Abstract
Varicella-zoster virus (VZV) is a medically important human herpesvirus that causes chickenpox and shingles, but its cell-associated nature has hindered structure studies. Here we report the cryo-electron microscopy structures of purified VZV A-capsid and C-capsid, as well as of the DNA-containing capsid inside the virion. Atomic models derived from these structures show that, despite enclosing a genome that is substantially smaller than those of other human herpesviruses, VZV has a similarly sized capsid, consisting of 955 major capsid protein (MCP), 900 small capsid protein (SCP), 640 triplex dimer (Tri2) and 320 triplex monomer (Tri1) subunits. The VZV capsid has high thermal stability, although with relatively fewer intra- and inter-capsid protein interactions and less stably associated tegument proteins compared with other human herpesviruses. Analysis with antibodies targeting the N and C termini of the VZV SCP indicates that the hexon-capping SCP—the largest among human herpesviruses—uses its N-terminal half to bridge hexon MCP subunits and possesses a C-terminal flexible half emanating from the inner rim of the upper hexon channel into the tegument layer. Correlation of these structural features and functional observations provide insights into VZV assembly and pathogenesis and should help efforts to engineer gene delivery and anticancer vectors based on the currently available VZV vaccine.
Varicella-zoster virus (VZV) is a ubiquitous and highly contagious human α-herpesvirus. Upon primary infection, VZV causes chickenpox (varicella) in susceptible individuals and establishes lifelong latency in sensory ganglia1–3. Chickenpox exerts a considerable disease burden worldwide, with an estimated annual minimum of 140 million new cases, 4.2 million severe complications leading to hospitalization, and 4,200 deaths4. VZV reactivates from latency in almost one-third of infected individuals later in life to cause shingles (herpes zoster), which is often characterized by severe pain and followed by post-herpetic neuralgia, substantially affecting patients’ quality of life5–7. Both chickenpox and shingles can lead to severe life-threatening complications7–10. VZV is the only human herpesvirus for which a licensed live attenuated vaccine, vOka, has been developed and widely administrated in several countries for decades11. VZV has also been used as an experimental vaccine vector against various pathogens12, including human immunodeficiency virus (HIV)13–16, and for the treatment of cancer17,18.
VZV has a double-stranded DNA (dsDNA) genome with a size of approximately 125 kb, which is the smallest among all known human herpesviruses and is about half that of human cytomegalovirus (HCMV). VZV is known for its extremely low titre in cultured cells and its cell-associated nature—that is, lack of secretion of mature virions to the culture supernatant19–22. These characteristics of VZV have been attributed to the diversion of newly formed virions to late endosomal or pre-lysosomal compartments, where they are degraded before exocytosis, producing a large proportion of coreless and pleomorphic-enveloped particles22,23. Therefore, growing and isolating mature VZV virions for high-resolution structural studies is challenging and, to our knowledge, no structure information has been reported for VZV particles. This is in contrast to the recently reported near-atomic structures of other members of the human herpesvirus family, including the α-herpesviruses herpes simplex virus type 1 (HSV-1)24,25 and HSV-226,27, the β-herpesviruses HCMV28 and human herpesvirus 6B (HHV-6B)29, and the γ-herpesvirus Kaposi’s sarcoma-associated herpesvirus (KSHV)30,31.
In this study, we used a sub-particle reconstruction method to work with minimally purified VZV sample and determined the structures of VZV capsids at near-atomic resolution by cryo-electron microscopy (cryo-EM). Atomic models built on the basis of these structures show that, compared with other human herpesviruses, VZV has a similarly sized capsid consisting of herpesvirus-conserved subunits, but with fewer intra- and inter-capsid protein interactions and less stably associated tegument proteins. VZV-specific structural features have been characterized and should offer insights into the mechanism of VZV capsid assembly and pathogenesis.
Results
Characterization and overall structure of VZV A- and C-capsids.
Uniquely among human herpesviruses, VZV is highly cell-associated and grows to very low titre in cell culture, presenting a major challenge for obtaining large amounts of mature virions for structural studies. We purified VZV capsids from whole-cell lysates of ARPE-19 cells infected by VZV (parental Oka (P-Oka) strain) (see a thin section examined by transmission electron microscopy in Extended Data Fig. 1a–c) in the presence of 1% Triton X-100 and 1 M KCl by sucrose-cushion ultracentrifugation. The purified capsids primarily consisted of naked A-capsids (empty particles, about 94%) and C-capsids (containing the viral genome, about 6%), and B-capsids (with a scaffold core) were barely observed (Extended Data Fig. 1d). From these particles, we determined the three-dimensional (3D) icosahedral reconstructions of the A- and C-capsids at resolutions of 4.3 and 5.3 Å, respectively. These reconstructions show nearly identical structures of the capsids (Fig. 1a,b, Supplementary Table 1 and Supplementary Video 1), but differ in the inner densities (Fig. 1c). Unexpectedly, the capsid-associated tegument complex (CATC) is absent on both A- and C-capsids, an observation confirmed by both structural (Fig. 1a,b) and biochemical (Extended Data Fig. 2) analyses.
Fig. 1 |. Cryo-EM reconstructions and atomic modelling of the VZV A- and C-capsids.
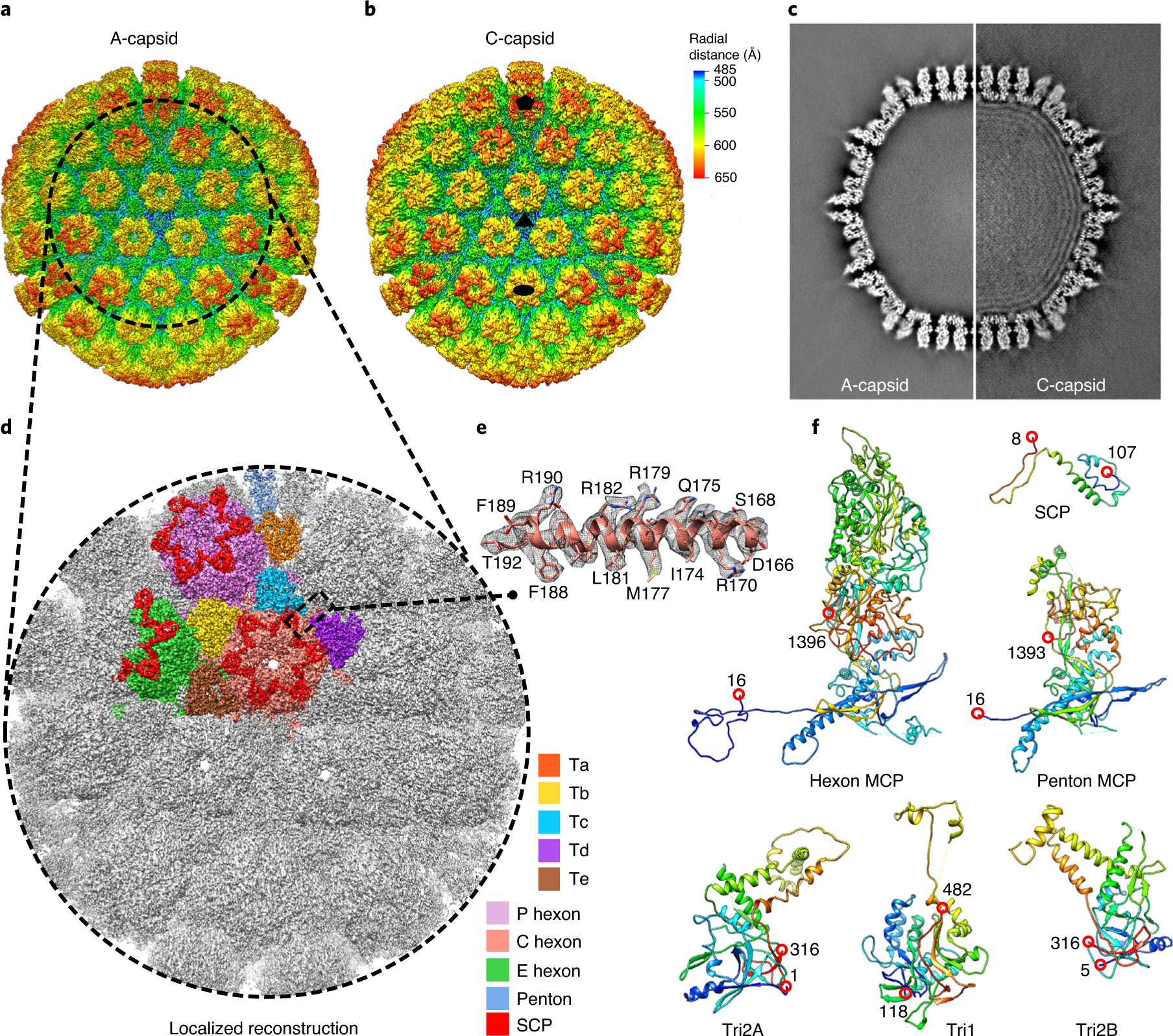
a,b, Radially coloured VZV A-capsid (a) and C-capsid (b) reconstructions at resolutions of 4.3 and 5.3 Å, respectively. Maps are viewed along a three-fold symmetry axis. The five-, three- and two-fold axes are denoted in the C-capsid reconstruction by a black pentagon, triangle and oval, respectively. c, Central slices of the VZV A-capsid (left) and C-capsid (right). The C-capsid slice shows interlayer dsDNA densities within the capsid shell. d,e, Localized reconstruction at 3.5 Å resolution of three-fold sub-particles in A-capsids (d) and a close-up view of the density map (grey mesh) of an α-helix in the MCP floor region, superposed with its model (e). f, Models of individual capsid proteins in ribbon representation in rainbow, from the N terminus (blue) to the C terminus (red).
To improve resolution, a sub-particle reconstruction strategy was used24,26,32. In brief, sub-particles of capsid areas around the three-fold symmetry axis were re-extracted and corrected for local defocus. Symmetry expansion and refinement of contrast transfer function were then applied to sub-particles for final reconstructions. The resolutions of sub-particle reconstructions of A-capsid and C-capsid were improved to 3.5 and 4.1 Å, respectively (Fig. 1d, Extended Data Fig. 3 and Supplementary Table 1). High-resolution features resolved in our cryo-EM maps, such as the polypeptide backbone and amino acid side chains (Fig. 1e and Extended Data Figs. 4 and 5), enabled us to build atomic models for both capsid types (Extended Data Fig. 6). Within an asymmetric unit, our models include 15 conformers of the MCP pORF40 in 2.5 hexons, one penton MCP, 15 conformers of the SCP pORF23 atop the 2.5 hexons, and 5 heterotrimeric triplexes (Ta, Tb, Tc, Td, and Te), each consisting of two conformers of the Tri2 pORF41 and one of the Tri1 pORF20 (Fig. 1f). The asymmetric unit also contains one-third of triplex Tf at the location of the three-fold symmetry axis, but the details of Tf were smeared by icosahedral averaging, precluding atomic modelling. The upper domain of the penton MCP was only resolved at a lower resolution, probably due to flexibility, and was not modelled (Fig. 1f). The models of asymmetric units of A- and C-capsids are similar, as indicated by their root mean square deviation of 0.76 Å over 25,884 aligned Cα positions of the two models (Extended Data Fig. 6).
Structure of the MCP.
The 1,396-residue MCP constitutes the penton and, together with SCP, all the hexons of VZV. Our model of the hexon MCP can be partitioned into seven domains following the nomenclature of HCMV28: upper (residues 495 to 1065), channel (424 to 494 and 1342 to 1380), buttress (1123 to 1351), helix–hairpin (205 to 241), dimerization (304 to 389), N-lasso (1 to 70) and Johnson fold (71 to 194, 238 to 294, 372 to 406 and 1048 to 1122) (Fig. 2a and Supplementary Video 2). The most prominent difference between VZV and other herpesviruses in hexon MCP is its flexibility at the dimerization domain (residues 349 to 374). Furthermore, the penton MCP of VZV is highly flexible for its upper and N-lasso domains (Fig. 2b). The VZV hexon has a larger internal diameter of ~17 Å (Fig. 2c) compared with those in other herpesviruses (~10 to 12 Å)24,26,28. Similar to other herpesviruses24,26,28,31,33, three types of MCP–MCP interactions (type I, II and III interactions) on the VZV capsid floor form a hierarchical level of network interactions (Extended Data Fig. 7) and a noncovalent chainmail. The interactions between hexon MCPs are similar to those in HSV-1 capsids. However, our structure shows that the penton–hexon interactions of VZV are much less extensive compared with those of HSV (strong helix–helical interactions)24, HCMV (buttress support reinforcement)28 and KSHV (six-stranded β-sheet interactions and buttress domain related interactions)31 (Fig. 2d,e). The helix pair interactions in HSV-1 have been suggested to stabilize pentons and serve to buttress the pentons against the internal pressure of packaged DNA24. The high flexibility in the dimerization domain and consequently, the lack of helical interactions (or type II and type III interactions), at the floor of VZV pentons probably accounts for their structural instability, and the relatively less stable binding of CATCs on the penton vertexes of VZV capsids (see below).
Fig. 2 |. MCP organization and pentonal MCP–hexonal MCP interactions.
