Abstract
Cells infected by influenza virus mount a large-scale antiviral response and most cells ultimately initiate cell-death pathways in an attempt to suppress viral replication. We performed a CRISPR/Cas9-knockout selection designed to identify host factors required for replication following viral entry. We identified a large class of presumptive antiviral factors that unexpectedly act as important pro-viral enhancers during influenza virus infection. One of these, IFIT2, is an interferon-stimulated gene with well-established antiviral activity but limited mechanistic understanding. As opposed to suppressing infection, we show here that IFIT2 is instead repurposed by influenza virus to promote viral gene expression. CLIP‐seq demonstrated that IFIT2 binds directly to viral and cellular mRNAs in AU‐rich regions, with bound cellular transcripts enriched in interferon‐stimulated mRNAs. Polysome and ribosome profiling revealed that IFIT2 prevents ribosome pausing on bound mRNAs. Together, the data link IFIT2 binding to enhanced translational efficiency for viral and cellular mRNAs and ultimately viral replication. Our findings establish a model for the normal function of IFIT2 as a protein that increases translation of cellular mRNAs to support antiviral responses and explain how influenza virus uses this same activity to redirect a classically antiviral protein into a pro-viral effector.
Cells express factors that selectively target and inhibit proteins and nucleic acids from invading pathogens1,2. Simultaneously, pathogens neutralize these inhibitors and co-opt other cellular factors for their own replication. The balance between these opposing forces influences the outcome of an infection and the course of disease.
Influenza virus is a serious public health threat causing significant morbidity and mortality. Humans are under continual assault by seasonal influenza virus outbreaks as well as sporadic pandemics with the potential for widespread infection and disease3,4. Like all viruses, influenza virus exploits, and in some cases subverts, cellular proteins and pathways to promote replication. These physical and genetic interactions have significant impacts on virus replication, innate antiviral responses and pathology. Moreover, the ability of influenza virus to productively engage the intracellular environment determines its host range and potential to transmit and cause disease in humans5.
Genome-wide knockout screens have provided a global view of the interface between influenza virus and its host6–10. Despite the success of these approaches, technical and biological limitations have created a strong bias toward steps involved in viral entry. Here we perform a screen to specifically query the post-entry role of pro-viral host factors during influenza virus infection. Counter to expectation, our screen revealed that several innate antiviral factors, especially IFIT2, enhanced influenza virus replication. Mechanistic studies showed that IFIT2 binds viral and host mRNAs and prevents ribosome pausing. This increases the translational efficiency of IFIT2-bound mRNAs, enhancing viral gene expression and replication. The combination of unbiased screening and mechanistic studies identified the process by which influenza virus hijacks IFIT2 to maximize replication, while also revealing how IFIT2 functions as part of the innate immune response.
Results
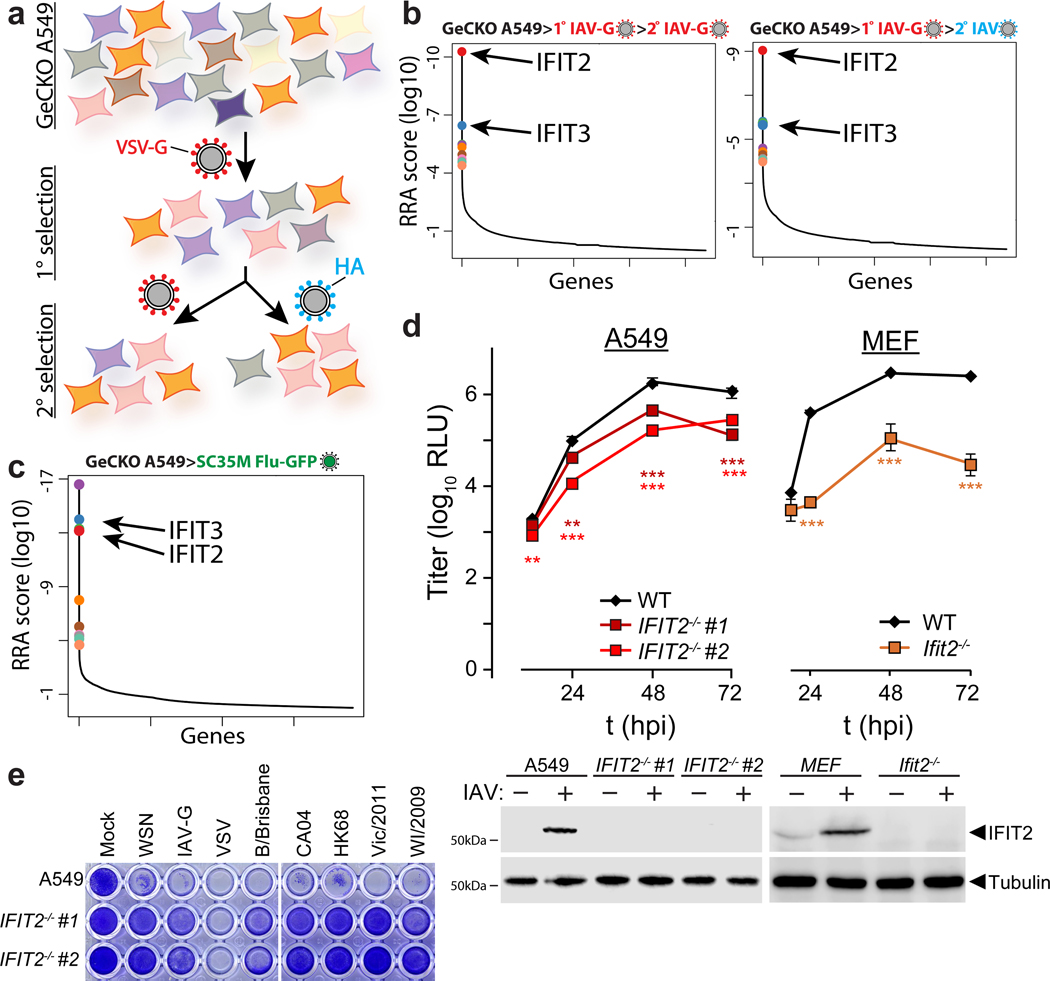
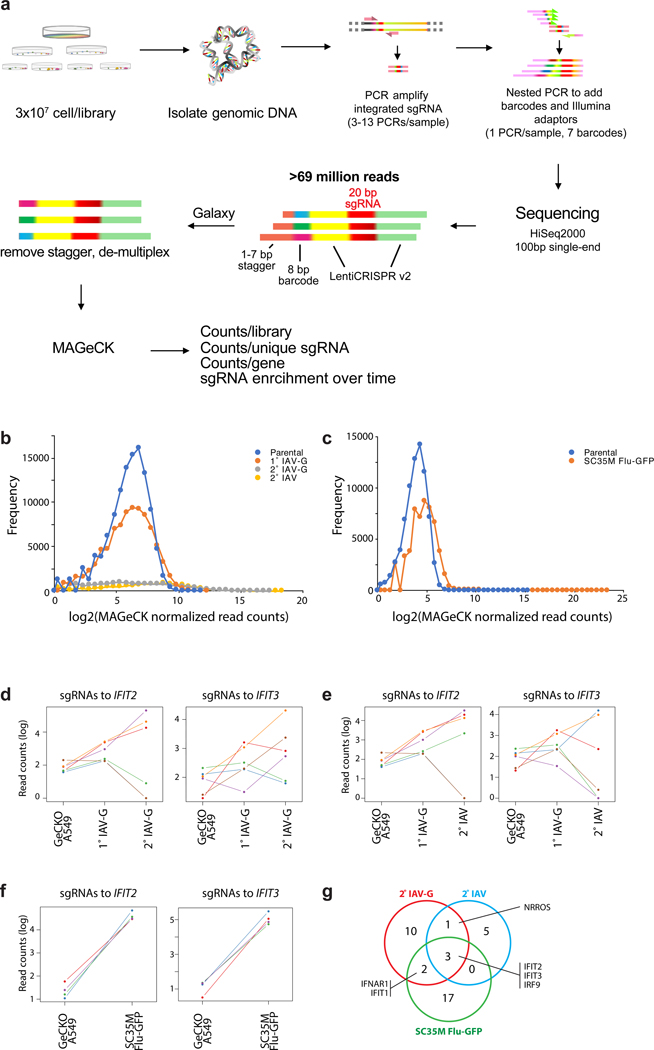
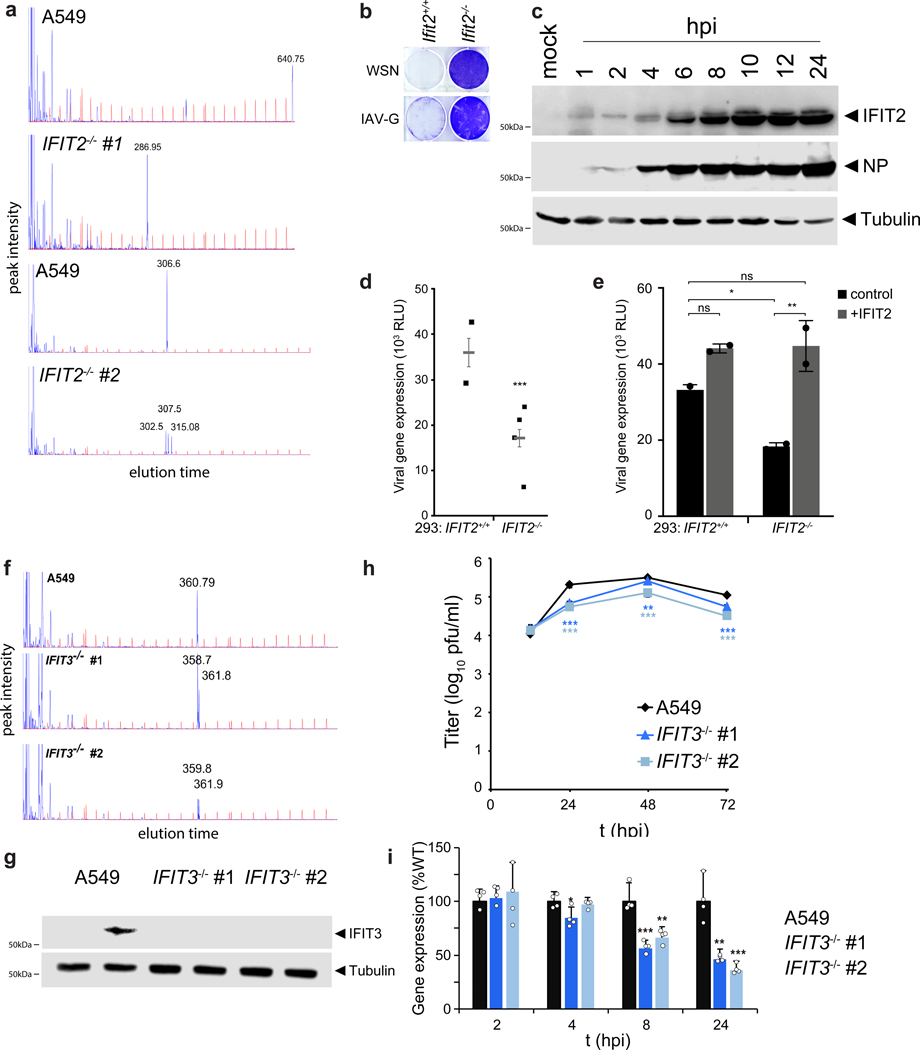
To identify cellular regulators of viral replication, we performed a genome-wide CRISPR-knockout selection (Fig. 1a, Extended Data Fig. 1a-c). Pooled gene-edited human A549 lung cells were challenged with influenza A virus (IAV) and surviving cells were isolated through sequential rounds of selection. Results from prior virus:host interaction screens identified key cellular co-factors, but also demonstrated a bias towards the entry process6–9. We therefore performed our selection with a IAV-G, a replication-competent influenza virus based on the A/WSN/1933 strain (H1N1; WSN) that is stably pseudotyped with the vesicular stomatitis virus glycoprotein (VSV-G) that bypasses this technical bottleneck11. Surviving cells were either re-challenged with IAV-G or were counter-selected with bona fide influenza virus. Both selection schemes enriched for cells with mutations in the genes coding for IFIT2 (interferon-induced protein with tetratricopeptide repeats 2) and IFIT3 (Fig. 1b, Supplementary Table 1). A separate selection was performed with an independently generated gene-edited pool of A549 cells. Cells were challenged with a different strain of IAV encoding GFP based on A/seal/Massachusetts/1-SC35M/1980 (SC35M Flu-GFP). IFIT2 and IFIT3 gene-edited cells were again among the most enriched cells in this selection (Fig. 1c, Supplementary Table 2). Thus, independent selections using different viruses showed that surviving cells were highly enriched for mutations in IFIT2 and IFIT3 (Extended Data Fig. 1d-g).
Figure 1. A CRISPR knockout screen identifies the antiviral proteins IFIT2 and IFIT3 as pro-viral regulators of influenza virus infection.
a, Schematic of the genome-scale CRISPR knockout (GeCKO) screen designed to specifically query host factors important for influenza virus infection after viral entry. b-c, MAGeCK (Model-based genome wide knock out)52 analysis for selection schemes involving sequential IAV-G selections (b, left), IAV-G followed by WSN (b, right), or seven rounds with SC35M Flu-GFP (c). d, Multicycle replication kinetics of WSN in wildtype or knockout cells. Western blotting confirmed loss of IFIT2 in two independent clonal human A549 cell lines and loss of Ifit2 in mouse embryo fibroblasts. Data are mean of n = 3 independent infections ± sd. (** p < 0.01, *** p < 0.001 when compared to WT; one-way ANOVA with post hoc Tukey’s HSD for multiple comparisons or a two-tailed Student’s t-test). e, Viability of parental or IFIT2 knockout A549 cells following challenge with diverse primary influenza isolates and VSV. VSV-G; vesicular stomatitis virus glycoprotein. IAV; influenza A virus. IAV-G; IAV stably encoding VSV-G. SC35M; A/seal/Mass/1-SC35M/1980. WSN; A/WSN/1933. B/Brisbane; B/Brisbane/60/2008. CA04; A/California/04/2009. HK68; A/Hong Kong/1/1968. Vic/2011; A/Victoria/361/2011. WI/2009; A/Wisconsin/15/2009.
IFITs are a family of proteins in humans (e.g. IFIT1, 2, 3 and 5) that are induced following type I interferon (IFN) treatment or pattern recognition receptor activation12,13. IFITs exert antiviral activity against a diverse collection of RNA and DNA viruses, although this activity varies depending on the virus, IFIT paralog, and the species of origin14–22; the molecular functions of IFIT2 are largely undefined. Thus, it was unexpected that our selection, which was designed to identify cellular factors necessary for influenza virus replication, instead identified IFN-stimulated genes (ISGs) with established antiviral activity for other viruses.
To validate the results of our selection, we evaluated the effects of IFIT2 on IAV replication. Multi-step IAV (WSN) replication was reduced by 1- to 2-log10 in two independently generated human IFIT2 knockout cell lines (Fig. 1d, Extended Data Fig. 2a). Similar results were obtained in mouse Ifit2 knockout embryo fibroblasts. Consistent with its role as an ISG, IFIT2 was largely undetectable in uninfected cells and upregulated upon infection. Knockout of IFIT2 or Ifit2 also conferred survival to cells challenged by diverse influenza viruses, including multiple IAV primary isolates, IAV-G, and influenza B virus (IBV) (Fig. 1e, Extended Data Fig. 2b). This phenotype was not a generic response, as knockout cells remained sensitive to infection and virally mediated cell death when infected with VSV (Fig. 1e). Additionally, we validated the role IFIT3 during IAV infection. Loss of IFIT3 also reduced IAV replication, although the magnitude was less than with loss of IFIT2 (Extended Data Fig. 2f-h). By contrast, loss of IFIT1 does not affect influenza virus replication21. The appearance of IFIT1 as a lower-confidence candidate in our screens could result from the ability of IFIT1, 2 and 3 to form hetero-oligomers and modulate each other’s activities15,23–25.
