Abstract
Several genes of the eut, pdu, and cob/cbi operons are responsible for the metabolism of ethanolamine (EA) and 1,2-propanediol (PD) and are essential during the pathogenic lifecycles of various enteric pathogens. Studies concerning EA and PD metabolism have primarily focused on bacterial genera from the family Enterobacteriaceae, especially the genus Salmonella. Listeria monocytogenes is a member of the Firmicutes phylum and is the causative agent of the rare but highly fatal foodborne disease listeriosis. The eut, pdu, and cob/cbi operons are organized as a single large locus collectively referred to as the cobalamin-dependent gene cluster (CDGC). The CDGC is well conserved in L. monocytogenes; however, functional characterization of the genes in this cluster and how they may contribute to Listeria virulence and stress tolerance in food production environments is highly limited. Previous work suggests that the degradation pathway of PD is essential for L. monocytogenes establishment in the gastrointestinal tract. In contrast, EA metabolism may be more important during intracellular replication. Other studies indicate that the CDGC is utilized when L. monocytogenes is exposed to food and food production relevant stress conditions. Perhaps most noteworthy, L. monocytogenes exhibits attenuated growth at cold temperatures when a key EA utilization pathway gene was deleted. This review aims to summarize the current knowledge of these pathways in L. monocytogenes and their significance in virulence and stress tolerance, especially considering recent developments.
Keywords: Listeria monocytogenes, food safety, virulence, stress survival, cobalamin, ethanolamine, 1, 2-propanediol, Rli47
Introduction
Foodborne pathogens must adapt to various stress conditions to survive in food production and host environments. The capacity to metabolize alternative substrates for energy is imperative for the survival of pathogens when more efficient nutrients are unavailable. Such environments include – among others – the nutrient-limited conditions within the phagosomes of macrophages. The metabolism of 1,2-propanediol (PD) and ethanolamine (EA) critically impacts the virulence of various enteric pathogens, including Salmonella enterica, Enterococcus faecalis, Escherichia coli, and Listeria monocytogenes. Enzymes required for the metabolism of PD and EA are dependent on cobalamin (vitamin B12) derivatives as cofactors. Cobalamin is synthesized de novo via the cob and cbi genes and recycled for use in these enzymatic reactions. Studies concerning the functional characterization of EA and PD metabolic genes and how they contribute to virulence have been, for the most part, conducted in Salmonella, E. coli, and Enterococcus. However, the ability to utilize EA and PD has been demonstrated in other distantly related genera, including Mycobacterium, Corynebacterium, Lactococcus, and Listeria as well as non-pathogenic bacteria (Nandedkar, 1974; Blackwell et al., 1976; Hartmans and Bont, 1986; Rodionov et al., 2003; Xue et al., 2008; Tsoy et al., 2009; Kutzner et al., 2016; Zeng et al., 2019).
Listeria monocytogenes is the causative agent of the foodborne illness listeriosis, a rare but severe disease with a high mortality rate in the immunocompromised, very young, and elderly populations. Furthermore, listeriosis also causes abortions in pregnant women (Allerberger and Wagner, 2010; Scallan et al., 2011; Radoshevich and Cossart, 2018). L. monocytogenes is also of considerable concern because of its long-term survival (also called persistence) in food production environments (Keto-Timonen et al., 2007; Carpentier and Cerf, 2011; Ferreira et al., 2014). L. monocytogenes may persist in these environments for up to decades due to increased tolerance to stresses associated with such environments (Bucur et al., 2018). Some L. monocytogenes strains exhibit increased tolerance toward quaternary ammonia disinfectants (Ferreira et al., 2014; Muller et al., 2014), acidic, oxidative, high or low temperatures, and high salt stress conditions (Bucur et al., 2018). Much is known concerning EA and PD metabolism genes and their contribution to L. monocytogenes virulence and stress conditions; however, a review that summarizes this information and highlights their comprehensive significance is absent in the scientific literature. Therefore, this review aims to summarize how EA and PD metabolism influences stress survival and virulence of L. monocytogenes.
1,2-Propanediol and Ethanolamine Metabolism
Bacterial microcompartments (BMCs) are organelle-like structures surrounded by semi-permeable protein shells encapsulating metabolic enzymes that often yield volatile toxic intermediates, thus protecting the cell from damage by mediating reactions within the BMC (Chowdhury et al., 2014; Kerfeld et al., 2018). The first BMCs, known as carboxysomes, were discovered by investigations of carbon fixation and contained high levels of ribulose-1,5-bisphosphate carboxylase oxygenase (Shively et al., 1973). Subsequent studies functionally characterized BMCs to mediate PD (Bobik et al., 1999; Sampson and Bobik, 2008) and EA (Garsin, 2010; Chowdhury et al., 2014) metabolism. Recently, Zeng et al. (2019) showed, for the first time, that BMCs form in L. monocytogenes, and their findings further indicated that BMCs mediate the metabolism of PD and that the addition of PD to minimal media stimulated growth in anaerobic conditions. The ability to metabolize EA in L. monocytogenes has been established (Kutzner et al., 2016); however, evidence of the formation of EA associated BMCs has yet to be elucidated.
