Abstract
The membrane-embedded protein rhodopsin is widely produced in organisms as a photoreceptor showing a variety of light-dependent biological functions. To investigate its molecular features, rhodopsin is often extracted from cellular membrane lipids by a suitable detergent as “micelles.” The extracted protein is purified by column chromatography and then is often reconstituted into “liposomes” by removal of the detergent. The styrene-maleic acid (“SMA”) copolymer spontaneously forms nanostructures containing lipids without detergent. In this study, we applied SMA to characterize two microbial rhodopsins, a thermally stable rhodopsin, Rubrobacter xylanophilus rhodopsin (RxR), and an unstable one, Halobacterium salinarum sensory rhodopsin I (HsSRI), and evaluated their physicochemical properties in SMA lipid particles compared with rhodopsins in micelles and in liposomes. Those two rhodopsins were produced in Escherichia coli cells and were successfully extracted from the membrane by the addition of SMA (5 w/v %) without losing their visible color. Analysis by dynamic light scattering revealed that RxR in SMA lipid particles (RxR-SMA) formed a discoidal structure with a diameter of 54 nm, which was 10 times smaller than RxR in phosphatidylcholine liposomes. The small particle size of RxR-SMA allowed us to obtain scattering-less visible spectra with a high signal-to-noise ratio similar to RxR in detergent micelles composed of n-dodecyl-β-D-maltoside. High-speed atomic force microscopy revealed that a single particle contained an average of 4.1 trimers of RxR (12.3 monomers). In addition, RxR-SMA showed a fast cyclic photoreaction (k = 13 s−1) comparable with RxR in phosphatidylcholine liposomes (17 s−1) but not to RxR in detergent micelles composed of n-dodecyl-β-D-maltoside (0.59 s−1). By taking advantage of SMA, we determined the dissociation constant (Kd) of chloride for HsSRI as 34 mM. From these results, we conclude that SMA is a useful molecule forming a membrane-mimicking assembly for microbial rhodopsins having the advantages of detergents and liposomes.
Significance
Membrane-embedded proteins play essential roles in a variety of biological phenomena in organisms. To investigate their structures and functions, membrane proteins are usually extracted from cellular membrane lipids by a suitable detergent as micelles, after which the extracted proteins are reconstituted into liposomes. Styrene-maleic acid (SMA) copolymer, which spontaneously forms nanostructures containing cellular membrane lipids without detergent, was used to characterize two microbial rhodopsins, Rubrobacter xylanophilus rhodopsin and Halobacterium salinarum sensory rhodopsin I. Of note, those two microbial rhodopsins were successfully extracted using SMA keeping the advantages of the detergents and liposomes. We conclude that SMA is a useful molecule forming a membrane-mimicking assembly for microbial rhodopsins.
Introduction
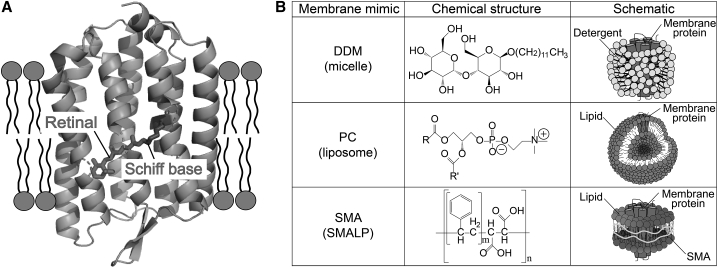
Membrane-embedded proteins are localized in cell membranes and play essential roles in a variety of biological functions, such as mass transfer and signal transduction across those membranes (1,2). Microbial rhodopsins are proteins containing seven-transmembrane α-helices with a chromophore retinal (vitamin A aldehyde) and form one of the largest phylogenetic groups among the membrane proteins (Fig. 1 A; (3)). The retinal is bound to an absolutely conserved Lys residue of the protein moiety (opsin) via a protonated retinylidene Schiff base linkage (Fig. 1 A). By differences in the interaction modes between the cognate opsin and the retinal, microbial rhodopsins absorb a wide range of visible wavelengths from 400 to 700 nm (3). In general, the light absorption of microbial rhodopsins causes the all-trans to 13-cis photoisomerization of the retinal (3), after which it returns to the original state through various spectroscopically distinctive photointermediates, such as K, L, M, N, and O (3,4). During a cyclic series of reactions called the photocycle, microbial rhodopsins show various light-dependent biological functions, such as photoelectric sensors, ion transporters, and enzymes within organisms (3,5). Thus, we can precisely control the function of microbial rhodopsins by visible light and can monitor the formation and decay of each intermediate with a high spatiotemporal resolution. It is worth noting that microbial rhodopsins are distributed in a variety of microorganisms, including three biological domains (i.e., eukaryotes, bacteria, and archaea) and viruses (5, 6, 7), indicating their biological significance in the host organisms and viruses. In addition, microbial rhodopsins are applicable for optogenetics technology, a method to control biological activity with light-activatable and genetically encoded molecules such as microbial rhodopsins and other photoreactive proteins (7,8). These features make microbial rhodopsins a valuable model both for membrane proteins and for photoactive proteins (6).
Figure 1.
Crystal structure of RxR and three membrane-mimicking systems. (A) Shown is the crystal structure of RxR (Protein Data Bank, PDB: 6KFQ (42)). The chromophore retinal binds to the seven-transmembrane domain via a protonated Schiff base linkage. (B) Shown are chemical structures and schematics of three membrane-mimicking systems. The characters m and n in the chemical structure of SMA represent the repeated numbers m, which correspond to 2 and ∼10 in this study. The number n was estimated from the molecular weight of styrene-maleic anhydride (SMAnh).
To investigate the molecular properties of microbial rhodopsins, they are often extracted from cellular membrane lipids by a suitable detergent as “micelles,” in which microbial rhodopsins are surrounded by amphipathic detergent molecules. The micelle system enables protein purification and makes it possible to perform biochemical, biophysical, and spectroscopic studies with the suppression of the scattering of the samples. Many microbial rhodopsins have been successfully solubilized by a class of detergents composed of a hydrophilic saccharide and a hydrophobic alkyl chain, such as n-dodecyl-β-D-maltoside (DDM) and n-octyl-β-D-glucoside, without losing their visible color (Fig. 1 B, upper panel; (9, 10, 11)). However, it is difficult to apply the detergent system to some unstable rhodopsins. In addition, because microbial rhodopsins function naturally in cell membranes, it is often difficult to investigate their biological activities in detergent micelles. In fact, it has been reported that the functional properties of several membrane proteins in micelles are significantly different from those in lipid bilayers (12). Therefore, detergent-solubilized rhodopsin is often reconstituted into “liposomes” by removal of the detergent and the addition of lipid molecules (13, 14, 15). Many microbial rhodopsins have been successfully reconstituted into a class of phospholipids such as phosphatidylcholine (PC) and phosphatidylglycerol (PG) without loss of their visible color (Fig. 1 B, middle panel; (14,15)). In general, the reconstitution of rhodopsins into liposomes has been performed in solutions with low salt concentrations (≤500 mM NaCl) because charges of the salt hamper the formation of liposomes because of the electric repulsion between salts and lipids. It should be noted that some microbial rhodopsins, especially those obtained from hyperhalophilic organisms such as Halobacterium salinarum sensory rhodopsin I (HsSRI), rapidly lose their visible absorption in low salt conditions within several seconds because of denaturation (16). Therefore, such unstable rhodopsins are not suitable for reconstitution into liposomes. Besides, detergent-solubilized rhodopsin is reconstituted into nanoscale phospholipids bilayers surrounded by amphipathic membrane scaffold protein(s) (MSP(s)), called MSP nanodiscs, by removing the detergent (17,18). Because it is necessary to solubilize and purify microbial rhodopsins in the detergent micelles before the reconstitution into the MSP nanodiscs, the rhodopsins that are unstable in the detergent micelles are not suitable for this system. Thus, in contrast to soluble proteins, membrane proteins face several problems for characterizing their molecular properties especially in vitro.
Styrene-maleic acid (SMA) copolymer spontaneously converts membranes into water-soluble nanodiscs (Fig. 1 B, lower panel). When mixed with cellular membranes, the styrene moieties of SMA spontaneously bind to the lipid acyl chain by hydrophobic interactions, leading to the formation of nanostructures called SMA lipid particles (SMALPs) (Fig. 1 B, lower panel; (19)). These characteristics make SMA a useful membrane mimic for solubilizing membrane proteins and for reconstituting them into lipid-based nanodiscs (20). Thus, by using SMA, membrane-embedded proteins can be extracted from the membrane and reconstituted into SMALPs without the use of detergents and extrinsic lipids (21). In fact, SMA has been successfully applied to various types of membrane proteins, including potassium channels (22), ATP-binding cassette (ABC) transporters (23), G-protein-coupled receptors (24), cytochrome b6f complex (25), and photosystem II complex (25), which were produced in various hosts such as bacteria, yeast, insect cells, and mammalian cells.