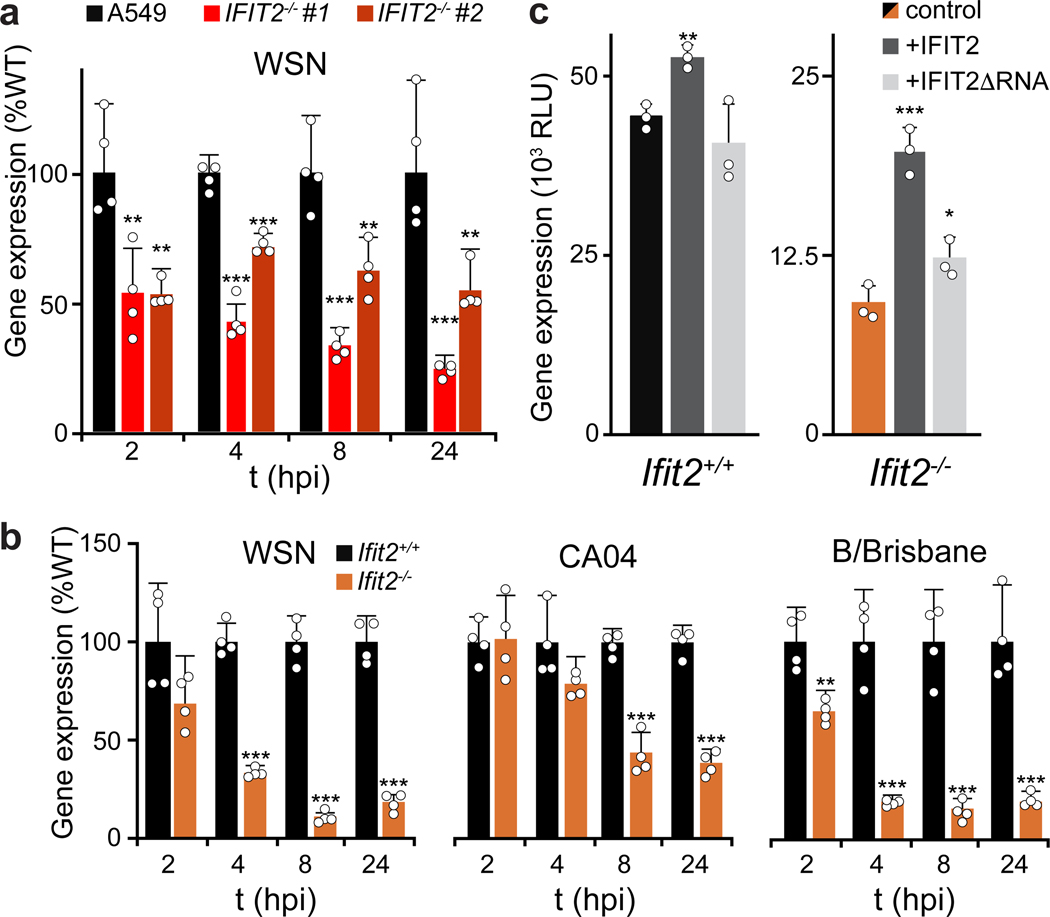
Cells lacking IFIT2 survive challenge by viruses encoding native HA or pseudotyped with VSV-G, suggesting that the phenotype is independent of the influenza entry process (Fig. 1b and 1e). Given that IFIT2 is rapidly induced upon IAV infection (Extended Data Fig. 2c and 26), we used highly sensitive reporter viruses to query primary transcription at early times post-inoculation and subsequent gene expression throughout infection27,28. Human A549 and 293 IFIT2−/− cell lines displayed ~50% decrease in viral gene expression beginning 2 hpi and continuing through 24 hpi (Fig. 2a, Extended Data Fig. 2d). Similar trends were obtained in mouse Ifit2−/− cells infected with reporter viruses based on WSN, the primary IAV isolate from the 2009 pandemic (CA04), or a primary IBV isolate (B/Brisbane) (Fig. 2b). IFIT3−/− cell lines also exhibit defects in gene expression (Extended Data Fig. 2i).
Figure 2. IFIT2 promotes IAV gene expression and progression through the viral life cycle.
a-b, Viral gene expression measured in parental and IFIT2 (a) or Ifit2 (b) knockout cells infected with IAV or IBV reporter viruses (MOI = 0.05). c, WSN viral gene expression was measured 12 hr after infection of MEFs transiently complemented with IFIT2 or the RNA-binding mutant IFIT2ΔRNA. For all panels, reporter activity is a proxy for gene expression and data are shown as mean of n = 3–4 independent infections + sd (*p < 0.05, **p < 0.01; *** p < 0.001 when compared to WT or control conditions; Two-tailed Student’s t-test for pairwise comparisons between control and knockout cells (b) or one-way ANOVA with post hoc Tukey’s HSD for multiple comparisons to control conditions (a and c).
Defects in viral gene expression in seven independently generated knockout lines from three different cell types strongly implicate mutations in the IFIT2 or Ifit2 loci as opposed to background effects or off-target edits (Fig. 2a-b, Extended Data Fig. 2d). To confirm the pro-viral role of IFIT2, we transiently complemented knockout cells. Ectopic expression of IFIT2 in MEFs or 293 cells more than doubled viral gene expression in the knockout cells, but exhibited modest enhancement in WT cells (Fig. 2c, Extended Data Fig. 2e). IFIT2 binds model RNAs in vitro29, although the RNAs bound by IFIT2 in cells are not known. To determine if RNA binding was important for the pro-viral function of IFIT2, cells were complemented with an RNA-binding IFIT2 mutant (R292E/K410E, IFIT2ΔRNA)29. The enhanced gene expression detected in cells complemented with IFIT2 was lost in cells complemented with IFIT2ΔRNA (Fig. 2c), suggesting a role for RNA binding by IFIT2 in promoting viral gene expression. Collectively, these data confirm that IFIT2 and IFIT3 have pro-viral activities and regulate expression of viral proteins during IAV infection.
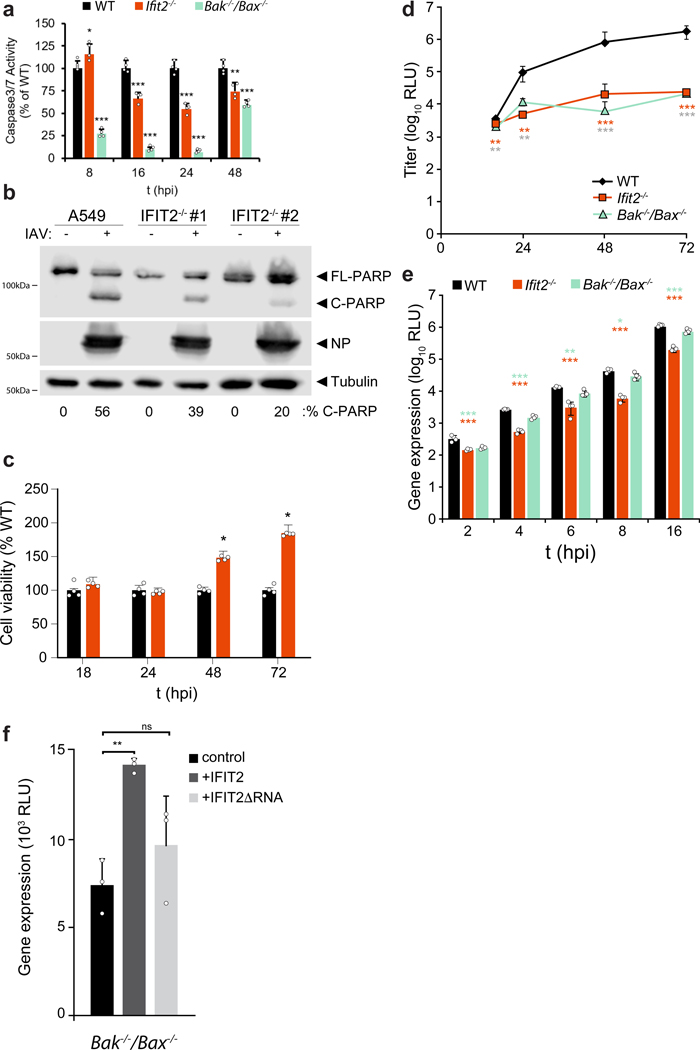
IFIT2 plays a role in inducing apoptosis via the intrinsic mitochondrial pathway and the proteins Bak and Bax23. Whereas apoptosis is a primary mechanism of cell death during IAV infection, it is also a key cellular process required for appropriate progression of the viral life cycle30–32. We therefore assessed whether changes in viral gene expression were related to IFIT2-induced apoptosis. Loss of IFIT2 reduced apoptosis and increased cell viability at later stages of infection (Extended data Fig. 3a-c). Lower viral replication in IFIT2-lacking cells may have contributed to reduced apoptosis. Interestingly, Bak−/−/Bax−/− cells phenocopied Ifit2−/− cells in multi-step replication assays, but showed minimal changes in gene expression early during infection, unlike the significant defects in gene expression in cells lacking Ifit2 (Extended data Fig. 3d-e). Moreover, expression of IFIT2 in Bak−/−/Bax−/− cells, but not IFIT2ΔRNA, enhanced viral gene expression during infection (Extended data Fig. 3f). Thus, IFIT2 still stimulates viral gene expression independent of its ability to stimulate apoptosis, indicating that these are separable functions.
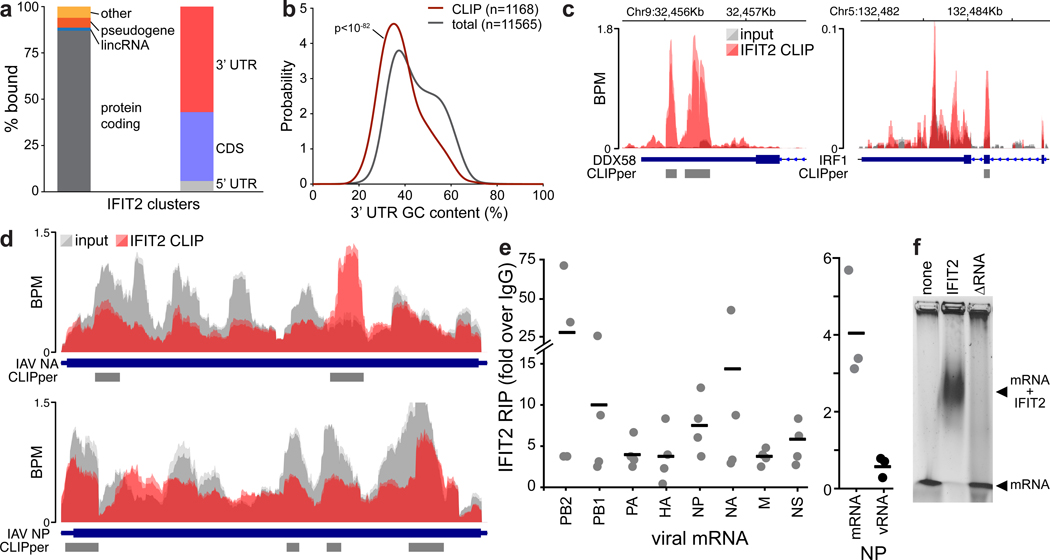
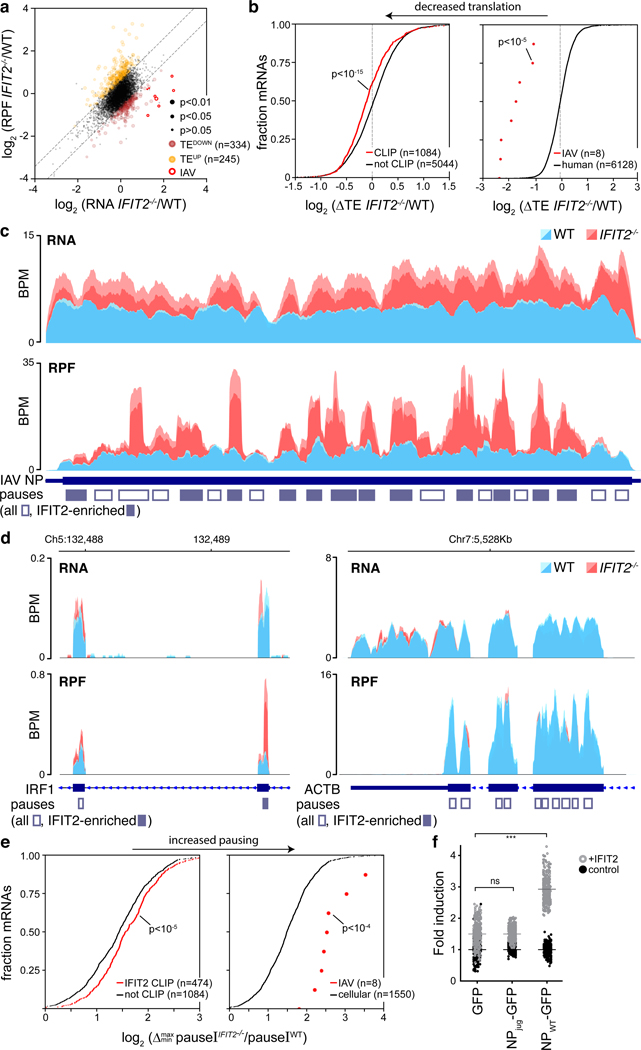
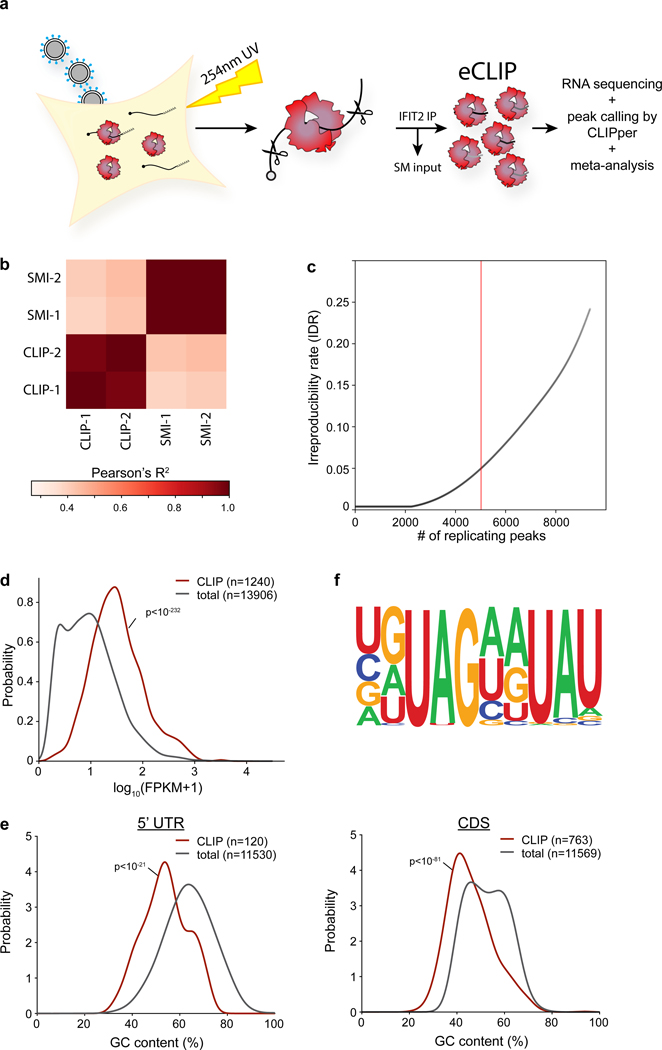
RNA binding by IFIT2 was important for enhanced viral gene expression. We therefore performed UV crosslinking and immuno-precipitation followed by sequencing (CLIP-Seq) to identify all RNAs bound by IFIT2 during infection. IFIT2 preferentially binds protein-coding RNAs, with 1804 specific binding sites in 1247 mRNAs (Fig. 3a, Extended Data Fig. 4a-c, Supplemental Table 3). Meta-analysis mapped most IFIT2 binding sites to the 3’ UTR of cellular mRNAs, although abundant binding in the coding sequence was also detected. Binding was not solely dependent on RNA levels as both abundant and lowly expressed transcripts were bound (Extended Data Fig. 4d). Motif discovery algorithms failed to find a strict sequence that defines IFIT2 binding sites. However, IFIT2-bound sites showed a clear skew in their nucleotide content and are AU rich, with ~20% of the sites containing the degenerate UAGnnUAU motif (Fig. 3b, Extended Data Fig. 4e-f). Gene ontology analysis revealed that IFIT2-bound mRNAs are highly enriched for genes involved in viral processes (Supplemental Table 3). ISGs were significantly enriched in the bound fraction (χ2 p<10−23), including potent regulators of viral infection like DDX58 (RIG-I), OAS, STAT1 and IRF1; IFIT2 even bound its own message as well as that of IFIT1 and IFIT3 (Fig. 3c, Supplemental Table 3).