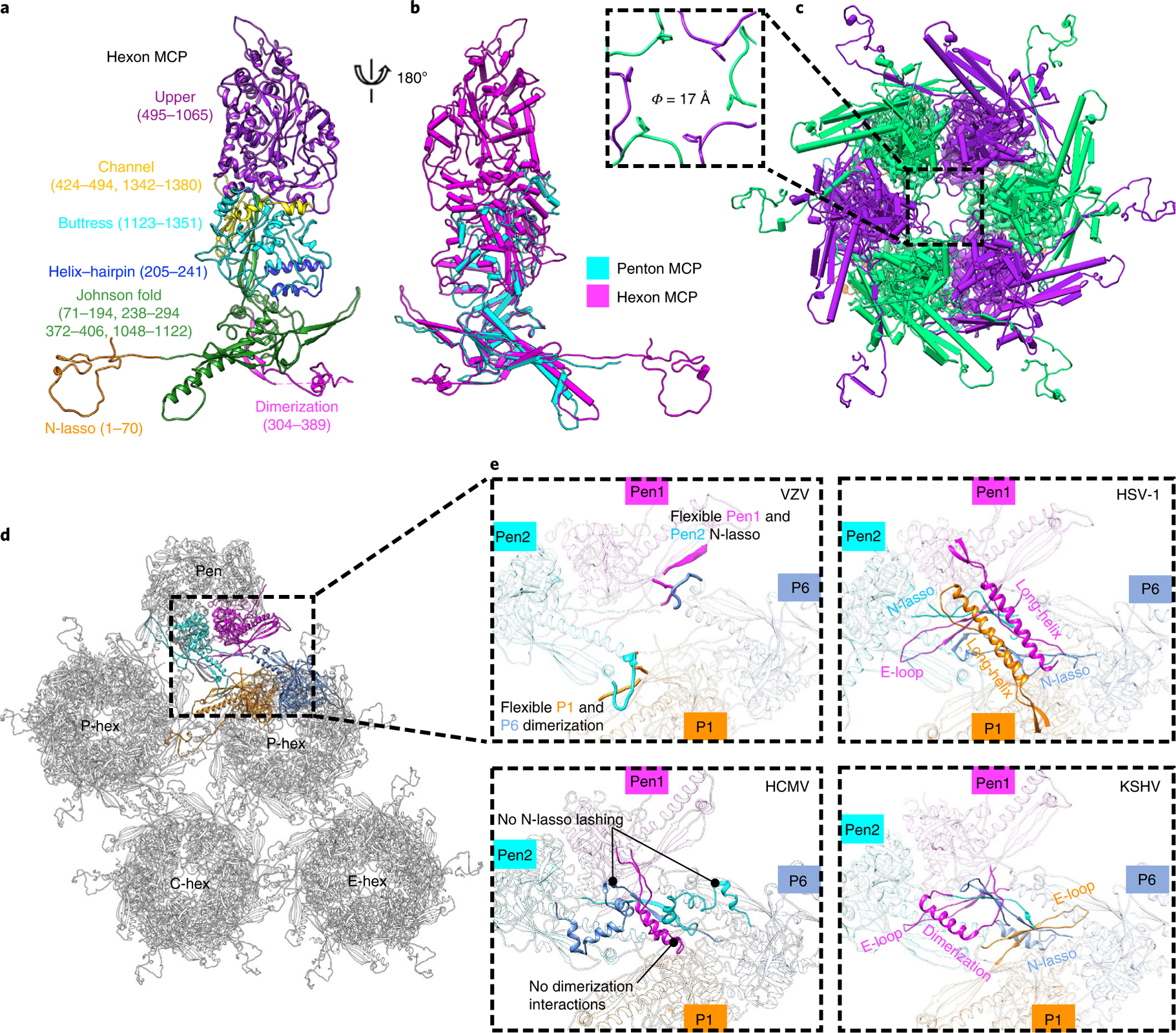
a, Canonical hexon MCP coloured and labelled by domain. b, Superposition of hexon and penton MCPs. c, Pipe-and-plank depiction of an E hexon showing the central channel (dashed square). The narrowest region has a dimension (Φ) of about 17 Å. d, Part of the MCP–MCP interaction network viewed from the outside. e, Comparisons of MCP interactions between hexons and pentons in the capsids of VZV, HSV-1, HCMV and KSHV.
Structure of the triplex and its interactions with MCPs.
Amid the canyons formed by three neighbouring capsomeres is a triplex, consisting of one Tri1 and two Tri2 conformers (denoted as Tri2A and Tri2B) (Fig. 3a–d). The VZV triplex, which adopts a trimeric arrangement in the quasi-three-fold symmetrical environment (Fig. 3a), shares common structural features with triplexes of other herpesviruses but possesses more flexible regions (Tri1 residues 1 to 118 and 371 to 385, Tri2A 162 to 178, and Tri2B 1 to 3, 164 to 172 and 250 to 267). The less rigid structure of the VZV triplex may result from its lack of inter- or intra-molecular disulfide bond formation compared with those reported in HSV-124 (Supplementary Fig. 1). Tri2A and Tri2B both contain two domains—trunk (142 to 288) and embracing arm (1 to 141 and 289 to 316)—but the embracing arms, which interact with each other, exhibit structural diversity between Tri2A and Tri2B (Fig. 3b–d). The N-anchor domain (1 to 117) of Tri1 is highly flexible, and only smeared densities are observed, which are inserted inside the capsid with a tri-lobed structure, anchoring the entire triplex (Fig. 3a,e,f). Extensive interactions exist between the triplex and three buttress arms extending from the three surrounding MCPs (Fig. 3g,h). Similar buttress arms involving triplex–MCP interactions have also been observed in HCMV, which appear to clamp the triplex more stably because the distal extremity of the buttress arm of HCMV contains a helix (which is a loop in VZV), thus forming stronger helix–helical interactions28.
Fig. 3 |. Structure of the triplex and its interactions with the surrounding MCPs.
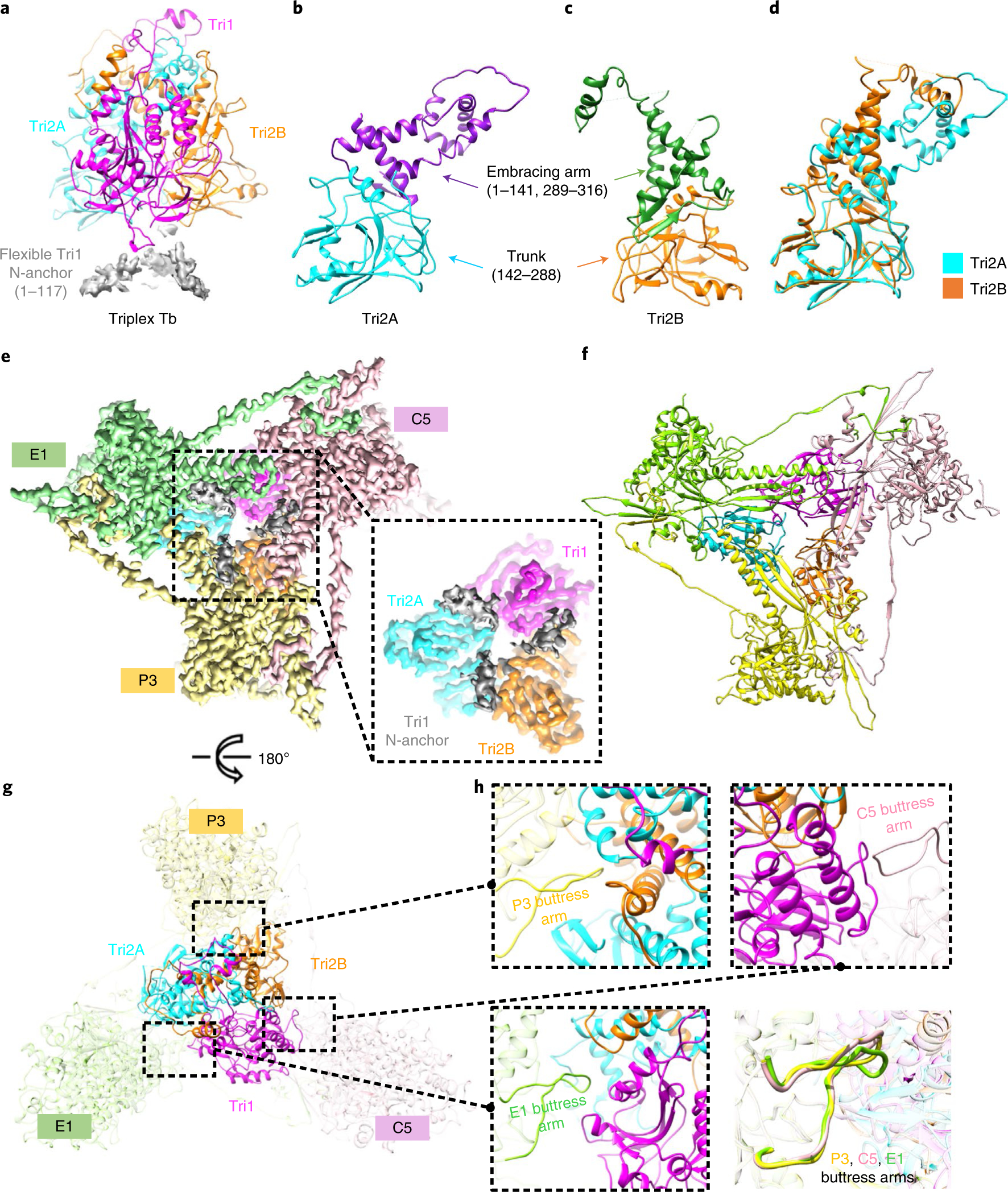
a–c, Detailed structures of triplex Tb (a) and its components Tri2A (b) and Tri2B (c). d, Superposition of Tri2A and Tri2B shows constant trunk domains and conformational differences in their embracing arm domains. e,f, Density map (e) and models (f) of triplex Tb and its surrounding MCPs. A close-up view of the density map of triplex Tb with the tri-lobed density of its flexible Tri1 N-anchor (grey) is shown in the dashed box. g,h, Interactions of the triplex with its surrounding MCPs. The buttress arms of MCP P3, E1 and C5 consistently insert into and interact with triplex Tb mainly through the loop containing residues 1176–1192. The interacting regions between the triplex and MCPs in g are highlighted with magnified views in the dashed boxes (h). h, Bottom right: the superimposed interacting regions with buttress arms of MCPs P3, E1 and C5 in the same orientation.
VZV-specific features in the CATC and packaged genome.
For HSV, the hetero-pentameric CATCs, which form extensive interaction networks with the capsid proteins, have been implicated in the stabilization and intracellular transport of the capsids24,27,34,35. However, CATCs were not observed in the purified VZV C-capsids, which raises the question of whether CATCs bind the capsid in intact VZV virions. Despite extreme difficulties due to association of VZV with its host cells, we attempted to purify intact VZV virions for structural analysis. We obtained 3,447 particle images of intact VZV virions from more than 16,000 cryo-EM micrographs (Extended Data Fig. 1e) and, from these images, computed a final icosahedral reconstruction at a resolution of 6.3 Å (Extended Data Fig. 3 and Supplementary Table 1). This virion capsid reconstruction at an intermediate resolution reveals unlike in the naked C-capsid, CATCs are present on the five-fold vertex of virion capsids (Fig. 4a,b). However, the CATC densities are much lower than for other capsid proteins (Fig. 4b), indicating that the binding sites of CATCs on VZV capsids are not fully saturated. Further sub-particle classification focused on penton vertexes shows that only about half (50.5%) of pentons are bound with CATCs (Fig. 4c). These results suggest low occupancy or relative flexibility of CATCs in VZV virions or a combination of both.
Fig. 4 |. Structure of the VZV virion showing the CATC and dsDNA genome.
