Abstract
At the end of the last century, genetic studies reported that genetic information is not transmitted solely by DNA, but is also transmitted by other mechanisms, named as epigenetics. The well-described epigenetic mechanisms include DNA methylation, biochemical modifications of histones, and microRNAs. The role of altered epigenetics in the biology of various fibrotic diseases is well-established, and recent advances demonstrate its importance in the pathogenesis of pulmonary fibrosis—predominantly referring to idiopathic pulmonary fibrosis, the most lethal of the interstitial lung diseases. The deficiency in effective medications suggests an urgent need to better understand the underlying pathobiology. This review summarizes the current knowledge concerning epigenetic changes in pulmonary fibrosis and associations of these changes with several cellular pathways of known significance in its pathogenesis. It also designates the most promising substances for further research that may bring us closer to new therapeutic options.
Key Points
| Epigenetic information is crucial for appropriate proliferation, maturation, and functioning of pulmonary fibroblasts and epithelial cells. |
| Lung tissue affected by fibrosis presents numerous alterations in epigenetic code, many of them associated with regions that code proteins of known contribution to the pathogenesis of fibrosis. |
| Specific agents that can modify the epigenome arise as potential medications for pulmonary fibrosis. |
Introduction
Interstitial lung diseases (ILDs) are a group of entities that predominantly affect lung interstitium. These diseases correspond to at least 15% of all known respiratory diseases. These are classified into subgroups, including idiopathic interstitial pneumonias (e.g., idiopathic pulmonary fibrosis [IPF]) and ILD with granuloma formation (e.g., sarcoidosis). Importantly, some ILDs can be induced by drugs or occupational factors (asbestos, beryllium, etc.), while others are associated with connective tissue disease (CTD)—mainly rheumatoid arthritis, scleroderma, polymyositis, or dermatomyositis.
The diagnosis and management of these diseases rely on clinical, radiological, and histopathological findings. The patient’s smoking status, exposure to toxins, coexistent diseases, and drug history should be noted carefully during an examination. The vast majority of symptoms of ILDs are limited to the respiratory tract (these would include dyspnea, cough, and less frequently wheezing and hemoptysis), but extrapulmonary symptoms may be present, and in many cases they are associated with underlying CTD (muscle weakness, arthritis, gastroesophageal reflux, skin lesions, digital clubbing, etc.). High-resolution CT (HRCT) of the chest plays a significant role in the diagnosis and follow-up of patients with ILDs [1]. Lung function tests may be helpful to estimate the severity of the ongoing disease, and blood tests can indicate the presence of an underlying autoimmunization. In some cases, HRCT combined with lung function test may be used to predict disease prognosis and to assess current staging [2].
When the diagnosis is uncertain, bronchoscopy with bronchoalveolar lavage (BAL) may be beneficial to support initial diagnosis or indicate other conditions. Differential cell count is recommended to be performed with the BAL fluid [3], as well as microbiological assessment and cytopathology in specific situations. During bronchoscopy, a transbronchial biopsy may be needed to evaluate granulomatous conditions (e.g., sarcoidosis). Finally, surgical lung biopsy is the essential last step when other results are equivocal and the data are not highlighting a specific disease. Finally, a multidisciplinary team meeting is a reasonable step in demanding cases, as this will augment confidence in the final diagnosis.
What escalates the complexity is that the late stages of many ILDs present with pulmonary fibrosis and the prognosis and treatment differ between specific diagnoses [4, 5].
With our increasing knowledge, researchers have discovered more and more alterations in gene expression among the ILD-lung or animal models; therefore, treating ILDs as a genetic disease like cancer is not an exaggeration. Targeted therapies already exist in pneumology, e.g., sirolimus in lymphangioleiomyomatosis (LAM), which is known to be caused by a fault in the mTOR signaling pathway—though it is not a fibrotic ILD.
Basics of Epigenetics
Every somatic cell in the human body has the same unique DNA sequence, and major genomic alterations that occur during ILD are described in the review by Furukawa et al. [6].
Epigenetics, on the other hand, is defined as a DNA-independent carrier of genetic information that can be modified by stress, nutrition, and other environmental factors. Moreover, epigenetics is the language of the cell; it can activate or silence gene expression via various mechanisms, and it plays a fundamental role both in the human physiology and pathology [7]. Pathogenesis of other pulmonary diseases like cancer [8], asthma [9], and chronic obstructive pulmonary disease (COPD) [10] have been already linked to epigenetics. Below we briefly describe particular epigenetic mechanisms.
DNA Methylation
Human DNA contains many CpG sites where cytosine occurs next to guanine, and these regions are often concentrated in gene promoters [11]. The methylation process occurs when a methyl group is added to the cytosine in CpG dinucleotides. Methylation of the CpG island via DNA methyltransferases (DNMTs) blocks RNA polymerase complex from binding to the promoter region and therefore suppresses gene expression. The majority of human CpG sites are methylated [12, 13]. Hypomethylated gene promoters should result in increased gene expression, while hypermethylated conducts decreased gene expression—a general rule discussed later in this review.
Histone Modifications
The activity of cells depends on nucleosomes, which consist of DNA strands and histone core proteins (H2A, H2B, H3, H4) [14]. The H1 family is the most alkaline histone and wraps the whole nucleosome together [15]. The discovery of histone post-translational modifications and rapid progress in our knowledge of the role of histones has provided an opportunity to create histone code theory—another crucial hypothesis in epigenetic inheritance [16]. These modifications have considerable influence on chromatin condensation level and consequently on gene expression rate.
To the present day, a couple of histone modifications have been described. The best elucidated modifications include acetylation by histone acetyltransferases (HATs) and deacetylation by histone deacetylase (HDAC), methylation by histone methyltransferase (HMT), demethylation, phosphorylation, ubiquitination, sumoylation, and polyADP-ribosylation. According to current knowledge, acetylation activates cell transcription by decompression of the chromatin. The function of other modifications is more complex, e.g., methylation may cause either activation or inhibition of gene expression because functional impact depends on the modified region [13, 17].
MicroRNA (miRNA)
MicroRNAs (miRNAs) belong to the family of non-coding RNA and contain 17–25 nucleotides each. They interact with mRNA’s 3′ untranslated region (3′-UTR) and recruit an miRNA-induced silencing complex (miRISC) [18], which leads to consequent mRNA inhibition or degradation—thereby controlling various biological processes. Approximately 2650 human miRNAs have been described [19]. Each miRNA molecule can affect a variety of genes, and each single gene can be controlled by multiple different miRNAs [20].
Long Non-coding RNA (lnc-RNA)
Long non-coding RNAs (lnc-RNAs) are poorly understood in terms of their regulatory function [21], in opposition to extensively studied short miRNAs. The first promising research has already shown up [22–24], but their role in pulmonary fibrosis still needs to be established. Therefore, we do not describe them in detail in this review.
Epigenetics and Interstitial Lung Diseases
DNA Methylation
Whole-Genome Studies
In search of differences in DNA methylation and RNA expression between lung samples from patients suffering IPF and normal tissue, Sanders et al. used microarrays [25]. The researchers merged the methylation data from almost 15,000 genes and data from gene expression microarrays, and managed to identify 16 genes showing inverse DNA methylation and gene expression levels—assuming that methylation of CpG dinucleotides selected by this method plays a crucial role in their regulation. Eight of these candidate genes had been previously reported as associated with lung fibrosis, e.g., matrix metalloproteinase 7 (MMP7) and collagen 3α1 (COL3A1) associated with extra-cellular matrix (ECM). A detailed validation of four genes (ZNF465, CLDN5, TP53INPI, and DDAH1) not previously associated with IPF has also been performed, suggesting their possible role in its pathogenesis [25].
Rabinovich et al. sought a global methylation pattern in IPF lung tissue and analyzed the methylation of 25,406 human CpG sites, finding 625 of these to be differentially methylated compared to control lung tissue [26]. Hypomethylation of the promoter region associated with increased transcription was proven by the examples of the STK17B and HIST1H2AH genes; however, only 8.8% of differentially methylated CpG islands were located in promoter regions. It has been shown that 65% of the CpG islands with an altered methylation pattern in IPF lung samples are also modified in lung cancer samples, which advocates for the similarity between IPF and cancer [26]. Unfortunately, small study groups and methodological differences may be the cause of the limited overlap between the differently methylated regions (DMRs) in the two studies mentioned above [27].
Huang et al. also analyzed the methylation profiles of lung fibroblasts from IPF patients and compared them to fibroblasts from histologically normal lung regions from patients undergoing resection for lung nodules and commercial fibroblast lines from the lungs of healthy individuals. Comparison revealed several hundred differently methylated loci (787 in IPF vs normal lung group and 333 in IPF vs human fibroblasts) and an overlap of 125 genes between these groups [28]. Most of the differentially methylated loci were located outside promoter regions in this study. Unfortunately, IPF fibroblasts from different cell lines showed significant heterogeneity in their individual methylation profiles.
Yang et al. identified 2130 DMRs in the IPF lung tissue genome (71% of these within gene bodies, 10% in promoter regions), the majority located in CpG island shores (60%; only 5% in CpG islands themselves). Among the multiple DMRs identified, some had been previously associated with IPF, namely CXCR4, thrombin, Wnt/β-catenin, vascular endothelial growth factor (VEGF), and epithelial adherens junction signaling [29]. Methylation changes were consistent with IPF genome-wide association studies (GWASs) [30, 31] in five out of the ten IPF GWAS loci. Five DMRs were influenced by combined use of corticosteroids and immunosuppressants. A total of 172 out of 1315 differently methylated genes showed a significant relationship between methylation and expression. Further analysis revealed the ten most affected networks [29]. Intriguingly, while mice deficient in amyloid protein precursor (APP) (presenilin 2) spontaneously develop lung fibrosis [32], the most significant network centers were located around APP [29].
Investigation of global methylation patterns in rat fibroblasts co-cultured in vitro with silica-exposed alveolar macrophages has shown 2842 DMRs in genes representing many of the pathways leading to myofibroblast differentiation [33]. Noteworthy clusters are associated with ‘regulation of actin skeleton’ characteristic for these cells, ‘focal adhesion’ that has its role in cell migration, ‘extracellular matrix’, and the ‘MAPK (mitogen-associated protein kinase) signaling pathway’ that is activated via transforming growth factor beta (TGFβ) [34]. The increased expression of MAPK9, one of the MAPK terminal kinases associated with negative regulation of a cell cycle, was confirmed with increased mRNA and translation product levels [33]. Intriguingly, elevated protein levels were not observed in reference to DMRs in the TGFβ1 pathway despite the increased mRNA levels. These findings most likely result from the overlap of different gene expression regulatory mechanisms.
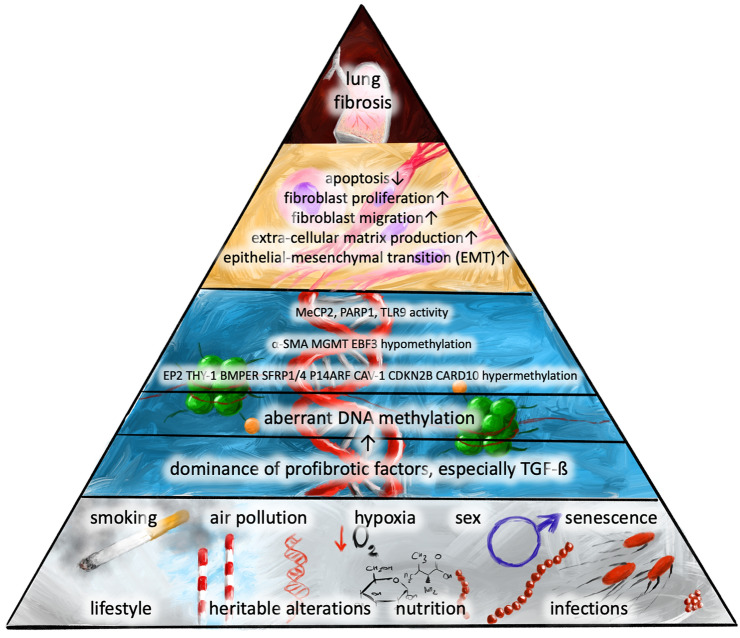
The studies described above show that DMRs are only partially located in promoter regions, while many are found, for example, in introns. Further studies are required to determine the significance of these alterations. Below, we describe the actual data according to the specific pathways known to play significant roles in pulmonary fibrosis. The order of events involving DNA methylation that leads to developed pulmonary fibrosis is presented in Fig. 1.