Figure 3. IFIT2 selectively binds AU-rich regions in viral and host mRNAs.
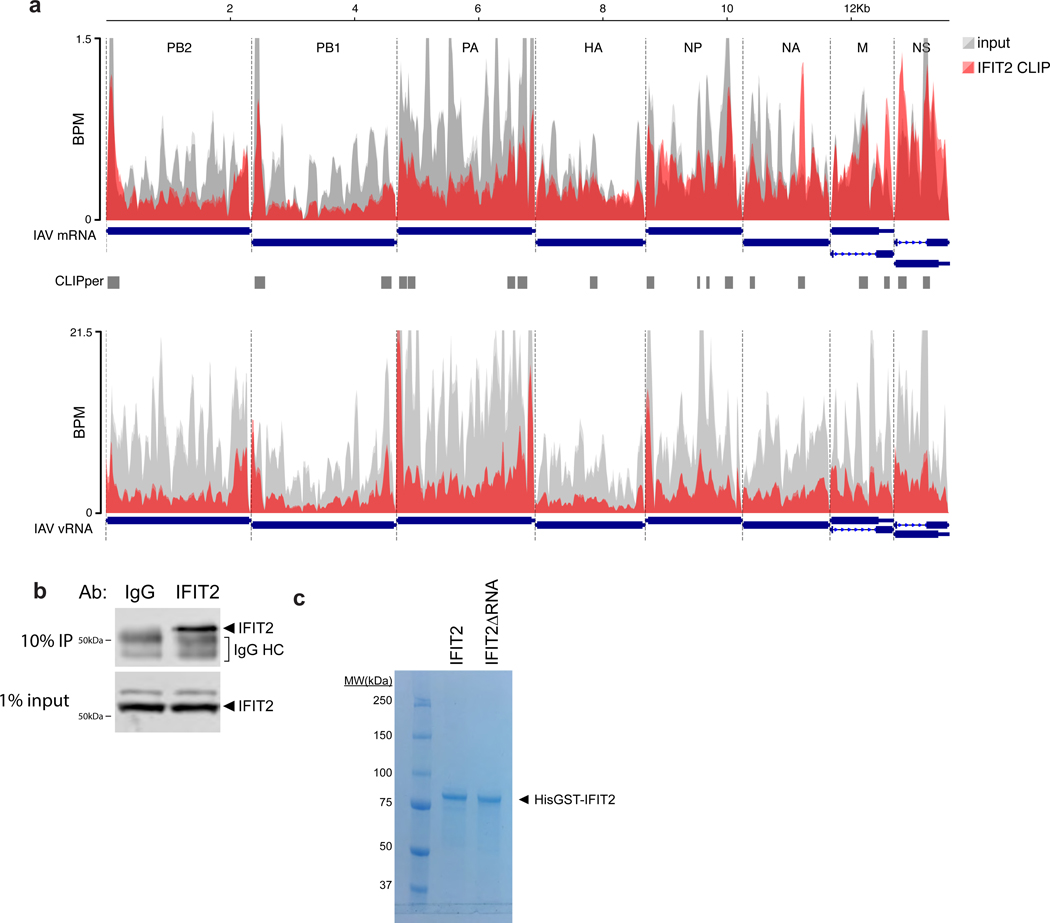
CLIP was performed to identify RNAs directly bound by IFIT2 during IAV WSN infection. a, RNA biotypes bound by IFIT2 and binding locations within protein-coding transcripts were determined. b, Meta-analysis of GC content for peaks identified in the 3’ UTR of bound mRNAs. IFIT2-bound sites were compared to all expressed 3’ UTRs by a Mann-Whitney U-test. c-d, Relative CLIP intensity for IFIT2-bound and size-matched input control RNAs for host (c) and viral (d) transcripts. Data from replicate experiments are plotted as mean and standard deviation using dark and light shades of the same color, respectively. CLIPper-identified peaks are shown below. BPM = bins per million. e, qRT-PCR of viral mRNAs or genomic vRNA in IFIT2 RNA-immunoprecipitation (RIP) samples from infected cells. All replicates are shown (n = 4) along with a bar indicating the mean. f, Electromobility shift assay of viral NP mRNA with recombinant IFIT2 or IFIT2ΔRNA.
Remarkably, IFIT2 bound transcripts from all 8 influenza virus genes, but did not bind genomic minus-sense vRNA (Fig. 3d, Extended Data Fig. 5a). The number of called peaks on viral mRNAs was limited due to high background levels of viral sequence in the input control. This is due to the fact that influenza virus NP, whose function is to bind viral RNAs, co-migrates with IFIT2 during gel electrophoresis. This causes an artificial enrichment of viral RNAs in the size-matched input control used to establish baseline RNA levels in CLIP-Seq. Hence, the peak calling here is likely an under-estimation of the intensity of IFIT2 binding to viral RNAs. To confirm interactions with viral RNAs, we performed RNA immunoprecipitations (RIPs) coupled with RT-qPCR. Again, IFIT2 selectively bound RNA from all influenza virus genes, showing ~3- to 25-fold enrichment over IgG controls (Fig. 3e, left, Extended Data Fig. 5b). IFIT2 RNA binding was specific for plus-sense RNA as genomic vRNA was not enriched by IFIT2 RIP (Fig. 3e, right). Electromobility shift assays using recombinant IFIT2 and in vitro transcribed NP mRNA demonstrated that IFIT2 directly binds viral mRNAs, further confirming the CLIP-Seq data (Fig. 3f, Extended Data Fig. 5c). This binding was lost with recombinant IFIT2ΔRNA. These data show that IFIT2 binds AU-rich regions on viral and host mRNAs during infection.
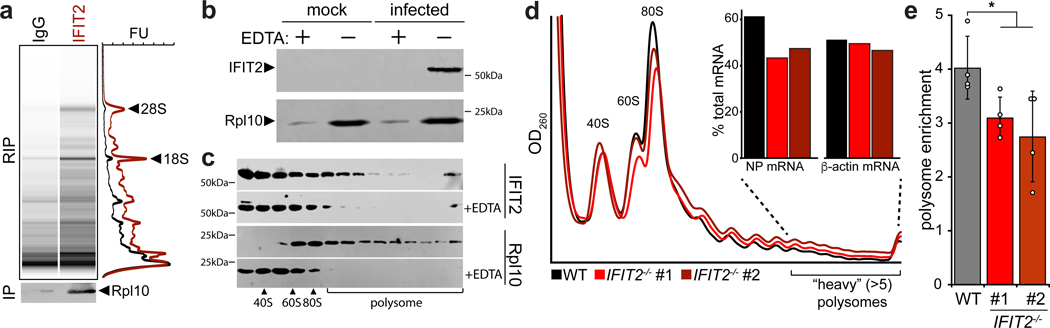
We next sought to establish a mechanism by which IFIT2 enhances viral gene expression. IFIT2 preferentially bound mRNAs, therefore we asked whether IFIT2 associated with mRNAs undergoing translation. Capillary electrophoresis of RNAs present in IFIT2 RIPs from infected cells demonstrated strong enrichment of ribosomal RNAs (rRNAs), including 28S, 18S and 5/5.8S (Fig. 4a). Parallel immunoprecipitations of IFIT2 showed specific co-precipitation of the ribosome, as measured by blotting for the large subunit protein Rpl10. IFIT2-mRNA interactions were further characterized by performing polysome pelleting to isolate actively translating ribosomes (Fig. 4b). IFIT2 co-pelleted with ribosomes from infected cell lysate. EDTA treatment dissociated polysomes, which dramatically reduced the amount of pelleted Rpl10 and completely eliminated co-pelleting IFIT2.
Figure 4. IFIT2 associates with active ribosomes and increases the translational efficiency of viral mRNAs.
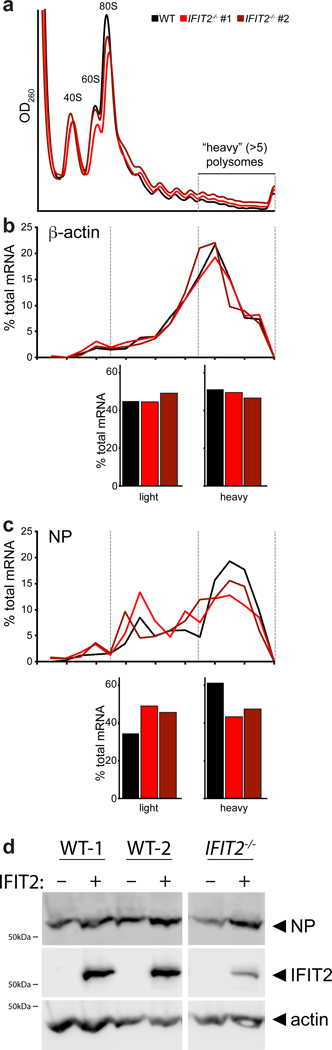
a, Capillary electrophoresis traces for RNAs that co-precipitate with IFIT2 or an IgG control from WSN-infected cell lysate. Ribosomal RNAs were identified by their size and are indicated. Simulated gels and paired chromatographs quantified by fluorescence units (FU) are shown. In a separate IFIT2 immuno-precipitation, co-precipitating ribosomal protein (Rpl10) was detected by western blot. b-c, Translationally active ribosomes were separated from infected or mock cell lysate by polysome pelleting (b) or profiling (c). Ribosome were dissociated with EDTA where indicated. Proteins in polysome pellets or fractions were detected by western blot. d, Polysome profiles from WT and IFIT2−/− A549 cells infected with IAV WSN. mRNA abundance amongst highly translated messages (>5 ribosomes/mRNA) was determined by qRT-PCR (inset). e, Enrichment of NP mRNA in the polysome relative to total RNA was determined by qRT-PCR for samples from WT and IFIT2/- cells infected with IAV. Data are mean of n = 4 biologically independent samples +/− sd. (* p < 0.05, one-way ANOVA with post hoc Tukey’s HSD for multiple comparisons to control conditions).
Polysome profiling was then performed to gain more resolution on the ribosome complexes with which IFIT2 interacts (Fig. 4c). IFIT2 co-migrated with initiating ribosomes and with the polysome fraction, indicating that IFIT2 is present with highly translated mRNAs containing multiple ribosomes. EDTA disrupted polysomes and IFIT2 no longer sedimented in the gradient. The abundance of 40S, 60S and 80S peaks as well as the polysome profiles were indistinguishable between WT and both IFIT2−/− A549 cell lines (Fig. 4d). Thus, knockout of IFIT2 did not cause global changes in translation during infection. The distribution of viral mRNAs in the polysome profile was then measured. Approximately 60% of total NP mRNA from infected cells was present in heavy polysome fractions, which represents highly translated messages with more than five ribosomes per mRNA. The abundance of viral NP mRNA amongst highly translated messages was reduced by approximately one third in the IFIT2−/− cell lines (Fig. 4d), and these mRNA were redistributed to fractions containing less efficiently translated messages (Extended Data Fig. 6a-c). The distribution of β-actin mRNA, a transcript that is not bound by IFIT2, was unchanged in its absence (Fig. 4d and Extended Fig. 6b). These findings were confirmed with a different separation strategy, measuring NP mRNA levels in polysome pellets from WT and IFIT2−/− cells lines (Fig. 4e). The proportion of total NP mRNA present in the polysome pellet was significantly decreased in both IFIT2−/− lines as compared to the WT control.
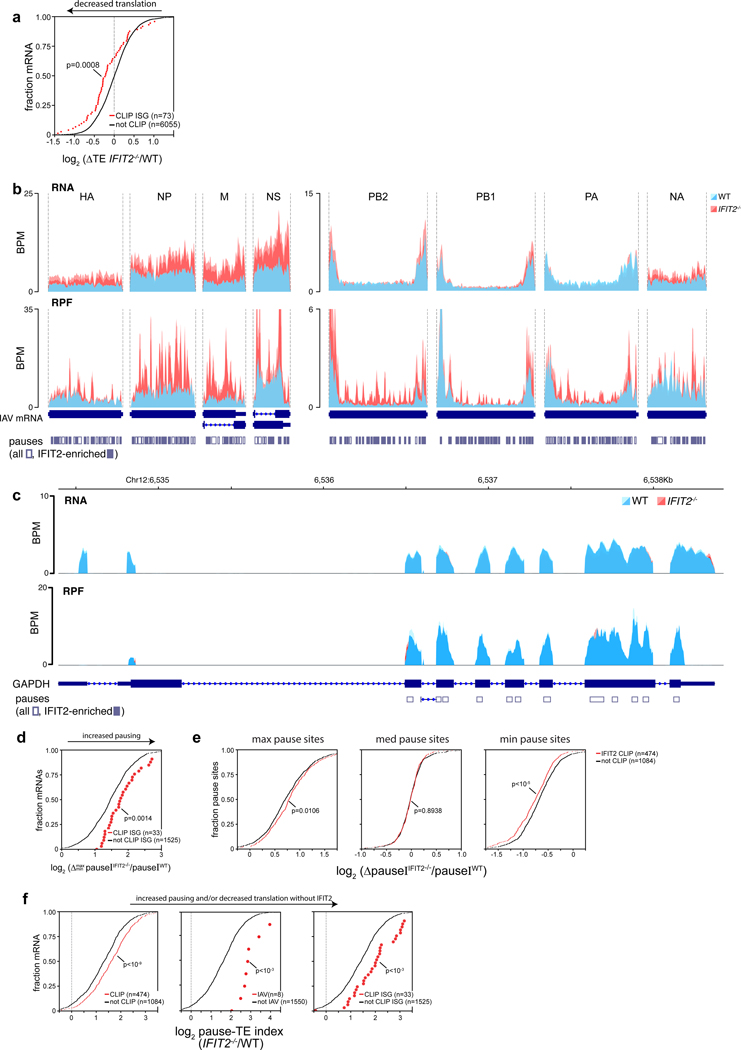
To specifically assess changes in translation to all IFIT2-bound mRNAs, we performed ribosome profiling of infected cells. Translational efficiency was measured as the number of ribosome protected fragments (RPFs) for an mRNA relative to its abundance in the cell33. The translational efficiencies of IFIT2-bound viral mRNAs, cellular mRNAs, and ISG mRNAs were all reduced in IFIT2−/− cells compared to WT, whereas translation of most unbound mRNAs was largely unchanged (Fig. 5a-b, Extended Data Fig. 7a, Supplemental Table 4). Viral mRNA levels increased ~2–4 fold in IFIT2−/− cells, possibly due to decreased translation of ISGs under these conditions. Whereas translation efficiency decreased for viral mRNAs in IFIT2−/− cells compared to WT, the number of RPFs, used as a proxy for absolute translation levels, was largely unchanged. Nonetheless, western blotting revealed viral NP levels were lower in infected IFIT2−/− cells and restored upon ectopic IFIT2 expression (Extended Data Fig. 6d). This agrees with prior results showing defects in viral gene expression in cells lacking IFIT2/Ifit2 (Fig. 2, Extended Data Fig. 2d-e). To understand this apparent disconnect, RPFs were mapped to their respective transcripts (Fig. 5c, Extended Data Fig. 7b). Ribosome footprint density was distributed across the coding sequence in viral mRNAs during infection of WT cells, with minor local variations, reflecting efficiently translated messages33. In contrast, strong peaks of ribosome density selectively appeared on viral mRNAs during infection of IFIT2−/− cells. Many of the peaks spanned ~28–32 nt, the predicted footprint of a single ribosome34. The appearance of highly aligned footprints in the absence of IFIT2 is indicative of extensive ribosome pausing on viral mRNAs33. Thus, many of the RPFs on viral mRNAs in IFIT2−/− cells may result from non-productive pauses, explaining the decrease in protein production without a corresponding reduction in RPFs.
Figure 5. IFIT2 modulates translational efficiency by preventing ribosome pausing.