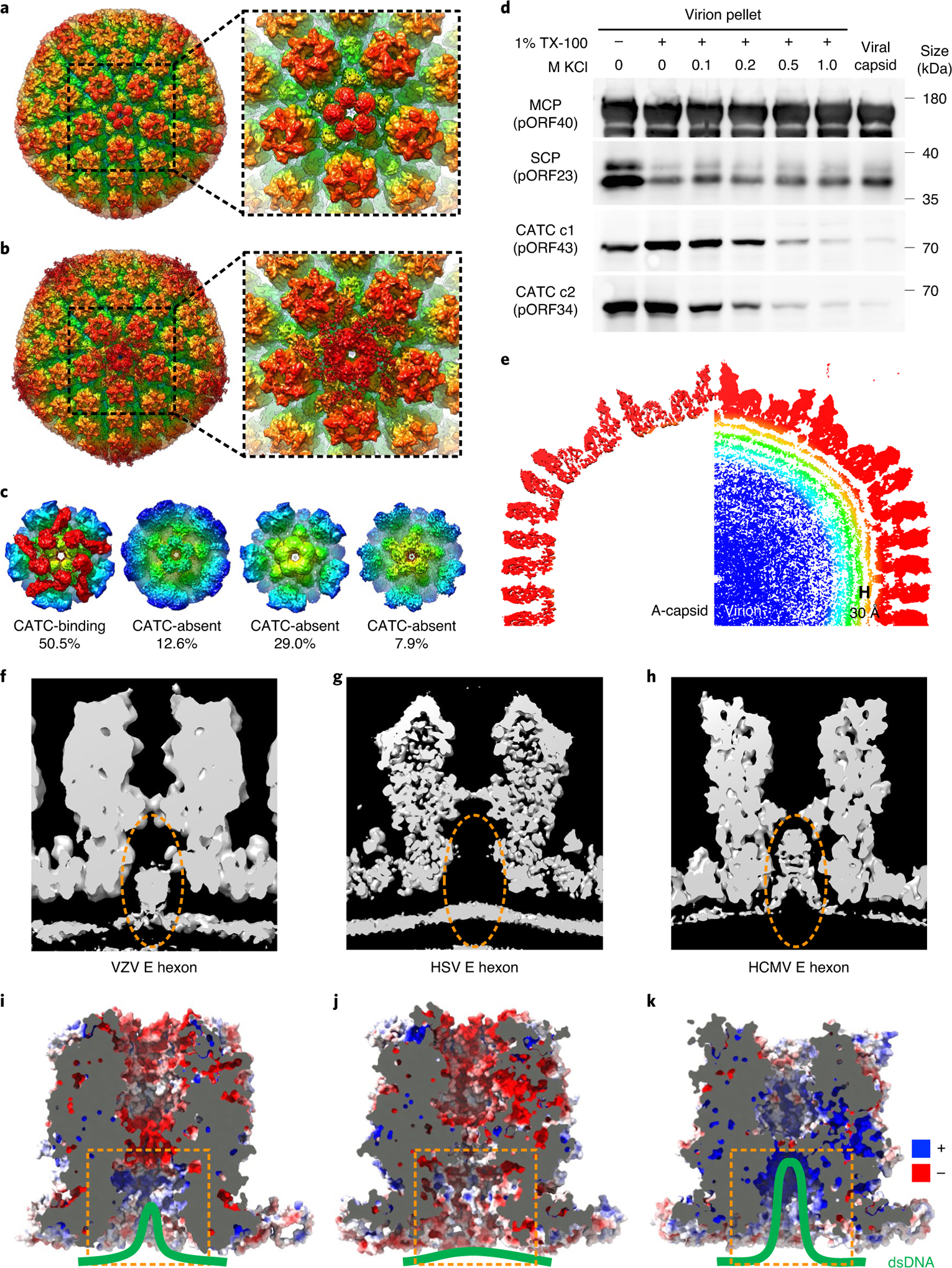
a,b, Radially coloured reconstructions of the VZV C-capsid (a) and virion (b). CATC is absent on VZV naked C-capsids (a) but present on capsids inside virions (b). c, Sub-particle classification of virion capsid penton vertexes shows that CATCs bind only half (class 1), while the rest (class 2–4) lack bound CATCs. d, Western blot showing a gradual loss of VZV CATC components (c1 and c2) from the virion pellets by detergent lysis (Triton X-100 (TX-100)) in the presence of increasing KCl concentrations (0, 0.1, 0.2, 0.5 and 1M). Experiments were performed three times independently with similar results. e, Radially coloured central slab of the VZV A-capsid (without internal density) and virion (with dsDNA density inside). At least three layers of dsDNA duplex with a 30 Å inter-duplex distance are recognizable, and part of the outer layer density bends and reaches into the hexon channels. f–h, Central sections of an E hexon from the density map of VZV (f), HSV26 (g) and HCMV28 (h), showing partial insertion, no insertion and deep insertion, respectively, of genomic DNA into the space (marked by ellipses) inside the capsid hexon channels. i–k, Central sections showing electrostatic surface potential of the capsid hexon channels of VZV (i), HSV (j) and HCMV (k). Blue and red denote positive and negative charges, respectively. Orange dashed circles mark the inner entrance to the hexon channel. The viral dsDNA at the hexon channel entrance is schematically indicated by the green solid line.
To characterize the binding of CATCs biochemically, VZV virions were treated with 1% Triton X-100 and different concentrations of KCl and then ultracentrifuged for western blot analysis (Fig. 4d and Extended Data Fig. 8a). Following Triton X-100 treatment, virions still contained substantial amounts of tegument, while increasing KCl from 0.1 to 0.5 or 1 M extracted several tegument proteins from viral capsids, including the putative CATC components pORF43 and pORF34, which are homologues of pUL17 and pUL25 of HSV-1, respectively. By contrast, the CATC of HSV-1 (exemplified by pUL17) was resistant to the same treatment and remained associated with capsids (Extended Data Fig. 8b), as also reported previously36,37. These data suggest that the CATC of VZV may bind to the capsid via relatively weaker interactions, and is easily discarded during the capsid purification process involving detergent and KCl treatment.
dsDNA in the virion capsid is packaged as regularly spaced dense shells with a larger interlayer distance of ~30 Å, distinguishing them from those of HSV (~25–26 Å) and HCMV (~23 Å) (Fig. 4e). The VZV genome densities show less ordered packing than those of HSV and HCMV and have only three to four recognizable layers, probably due to its small genomic size (Fig. 4e). Unexpectedly, the outmost layer of the VZV genome seems to partially occupy the narrow interior confines of the capsid’s hexon channel (Fig. 4f), similar to that of HCMV with a 235 kb genome, but unlike that of HSV (Fig. 4g,h). This observation appears to counter the argument for more space being needed to accommodate the large HCMV genome, as VZV has a much smaller genome. A comparison of the surface charge properties of hexon channels among these herpesviruses indicates that, although they are mainly neutral for the HSV hexon channel, residues lining the hexon channel entrance of both VZV and HCMV are positively charged (Fig. 4i–k). Moreover, those deep inside the channel become negatively charged in VZV (Fig. 4i), but remain positive in HCMV (Fig. 4k). Thus, the reach of surface residues with a positive charge, complementary to that of the dsDNA genome, is correlated with the extent of the DNA inserted into these channels (Fig. 4f–k), suggesting that electrostatic interactions rather than genome sizes influence DNA distribution inside the hexon channels.
VZV-specific features in the SCP.
Ranging from 8 to 35 kDa in molecular mass, the smallest capsid proteins vary the most in size across herpesviruses, whereas the largest capsid proteins, the 150 kDa MCPs, vary the least. The 235-residue SCP of VZV is the largest among human herpesviruses, with a unique C terminus (residues 108 to 235; Supplementary Fig. 1), and is essential for VZV infection of human skin tissues in vivo38. This prompted us to investigate the role of the SCP in VZV. Unlike those in β- and γ-herpesviruses but similar to those in the α-herpesviruses HSV-139, HSV-240 and pseudorabies virus (PrV)41, VZV SCP caps hexons but not pentons (Fig. 1d), interfacing between the capsid and the tegument. To investigate its role in VZV assembly, we prepared antibodies targeted to the N terminus (Ab-SCPn, targeting residues 1–15) and C terminus (Ab-SCPc, targeting residues 108–235) of the VZV SCP. Western blot analysis using these antibodies showed that purified VZV capsids contained the full-length SCP (~35 kDa; Fig. 5a). However, our cryo-EM structure only shows ordered densities of the N-terminal region of SCP, and the densities of its C terminus are smeared (Fig. 5b). To our knowledge, these disordered C-terminal densities—which emanate from the inner rim of the upper hexon channel and project into the tegument layer—have not been observed in other herpesviruses, although they disappear when increasing the threshold of the map (σ from 0.25 to 3.00; Fig. 5c). These observations suggest that the C-terminal segment of SCP (residues 108 to 235) is highly flexible. The model of the N terminus (residues 3 to 107) of SCP was then built out; it exhibits structural similarity to HSV-1 VP26, with a helix-rich domain (residues 8 to 75) involved in MCP binding, along with a long insertion hairpin-loop domain (residues 76 to 107) to stabilize MCP binding (Fig. 5d,e). Similar to HSV, six SCP subunits form a gear-shaped hexameric ring crowning each hexon, with their hairpin-loop domains filling in the gap between adjacent hexon MCP upper domains (Fig. 5f–h). The SCPs cross-link each other and cap hexons via two types of interactions: (1) the insertion loop extends into the groove formed by the upper domains of two adjacent hexon SCPs, forming hydrophobic interactions. The N terminus of the adjacent SCP also interacts with the insertion loop and stabilizes the insertion (Fig. 5h); (2) residues of the connection loop (residues 76 to 79), together with a part of the stem helix (residues 67 to 75), are positively charged and electrostatically interact with the negatively charged MCP upper region (residues 865 to 883 and 908 to 915; Fig. 5i). These noncovalent cross-linking and capping interactions of SCPs and MCP–SCP resemble those of the 112-amino-acid SCP homologue VP26 in HSV-1 and HSV-2, and may have a similar function in stabilizing the hexons.
Fig. 5 |. Characterization and structure of the SCP and its interactions with MCPs.
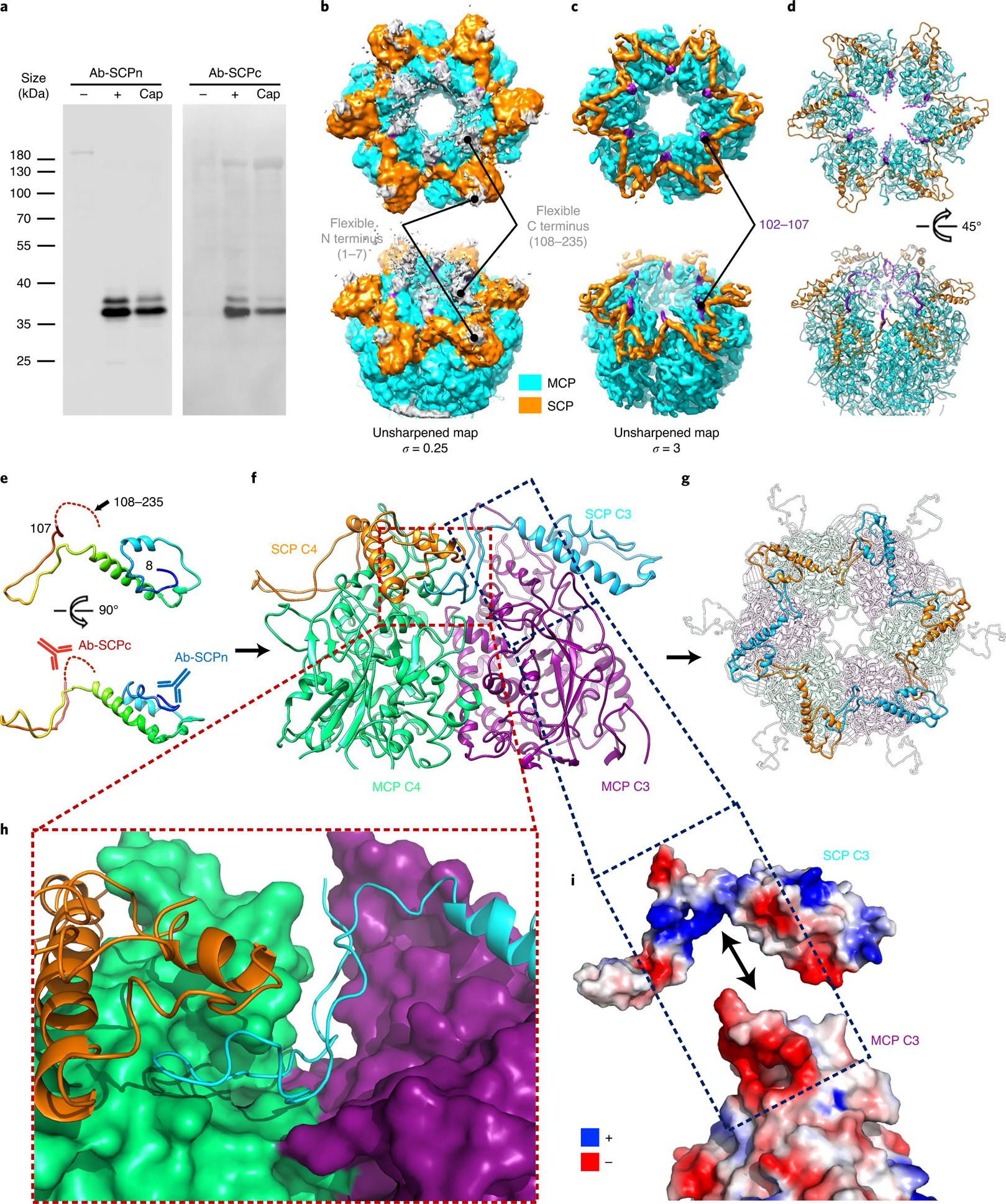
a, Western blot analysis of SCP on VZV capsids (cap) using antibodies raised to target either the N terminus (Ab-SCPn) or C terminus (Ab-SCPc) of pORF23. Lysates of uninfected (−) or VZV-infected (+) cells were used as negative and positive controls, respectively. Molecular mass markers (in kDa) are shown on the left. Experiments were performed two times independently with similar results. b–d, Localized reconstruction of an E hexon of the A-capsid shown with different density thresholds. MCPs and SCPs are coloured in cyan and yellow, respectively. b, The lower threshold display of the map shows an extra density blob (grey) emanating from the last modelled residue of both the N-terminal end (residue 8) and C-terminal end (residue 107) of each SCP subunit. c, When the map is displayed at the higher threshold of three standard deviations (σ) above the mean, both density blobs disappear, suggesting flexibility of the N-terminal 7 residues and C-terminal 127 residues. d, Purple dashed lines in the model simulate disordered C termini of SCP on the basis of the extra densities as shown in b. e, Atomic model of an SCP monomer shown as ribbons, rainbow-coloured from the N terminus (blue) to the C terminus (red). The flexible C-terminal extension is depicted as a dashed line, and the binding sites of antibodies are indicated by the schematic drawing of Y-shaped antibodies (Ab-SCPn, blue; Ab-SCPc, red). f, The SCP bridging two underlying MCPs (MCP C3 and C4). g, Cross-linking of six SCP subunits forms a gear-shaped hexameric ring crowning each hexon. h,i, Interaction details of SCPs with MCPs. h, The hairpin-loop of SCP C3 inserts into a groove formed by MCP C4 and SCP C4. i, Open-book view of the electrostatic potential surfaces of SCP C3 and MCP C3 shows their complementary electrostatic surface charges.