On the basis of that background, in this study, we used SMA with two microbial rhodopsins, a thermally stable rhodopsin, Rubrobacter xylanophilus rhodopsin (RxR) (11,26), and an unstable one, Halobacterium salinarum sensory rhodopsin I (HsSRI) (27), and quantitatively investigated their physicochemical properties in SMA and compared those with DDM micelles and PC liposomes. RxR is produced in the eubacterium R. xylanophilus, which lives in high temperatures (∼60°C) as a light-driven outward proton pump (11,26). The proton gradient generated across the membrane is thought to be utilized for the production of ATP through ATP synthase. HsSRI is produced in the hypersaline archaeon H. salinarum, which lives in extremely high salt environments (i.e., 4 M NaCl). HsSRI functions as a dual photosensor both for negative and for positive phototaxis (28). Of note, HsSRI is unstable, especially in the detergent-solubilized state at low salt conditions (≤500 mM NaCl) (29). In this study, we used RxR and HsSRI to evaluate the applicability of SMA forming a membrane-mimicking assembly for microbial rhodopsins. Previous studies have applied SMA so far to several microbial rhodopsins, such as H. salinarum bacteriorhodopsin, Natronomonas pharaonis sensory rhodopsin II (NpSRII), and proteorhodopsin (30, 31, 32, 33). We will fully discuss the results and applicability of SMA estimated in this study in comparison with the previous studies.
Materials and Methods
Sample preparation
For the preparation of SMA, a styrene-maleic anhydride (SMAnh) copolymer (SMA2000, in which the ratio of styrene and maleic anhydride is ∼2:1; Cray Valley, Exton, PA) was kindly provided by Kawahara Petrochemical (Osaka, Japan). The copolymer has been widely used to characterize several membrane proteins in the previous studies (19,33). The number average molecular weight, weight average molecular weight, and polydispersity index were 3000, 7500, and 2.5, respectively. We converted the hydrophobic SMAnh into the water-soluble, amphiphilic, membrane-active acid derivative SMA in a hydrolysis reaction according to a previously published method (34). In short, SMAnh was solubilized in 1 M NaOH and refluxed for 2 h while heating the suspension to 100°C. After being cooled to room temperature, the polymer was precipitated by adding HCl and washed six times with deionized water to remove Na+ ions. The sample was dissolved in 0.6 M NaOH and adjusted to pH 8 to prepare the stock solutions. The solutions were lyophilized and then stored at −20°C until the use.
For DNA manipulation and the production of RxR and HsSRI, the Escherichia coli strains, DH5α and BL21(DE3), were used as hosts, respectively. Those expression plasmids encoded a hexahistidine motif (His-tag) at the C-terminus as described previously (26,29). E. coli cells harboring the expression plasmids were incubated in 50 mL LB medium containing 50 μg/mL ampicillin and were grown at 37°C for 15 h. After the preculture, the growth medium was directly transferred to 1.0 L LB medium containing 50 μg/mL ampicillin, and the cells were grown at 37°C until the optical density at 660 nm reached 1.4–1.6. Isopropyl-D-1-thiogalactopyranoside (final concentration = 1 mM) and all-trans retinal (final concentration = 10 μM) were then added to the culture medium and were further incubated at 37°C for 3 h to induce protein production, after which the cells were collected by centrifugation (5535 × g for 10 min at 4°C). The crude membranes of rhodopsin-producing cells were solubilized with a detergent 1 w/v % DDM (RxR, for 45 min at room temperature; HsSRI, for 30 min at 4°C) (Dojindo, Kumamoto, Japan) or with 2.5 and 5 w/v % SMA (RxR, for 2 h at room temperature; HsSRI, for 2 h at 4°C) (20,23,24,32,33,35,36). The suspension of the membranes was centrifuged (103,900 × g for 30 min at 4°C) and then the supernatant was collected as the solubilized fraction. The red color of the supernatant extracted with 5 w/v % SMA was brighter than that with 2.5 w/v % SMA (Fig. S1), which indicates that the rhodopsins were more efficiently extracted from the membrane with 5 w/v % SMA. The solubilized fraction was purified by Ni2+ affinity column chromatography utilizing the His-tag attached to RxR and HsSRI. The purified proteins were concentrated and replaced with the appropriate buffer solution by centrifugation using an Amicon Ultra Filter at 4°C. The yield of the purified RxR was ∼5 mg per 1 L culture medium for 5 w/v % SMA, which was 2.5-fold higher than that (i.e., ∼2 mg per 1 L culture medium) for 2.5 w/v % SMA. Therefore, we decided to use 5 w/v % SMA for extracting a large amount of the samples to be used in their detailed analysis in SMALPs. The RxR purified by DDM was reconstituted into egg PC liposomes (Sigma-Aldrich, Tokyo, Japan), from which DDM was removed with Bio-Beads SM-2 (Bio-Rad Laboratories, Hercules, CA). The molar ratio of RxR and PC was adjusted to 1:30. The PC-reconstituted RxR was collected by centrifugation (20,631 × g for 20 min at 4°C) and was washed several times with buffer containing 50 mM Tris-HCl (pH 8), 150 mM NaCl, and 10 v/v % glycerol. The samples were then resuspended in the same buffer for the measurements.
Dynamic light scattering and high-speed atomic force microscopy experiments
Dynamic light scattering (DLS) signals were recorded using a Zetasizer uV (Malvern Panalytical, Worcestershire, UK) in a buffer solution containing 50 mM Tris-HCl (pH 8), 150 mM NaCl, and 10 v/v % glycerol at room temperature (∼25°C). The particle diameters were calculated by the Einstein Stokes equation as follows (Eq. 1):
| (1) |
where kB, T, η0, and a represent Boltzmann’s constant, absolute temperature, dynamic viscosity, and the radius of the discoidal particle, respectively.
High-speed atomic force microscopy (HS-AFM) images of RxR in SMALPs (RxR-SMA) were obtained as described previously (37,38). Briefly, HS-AFM was operated in the tapping mode. Deflections of the cantilever were detected with an optical beam deflection detector using an infrared laser (0.8 mW, 780 nm) focused through a ×60 objective lens (CFI S Plan Fluor ELWD 60×; Nikon, Tokyo, Japan) onto the back side of a cantilever (BL-AC7DS-KU4; Olympus, Tokyo, Japan) covered with a gold film, and the reflected infrared laser was detected by a two-segmented p-intrinsic-n (PIN) photodiode. The free oscillation amplitude was ∼1 nm, and the set-point amplitude was ∼90% of the free amplitude for feedback control of the HS-AFM. An amorphous carbon tip generated by electron beam deposition was used as an atomic force microscopy (AFM) probe. The experiments were performed on a mica substrate in a buffer solution of 20 mM Tris-HCl (pH 7.4) and 150 mM KCl at room temperature (∼25°C).
Ultraviolet visible spectra, high-performance liquid chromatography, and time-resolved flash-photolysis experiments
Ultraviolet (UV) visible spectra were recorded using a UV-2450 spectrophotometer (Shimadzu, Kyoto, Japan) at 25°C in the appropriate buffer solution. The retinal composition was analyzed by high-performance liquid chromatography (HPLC) as reported previously (39). In short, the protein concentration was adjusted to 5.6 μM, and then the retinal was extracted with hexane as retinal oxime by the addition of 1 M hydroxylamine (final concentration = 100 mM) with 99.7 v/v % methanol (final concentration = 66.7 v/v %) at room temperature (∼25°C). The extraction was carefully carried out under dim red light (>580 nm) to avoid the photoactivation of the proteins. Each sample was kept in the dark for more than 1 week before analysis. The molar composition of each retinal isomer was calculated from the areas of the peaks in HPLC patterns monitored at 360 nm. Upon calculation, the reported absorption coefficients of retinal isomers were used (51,600, 54,600, and 47,900 cm−1 M−1 for all-trans 15-anti, all-trans 15-syn, and 13-cis retinal oximes, respectively) (40). The pH titration experiments were performed using essentially the same method described previously (39). In short, the experiments were performed at room temperature using RxR reconstituted into PC liposomes and solubilized by DDM and SMA in a solution containing 150 mM NaCl and Good’s buffer (0.89 mM citric acid, 0.89 mM MES, 1.1 mM TES, 0.78 mM TAPS, 1.1 mM CHES, and 0.33 mM CAPS) with 0.05% DDM. The buffer composition had the same buffer capacity over a wide range of pH-values. The pH was then adjusted to the desired value by the addition of NaOH. The data were fitted to the Henderson-Hasselbalch equation with single pKa as follows (Eq. 2):
| (2) |
where a represents the amplitude of the change of absorption differences, and b is an offset. After the measurements, the reversibility of the pH changes of the sample was checked to confirm that the sample was not denatured during the measurements.