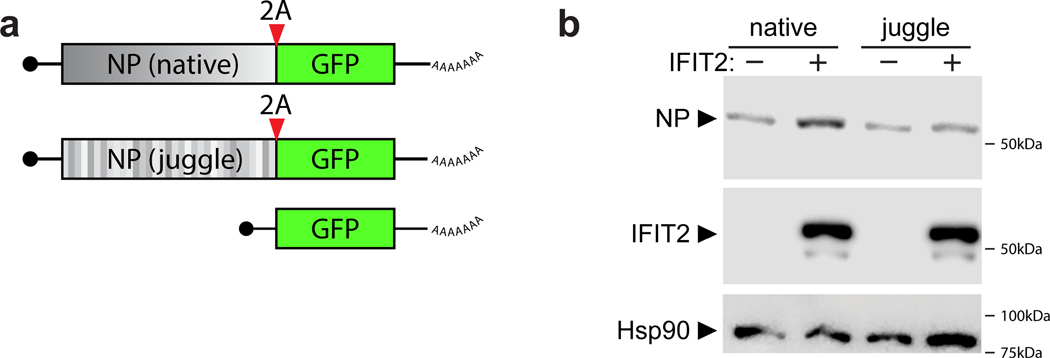
Differentially translated mRNAs were identified by ribosome profiling of IAV WSN-infected WT and IFIT2−/− A549 cells. a, The relative contribution of transcription and translational were assessed by plotting fold changes between IFIT2−/− and WT cells for input RNA versus ribosome protected fragments (RPF) from each transcript. Dotted lines represent a 1.5-fold (log2 0.58) threshold. Transcripts that are differentially translated in IFIT2−/− cells have been colored, with IAV mRNAs highlighted. p values derived from a two-sided F-test (see Supplemental Table 4). b, The translational efficiency of all IFIT2-bound mRNAs (left) or viral mRNAs (right) is decreased in the absence of IFIT2. Bound transcripts were compared to unbound transcripts via a Mann-Whitney U-test. c-d, Accumulation of paused ribosomes in the absence of IFIT2. Normalized read density for total RNA (top) and RPFs (bottom) mapping to IFIT2-bound NP (c) and IRF1 (d, left) or unbound β-actin (d, right) mRNAs in infected WT and IFIT2−/− cells. Data from replicate experiments are plotted as mean and standard deviation using dark and light shades of the same color, respectively. Pause sites are shown below. Pause sites enriched in IFIT2−/− cells >1.5-fold are filled in. BPM = bins per million. e, Ribosome pausing increases during infection of IFIT2−/− cell. Changes in pause intensity (pauseI) at the maximum and minimum pauseI sites on each transcript were calculated during infection of WT and IFIT2−/− cells. All IFIT2-bound mRNAs (left) or viral mRNAs (right) were compared to remaining transcripts via a two-sided Mann-Whitney U-test. f, Sequence-dependent enhancement of NP translation in the presence of IFIT2 or control. NP was encoded as a GFP polyprotein using WT or juggled codons. Expression was quantified by fluorescence imaging of live cells. n = 2 biological replicates quantifying 177 fields of view each. IFIT2 induction of NP-GFP variants was compared to the GFP-only control using a one-way ANOVA with post hoc Tukey’s HSD (*** p < 0.001).
Global changes in translation kinetics were analyzed by identifying peaks in ribosome density, quantifying the intensity of pausing at the peak, and then comparing pause intensity (pauseI) between infected WT and IFIT2−/− cells. Ribosome pauses were present on viral and host mRNAs in WT cells, consistent with known position-specific variations in the speed of the ribosome (Fig 5c-d, Extended Data Fig. 7b-c)33,34. However, pausing was consistently increased in IFIT2−/− cells for cellular mRNAs, ISG mRNAs and viral mRNAs that are normally bound by IFIT2 (Fig 5e, Extended Data 7d-e). Pausing increased at many sites on viral mRNAs and at more limited positions in IFIT2-bound cellular transcripts (Fig. 5c-d). Changes in pause intensity at these sites were not the result of global changes in ribosome processivity, as polysome profiles were indistinguishable in WT and IFIT2−/− cells and unbound control transcripts like ACTB and GAPDH showed no differential pausing (Fig. 4d, 5d, Extended Data Fig. 7c).
These data suggest that decreased translational efficiency for viral mRNA and cellular IFIT2-bound mRNAs may be due to ribosome pausing that occurs when IFIT2 is absent. We tested this possibility by correlating IFIT2-dependent changes in pause intensity and translational efficiency. IFIT2-bound viral mRNAs, cellular mRNAs, and ISG mRNA all showed a stronger correlation than unbound transcripts (Extended Data Fig. 7f). Finally, we used a functional assay to connect IFIT2 binding to changes in translation. IFIT2 was co-expressed in cells with an influenza virus NP-GFP polyprotein where NP was encoded by the native transcript, or one where silent mutations juggled the RNA sequence without changing the coding potential (NPjug) (Extended Data Fig. 8a). NP and NPjug contain a similar overall AU content, but the codon juggling eliminated clustered AU-rich regions found in the native NP mRNA. Changing the coding sequence did not alter expression of NP or NPjug when they were expressed on their own (Fig. 5f). IFIT2 co-expression caused a ~3-fold increase in natively coded NP, following patterns observed in infection. By contrast, IFIT2 caused background-level changes in NPjug expression that mirrored the GFP control. Similar results were obtained when blotting NP itself (Extended Data 8b). Combined, these data support a model where IFIT2 binds to and enhances the translational efficiency of host mRNAs by suppressing ribosome pausing.
Discussion
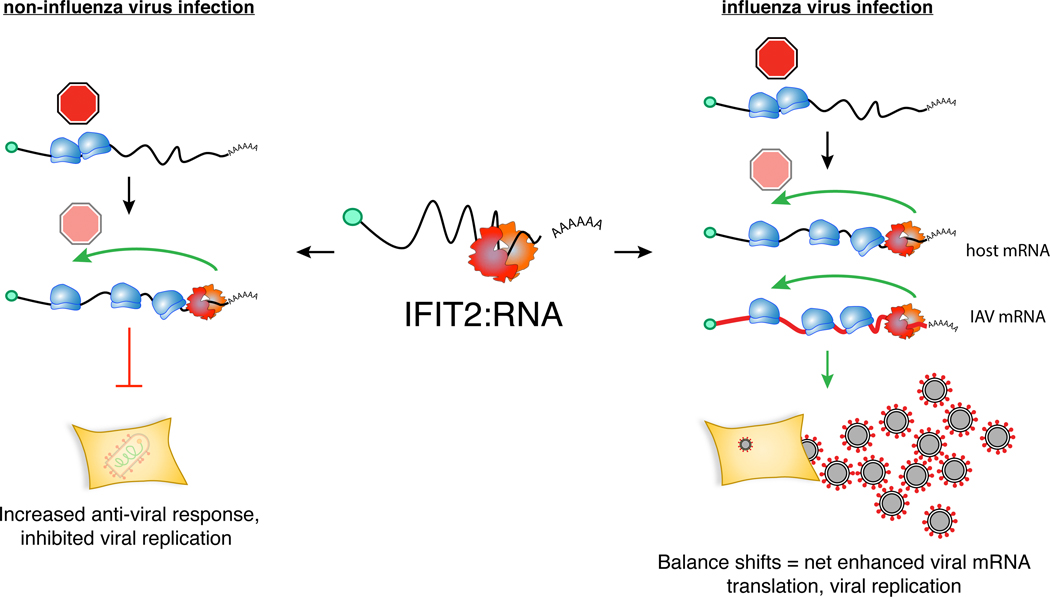
Viruses must overcome cellular defense to enable successful replication, often by evading detection or deploying viral countermeasures. Data presented here show that influenza virus utilizes an additional strategy – exploiting cellular defenses to stimulate viral gene expression and replication. IFIT2 binds AU-rich mRNAs and increases their translational efficiency by decreasing ribosome pausing. Many of the transcripts bound by IFIT2 are important for antiviral responses, perhaps explaining the broad antiviral activity of IFIT212,13. However, influenza virus co-opts this strategy, where viral mRNAs are also bound by IFIT2 resulting in increased production of viral gene products and ultimately tipping the balance to higher levels of viral replication (Extended Data Fig. 9).
The strategy of repurposing antiviral functions to support infection may be part of a more common tactic exploited by viruses: the entry of multiple enveloped viruses is enhanced by LY6E, the product of an ISG35; human cytomegalovirus virus has usurped the antiviral viperin and IFN-induced transmembrane (IFITM) proteins to facilitate infection36,37; and, HIV and BK virus may exploit the mutator function of APOBEC3 to acquire beneficial changes that accelerate escape from immune pressure or drug treatment38–40. Thus, the potent innate immune response creates strong pressures driving viruses to adopt multiple strategies to counteract these defenses, including converting antiviral proteins into pro-viral effectors.
Methods
Cells
Human alveolar epithelial (A549), Madin-Darby canine kidney (MDCK), Madin-Darby bovine kidney (MDBK), baby hamster kidney (BHK), and HEK293T cells were acquired from ATCC cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% Penicillin/Streptomycin, and Corning GlutaGro supplement (Mediatech). Line were authenticated by the supplier. Murine embryonic fibroblasts (MEFs) derived from wildtype (WT), Ifit2−/− C57BL/6 embryos41, Bax−/−/Bak−/− double knockout embryos (a gift from Chi Li, University of Louisville) were maintained in DMEM supplemented with 10% FBS and nonessential amino acids. Transduced A549 cells were selected and maintained in cell growth medium containing 0.5 μg/ml Puromycin (GenDepot). All cells were grown at 37°C in 5% CO2. Cells are routinely tested for mycoplasma contamination using MycoAlert (Lonza).
Plasmids
The human LentiCRISPR Genome-wide CRISPR Knock Out (GeCKO) v2 (#1000000048) and Brunello (#73178) libraries were obtained from Addgene and prepared according to published protocols42–44. Briefly, each library was electroporated into E. cloni 10G Elite electrocompetent cells (Lucigen) in quadruplicate. After recovery, cells were plated and grown at 30°C for 14 h. Colonies were harvested by scraping plates. GeCKOv2 plasmid libraries were isolated using the QIAFilter Maxi Kit (Qiagen). Plasmids expressing the viral polymerase proteins, viral NP, and a genomic reporter vRNA have been described45. Plasmids encoding V5-tagged human IFIT2 and IFIT3 were a kind gift from N. Reich (Stony Brook University)23. The IFIT2 RNA binding mutant DM2 R292E/K410E (IFIT2ΔRNA)29 was generated by sequential inverse PCR and confirmed by sequencing. Primers to generate the RNA binding mutant are listed below:
| hIFIT2 R292E For | GTGCTGCTATgagGCAAAAGTCTTC |
| hIFIT2 R292E Rev | CCAATTTGGCAATGCAGG |
| hIFIT2 K410E For | GGAGAAAGAAgagATGAAAGACAAAC |
| hIFIT2 K410E Rev | CTTGATTTCTGGTTTATTTTTACAC |
For bacterial expression, both WT and IFIT2ΔRNA were cloned into pENTR and moved into pHGGWA by Gateway recombination (Invitrogen).
The coding sequence of NP was altered with synonymous changes using GeneDesign to exchange the original codons with random selections from the same family46, while avoiding rare codons and maintaining a similar overall codon adaptation index. Juggled NP sequence is:
>NP_juggle
ATGGCAACTAAGGGTACTAAGAGGAGCTATGAGCAAATGGAAACCGACGGCGAGCGTCAAAACGCTACAGAGATAAGGGCCAGTGTGGGCAAGATGATCGACGGGATCGGGCGGTTTTATATACAGATGTGTACTGAGTTGAAGCTTTCTGACTACGAAGGGAGACTAATCCAAAATAGTTTGACCATTGAACGTATGGTACTATCGGCGTTCGATGAACGGCGCAACAAGTACCTCGAAGAGCACCCTTCTGCCGGAAAGGACCCAAAAAAGACAGGTGGCCCGATTTATCGCCGCGTGGACGGTAAATGGCGTCGTGAGCTTATTTTGTACGATAAGGAGGAGATCCGTAGGATATGGCGTCAGGCAAACAACGGCGATGACGCGACCGCCGGACTCACCCATATGATGATATGGCATTCGAACCTGAACGACGCGACGTATCAACGAACTAGAGCACTGGTACGTACTGGTATGGACCCACGGATGTGTTCCTTGATGCAAGGATCTACACTACCACGACGTAGTGGAGCAGCCGGCGCGGCGGTAAAGGGTGTCGGGACTATGGTAATGGAGCTGATAAGGATGATAAAGCGGGGCATAAACGACAGAAATTTTTGGCGTGGCGAAAACGGGAGACGGACCCGCATAGCATACGAGCGTATGTGTAATATACTGAAGGGTAAGTTCCAGACCGCGGCCCAGCGCACGATGGTTGACCAGGTAAGGGAATCGAGGAACCCGGGCAACGCGGAATTTGAGGACCTGATTTTCCTTGCGAGATCGGCTCTAATTCTCCGGGGTTCCGTCGCACATAAAAGCTGTCTCCCAGCTTGCGTTTACGGTTCGGCTGTTGCAAGCGGGTATGATTTCGAGCGCGAAGGTTATTCACTCGTAGGGATCGATCCCTTTCGCTTACTGCAGAATTCCCAGGTCTATTCGCTCATTCGTCCTAACGAAAACCCGGCCCATAAATCTCAGTTGGTTTGGATGGCTTGTCACTCAGCGGCGTTCGAGGACCTCCGAGTCTCCTCCTTTATTCGAGGAACAAAGGTTGTACCGCGGGGCAAACTGTCTACCCGGGGCGTCCAGATAGCGTCAAACGAGAATATGGAAACCATGGAGTCGTCAACGTTAGAGTTGCGCTCACGGTATTGGGCAATCCGAACTCGTTCGGGTGGCAATACAAACCAGCAACGCGCCTCTTCCGGACAGATATCTATTCAGCCCACTTTTTCTGTGCAACGGAACTTGCCCTTCGATCGGCCCACGATCATGGCGGCGTTTACAGGCAACACTGAAGGACGAACCTCCGATATGCGTACTGAGATTATCCGCCTTATGGAGTCTGCTCGGCCTGAGGACGTCAGTTTTCAAGGACGCGGCGTTTTTGAACTAAGCGATGAGAAAGCTACCTCTCCCATAGTTCCGTCGTTCGATATGTCCAACGAGGGTTCGTACTTTTTTGGCGATAACGCGGAAGAATATGATAACTAA
The global GC content of the native and juggled NP are 47% and 52%, respectively. The gene was synthesized (GenScript) and cloned to created pCDNA3-jugNP. WT and juggled NP were also cloned into pCDNA3 as fusion proteins with a C-terminal GFP separated by 2A peptide from porcine teschovirus-1 polyprotein to create pCDNA3-NP-2A-GFP or pCDNA3-jugNP-2A-GFP. All plasmids were verified by sequencing.
Viruses and multicycle replication
Viral infections were performed using A/WSN/1933 (WSN), WSN stably encoding the VSV G glycoprotein (VSV-G) and GFP (IAV-G)11, WSN encoding PB2-FLAG47, A/California/04/2009 (CA04), and primary isolates obtained from BEI Resources: A/Hong Kong/1/1968 (H3N2) (HK68; Cat# NR-28620), A/Victoria/361/2011 (H3N2) (Vic/2011; Cat# NR-44022), and A/Wisconsin/15/2009 (H3N2) (WI/2009; Cat# NR-42007). Reporter viruses encoding the NanoLuc (Nluc) reporter gene as a polyprotein with PA have been previously described for the WSN (PASTN) and A/California/04/2009 (H1N1) strains (CA04-PASTN)27,48. A similar strategy was used to produce the influenza B reporter virus in the B/Brisbane/60/2008 background (B-PASTN). Replication kinetics of PASTN-B and the parental recombinant virus are shown (Extended Data Fig. 10). Recombinant SC35M NS1–2A-GFP-2A-NEP (SC35M Flu-GFP) is based on A/Seal/Massachusetts/1-SC35M/80 (H7N7) and was prepared as previously described plasmids49. VSV-GFP was generously provided by P. Bates (University of Pennsylvania). Viruses were propagated in MDCK or MDBK cells in virus growth medium (VGM; DMEM, 0.3% bovine serum albumin, 25 mM HEPES, and 0–0.5 μg/ml of L-1-tosylamido-2-phenylethyl chloromethyl ketone [TPCK] trypsin) or in 9–11 day old embryonated chicken eggs. Viral stocks were titered on MDCK cells. VSV and FVG-G viruses were titered on BHK cells.
Multicycle replication experiments were performed in triplicate and infected at multiplicities of 0.01–0.1. For infections of MEFs, viruses were diluted and grown in DMEM containing 1% FBS and Penicillin/Streptomycin. All other infections were performed in VGM. Aliquots were removed at the indicated time points and titers were determined by plaque assay or by NanoGlo Titer Assay28.