Discussion
The lack of structural information about the VZV particles represents both a knowledge gap in understanding VZV capsid assembly and an untapped opportunity for structure-based rational design of VZV-based vectors for prophylactic and therapeutic applications. We present here the near-atomic-resolution structure of the VZV capsid. The overall structure is highly similar to those of HSV-1 and HSV-2 but has some specific features, such as more regions with flexibility in pentons and triplexes, as well as SCP C terminus densities. Furthermore, the structure shows that the VZV capsid is organized by weaker or less extensive interactions compared with capsids of other herpesviruses, including penton–hexon, triplex–MCP, and even CATC–capsid interactions. Unlike the numerous disulfide bonds observed in HSV-1 capsids, there is a general lack of disulfide bonding in the VZV capsid. On the basis of these observations, it is tempting to speculate that the VZV capsid has relatively lower stability. Nonetheless, the VZV capsid is sufficiently stable, as indicated by our thermal stability assay (Extended Data Fig. 9) showing that VZV capsids withstood high temperatures (up to 87 °C), similar to HSV-1 capsids (up to 86 °C) and by the fact that high-resolution structures were obtained by averaging many particles.
Herpesviruses assemble a similarly sized capsid but have diverse capsid-fortifying strategies to ensure encapsidation of their differently sized genomes, overcoming up to tens of atmospheres of internal pressure due to compacted dsDNA genome42,43. Our structures reveal that, although the VZV capsid tends to have fewer links and more flexibility, most of its structure and the capsomer–capsomer interaction networks within the structure generally resemble those of other herpesviruses. Thus, we speculate that the high stability of VZV capsids is derived from the intrinsic structural features of herpesvirus capsids. Moreover, due to the smaller genomic DNA and therefore lower internal pressure, the VZV capsid may enable fewer or weaker intra-subunit and inter-subunit interactions to maintain structural stability. Conversely, we have identified increased structural flexibility in pentons, triplexes and the SCP C terminus of VZV capsids compared with solved structures of mostly tegumented capsids from the mature virions of other herpesviruses. Whether these features are unique to VZV or associated with the structural perturbation caused by purification steps should be further confirmed by solving high-resolution structures of VZV capsids in intact virions, which remains challenging.
CATCs binding at capsid vertices of herpesviruses could enhance capsid stability27,28,44,45. Similar to their homologues in HSV, all three subunits of VZV CATC are essential for viral replication46. Occupancy of CATC binding sites on virion capsids is ~50% in VZV (Fig. 4c), compared with near 100% in the other α-herpesviruses HSV-124, HSV-227 and PrV47. Moreover, VZV CATC dissociated from the capsid when treated with detergent and at high KCl concentration (Fig. 4d); by contrast, HSV-1 CATC was resistant to the same treatment (Extended Data Fig. 8). Our results also showed high thermal stability of the VZV C-capsids without CATCs (Extended Data Fig. 9). Together, these results suggest a diminished role, if any, for VZV CATC in stabilizing the pressurized DNA-containing capsids. Interestingly, similar low occupancy of peri-penton CATC binding sites has been observed in the virions of the γ-herpesviruses KSHV31,44 and Epstein–Barr virus (EBV) (see also the accompanying Article48), although their peri-portal CATC binding sites are fully occupied, pointing to conserved functions of CATC in facilitating DNA packaging and ejection.
The capsids of all herpesviruses have the same protein compositions for their capsid shell: MCP, SCP, Tri1 and Tri2. Among them, SCP is the smallest and the most divergent, both structurally and functionally, across different subfamilies of the Herpesviridae, and even within the α-herpesvirus subfamily. The VZV SCP is 2.1 times the size of the SCP homologues in the α-herpesviruses HSV-1 and HSV-2, about 3 times that of the β-herpesviruses HCMV28 and HHV-6B29, and about 1.4 times that of the γ-herpesviruses KSHV30,31 and EBV (Supplementary Fig. 1). Similar to SCP homologues in HSV-1 and HSV-224,26, six copies of VZV SCP form a ring crowning only hexons, compared with those in β- and γ-herpesviruses, which bind both hexons and pentons28,31. Relevant to chickenpox and shingles, SCP was shown to be essential for VZV growth in human fetal skin xenografts in a SCID humanized mice model in vivo, and in melanoma cells in vitro38. By contrast, SCPs were proved to be nonessential for replication of HSV-1 and PrV49,50. Similar to that of VZV, SCPs of HCMV and KSHV are required for viral growth51,52. Further studies on structure–function relationships should provide insights into the molecular basis for the specific roles of SCPs in the replication of different herpesviruses. For VZV, SCP interacts with MCP to facilitate its nuclear translocation, suggesting a role in the process of capsid assembly38. The MCP-bound N-terminal region of SCP (residues 8 to 107) appears to serve as the capsid anchor of SCP’s flexible C-terminal segment (residues 108 to 235), which is unique among human herpesviruses. With this long C terminus projecting up from the capsid surface (Fig. 5b–d), the VZV SCP is probably involved in functional interactions with other viral or host factors. Indeed, previous yeast two-hybrid studies have shown that VZV SCP interacts with components of the nuclear egress complex, including pORF24 and pORF27 (homologues of pUL34 and pUL31 of HSV-1, respectively), as well as the tegument proteins pORF7, pORF9 and pORF21 (homologues of pUL51, VP22 and pUL37 of HSV-1, respectively)53–55. These findings suggest possible alternative roles for SCP in the nuclear egress and secondary envelopment of VZV particles.
Overall, the near-atomic structures of VZV capsids reported here, together with the previously reported cryo-EM structures of HSV capsids, reveal structural similarities and differences among these human α-herpesviruses and provide insights into their capsid-assembly mechanisms. Interaction hotspots revealed in our structures could be targeted either for designing competitive inhibitors as antivirals or for engineering live attenuated VZV strains as vaccines against chickenpox and shingles. Moreover, as it has the smallest genome size among human herpesviruses and high capsid stability, safe VZV vectors based on the commercially available live VZV vaccine could be engineered to carry exogenous DNA fragments as platforms for development of multivalent vaccines and anticancer agents.
Methods
Cells.
Human retinal pigment epithelial ARPE-19 cells (CRL-2302) and African green monkey Vero kidney cells (CCL-81) were obtained from the American Type Culture Collection and maintained in DMEM containing 10% fetal bovine serum, penicillin/streptomycin and l-glutamine. Mouse SP2/0-Ag14 myeloma cells (GDC0019) were obtained from the China Center for Type Culture Collection and maintained in RPMI 1640 medium containing 10% fetal bovine serum, penicillin/streptomycin and l-glutamine. The cell lines were authenticated by the providers using morphology, karyotyping and PCR-based approaches and were not further authenticated in our laboratory. Cells tested negative for Mycoplasma contamination by PCR before use in experiments.
VZV particle preparation.
ARPE-19 cells were infected with the wild-type parental Oka strain (P-Oka) of VZV at a multiplicity of infection (MOI) of 0.3 when reaching ~80% confluence. For VZV capsid purification, infected cells were collected at 3 d post infection (dpi) with the cytopathic effect reaching >80%, resuspended in ice-cold lysis buffer (PBS containing 1% Triton X-100, 1 M KCl and protease inhibitor) and incubated on ice for 30 min. After lysis, large cell debris was removed from the solution by centrifugation at 1,500g at 4 °C for 15 min. Then, the capsids were pelleted through a 30% (w/v in PBS) sucrose cushion by centrifugation at 110,000g at 4 °C for 15 min in a Beckman SW28 rotor and resuspended in PBS for cryo-EM sample preparation and other experiments.
For VZV virion purification, VZV-infected ARPE-19 cells were collected when showing >80% cytopathic effect and were homogenized with a Dounce homogenizer for 20–30 strokes. The clarified cytoplasmic extract was centrifuged at 1,500g at 4 °C for 15 min to remove large cell debris. The resulting solution was centrifuged through a linear 20% to 50% (w/v in PBS) sucrose gradient at 110,000g at 4 °C for 1 h in a Beckman SW28 rotor. The lower visible band was collected, diluted in PBS and centrifuged through a 30% (w/v in PBS) sucrose cushion at 110,000g at 4 °C for 15 min. The pelleted crude VZV virions were resuspended in PBS and subsequently subjected to cryo-EM sample preparation and other experiments.
HSV-1 particle preparation.
Vero cells were infected with the KOS strain of HSV-1 at an MOI of 0.01. At 3 dpi, the cells and culture medium were collected for purification of naked capsids and intact virions, respectively. HSV-1 capsids were purified from whole-cell lysate using the same method as for VZV capsid purification. HSV-1 virions were purified using the method described previously, with slight modification24. In brief, after removal of large cell debris, the culture medium was centrifuged at 80,000g for 1 h. Pellets were resuspended in PBS and then centrifuged through a linear 20% to 50% (w/v) sucrose density gradient at 80,000g for 1 h. The upper band was collected, diluted with PBS and pelleted by centrifuging at 80,000g for 1 h. The pelleted HSV-1 virions were resuspended in PBS and frozen at −80 °C before use.
Antibodies.
To obtain antibodies against the capsid proteins (pORF40, pORF41, pORF20 and pORF23 of VZV and HSV-1 VP5), the portal protein (VZV pORF54) and the CATC proteins (pORF34 and pORF43 of VZV, and HSV-1 pUL25), BALB/c mice were immunized subcutaneously with the corresponding recombinant viral proteins (100 μg) or synthetic peptides conjugated to bovine serum albumin (BSA, 100 μg) emulsified in complete Freund’s adjuvant and boosted twice, at two week intervals, with the same doses of antigen in incomplete Freund’s adjuvant. Two weeks after the final boost, splenic cells from immunized mice were fused with SP2/0-Ag14 myeloma cells and the hybridoma supernatants were screened using an enzyme-linked immunosorbent assay (ELISA) against each antigen. Finally, antibodies were purified from mouse ascitic fluid using protein A affinity columns. All animal experimental protocols were approved by the Xiamen University Laboratory Animal Center and were conducted in strict compliance with the guidelines of the Xiamen University Institutional Committee for Care and Use of Laboratory Animals (approval code: XMULAC20160049). Additionally, antibodies against pORF9, pORF62 and gE of VZV were prepared previously56,57, and antibodies against ICP4, VP16, and gD of HSV-1 were purchased from Santa Cruz Biotechnology.
Western blot analysis.
Purified viral particles and lysates prepared from mock-infected or VZV or HSV-1-infected cells were separated by 10% SDS–PAGE, and proteins were then transferred to nitrocellulose membranes. After blocking for 1 h, membranes were incubated for 1 h with primary mouse antibodies against the capsid proteins (pORF40, pORF41, pORF20 and pORF23 of VZV, and HSV-1 VP5), the portal protein (VZV pORF54), the CATC proteins (pORF34 and pORF43 of VZV, and HSV-1 pUL25), the viral glycoproteins (VZV gE and HSV-1 gD), and other tegument proteins (pORF9 and pORF62 of VZV, and ICP4 and VP16 of HSV-1) at a concentration of 1 μg ml. Then, the membranes were incubated with horseradish peroxidase-conjugated goat anti-mouse IgG secondary antibody at a dilution of 1:5,000 for 30 min, and finally protein bands were visualized by chemiluminescence. All procedures were conducted at room temperature.
Particle-stability thermal-release assay.
Thermofluor experiments were performed with a CFX86 PCR instrument (Bio-rad). The fluorescent dyes SYTO-Green and SYTO9 (Thermo Fisher Scientific) were used to detect the presence of DNA and RNA, respectively. Each 50 μl reaction was set up in a thin-walled PCR plate containing 1.0 μg of purified capsids of VZV or HSV-1 and 5 μM SYTO-Green or SYTO9 in PBS. The temperature was ramped from 45 to 99 °C and the fluorescence was recorded in triplicate at 0.5 °C intervals. The negative first derivatives of the fluorescence change (−d(RFU)/dT, where RFU is relative fluorescence units and T represents time) were plotted against temperature.
Transmission electron microscopy.
ARPE-19 cells infected with VZV P-Oka strain (at an MOI of 0.3) were collected at 3 dpi and prepared for transmission electron microscopy analysis as previously described58,59. In brief, samples were fixed in 2.5% glutaraldehyde in phosphate buffer (0.1 M, pH 7.4), post-fixed with 1% osmium tetroxide, incubated in 1% aqueous uranyl acetate overnight, dehydrated through graded ethanol and embedded with an Embed 812 kit (Electron Microscopy Sciences). Ultra-thin sections were stained with 3.5% aqueous uranyl acetate and 0.2% lead citrate. Images were recorded using a Tecnai G2 Spirit transmission electron microscope (FEI).
Cryo-EM imaging.