The Cl− titration experiments were performed using essentially the same method described previously (41). In short, the experiments were performed on ice using HsSRI solubilized by SMA in a buffer containing 50 mM Tris-H2SO4 (pH 8.5) and 10 v/v % glycerol. The chloride (Cl−) ions were supplied and adjusted to the desired value using a concentrated NaCl solution in which the following two buffers were used in the appropriate ratio to maintain the ionic strength: 1) 4 M NaCl, 50 mM Tris-H2SO4 (pH 8.5), and 10 v/v % glycerol; and 2) 1 M Na2SO4, 50 mM Tris-H2SO4 (pH 8.5), and 10 v/v % glycerol. The NaCl-induced absorbance changes of HsSRI were plotted against the NaCl concentration and were analyzed by fitting the data of the Hill equation as follows (Eq. 3):
| (3) |
where ΔAmax represents the maximal absorbance change, and [X] is the NaCl concentration. The Hill coefficient was fixed as 1.
The apparatus and the procedure for time-resolved laser flash photolysis were essentially the same as described previously (39). In short, absorption spectra from 380 to 700 nm at 5-nm intervals were obtained using a homemade computer-controlled flash-photolysis system equipped with an Nd:YAG laser as an actinic light source. The wavelength of the actinic pulse was tuned at 545 nm (4 ns) using an optical parametric oscillator (Surelite OPO Plus; Continuum, San Jose, CA). The pulse intensity was adjusted to 2 mJ/pulse. The data before the laser flash were adopted as a baseline. All measurements were conducted at 25°C using a thermostat. The data were analyzed by fitting the data of the multiexponential equation with the linear combination as follows (Eq. 4):
| (4) |
where t represents each time constant, and A represents the rate of each intermediate at each time constant. After the measurements, the reproducibility of the data was checked to confirm that the sample was not denatured during the measurements.
Results
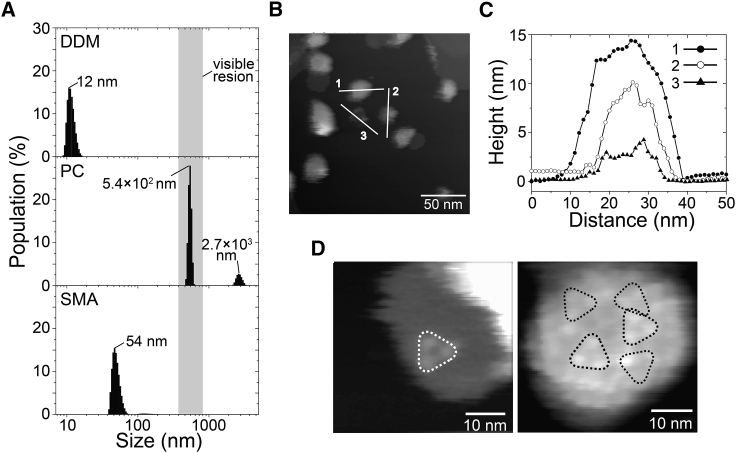
Firstly, to investigate the applicability of SMA for RxR, we analyzed the particle structures of RxR-SMA and then quantitatively compared them with those of RxR in DDM micelles (RxR-DDM) and RxR in PC liposomes (RxR-PC). Fig. 2 A shows the DLS patterns of RxR-DDM, RxR-PC, and RxR-SMA in solution. The population of the samples was plotted against the cognate diameters. As seen, sharp peaks were observed at 12 ± 1.5, 54 ± 6.9, and 5.4 × 102 ± 23 nm for RxR-DDM, RxR-SMA, and RxR-PC, respectively (Table 1). The particle size for RxR-SMA (54 ± 6.9 nm) was much smaller than visible wavelengths (400–800 nm), suggesting relatively scattering-less visible spectra. For RxR-PC, in addition to the main peak, a subpeak was also observed at 2.7 × 103 ± 2.2 × 102 nm. The areas of the peaks in Fig. 2 A reflect the population of the cognate particles. From the area of the signals, the ratio of small and large particles was estimated at 86 and 14%, respectively, indicating the heterogeneity of RxR-PC.
Figure 2.
Structural information revealed by DLS and HS-AFM. (A) Shown are the DLS patterns of RxR-DDM (upper panel), RxR-PC (middle panel), and RxR-SMA (lower panel) with averaged sample diameters. The integration of the areas of the peaks is 100% in each sample. The visible wavelength region (400–800 nm) is colored gray in the background. (B) Shown are the HS-AFM images of RxR-SMA on the mica surface. Scale bars, 50 nm. (C) The cross section profiles of the particles are shown in (B) as white lines 1, 2, and 3. (D) Shown are snapshots of the AFM video. Scale bars, 10 nm. The estimated RxR trimers are shown as black and white triangles.
Table 1.
Comparison of the Physicochemical Properties of RxR-DDM, RxR-PC, and RxR-SMA
| Membrane Mimic | Particle Diameter (nm) | All-Trans Retinal (%) | pKa of the Schiff Base | Photocycling Rate (s−1) |
|---|---|---|---|---|
| DDM (micelle) | 12 ± 1.5 | 98.3 | 10.4 ± 0.021 | 0.59 |
| PC (liposome) | 5.4 × 102 ± 23 (86%) | 98.4 | 11.7 ± 0.049 | 17 |
| 2.7 × 103 ± 2.2 × 102 (14%) | ||||
| SMA (SMALP) | 54 ± 6.9 | 99.0 | 12.2 ± 0.10 | 13 |
It has been reported that SMA forms a discoidal structure with a Stokes diameter of ∼10 nm (20,23,27), which is significantly smaller than RxR-SMA (54 ± 6.9 nm) observed here (Fig. 2 A). Several previous studies reported that the SMALPs formed multiple-layered particles by AFM and DLS measurements (31,36). The multiple-layered structure of the SMALPs could lead to the increased Stokes diameter observed in this study. To investigate the reason for that, we performed HS-AFM (Fig. 2, B–D). As seen in Fig. 2, B and C, discoidal particles with diameters of several tens of nanometers and heights of ∼5, 10, and 15 nm were observed on a mica surface in which the size roughly matched well with RxR-SMA. The average diameter of the particles was estimated as 30 ± 6.4 nm (n = 53) (Fig. S2 A). The ratio of particles with heights of ∼5, 10, and 15 nm was estimated as 40, 20, and 40%, respectively (n = 214). Because the heights of particles with 5, 10, and 15 nm matched well with the single, double, and triple values of the thickness of a single lipid bilayer (i.e., ∼5 nm), the RxR-SMA should have consisted of single-, double-, and triple-layered particles. The large diameter and multilayered structure of RxR-SMA would lead to the increased Stokes diameter in DLS measurements compared with the previously reported size (20,35). An expanded view of the AFM image of RxR-SMA and its video showed that RxR forms a trimer in the particle (Fig. 2 D; Video S1). We directly counted the numbers of RxR trimers from the AFM images (n = 8) (Fig. S2 B) and estimated the average number of RxR trimers per particle as 4.1 ± 1.4 (12.3 monomers). Because the average diameter of the particles was estimated as 30 ± 6.4 nm, the average number of lipids contained in an empty particle with a diameter of 30 nm was estimated as 1125∼1250 lipids, assuming that an empty particle with a diameter of 12 nm contains 180∼200 lipids (35). The volume of 4.1 trimers of RxR corresponds to 492 lipids, assuming that the volume of crystal structure of monomeric RxR (∼37 nm3) corresponds to 40 lipids (42). By subtracting 492 lipids from 1125 to ∼1250 lipids, we calculated the number of lipids as 635∼760 in the particle with a diameter of 30 nm containing 4.1 timers of RxR. Thus, the ratio of RxR molecules to lipids was estimated as 1:52∼62 in this study. In general, the diameter of the particle was around 10 nm, and the ratio of protein molecules to lipids was estimated as 1:10∼170 (20,35). Although the particle diameter in this study was much larger than those of the typical particles, the ratio of protein molecules to lipids in this study was within the reported range of the ratio (i.e., 10∼170).
HS-AFM video of RxR-SMA on the mica surface with 3.3 fps.