Viral gene expression assays
Viral gene expression was measured using PASTN-based reporter following previous protocols27,28. This virus encodes the NanoLuc (NLuc) reporter in a polycistronic version of the PA gene. Briefly, cells were grown to confluence in a white 96-well plate and infected at MOI = 0.01 at least in triplicate. Cells were washed after 1 hr to remove the inoculum and any residual NLuc from the viral stocks. Infections were allowed to proceed for the indicated times and NLuc activity was quantified as a proxy for viral gene expression. NLuc activity was measured in situ using the Nano-Glo assay system (Promega) and a plate reader.
GeCKOv2 lentivirus library packaging and A549 transduction
For lentiviral packaging, 293T cells (2.4 × 108 cells) were grown in 15 cm dishes to 40% confluence. Cells were transfected with equivalent concentrations of psPAX2, pVSV-G, and LentiCRISPRv2 library A and B using Transit-2020 (Mirus). After 6 h, the medium was replaced with DMEM containing 10% FBS and 1% BSA. After 60 h post transfection, the supernatant was harvested, pooled, centrifuged for 10 min at 4°C, and filtered through a 0.45 μm low protein binding membrane. Lentiviral libraries were then concentrated using ultracentrifugation and resuspended in DMEM containing 10% FBS and 1% BSA. The LentiCRISPRv2 lentivirus library was then titered by transducing A549 cells and counting puromycin-resistant colonies three weeks post-selection and by determining percent transduction by counting viable cells following 4 days of puromycin selection. To generate the GeCKO A549 knockout library, 108 early passage A549 cells were transduced in the presence of 8 μg/ml polybrene at a multiplicity of 0.3 for ~287-fold library coverage for selection with IAV-G and WSN. A separate GeCKO A549 knockout library was prepared for selection with SC35M Flu-GFP using an independently generated lentiviral library. Cells were selected with puromycin containing media (0.5 μg/ml) 24–48 h post-transduction. Transduced cells were frozen 10–11 days following antibiotic selection or used immediately.
Genome-wide GeCKO selection with IAV-G and WSN and sequencing
IAV-G and WSN selections: The experimental workflow is diagrammed in Fig. 1 and Extended Data Fig. 1. GeCKO-A549 cells were created with the GeCKO genome-wide sgRNA library43 and plated at a presumed 530X library coverage. GeCKO-A549 cells were infected at an MOI of 0.3 with IAV-G50, which was determined to be the minimal multiplicity of infection necessary for complete killing of non-transduced A549 cells. Infected cells were incubated for 20–21 days with media changes every 3–4 days. Following the primary IAV-G selection, surviving cells were re-plated and a fraction was frozen. Re-plated cells were subject to a secondary challenge with IAV-G or WSN to enrich for resistant cells. Surviving cells were expanded for an additional 20–21 days. Even though this approach contained multiple rounds of selection and a counter-screen, we consider it a single biological replicate as the selection began with a single pool of GeCKO-A549 cells. A completely independent selection was performed using a uniquely generated knockout cell pool and IAV-SC35M (see below).
Deep sequencing and data analysis of selected gRNAs: Genomic DNA was extracted from frozen cell pellets using the Blood & Cell Culture Midi kit (Qiagen). To ensure sufficient coverage of the library, genomic DNA was extracted from 3×107 cells from the GeCKO-A549 parental library and selected cells from the primary selections. For each sample, sgRNA sequences were amplified by nested PCR in 3–13 separate reactions containing 10 μg of genomic DNA in each reaction. Previously published primers were used to amplify the LentiCRISPRv2 sgRNAs42:
| Primer Name | Sequence |
|---|---|
| v2Adaptor_F | AATGGACTATCATATGCTTACCGTAACTTGAAAGTATTTCG |
| v2Adaptor_R | TCTACTATTCTTTCCCCTGCACTGTtgtgggcgatgtgcgctctg |
| F01 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTtAAGTAGAGtcttgtggaaaggacgaaacaccg |
| F02 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTatACACGATCtcttgtggaaaggacgaaacaccg |
| F03 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTgatCGCGCGGTtcttgtggaaaggacgaaacaccg |
| F04 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTcgatCATGATCGtcttgtggaaaggacgaaacaccg |
| F05 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTtcgatCGTTACCAtcttgtggaaaggacgaaacaccg |
| F06 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTatcgatTCCTTGGTtcttgtggaaaggacgaaacaccg |
| F07 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTgatcgatAACGCATTtcttgtggaaaggacgaaacaccg |
| F08 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTcgatcgatACAGGTATtcttgtggaaaggacgaaacaccg |
Resulting amplicons were gel-quantified and equimolar amounts were pooled, gel extracted using the MinElute gel extraction kit (Qiagen), and sequenced using a HiSeq 2500 (Illumina). Raw FASTQ files were demultiplexed and trimmed using the FASTX-Toolkit in Galaxy and then processed by Galaxy to contain only the unique sgRNA sequence51. MAGeCK was then used to align the reads to the GeCKO library, calculate the number of aligned reads for each sequence, and identify positively selected hits52 (Supplemental Table 1).
Genome-wide Brunello selection with SC35M Flu-GFP and sequencing
SC35M Flu-GFP selections: Brunello A549 knockout library cells were created using the two-vector Brunello system by transducing A549-Cas9 cells with the sgRNA library44. Cells were plated at a presumed 1000-fold coverage and infected with SC35M Flu-GFP at an MOI of 0.1. In parallel, mock-infected cells were harvested. Surviving cells were expanded and re-infected with SC35M Flu-GFP under the same conditions for 7 rounds of selection and harvested on day 63 post infection. The SC35M Glu-GFP selection was performed as a single biological replicate. A completely independent selection was performed using a uniquely generated knockout cell library and IAV-WSN (see above).
Deep sequencing and data analysis of selected gRNAs: Genomic DNA (gDNA) of cell pellets was isolated using Quick-DNA™ Miniprep Plus Kit (Zymo Research). PCR was performed on gDNA to construct Illumina sequencing libraries with each containing 5 μg gDNA following the Broad Institute protocol PCR of sgRNAs for Illumina sequencing. Briefly, gDNA was aliquoted into PCR tubes in a 50 μl total volume. A PCR mastermix containing Extaq DNA polymerase (Clontech), ExTaq buffer, dNTP, P5 stagger primers (see below) and water were prepared. 40 μl of PCR mixture and 10 μl of a barcode P7 primer (see below) were added to each tube containing 50 μl of gDNA. Primers used for Brunello amplicons are:
| Name | Sequence |
|---|---|
| P5–0 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTTTGTGGAAAGGACGAAACACCG |
| P5–1 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTCTTGTGGAAAGGACGAAACACCG |
| P5–2 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTGCTTGTGGAAAGGACGAAACACCG |
| P5–3 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTAGCTTGTGGAAAGGACGAAACACCG |
| P5–4 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTCAACTTGTGGAAAGGACGAAACACCG |
| P5–6 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTTGCACCTTGTGGAAAGGACGAAACACCG |
| P5–7 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTACGCAACTTGTGGAAAGGACGAAACACCG |
| P5–8 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTGAAGACCCTTGTGGAAAGGACGAAACACCG |
| P7-A01 | CAAGCAGAAGACGGCATACGAGATCGGTTCAAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTCTACTATTCTTTCCCCTGCACTGT |
| P7-A02 | CAAGCAGAAGACGGCATACGAGATGCTGGATTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTTCTACTATTCTTTCCCCTGCACTGT |
PCR samples were amplified as follows: 95°C for 1 min, followed by 28 cycles of 95 °C for 30 s, 53 °C for 30 s, 72 °C for 30 s with a final 10 min. at 72 °C. The PCR products were then purified from 2% agarose gels using a gel extraction kit (Zymo Research). PCR amplicons were purified via double size-selection with Sera-Mag SpeedBeads (GE). Cleanup was verified by visualizing samples on 2% agarose gels. Concentrations of each sample were determined using a Qubit fluorometer with dsDNA high-sensitivity assay reagents. Samples were then sequenced on an Illumina NextSeq 500. The resulting sequencing reads were trimmed to 20-nt potential sgRNA sequences using cutadapt53. The MAGeCK software suite was used to assign these sequences to targets and genes52. Comparisons between survivor and control samples were carried out using MAGeCK’s robust rank aggregation method for significance testing (Supplemental Table 2).
Knockout clone generation and genotyping by Indel Detection by Amplicon Analysis
IFIT2 and IFIT3 knockout A549 cells were generated by cloning sgRNA oligos into pLentiCRISPRv2 at the BsmBI site. The oligo sequences are:
| IFIT2.1 sgRNA For | caccgAGAACGCCATTGACCCTCTG |
| IFIT2.1 sgRNA Rev | aaacCAGAGGGTCAATGGCGTTCTc |
| IFIT2.2 sgRNA For | caccgGGCCAGTAGGTTGCACATTG |
| IFIT2.2 sgRNA Rev | aaacCAATGTGCAACCTACTGGCCc |
| IFIT3.1 sgRNA For | caccgCACTGCGGAGGACATCTGTT |
| IFIT3.1 sgRNA Rev | aaacAACAGATGTCCTCCGCAGTGc |
Lentivirus particles were produced and used to transduce A549 cells as above. Following 11–14 days of antibiotic selection, transduced cells were sorted into 96-well plates to generate clonal cell lines. Each 96-well plate was incubated for 3–4 weeks and colonies were amplified. Indel Detection by Amplicon Analysis was used as described to genotype the edited clones54. Briefly, genomic DNA was isolated in each 96-well plate and A549 control cells. Amplicons were fluorophore labelled by tri-primer PCR using a universal 6-FAM M13 5’-labelled primer and primers flanking the sgRNA target site in the genomic DNA with the sense primer containing a M13 extension. Primers are listed below:
| 6-FAM M13 Uni | GTAAAACGACGGCCAGTG |
| IFIT2.1 IDAA M13 For | GTAAAACGACGGCCAGTGgtccctatagaattgagagtccag |
| IFIT2.1 IDAA Rev | GAACATCTGTTACACCTGGGG |
| IFIT2.2 IDAA M13 For | GTAAAACGACGGCCAGTGgaattccttggagagcagcctac |
| IFIT2.2 IDAA Rev | CAGGCATAGTTTCCCCAGGTG |
| IFIT3.1 IDAA M13 For | GTAAAACGACGGCCAGTGgccttgtagcagcacccaatc |
| IFIT3.1 IDAA Rev | CTGGACTGGCAATTGCGATGTAC |
PCRs were diluted 1:5, mixed with the Geneflo Rox-625 size standard (ABI/Life Technology), and analyzed by capillary gel electrophoresis. Data were analyzed using Peak Scanner Software V2.0 (ABI/Life Technology). Positive clones were selected by both IDAA and confirmed by western blotting.
Knockout 293 cells were generated by Synthego. Homozygous knockouts clones were created by limiting dilution, identified by western blotting lysate from infected or IFN-treated cells, and verified by Sanger sequencing across the edited site.
Viral clearance assay
To compare resistance of cells to different viruses, A549 or edited cells were seeded in a 96-well plate at 40,000 cells per well and treated with different concentrations of viruses that were the lowest infectious dose shown to produce extensive cytopathic effects in pilot experiments (~1 TCID50). Approximately seven days after treatment, cells were rinsed with phosphate-buffered saline (PBS), fixed with ethanol, and stained with crystal violet.
Western blotting
Cells were harvested and lysed in RIPA buffer containing protease inhibitor cocktail (Roche Life Science). Protein concentration was measured by Bradford Assay. Lysates were separated on 10–15% PAGE gel and transferred to a PVDF membrane for blocking and antibody incubation. The antibodies used were as follows: anti-IFIT2 (1:300, Proteintech 126041AP), anti-RNP (1:1000, BEI Resources Repository, NR-3133), anti-V5-HRP (1:10,000, Sigma, V2260), anti-Tubulin (1:5000, Sigma, T6199), anti-Rpl10 (1:500, Santa Cruz sc-768), PARP-1 (1:1000, Cell Signaling Technologies, 9542S), M2 anti-FLAG (1:1000, Sigma F1804), anti-NP (1μg/ml, clone H16-L10–4R555) and a custom mouse anti-IFIT3 monoclonal (1:5 of hybridoma supernatant).
Complementation assay
WT and Ifit2−/− MEFs were complemented with IFIT2, IFIT2ΔRNA, or empty vector control by nucleofection. 2×106 MEFs were nucleofected with 5 μg total DNA using the MEF nuclefector kit 1 (Lonza VPD-1004) and setting T-020 in the Nucleofector 2b device. Cells were plated allowed to recover prior to inoculation with PASTN at an MOI of 0.01. Viral gene expression was measured 12 hpi using the Nano-Glo assay system (Promega).
Knock-in of epitope tag at IFIT2 locus in A549 cells
CRISPR/Cas9-mediated knock-in of a V5 epitope tag was performed by the Genome Engineering & iPSC Center at Washington University School of Medicine. An sgRNA compatible with S. pyogenes Cas9 was designed to target TTCAGCATCAAGCTGGAATG at the 3’ end of the IFIT2 open reading frame. Donor DNA was synthesized encoding a linker sequence followed by the V5–6xHis epitope tag, blocking mutations to prevent re-cutting, and 150 bp homology arms on both sides.
>Donor DNA
AAATGCGACTTTCTAAAAATGGAGCAGATTCTGAGGCTTTGCATGTCTTGGCATTCCTTCAGGAGCTGAATGAAAAAATGCAACAAGCAGATGAAGACTCTGAGAGGGGTTTGGAGTCTGGAAGCCTCATCCCTTCAGCATCAAGCTGGAACGGTGAAGGATCCACTAGTCCAGTGTGGTGGAATTCTGCAGATATCCAGCACAGTGGCGGCCGCTCGAGTCTAGAGGGCCCGCGGTTCGAAGGTAAGCCTATCCCTAACCCTCTCCTCGGTCTCGATTCTACGCGTACCGGTCATCATCACCATCACCATTGAAGAATAGAGATGTGGTGCCCACTAGGCTACTGCTGAAAGGGAGCTGAAATTCCTCCACCAAGTTGGTATTCAAAATATGTAATGACTGGTATGGCAAAAGATTGGACTAAGACACTGGCCATACCACTGGACAGGGTTATGTTAACACCT
A549 cells were edited with Cas9/sgRNA in the presence of donor DNA to favor homology directed repair. The pool of edited cells was single cell cloned and homozygous knock-in clones were identified by high-throughput sequencing.
IFIT2 eCLIP
IFIT2 eCLIP protocol was adapted from previously published work56–58 and performed on two independent biological replicates for each experimental condition. Briefly, 107 A549-IFIT2 knock-in cells were plated the day prior to infection for 24 hr (MOI = 0.02 WSN). To form RNA:protein adducts, cells were washed in PBS, placed on ice for UV crosslinking with 254 nm light at 400 mJ/cm2, exposed again at 200 mJ/cm2, collected by scraping, pelleted and snap frozen in liquid nitrogen. Cells were thawed on ice, lysed in CLIP lysis buffer (50 mM HEPES [pH 7.5], 150 mM KCl, 2 mM EDTA, 0.5% [v/v] NP-40, 1 mM dithiothreitol [DTT], 0.5% DOC w/v, 1× EDTA-free protease inhibitor cocktail [Roche]), and clarified by centrifugation. RNase T1 (ThermoFisher) was added to 0.1 units/μl and lysates were incubated at room temperature for 10 min. Digestion was halted by placing samples on ice. Aliquots were removed for generation of the size-matched input control. IFIT2:RNA complexes were captured by adding Dynabeads protein A (Invitrogen) with pre-bound anti-V5 antibody and mixing for 3 hr at 4˚C. Beads were magnetically isolated and washed extensively in lysis buffer, followed by a high salt buffer (50 mM Tris pH [7.4], 1 M NaCl, 1 mM EDTA, 0.1% SDS, 0.5% Na-deoxycholate, 1% NP-40), and ultimately exchanged into a dephosphorylation buffer (50 mM Tris-HCl [pH 7.9], 100 mM NaCl, 10 mM MgCl2, 1 mM DTT). Samples were treated with calf intestinal phosphatase (NEB), washed with dephosphorylation buffer and exchanged into PNK buffer (50 mM Tris-HCl [pH 7.5], 50 mM NaCl, 10 mM MgCl2, 5 mM DTT). RNA was phosphorylated using [γ−32P]-ATP and polynucleotide kinase (NEB). Samples were washed sequentially in PNK buffer, high salt buffer, and finally lysis buffer.