A 3 μl aliquot of purified VZV capsids or virion was applied to freshly glow-discharged holey carbon Quantifoil Cu grids (R2/2, 200 mesh, Quantifoil Micro Tools), and then blotted for 6 s before plunge-freezing the grids into liquid ethane cooled by liquid nitrogen inside a Vitrobot Mark IV (Thermo Fisher Scientific) at 100% humidity and 4 °C. The cryo-grids were initially checked at 300 kV with the FEI Tecnai F30, with only good quality grids selected for data collection. Cryo-EM images of naked capsids were acquired with the FEI Titan Kiros (Thermo Fisher Scientific) with a Gatan K2 Summit direct electron detector at a nominal ×130,000 magnification, corresponding to a calibrated physical pixel size of 1.305 Å (0.6525 Å for superresolution pixel size). Each movie had 40 frames, with a total dose of 56 e− Å−2 with an exposure time of 8 s. Data were semi-automatically collected using SerialEM60. Cryo-EM images of virions were acquired with the FEI Tecnai F30 with a Falcon III direct electron detector (Thermo Fisher Scientific) at a nominal magnification of ×59,000, corresponding to a pixel size of 1.76 Å. A total electron dose of about 40 e− Å−2 was fractionated into 39 frames in every movie at an exposure time of 1 s. Data were automatically collected with Thermo Fisher EPU software.
Image processing and 3D reconstruction.
Drift and beam-induced motion correction was performed with MotionCor261 to produce a micrograph from each movie. Contrast transfer function fitting and phase-shift estimation were conducted with Gctf62. Micrographs with astigmatism, obvious drift or contamination were discarded before reconstruction. Particles were automatically picked and screened using cisTEM63. The initial 3D models for each dataset were generated with a random model method using AUTO3DEM64. Several rounds of reference-free 2D classifications and unsupervised 3D classifications were executed using Relion 2.165. Sorted particles were then subjected to final homogenous refinement using Relion 2.1 and cisTEM. The resolution of all density maps was determined by the gold-standard Fourier shell correlation curve, with a cut-off of 0.14366. Local map resolution was estimated with ResMap67.
To improve the resolution, we used Relion symmetry expansion to exact and perform sub-particle reconstruction as previously described32. In brief, after 3D refinement with imposition of icosahedral symmetry, we extracted sub-particles from the three-fold region in a box-size of 540 × 540 pixels and expanded the sub-particles with I1 symmetry. The extracted sub-particles were used to generate the initial model using Relion relion_reconstruct command. Further 3D classification and refinement were performed by Relion 2.1 and cisTEM. The resolution was assessed by Fourier shell correlation curves with a cut-off at 0.143 from two independent half-sets of the sub-particles.
Atomic model building, refinement and 3D visualization.
As the upper domain of the pentonal MCP exhibited very weak densities, we built the model only for the middle and floor regions. The initial VZV capsid model was generated from homology modelling on the basis of the atomic model of HSV-124 using Accelrys Discovery Studio software (https://www.3dsbiovia.com). We initially fitted the templates into the corresponding segmented volume (enclosing an asymmetric unit) of the final cryo-EM maps using Chimera68, and further corrected and adjusted them manually by real-space refinement in Coot69. The resulting models were then refined with phenix.real_space_refine in PHENIX70. These operations were executed iteratively until the problematic regions, Ramachandran outliers and poor rotamers were either eliminated or moved to the favoured regions. After several cycles of refinement, the resulting models were fitted into the map of six neighbouring asymmetric units. The totality of these seven asymmetric units were subjected to further real-space refinement to optimize the clashes. The final atomic models were validated with Molprobity71. Structure-based sequence alignment was performed with Clustal Omega on the EBI server (https://www.ebi.ac.uk/Tools/msa/clustalo/). For the intermolecular interactions, maximum cut-off distances of 4.0 Å were used for hydrogen bonding, salt-bridge formation and other contacts. All figures were generated with Chimera or Pymol software (http://www.pymol.org).
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Structure coordinates have been deposited in the Protein Data Bank under accession numbers 6LGL (A-capsid) and 6LGN (C-capsid). The corresponding electron microscopy density maps have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-0880 (icosahedral reconstruction of the A-capsid), EMD-0881 (icosahedral reconstruction of the C-capsid), EMD-30251 (icosahedral reconstruction of the virion capsid), EMD-30248 (localized reconstruction of the A-capsid) and EMD-30250 (localized reconstruction of the C-capsid). Source data are provided with this paper.
Extended Data
Extended Data Fig. 1 |. Electron microscopic imaging of VZV particles.
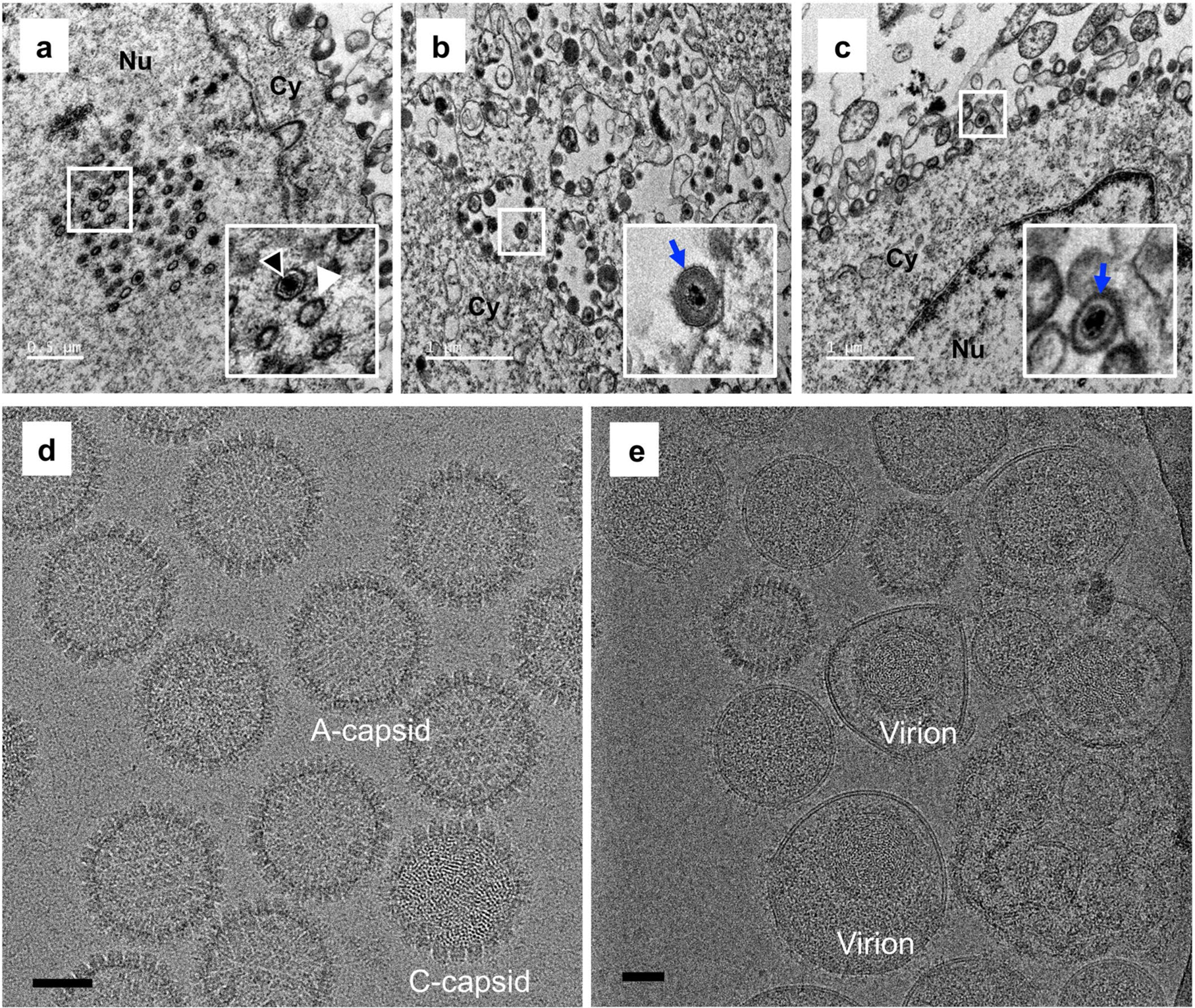
a–c, Representative electron microscopic images of VZV-infected ARPE-19 cells recorded at 3 days post-infection (dpi) using a Tecnai G2 Spirit transmission electron microscope. Panel a shows VZV nucleocapsids in the nucleus of an infected cell. Intact capsids are indicated by black arrow heads, and capsids without an electron-dense core are indicated by white arrow heads. Panel b,c show enveloped viral particles (blue arrows) within cytoplasmic vacuoles and on the surface of infected cells, respectively. Insets show enlarged images of the boxed areas. Experiments were performed two times independently with similar results. Nu and Cy indicate the nucleus and cytoplasm, respectively. Scale bars in a–c denote 0.5 μm, 1 μm and 1 μm, respectively. d, Purified VZV naked particles were recorded using a Gatan K2 Summit direct electron detector, showing two types of particles in our sample preparation: DNA-containing C-capsid and DNA-devoid A-capsid. Experiments were performed two times independently with similar results. e, Purified VZV virions were recorded using a Falcon3 direct electron detector. Experiments were performed two times independently with similar results. Scale bars in d,e denote 50nm.
Extended Data Fig. 2 |. Immunoblot characterization of purified VZV capsids.
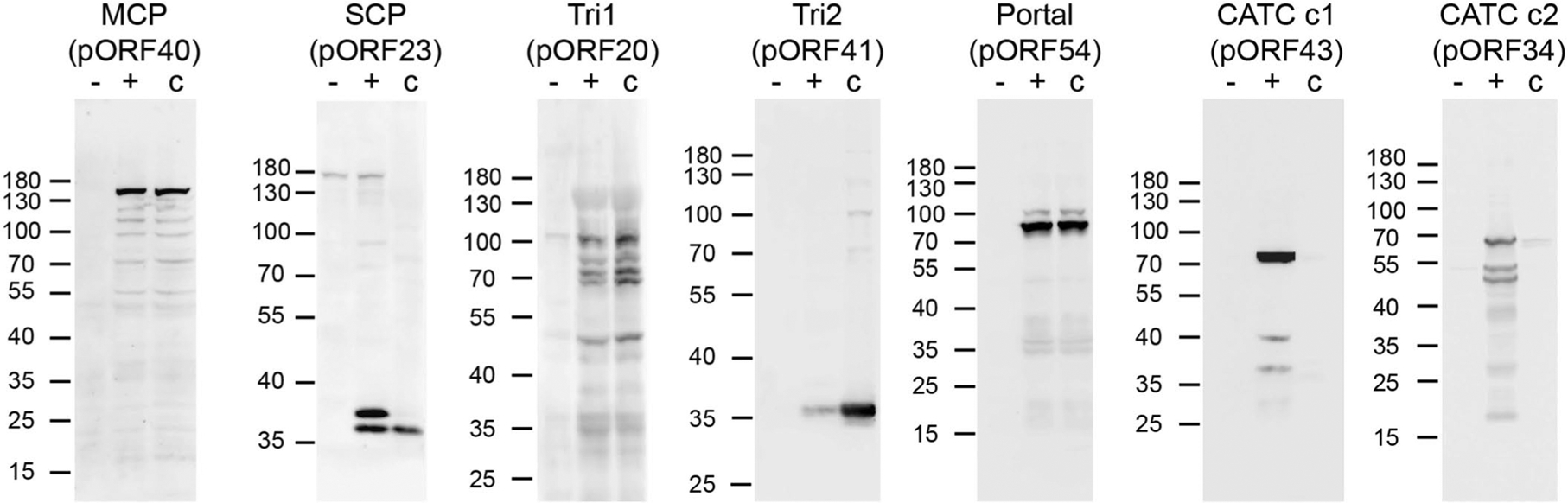
VZV capsids (c) were purified from the infected ARPE-19 cells at 3 dpi and were analyzed by western blot using antibodies against the indicated viral proteins. Lysates of uninfected (−) and VZV-infected (+) cells were used as negative and positive controls, respectively. Molecular mass markers in kilodaltons (kDa) are shown on the left. Experiments were performed two times independently with similar results. pORF43 and pORF34 of VZV are homologs of pUL17 and pUL25 of HSV-1, respectively, and represent two putative components (c1 and c2) of VZV CATC.
Extended Data Fig. 3 |. Global and local resolution analysis of 3D reconstructions of VZV A-capsid and C-capsid.