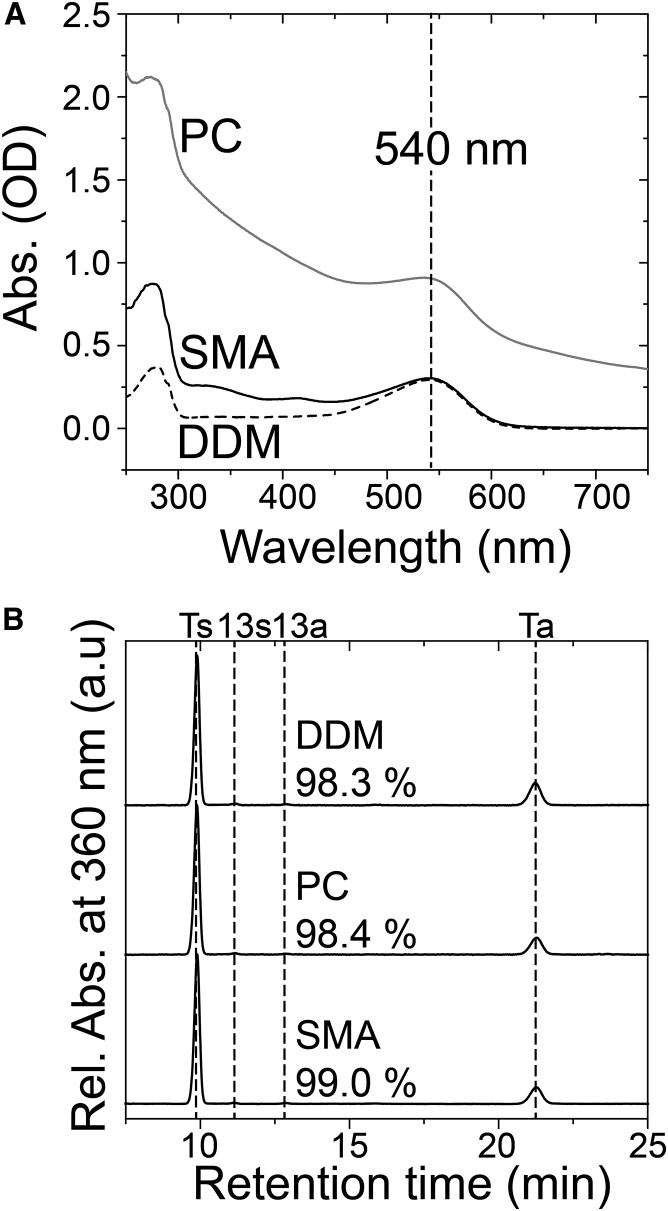
Secondly, we analyzed the absorption spectra and retinal configurations of RxR-DDM, RxR-PC, and RxR-SMA (Fig. 3). In the visible region, an absorption maximum (λmax) was commonly located at 540 nm for RxR-DDM, RxR-PC, and RxR-SMA, suggesting a less significant alteration in the protein structure by their membrane mimics (Fig. 3 A). These data demonstrated that we were able to purify RxR as a photoactive protein using SMA without a detergent. Fig. 3 B and Table 1 show the HPLC patterns of the retinal oxime extracted from the cognate samples. As shown in Fig. 3 B, the isomeric state of retinal is predominantly all-trans (>98.3%, the sum of the area of Ts and Ta), with a small proportion of 13-cis (<1.7%, the sum of the area of 13s and 13a). As well as other microbial rhodopsins, RxR shows a function (i.e., outward proton pumping activity) in collaboration only with the all-trans retinal chromophore but not with the 13-cis form. From these results, we concluded that SMA shows no significant structural change either in the protein or in the retinal.
Figure 3.
Absorption spectra and HPLC patterns of RxR-DDM, RxR-PC, and RxR-SMA. (A) Shown are the UV-Vis spectra of RxR-DDM (broken line), RxR-PC (gray solid line), and RxR-SMA (black solid line). (B) Shown are the HPLC patterns of the retinal oxime of RxR-DDM, RxR-PC, and RxR-SMA in the dark (∼25°C). Ts, Ta, 13s, and 13a represent all-trans-15-syn, all-trans-15-anti, 13-cis-15-syn, and 13-cis-15-anti retinal oxime, respectively. The molar compositions of retinal isomers were calculated from the areas of the peaks in the HPLC patterns.
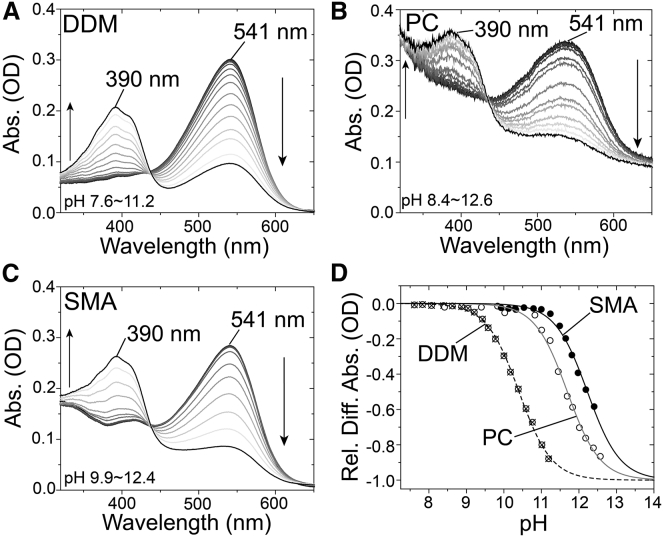
Thirdly, we analyzed the pKa of the protonated Schiff base of the retinal chromophore. For membrane-embedded proteins, including microbial rhodopsins, charged residues such as Lys, Arg, Asp, and Glu play essential roles in protein functions (26). In particular, a Lys residue located in the seventh helix is absolutely conserved among microbial rhodopsins and forms a protonated Schiff base with the retinal aldehyde. It is known that the deprotonation of the Schiff base nitrogen under alkaline condition leads to an increase in the energy gap between the electronic ground and excited states and then a spectral blue shift from the visible region to the UV region (43). In general, other titratable groups are not seen in this region. We performed pH titration experiments to estimate the pKa-value of the protonated Schiff base (Lys209 in RxR) by monitoring the spectral blue shift due to the deprotonation of the Schiff base at an alkaline pH. Fig. 4 shows the absorption changes of RxR-DDM (Fig. 4 A), RxR-PC (Fig. 4 B) and RxR-SMA (Fig. 4 C) at varying pH-values from 7.6 to 12.6. Decreases and increases in absorption at 541 nm and 390 nm, respectively, were observed with an isosbestic point at around 436 nm, indicating the presence of an equilibrium between the protonated and deprotonated forms of the Schiff base in RxR without the protonated and deprotonated states of other titratable groups. When the absorption changes at 541 nm were plotted against the environmental pH, the titration curves were fitted well by the Henderson-Hasselbalch equation (see Eq. 2; Fig. 4 D). From these data, the pKa-values of Lys209 of RxR-DDM, RxR-PC, and RxR-SMA were estimated to be 10.4 ± 0.021, 11.7 ± 0.049 and 12.2 ± 0.10, respectively.
Figure 4.
pH titration experiments of RxR-DDM, RxR-PC, and RxR-SMA. (A–C) Shown are the absorption spectra of RxR-DDM (A), RxR-PC (B), and RxR-SMA (C) obtained at pH-values ranging from 7.6 to 12.6. The pH was adjusted to the desired values by the addition of NaOH. (D) Shown is the estimation of the pKa of the Schiff base. Absorption changes at 541 nm were plotted against respective pH-values. The data were fitted by the Henderson-Hasselbalch equation (Eq. 2, see Materials and Methods).