Purified IFIT2:RNA adducts were separated by SDS-PAGE, transferred to nitrocellulose and visualized by phosphor-imaging. Size-matched input controls were separated in parallel on the same gel. Standard western blotting on lysates was also performed separately to confirm the position of IFIT2. Regions corresponding to ~10–200 kDa larger than IFIT2 were cut from the RNA-bound membrane and processed into 2 mm strips. Membranes were placed in Eppendorf tubes and RNA was released by treating samples with proteinase K (Dot Scientific) in digestion buffer (100 mM Tris pH 7.5, 10 mM EDTA, 50 mM NaCl, 1% SDS). An equal volume of phenol (pH 6.8):chloroform:isoamyl alcohol (250:49:1 ratio) was added and RNA extracted into the aqueous layer. RNA was purified on column (Zymo Research) and eluted in water.
RNAs were converted into sequencing libraries. The 3’ ends of RNA fragments were healed by treating with polynucleotide kinase in the absence of ATP and purified with Dynabeads MyOne silane (ThermoFisher). A pre-adenylated DNA adapter (NEB S1315) was ligated onto the 3’ end using a truncated T4 RNA ligase 2 (NEB). Ligation was terminated and free adapter was removed by purifying product with Dynabeads MyOne silane beads. Product was used for reverse transcription by SuperScript III (Invitrogen) in First Strand buffer using one of the following primers which contain indices to allow for multiplexing (index lowercase) and unique molecular identifies (UMI, N’s) to allow for accurate quantification:
5’-NNaaccNNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNacaaNNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNgccaNNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNgaccNNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNtccgNNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNtgccNNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNccggNNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNttaaNNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNattgNNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNaggtNNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNcgccNNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNctaaNNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNcattNNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNggttNNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNgtggNNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNtattNNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
The reaction was terminated with ExoSap-IT (ThermoFisher), RNA was hydrolyzed with NaOH and neutralized with HCl. cDNA was purified with Dynabeads MyOne silane, 5’ phosphorylated with polynucleotide kinase, and purified once more with Dynabeads MyOne silane.
cDNA was circularized with CircLigase II (Lucigen) annealed to an oligo (5’-ATCAGGGATCCACGACGCTCTTCAAAA-3’) and digested with BamHI. Linearized product was purified with Dynabeads MyOne silane. qPCR was performed to determine the appropriate cycle number for preparative amplification. Library was amplified with sequencing adapters using Q5 DNA polymerase (NEB) and the following primers:
p5 5’-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT-3’
p3 5’-CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGATTGATGGTGCCTACAG-3’
PCR product was size-selected on an agarose gel and purified with on a gel extraction kit (Zymo Research). The library was quantified with the NEBNext Library Quant Kit for Illumina (NEB).
Each library was pooled and sequenced on an Illumina HiSeq4000 (Novogene). Summary statistics and accession numbers are in Supplementary Table 6. Data quality control filtering was done with FastQC v0.11.5, de-multiplexed, adapters were trimmed with BBMAP 37.56, and sequences were aligned using HISAT2 v2.0.5 to a custom hg38 concatenated to the influenza A virus genome (A/WSN/33). PCR duplicates were removed by collapsing reads based on UMIs to yield a unique number of RNAs mapping to each position. The results were used as input for CLIPPER 0.1.4 to identify CLIP peaks statistically enriched over the size-matched input controls. Correlation analysis showed a high degree of overlap between replicates (Extended Data Fig. 5b). Data were analyzed with IDR 2.0.03 to measure peak reproducibility across replicates (Extended Data Fig. 5c). Bedtools v2.26.0 was used for genomic data manipulation and pyGenomeTracks 2.1 was used for bigwig visualization. Motif searching and logo generation were performed on CLIP peaks with HOMER v4.1159. Detailed analytical pipeline is shown below.
- Trim fastq with BBDuk in the BBMap suite60.
- bbduk_runner_CLIP.py
- Demultiplex and slice UMIs with custom python script.
- debarcode.py
- Retrim with BBDuk.
- bbduk_runner2.py
- Map to human and WSN genomes.
- hisat2_runner.py
- Filter out unmapped reads.
- preparesamforpcrdedup.py
- PCR-deduplicate with UMIs and read-starts using custom python script.
- PCR_dup_remover.py
- Call peaks with CLIPper using human and WSN genomes61.
- CLIPPER_runner.py
- Concatenate adjacent and overlapping peaks with custom python script.
- merge_adjacent_peaks.py
Count reads in CLIP peaks and size-matched input using bedtools coverage62.
- Perform input normalization with a Chi2 or Fisher’s exact test (depending on number of counts) with custom python script.
- 9+10 wrapped together in one script, input_norm.py
- then, mergetranscriptpeaks.py
- Perform IDR on rep1 and rep2 to filter out un-replicating peaks. Ranking based on p value from Chi2 or Fisher’s exact test. Set IDR cutoff at 540 (p value < 0.05).
- run_IDR_analysis.py, https://github.com/nboley/idr
- Cluster biotype analysis with custom python script.
- Intersect bed with gene_biotype.bed derived from GRCh38 gtf
- count_CLIP_metabiotype_new.py
Filter on protein-coding genes with custom python script.
- Filter on 2 log fold-change (CLIP/input) and relative proportion of dataset (>10^−18 average coverage) with custom python script.
- plot_coverage_CLIPvexpression.py
- Intersect and segregate clusters with protein coding mRNAs with custom python script.
- Intersect with bed file split on 5’UTR,3’UTR, and CDS derived from GRCh38 gtf
- split_CLIP_meta.py
- Calculate FPKM of CLIP origin mRNAs compared to expressed mRNAs with custom python script.
- fpkm_distribution.py
- Calculate GC-content of clusters compared to all corresponding sub-mRNA regions that are expressed with custom python script.
- fasta_peak_to_gc_kde.py
- Calculate length of sub-mRNA regions on CLIPped transcripts and compare to the same regions in all expressed mRNAs with custom python script.
- bed12_bed6_length_kde.py
- Perform saturation analysis with custom python script by testing peak recall on sub-sampled bams with CLIPper.
- Sub-sample 50% of bam, determine % of peaks that can be re-discovered with half of the input data.
Visualization with bedtools coverage, then normalization with custom python scripts and plotting with pyGenomeTracks63.
GO analysis performed with ShinyGO64
Electromobility shift assays
His-GST-IFIT2 and His-GST-IFIT2ΔRNA were purified based on prior approaches29. Briefly, proteins were expressed in E. coli at 16 ˚C overnight. Bacteria was lysed by sonication in high salt buffer (20 mM HEPES pH 7.4, 1 M NaCl) supplemented with 0.5 mg/ml lysozyme and 0.2 mM PMSF. Lysates were clarified by centrifugation and applied to glutathione sepharose FF (GE Life Sciences). Resin was washed extensively with high salt buffer followed by low salt buffer (20 mM HEPES pH 7.4, 150 mM NaCl). Protein was eluted with low salt buffer containing 10 mM reduced glutathione. Protein was dialyzed against an excess of low salt buffer containing 10% glycerol. Proteins were quantified by Bradford assay, assessed by gel electrophoresis, aliquoted, flash frozen in liquid nitrogen and stored at −80 ˚C until use.
Capped and poly-adenylated NP mRNA was synthesized from a PCR-generated template using the HiScribe T7 ARCA (with tailing) kit as recommended (NEB E2060S). The template was designed to mimic virally synthesized mRNA by initiating transcription at the extreme 3’ end of NP vRNA and terminating at the poly-U tract. Products were verified by formaldehyde denaturing agarose gel electrophoresis.
Protein:RNA binding reactions were performed by assembling 20 μl reactions containing ~25 nM mRNA and 0 or 2.5 μM protein in 20 mM HEPES, 150 mM NaCl, 2.5 mM MgCl2, 1 mM DTT and 4U RNasin. binding was performed at room temperature for 45 min. Samples were separated on a 0.8% agarose gel at 4 ˚C. The gel was prepared and run in 1x TBE with 5 mM MgCl2. After separation, the gel was stained with Sybr Gold and RNA was visualized.
qRT-PCR
RNA was isolated using TRIzol (ThermoFisher) and reverse-transcribed essentially as described65 with the following modifications. Influenza mRNAs were reverse-transcribed with in-house produced MMLV-RT at 42˚C for one hour with a uniquely-tagged poly-dT primer (tag in bold, CGCACCAGATCGTTCGAGTCGTTTTTTTTTTTTTTTTTTTT). Influenza vRNAs were reverse-transcribed at 42˚C in independent reactions with uniquely-tagged primers specific for the NP segment (GGCCGTCATGGTGGCGAATACCATAATGACCGATGGCCCAAGT). For normalization, 18S rRNA was reverse-transcribed in the same reactions with the uniquely-tagged primer (CGGTCATGGTGGCGAATAACCAAGATCCAACTACGAGCTT)66. Resulting cDNA was diluted 1:10 in water, and qPCR was performed in technical duplicate or triplicate with iTaq Universal SYBR Green Supermix (Bio-Rad) in a Step One Plus RT-PCR instrument (Applied Biosystems) using the following sets of forward primers and a reverse primer identical in sequence and direction to the bolded tags described in the reverse-transcription primers:
PB2_mRNAF: AGGAGATATGGACCAGCATTAAG
PB1_mRNAF: GTATGGTGGAGGCTATGGTTT
PA_mRNAF: GAACCTGGGACCTTTGATCTT
HA_mRNAF: GTATCAGATTCTGGCGATCTACTC
NP_mRNAF: AGACCAGAAGATGTGTCTTTCC
NA_mRNAF: CTGTATGAGGCCTTGCTTCT
M_mRNAF: CTCTCGTCATTGCAGCAAATATC
NS_mRNAF: GCGGGAACAATTAGGTCAGA
NA_vRNAF: ACATCACTTTGCCGGTATCAGGGT
NP_vRNAF: CTCAATATGAGTGCAGACCGTGCT
NA_cRNAF: TGAATAGTGATACTGTAGATTGGTCT
NP_cRNAF: CGATCGTGCCCTCCTTTG
18S_F: GGCCCTGTAATTGGAATGAGTC.
Specific forward and reverse primers were used for the amplification of GAPDH and β-Actin cDNA primed by the tagged poly-dT primer:
GAPDH_F: ACCGTCAAGGCTGAGAACGG
GAPDH_R: GTGGTGAAGACGCCAGTGGA
β-Actin_F: AGAGCTACGAGCTGCCTGAC
β -Actin_R: AGCACTGTGTTGGCGTACAG
IFIT2-RIP qRT-PCR and Bioanalyzer
A549 cells were infected with WSN (MOI = 0.02) for 24 h. Cells were lysed in co-immunoprecipitation buffer (50 mM Tris pH 7.5, 150 mM NaCl, 0.5% NP40, 1x protease inhibitor cocktail (Sigma), and 20U/mL RNasin (Promega)). Clarified lysates were precleared and incubated for 16 h with 3 μg of IFIT2 antibody or rabbit IgG (Cell Signaling Technologies 2729S) in the presence of 1mg/mL BSA. Immuno-complexes were captured with protein A agarose resin (Sigma) for one hour and washed 6 times with co-immunoprecipitation buffer. 10% of the resin was reserved for western blot analysis and RNA was extracted from the remainder with TRIzol, precipitated and resuspended in 10 μl of water. An equal volume of IFIT2-bound RNA and IgG-bound RNA was used for qRT-PCR analysis. RT was performed as above and the sample was subsequently split into 8 PCRs to separately measure each viral gene. IFIT2-bound samples were normalized to IgG-bound samples using the ΔCt method. RNA was visualized on an Agilent 2100 Bioanalyzer with an RNA 6000 Pico Chip following the manufacturer’s instructions.
IFIT2 reporter assays
The impact of IFIT2 on NP translation was measured using the NP-2A-GFP fusion proteins. 293T cells were transfected in triplicate with constructs expressing either NP-2A-GFP, jugNP-2A-GFP or GFP alone in the presence or absence of IFIT2. Cells were stained with Hoescht 33342 48 hr post-transfection and fluorescence was quantified using a Cytation 5 spectrophotometer (BioTek). GFP and Hoescht fluorescence were measured from a 15 × 15 point grid in each well. Fold induction was calculated relative to the IFIT2-minus controls and average across all replicates.
Ribosome Profiling
Ribosome profiling was based on previous protocols and performed on two independent biological replicates67,68. 5×106 A549 cells were plated the day prior and subsequently infected for 24 hr (MOI = 0.02 WSN). Following treatment cells were rapidly frozen in situ in liquid nitrogen. Cells were thawed, lysed in buffer (10 mM HEPES [pH 7.5], 100 mM KCl, 5 mM MgCl2, 0.5% NP-40, 0.5% deoxycholate, 4 mM DTT, 20 U/mL RNasin, 1x protease inhibitor without EDTA), and disrupted by passage through a 23 G needle. Lysates were clarified by centrifugation and a sample was removed for total RNA analysis. The remainder was digested with RNase I (ThermoFisher AM2294) at 0.5 U/μl for 30 m at 24˚C with mixing. Reaction was quenched with RNasin and layered onto pre-formed 10–50% sucrose gradient prepared in 20mM HEPES (pH 7.5), 75 mM KCl, 5 mM MgCl2, 20 U/mL RNasin, 4 mM DTT and 1x protease inhibitor without EDTA. Gradients were centrifuged in a swinging bucket rotor at 150,000 x g for 2 hrs at 4˚C. Gradients were fractionated. Monosome-containing fractions were identified by western blot, pooled and concentrated by centrifugation on a 100,000 MWCO filter. Monosomes were disrupted into large and small subunits by dilution into buffer supplemented with 50mM EDTA. Samples were centrifuged again through a 100,000 MWCO filter, this time recovering ribosome-protected RNA fragments present in the flow through. RNA was extracted with an equal volume of phenol (pH 6.8):chloroform:isoamyl alcohol (250:49:1 ratio) and precipitated from the aqueous phase following resuspension in water.
Total RNA was extracted from cell lysates using Trizol and poly A selection was performed on 10μg total RNA using magnetic oligo-dT25 beads (NEB S1419). Poly A RNA was fragmented with Mg2+ by heating in First Strand buffer at 94˚C for 24 m. Fragments were precipitated and resuspended in water. Fragmented poly A RNA and the ribosome protected fragments were separated on a 15% TBE-UREA gel using 0.5x TBE. Gels were stained with SYBR gold and RNA corresponding to 27–32 nt was excised from ribosome protected samples and 25–48 nt was excised from fragmented poly A RNA. RNA was eluted from the gel, extracted with Trizol and resuspended in water.
RNAs were converted into sequencing libraries following the same approach used in the eCLIP protocol above, except with 24 unique 5-nt barcodes.
5’-NNTGTCTNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNGACAGNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNTTGGANNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNCCGTTNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNAACGANNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNACTTCNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNACTCGNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNTGAGANNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNCTCAGNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNGAGACNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNCGATCNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNGTACTNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNAGAGGNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNGCACANNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNTCTGGNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNCTCTTNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNCTGTANNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNCACATNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNATCCANNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNTGTTCNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNTGGACNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNGATCCNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNGCAAGNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
5’-NNAAGGTNNNNAGATCGGAAGAGCGTCGTGGATCCCTGATTGATGGTGC-3’
Each library was pooled and sequenced on an Illumina NextSeq500 (NUSeq Core, Northwestern University). Ribosome protected fragments from all conditions were ~32nt long, consistent with protection afforded by a monosome. Summary statistics and accession numbers are in Supplementary Table 6. Data were analyzed with the following pipeline.
Ribosome profiling analysis
- Trim fastq with BBDuk60.
- bbduk_runner_ribo.py
- Demultiplex and slice UMIs with custom python script.
- debarcode_ribo.py
- Retrim with BBDuk.
- bbduk_runner2.py
- Map to human rRNA contig using HISAT269.
- hisat2_runner_rRNA.py
- Filter and retain unmapped reads.
- rRNAalign_refastq.py
- Map to human and WSN genomes with splice database.
- Generate splice database with hisat2_extract_splice_sites.py from GRCh38 gtf
- hisat2_runner.py
- Filter out unmapped reads.
- preparesamforpcrdedup.py
- PCR-deduplicate with UMIs and read-starts using custom python script.
- PCR_dup_remover.py
Phase ribosomes with non-overlapping ORFs using plastid70.