Fig. 1.
The sequence of events including impaired DNA methylation that leads to pulmonary fibrosis. α-SMA α-smooth muscle actin, BMP bone morphogenetic protein, BMPER BMP endothelial cell precursor-derived regulator, CARD10 caspase recruitment domain-containing protein 10, Cav-1 caveolin 1, CDKN2B a cyclin-dependent kinase 4 inhibitor B, EBF3 early B-cell factor 3, EP2 prostaglandin E2 receptor subtype EP2, MeCP2 methyl CpG binding protein, MGMT O6-alkylguanine DNA alkyltransferase, PARP1 poly(ADP-ribose)polymerase 1, SFRP secreted fizzled protein, TGF-β transforming growth factor β, Thy-1 thymocyte differentiation antigen 1, TLR9 toll-like receptor 9
Prostaglandin E2 (PGE2) Pathway
Prostaglandin E2 (PGE2) participates in maintaining fibroblasts’ suppression and their proper apoptosis. The decreased levels and tissue effects of PGE2 are simultaneous mechanisms in IPF pathogenesis [35]. Huang et al. sought to determine whether decreased EP2 (PGE2 receptor subtype) expression in fibroblasts is conditioned by DNA hypermethylation of its promoter, PTGER2 [36], both in the murine model and humans with pulmonary fibrosis. After proving this fact, the attempt to restore EP2 gene expression with the use of DNA methylation inhibitors (DNMTi) was made, resulting in an increase in EP2 expression in fibrotic fibroblasts and no influence on non-fibrotic fibroblasts. A rise in EP2 expression resulted in the restored ability of PGE2 to inhibit collagen synthesis and cell proliferation in these cells [36]. DNMTi in this study were 5-aza-2′-deoxycytidine and zebularine, the first of which is already approved for use in myelodysplasia and acute myeloid leukemia [37, 38]. To avoid confusion, the agent 5-azacytidine and its derivate 5-aza-2′-deoxycytidine are called 5-aza later in this review.
With regard to the underlying cause of reduced PGE2 levels, other researchers have found that not only are cyclooxygenase-2 (COX-2) mRNA basal levels reduced in IPF and systemic sclerosis (SSc) lung fibroblasts, but TGFβ1 induction is also restricted [39]. A significant reversion of this state has been shown after the treatment of IPF- and SSc-derived cells with 5-aza. The intervention partially corrected aberrant PGE2 and collagen 1α expression and restored sensitivity to FasL-induced apoptosis [39]. As a whole, these results advocate a positive change in cell phenotype assigned to 5-aza treatment. Interestingly, CpG promoter methylation did not show significant differences. Looking for a potential mechanism, investigators have found five DMRs in the c8orf4 promoter, while c8orf4 itself was further proven to interact with COX-2 promoter and regulate its expression [39]—forming a probable direct connection between DNA methylation and impaired COX-2 expression.
On the other hand, human fetal and adult lung fibroblasts, when incubated with PGE2, tend to increase transcription factors for DNMT3a [40] along with increased DNMT expression and elevated overall DNA methylation level. This effect might be unique for fibroblasts and pathogenesis of lung fibrosis, as it stands opposite to lowering DNMT1 and DNMT3a levels in macrophages.
Thymocyte Differentiation Antigen 1 (Thy-1)
Several studies refer to thymocyte differentiation antigen 1 (Thy-1), a cell outer membrane glycoprotein also known as CD90—a thymocyte differentiation antigen. It has long been known that Thy-1 knockout mice demonstrate enhanced lung fibrosis when treated with intratracheal bleomycin [41]. Additionally, cells have rarely shown Thy-1 expression in fibrotic areas, as opposed to the diffuse presence of phosphorylated Smad2/3, active TGFβ, and α-smooth muscle actin (α-SMA).
The absence of Thy-1 [42] along with increased α-SMA has been confirmed in human lung specimens from patients with fibrotic lung disease. Human lung fibroblasts that have lost Thy-1 expression via incubation with tumor necrosis factor (TNF) and interleukin-1 (IL-1) also demonstrated elevated levels of α-SMA, pSMAD2/3, and overall SMAD—consistent with the fibrogenic phenotype identified in murine models [41]. These experiments suggest that suppression of Thy-1 expression promotes myofibroblast differentiation.
How is DNA methylation and altered Thy-1 gene expression associated? The Thy-1 promoter hypermethylation was proven in Thy-1 non-expressing cells and IPF-derived fibroblasts, in opposition to Thy-1 expressing cells [43]. Another interesting observation has been made by Robinson et al. who showed that hypoxia results in escalated methylation and reduced Thy-1 transcription in normal human pulmonary fibroblasts [44]. The study also suggests that hypoxia promotes myofibroblast differentiation, as it increases α-SMA, collagen 1 and 3 mRNA levels, and α-SMA protein level—specific for this cell phenotype. As hypoxia is a result of smoking and oxidative stress, strongly associated with lung fibrosis, these findings might be an important part of the disease’s pathogenesis.
Coal and silica exposure induces a type of lung fibrosis commonly known as pneumoconiosis. In RLE-6TN cells (rat alveolar epithelial cells [AECs]) and rat lungs, coal and silica exposure resulted in accumulation of collagen 1 and 3, indicating fibrotic changes. At the same time, DNMT1 levels were raised, which corresponds to Thy-1 promoter hypermethylation and a decrease in Thy-1 protein expression. 5-Aza administration 24 h after coal and silica exposure significantly inhibited pneumoconiosis [45].
An increase in Thy-1 expression after demethylation with 5-aza was proved in primary mouse lung fibroblasts [46] and rat and human lung fibroblasts [42]. Similar results were obtained after transfecting mouse fibroblasts with DNMT1 silencing RNA [46]. These interventions reversed the effect of TGFβ1, consistently with lowered DNMT1 activity and collagen type 1, alpha 1 (Col1α1) expression.
Thy-1 was also upregulated in Thy-1 age-deficient cells after incubation with 5-aza. Unfortunately, a study by Sueblinvong et al. despite showing a decrease in Thy-1 mRNA expression of fibroblasts from old mice lungs compared to young ones, did not show an increase in mRNA levels after treatment with this DNMTi [47].
Nevertheless, the findings strongly suggest an attenuation of the profibrotic cell phenotype with the use of a demethylation agent, which provides an opportunity for clinical studies and, potentially, for the development of a new treatment approach.
Sanders et al. stated that treatment with DNMTi and, alternatively, histone deacetylase inhibitors (HDACi) restored expression of Thy-1 gene and inhibited myosin expression [43]. Regulation of Thy-1 expression in the context of histone acetylation is reviewed in the following chapter.
Myofibroblast Differentiation
Fibroblast to myofibroblast differentiation has usually been measured by the level of α-SMA. α-SMA gene methylation has shown DMRs in at least four CpG sites in rat fibroblasts and AECs. When compared to epithelial type II cells that do not express α-SMA, fibroblasts exhibit significantly lower α-SMA gene methylation and DNMT levels. During inhibition of methylation activity via 5-aza or short interfering RNA (siRNA) specific for DNMT1, DNMT3a, and DNMT3b, α-SMA expression increased, while transfection with plasmids containing DNMT genes did the opposite [48], underlying the importance of these mechanisms in epithelial–mesenchymal transition (EMT) regulation. Other methyltransferase inhibitors such as arginine N-methyltransferase inhibitor-1 (AMI1) and sinefungin also present the potential to disrupt the ECM architecture generated by IPF fibroblasts, as recently discovered by Sala et al. [49].
While TGFβ is the most recognizable fibrosis-leading factor, the balance between TGFβ and bone morphogenetic proteins (BMPs), especially BMP4, is crucial for lung repair mechanisms, fibroblast proliferation, and EMT [50–52]. BMP endothelial cell precursor-derived regulator (BMPER), which binds BMPs, is overexpressed along with α-SMA in the fibrotic foci of IPF lung tissue. In a series of experiments, BMPER revealed its influence on migration and invasion of fibroblasts and matrix production measured by hyaluronan levels [53]. It also affects TGFβ/BMP signaling in favor of the profibrotic TGFβ pathway. Elevated levels of BMPER were attenuated via administration of 5-aza, which indicates DNA methylation is the main regulatory mechanism. However, in this study, even more 5-aza effects have been shown, including reduced matrix production and fibroblast migration in IPF fibroblasts, as well as lowering the degree of lung fibrosis in bleomycin-treated mice. The phenotypic changes in primary and IPF lung fibroblasts after diminishing BMPER via siRNA and via 5-aza are convergent [53].
Toll-like receptor 9 (TLR9), which is excessively expressed in histological specimens of UIP and NSIP patterns characteristic for ILDs, appears to be a significant factor that drives myofibroblast differentiation dependent on type 2 T helper cell (TH2) lymphocyte/IL-4 expression [54]. Hypomethylation of fibroblastic DNA through the TLR9-associated pathway leads to defective miRNA processing (via the loss of Argonaute proteins—AGO1 and AGO2), causing the progressive phenotype of IPF [55].
Hu and colleagues studied the role of poly(ADP-ribosyl)ation in IPF. The reaction of poly(ADP-ribosyl)ation is performed by the poly(ADP-ribose)polymerase (PARP) enzyme family, mainly by PARP1. They revealed that PARP1 promotes myofibroblast differentiation via the upregulation of α-SMA expression, while the lack of PARP1 causes hypermethylation of CpG islands of the α-SMA gene. The authors also reported higher expression of PARP1 in the IPF fibroblasts and reduced fibrotic response in bleomycin-induced lung fibrosis (BILF) mice with PARP1 deficiency [56].
Methyl CpG binding protein 2 (MeCP2) binds to the α-SMA gene, increases its expression [57], and drives fibroblast differentiation into myofibroblasts, which is in opposition to its known function as a gene repressor. Mice deficient in MeCP2 surprisingly presented with much less fibrosis, as measured by hydroxyproline and collagen type 1 [58]. Upregulation of α-SMA via MeCP2 association to the α-SMA gene can be significantly reversed with 5-aza in human lung fibroblasts (IMR-90 line).
The Wnt signaling pathway that plays a crucial role in fibroblast activation has a significant role in pulmonary fibrosis [59]. Secreted fizzled proteins (SFRPs) bind to the Wnt ligand, forming non-functioning complexes, and their downregulation is associated with excessive fibrosis in various organs. Zhou et al. showed that SFRP1 and SFRP4 were downregulated in the mouse model of pulmonary fibrosis, that their promoter regions were progressively hypermethylated, along with the duration of the disease, and that this effect was attenuated via incubation of fibroblasts with 5-aza [60]. Intraperitoneal injection of 5-aza has also reduced the levels of COL1α1 and α-SMA, corresponding to the clinical effect of decreasing pulmonary density and pulmonary fibrosis scores in this study.
CDKN2B (a cyclin-dependent kinase 4 inhibitor B considered to be a tumor suppressor), CARD (caspase recruitment domain-containing protein 10, involved in apoptosis), and MGMT (O6-alkylguanine DNA alkyltransferase, a crucial DNA repair enzyme) were the focus of interest in a study by Huang et al. CDKN2B and CARD10 genes were proven to be hypermethylated, with an accompanying reduced expression, and MGMT was hypomethylated with overexpression in IPF fibroblasts [28]. Treatment of these cells with 5-aza resulted in a significant decrease in CARD10 methylation and a modest decrease in CDKN2B methylation, with increased protein expression of both gene products.
The effect of silencing CDKN2B expression via siRNA in normal fibroblasts is not yet clear; it resulted in increased cell proliferation among unstimulated cells [28], but this appeared to decrease with growth factor stimuli [61], which is opposite to the reaction of epithelial cells. In IPF, a similar paradox is described in the context of higher sensitivity of epithelial cells and lower sensitivity of fibroblasts to FasL-mediated apoptosis [62]. The authors suggest that the loss of CDKN2B in fibroblasts promotes EMT rather than proliferation, as both the mRNA and protein levels of collagen 1 and α-SMA increase, along with the transcription factors associated with myofibroblast differentiation such as serum response factor (SRF) and myocardin-related transcription factor A (MRTF-A).