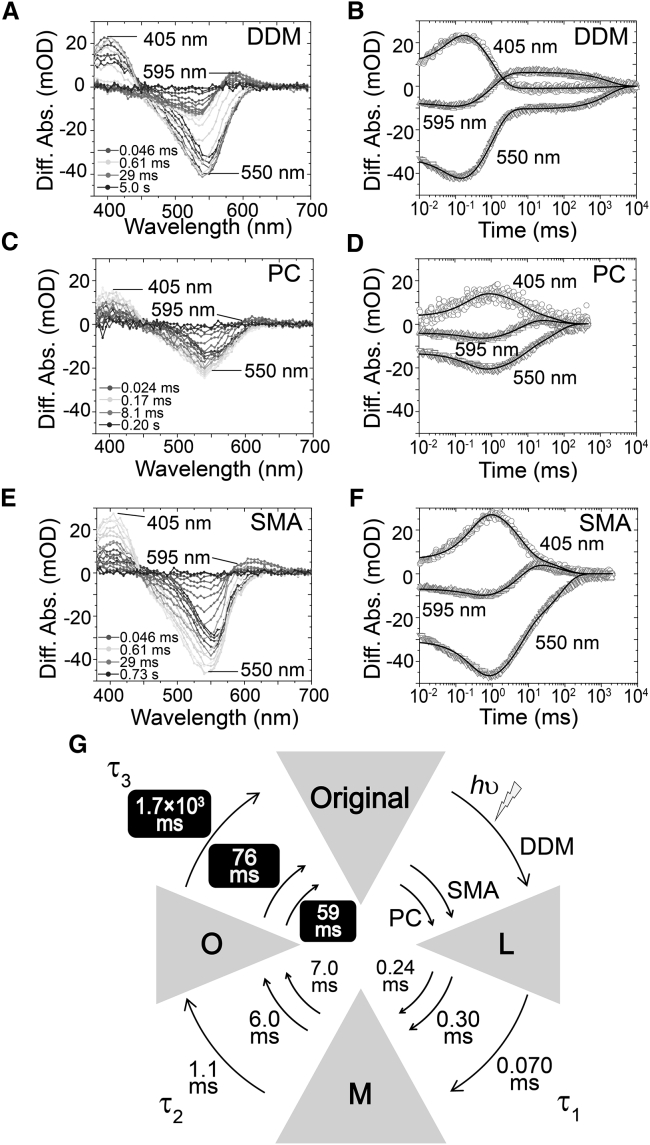
Fourthly, we performed time-resolved flash-photolysis experiments to examine the photoreactions of RxR-DDM, RxR-PC, and RxR-SMA. Microbial rhodopsin shows a cyclic series of reactions (called the photocycle) upon illumination that returns to the original state through several intermediates having distinctive absorption maxima. During the photocycle, cognate rhodopsin function is achieved. Therefore, it is important to monitor the formation and decay of each intermediate to understand the protein function. Fig. 5 shows the flash-induced difference spectra of RxR-DDM (Fig. 5, A and B), RxR-PC (Fig. 5, C and D), and RxR-SMA (Fig. 5, E and F) (pH 8). Flash-induced bleaching and recovery of the original pigment were observed at around 550 nm, a value similar to the absorption maximum of RxR, as shown in Fig. 3. At 405 nm, an increase and a decrease in absorbance were observed, implying the formation and decay of a blue-shifted M intermediate of RxR (RxRM). During the decay of RxRM, a red-shifted O intermediate of RxR was sequentially produced at around 595 nm. The time courses of the absorbance change at selected wavelengths (405 nm for RxRM, 540 nm for the unphotolyzed state, and 595 nm for O intermediate of RxR) were fitted well to the multiexponential equation (see Eq. 4) with three time constants, τ1, τ2, and τ3 (solid black lines in Fig. 5, B, D, and F), which can be assigned to the M formation (τ1), O formation (τ2), and O decay (τ3), respectively. As seen in Fig. 5 G, the three time constants of RxR-SMA (τ1 = 0.30 ms, τ2 = 6.0 ms, and τ3 = 76 ms) and of RxR-PC (τ1 = 0.24 ms, τ2 = 7.0 ms, and τ3 = 59 ms) were almost identical, but they were dramatically different from those of RxR-DDM (τ1 = 0.070 ms, τ2 = 1.1 ms, and τ3 = 1.7 × 103 ms), especially τ3. On the basis of these results, we propose a putative model of the photochemical reaction cycle of RxR-DDM, RxR-PC, and RxR-SMA with the time constants τ1, τ2 and τ3 (Fig. 5 G).
FIGURE 5.
Photoreaction kinetics of RxR-DDM, RxR-PC, and RxR-SMA. (A–F) Shown are the flash-induced difference absorption spectra of RxR-DDM (A and B), RxR-PC (C and D), and RxR-SMA (E and F) at 25°C. The data were fitted using the multiexponential equation (Eq. 4, see Materials and Methods) with three time constants, τ1, τ2, and τ3 (solid lines in B, D, and F). (G) Given is a putative photocycle model of RxR with the cognate time constants.
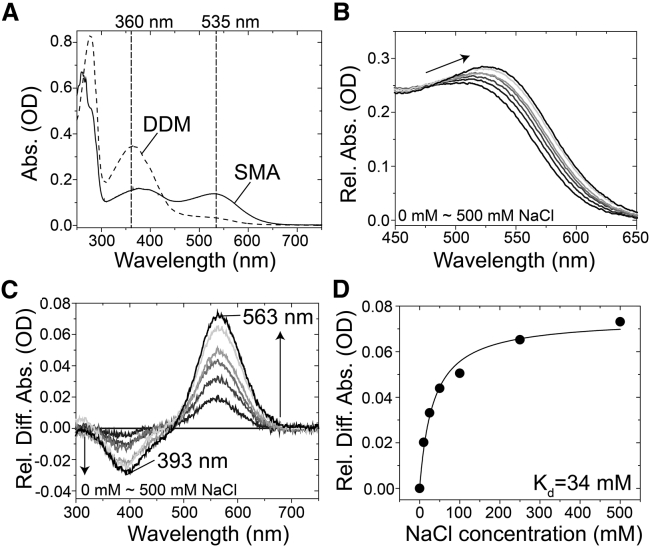
Finally, to further investigate the applicability of SMA for microbial rhodopsins, we employed HsSRI as a model for unstable microbial rhodopsins. As seen in Fig. S3, HsSRI can be produced in E. coli cells as a recombinant protein with a blue color (29,44), but its color becomes yellow (λmax = ∼360 nm) when solubilized with the detergent DDM in low salt conditions (500 mM NaCl) because of the protein denaturation (Fig. 6 A, broken line; Fig. S3). On the other hand, when SMA was used for E. coli cells producing HsSRI, visible absorption at 535 nm was observed in addition to the protein denaturation signal at 360 nm (Fig. 6 A, solid line; Fig. S3) even in low salt conditions (500 mM NaCl). So far, we have identified two relatively stable sensory rhodopsin I (SRI) proteins, SRI from the eubacterium Salinibacter ruber (SrSRI) and SRI from the halophilic archaeon Haloarcula vallismortis (29). Using those proteins, we reported chloride binding to SrSRI with an affinity of 307 mM (41). The visible absorption of HsSRI in SMALPs (HsSRI-SMA) was kept more than 1 h even in the absence of NaCl, and its high stability allowed us to investigate the chloride binding to HsSRI-SMA. As seen for SrSRI and SRI from the halophilic archaeon H. vallismortis, an NaCl-induced spectral red shift from 521 to 535.5 nm was observed for HsSRI-SMA (Fig. 6 B). The difference spectra showed increases and decreases in absorbance at 563 and 393 nm, respectively, with an isosbestic point at around 485 nm (Fig. 6 C) indicating an equilibrium between the chloride-bound and -unbound forms of HsSRI-SMA. The absorbance changes at 563 nm were plotted against the NaCl concentration (Fig. 6 D) and the dissociation constant (Kd) estimated using the Hill equation (see, Eq. 3) was 34 ± 5.1 mM.
Figure 6.
Applicability of SMA to HsSRI. (A) Shown are the absorption spectra of HsSRI-DDM (broken line) and HsSRI-SMA (solid line). (B) Shown are the changes in the absorption spectra of HsSRI-SMA by adding NaCl to the desalted buffer over a spectral range from 450 to 650 nm. The NaCl concentrations are 0, 10, 25, 50, 100, 250, and 500 mM NaCl. (C) Shown are the difference absorption spectra by adding NaCl to the desalted HsSRI-SMA sample. Each spectrum corresponds to 10, 25, 50, 100, 250, and 500 mM NaCl. (D) The absorbance changes at 563 nm were plotted against NaCl concentrations, and the dissociation constant (Kd) value of Cl− was calculated using the Hill equation (Eq. 3, see Materials and Methods) as 34 ± 5.1 mM.