Count RPFs and RNAs in coding sequences using featureCounts71. Filter out mitochondrial mRNAs and histones.
- Run differential expression analysis on RNA and RPF using edgeR in R script72.
- The design matrix was used to program the negative binomial general linear models in EdgeR to model WT and knockout cells as two different cell types (two-factor design).
- Model as a full factorial experiment, then contrast the difference between IFIT2KO infection/IFIT2KO mock vs WT infection/WT mock
- run_edgeR.R
- Run analysis on RNA-normalized RPFs using custom R script. Analyze these data with edgeR, modeling data from WT and knockout cells as two different cell types.
- dispersion factors for each gene for RNA and RPF are modeled separately and then recombined, to better look at situations in which only RNA or only RPF change with treatment.
- data were modeled as a full factorial experiment similar to above. This allowed the comparison of IFIT2KO infection TE/ IFIT2KO mock TE vs WT infection TE/WT mock TE, i.e. normalizing for differences in cell type before looking at the effect of IFIT2 on infection.
- Prepare cumulative distribution frequency curves with custom python scripts, using a log cpm cutoff to 5.
- all listed as cumulative_distribution_CLIP*.py
Visualization with bedtools coverage62, then normalization with custom python scripts and plotting with pyGenomeTracks63
Pause site analysis:
Identification of pause peaks:
Extract coverage over all expressed CDS from all RPF files using bedtools coverage62.
To prevent confounding effects of alternative splicing, filter out genes that have <75% of sequence covered in RPFs and those that have average RPF coverage < 8 (~roughly corresponds to RPKM of ~10 in our datasets).
Remove regions where there is 0 RPF coverage.
Median-normalize coverage on a per-gene basis
Calculate the maximum change in slope over a gapped interval representing boundaries consistent with a pause ribosome (50 nt interval blocking internal 28 nt). Ascending slope is positive (m_a), descending slope is negative (m_d), max_delta_slope (m_max) = m_a-m_d.
Call peaks using scipy.signal find_peaks algorithm33. Peaks are plateau-shaped and peaks are called in the middle of plateau.
Combine peaks from replicate samples. Re-center peaks that overlap.
Combine peaks from the same conditions. Re-center overlapping peaks.
Extend peaks 17-nt either side to generate the pause site and record as bed12.
Quantify peaks:
Count reads overlapping each pause in all datasets with bedtools coverage62.
Analyze differential expression analysis on peaks using edgeR and R script72. Libraries were re-scaled to the size of original RPF libraries – calculate the library size and normalization factor of RPF library and replace original in DGEList with these. Each pause was modeled as an independent mRNA.
- To produce an adjusted log2 fold change for each pause, a custom offset matrix was used where each row of the original offset matrix (loge(library_size) from 2) was transformed by taking the base e exponential to restore untransformed library size. This was multiplied by a vector containing 1’s (for non-IFIT2KOixn libraries) or the fold change from log2FC_IFIT2KO/WTinfection (for IFIT2KOixn libraries). This new matrix was loge transformed and replaced the offset matrix after dispersion estimation. This effectively scales the library size of the IFIT2KO infection libraries with log2FC of the RPF libraries, preventing changes in RPF density from being called as differential pausing.
- Same full factorial design as RPF analysis 11+12 above
- In addition, use an additional contrast in glmQLFTest as a second coefficient, KO/WT infection w/o mock normalization (or model as the same cell type). This outputs a second log2FC (simply KO/WT infection) and a p-value after testing for differential expression of the combined effects of both coefficients. This can be used as an additional filter when looking for differential pausing.
Catalog these log2 fold changes as pauseI and separate sub-populations based on the maximum, median and minimum intensity pause sites present on each transcript, using normalized log2 fold changes from 3a above.
Determine changes in pauseI during infection of WT and IFIT2−/− cells by measuring changes in intensity of the maximum and minimum pause sites on each transcript.
Conservative comparisons of changes in pauseI at each site in WT or IFIT2−/− cells were made using mock-normalized data, and more general changes in pauseI were assessed using only data from infected cells. Pause sites with log2 fold change of at least 0.58 under both comparisons were considered (Supplementary Table 5).
CLIP-ribo cross analysis via pause-TE index:
Calculate pause-TE index as pauseI/TE using the maximum pause site for each transcript.
- Plot cumulative distribution frequency curves with custom python scripts, filtering on RPKM > 10 in RPF libraries.
- all cumulative_distribution_CLIP*.py files
Polysome Pelleting
Polysome pelleting was performed essentially as described73 with the following modifications. A549 cells were mock-infected or infected with WSN at an MOI of 1 for 6 h. Before harvest, cells were treated with 100 ug/mL cycloheximide for 5 min at 37˚C. Cells were lysed in modified polysome extraction buffer (10 mM HEPES pH 7.5, 100 mM KCl, 5 mM MgCl2, 4 mM DTT, 0.5% NP-40, 100 ug/mL cycloheximide, 20 U/mL RNasin, 10%[W/V] sucrose, and 1x protease inhibitor cocktail without EDTA). Clarified lysate was split in half and one half was treated with 30 mM EDTA to dissociate ribosomes.
Western blot of polysome: Polysomes were pelleted by layering lysate onto 2.5 mL of 25% (W/V) sucrose prepared with polysome cushion buffer (20 mM HEPES pH 7.5, 75 mM KCl, 5 mM MgCl2, 4 mM DTT, 20 U/mL RNasin, 100 μg/mL cycloheximide, 1x protease inhibitor cocktail without EDTA, and +/− 30 mM EDTA) followed by centrifugation in an SW41 rotor at 150,000 x g (29,500 rpm) for 4 h at 4˚C. The supernatant was removed and the pelleted protein was dissolved in 1x SDS-PAGE sample buffer with gentle shaking at 60˚C for 30 mins. Samples were analyzed by western blot.
qRT-PCR of RNA from high molecular weight polysomes: A fraction of the cell lysate was reserved as an input control. High molecular weight polysomes were pelleted by layering cell lysates onto 2.5 mL of 37.5% (W/V) sucrose prepared with polysome cushion buffer followed by centrifugation a in an SW41 rotor at 150,000 x g (29,500 rpm) for 2 h at 4˚C. The supernatant was removed and RNA was extracted from pelleted polysomes and the corresponding input samples with TRIzol, pelleted and re-dissolved in water. The abundance of NP mRNA in the polysome pellet relative to total RNA was calculated using the ΔΔCt method with18S rRNA as reference control. Data were then presented as fold-enrichment in the polysome pellet.
Polysome profiling
Polysome profiling was based on published protocols74,75. A549 cells were mock-infected or infected with WSN at an MOI of 1 for 6 h. Cells were treated with 100 μg/mL cycloheximide for 5 min at 37˚C prior immediately prior to lysing in polysome extraction buffer. Clarified lysate was split in half and one half was treated with 30 mM EDTA to dissociate ribosomes. Lysate corresponding to a cell equivalent of ~10 OD260 was layered on 12.5 mL linear 10–50% (W/V) sucrose gradients prepared with polysome cushion buffer. Samples were centrifuged in an SW41 rotor at 210,000 x g (35,000 rpm) for 2 h at 4˚C.
Gradients were separated on a Teledyne ISCO gradient fractionation system outfitted with a Foxy JR fraction collector. Sixteen equal fractions were collected using the “time” setting on the fraction collector while continuously monitoring absorbance of the gradient using a UA-5 detector and a Type-6 optical unit equipped with a 260 nm filter. The analog voltage output of the detector was converted to a digital signal and recorded every 10 milliseconds to plot an absorbance trace.
RNA was isolated from polysome profiles by extracting 200 μl of each fraction with TRIzol and dissolving the RNA in 20 μl of water. qRT-PCR analysis was performed for NP and β-actin mRNA using the percent-total normalization technique76. Western-blots were directly performed on all gradient fractions after addition of 5x SDS-PAGE sample buffer.
Caspase activation
Induction of apoptosis was quantified by measuring caspase 3 and caspase 7 activity with the Caspase-Glo 3/7 Assay (Promega). For virally induced apoptosis, WT or knockout MEFs were infected at an MOI of 0.01 and measured at the indicated times. For IFIT2-mediated apoptosis, IFIT2 was expressed in 293T cells for 72 h prior to measuring caspase activity. To measure virus production when caspase activity was inhibited, A549 cells were inoculated with WSN at an MOI of 0.3 and the infection was allowed to proceed for 24 h to establish robust replication. Media and any virus produced during the first 24 h were removed and replaced with fresh media containing increasing concentrations of Z-VAD-FMK (EMD Millipore) or the DMSO solvent. Cells were incubated for an additional 8 h. Caspase activity was measured and virus produced during this period was quantified by plaque assay.
Statistics and Reproducibility
Unless otherwise noted, data represent means ± standard deviations (n=3 or more technical replicates) and are representative of at least three independent biological replicates. Excel (16.15) and Prism 8 were used for graphing and statistical analyses. Pairwise comparisons were made using a Student’s T test. Multiple comparison were performed by a one-way ANOVA with post hoc Tukey’s HSD. Significance was defined as P< 0.05 and is indicated when necessary with an asterisk (*). Double (**) or triple (***) asterisks are denoted where P <0.01 or P < 0.001, respectively. Analyses used for eCLIP and ribosome profiling are indicated within their respective protocols. Cumulative distribution frequencies were compared using a Mann-Whitney U test.
Data availability
Data supporting the findings of this study are available within the paper and its Supplementary Information and Tables, with the exception of raw data for eCLIP and ribosome profiling which are accessible as BioProject PRJNA633047 containing BioSample accessions SAMN14931303- SAMN14931312 and SRA accession SRR11794832-SRR11794851 (Supplementary Table 6). All other data are available from the corresponding author upon reasonable request.
Code Availability
Existing code used for data analysis are described above. Custom code and pipelines are described above, available at https://github.com/mehlelab/ or from the corresponding author upon request.
Extended Data
Extended Data Fig. 1. Enrichment of IFIT2 and IFIT3 over sequential influenza virus selections.
a, Schematic for hit analysis and selection. b-c, Frequency distribution of sgRNAs present in the parental libraries and those following selection with WSN (b) or SC35M (c). d-f, sgRNA enrichment trajectories following (d) sequential IAV-G selections, (e) IAV-G followed by bona fide influenza virus (WSN) infection or (f) selection with SC35M-GFP. The abundance of each sgRNA targeting the indicated gene is shown. g, Overlap between top hits from each screen. A conservative cutoff of RRA<10−5 was used to define top hits.
Extended Data Fig. 2. Characterization of replication and protein expression in the presence or absence of IFIT2 or IFIT3.
a, Two IFIT2−/− clones were created in A549 cells by CRISPR/Cas9 mutagenesis using distinct sgRNAs. Homozygous knockouts were identified by IDAA (indel detection by amplicon analysis). Chromatographs are shown for WT and mutant amplicons from IFIT2 knockout line #1 targeting AGAACGCCATTGACCCTCTG and IFIT2 knockout line #2 targeting GGCCAGTAGGTTGCACATTG. Size standards appear in red and amplicon peaks are shown in blue with calculated sizes in nucleotides listed above each peak. b, Ifit2−/− MEFS survive infection. WT and knockout cells were challenged with WSN or IAV stably encoding VSV-G (IAV-G). Surviving cells were visualized by staining with crystal violet. c, Kinetics of IFIT2 and NP expression were determined by infecting A549 cells with WSN (MOI = 0.2) and measuring protein expression by western blotting samples acquired at the indicated times post-infection. d, Clonal WT or IFIT2−/− 293 cells were infected with an IAV (WSN) reporter virus at an MOI = 0.01 and viral gene expression was measured 8 hpi. Each point represents mean gene expression (n = 4) from 4 independent clonal knockout lines. The mean of all lines is shown as a gray bar ± SEM. e, IFIT2 was expressed in WT 293 cells (clonal line 1B8) or IFIT2−/− knockout 293 cells (clonal line 3B9) for 12 hr followed by infection with an IAV (WSN) reporter virus (MOI = 0.1) for 8 hr. n = 2 independent infections. f, Two homozygous IFIT3−/− clones were identified by IDAA. Chromatographs are shown for WT and mutant amplicons from IFIT3 knockout lines #1 and #2 targeting CACTGCGGAGGACATCTGTT. Size standards appear in red and amplicon peaks are shown in blue with calculated sizes in nucleotides listed above each peak. g, Infection-induced IFIT3 expression was monitored in WT and IFIT3 knockout cells by western blotting. h, Multicycle replication of WSN in wildtype or IFIT3 knockout A549 human lung cells. Data represent mean ± standard deviation (n = 3 independent infections). i, Viral gene expression in wildtype or IFIT3 knockout cells infected with an influenza reporter virus for the indicated times. Data represent mean + sd (n = 4 independent infections). (*** p < 0.001; two-tailed Student’s t-test for pairwise comparisons between control and knockout clonal lines in d; * p < 0.05; ** p < 0.01 one-way ANOVA with post hoc Tukey’s HSD for e, h and i).
Extended Data Fig. 3. Human IFIT2 and murine Ifit2 promote virus-induced apoptosis.
a, Caspase 3/7 activity was measured in wildtype, Ifit2−/−, and Bak/Bax−/− MEFs infected with WSN at an MOI of 0.01 for the indicated times. n = 4 independent infections ± standard deviation. b, Expression of cleaved (C-PARP) versus full-length (FL-PARP) PARP-1 in IAV infected A549 and IFIT2 knockout cell lines. Protein bands were quantified and the percent C-PARP of total PARP signal normalized to a tubulin loading control is indicated below. c, Viability of Ifit2−/− cells during infection with WSN assessed by CellTiter Glo Assay at the indicated time points. n = 4 independent infections ± standard deviation. d, Multicycle replication of WSN reporter virus in in WT, Ifit2−/−, and Bak/Bax−/− MEFs. n = 3 independent infections ± standard deviation. e, Single-cycle WSN reporter virus gene expression at early time points post-infection in WT, Ifit2−/−, and Bak/Bax−/− MEFs. n = 4 independent infections ± standard deviation. f, WSN reporter virus gene expression in Bak/Bax−/− cells ectopically expressing an empty vector, hIFIT2, or hIFIT2-ΔRNA. n = 3 independent infections ± standard deviation. *p < 0.05, **p < 0.01; *** p < 0.001; Two-tailed Student’s T-test for pairwise comparisons (c) or one-way ANOVA with post hoc Tukey’s HSD for multiple comparisons to WT cells (a, d, e and f). All data are plotted as mean ± sd.
Extended Data Fig. 4. Analysis of IFIT2 CLIP-Seq data.
a, Experimental workflow for eCLIP of IFIT2 from IAV WSN-infected A549 cells. RNA-protein adducts were formed in infected cells by UV cross-linking. Lysates were prepared and RNA was partially digested with RNase. A fraction of lysate was removed to serve as the size-matched input control (SM input). IFIT2 was immunopurified and bound RNAs were processed via eCLIP followed by sequencing and analysis. b, CLIP-Seq was performed in duplicate and compared for IFIT2 CLIP or size-matched input (SMI) controls. Pearson’s correlation coefficient is shown as a heatmap. c, Concordance between ranked peaks in two independent biological replicates of IFIT2 CLIP-Seq was assessed by calculating an irreproducible discover rate (IDR). A conservative threshold of 5% was used to identify significant peaks. d, Abundance distribution in the size-matched input control for all transcripts and those with IFIT2 CLIP peaks. Populations were compared by a two-sided Mann-Whitney U-test. e, Meta-analysis of GC content for peaks identified in the 5’ UTR and coding sequence (CDS) of bound transcripts. IFIT2-bound RNAs were compared to all expressed 5’ UTRs or CDS via a two-sided Mann-Whitney U-test. f, A degenerate UAGnnUAU motif was found in ~20% of IFIT2 CLIP peaks (p = 10−279 compared to background).
Extended Data Fig. 5. IFIT2 binds viral mRNA but not genomic vRNA.
a, IFIT2 CLIP-Seq was performed on WSN infected cells. Reads were mapped to viral mRNA (top) or genomic vRNA (bottom) and analyzed with CLIPper. No CLIP peaks were identified on the genomic RNA. Data from biological duplicates of size-matched input controls or IFIT2 CLIP are plotted as mean and standard deviation using dark and light shades of the same color, respectively. BPM = bins per million. b, Western blot of immunoprecipitated IFIT2 or IgG controls used for RIP-qRT-PCR shown in Figure 3e. c, Recombinant HisGST-IFIT2 used for EMSA analysis in Figure 3f was expressed and purified from E. coli. Protein integrity and purity was verified by gel electrophoresis and Coomassie staining. IFIT2∆RNA = R212E/K410E mutant.