Finally, a positive correlation between pulmonary fibrosis among pigeon breeder’s lung patients and EBF3 (early B-cell factor 3, a B-cell maturation factor) hypomethylation has been shown. TH2 dominance and excessive B-cell activation are thought to be important factors in fibrosis that complicate chronic stages of hypersensitivity pneumonitis [63]. The methylation rate presented statistically significant differences between patients who were breeding pigeons with and without consequent ILD [64]. The EBF3 mRNA and IL-10 levels were increased in the study group, negatively correlating with EGF3 gene methylation levels.
Apoptosis
Regarding the reduced apoptotic activity of fibroblasts in the fibroblastic foci of IPF lungs, Cisneros et al. evaluated promoter methylation of the proapoptotic (tumor suppressor) gene p14ARF in fibroblasts obtained from IPF and normal lungs. Half (four) of the fibroblast cell lines from IPF patients presented hypermethylation of p14ARF promoter, while the other four IPF fibroblast and four normal cell lines remained unmethylated. The hypermethylation resulted in the underexpression of p14ARF, which consequently reduced the apoptosis rate and p53 levels in fibroblasts [65]. The researchers demonstrated these effects were reversible via incubating the cell lines in, once more, 5-aza.
Histone Modifications
Epithelial–Mesenchymal Transition
In search of epigenetic changes associated with epithelial-to-mesenchymal transition, Noguchi et al. performed RNA sequencing of A549 cells (alveolar epithelial type II cells) stimulated with the profibrotic factor TGFβ1, finding several up- and downregulated genes [66]. Furthermore, valproic acid (VPA), known to inhibit histone deacetylase 1 (HDAC1) activity, was administered, resulting in partial reversal of the downregulation, especially in CDH1 (E-cadherin; epithelium-specific gene) and GATSL2 (GATS protein-like 2) genes. The H3K27 histone in the CDH1 loci has been shown to be deacetylated by TGFβ1 and restored under VPA stimulation, which strongly corresponds with expression changes. In this study, there was almost no such effect on TGFβ1-upregulated genes, with the exception of COL1A1 [66].
Ota et al. also examined A549 cells exposed to TGFβ1 and EMT reversibility via HDACi—trichostatin A (TSA). The histological phenotype did not alter after the intervention, but TSA treatment increased mRNA expression of surfactant protein C (Sftpc), histone H4 acetylation in its promoter region, and propeptide of surfactant C protein (pro-Sp-C) in both control and TGFβ1-induced cells in vitro. Surfactant protein C (Sp-C) is necessary for alveolar homeostasis and was reported to be reduced in the lungs of bleomycin-treated mice. In the lung tissue from these mouse models, TSA treatment elevated the overall histone H3 and H4 acetylation levels and, especially, pro-SP-C expression. Among magnetically separated alveolar type II cells from BILF mice, the Sftpc and Cdh-1 mRNA levels were restored after TSA treatment [67]. TSA also inhibited TGFβ-induced expression of α-SMA and collagen type 1 and blocked contractile function of normal human lung fibroblasts [68].
The expression of one of the best known transcription factors that mediates EMT, zinc finger E-box-binding homeobox 1 (ZEB1), is highly dependent on H3K4me3 and H3K9me3 balance [69]. ZEB1 has a known effect of suppressing epithelial marker expression, E-cadherin [70], thus allowing EMT, cell migration, and invasion. Challenging A549 cells with TGFβ1 results in a loss of H3K9me3, increased ZEB1 transcription, and switching to the mesenchymal phenotype. The addition of Schisandrin B, a substance obtained from a fruit used in Chinese medicine, restored H3K9me3 and reverted EMT of A549 cells, revealing a strong epigenetic influence [69].
HDACi have also been tested for a direct in vitro effect on fibroblasts. Davies et al. examined the effect of spiruchostatin A (SpA), an HDACi specific for class I histone deacetylase, on IPF fibroblasts. The presence of SpA resulted in significant inhibition of proliferation after 144 h of incubation, irrespective of the presence of TGFβ1, with an effect that was still evident after 96 h post-treatment, with little cytotoxicity at effective doses. Histone H3 acetylation was accelerated after treatment and P21WAF1 (cell-cycle regulator) expression level increased, thus revealing the possible antiproliferative effect of SpA. Treatment with SpA suppressed α-SMA (mRNA and protein), collagen 1 and 1 overexpression caused by TGFβ, as well as the formation of actin filaments—characteristic for myofibroblast differentiation and fibrosis [71].
Lysine-specific demethylase 1 (LSD1), known to mediate tumor cell behavior via epigenetic mechanisms, was recently investigated in a BILF mouse model. Its expression is increased in BILF mice lungs, while its knockdown alleviates lung fibrosis in vivo [72]. Lung coefficient and histological scores, as well as ECM accumulation measured with COL1α levels, were significantly in favor of the tested group.
When Pan et al. tried to achieve similar results in vitro, they found that TGFβ-incubated primary mouse lung fibroblasts slowed their migration and were less likely to differentiate into α-SMA positive cells after LSD1 knockdown. The effects are dependent on the TGFβ/Smad3 signaling pathway, which was probably downregulated due to high repressive H3K9me1/2 expression [72].
Thy-1
Mouse lung fibroblasts challenged with lipopolysaccharide (LPS) resulted in a significant increase in proliferation in comparison to the control group. Further investigation revealed a rapid decrease in Thy-1 (CD90) mRNA and protein expression, which indicates a profibrotic transformation phenotype. Global histone H3 and H4 acetylation levels were lowered in the LPS-challenged cells, as well as Ace-H4 level in the Thy-1 promoter region, suggesting epigenetic silencing of the Thy-1 gene. Furthermore, pre-challenge depletion of TLR4 (receptor for LPS) via the effect of specific siRNA resulted in a lack of Thy-1 expression inhibition and significantly higher Ace-H3 and Ace-H4 levels [73].
In another experiment, the researchers managed to partially restore Thy-1 expression in Thy-1 (−) rat lung fibroblasts using TSA, while open chromatin markers associated with Thy-1 promoter raised their levels. The HDACi also partially increased CpG island methylation in the Thy-1 promoter region, consistently raising overall DNMT levels. Thy-1 re-expression was associated with decreased expression of α-SMA, suggesting the cells were undergoing a phenotypic alteration [74].
Apoptosis
Huang et al. have shown resistance to Fas-mediated apoptosis and impaired Fas expression in fibroblasts acquired from lungs of bleomycin-injured mice [75]. Decreased histone H3 and H4 acetylation in the Fas promoter region and increased overall HDAC2/4 expression were detected, attributing histone deacetylation to phenotypic changes. Treatment with HDACi (TSA and suberoylanilide hydroxamic acid [SAHA]) increased Fas mRNA and surface expression levels significantly, partially restoring the apoptotic activity of fibroblasts. SAHA treatment restored histone H3 acetylation levels in BILF fibroblasts, while TSA treatment elevated the susceptibility to Fas-mediated apoptosis in this cell group, with no such effect in control cells [75]. Increased trimethylation of H3K9 histone was an additional finding of no known significance. SAHA, also known as vorinostat, is a non-specific histone deacetylase inhibitor (HDI) registered for treatment of cutaneous T-cell lymphoma.
Reduced histone H3 acetylation and increased histone H3K9 methylation at the Fas promoter region, along with restoration of Fas mRNA and protein expression after TSA treatment, has been confirmed in human IPF fibroblasts in a subsequent experiment [75].
Cav-1 is a tumor suppression gene. Reduced levels of caveolin 1 (Cav-1) have been reported in IPF lung fibroblasts. In the mouse model, Cav-1 is experimentally suppressed along with elevated collagen 1α1 in response to bleomycin lung injury. Sanders et al. have found that it is possibly a result of decreased association of Cav-1 with H3K4Me3 (an active histone mark), as this differed significantly when compared to control cells. Additionally, the inhibition of the p38 MAPK pathway prevented downregulation of Cav-1 gene associated with H3K4Me3 [76].
In contrast, no significant differences in DNA methylation were proven between IPF and control fibroblasts, as well as between normal fibroblasts with and without TGFβ1 stimulation, suggesting that DNA methylation is irrelevant to Cav-1 regulation.
Wang et al. assessed the antifibrotic effect of SAHA on human fibroblasts and its anti-inflammatory effect on peripheral blood morphonuclear cells (PBMCs) and lymphocytes. It emerged that SAHA induces acetylation of histone 3 and α-tubulin in both TGFβ1 (+) and TGFβ1 (−) conditions. It mitigates the myofibroblast differentiation and collagen I deposition and reduces both the elevated matrix metalloproteinase-1 (MMP1) and tissue inhibitor of metalloproteinases 1 (TIMP1) levels in IPF lung fibroblasts. SAHA also inhibited cell proliferation with no effect on apoptosis. HDACi affected cytokine release in PBMCs and lymphocytes, with TNFα, IL-8, IL-13, and IL-10 being decreased, while IL-6 and IL-10 levels, respectively, were diminished in lymphocytes and monocytes alone [77].
Another research team reported decreased COL3A1 gene expression and collagen 3 protein after the SAHA treatment. It reduced the high acetylation rate of histone H3 and H4 observed in the IPF group [78]. Moreover, SAHA promoted apoptosis of IPF myofibroblasts by stimulating apoptotic function of Bcl2-antagonist/killer (Bak), while inhibiting Bcl-xl’s (one of the B-cell lymphoma 2, Bcl2, protein family) anti-apoptotic activity [79].
A recent study by Bai et al. showed that IPF fibroblasts cultured without glutamine (Gln) exhibit an increased apoptosis rate. A similar result of increased apoptosis was observed after glutaminase (GLS1), Gln-glutamate conversion enzyme, depletion [80]. GLS1 inhibition in BILF mice attenuates pulmonary fibrosis in vivo [81]. The effect seems to be associated with the lack of Gln metabolites. Molecules from the inhibitor of apoptosis protein (IAP) family such as X-linked inhibitor of apoptosis (XIAP) and survinin exhibit increased expression in IPF lung fibroblasts, but their levels are highly dependent on Gln-glutamate balance [80]. It also appears that H3K27me3 rate increases during GLS1 gene knockdown and when incubating IPF fibroblasts in a medium deficient in Gln. To sum up, Gln emerges as a key factor that prevents apoptosis and mediates survival of fibroblasts, possibly via epigenetic silencing of IAPs.
PGE2 Regulation
As discussed in Sect. 2.1.2., low COX-2 expression has been linked to hypoacetylation of histone H3 and H4 caused by decreased activity of HATs [82]. The same research group revealed histone H3 and H4 deacetylation, as well as histone H3 hypermethylation of gamma interferon induced protein 10 gene (IP-10/CXCL10). These abnormalities were caused by reduced recruitment of HATs, increased activity of HDAC-containing repressor complexes (HMTs G9a and SUV39H1), and the presence of heterochromatin protein 1 at the IP-10 promoter region [83].
When inhibiting or knocking out G9a or EZH2 (H3K9 and H3K27-specific HMT, respectively) in IPF fibroblasts, the histone H3 and H4 acetylation at the IP-10 promoter region increases in IL-1β-stimulated cells, and the previously reduced expression of IP-10 seems to be restored [84]. H3K9me3 and H3K27me3 levels are lowered under similar conditions, which means less repressive chromatin in this region after the intervention.
On the other hand, the knockdown of either G9a or EZH2 in normal lung fibroblasts reverses the excessive (TGFβ-induced) association of EZH2 to the IP-10 promoter [84]. It appears that they physically interact with each other and both are required for gene expression regulation. The intervention mentioned above also affects H3K9me3 and H3K27me3 levels. Moreover, histone and DNMTs cooperate in the case of epigenetic changes, as inhibition of either G9a or EZH2 alone significantly reduces DNA methylation in the COX-2 promoter region [85]. Taken together, results are suggestive for a complex epigenetic regulatory mechanism.
What is the association between G9a, EZH2, and COX-2? Coward et al. investigated G9a- and EZH2-associated histone and DNA methylation at the COX-2 promoter in IPF fibroblasts [85]. The reduction of repressive histone H3 methylation and DNA methylation using G9a, EZH2, and DNMT1 inhibitors, as well as specific siRNAs, has been confirmed; the expected variability between experiments that is also partially dependent on stimulation from IL-β1 was also shown. Both inhibition and disruption of these factors increased COX-2 mRNA and PGE2 levels [85], including cells previously stimulated with IL-β1.
