Dear Editor,
Acinetobacter baumannii is a prevalent nosocomial pathogen that causes serious threat in health care institutions1. A. baumannii has demonstrated resistance to a wide array of antibiotics, including the last-resort colistin or polymyxin B2–4. The MlaFEDB-mediated glycerophospholipid (PL) transport was reported to play an important role in maintaining the integrity of the lipid membrane2,3. The MlaFEDB complex is an ATP-binding cassette transporter to actively translocate the phospholipids between the inner membrane and the periplasmic protein MlaC4,5. Despite the progress that has been made in understanding the function of the MlaFEDB complex by genetic and biochemical strategies in A. baumannii, questions still remain open surrounding the directionality in glycerophospholipids transport via MlaFEDB in both retrograde and anterograde transport6,7. The low-resolution structure of the MlaFEDB complex from A. baumannii at 8.7 Å has provided initial structural insights into this complex8. However, the molecular details of the transport complex assembly, the interactions with glycerophospholipids, and the transport mechanisms of the MlaFEDB in A. baumannii remain enigmatic.
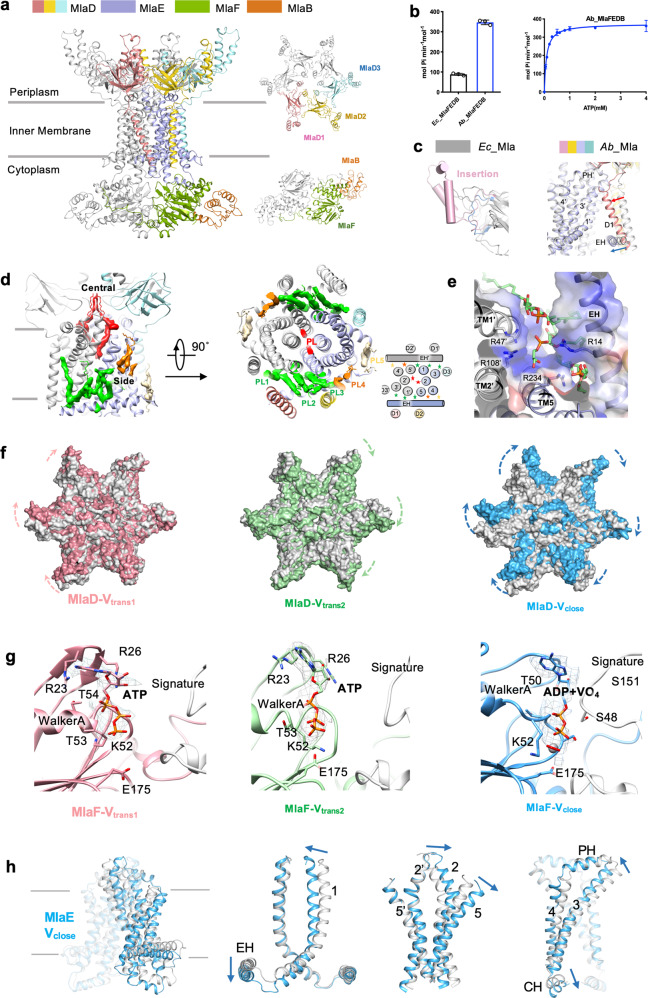
To chase the high-resolution structure of MlaFEDB, we overexpressed the A. baumannii mlaFEDCB operon in Escherichia coli BL21(DE3) pLyS. The MlaFEDB proteins formed a stable complex and were reconstituted into lipid nanodisc (Fig. 1a and Supplementary Fig. S1a–c). The ATPase activity of A. baumannii MlaFEDB is approximately three times that of the E. coli MlaFEDB observed in our previous study9 (Fig. 1b, left). The ATPase activity exhibits the Km around 0.085 ± 0.005 mM and the Vmax around 373.7 ± 4.08 mol ATP per min per mol MlaFEDB (Fig. 1b, right), suggesting a high ATP-binding affinity during the transportation cycle.