Discussion
Schematics of RxR-SMA
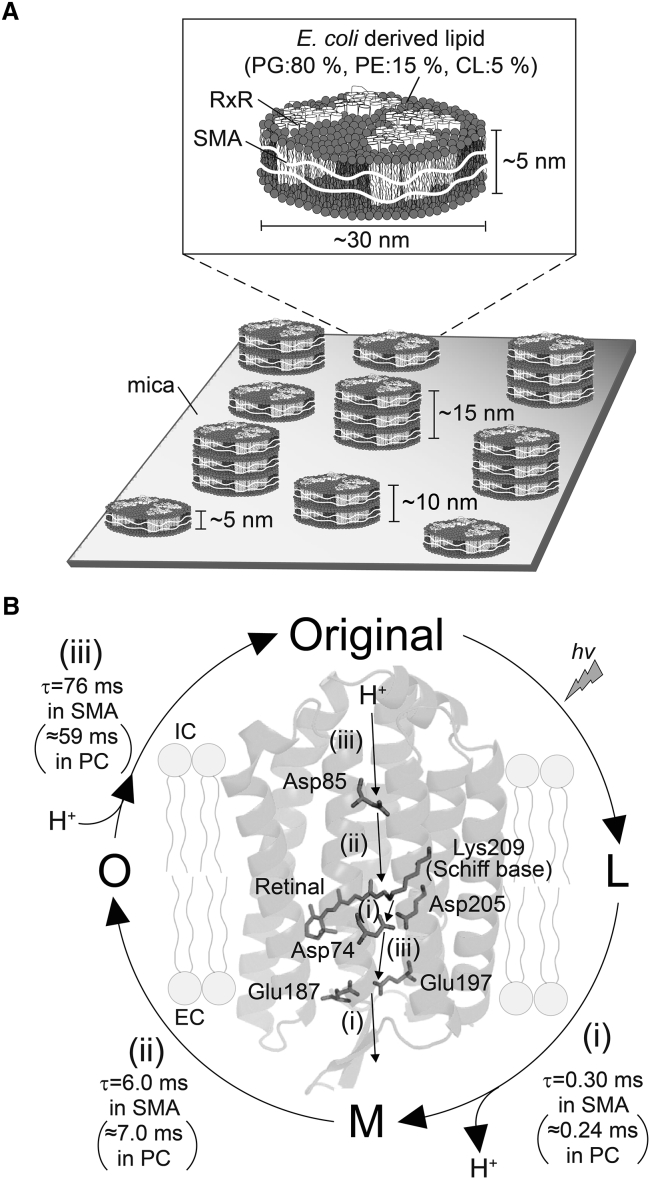
On the basis of our results and the previous report in which the styrene groups of the polymer were shown to interact with the lipid acyl chains (45), we propose that RxR-SMA formed single-, double-, and triple-layered discoidal particles with diameters of 30 nm shown schematically in Fig. 7 A. Many previous studies indicated that SMALPs formed discoidal particles with diameters of ∼10 nm (20,23,27), which is significantly smaller than that of RxR-SMA observed here. We speculate that the cause of such the large difference in particle sizes may have come from several differences in experimental procedures to prepare the SMALPs between this study and the previous studies. Firstly, we solubilized cellular membrane lipids with 5 w/v % SMA, which was higher than the concentration (i.e., 2.5 w/v %) reported in the previous studies (20,23,27). Secondly, we solubilized cellular membrane lipids by SMA without adding exogenous lipids, whereas the exogenous lipids (e.g., dimyristoyl PC) were added during the solubilization process in the previous studies (20,23). Thus, the ratio of SMA to lipids was higher in this study than those in the previous studies. Because it has been reported that the diameters of the SMALPs were increased according to the higher ratio of SMA to lipids (46), we speculate that the higher ratio of SMA to lipids led to the formation of the extremely large SMALPs observed in this study. On the other hand, it may be possible that the stacked multilayers of RxR-SMA influence the properties of the rhodopsins while we analyzed them using the samples consisting of single-, double-, and triple-layered particles with populations of 40, 20 and 40%, respectively, in this study (Fig. 7 A). It is assumed that the single-, double-, and triple-layered states form the equilibrium in the SMALPs samples. To elucidate the effect of the stacked multilayers, it will be necessary to shift the equilibrium to each state by changing several experimental conditions, such as temperature, pH, pressure ionic composition, and ionic strength, and then quantitatively analyze the properties (e.g., photocycle and estimation of pKa of the Schiff base) of each state as future works. Besides, the results of AFM measurements could not completely deny the possibilities that the SMALPs samples form swollen single lipid bilayers (e.g., the phospholipid headgroups of the lower leaflet are not in immediate contact with the solid support and that the protein protrudes from at least one side of the nanodiscs), complex aggregates, or copolymer micelles rather than the multiple-layered structure. It will be necessary to observe the detailed morphology of the SMALPs by electron microscopy in the future.
Figure 7.
Schematic structure of RxR-SMA and proton transport mechanism in RxR-SMA. (A) Shown are the schematics of RxR-SMA. The RxR-SMA samples consisted of single-, double-, and triple-layered particles having diameters of several tens of nanometers with the heights of ∼5, 10, and 15 nm, respectively. A single particle contains 4.1 RxR trimers (i.e., 12.3 RxR monomers) on average, with intrinsic lipids from E. coli cells. The native E. coli membrane consists of PG, PE, and CL (phosphatidylglycerol, phosphatidylethanolamine, and cardiolipin, respectively). (B) Shown is the proton transport mechanism in RxR-SMA during the photocycle. The proton transport pathway and key carboxylates are indicated on the crystal structure of RxR (PDB: 6KFQ (42)). A proton is transported from the intracellular side (IC) to extracellular side (EC) through several carboxylates during the photocycle.
Advantages of SMA in comparison with DDM and PC
Membrane-embedded proteins are often extracted and purified from lipid membranes using detergents to investigate their structures and functions. When necessary, the purified proteins are reconstituted into phospholipids such as PC and PG (13, 14, 15,47,48). Instead of detergents and lipids, in this study, we successfully used SMA as a molecule forming a membrane-mimicking assembly to characterize two microbial rhodopsins, RxR and HsSRI. Here, we discuss the applicability of SMA for RxR and HsSRI and compare that with DDM and PC.
From the viewpoint of RxR
The physicochemical and photochemical properties of RxR-SMA are listed in Table 1 and are compared with RxR-DDM (in detergent) and RxR-PC (in lipids). So far, the applicability of SMA has been reported for several membrane proteins, such as ABC transporters and KcsA (22,23), but its comparison with micelles and liposomes at the molecular level has not been reported. As seen in Figs. 2 and 3 and Table 1, the absorption maximum and retinal configuration of RxR-SMA (540 nm and 99.0%, respectively) are almost identical to those of RxR-DDM and RxR-PC, indicating that the overall protein and retinal structures are roughly identical among them. Thus, these three membrane mimic molecules (i.e., SMA, DDM, and PC) are generally suitable for investigating the structure and function of RxR. On the other hand, the DLS analysis revealed that the size of RxR-PC particles (5.4 × 102 ± 23 nm) matched well with the visible wavelength region (400–800 nm), suggesting that scattering would be observed for the visible spectroscopy of RxR-PC. In fact, RxR-PC showed a largely scattered spectrum, whereas RxR-SMA showed a scattering-less spectrum as well as RxR-DDM (Fig. 3). Thus, RxR-SMA is more suitable for spectroscopic analysis compared with RxR-PC. The pH titration experiments for RxR-DDM revealed that the pKa of the protonated Schiff base nitrogen (Lys209), which is a crucial residue for all rhodopsins because of its linkage with the retinal chromophore, was slightly (1.3–1.8 unit) smaller than the others (RxR-PC and RxR-SMA). RxR-DDM is surrounded by the detergent DDM in solution, whereas the others (RxR-PC and RxR-SMA) are surrounded by lipids (Fig. 1). Therefore, differences in the pKa of the Schiff base can be explained by differences in the interactions between RxR and surrounding molecules. Thus, these results suggest that two membrane mimics, SMA and PC, are more suitable for investigating the function and structure of RxR than is DDM. This feature is also shown in the reaction kinetics of RxR upon illumination. Time-resolved flash photolysis revealed that the reaction kinetics for RxR-SMA are similar to those of RxR-PC but are quite different from those of RxR-DDM (Fig. 5; Table 1). The mutational and theoretical analysis of RxR in our previous study revealed its proton transport steps as follows (Fig. 7 B): 1) a proton of the Schiff base (Lys209) is transferred to the counterion Asp74 during the transition from the L to M intermediate, and simultaneously, a respective proton is released from the proton releasing group (Glu187/Glu197) to the extracellular side, 2) a proton of Asp85 is transferred to the Schiff base during the transition from the M to O intermediate, and 3) a proton is taken up from the intracellular side to Asp85 during the transition from O intermediate to original state (26). All three time constants (i.e., τ1, τ2, and τ3) reflect the rates of the cognate proton transport steps and are physiologically important in the proton pumping function of RxR. The similarity of the reaction kinetics for RxR-SMA and RxR-PC suggests that RxR-SMA shows the equivalent proton pumping function with RxR-PC. Similar to the pKa of the Schiff base, the differences in rate constants between RxR-SMA, RxR-PC, and RxR-DDM can be explained by differences in the interactions between RxR and surrounding molecules. Microbial rhodopsins including RxR show cognate biological functions during the photocycle, indicating their biological importance. In outward proton pumps, including H. salinarum bacteriorhodopsin and RxR, one proton moves from the intracellular side to the extracellular side during a single photocycle, and therefore, the photocycling rate is tightly coupled with their proton pumping function. In this study, the photocycling rate of RxR-DDM was ∼30-fold smaller than RxR-SMA and RxR-PC, indicating its significantly lower proton pumping activity. Thus, these results suggest that two membrane mimics, SMALPs and PC liposome, are more suitable for investigating the function and structure of RxR compared with the detergent DDM. In contrast, a previous study reported that NpSRII showed 10-fold faster M decay in SMALPs (diameter: ∼10 nm) as compared with that in liposomes and MSP nanodiscs (32). The reason for the difference was proposed to be that the high local proton concentration around NpSRII in SMALPs is formed by the negative charge of carboxylic groups of SMA and affects the rate of M decay. We speculate that the larger distance between carboxylic groups of SMA and RxR due to the larger SMALPs (diameter: ∼30 nm) in this study prevents the high local proton concentration around RxR and effects on the rate of the photocycle. Thus, the large SMALPs allows us to reconstitute microbial rhodopsins into lipids without the undesired charge effects SMA.