Nevertheless, the study on fibroblasts from non-fibrotic lung tissue did not show the same effect on COX-2 mRNA levels via G9a and EZH2 inhibition. In this study, Pasini et al. identified a significant increase in COX-2 protein expression without an increase in COX-2 mRNA levels in response to SAHA, which may be mediated via other factors, e.g., the suppression of a translational silencer TIA-1 or via changes in miRNA expression [86], which requires validation by further experiments.
Other
While H3K9 dimethylation is overrepresented in fibrotic regions of both BILF mice and IPF lungs [87], G9a specifically methylates H3K9, allowing binding of chromobox homolog 5 (CBX5) and a consequent assembly of a transcriptional repressor complex [88]. Consequent chromatin modifications inhibit transcription of PPARγ coactivator 1α (PPARGC1A—peroxisome proliferator-activated receptor gamma coactivator 1-alpha) [87] that encodes peroxysmal protein PGC1α. PGC1α acts as a regulator of mitochondrial biogenesis and fatty acid oxidation [89], probably playing an important fibrosis-inhibiting role in BILF [90] and IPF.
This study was first to identify the roles of the G9a/CBX5 pathway that both directly and via H3K9 methylation depresses PGC1α expression in its gene promoter region and leads to an uncontrolled IPF fibroblast activation. Administration of BIX01294, a G9a inhibitor, prevents PGC1α depression and mitigates profibrotic effects of TGFβ on IPF fibroblasts; knockdown of either CBX5 or G9a reduces the profibrotic ACTA2 gene expression in IPF lung fibroblasts [87].
Recent research by Jones et al. presents the effects of another HDAC inhibitor, pracinostat, on IPF primary lung fibroblasts [91]. The significant reduction of α-SMA, fibronectin, and collagen 1 levels after TGFβ stimulation was proved, which means alleviation of myofibroblast differentiation and ECM production. Intervention altered fibroblast contractile functions as well. Pracinostat increased global histone acetylation rate; it prevented TGFβ from repressing various antifibrotic genes [91]. The authors investigated the effects of HDAC knockdown on lung fibroblast activation, and HDAC7 emerges as the most important HDAC that mediates TGFβ-induced fibroblast activation [91].
miRNA
miRNAs have been extensively studied in the pathogenesis of pulmonary fibrosis. The comprehensive study conducted by Xie et al. revealed different expression of 161 miRNAs in BILF. The altered miRNAs were involved in a number of pathways and regulatory processes, including cell apoptosis and the TGFβ and Wnt signaling pathway [92]. Abnormal miRNA expression in BILF may also be associated with insulin-like growth factor signaling according to increased expression of Igf1 in lungs of these mice [93].
One paper has identified circulating miRNAs in patients’ sera [94]. Forty-seven miRNAs differed between IPF and a control group (21 upregulated vs 26 downregulated). In the second cohort, authors reported enhanced expression of miR-21, miR-199a-5p, and miR-200c, accompanied by reduced expression of miR-31, let-7a, and let-7d in IPF patients. miRNAs in this case have been assigned to disrupted molecular pathways such as TGFβ, MAPK, PI3K-Akt, Wnt, HIF-1, Jak-STAT [94].
Regarding the high variability of disease progression, the study conducted by Oak et al. showed different expression of miRNAs in patients with progressive IPF [95]. miRNAs were assessed in surgical lung biopsy specimens, and overexpression of five miRNAs (miR-302c, miR-423-5p, miR-210, miR-376c, and miR-185) has been linked to rapid progression, while only one miRNA level (miR-423-3p) was decreased when compared to more stable disease. Intriguingly, miR-302c was downregulated in lung tissue in sarcoidosis, opposite to that seen in IPF, a fact possibly useful for establishing the final diagnosis [96]. mRNA profiling revealed promotion of EMT via downregulation of specific miRNAs. The vital components of the miRISC complex that process miRNA, ARO1 and ARO2, were also shown to have lower expression in subjects with IPF [95]. The results strongly relate miRNA abnormalities to IPF.
Profibrotic miRNA
miR-21 presumably plays a key role in the pathogenesis of IPF. Enhanced production of TGFβ stimulates miR-21 expression. It is supposed that miR-21 induces EMT and promotes the fibrotic processes via inhibition of Smad7 [97]. High expression levels of miR-21 were found in myofibroblasts from patients with IPF and also in an experimental model of pulmonary fibrosis [98]. Yang and colleagues found that the presence of miR-145 leads to α-SMA expression, which is a characteristic of fibroblast–myofibroblast transition. Additionally, miR-145 activates the latent TGFβ, inducing fibrosis [99]. miR-424 seems to promote myofibroblast differentiation during EMT through targeting Smurf2 (TGFβ pathway inhibitor) and thereby enhancing TGFβ production [100]. Another group found that increased expression of miR-96 in response to a collagen-rich environment suppressed forkhead box O3a (FoxO3a), which made IPF fibroblasts capable of abnormal expansion in the diseased lung. This action can be explained by impaired tumor suppressor factors p27, p21, and Bim activity [101].
Relaxin-deficient mice develop age-related lung fibrosis, and treatment with relaxin reverses collagen deposition in their lungs [102]. The expression of relaxin receptor, relaxin/insulin-like family peptide receptor 1 (RXFP1), is downregulated in lungs from IPF and SSc patients, which limits the antifibrotic effect of endogenous relaxin and its potential therapeutic use [103]. Additionally, lowest RXFP1 levels are associated with highest predicted mortality. miR-144-3p is an miRNA upregulated over 70-fold in IPF fibroblasts, and its mimic has been recently proven to downregulate RXFP1 in IPF lung fibroblasts [104]. Decrease in RXFP1 accompanies increased α-SMA levels and implies a direct profibrotic effect of miR-144-3p.
Milosevic et al. revealed the profibrotic function of miR-154 in IPF through activation of the WNT pathway. miR-154 regulates fibroblast migration and proliferation, additionally inhibiting p15 expression [105]. Another molecule, the miR-23a cluster, acts together with ZEB1 transcription factor to reinforce the presence of mesenchymal cells in pulmonary fibrosis [106], in opposition to miR-200c. Lino Cardenas et al. found increased expression of miR-199a-5p in IPF samples, particularly in injured murine myofibroblasts and fibroblastic foci. The synthesis of miR-199a-5p was enhanced with TGFβ, but addition of TGFβ was not required to activate pulmonary fibroblasts. The authors suggest that miR-199a-5p regulates Cav-1 and has an important role in the mouse model of liver and renal fibrosis [107]. Bodempudi and co-workers reported that hypoxia stimulates fibroblast proliferation via miR-210 expression. HIF-2α, a protein induced by hypoxia, targeted miR-210 and upregulated its expression [108]. Abnormal expression of miR-21, miR-101-3p, and miR-155 were matched with impaired forced vital capacity and radiological patterns in IPF [109].
Antifibrotic miRNA
Liang et al. described low expression of miR-26a in a mouse model of pulmonary fibrosis and consequently sought the antifibrotic effect of miR-26. TGFβ1 inhibits expression of miR-26a by phosphorylation of Smad3 in both a mouse model of pulmonary fibrosis and IPF. Admission of the exogenous miR-26 reduces collagen and connective tissue growth factor (CTGF) expression, as well as the overall level of lung fibrosis in BILF mice [110] to a degree dependent on the time from symptom onset. Additionally, Li et al. revealed that miR-26a targets TGFβ fibrotic response via regulation of cyclin D2, TGFβ2, and the receptor for TGFβ1 [111]. miR-26a targets high-mobility group AT-hook 2 (HMGA2), a transcriptional factor which plays a significant role in EMT, and downregulation of miR-26a boosts this process [112]. A number of other miRNAs are suppressed in pulmonary fibrosis via the influence of TGFβ. Graham and colleagues found that low expression of miR-27b is associated with increased collagen expression. miR-27b binds directly to the 3′-UTR of Gremlin 1 and blocks the synthesis of its mRNA; decrease in its function causes enhanced Gremlin 1 expression [113]. The miR-200 family was markedly decreased in IPF patients and BILF mice, while its overexpression plays a crucial role in the inhibition of EMT [114]. miR-326 inhibits profibrotic genes such as Ets1, Smad3, and matrix metalloproteinase 9 (MMP9) and stimulates the antifibrotic Smad7 [115]. miR-375’s function is the regulation of AEC type II trans-differentiation into AEC type I cells, which occurs after lung injury, via the WNT/b-catenin pathway [116]. miR-30a and miR-92a levels were predicted to target and mitigate WNT1-inducible signaling pathway protein 1 (WISP1) expression, which is upregulated in response to TGFβ1 and is probably associated with de novo collagen production in BILF [117]. One of the targets of let-7d is HMGA2, which becomes overexpressed in fibrotic lung. The loss of let-7d in IPF fibroblasts impairs epigenetic gene silencing via the MiCEE complex required for normal lung repair; it also interferes with HDAC1 and HDAC2 that are part of this complex [118]. The inhibition of let-7d leads to EMT and promotes collagen deposition in mouse lungs [119]. Let-7d interacts during the EMT process; its administration diminishes mesenchymal transition by reducing α-SMA and other markers involved in this process [120].
ECM deposition is highly dependent on miR-29 regulation. Cushing et al. suggest that miR-29 is a controller of many genes associated with complex fibrotic response such as laminins, integrins, ADAM metallopeptidases, MMPs, and collagen and that its downregulation enhances fibrosis and sensitizes cultured lung fibroblasts to TGFβ effects [121, 122]. In addition, miR-29 reduces expression of COL1α via phosphorylation of PI3K-Akt [123]; however, its expression is downregulated by TGFβ and SMAD3 [124]. miR-29c also mitigates the abnormal mRNA levels of LOXL2, COL3A1, and SPARC, which are associated with extracellular matrix; at the same time, it accelerates the expression of Fas mRNA and protein, increasing the Fas-mediated apoptosis in human lung fibroblasts (HFL1 cells) [125]. It also reverses the apoptotic resistance in cells treated with TGFβ. To sum up, overexpression of miR-29 limits ECM production and deposition [126]. Inhibition of miR-29c had the opposite biological effects in the abovementioned studies, and expression of miR-29 is known to be reduced in lung fibrosis. There is evidence that its biological effects are associated with the regulation of DNA methylation, as miR-29c directly decreases DNMT3A and 3B levels and there was no change in Fas levels after treatment with miR-29c inhibitor in the environment containing 5-aza [125].
The experiment performed by Xiao et al. deserves even more attention. Firstly, the researchers confirmed, using the mouse model, that bleomycin-induced pulmonary fibrosis, as well as the loss of the miR-29 family, is associated with Smad3 expression, possibly due to induction from TGFβ1. Secondly, they showed that transfection of pre-miR-29b to the lung tissue is possible, efficient, and—most importantly—that it restores miR-29b to the normal levels and blocks collagen 1, collagen 3, and fibronectin expression, as well as macrophage infiltration [124]. When similar tests were performed after 14 consecutive days, the miR-29 transfection similarly inhibited progressive disease. It did not modify the established lung fibrosis, which is consistent with expectations. Fibrogenetic factors, such as TGFβ1, CTGF, and phospho-Smad3, were also negatively affected by the intervention described above.
Other data considering miR-29b efficacy in vivo come from the paper from Montgomery et al. [127]. Histological specimens of BILF mice lungs after miR-29b administration presented diminished fibrotic and inflammatory reaction. IL-12, IL-4, and granulocyte colony-stimulating factor (G-CSF) levels dropped in broncho-alveolar lavage fluids (BALF) from miR-29b-treated BILF mice, along with neutrophil, lymphocyte, and macrophage counts. Once more, the alleviation of ECM production has been confirmed to some extent, even when miR-29b administration started 10 days after bleomycin supply. The researchers also confirmed the effect on Col1α1 and Col3α1 expression in vitro on human IPF fibroblasts and human AEC type II cells (A459 line) [127].
miR-155 levels in normal lung fibroblasts change with cytokine stimulation. Treatment with TNFα, interferon-γ (IFNγ), IFNβ, and poly (I:C) inducts [128] and TGFβ [129] represses its levels. The instillation of bleomycin that provokes lung fibrosis elevates miR-155 in mice lung fibroblasts, in a scale dependent on the mice strain [129]. To test the functional consequences, HLF1 cells (human lung fibroblasts) have been transfected with pre-miR-155 and control pre-miRNA, and pangenomic assays revealed changes in expression of 474 and 1404 genes at 24/48 h respectively; all of them associated with cell signaling, death, and movement. These findings were somehow confirmed via testing for the increased levels of caspase 3—possibly indicating an enhanced apoptosis—and for the elevated mobility of these cells, especially on the base of collagen 1. Last but not least, it was proved to directly alleviate IL-1β-induced keratinocyte growth factor/fibroblast growth factor 7 (KGF/FGF7) expression, which is an EMT-involved mitogen produced by mesenchymal cells [129].
miR-200c interferes with ZEB1 transcription factor to inhibit the EMT of AECs. Downregulation of miR-200c was revealed in BILF and IPF lungs [114], which is possibly via a novel lnc-RNA-ATB mechanism [23]. In recent research, miR-200c agomir attenuated silica-induced lung fibrosis in mice [23].