Fig. 1. Cryo-EM structures of glycerophospholipid transporter complex MlaFEDB from A. baumannii.
a Cryo-EM structure of the MlaFEDB complex. MlaD, MlaE, MlaF and MlaB are colored separately. b The ATPase activities of MlaFEDB from A. baumannii and E. coli. Each point represents mean ± SD. c Overlay of the MCE domain of MlaD (left) and TMs of MlaED between A. baumannii (color) and E. coli (gray, PDB 7CGE). d Cross-sectional view (left) and periplasmic view (middle) of the model with phospholipid binding. Twelve glycerophospholipids (PLs) are modeled and are shown as stars in the cartoon view (right). Two PLs bound in the central cavity are colored red. PL1–3, PL4, and PL5 bound in one side cavity are shown and colored green, orange, and yellow, respectively. e Electrostatic surface view of the interaction between PL1–3 and MlaE, indicating areas of positive charge (blue). f Surface overlay of the MCE domain of MlaD between the nucleotide-free (gray) and Vtrans1, Vtrans2, and Vclose conformations. The arrows indicate the movement of MCE. g Cryo-EM densities for the nucleotides bound at the ATP-binding sites of two subunits of MlaF in Vtrans1 (raspberry), Vtrans2 (green), and Vclose (blue). ATP or ADP-VO4 binds to the Walker A and Signature motifs. R23, R26, K52, T53, T54, and E175 participate in the binding. h Structure overlay of MlaE in the nucleotide-free (gray) and Vclose (blue) conformations. Blue arrow indicates the movement of TMD of MlaE.
The structure of MlaFEDB complex in nanodisc in nucleotide-free form was determined by cryogenic electron microscopy (cryo-EM). The two-dimensional class averages of MlaFEDB showed clear structural features (Supplementary Fig. S2a). In our previous studies, E. coli MlaFEDB exhibited twofold symmetry, with bound glycerophospholipid densities along the axis of symmetry9 (PDB 7CGE). To improve and confirm densities along the central axis, no symmetry was applied during the reconstruction of A. baumannii MlaFEDB (Fig. 1a and Supplementary Fig. S2b). The final map of the MlaFEDB complex was well defined at 3.1 Å resolution, allowing us to build a nearly complete model for MlaFEDB, which includes a high-resolution structure of transmembrane helices (TMs) of MlaE and MlaD (Supplementary Fig. S3 and Table S1). Consistent with the previous reported low-resolution A. baumannii MlaFEDB map8 and our E. coli MlaFEDB map9 (EMD-30355), the MlaF (cytoplasmic nucleotide-binding domain, NBD), MlaE (transmembrane domain), MlaD (periplasmic phospholipid-binding domain), and MlaB (cytoplasmic auxiliary domain) subunits are assembled in a stoichiometry of 2:2:6:2 (Fig. 1a).
On the periplasmic side, the mammalian cell entry (MCE) domain of MlaD is assembled as a hexameric ring (Fig. 1a). The MlaD of A. baumannii shares 27.2% identity with the MlaD of E. coli. The MCE domain is composed of a seven-stranded β-barrel fold. Extra density corresponds to a 47 amino acid α-helices insert between the fourth and fifth strand β-sheet (Fig. 1c left and Supplementary Fig. S4). The function of the insertion is unclear. On the cytoplasmic side, two subunits of MlaF interact with two subunits of MlaB (Supplementary Fig. S5a) and the C terminus of one MlaF subunit extends to wrap around the opposite subunit of MlaF and the neighboring MlaB, forming close contacts (Fig. 1a and Supplementary Fig. S5b).
In the transmembrane region, the MlaFEDB complex consists of a total of 18 long helices in the inner membrane, formed by 2 subunits of MlaE and 6 subunits of MlaD. Three protomers of the MlaD hexamer (MlaD1, MlaD2, and MlaD3) incorporate with one subunit of MlaE (Fig. 1a and Supplementary Fig. S6a). Two subunits of MlaE form a homodimer and each subunit contains one elbow helix (EH), five TMs, and one coupling helix and one periplasmic helix (Supplementary Fig. S6a, right). The EH is amphipathic and runs parallel to the inner membrane representing the novel feature of MlaFEDB complex. The TM of MlaD1 (D1) and MlaD2 (D2) interact tightly with EH of MlaE (Supplementary Fig. S6b, c). The TM of MlaD3 (D3) interacts with TM1 and TM3 (Supplementary Fig. S6a). The assembly of TM1–5 in MlaE are essentially identical to that in E. coli MlaE9 (PDB 7CGE). Notably, the EH moves inward to the TMs of MlaE coordinating inward movement of D1 and D2, and forming a smaller side cavity in comparison to the cavity formed by MlaE and MlaD in E. coli9 (Fig. 1c right and Supplementary Fig. S6d).