These comparisons of spectroscopic properties revealed that RxR-SMA has advantages compared with RxR-DDM (i.e., small scattering) and RxR-PC (i.e., high pKa of the Schiff base and fast photocycle). Therefore, SMA will be a useful tool to study microbial rhodopsins instead of detergents such as DDM and lipids such as PC.
From the viewpoint of HsSRI
The applicability of using SMA for microbial rhodopsins was also revealed for HsSRI. For membrane-embedded proteins, reconstitution of the purified proteins into lipids such as PC is effective for their functional and structural studies because they work in the lipid bilayer in living organisms. On the other hand, membrane proteins are often denatured during purification with detergents because of their instability in micelles. To avoid denaturation, membrane proteins are produced in cell-free systems in which they are synthesized in lipids and simultaneously reconstituted into them (49). In addition, some membrane proteins from halophilic organisms are often unstable without salts (29). Because of the electrostatic interactions between salts and lipids, it is quite difficult to reconstitute membrane proteins into lipids in high salt conditions. Historically, microbial rhodopsins were first discovered from hypersaline environments; therefore, some of them are unstable in low salt conditions (≤500 mM NaCl). As reported so far, HsSRI can be successfully produced both in the archaeon H. salinarum (27) and in the eubacterium E. coli cells (29) with blue color. Fortunately, HsSRI can be purified using the detergent DDM in the presence of high salt conditions (>4 M NaCl) without loss of its color (29,44). However, its instability in low salt conditions hampers its reconstitution into lipids. In fact, HsSRI rapidly lost its visible color when DDM-solubilized HsSRI samples were suspended in a solution containing 500 mM NaCl (Fig. 6 A, broken line; Fig. S3). On the other hand, using SMA, HsSRI can be extracted and reconstituted into cellular membrane lipids and retains its visible color (Fig. 6 A, solid line; Fig. S3). It has been reported that SMALPs contain a central lipid bilayer supported by an outer annulus of SMA polymer in which the structure is stabilized by the intercalation of hydrophobic styrene groups between the acyl chains of the lipid bilayer (20). In this case, the hydrophilic maleic acid groups face the solvent. Our AFM images are consistent with that assumption (Fig. 2). Thus, the use of SMA is a universally applicable method that could extract active membrane proteins, including microbial rhodopsins, without the need for detergent.
Conclusions
The extraction of membrane proteins from lipids and their reconstitution into lipids are important steps for understanding the functions and structures of membrane proteins. In this study, we successfully used SMA to purify two seven-transmembrane photoactive proteins, RxR and HsSRI. As a result, those microbial rhodopsins were obtained as SMALPs without the need for detergent and without changing their physicochemical properties. Thus, SMA is a universally applicable method for solubilizing and characterizing membrane proteins.
Author Contributions
Y.S. designed the research. T.U. and M.S. performed the research. T.H. and S.N. contributed analytic tools. T.U., K.K., M.S., and Y.S. analyzed the data. T.U., K.K., T.H., M.S., and Y.S. wrote the manuscript.
Acknowledgments
We thank Ms. Kanae Kanehara, Mr. Shinichiro Yonekura, and Dr Atsushi Shibukawa for assistance with sample preparation and DLS experiments and invaluable discussion, respectively.
This work was financially supported by JSPS KAKENHI grant numbers JP19K16090 to K.K., JP18K06156 to T.H., JP18H01836, JP19H05257 to M.S., and JP18H02411, JP19H04727, and JP19H05396 to Y.S. This research was partially supported by CREST-JST (JPMJCR1656) and AMED (18dm0207060h0002) to Y.S.
Editor: Charles Deber.
Footnotes
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.09.026.
Supporting Material
References
- 1.Scaltriti M., Baselga J. The epidermal growth factor receptor pathway: a model for targeted therapy. Clin. Cancer Res. 2006;12:5268–5272. doi: 10.1158/1078-0432.CCR-05-1554. [DOI] [PubMed] [Google Scholar]
- 2.McCudden C.R., Hains M.D., Willard F.S. G-protein signaling: back to the future. Cell. Mol. Life Sci. 2005;62:551–577. doi: 10.1007/s00018-004-4462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ernst O.P., Lodowski D.T., Kandori H. Microbial and animal rhodopsins: structures, functions, and molecular mechanisms. Chem. Rev. 2014;114:126–163. doi: 10.1021/cr4003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lanyi J.K. Bacteriorhodopsin. Annu. Rev. Physiol. 2004;66:665–688. doi: 10.1146/annurev.physiol.66.032102.150049. [DOI] [PubMed] [Google Scholar]
- 5.Kurihara M., Sudo Y. Microbial rhodopsins: wide distribution, rich diversity and great potential. Biophys. Physicobiol. 2015;12:121–129. doi: 10.2142/biophysico.12.0_121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kojima K., Shibukawa A., Sudo Y. The unlimited potential of microbial rhodopsins as optical tools. Biochemistry. 2020;59:218–229. doi: 10.1021/acs.biochem.9b00768. [DOI] [PubMed] [Google Scholar]
- 7.Govorunova E.G., Sineshchekov O.A., Spudich J.L. Microbial rhodopsins: diversity, mechanisms, and optogenetic applications. Annu. Rev. Biochem. 2017;86:845–872. doi: 10.1146/annurev-biochem-101910-144233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang F., Vierock J., Deisseroth K. The microbial opsin family of optogenetic tools. Cell. 2011;147:1446–1457. doi: 10.1016/j.cell.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Michel H., Oesterhelt D. Three-dimensional crystals of membrane proteins: bacteriorhodopsin. Proc. Natl. Acad. Sci. USA. 1980;77:1283–1285. doi: 10.1073/pnas.77.3.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hohenfeld I.P., Wegener A.A., Engelhard M. Purification of histidine tagged bacteriorhodopsin, pharaonis halorhodopsin and pharaonis sensory rhodopsin II functionally expressed in Escherichia coli. FEBS Lett. 1999;442:198–202. doi: 10.1016/s0014-5793(98)01659-7. [DOI] [PubMed] [Google Scholar]
- 11.Kanehara K., Yoshizawa S., Sudo Y. A phylogenetically distinctive and extremely heat stable light-driven proton pump from the eubacterium Rubrobacter xylanophilus DSM 9941 T. Sci. Rep. 2017;7:44427. doi: 10.1038/srep44427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kojima K., Imamoto Y., Shichida Y. Rod visual pigment optimizes active state to achieve efficient G protein activation as compared with cone visual pigments. J. Biol. Chem. 2014;289:5061–5073. doi: 10.1074/jbc.M113.508507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miranda M.R., Choi A.R., Brown L.S. The photocycle and proton translocation pathway in a cyanobacterial ion-pumping rhodopsin. Biophys. J. 2009;96:1471–1481. doi: 10.1016/j.bpj.2008.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shionoya T., Mizuno M., Mizutani Y. High thermal stability of oligomeric assemblies of thermophilic rhodopsin in a lipid environment. J. Phys. Chem. B. 2018;122:6945–6953. doi: 10.1021/acs.jpcb.8b04894. [DOI] [PubMed] [Google Scholar]
- 15.Sudo Y., Okada A., Homma M. Characterization of a signaling complex composed of sensory rhodopsin I and its cognate transducer protein from the eubacterium Salinibacter ruber. Biochemistry. 2009;48:10136–10145. doi: 10.1021/bi901338d. [DOI] [PubMed] [Google Scholar]
- 16.Yagasaki J., Suzuki D., Sudo Y. Spectroscopic studies of a sensory rhodopsin I homologue from the archaeon Haloarcula vallismortis. Biochemistry. 2010;49:1183–1190. doi: 10.1021/bi901824a. [DOI] [PubMed] [Google Scholar]