Hybrid Approach
Another study has investigated the role of the miR-17–92 cluster in the regulation of DNA methylation in IPF. The miR-17–92 cluster targets specific genes involved in fibrosis, including collagen, TGFβ, and metalloproteinases. The authors found that the miR-17–92 cluster has an overall reduced expression in IPF. Moreover, the promoter CpG islands of this cluster were severely methylated and there was evidence of DNMT1 overexpression, which may explain excessive methylation [130]. Mice with bleomycin-induced fibrosis were treated with 5-aza-2′-deoxycytidine and exhibited raised expression of miR-17–92, presumably via demethylation of its promoter. Treatment had not induced a clearance of collagen, but inhibited its further deposition [130].
There is evidence that miRNA biological effects are associated with the regulation of DNA methylation, e.g., miR-29c directly decreases DNMT3A and 3B levels and there was no change in Fas levels after treatment with miR-29c inhibitor in the environment containing 5-aza [125].
Another molecule, miR-30a, is decreased in lung fibrosis animal models, HFL fibroblasts after H2O2 stimulation, and IPF patients’ blood [131]. In the search for its target gene, tet methylcytosine dioxygenase 1 (TET1) seems most promising. It is a protein that converts 5-methylcytosine to 5-hydroxymethylcytosine, an epigenetic mark for actively expressed genes, and a DNA demethylation/activation promoter. In this study, an miR-30a designed agomir was sprayed across BILF mice lungs, resulting in fewer histological changes compared to the control BILF group, with fewer collagen fibers; which is suggestive of an antifibrotic effect in vivo [131]. The expression of hydroxyproline, α-SMA, and vimentin was reduced, while E-cadherin levels increased, indicating alleviation of EMT and enhancement of apoptosis. The miR-30a level was raised, while the TET1 level dropped—reversing the toxic effect of bleomycin. Other experiments suggest it can also alleviate the bleomycin-induced levels of dynamin-related protein 1 (Drp-1) that lead to epithelial cell apoptosis and which is correlated with higher p53 rates [132].
Drugs and miRNA
Arsenic trioxide was used by Chinese investigators to study its role in BILF rats. The treatment prevented pulmonary fibrosis via high expression of miR-98 and decreased the signal transducer and activator of transcription 3 (STAT3) signaling pathway, inhibiting the production of collagen 1 and hydroxyproline [133]. Another group has explored a sulindac (a nonsteroidal anti-inflammatory drug [NSAID]) mechanism in BILF. Sulindac diminished the severity of fibrosis measured by collagen deposition, thickening of the alveolar interval, and wet lung to body weight ratios. Moreover, sulindac inhibits EMT by restoring E-cadherin and SMA levels. Authors suggest that sulindac might decrease STAT3 expression with IFNγ and miR-21 involvement [134]. Wang et al. found that administration of paclitaxel induces miR-140 expression and ameliorates pulmonary fibrosis in BILF rats via EMT reduction [135]. Tectorigenin, a type of isoflavone, reduces fibroblasts’ ability to proliferate in BILF rats via increased miR-338 expression and, in turn, via downregulation of its plausible target gene—lysophosphatidic acid receptor 1 (LPA1) [136]. Another flavone that showed the antifibrotic effect is baicalein; lowered expression of miR-21 in BILF rats treated with this agent was proven [137].
Conclusions
There is a continuous deficiency of therapeutic options in fibrotic lung diseases. With regard to countering IPF, the fatal disease this paper primarily discusses, there are only two antifibrotic medications that slow down progression, but adverse effects limit their use [138]. Better understanding of the epigenetics of ILDs may potentially increase our knowledge about their natural history and, eventually, contribute to the development of new therapeutic strategies (Table 1). The use of drugs that alter oligonucleotide activity, methylation pattern, or histone structure is a tempting possibility yet to be verified in clinical trials. Several substances are already registered for malignant lesions, and they seem to be obvious candidates for further experiments, especially considering their efficacy in alleviating BILF in mice. However, the heterogeneity of methodologies used in particular studies should be considered as a major limitation and indicates a strong need for further, intensive research in this field.
Table 1.
List of the substances used in experimental studies cited in the review and their discovered in vitro and in vivo effects
| Substance | Phenotypic effect | Association | Model |
|---|---|---|---|
| DNA methylation modifiers | |||
| 5-Aza-2′-deoxycytidine and zebularine | Inhibition of collagen synthesis and cell proliferation | PGE2 | Murine BILF and human IPF fibroblasts |
| 5-Aza-2′-deoxycytidine | Reduced collagen 1α expression; increased sensitivity to FasL apoptosis | PGE2 | Human IPF and SSc fibroblasts |
| 5-Aza-2′-deoxycytidine | Alleviation of stress fiber formation and EMT in response to TGFβ1 | Thy-1 | Murine primary lung fibroblasts |
| 5-Azacytidine | Alleviation of matrix production and migration of fibroblasts | BMPER | Human IPF fibroblasts |
| 5-Azacytidine | Attenuation of matrix accumulation and lung fibrosis | BMPER | Murine BILF lungs in vivo |
| 5-Aza-2′-deoxycytidine | Reduction of collagen 1α1 and α-SMA levels; attenuation of histological alveolitis and fibrosis scores, as well as radiological density and fibrosis scores | SFRPs and Wnt pathway | Murine BILF lungs in vivo |
| AMI1 and sinefungin | Disruption of fibroblast-derived ECM architecture | Snail1 | Human IPF fibroblasts |
| Histone structure modifiers | |||
| Trichostatin A | No changes of histological phenotype | – | Human AEC type II A549 |
| Trichostatin A | Mitigation of α-SMA production in response TGFβ; less cell contraction | Akt/SMAD | Normal human lung fibroblasts |
| Spiruchostatin A | Inhibition of fibroblast proliferation | P21WAF1 | Human IPF fibroblasts |
| Spiruchostatin A | Reduced expression of α-SMA and actin filament formation | TGFβ | Human IPF fibroblasts |
| Trichostatin A | Mitigation of α-SMA production and Thy-1 expression restoration | Thy-1 | Primary rat lung fibroblasts |
| Trichostatin A | Increased susceptibility to Fas-mediated apoptosis | Fas | Murine BILF fibroblasts |
| Trichostatin A | Restoration of Fas expression | Fas | Human IPF fibroblasts |
| SAHA | Lower expression of α-SMA and collagen 1 deposition; reduced cell proliferation | TGFβ1 | Human IPF, adult and fetal fibroblasts |
| SAHA | Reduced collagen 3α1 expression | – | Human IPF fibroblasts |
| SAHA | Reduced collagen 3α1 expression, less aberrated lung structure | – | Murine BILF lungs in vivo |
| BIX01294 | Mitigation of profibrotic TGFβ actions | G9a, PGC1α | Human IPF fibroblasts |
| Schisandrin B | Reversion of EMT among lung epithelial cells | ZEB1 | Human AEC type II A549 |
| Pracinostat | Inhibition of α-SMA, fibronectin and collagen 1 production in response to TGFβ | HDAC7 | Human IPF fibroblasts |
| Engineered miRNA | |||
| agomiR-26a | Attenuation of collagen, CTGF expression and lung fibrosis | p-SMAD3 | Murine BILF lungs in vivo |
| let-7d | EMT alleviation (α-SMA and N-CAD reduction); slowing of fibroblast migration and proliferation | HMGA2, Myc, cyclin D2 | Human FLF and NHLF fetal and normal lung fibroblasts |
| Pre-miR-29c | Mitigation of ECM-related gene expression (LOXL2, COL3A1, SPARC) and Fas-related apoptosis resistance | – | Human HFL1 fetal lung fibroblasts |
| Pre-miR-29b | Alleviation of matrix production and macrophage infiltration | Smad3, CTGF | Murine BILF lungs in vivo |
| miR-29b mimic | Mitigation of lung fibrosis and tissue inflammation (reduced collagen 1α1, hydroxyproline, IL-12, IL-4, G-CSF) | IGF1 | Murine IPF fibroblasts in vivo |
| miR-29b mimic | Mitigation of collagen induction | – | Human IPF fibroblasts, A549 AEC II |
| Has-miR29c | Mitigation of ECM-related gene expression, especially COL12A1 | – | Human IPF fibroblasts |
| Pre-miR-155 | Increased apoptosis (caspase-3) and motility | – | Human HFL1 fetal lung fibroblasts |
| agomiR-30a | Fibrosis and EMT alleviation; apoptosis enhancement | TET1, Drp-1 | Murine BILF fibroblasts |
| miR-200c agomir | Decrease in α-SMA expression and EMT | ZEB1, lnc-RNA-ATB | Murine silica-induced pulmonary fibrosis fibroblasts |
| Other antifibrotic factors with known miRNA associations | |||
| 5-Aza-2′-deoxycytidine | Inhibition of collagen deposition (P > 0.05) and expression of VEGF, CTGF, DNMT1 | miR17–92 | Murine BILF lungs in vivo |
| Arsenic trioxide | Mitigation of lung fibroblasts, collagen deposition and lung density | STAT3 and miR-98 | Rats’ BILF lungs in vivo |
| Sulindac | Diminished severity of fibrosis—reduced collagen deposition and alveolar interval thickening | STAT3/IFNγ; miR-21 | Rats’ BILF lungs in vivo |
| Sulindac | EMT alleviation | STAT3/IFNγ; miR-21 | A459 AEC II |
| Paclitaxel | EMT alleviation | TGFβ1/SMAD3 and miR-140 | A459; RLE-6TN AEC II cells |
| Paclitaxel | Less interalveolar collagen 1 deposition and alveolar disruption | TGFβ1/SMAD3 and miR-140 | Rats’ BILF lungs in vivo |
| Tectorigenin | Reduction of fibroblast proliferation | LPA1; mir-338 | Rats’ BILF fibroblasts |
α-SMA α-smooth muscle actin, AEC alveolar epithelial cell, AMI1 arginine N-methyltransferase inhibitor-1, BILF bleomycin-induced lung fibrosis, BMP bone morphogenetic protein, BMPER BMP endothelial cell precursor-derived regulator, COL3A1 collagen 3α1, COL12A1 collagen 12α1, CTGF connective tissue growth factor, DNMT DNA methyltransferase, Drp-1 dynamin-related protein, ECM extra-cellular matrix, EMT epithelial-mesenchymal transition, FLF fetal lung fibroblasts, G-CSF granulocyte colony-stimulating factor, HDAC histone deacetylase, HFL1 human lung fibroblasts 1, HMGA2 high-mobility group AT-hook 2, IFNγ interferon-γ, IGF1 insulin-like growth factor 1, IL interleukin, LOXL2 lysyl oxidase homolog 2, LPA1 lysophosphatidic acid receptor 1, IPF idiopathic pulmonary fibrosis, lncRNA long non-coding RNA, N-CAD N-cadherin, NHLF normal human lung fibroblasts, PGC1α peroxisome proliferator-activated receptor gamma coactivator 1-alpha, PGE2 prostaglandin E2, SAHA suberoylanilide hydroxamic acid, SFRP secreted fizzled protein, SPARC secreted protein acidic and rich in cysteine, SSc systemic sclerosis, STAT3 signal transducer and activator of transcription 3, TET1 tet methylcytosine dioxygenase 1, TGF transforming growth factor, Thy-1 thymocyte differentiation antigen 1, VEGF vascular endothelial growth factor, ZEB1 zinc finger E-box-binding homeobox 1
Declarations
Funding
The Medical University of Lodz. Open access funding was provided through an agreement between Springer Nature and the Polish Ministry of Science and Higher Education.
Conflict of Interest
The authors declare no conflict of interest.
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Availability of Data and Material
Not applicable.