Cryo-EM structure of MlaFEDB in nucleotide-free state also reveals glycerophospholipids bound inside the MlaE and MlaD, suggesting that the overexpressed A. baumannii MlaFEDB can bind glycerophospholipids from E. coli. Clusters of diacyl glycerophospholipid densities were resolved to be located in the central cavity and side cavities symmetrically (Fig. 1d). Two glycerophospholipids (PL center) are bound in the central cavity (Fig. 1d). The well-defined densities are resolved with one acyl chain reaching down into the MlaE hydrophobic cavity and the other reaching upwards into the cavity formed by MCE of MlaD (Supplementary Fig. S7a, d), which is consistent with the lipids found in E. coli (PDB 7CGE). Glycerophospholipid densities are also found on the two side cavities formed by MlaE and MlaD (Fig. 1d). Five diacyl glycerophospholipids (PL1–5) are built in one side cavity (Fig. 1d, right), which is formed by EH, TM3’ TM1’, TM5, and TM4 from MlaE and D3’, D1, and D2 from MlaD. The head groups of PL1–3 are pointing down and are accommodated by a cluster of positively charged residues from MlaD, which include Arg47, Arg108, Arg14, and Arg234 (Fig. 1e and Supplementary Fig. S7d, e). PL4 is trapped in a position close to the outer leaflet of the inner membrane and PL5 locates close to the end of EH (Fig. 1d and Supplementary Fig. S7c). Extra trace of glycerophospholipid’s tail densities exist in the area close to the PL5. The bound glycerophospholipids in the side cavity may play a role in the regulation of glycerophospholipid translocation or structural integrity of the complex.
To understand the mechanism of glycerophospholipid translocation coupled with the conformational change, distinct conformations of MlaFEDB were trapped using sodium ortho-vanadate10. Vanadate acts as a potent inhibitor of ATPase by forming an ADP–vanadate complex that mimics the intermediate state formed during hydrolysis. In the presence of 1 mM vanadate, the ATPase activity of MlaFEDB is reduced by ~50% (Supplementary Fig. S1d), indicating an incomplete inhibition state. Three individual conformations were classified and refined to 4.0, 3.6, and 4.0 Å, and were named Vtrans1, Vtrans2, and Vclose, respectively (Supplementary Figs. S8,S9 and Table S1). All three structures display bound nucleotides at the ATP-binding sites (Fig. 1g). However, the maps are of insufficient quality to distinguish between ADP, ATP, and ADP–vanadate.
Comparison of Vtrans1 and Vtrans2 to nucleotide-free structure shows the conformational rearrangement of the MlaD. MlaD hexametric ring rotates clockwise at different levels (Fig. 1f and Supplementary Movies S1 and S2), whereas MlaFEB remains unchanged. The structure of Vclose shows the tight dimerization and close proximity of the NBD of MlaF subunits, caused by the trapping of ADP–vanadate or ATP at the ATP-binding sites comprising the Walker A, Walker B motifs, and the signature motif (Fig. 1g). The dimerization of the NBDs engages the coupled structural rearrangements of MlaE and MlaD, respectively. The MlaD ring rotates clockwise and the two subunits of MlaE squeeze towards each other to collapse the central phospholipid-binding cavity to extrude lipids (Fig. 1f, h and Supplementary Movie S3). The conformational changes from nucleotide-free to Vclose in A. baumannii are consistent with the changes captured from nucleotide-free (PDB 7CGE) to EQclose (PDB 7CH0) in E. coli9. No bound glycerophospholipid densities were observed in the central cavity in Vclose.
In summary, we characterize four structures of the A. baumannii MlaFEDB complex in nucleotide-free and vanadate-trapped conformations using single-particle cryo-EM. The 3.1 Å resolution structure of nucleotide-free MlaFEDB reveals molecular details of the whole complex assembly and the glycerophospholipid binding, suggesting the unique and conserved features as compared to the E. coli MlaFEDB. The three vanadate-trapped structures of MlaFEDB reveal different levels of conformational rearrangement of MlaD and MlaE coupled with nucleotide binding and glycerophospholipid translocation. Our findings uncover the binding sites and potential pathway of glycerophospholipid translocation and provide structural insights into the glycerophospholipid transport mechanism mediated by MlaFEDB in A. baumannii.