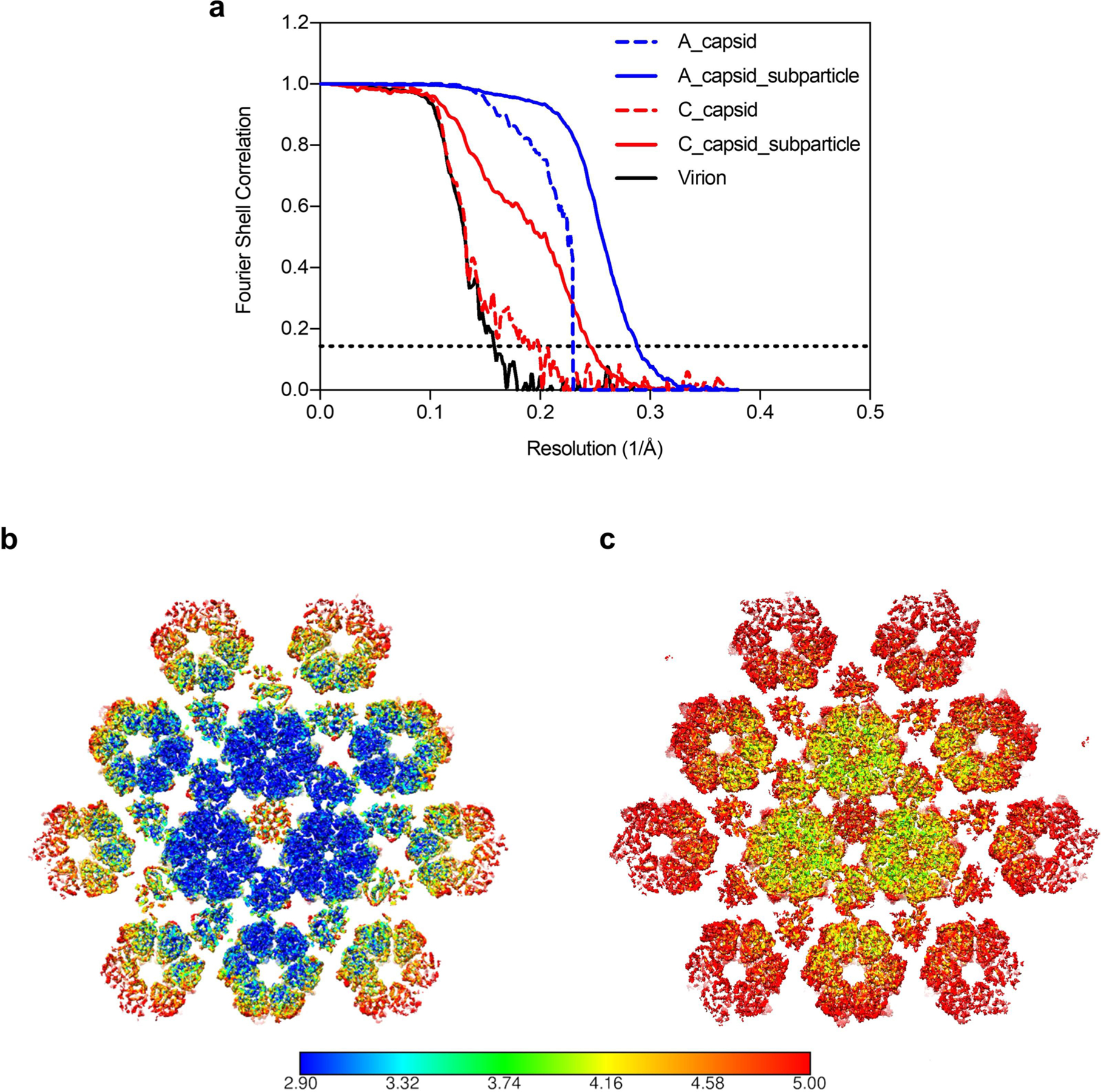
a, Fourier shell correlation (FSC) of global and localized reconstructions of three kinds of purified VZV particles. Resolutions were determined using a reference-based FSC coefficient criterion of 0.143. b,c, Maps from localized reconstructions of A- (b) and C-capsid (c) were analyzed by Resmap.
Extended Data Fig. 4 |. Density maps and atomic models of MCP and SCP.
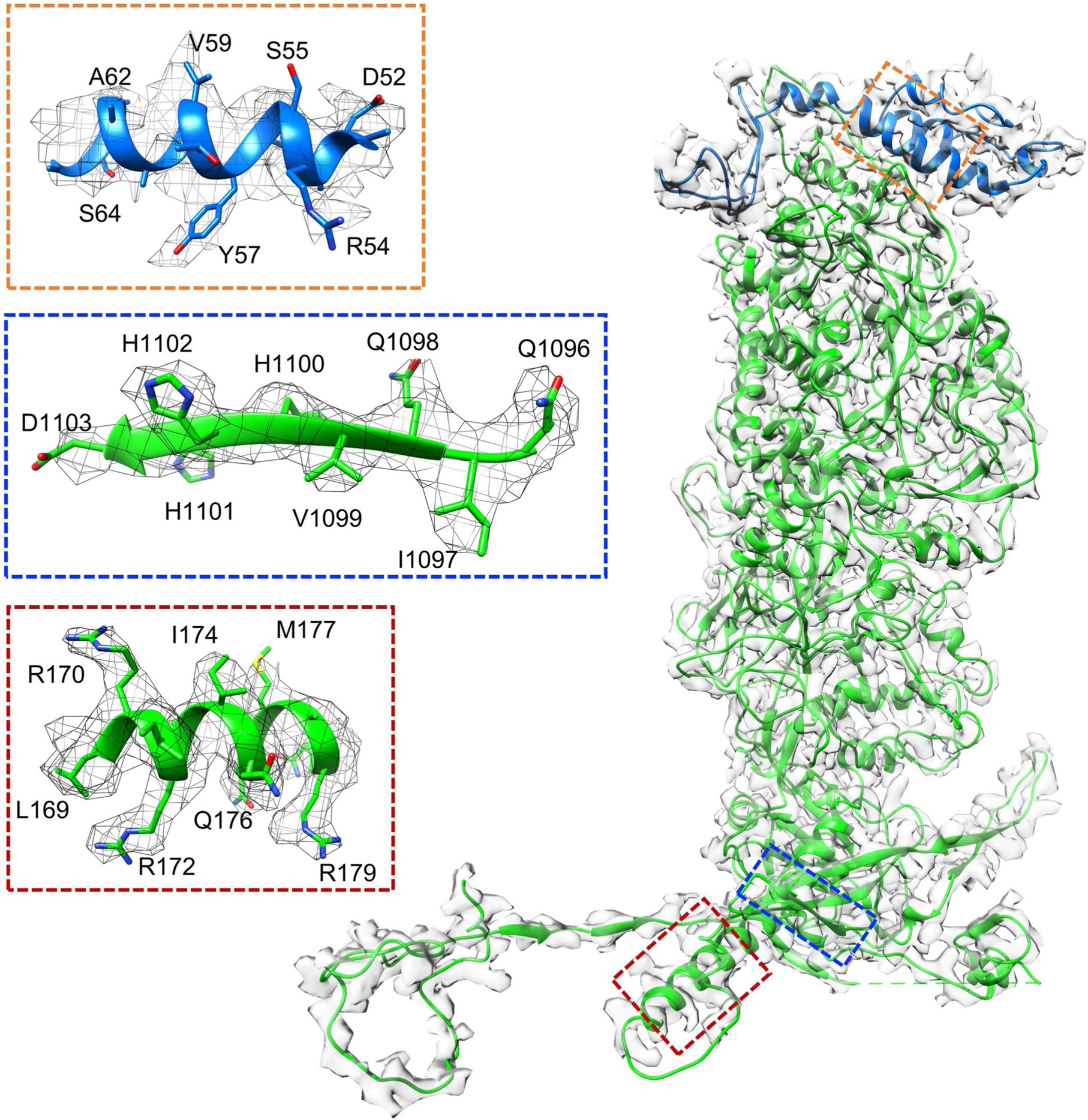
The fitted models of MCP and SCP are colored in green and blue, respectively. Insets correspond to zoomed-in views of boxed regions and illustrate residue features in the density map (mesh).
Extended Data Fig. 5 |. Density maps and atomic models of Tri1 (a), Tri2a (b) and Tri2b (c).
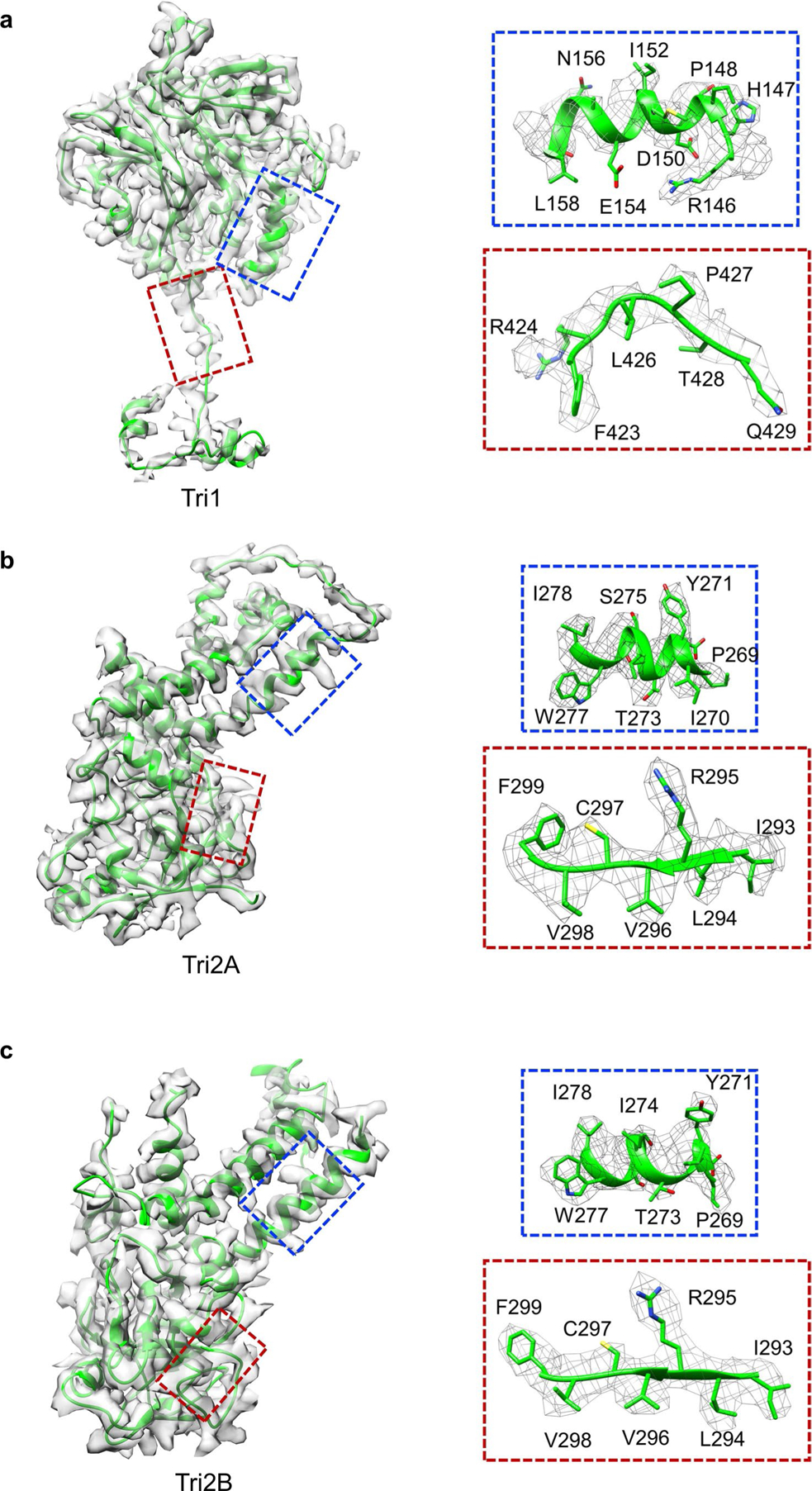
Insets correspond to zoomed-in views of boxed regions and illustrate residue features in the density map (mesh).
Extended Data Fig. 6 |. Structural comparison of A-capsid and C-capsid.