Code Availability
Not applicable.
References
- 1.Raghu G, Remy-Jardin M, Myers JL, Richeldi L, Ryerson CJ, Lederer DJ, et al. Diagnosis of idiopathic pulmonary fibrosis. An Official ATS/ERS/JRS/ALAT Clinical Practice Guideline. Am J Respir Crit Care Med. 2018;198:e44–e68. doi: 10.1164/rccm.201807-1255ST. [DOI] [PubMed] [Google Scholar]
- 2.Walsh SLF, Hansell DM. High-resolution CT of interstitial lung disease: a continuous evolution. Semin Respir Crit Care Med. 2014;35:129–144. doi: 10.1055/s-0033-1363458. [DOI] [PubMed] [Google Scholar]
- 3.Meyer KC, Raghu G, Baughman RP, Brown KK, Costabel U, du Bois RM, et al. An official American Thoracic Society clinical practice guideline: the clinical utility of bronchoalveolar lavage cellular analysis in interstitial lung disease. Am J Respir Crit Care Med. 2012;185:1004–1014. doi: 10.1164/rccm.201202-0320ST. [DOI] [PubMed] [Google Scholar]
- 4.Meyer KC. Diagnosis and management of interstitial lung disease. Transl Respir Med. 2014;2:4. doi: 10.1186/2213-0802-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gulati M. Diagnostic assessment of patients with interstitial lung disease. Prim Care Respir J J Gen Pract Airw Group. 2011;20:120–127. doi: 10.4104/pcrj.2010.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furukawa H, Oka S, Shimada K, Tsuchiya N, Tohma S. Genetics of interstitial lung disease: Vol de Nuit (Night Flight) Clin Med Insights Circ Respir Pulm Med. 2015;9:1–7. doi: 10.4137/CCRPM.S23283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang B, Jiang C, Zhang R. Epigenetics: the language of the cell? Epigenomics. 2014;6:73–88. doi: 10.2217/epi.13.72. [DOI] [PubMed] [Google Scholar]
- 8.Brzeziańska E, Dutkowska A, Antczak A. The significance of epigenetic alterations in lung carcinogenesis. Mol Biol Rep. 2013;40:309–325. doi: 10.1007/s11033-012-2063-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kabesch M. Epigenetics in asthma and allergy. Curr Opin Allergy Clin Immunol. 2014;14:62–68. doi: 10.1097/ACI.0000000000000025. [DOI] [PubMed] [Google Scholar]
- 10.Schamberger AC, Mise N, Meiners S, Eickelberg O. Epigenetic mechanisms in COPD: implications for pathogenesis and drug discovery. Expert Opin Drug Discov. 2014;9:609–628. doi: 10.1517/17460441.2014.913020. [DOI] [PubMed] [Google Scholar]
- 11.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 13.Rothbart SB, Strahl BD. Interpreting the language of histone and DNA modifications. Biochim Biophys Acta. 2014;1839:627–643. doi: 10.1016/j.bbagrm.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 28 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 15.Thomas JO, Stott K. H1 and HMGB1: modulators of chromatin structure. Biochem Soc Trans. 2012;40:341–346. doi: 10.1042/BST20120014. [DOI] [PubMed] [Google Scholar]
- 16.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 17.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Brien J, Hayder H, Zayed Y, Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol [Internet]. 2018. https://www.frontiersin.org/articles/10.3389/fendo.2018.00402/full. Accessed 03 Aug 2020. [DOI] [PMC free article] [PubMed]
- 19.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 21.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Luo G, Zhang Y, Zhang M, Zhou J, Gao W, et al. Critical effects of long non-coding RNA on fibrosis diseases. Exp Mol Med. 2018;50:e428. doi: 10.1038/emm.2017.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Li Y, Xu Q, Yao W, Wu Q, Yuan J, et al. Long non-coding RNA-ATB promotes EMT during silica-induced pulmonary fibrosis by competitively binding miR-200c. Biochim Biophys Acta Mol Basis Dis. 2018;1864:420–431. doi: 10.1016/j.bbadis.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Hadjicharalambous MR, Roux BT, Csomor E, Feghali-Bostwick CA, Murray LA, Clarke DL, et al. Long intergenic non-coding RNAs regulate human lung fibroblast function: implications for idiopathic pulmonary fibrosis. Sci Rep. 2019;9:6020. doi: 10.1038/s41598-019-42292-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanders YY, Ambalavanan N, Halloran B, Zhang X, Liu H, Crossman DK, et al. Altered DNA methylation profile in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2012;186:525–535. doi: 10.1164/rccm.201201-0077OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rabinovich EI, Kapetanaki MG, Steinfeld I, Gibson KF, Pandit KV, Yu G, et al. Global methylation patterns in idiopathic pulmonary fibrosis. PloS One. 2012;7:e33770. doi: 10.1371/journal.pone.0033770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rabinovich EI, Selman M, Kaminski N. Epigenomics of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2012;186:473–475. doi: 10.1164/rccm.201208-1350ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang SK, Scruggs AM, McEachin RC, White ES, Peters-Golden M. Lung fibroblasts from patients with idiopathic pulmonary fibrosis exhibit genome-wide differences in DNA methylation compared to fibroblasts from nonfibrotic lung. PLoS One. 2014;9:e107055. doi: 10.1371/journal.pone.0107055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang IV, Pedersen BS, Rabinovich E, Hennessy CE, Davidson EJ, Murphy E, et al. Relationship of DNA methylation and gene expression in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2014;190:1263–1272. doi: 10.1164/rccm.201408-1452OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fingerlin TE, Murphy E, Zhang W, Peljto AL, Brown KK, Steele MP, et al. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat Genet. 2013;45:613–620. doi: 10.1038/ng.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noth I, Zhang Y, Ma S-F, Flores C, Barber M, Huang Y, et al. Genetic variants associated with idiopathic pulmonary fibrosis susceptibility and mortality: a genome-wide association study. Lancet Respir Med. 2013;1:309–317. doi: 10.1016/S2213-2600(13)70045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herreman A, Hartmann D, Annaert W, Saftig P, Craessaerts K, Serneels L, et al. Presenilin 2 deficiency causes a mild pulmonary phenotype and no changes in amyloid precursor protein processing but enhances the embryonic lethal phenotype of presenilin 1 deficiency. Proc Natl Acad Sci USA. 1999;96:11872–11877. doi: 10.1073/pnas.96.21.11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Yao W, Zhang L, Bao L, Chen H, Wang D, et al. Genome-wide DNA methylation analysis in lung fibroblasts co-cultured with silica-exposed alveolar macrophages. Respir Res. 2017;18:91. doi: 10.1186/s12931-017-0576-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stempien-Otero A, Kim D-H, Davis J. Molecular networks underlying myofibroblast fate and fibrosis. J Mol Cell Cardiol. 2016;97:153–161. doi: 10.1016/j.yjmcc.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bozyk PD, Moore BB. Prostaglandin E2 and the pathogenesis of pulmonary fibrosis. Am J Respir Cell Mol Biol. 2011;45:445–452. doi: 10.1165/rcmb.2011-0025RT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang SK, Fisher AS, Scruggs AM, White ES, Hogaboam CM, Richardson BC, et al. Hypermethylation of PTGER2 confers prostaglandin E2 resistance in fibrotic fibroblasts from humans and mice. Am J Pathol. 2010;177:2245–2255. doi: 10.2353/ajpath.2010.100446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ghoshal K, Bai S. DNA methyltransferase as targets for cancer therapy. Drugs Today. 2007;43:395. doi: 10.1358/dot.2007.43.6.1062666. [DOI] [PubMed] [Google Scholar]
- 38.Estey EH. Epigenetics in clinical practice: the examples of azacitidine and decitabine in myelodysplasia and acute myeloid leukemia. Leukemia. 2013;27:1803–1812. doi: 10.1038/leu.2013.173. [DOI] [PubMed] [Google Scholar]
- 39.Evans IC, Barnes JL, Garner IM, Pearce DR, Maher TM, Shiwen X, et al. Epigenetic regulation of cyclooxygenase-2 by methylation of c8orf4 in pulmonary fibrosis. Clin Sci. 2016;130:575–586. doi: 10.1042/CS20150697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang SK, Scruggs AM, Donaghy J, McEachin RC, Fisher AS, Richardson BC, et al. Prostaglandin E2 increases fibroblast gene-specific and global DNA methylation via increased DNA methyltransferase expression. FASEB J. 2012;26:3703–3714. doi: 10.1096/fj.11-203323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hagood JS, Prabhakaran P, Kumbla P, Salazar L, MacEwen MW, Barker TH, et al. Loss of fibroblast Thy-1 expression correlates with lung fibrogenesis. Am J Pathol. 2005;167:365–379. doi: 10.1016/S0002-9440(10)62982-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanders YY, Pardo A, Selman M, Nuovo GJ, Tollefsbol TO, Siegal GP, et al. Thy-1 promoter hypermethylation: a novel epigenetic pathogenic mechanism in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2008;39:610–618. doi: 10.1165/rcmb.2007-0322OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanders YY, Cisneros J, Nuovo G, Selman M, Hagood JS. Epigenetic regulation mediates Thy-1 gene silencing and induction of the myofibroblast phenotype. Proc Am Thorac Soc. 2008;5:363–364. [Google Scholar]
- 44.Robinson CM, Neary R, Levendale A, Watson CJ, Baugh JA. Hypoxia-induced DNA hypermethylation in human pulmonary fibroblasts is associated with Thy-1 promoter methylation and the development of a pro-fibrotic phenotype. Respir Res. 2012;13:74. doi: 10.1186/1465-9921-13-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang N, Liu K, Wang K, Zhou C, Wang H, Che S, et al. Dust induces lung fibrosis through dysregulated DNA methylation. Environ Toxicol. 2019;34:728–741. doi: 10.1002/tox.22739. [DOI] [PubMed] [Google Scholar]
- 46.Neveu WA, Mills ST, Staitieh BS, Sueblinvong V. TGF-β1 epigenetically modifies Thy-1 expression in primary lung fibroblasts. Am J Physiol Cell Physiol. 2015;309:C616–C626. doi: 10.1152/ajpcell.00086.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sueblinvong V, Neujahr DC, Todd Mills S, Roser-Page S, Guidot D, Rojas M, et al. Predisposition for disrepair in the aged lung. Am J Med Sci. 2012;344:41–51. doi: 10.1097/MAJ.0b013e318234c132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu B, Gharaee-Kermani M, Wu Z, Phan SH. Epigenetic regulation of myofibroblast differentiation by DNA methylation. Am J Pathol. 2010;177:21–28. doi: 10.2353/ajpath.2010.090999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sala L, Franco-Valls H, Stanisavljevic J, Curto J, Vergés J, Peña R, et al. Abrogation of myofibroblast activities in metastasis and fibrosis by methyltransferase inhibition. Int J Cancer. 2019;145:3064–3077. doi: 10.1002/ijc.32376. [DOI] [PubMed] [Google Scholar]
- 50.Koli K, Myllärniemi M, Vuorinen K, Salmenkivi K, Ryynänen MJ, Kinnula VL, et al. Bone morphogenetic protein-4 inhibitor gremlin is overexpressed in idiopathic pulmonary fibrosis. Am J Pathol. 2006;169:61–71. doi: 10.2353/ajpath.2006.051263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Masterson JC, Molloy EL, Gilbert JL, McCormack N, Adams A, O’Dea S. Bone morphogenetic protein signalling in airway epithelial cells during regeneration. Cell Signal. 2011;23:398–406. doi: 10.1016/j.cellsig.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 52.Rosendahl A, Pardali E, Speletas M, ten Dijke P, Heldin C-H, Sideras P. Activation of bone morphogenetic protein/Smad signaling in bronchial epithelial cells during airway inflammation. Am J Respir Cell Mol Biol. 2002;27:160–169. doi: 10.1165/ajrcmb.27.2.4779. [DOI] [PubMed] [Google Scholar]