- 17.Bayburt T.H., Grinkova Y.V., Sligar S.G. Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch. Biochem. Biophys. 2006;450:215–222. doi: 10.1016/j.abb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Wang J., Sasaki J., Spudich J.L. HAMP domain signal relay mechanism in a sensory rhodopsin-transducer complex. J. Biol. Chem. 2012;287:21316–21325. doi: 10.1074/jbc.M112.344622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dörr J.M., Scheidelaar S., Killian J.A. The styrene-maleic acid copolymer: a versatile tool in membrane research. Eur. Biophys. J. 2016;45:3–21. doi: 10.1007/s00249-015-1093-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knowles T.J., Finka R., Overduin M. Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J. Am. Chem. Soc. 2009;131:7484–7485. doi: 10.1021/ja810046q. [DOI] [PubMed] [Google Scholar]
- 21.Swainsbury D.J., Scheidelaar S., Jones M.R. Bacterial reaction centers purified with styrene maleic acid copolymer retain native membrane functional properties and display enhanced stability. Angew. Chem. Int.Engl. 2014;53:11803–11807. doi: 10.1002/anie.201406412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dörr J.M., Koorengevel M.C., Killian J.A. Detergent-free isolation, characterization, and functional reconstitution of a tetrameric K+ channel: the power of native nanodiscs. Proc. Natl. Acad. Sci. USA. 2014;111:18607–18612. doi: 10.1073/pnas.1416205112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gulati S., Jamshad M., Rothnie A.J. Detergent-free purification of ABC (ATP-binding-cassette) transporters. Biochem. J. 2014;461:269–278. doi: 10.1042/BJ20131477. [DOI] [PubMed] [Google Scholar]
- 24.Logez C., Damian M., Banères J.L. Detergent-free isolation of functional G protein-coupled receptors into nanometric lipid particles. Biochemistry. 2016;55:38–48. doi: 10.1021/acs.biochem.5b01040. [DOI] [PubMed] [Google Scholar]
- 25.Swainsbury D.J.K., Proctor M.S., Hunter C.N. Probing the local lipid environment of the Rhodobacter sphaeroides cytochrome bc 1 and Synechocystis sp. PCC 6803 cytochrome b 6 f complexes with styrene maleic acid. Biochim. Biophys. Acta Bioenerg. 2018;1859:215–225. doi: 10.1016/j.bbabio.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kojima K., Ueta T., Sudo Y. Vectorial proton transport mechanism of RxR, a phylogenetically distinct and thermally stable microbial rhodopsin. Sci. Rep. 2020;10:282. doi: 10.1038/s41598-019-57122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Losi A., Braslavsky S.E., Spudich J.L. Time-resolved absorption and photothermal measurements with sensory rhodopsin I from Halobacterium salinarum. Biophys. J. 1999;76:2183–2191. doi: 10.1016/S0006-3495(99)77373-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoff W.D., Jung K.H., Spudich J.L. Molecular mechanism of photosignaling by archaeal sensory rhodopsins. Annu. Rev. Biophys. Biomol. Struct. 1997;26:223–258. doi: 10.1146/annurev.biophys.26.1.223. [DOI] [PubMed] [Google Scholar]
- 29.Kitajima-Ihara T., Furutani Y., Sudo Y. Salinibacter sensory rhodopsin: sensory rhodopsin I-like protein from a eubacterium. J. Biol. Chem. 2008;283:23533–23541. doi: 10.1074/jbc.M802990200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gakhar S., Risbud S.H., Longo M.L. Structure retention of silica gel-encapsulated bacteriorhodopsin in purple membrane and in lipid nanodiscs. Colloids Surf. B Biointerfaces. 2020;186:110680. doi: 10.1016/j.colsurfb.2019.110680. [DOI] [PubMed] [Google Scholar]
- 31.Bagrov D.V., Voskoboynikova N., Shaitan K.V. Characterization of lipodisc nanoparticles containing sensory rhodopsin II and its cognate transducer from Natronomonas pharaonis. Biophys. 2016;61:942–949. [Google Scholar]
- 32.Mosslehy W., Voskoboynikova N., Steinhoff H.J. Conformational dynamics of sensory rhodopsin II in nanolipoprotein and styrene-maleic acid lipid particles. Photochem. Photobiol. 2019;95:1195–1204. doi: 10.1111/php.13096. [DOI] [PubMed] [Google Scholar]
- 33.Ganapathy S., Opdam L., de Grip W.J. Membrane matters: the impact of a nanodisc-bilayer or a detergent microenvironment on the properties of two eubacterial rhodopsins. Biochim. Biophys. Acta Biomembr. 2020;1862:183113. doi: 10.1016/j.bbamem.2019.183113. [DOI] [PubMed] [Google Scholar]
- 34.Lee S.C., Knowles T.J., Dafforn T.R. A method for detergent-free isolation of membrane proteins in their local lipid environment. Nat. Protoc. 2016;11:1149–1162. doi: 10.1038/nprot.2016.070. [DOI] [PubMed] [Google Scholar]
- 35.Orwick-Rydmark M., Lovett J.E., Watts A. Detergent-free incorporation of a seven-transmembrane receptor protein into nanosized bilayer Lipodisq particles for functional and biophysical studies. Nano Lett. 2012;12:4687–4692. doi: 10.1021/nl3020395. [DOI] [PubMed] [Google Scholar]
- 36.Broecker J., Eger B.T., Ernst O.P. Crystallogenesis of membrane proteins mediated by polymer-bounded lipid nanodiscs. Structure. 2017;25:384–392. doi: 10.1016/j.str.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 37.Shibata M., Yamashita H., Ando T. High-speed atomic force microscopy shows dynamic molecular processes in photoactivated bacteriorhodopsin. Nat. Nanotechnol. 2010;5:208–212. doi: 10.1038/nnano.2010.7. [DOI] [PubMed] [Google Scholar]
- 38.Shibata M., Inoue K., Uchihashi T. Oligomeric states of microbial rhodopsins determined by high-speed atomic force microscopy and circular dichroic spectroscopy. Sci. Rep. 2018;8:8262. doi: 10.1038/s41598-018-26606-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inoue S., Yoshizawa S., Sudo Y. Spectroscopic characteristics of Rubricoccus marinus xenorhodopsin (RmXeR) and a putative model for its inward H + transport mechanism. Phys. Chem. Chem. Phys. 2018;20:3172–3183. doi: 10.1039/c7cp05033j. [DOI] [PubMed] [Google Scholar]
- 40.Chan S.K., Kawaguchi H., Kouyama T. Crystal structure of the 11-cis isomer of pharaonis halorhodopsin: structural constraints on interconversions among different isomeric states. Biochemistry. 2016;55:4092–4104. doi: 10.1021/acs.biochem.6b00277. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki D., Furutani Y., Sudo Y. Effects of chloride ion binding on the photochemical properties of salinibacter sensory rhodopsin I. J. Mol. Biol. 2009;392:48–62. doi: 10.1016/j.jmb.2009.06.050. [DOI] [PubMed] [Google Scholar]
- 42.Hayashi T., Yasuda S., Kinoshita M. How does a microbial rhodopsin RxR realize its exceptionally high thermostability with the proton-pumping function being retained? J. Phys. Chem. B. 2020;124:990–1000. doi: 10.1021/acs.jpcb.9b10700. [DOI] [PubMed] [Google Scholar]
- 43.Katayama K., Sekharan S., Sudo Y. Color tuning in retinylidene proteins. In: Yawo H., Kandori H., Koizumi A., editors. Optogenetics: Light-Sensing Proteins and Their Applications. Springer Japan; 2015. pp. 89–107. [Google Scholar]
- 44.Sudo Y., Yamabi M., Kamo N. Importance of specific hydrogen bonds of archaeal rhodopsins for the binding to the transducer protein. J. Mol. Biol. 2006;357:1274–1282. doi: 10.1016/j.jmb.2006.01.061. [DOI] [PubMed] [Google Scholar]
- 45.Jamshad M., Grimard V., Dafforn T.R. Structural analysis of a nanoparticle containing a lipid bilayer used for detergent-free extraction of membrane proteins. Nano Res. 2015;8:774–789. doi: 10.1007/s12274-014-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Esmaili M., Overduin M. Membrane biology visualized in nanometer-sized discs formed by styrene maleic acid polymers. Biochim. Biophys. Acta Biomembr. 2018;1860:257–263. doi: 10.1016/j.bbamem.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rigaud J.L., Lévy D. Reconstitution of membrane proteins into liposomes. Methods Enzymol. 2003;372:65–86. doi: 10.1016/S0076-6879(03)72004-7. [DOI] [PubMed] [Google Scholar]
- 48.Oiki S. Channel function reconstitution and re-animation: a single-channel strategy in the postcrystal age. J. Physiol. 2015;593:2553–2573. doi: 10.1113/JP270025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sachse R., Dondapati S.K., Kubick S. Membrane protein synthesis in cell-free systems: from bio-mimetic systems to bio-membranes. FEBS Lett. 2014;588:2774–2781. doi: 10.1016/j.febslet.2014.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HS-AFM video of RxR-SMA on the mica surface with 3.3 fps.