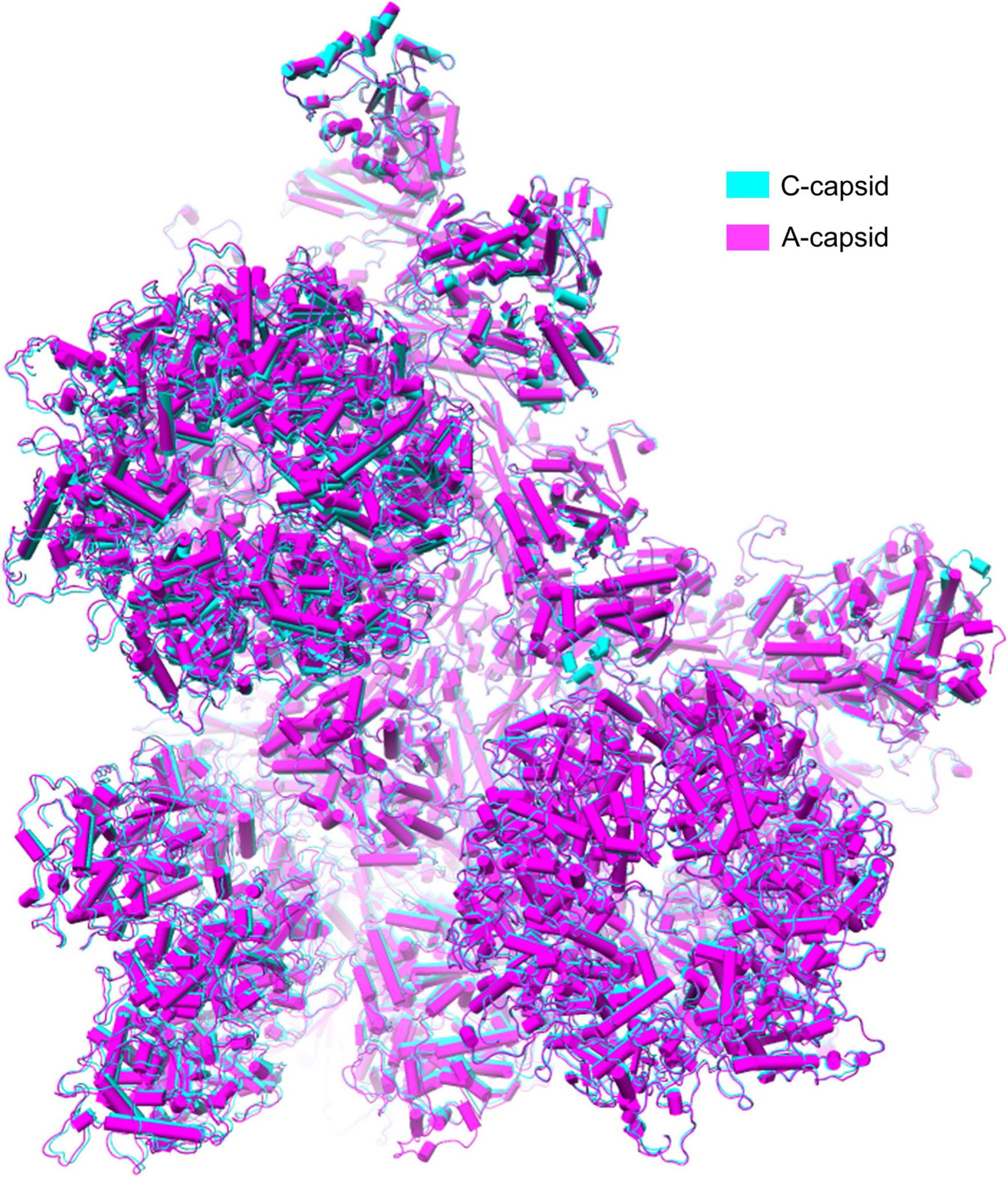
Superposition of the asymmetric units of A-capsid (magenta) and C-capsid (cyan) shows highly structural similarity. The models of asymmetric units of A- and C-capsid show high similarity with root mean square deviation of 0.76 Å over 25,884 aligned Cα positions.
Extended Data Fig. 7 |. Hexon MCP-involved network interactions.
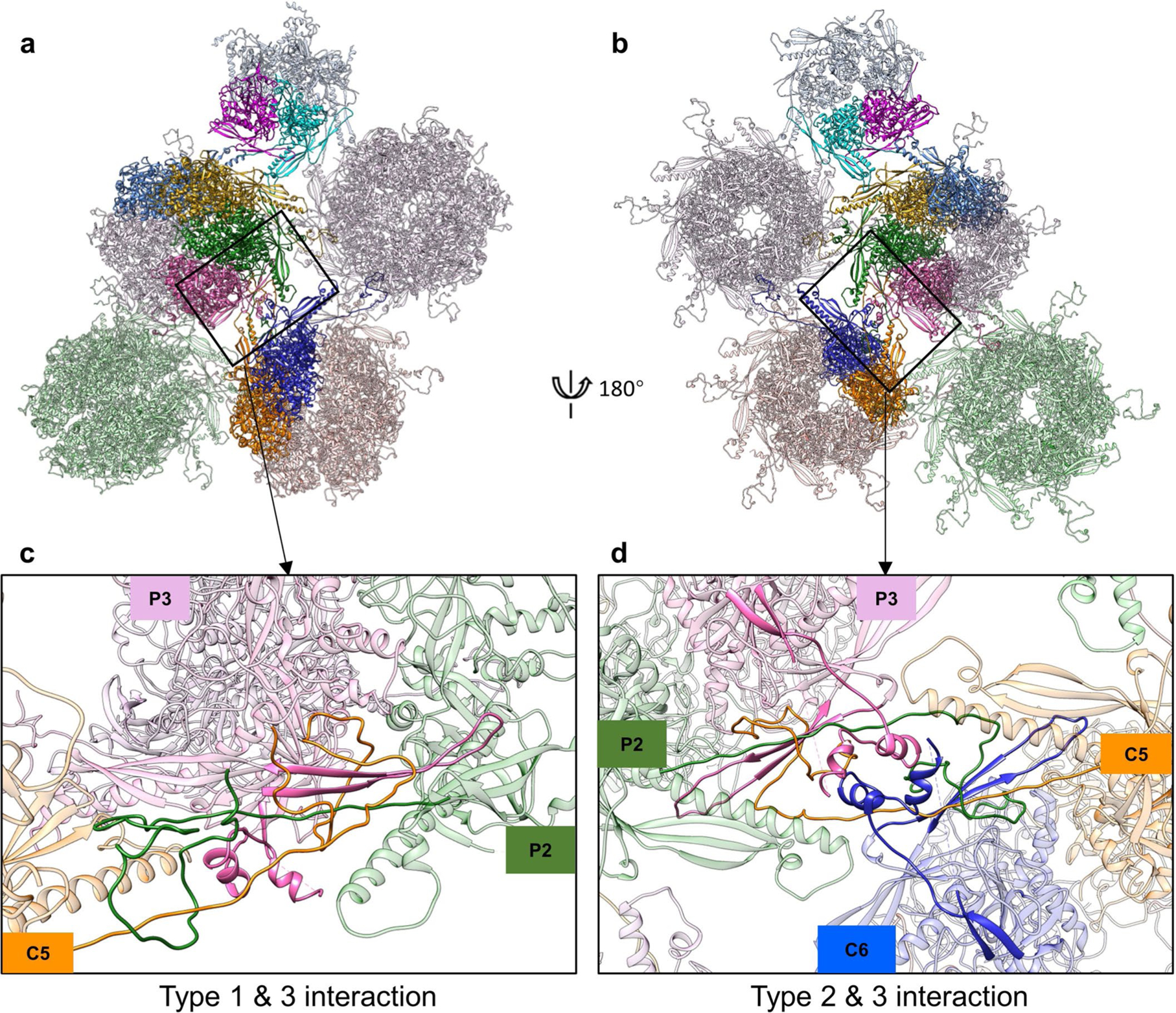
a,b, Part of the MCP-MCP interaction network viewed from the outside (a) or inside (b) of VZV A-capsid. c,d, Three types of interactions among hexon MCPs are shown. Type I interactions are intracapsomeric augmentations of β-strands from adjacent MCPs (P2 and P3 in c). Type II and type III interactions are intercapsomeric interactions among two pairs of MCPs (P2–P3 and C5–C6 in d).
Extended Data Fig. 8 |. Immunoblot characterization of virion capsids of VZV and HSV-1 after Triton X-100 lysis and KCl treatment.
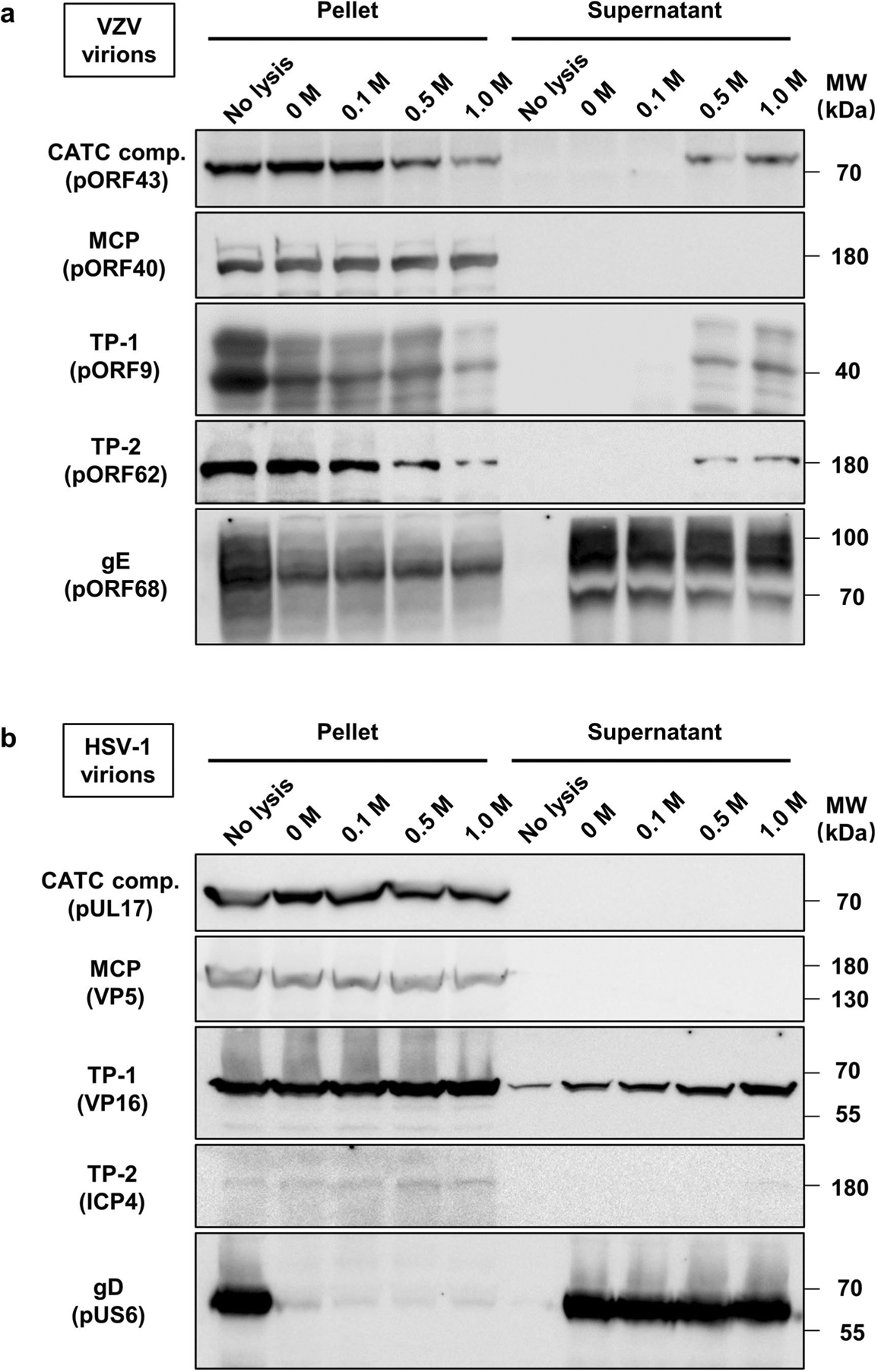
Purified virions of VZV (a) and HSV-1 (b) were not treated (no lysis) or treated with 1% Triton X-100 in the presence of 0, 0.1, 0.5, or 1M KCl. Viral lysates were then centrifuged through a 30% sucrose cushion. Pelleted (pellet) and released (supernatant) fractions were analyzed by SDS-PAGE and immunoblotting with antibodies targeting the CATC component (comp.) (VZV pORF43 and its HSV-1 counterpart, pUL17), MCP (VZV pORF40 and its HSV-1 counterpart, VP5), glycoproteins (VZV gE and HSV-1 gD), and other tegument proteins (TP; VZV pORF62 and pORF9, HSV-1 VP16 and ICP4). Markers of protein molecular mass (MW) are indicated in kDa on the right side of the panel. Experiments were performed two times independently with similar results.
Extended Data Fig. 9 |. Particle stability thermal release assay of purified VZV and HSV-1 capsids.