- 53.Huan C, Yang T, Liang J, Xie T, Cheng L, Liu N, et al. Methylation-mediated BMPER expression in fibroblast activation in vitro and lung fibrosis in mice in vivo. Sci Rep. 2015;5:14910. doi: 10.1038/srep14910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meneghin A, Choi ES, Evanoff HL, Kunkel SL, Martinez FJ, Flaherty KR, et al. TLR9 is expressed in idiopathic interstitial pneumonia and its activation promotes in vitro myofibroblast differentiation. Histochem Cell Biol. 2008;130:979–992. doi: 10.1007/s00418-008-0466-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hogaboam CM, Murray L, Martinez FJ. Epigenetic mechanisms through which Toll-like receptor-9 drives idiopathic pulmonary fibrosis progression. Proc Am Thorac Soc. 2012;9:172–176. doi: 10.1513/pats.201201-002AW. [DOI] [PubMed] [Google Scholar]
- 56.Hu B, Wu Z, Hergert P, Henke CA, Bitterman PB, Phan SH. Regulation of myofibroblast differentiation by poly(ADP-ribose) polymerase 1. Am J Pathol. 2013;182:71–83. doi: 10.1016/j.ajpath.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiang Z, Zhou Q, Hu M, Sanders YY. MeCP2 epigenetically regulates alpha-smooth muscle actin in human lung fibroblasts. J Cell Biochem. 2020;121:3616–3625. doi: 10.1002/jcb.29655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu B, Gharaee-Kermani M, Wu Z, Phan SH. Essential role of MeCP2 in the regulation of myofibroblast differentiation during pulmonary fibrosis. Am J Pathol. 2011;178:1500–1508. doi: 10.1016/j.ajpath.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baarsma HA, Königshoff M. ‘WNT-er is coming’: WNT signalling in chronic lung diseases. Thorax. 2017;72:746–759. doi: 10.1136/thoraxjnl-2016-209753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou J, Yi Z, Fu Q. Dynamic decreased expression and hypermethylation of secreted frizzled-related protein 1 and 4 over the course of pulmonary fibrosis in mice. Life Sci. 2019;218:241–252. doi: 10.1016/j.lfs.2018.12.041. [DOI] [PubMed] [Google Scholar]
- 61.Scruggs AM, Koh HB, Tripathi P, Leeper NJ, White ES, Huang SK. Loss of CDKN2B promotes fibrosis via increased fibroblast differentiation rather than proliferation. Am J Respir Cell Mol Biol. 2018;59:200–214. doi: 10.1165/rcmb.2017-0298OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maher TM, Evans IC, Bottoms SE, Mercer PF, Thorley AJ, Nicholson AG, et al. Diminished prostaglandin E2 contributes to the apoptosis paradox in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2010;182:73–82. doi: 10.1164/rccm.200905-0674OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mitaka K, Miyazaki Y, Yasui M, Furuie M, Miyake S, Inase N, et al. Th2-biased immune responses are important in a murine model of chronic hypersensitivity pneumonitis. Int Arch Allergy Immunol. 2011;154:264–274. doi: 10.1159/000321114. [DOI] [PubMed] [Google Scholar]
- 64.Wu C, Luo Z, Pang B, Wang W, Deng M, Jin R, et al. Associations of pulmonary fibrosis with peripheral blood Th1/Th2 cell imbalance and EBF3 gene methylation in Uygur Pigeon Breeder’s lung patients. Cell Physiol Biochem. 2018;47:1141–1151. doi: 10.1159/000490208. [DOI] [PubMed] [Google Scholar]
- 65.Cisneros J, Hagood J, Checa M, Ortiz-Quintero B, Negreros M, Herrera I, et al. Hypermethylation-mediated silencing of p14(ARF) in fibroblasts from idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2012;303:L295–L303. doi: 10.1152/ajplung.00332.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Noguchi S, Eitoku M, Moriya S, Kondo S, Kiyosawa H, Watanabe T, et al. Regulation of gene expression by sodium valproate in epithelial-to-mesenchymal transition. Lung. 2015;193:691–700. doi: 10.1007/s00408-015-9776-9. [DOI] [PubMed] [Google Scholar]
- 67.Ota C, Yamada M, Fujino N, Motohashi H, Tando Y, Takei Y, et al. Histone deacetylase inhibitor restores surfactant protein-C expression in alveolar-epithelial type II cells and attenuates bleomycin-induced pulmonary fibrosis in vivo. Exp Lung Res. 2015;41:422–434. doi: 10.3109/01902148.2015.1060275. [DOI] [PubMed] [Google Scholar]
- 68.Guo W, Shan B, Klingsberg RC, Qin X, Lasky JA. Abrogation of TGF-beta1-induced fibroblast–myofibroblast differentiation by histone deacetylase inhibition. Am J Physiol Lung Cell Mol Physiol. 2009;297:L864–L870. doi: 10.1152/ajplung.00128.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhuang W, Li Z, Dong X, Zhao N, Liu Y, Wang C, et al. Schisandrin B inhibits TGF-β1-induced epithelial-mesenchymal transition in human A549 cells through epigenetic silencing of ZEB1. Exp Lung Res. 2019;45:157–166. doi: 10.1080/01902148.2019.1631906. [DOI] [PubMed] [Google Scholar]
- 70.Ohira T, Gemmill RM, Ferguson K, Kusy S, Roche J, Brambilla E, et al. WNT7a induces E-cadherin in lung cancer cells. Proc Natl Acad Sci. 2003;100:10429–10434. doi: 10.1073/pnas.1734137100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davies ER, Haitchi HM, Thatcher TH, Sime PJ, Kottmann RM, Ganesan A, et al. Spiruchostatin A inhibits proliferation and differentiation of fibroblasts from patients with pulmonary fibrosis. Am J Respir Cell Mol Biol. 2012;46:687–694. doi: 10.1165/rcmb.2011-0040OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pan X, Li J, Tu X, Wu C, Liu H, Luo Y, et al. Lysine-specific demethylase-1 regulates fibroblast activation in pulmonary fibrosis via TGF-β1/Smad3 pathway. Pharmacol Res. 2020;152:104592. doi: 10.1016/j.phrs.2019.104592. [DOI] [PubMed] [Google Scholar]
- 73.He Z, Wang X, Deng Y, Li W, Chen Y, Xing S, et al. Epigenetic regulation of Thy-1 gene expression by histone modification is involved in lipopolysaccharide-induced lung fibroblast proliferation. J Cell Mol Med. 2013;17:160–167. doi: 10.1111/j.1582-4934.2012.01659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sanders YY, Tollefsbol TO, Varisco BM, Hagood JS. Epigenetic regulation of thy-1 by histone deacetylase inhibitor in rat lung fibroblasts. Am J Respir Cell Mol Biol. 2011;45:16–23. doi: 10.1165/rcmb.2010-0154OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang SK, Scruggs AM, Donaghy J, Horowitz JC, Zaslona Z, Przybranowski S, et al. Histone modifications are responsible for decreased Fas expression and apoptosis resistance in fibrotic lung fibroblasts. Cell Death Dis. 2013;4:e621. doi: 10.1038/cddis.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sanders YY, Liu H, Scruggs AM, Duncan SR, Huang SK, Thannickal VJ. Epigenetic regulation of caveolin-1 gene expression in lung fibroblasts. Am J Respir Cell Mol Biol. 2017;56:50–61. doi: 10.1165/rcmb.2016-0034OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Z, Chen C, Finger SN, Kwajah S, Jung M, Schwarz H, et al. Suberoylanilide hydroxamic acid: a potential epigenetic therapeutic agent for lung fibrosis? Eur Respir J. 2009;34:145–155. doi: 10.1183/09031936.00084808. [DOI] [PubMed] [Google Scholar]
- 78.Zhang X, Liu H, Hock T, Thannickal VJ, Sanders YY. Histone deacetylase inhibition downregulates collagen 3A1 in fibrotic lung fibroblasts. Int J Mol Sci. 2013;14:19605–19617. doi: 10.3390/ijms141019605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sanders YY, Hagood JS, Liu H, Zhang W, Ambalavanan N, Thannickal VJ. Histone deacetylase inhibition promotes fibroblast apoptosis and ameliorates pulmonary fibrosis in mice. Eur Respir J. 2014;43:1448–1458. doi: 10.1183/09031936.00095113. [DOI] [PubMed] [Google Scholar]
- 80.Bai L, Bernard K, Tang X, Hu M, Horowitz JC, Thannickal VJ, et al. Glutaminolysis epigenetically regulates antiapoptotic gene expression in idiopathic pulmonary fibrosis fibroblasts. Am J Respir Cell Mol Biol. 2019;60:49–57. doi: 10.1165/rcmb.2018-0180OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Choudhury M, Yin X, Schaefbauer KJ, Kang J-H, Roy B, Kottom TJ, et al. SIRT7-mediated modulation of glutaminase 1 regulates TGF-β-induced pulmonary fibrosis. FASEB J. 2020;34:8920–8940. doi: 10.1096/fj.202000564R. [DOI] [PubMed] [Google Scholar]
- 82.Coward WR, Watts K, Feghali-Bostwick CA, Knox A, Pang L. Defective histone acetylation is responsible for the diminished expression of cyclooxygenase 2 in idiopathic pulmonary fibrosis. Mol Cell Biol. 2009;29:4325–4339. doi: 10.1128/MCB.01776-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Coward WR, Watts K, Feghali-Bostwick CA, Jenkins G, Pang L. Repression of IP-10 by interactions between histone deacetylation and hypermethylation in idiopathic pulmonary fibrosis. Mol Cell Biol. 2010;30:2874–2886. doi: 10.1128/MCB.01527-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Coward WR, Brand OJ, Pasini A, Jenkins G, Knox AJ, Pang L. Interplay between EZH2 and G9a regulates CXCL10 gene repression in idiopathic pulmonary fibrosis. Am J Respir Cell Mol Biol. 2018;58:449–460. doi: 10.1165/rcmb.2017-0286OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Coward WR, Feghali-Bostwick CA, Jenkins G, Knox AJ, Pang L. A central role for G9a and EZH2 in the epigenetic silencing of cyclooxygenase-2 in idiopathic pulmonary fibrosis. FASEB J Off Publ Fed Am Soc Exp Biol. 2014;28:3183–3196. doi: 10.1096/fj.13-241760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pasini A, Brand OJ, Jenkins G, Knox AJ, Pang L. Suberanilohydroxamic acid prevents TGF-β1-induced COX-2 repression in human lung fibroblasts post-transcriptionally by TIA-1 downregulation. Biochim Biophys Acta. 2018;1861:463–472. doi: 10.1016/j.bbagrm.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ligresti G, Caporarello N, Meridew JA, Jones DL, Tan Q, Choi KM, et al. CBX5/G9a/H3K9me-mediated gene repression is essential to fibroblast activation during lung fibrosis. JCI Insight. 2019;5:12. doi: 10.1172/jci.insight.127111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eissenberg JC, Elgin SCR. HP1a: a structural chromosomal protein regulating transcription. Trends Genet. 2014;30:103–110. doi: 10.1016/j.tig.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mora AL, Bueno M, Rojas M. Mitochondria in the spotlight of aging and idiopathic pulmonary fibrosis. J Clin Investig. 2017;127:405–414. doi: 10.1172/JCI87440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu G, Tzouvelekis A, Wang R, Herazo-Maya JD, Ibarra GH, Srivastava A, et al. Thyroid hormone inhibits lung fibrosis in mice by improving epithelial mitochondrial function. Nat Med. 2018;24:39–49. doi: 10.1038/nm.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jones DL, Haak AJ, Caporarello N, Choi KM, Ye Z, Yan H, et al. TGFβ-induced fibroblast activation requires persistent and targeted HDAC-mediated gene repression. J Cell Sci. https://jcs.biologists.org/content/132/20/jcs233486. [DOI] [PMC free article] [PubMed]
- 92.Xie T, Liang J, Guo R, Liu N, Noble PW, Jiang D. Comprehensive microRNA analysis in bleomycin-induced pulmonary fibrosis identifies multiple sites of molecular regulation. Physiol Genom. 2011;43:479–487. doi: 10.1152/physiolgenomics.00222.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Honeyman L, Bazett M, Tomko TG, Haston CK. MicroRNA profiling implicates the insulin-like growth factor pathway in bleomycin-induced pulmonary fibrosis in mice. Fibrogenesis Tissue Repair. 2013;6:16. doi: 10.1186/1755-1536-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang G, Yang L, Wang W, Wang J, Wang J, Xu Z. Discovery and validation of extracellular/circulating microRNAs during idiopathic pulmonary fibrosis disease progression. Gene. 2015;562:138–144. doi: 10.1016/j.gene.2015.02.065. [DOI] [PubMed] [Google Scholar]