Supplementary information
Acknowledgements
We are grateful to the technical assistance from Westlake cryo-EM facility. This work was supported by the Zhejiang Provincial Natural Science Foundation of China (LR20C050001) and foundation of Westlake University.
Author contributions
Y.L. and Q.Z. conceived and designed the project. Q.F. performed cloning, expression, purification, and collected the cryo-EM data. Y.Z. processed cryo-EM data and built the atomic models. Y.Z., X.C., and Y.L. analyzed the data and made the figures. Y.L. wrote the manuscript.
Data availability
Four three-dimensional cryo-EM density maps of A. baumannii MlaFEDB in nanodiscs have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-30525 (nucleotide-free state of MlaFEDB), EMD-30526 (Vtrans1 state of MlaFEDB), EMD-30527 (Vtrans2 state of MlaFEDB), and EMD-30528 (Vclose state of MlaFEDB). Four atomic coordinates for the atomic models have been deposited in the Protein Data Bank under accession numbers 7D06 (nucleotide-free state of MlaFEDB), 7D08 (Vtrans1 state of MlaFEDB), 7D09 (Vtrans2 state of MlaFEDB), and 7D0A (Vclose state of MlaFEDB).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Qiang Zhou, Email: zhouqiang@westlake.edu.cn.
Yanyan Li, Email: liyanyan@westlake.edu.cn.
Supplementary information
Supplementary Information accompanies the paper at (10.1038/s41421-020-00230-5).
References
- 1.World Health Organization. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Geneva, Switzerland: World Health Organization; 2017. [Google Scholar]
- 2.Henderson JC, et al. The power of asymmetry: architecture and assembly of the Gram-negative outer membrane lipid bilayer. Annu. Rev. Microbiol. 2016;70:255–278. doi: 10.1146/annurev-micro-102215-095308. [DOI] [PubMed] [Google Scholar]
- 3.Leung LM, et al. A prospective study of acinetobacter baumannii complex isolates and colistin susceptibility monitoring by mass spectrometry of microbial membrane glycolipids. J. Clin. Microbiol. 2019;57:1–9. doi: 10.1128/JCM.01100-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thong S, et al. Defining key roles for auxiliary proteins in an ABC transporter that maintains bacterial outer membrane lipid asymmetry. Elife. 2016;5:e19042. doi: 10.7554/eLife.19042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powers MJ, Trent MS. Expanding the paradigm for the outer membrane: Acinetobacter baumannii in the absence of endotoxin. Mol. Microbiol. 2018;107:47–56. doi: 10.1111/mmi.13872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Powers MJ, Trent MS. Phospholipid retention in the absence of asymmetry strengthens the outer membrane permeability barrier to last-resort antibiotics. Proc. Natl. Acad. Sci. USA. 2018;115:E8518–E8527. doi: 10.1073/pnas.1806714115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Powers MJ, Trent MS. Intermembrane transport: glycerophospholipid homeostasis of the Gram-negative cell envelope. Proc. Natl. Acad. Sci. USA. 2019;116:17147–17155. doi: 10.1073/pnas.1902026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamischke C, et al. The Acinetobacter baumannii Mla system and glycerophospholipid transport to the outer membrane. Elife. 2019;8:e40171. doi: 10.7554/eLife.40171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi X, et al. Structural mechanism of phospholipids translocation by MlaFEDB complex. Cell Res. 2020 doi: 10.1038/s41422-020-00404-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, et al. Structural basis of lipopolysaccharide extraction by the LptB2FGC complex. Nature. 2019;567:486–490. doi: 10.1038/s41586-019-1025-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Four three-dimensional cryo-EM density maps of A. baumannii MlaFEDB in nanodiscs have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-30525 (nucleotide-free state of MlaFEDB), EMD-30526 (Vtrans1 state of MlaFEDB), EMD-30527 (Vtrans2 state of MlaFEDB), and EMD-30528 (Vclose state of MlaFEDB). Four atomic coordinates for the atomic models have been deposited in the Protein Data Bank under accession numbers 7D06 (nucleotide-free state of MlaFEDB), 7D08 (Vtrans1 state of MlaFEDB), 7D09 (Vtrans2 state of MlaFEDB), and 7D0A (Vclose state of MlaFEDB).