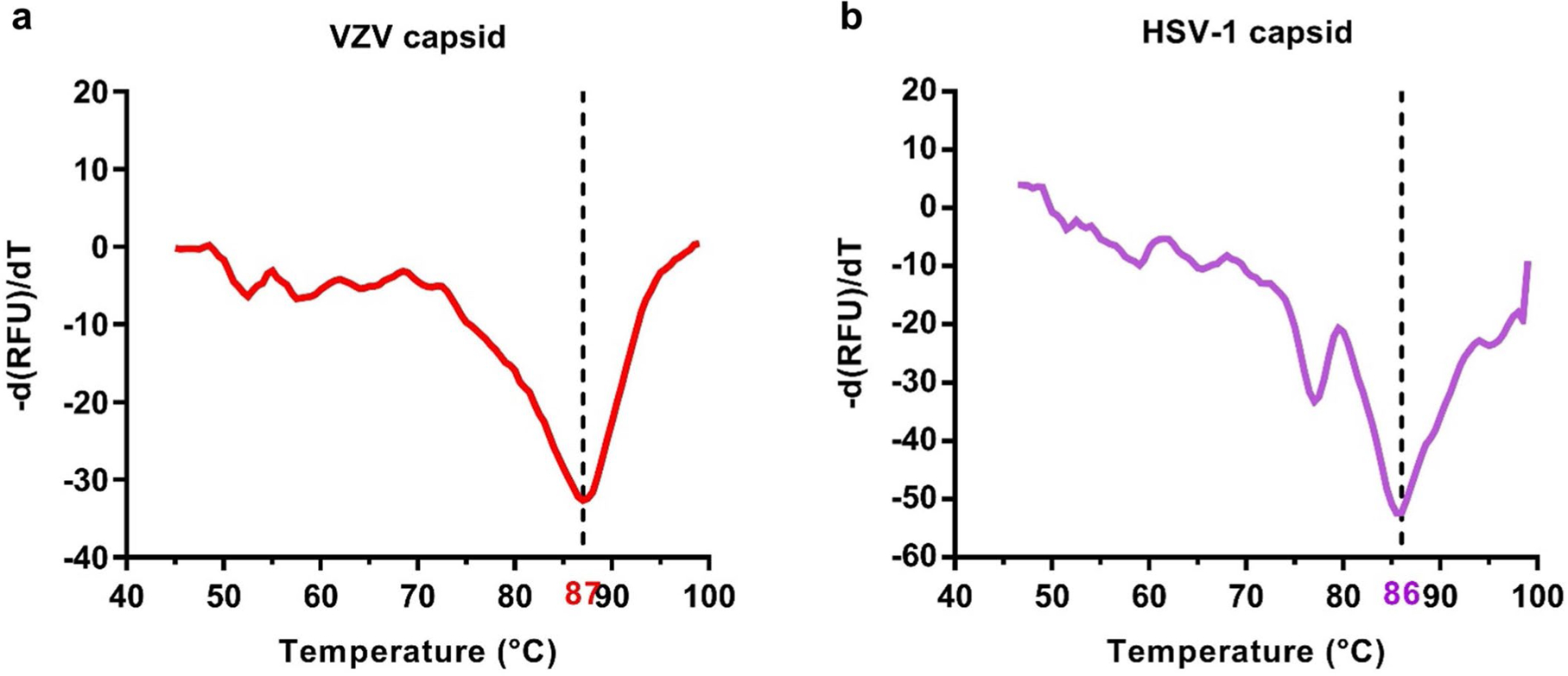
Purified capsids (a mixture of A-, B- and C-capsids) of VZV (a) and HSV-1 (b) were subjected to the thermofluor stability assay using the dyes SYTO-Green and SYTO9 to detect DNA and RNA exposures, respectively. The average negative first derivative of the fluorescence (measured in triplicate) with respect to temperature (−d(RFU)/dT) is shown.
Supplementary Material
Acknowledgements
Single-particle cryo-EM data were collected at the Center of Cryo-Electron Microscopy at Zhejiang University. We thank X. Zhang and S. Chang for support with facility access and data acquisition. This research was supported by grants from the National Science and Technology Major Projects for Major New Drugs Innovation and Development (no. 2018ZX09711003-005-003), the National Science and Technology Major Project of Infectious Diseases (no. 2017ZX10304402), the National Natural Science Foundation of China (no. 81871648, 81601762), the Research Unit of Frontier Technology of Structural Vaccinology of Chinese Academy of Medical Sciences (no. 2019RU022) and the US National Institutes of Health (DE025567/028583). The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
Competing interests
The authors declare no competing interests.
Supplementary information is available for this paper at https://doi.org/10.1038/s41564-020-0785-y.
References
- 1.Schmid DS & Jumaan AO Impact of varicella vaccine on varicella-zoster virus dynamics. Clin. Microbiol. Rev 23, 202–217 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arvin AM Varicella-zoster virus. Clin. Microbiol. Rev 9, 361–381 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross AH Modification of chicken pox in family contacts by administration of gamma globulin. N. Engl. J. Med 267, 369–376 (1962). [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization. Varicella and herpes zoster vaccines: WHO position paper, June 2014. Wkly. Epidemiol. Rec 89, 265–287 (2014). [PubMed] [Google Scholar]
- 5.Lukas K, Edte A & Bertrand I The impact of herpes zoster and post-herpetic neuralgia on quality of life: patient-reported outcomes in six European countries. Z. Gesundh. Wiss 20, 441–451 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson RW et al. The impact of herpes zoster and post-herpetic neuralgia on quality-of-life. BMC Med. 8, 37 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilden DH, Kleinschmidt-DeMasters BK, LaGuardia JJ, Mahalingam R & Cohrs RJ Neurologic complications of the reactivation of varicella-zoster virus. N. Engl. J. Med 342, 635–645 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Pergam SA, Limaye AP & AST Infectious Diseases Community of Practice. Varicella zoster virus in solid organ transplantation: guidelines from the American Society of Transplantation Infectious Diseases Community of Practice. Clin. Transplant 33, e13622 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Bozzola E & Bozzola M Varicella complications and universal immunization. J. Pediatr 92, 328–330 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Gnann JW Jr. Varicella-zoster virus: atypical presentations and unusual complications. J. Infect. Dis 186(Suppl 1), S91–S98 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Quinlivan M & Breuer J Clinical and molecular aspects of the live attenuated Oka varicella vaccine. Rev. Med. Virol 24, 254–273 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Cohen JI The varicella-zoster virus genome. Curr. Top. Microbiol. Immunol 342, 1–14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Staprans SI et al. Enhanced SIV replication and accelerated progression to AIDS in macaques primed to mount a CD4 T cell response to the SIV envelope protein. Proc. Natl Acad. Sci. USA 101, 13026–13031 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willer DO et al. Experimental infection of cynomolgus macaques (Macaca fascicularis) with human varicella-zoster virus. J. Virol 86, 3626–3634 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perciani CT et al. Protocol of a randomised controlled trial characterising the immune responses induced by varicella-zoster virus (VZV) vaccination in healthy Kenyan women: setting the stage for a potential VZV-based HIV vaccine. BMJ Open 7, e017391 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perciani CT et al. Live attenuated varicella-zoster virus vaccine does not induce HIV target cell activation. J. Clin. Invest 129, 875–886 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landmeier S et al. Gene-engineered varicella-zoster virus reactive CD4+ cytotoxic T cells exert tumor-specific effector function. Cancer Res. 67, 8335–8343 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Tanaka M et al. Vaccination targeting native receptors to enhance the function and proliferation of chimeric antigen receptor (CAR)-modified T cells. Clin. Cancer Res 23, 3499–3509 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carpenter JE, Hutchinson JA, Jackson W & Grose C Egress of light particles among filopodia on the surface of varicella-zoster virus-infected cells. J. Virol 82, 2821–2835 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cook ML & Stevens JG Labile coat: reason for noninfectious cell-free varicella-zoster virus in culture. J. Virol 2, 1458–1464 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harson R & Grose C Egress of varicella-zoster virus from the melanoma cell: a tropism for the melanocyte. J. Virol 69, 4994–5010 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gershon AA et al. Intracellular transport of newly synthesized varicella-zoster virus: final envelopment in the trans-Golgi network. J. Virol 68, 6372–6390 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen JJ, Zhu Z, Gershon AA & Gershon MD Mannose 6-phosphate receptor dependence of varicella zoster virus infection in vitro and in the epidermis during varicella and zoster. Cell 119, 915–926 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Dai X & Zhou ZH Structure of the herpes simplex virus 1 capsid with associated tegument protein complexes. Science 360, eaao7298 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu YT, Jih J, Dai X, Bi GQ & Zhou ZH Cryo-EM structures of herpes simplex virus type 1 portal vertex and packaged genome. Nature 570, 257–261 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan S et al. Cryo-EM structure of a herpesvirus capsid at 3.1 Å. Science 360, eaao7283 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Wang J et al. Structure of the herpes simplex virus type 2 C-capsid with capsid-vertex-specific component. Nat. Commun 9, 3668 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu X, Jih J, Jiang J & Zhou ZH Atomic structure of the human cytomegalovirus capsid with its securing tegument layer of pp150. Science 356, eaam6892 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y et al. Atomic structure of the human herpesvirus 6B capsid and capsid-associated tegument complexes. Nat. Commun 10, 5346 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong D et al. DNA-packing portal and capsid-associated tegument complexes in the tumor herpesvirus KSHV. Cell 178, 1329–1343 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dai X et al. Structure and mutagenesis reveal essential capsid protein interactions for KSHV replication. Nature 553, 521–525 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ilca SL et al. Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat. Commun 6, 8843 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou ZH et al. Four levels of hierarchical organization, including noncovalent chainmail, brace the mature tumor herpesvirus capsid against pressurization. Structure 22, 1385–1398 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cardone G et al. The UL36 tegument protein of herpes simplex virus 1 has a composite binding site at the capsid vertices. J. Virol 86, 4058–4064 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grunewald K et al. Three-dimensional structure of herpes simplex virus from cryo-electron tomography. Science 302, 1396–1398 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Wolfstein A et al. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic 7, 227–237 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Radtke K et al. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog. 6, e1000991 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaudhuri V, Sommer M, Rajamani J, Zerboni L & Arvin AM Functions of varicella-zoster virus ORF23 capsid protein in viral replication and the pathogenesis of skin infection. J. Virol 82, 10231–10246 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou ZH et al. Assembly of VP26 in herpes simplex virus-1 inferred from structures of wild-type and recombinant capsids. Nat. Struct. Biol 2, 1026–1030 (1995). [DOI] [PubMed] [Google Scholar]
- 40.Huet A et al. Extensive subunit contacts underpin herpesvirus capsid stability and interior-to-exterior allostery. Nat. Struct. Mol. Biol 23, 531–539 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu YT et al. A pUL25 dimer interfaces the pseudorabies virus capsid and tegument. J. Gen. Virol 98, 2837–2849 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bauer DW, Huffman JB, Homa FL & Evilevitch A Herpes virus genome, the pressure is on. J. Am. Chem. Soc 135, 11216–11221 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brandariz-Nunez A, Liu T, Du T & Evilevitch A Pressure-driven release of viral genome into a host nucleus is a mechanism leading to herpes infection. eLife 8, e47212 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dai X, Gong D, Wu TT, Sun R & Zhou ZH Organization of capsid-associated tegument components in Kaposi’s sarcoma-associated herpesvirus. J. Virol 88, 12694–12702 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snijder J et al. Vertex-Specific Proteins pUL17 and pUL25 Mechanically Reinforce Herpes Simplex Virus Capsids. J. Virol 91, e00123–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Z et al. Genome-wide mutagenesis reveals that ORF7 is a novel VZV skin-tropic factor. PLoS Pathog. 6, e1000971 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Homa FL et al. Structure of the pseudorabies virus capsid: comparison with herpes simplex virus type 1 and differential binding of essential minor proteins. J. Mol. Biol 425, 3415–3428 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu W et al. Structures of capsid and capsid-associated tegument complex inside the Epstein–Barr virus. Nat. Microbiol 10.1038/s41564-020-0758-1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Desai P, DeLuca NA & Person S Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology 247, 115–124 (1998). [DOI] [PubMed] [Google Scholar]
- 50.Krautwald M, Maresch C, Klupp BG, Fuchs W & Mettenleiter TC Deletion or green fluorescent protein tagging of the pUL35 capsid component of pseudorabies virus impairs virus replication in cell culture and neuroinvasion in mice. J. Gen. Virol 89, 1346–1351 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Borst EM, Mathys S, Wagner M, Muranyi W & Messerle M Genetic evidence of an essential role for cytomegalovirus small capsid protein in viral growth. J. Virol 75, 1450–1458 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perkins EM et al. Small capsid protein pORF65 is essential for assembly of Kaposi’s sarcoma-associated herpesvirus capsids. J. Virol 82, 7201–7211 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uetz P et al. Herpesviral protein networks and their interaction with the human proteome. Science 311, 239–242 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Stellberger T et al. Improving the yeast two-hybrid system with permutated fusions proteins: the varicella zoster virus interactome. Proteome Sci. 8, 8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang W et al. Varicella-zoster virus ORF7 interacts with ORF53 and plays a role in its trans-Golgi network localization. Virol. Sin 32, 387–395 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu J et al. Serological evaluation of immunity to the varicella-zoster virus based on a novel competitive enzyme-linked immunosorbent assay. Sci. Rep 6, 20577 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang HF et al. ORF7 of varicella-zoster virus is required for viral cytoplasmic envelopment in differentiated neuronal cells. J. Virol 91, e00127–00117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Selariu A et al. ORF7 of varicella-zoster virus is a neurotropic factor. J. Virol 86, 8614–8624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang W et al. Outer nuclear membrane fusion of adjacent nuclei in varicella-zoster virus-induced syncytia. Virology 512, 34–38 (2017). [DOI] [PubMed] [Google Scholar]
- 60.Mastronarde DN SerialEM: A program for automated tilt series acquisition on Tecnai microscopes using prediction of specimen position. Microsc. Microanal 9, 1182–1183 (2003). [Google Scholar]
- 61.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang K Gctf: real-time CTF determination and correction. J. Struct. Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grant T, Rohou A & Grigorieff N cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yan X, Sinkovits RS & Baker TS AUTO3DEM—an automated and high throughput program for image reconstruction of icosahedral particles. J. Struct. Biol 157, 73–82 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kimanius D, Forsberg BO, Scheres SH & Lindahl E Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scheres SH & Chen S Prevention of overfitting in cryo-EM structure determination. Nat. Methods 9, 853–854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kucukelbir A, Sigworth FJ & Tagare HD Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pettersen EF et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 69.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 70.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structure coordinates have been deposited in the Protein Data Bank under accession numbers 6LGL (A-capsid) and 6LGN (C-capsid). The corresponding electron microscopy density maps have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-0880 (icosahedral reconstruction of the A-capsid), EMD-0881 (icosahedral reconstruction of the C-capsid), EMD-30251 (icosahedral reconstruction of the virion capsid), EMD-30248 (localized reconstruction of the A-capsid) and EMD-30250 (localized reconstruction of the C-capsid). Source data are provided with this paper.