Extended Data Fig. 6. IFIT2 enhances the translational efficiency of influenza virus NP mRNA.
a-c, Full dataset underlying results presented in Fig 4d. Polysome profiling was performed on WT and IFIT2−/− A549 cells infected with WSN. a, The polysome profile is shown again for clarity. b, qRT-PCR quantification of β-actin or c, NP mRNA in each fraction. The sum of less efficiently translated mRNA present in “light” polysome fractions (5 or less ribosomes per message) or efficiently translated mRNA in “heavy” polysome fractions (>5 ribosomes per message) is shown under each trace. Loss of IFIT2 shifts NP mRNA towards the top of the gradient, indicating less efficient translation than in WT cells. d, IFIT2−/− 293 cells or two clones of WT cells were transfected with an IFIT2 expression vector or an empty vector control prior to infection with WSN. Proteins were detected by western blot.
Extended Data Fig. 7. Loss of IFIT2 results in ribosomal pausing that decreases translation and increases pausing of IFIT2-bound mRNAs.
Ribosome dynamics were monitored in cells infected with IAV WSN. a, The translational efficiency of IFIT2-bound ISG mRNAs is decreased in the absence of IFIT2. Bound transcripts were compared to unbound transcripts via a two-sided Mann-Whitney U-test. b-c, Accumulation of paused ribosomes in the absence of IFIT2. Normalized read density for total RNA (top) and ribosome-protected fragments (RPFs) (bottom) mapping to IAV mRNAs (b) or GAPDH (c) in infected WT and IFIT2−/−cells. A close-up view of NP is shown in Figure 5c. Data from replicate experiments are plotted as mean and standard deviation using dark and light shades of the same color, respectively. Pause sites are shown below. Pause sites enriched in IFIT2−/− cells >1.5-fold are filled in. High coverage at the 5’ and 3’ end of PB2, PB1 and PA is consistent with low levels of defective-interfering (DI) particles commonly found in laboratory viral stocks. Note that there are no IFIT2-dependent pause sites identified on GAPDH mRNA. BPM = bins per million. d, Ribosome pausing increases during infection of IFIT2−/− cells. Changes in pause intensity (pauseI) between the maximum and minimum pauseI sites on each transcript were calculated during infection of WT and IFIT2−/− cells. e, Transcripts contain multiple pause sites. The pause sites with the minimum, median and maximum pauseI were identified. The effect of IFIT2 on pausing for each of these classes was determined by comparing IFIT2-bound transcripts to unbound transcripts. In the absence of IFIT2, pauseI increases at the strongest pause sites (maximum) while it is reduced at the weakest pause sites (minimum). The median pause sites are unchanged. f, Increased pausing and decreased translation are correlated for IFIT2-bound mRNAs. The relationship between pausing and translation efficiency was assessed by calculating a pause-TE index (pauseImax/TE) for each transcript. Changes in the pause-TE index were assessed for IFIT2-bound mRNAs (left), IAV mRNAs (middle), and ISG mRNAs (right). Comparisons in (d-f) were performed via a two-sided Mann-Whitney U-test.
Extended Data Fig. 8. Sequence-dependent enhancement of NP translation in the presence of IFIT2.
a, Diagram of NP-2A-GFP polyprotein. For cell-based assays in Figure 5f, NP was expressed as a polyprotein where NP was separated from GFP by the 2A cleavage sequence from porcine teschovirus. NP is shaded to represent the codon juggling that introduced silent mutations to NPjug. See Methods for details. b, NP was expressed in 293T cells from a gene encoding WT or juggled codons for only NP, and not a polyprotein. IFIT2-V5 was co-expressed where indicated. Expression was detected by western blot, including Hsp90 as a loading control.
Extended Data Fig. 9. Classic “antiviral” proteins can be re-purposed during infection into pro-viral effectors.
A model for the dichotomous functions of IFIT2. IFIT2 enhances translation of host mRNAs, including ISGs, to exert antiviral activity to diverse viruses. Yet, as shown here, IFIT2 is re-purposed during influenza virus infection to increase translation of viral mRNAs and shift the balance resulting in a net increase in viral mRNA translation and viral replication.
Extended Data Fig. 10. Replication kinetics of recombinant B/Brisbane/60/2008 and the reporter virus B/Brisbane/60/2008-PASTN.
An influenza B reporter virus encoding NanoLuc in the PA gene segment (see Methods for details). Infections were performed in MDCK cells (MOI = 0.01) at 33 ˚C and viral titers were determined by plaque assay at the indicated time points. Data are mean of n = 3 independent infection ± standard deviation.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health R21AI125897 and R01AI125271, the UW2020:WARF Discovery Initiative, and a Shaw scientist award to A.M., an NIH National Service Award T32 GM07215 to V.T., a National Science Foundation grant GRFP DGE-1747503 to M.P.L., R01AI104972 to M.S.D, a R01AI118938 to A.C.B. the German Research Foundation (DFG; SFB 1160, project 13) to M.S., the Excellence Initiative of the German Research Foundation (GSC-4, Spemann Graduate School) and the Ministry for Science, Research and Arts of the State of Baden-Wuerttemberg to T.T., U19AI106754 to C.B, and HHSN272201400008C - Center for Research on Influenza Pathogenesis (CRIP), a NIAID-funded Center of Excellence for Influenza Research and Surveillance (CEIRS), and U19AI135972 A.G-S. A.M. holds an Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. We thank D. Poole for technical assistance, members of the Mehle laboratory, M. Harrison, C. Fraser, S. Floor and J.D. Sauer for valuable input and C. Li, N. Reich, P. Friesen, Y. Kawaoka, L. Ristow, and R. Welch for sharing reagents. We acknowledge F. Zheng for reagents deposited in Addgene and the University of Wisconsin Biotechnology Center DNA Sequencing Facility for sequencing and IDAA services.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Iwasaki A & Pillai PS Innate immunity to influenza virus infection. Nat. Rev. Immunol. 14, 315–28 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Downey J, Pernet E, Coulombe F & Divangahi M. Dissecting host cell death programs in the pathogenesis of influenza. Microbes Infect. (2018). doi: 10.1016/J.MICINF.2018.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krammer F et al. Influenza. Nat. Rev. Dis. Prim. 4, 3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D, Zhu W, Yang L & Shu Y. The Epidemiology, Virology, and Pathogenicity of Human Infections with Avian Influenza Viruses. Cold Spring Harb. Perspect. Med. (2020). doi: 10.1101/cshperspect.a038620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long JS, Mistry B, Haslam SM & Barclay WS Host and viral determinants of influenza A virus species specificity. Nat. Rev. Microbiol. 17, 67–81 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Carette JEE et al. Haploid genetic screens in human cells identify host factors used by pathogens. Science 326, 1231–5 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Heaton BE et al. A CRISPR Activation Screen Identifies a Pan-avian Influenza Virus Inhibitory Host Factor. Cell Rep. 20, 1503–1512 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li B et al. Genome-wide CRISPR screen identifies host dependency factors for influenza A virus infection. Nat. Commun. 11, 164 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han J et al. Genome-Wide CRISPR/Cas9 Screen Identifies Novel Host Factors Essential for Influenza Virus Replication. CellReports 23, 596–607 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schaack GA & Mehle A Experimental approaches to identify host factors important for influenza virus in Influenza: The Cutting Edge (eds. Neumann G & Kawaoka Y) In Press (Cold Spring Harbor Laboratory Press, 2019). doi: 10.1101/cshperspect.a038521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hao L et al. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454, 890–3 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diamond MS & Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 13, 46–57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fensterl V & Sen GC Interferon-Induced Ifit Proteins: Their Role in Viral Pathogenesis. J. Virol. 89, 2462–2468 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y et al. ISG56 is a negative-feedback regulator of virus-triggered signaling and cellular antiviral response. Proc. Natl. Acad. Sci. U. S. A. 106, 7945–7950 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pichlmair A et al. IFIT1 is an antiviral protein that recognizes 5’-triphosphate RNA. Nat. Immunol. 12, 624–30 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Daffis S et al. 2’-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468, 452–456 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schoggins JW et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szretter KJ et al. 2’-O methylation of the viral mRNA cap by West Nile virus evades Ifit1-dependent and -independent mechanisms of host restriction in vivo. PLoS Pathog. 8, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terenzi F, Saikia P & Sen GC Interferon-inducible protein, P56, inhibits HPV DNA replication by binding to the viral protein E1. EMBO J. 27, 3311–3321 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fensterl V et al. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathog. 8, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinto AK et al. Human and Murine IFIT1 Proteins Do Not Restrict Infection of Negative-Sense RNA Viruses of the Orthomyxoviridae, Bunyaviridae, and Filoviridae Families. J. Virol. 89, 9465–76 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daugherty MD, Schaller AM, Geballe AP & Malik HS Evolution-guided functional analyses reveal diverse antiviral specificities encoded by IFIT1 genes in mammals. Elife 5, 1–22 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stawowczyk M, Van Scoy S, Kumar KP & Reich NC The interferon stimulated gene 54 promotes apoptosis. J. Biol. Chem. 286, 7257–7266 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleith RC et al. IFIT3 and IFIT2/3 promote IFIT1-mediated translation inhibition by enhancing binding to non-self RNA. Nucleic Acids Res. 46, 5269–5285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson B et al. Human IFIT3 Modulates IFIT1 RNA Binding Specificity and Protein Stability. Immunity 48, 487–499 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benitez AA et al. In Vivo RNAi Screening Identifies MDA5 as a Significant Contributor to the Cellular Defense against Influenza A Virus. Cell Rep 11, 1714–1726 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tran V, Moser LA, Poole DS & Mehle A. Highly sensitive real-time in vivo imaging of an influenza reporter virus reveals dynamics of replication and spread. J. Virol. 87, 13321–13329 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran V et al. Multi-Modal Imaging with a Toolbox of Influenza A Reporter Viruses. Viruses 7, 5319–5327 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z et al. Crystal structure of ISG54 reveals a novel RNA binding structure and potential functional mechanisms. Cell Res. 22, 1328–1338 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takizawa T et al. Induction of programmed cell death (apoptosis) by influenza virus infection in tissue culture cells. J. Gen. Virol. 74 ( Pt 11, 2347–55 (1993). [DOI] [PubMed] [Google Scholar]
- 31.Mühlbauer D et al. Influenza Virus-Induced Caspase-Dependent Enlargement of Nuclear Pores Promotes Nuclear Export of Viral Ribonucleoprotein Complexes. J. Virol. 89, JVI.03531–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wurzer WJ et al. Caspase 3 activation is essential for efficient influenza virus propagation. EMBO J. 22, 2717–28 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ingolia NT, Lareau LF & Weissman JS Ribosome Profiling of Mouse Embryonic Stem Cells Reveals the Complexity and Dynamics of Mammalian Proteomes. Cell 147, 789–802 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolin SL & Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 7, 3559–3569 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mar KB et al. LY6E mediates an evolutionarily conserved enhancement of virus infection by targeting a late entry step. Nat. Commun. 9, 3603 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seo J, Yaneva R, Hinson ER & Cresswell P. Human cytomegalovirus directly induces the antiviral protein viperin to enhance infectivity. Science (80-. ). 332, 1097–1100 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Xie M et al. Human Cytomegalovirus Exploits Interferon-Induced Transmembrane Proteins To Facilitate Morphogenesis of the Virion Assembly Compartment. J. Virol. 89, 3049–3061 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peretti A et al. Characterization of BK Polyomaviruses from Kidney Transplant Recipients Suggests a Role for APOBEC3 in Driving In-Host Virus Evolution. Cell Host Microbe 23, 628–635.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim E-Y et al. Human APOBEC3 induced mutation of human immunodeficiency virus type-1 contributes to adaptation and evolution in natural infection. PLoS Pathog 10, e1004281 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mulder LCF, Harari A & Simon V. Cytidine deamination induced HIV-1 drug resistance. Proc. Natl. Acad. Sci. U. S. A. 105, 5501–5506 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho H, Shrestha B, Sen GC & Diamond MS A role for Ifit2 in restricting West Nile virus infection in the brain. J. Virol. 87, 8363–71 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanjana NE, Shalem O & Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shalem O et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doench JG et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mondal A et al. Influenza virus recruits host protein kinase C to control assembly and activity of its replication machinery. Elife 6, e26910 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richardson SM, Wheelan SJ, Yarrington RM & Boeke JD GeneDesign: Rapid, automated design of multikilobase synthetic genes. Genome Res. 16, 550–556 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dos Santos Afonso E, Escriou N, Leclercq I, van der Werf S & Naffakh N. The generation of recombinant influenza A viruses expressing a PB2 fusion protein requires the conservation of a packaging signal overlapping the coding and noncoding regions at the 5’ end of the PB2 segment. Virology 341, 34–46 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Karlsson EA et al. Visualizing real-time influenza virus infection, transmission and protection in ferrets. Nat. Commun. 6, 6378 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reuther P et al. Generation of a variety of stable Influenza A reporter viruses by genetic engineering of the NS gene segment. Sci. Rep. 5, 11346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Watanabe T, Watanabe S, Noda T, Fujii Y & Kawaoka Y. Exploitation of nucleic acid packaging signals to generate a novel influenza virus-based vector stably expressing two foreign genes. J. Virol. 77, 10575–83 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Afgan E et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. ahead of p, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li W et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 1–12 (2014). doi: 10.1186/s13059-014-0554-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10 (2011). [Google Scholar]
- 54.Yang Z et al. Fast and sensitive detection of indels induced by precise gene targeting. Nucleic Acids Res. 43, e59 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yewdell JW & Gerhard W. Antigenic characterization of viruses by monoclonal antibodies. Annu. Rev. Microbiol. 35, 185–206 (1981). [DOI] [PubMed] [Google Scholar]
- 56.Huppertz I et al. iCLIP: Protein-RNA interactions at nucleotide resolution. Methods 65, 274–287 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Nostrand EL et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 13, 508–514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kutluay SB et al. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell 159, 1096–1109 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heinz S et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bushnell B BBMap (version 37.75) [Software]. Available at https://sourceforge.net/projects/bbmap/ (2015). [Google Scholar]
- 61.Lovci MT et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat. Struct. Mol. Biol. 20, 1434–1442 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quinlan AR & Hall IM BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramírez F et al. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun. 9, 189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ge SX, Jung D & Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics (2019). doi: 10.1093/bioinformatics/btz931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kawakami E et al. Strand-specific real-time RT-PCR for distinguishing influenza vRNA, cRNA, and mRNA. J. Virol. Methods 173, 1–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kiselak EA et al. Transcriptional regulation of an axonemal central apparatus gene, sperm-associated antigen 6, by a SRY-related high mobility group transcription factor, S-SOX5. J. Biol. Chem. 285, 30496–505 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ingolia NT, Brar GA, Rouskin S, McGeachy AM & Weissman JS The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo H, Ingolia NT, Weissman JS & Bartel DP Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim D, Langmead B & Salzberg SL HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dunn JG & Weissman JS Plastid: nucleotide-resolution analysis of next-generation sequencing and genomics data. BMC Genomics 17, 958 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liao Y, Smyth GK & Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sheets MD, Fritz B, Hartley RS & Zhang Y. Polyribosome analysis for investigating mRNA translation in Xenopus oocytes, eggs and embryos. Methods 51, 152–156 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mayeur GL, Fraser CS, Peiretti F, Block KL & Hershey JWB Characterization of eIF3k: a newly discovered subunit of mammalian translation initiation factor elF3. Eur. J. Biochem. 270, 4133–9 (2003). [DOI] [PubMed] [Google Scholar]
- 75.Gantt KR, Jain RG, Dudek RW & Pekala PH HuB localizes to polysomes and alters C/EBP-beta expression in 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 313, 619–22 (2004). [DOI] [PubMed] [Google Scholar]
- 76.Panda AC, Martindale JL & Gorospe M. Polysome Fractionation to Analyze mRNA Distribution Profiles. Bio-protocol 7, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are available within the paper and its Supplementary Information and Tables, with the exception of raw data for eCLIP and ribosome profiling which are accessible as BioProject PRJNA633047 containing BioSample accessions SAMN14931303- SAMN14931312 and SRA accession SRR11794832-SRR11794851 (Supplementary Table 6). All other data are available from the corresponding author upon reasonable request.