- 95.Oak SR, Murray L, Herath A, Sleeman M, Anderson I, Joshi AD, et al. A micro RNA processing defect in rapidly progressing idiopathic pulmonary fibrosis. PLoS One. 2011;6:e21253. doi: 10.1371/journal.pone.0021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Crouser ED, Julian MW, Crawford M, Shao G, Yu L, Planck SR, et al. Differential expression of microRNA and predicted targets in pulmonary sarcoidosis. Biochem Biophys Res Commun. 2012;417:886–891. doi: 10.1016/j.bbrc.2011.12.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, et al. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 2010;207:1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yamada M, Kubo H, Ota C, Takahashi T, Tando Y, Suzuki T, et al. The increase of microRNA-21 during lung fibrosis and its contribution to epithelial–mesenchymal transition in pulmonary epithelial cells. Respir Res. 2013;14:95. doi: 10.1186/1465-9921-14-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang S, Cui H, Xie N, Icyuz M, Banerjee S, Antony VB, et al. miR-145 regulates myofibroblast differentiation and lung fibrosis. FASEB J Off Publ Fed Am Soc Exp Biol. 2013;27:2382–2391. doi: 10.1096/fj.12-219493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xiao X, Huang C, Zhao C, Gou X, Senavirathna LK, Hinsdale M, et al. Regulation of myofibroblast differentiation by miR-424 during epithelial-to-mesenchymal transition. Arch Biochem Biophys. 2015;566:49–57. doi: 10.1016/j.abb.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nho RS, Im J, Ho Y-Y, Hergert P. MicroRNA-96 inhibits FoxO3a function in IPF fibroblasts on type I collagen matrix. Am J Physiol Lung Cell Mol Physiol. 2014;307:L632–L642. doi: 10.1152/ajplung.00127.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Samuel CS, Zhao C, Bathgate RAD, Bond CP, Burton MD, Parry LJ, et al. Relaxin deficiency in mice is associated with an age-related progression of pulmonary fibrosis. FASEB J. 2003;17:121–123. doi: 10.1096/fj.02-0449fje. [DOI] [PubMed] [Google Scholar]
- 103.Tan J, Tedrow JR, Dutta JA, Juan-Guardela B, Nouraie M, Chu Y, et al. Expression of RXFP1 is decreased in idiopathic pulmonary fibrosis implications for relaxin-based therapies. Am J Respir Crit Care Med. 2016;194:1392–1402. doi: 10.1164/rccm.201509-1865OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bahudhanapati H, Tan J, Dutta JA, Strock SB, Sembrat J, Àlvarez D, et al. MicroRNA-144-3p targets relaxin/insulin-like family peptide receptor 1 (RXFP1) expression in lung fibroblasts from patients with idiopathic pulmonary fibrosis. J Biol Chem. 2019;294:5008–5022. doi: 10.1074/jbc.RA118.004910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Milosevic J, Pandit K, Magister M, Rabinovich E, Ellwanger DC, Yu G, et al. Profibrotic role of miR-154 in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2012;47:879–887. doi: 10.1165/rcmb.2011-0377OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cho J-H, Gelinas R, Wang K, Etheridge A, Piper MG, Batte K, et al. Systems biology of interstitial lung diseases: integration of mRNA and microRNA expression changes. BMC Med Genom. 2011;4:8. doi: 10.1186/1755-8794-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 107.Lino Cardenas CL, Henaoui IS, Courcot E, Roderburg C, Cauffiez C, Aubert S, et al. miR-199a-5p Is upregulated during fibrogenic response to tissue injury and mediates TGFbeta-induced lung fibroblast activation by targeting caveolin-1. PLoS Genet. 2013;9:e1003291. doi: 10.1371/journal.pgen.1003291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bodempudi V, Hergert P, Smith K, Xia H, Herrera J, Peterson M, et al. miR-210 promotes IPF fibroblast proliferation in response to hypoxia. Am J Physiol Lung Cell Mol Physiol. 2014;307:L283–L294. doi: 10.1152/ajplung.00069.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li P, Li J, Chen T, Wang H, Chu H, Chang J, et al. Expression analysis of serum microRNAs in idiopathic pulmonary fibrosis. Int J Mol Med. 2014;33:1554–1562. doi: 10.3892/ijmm.2014.1712. [DOI] [PubMed] [Google Scholar]
- 110.Liang H, Xu C, Pan Z, Zhang Y, Xu Z, Chen Y, et al. The antifibrotic effects and mechanisms of microRNA-26a action in idiopathic pulmonary fibrosis. Mol Ther J Am Soc Gene Ther. 2014;22:1122–1133. doi: 10.1038/mt.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li X, Liu L, Shen Y, Wang T, Chen L, Xu D, et al. MicroRNA-26a modulates transforming growth factor beta-1-induced proliferation in human fetal lung fibroblasts. Biochem Biophys Res Commun. 2014;454:512–517. doi: 10.1016/j.bbrc.2014.10.106. [DOI] [PubMed] [Google Scholar]
- 112.Liang H, Gu Y, Li T, Zhang Y, Huangfu L, Hu M, et al. Integrated analyses identify the involvement of microRNA-26a in epithelial–mesenchymal transition during idiopathic pulmonary fibrosis. Cell Death Dis. 2014;5:e1238. doi: 10.1038/cddis.2014.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Graham JR, Williams CMM, Yang Z. MicroRNA-27b targets Gremlin 1 to modulate fibrotic responses in pulmonary cells. J Cell Biochem. 2014;115:1539–1548. doi: 10.1002/jcb.24809. [DOI] [PubMed] [Google Scholar]
- 114.Yang S, Banerjee S, de Freitas A, Sanders YY, Ding Q, Matalon S, et al. Participation of miR-200 in pulmonary fibrosis. Am J Pathol. 2012;180:484–493. doi: 10.1016/j.ajpath.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Das S, Kumar M, Negi V, Pattnaik B, Prakash YS, Agrawal A, et al. MicroRNA-326 regulates profibrotic functions of transforming growth factor-β in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2014;50:882–892. doi: 10.1165/rcmb.2013-0195OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang Y, Huang C, Reddy Chintagari N, Bhaskaran M, Weng T, Guo Y, et al. miR-375 regulates rat alveolar epithelial cell trans-differentiation by inhibiting Wnt/β-catenin pathway. Nucleic Acids Res. 2013;41:3833–3844. doi: 10.1093/nar/gks1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Blaauboer ME, Emson CL, Verschuren L, van Erk M, Turner SM, Everts V, et al. Novel combination of collagen dynamics analysis and transcriptional profiling reveals fibrosis-relevant genes and pathways. Matrix Biol. 2013;32:424–431. doi: 10.1016/j.matbio.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 118.Rubio K, Singh I, Dobersch S, Sarvari P, Günther S, Cordero J, et al. Inactivation of nuclear histone deacetylases by EP300 disrupts the MiCEE complex in idiopathic pulmonary fibrosis. Nat Commun [Internet]. 2019. [cited 2020 Jul 16];10. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6527704/. [DOI] [PMC free article] [PubMed]
- 119.Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, Gibson KF, et al. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2010;182:220–229. doi: 10.1164/rccm.200911-1698OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Huleihel L, Ben-Yehudah A, Milosevic J, Yu G, Pandit K, Sakamoto K, et al. Let-7d microRNA affects mesenchymal phenotypic properties of lung fibroblasts. Am J Physiol Lung Cell Mol Physiol. 2014;306:L534–L542. doi: 10.1152/ajplung.00149.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, et al. miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol. 2011;45:287–294. doi: 10.1165/rcmb.2010-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cushing L, Kuang P, Lü J. The role of miR-29 in pulmonary fibrosis. Biochem Cell Biol. 2015;93:109–118. doi: 10.1139/bcb-2014-0095. [DOI] [PubMed] [Google Scholar]
- 123.Yang T, Liang Y, Lin Q, Liu J, Luo F, Li X, et al. miR-29 mediates TGFβ1-induced extracellular matrix synthesis through activation of PI3K-AKT pathway in human lung fibroblasts. J Cell Biochem. 2013;114:1336–1342. doi: 10.1002/jcb.24474. [DOI] [PubMed] [Google Scholar]
- 124.Xiao J, Meng X-M, Huang XR, Chung AC, Feng Y-L, Hui DS, et al. miR-29 inhibits bleomycin-induced pulmonary fibrosis in mice. Mol Ther J Am Soc Gene Ther. 2012;20:1251–1260. doi: 10.1038/mt.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Matsushima S, Ishiyama J. MicroRNA-29c regulates apoptosis sensitivity via modulation of the cell-surface death receptor, Fas, in lung fibroblasts. Am J Physiol Lung Cell Mol Physiol. 2016;311:L1050–L1061. doi: 10.1152/ajplung.00252.2016. [DOI] [PubMed] [Google Scholar]
- 126.Parker MW, Rossi D, Peterson M, Smith K, Sikström K, White ES, et al. Fibrotic extracellular matrix activates a profibrotic positive feedback loop. J Clin Investig. 2014;124:1622–1635. doi: 10.1172/JCI71386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Montgomery RL, Yu G, Latimer PA, Stack C, Robinson K, Dalby CM, et al. MicroRNA mimicry blocks pulmonary fibrosis. EMBO Mol Med. 2014;6:1347–1356. doi: 10.15252/emmm.201303604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pottier N, Maurin T, Chevalier B, Puisségur M-P, Lebrigand K, Robbe-Sermesant K, et al. Identification of keratinocyte growth factor as a target of microRNA-155 in lung fibroblasts: implication in epithelial–mesenchymal interactions. PLoS One. 2009;4:e6718. doi: 10.1371/journal.pone.0006718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dakhlallah D, Batte K, Wang Y, Cantemir-Stone CZ, Yan P, Nuovo G, et al. Epigenetic regulation of miR-17–92 contributes to the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med. 2013;187:397–405. doi: 10.1164/rccm.201205-0888OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang S, Liu H, Liu Y, Zhang J, Li H, Liu W, et al. miR-30a as potential therapeutics by targeting TET1 through regulation of Drp-1 promoter hydroxymethylation in idiopathic pulmonary fibrosis. Int J Mol Sci [Internet]. 2017 [cited 2020 May 20];18. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5372646/. [DOI] [PMC free article] [PubMed]
- 132.Mao C, Zhang J, Lin S, Jing L, Xiang J, Wang M, et al. MiRNA-30a inhibits AECs-II apoptosis by blocking mitochondrial fission dependent on Drp-1. J Cell Mol Med. 2014;18:2404–2416. doi: 10.1111/jcmm.12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gao S-Y, Zhou X, Li Y-J, Liu W-L, Wang P-Y, Pang M, et al. Arsenic trioxide prevents rat pulmonary fibrosis via miR-98 overexpression. Life Sci. 2014;114:20–28. doi: 10.1016/j.lfs.2014.07.037. [DOI] [PubMed] [Google Scholar]
- 134.Zhou X, Li Y-J, Gao S-Y, Wang X-Z, Wang P-Y, Yan Y-F, et al. Sulindac has strong antifibrotic effects by suppressing STAT3-related miR-21. J Cell Mol Med. 2015;19:1103–1113. doi: 10.1111/jcmm.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang C, Song X, Li Y, Han F, Gao S, Wang X, et al. Low-dose paclitaxel ameliorates pulmonary fibrosis by suppressing TGF-β1/Smad3 pathway via miR-140 upregulation. PLoS One [Internet]. 2013 [cited 2015 Nov 12];8. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3744547/. [DOI] [PMC free article] [PubMed]
- 136.Zhang H, Liu X, Chen S, Wu J, Ye X, Xu L, et al. Tectorigenin inhibits the in vitro proliferation and enhances miR-338* expression of pulmonary fibroblasts in rats with idiopathic pulmonary fibrosis. J Ethnopharmacol. 2010;131:165–173. doi: 10.1016/j.jep.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 137.Gao Y, Lu J, Zhang Y, Chen Y, Gu Z, Jiang X. Baicalein attenuates bleomycin-induced pulmonary fibrosis in rats through inhibition of miR-21. Pulm Pharmacol Ther. 2013;26:649–654. doi: 10.1016/j.pupt.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 138.Spagnolo P, Tzouvelekis A, Bonella F. The management of patients with idiopathic pulmonary fibrosis. Front Med [Internet]. 2018 [cited 2020 Jul 17];5. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6036121/. [DOI] [PMC free article] [PubMed]