PD metabolism is well documented in Listeria. PD, also known as propylene glycol, is a United States Food and Drug Administration approved additive to food and food containers. PD is biogenically produced from the catabolism of rhamnose or fucose and is often a fermentative end product of commensal bacteria. These sugars are highly abundant in plants, bacterial capsules, and eukaryotic glycoconjugates (Chowdhury et al., 2014). In L. monocytogenes, PD is metabolized by the enzymes of the pdu gene cluster. First, rhamnose or fucose is converted into L-lactaldehyde by aldolases that correspond to the sugar’s stereochemistry. Secondly, L-lactaldehyde is then oxidized by a propanediol oxidoreductase to form PD (Baldoma and Aguilar, 1988). The metabolism of PD produces the toxic intermediate propionaldehyde within the PD BMC, thus protecting L. monocytogenes from further damage. Propionaldehyde is then converted by a series of pdu enzymes to form propionate, propanol, or succinate (Baldoma and Aguilar, 1988; Garsin, 2010; Zeng et al., 2019).
Recent studies have revealed that L. monocytogenes can utilize EA as a carbon and nitrogen source. Phosphatidylethanolamine is the most abundant phospholipid in membranes of mammalian enterocytes (Kawai et al., 1974). Phosphatidylethanolamine is oxidized to glycerol and EA by non-specific phospholipases and then can freely diffuse across bacterial membranes. However, in acidic conditions, the EA transporter EutH is required for the translocation of protonated EA across cellular membranes (Anderson et al., 2018; Kaval and Garsin, 2018). Listeria may use EA as a carbon and nitrogen source by the further breakdown of EA to acetaldehyde and ammonia mediated by an EA ammonia-lyase known as the EutB/EutC complex (Kutzner et al., 2016; Kaval and Garsin, 2018). The EA utilization pathway genes (eut) can further metabolize acetaldehyde to ethanol or acetyl-CoA. Acetyl-CoA is subsequently oxidized to pyruvate or acetate to accommodate the current metabolic needs of the cell (Joseph et al., 2006; Garsin, 2010; Kaval and Garsin, 2018). For the remainder of this review, the pdu, eut, and cbi/cob genes will be collectively referred to as the CDGC.
Conservation
The CDGC is highly conserved within a single large locus (Supplementary Table 1) among Listeria sensu strictu species representing Listeria species capable of growth in the gastrointestinal tracts (GI) of animals (except for the loss of one gene, pduJ, in L. marthii). However, homologs are absent in Listeria sensu lato species, which are exclusively environmental isolates (Buchrieser et al., 2003; Chiara et al., 2015; Schardt et al., 2017; Zeng et al., 2019), further supporting the hypothesis that PD and EA metabolism is advantageous for Listeria species that colonize the mammalian GI tract. It is likely that an ancestor of Listeria sensu strictu acquired the CDGC through horizontal gene transfer from Bacillus cereus (Chiara et al., 2015).
Regulation
The regulation of PD and EA utilization has been well-characterized in Listeria sensu strictu species (Joseph et al., 2006; Xue et al., 2008; Mellin et al., 2013, 2014; Anderson et al., 2018; Costa and Escalante-Semerena, 2018; Zeng et al., 2019). In L. monocytogenes, both the pdu and eut operons require cobalamin as a cofactor for essential catabolic reactions, and PD or EA must be present for optimal expression of each respective metabolic pathway. The availability of cobalamin tightly regulates both pathways by binding to two Cis-acting riboswitches, Rli39 and Rli55 (Mellin et al., 2013, 2014).
The regulation of the eut operon in L. monocytogenes diverges from the well-studied pathways found in the family Enterobacteriaceae in that it lacks the classic EutR regulator and instead encodes a two-component response regulator (EutV/EutW) that is tightly regulated by a cobalamin-dependent riboswitch (Rli55) (Mellin et al., 2014). Many Firmicutes, including L. monocytogenes, have long eut operons, unlike the shorter operons found in many Proteobacteria, and Tsoy et al. (2009) also found that Enterobacteriaceae obtained the eut operon from Firmicutes and later acquired the EutR regulator by horizontal gene transfer.
The pdu regulon of L. monocytogenes shares many common traits with that of the well-characterized pdu regulon of Salmonella. PocR is the central regulator of the pdu genes in both organisms and is activated by PD in the absence of more efficient carbon sources. However, the pdu operon of Salmonella is controlled differently on a global scale through the mediation of a cyclic AMP receptor protein-cyclic AMP complex and the ArcA/ArcB system (Chowdhury et al., 2014). In L. monocytogenes, pocR gene expression is finely tuned by a cobalamin-dependent riboswitch (Rli39) that, in the absence of cobalamin, will generate a longer transcript that is antisense of pocR (aspocR), thereby hindering the translation of pocR mRNA. After translation, PocR is activated in the presence of PD. PocR acts as a positive regulator of the pocR gene (ensuring its production while PD is present) and will, in turn, activate the pdu and possibly the cob and cbi genes, thereby helping maintain production of cobalamin derived cofactors and the utilization of PD (Mellin et al., 2013). Several L. monocytogenes pdu and cob genes are regulated on a global scale directly by the stress regulator σB (Liu et al., 2017), and are directly or indirectly influenced by the non-coding RNA, Rli47 (Marinho et al., 2019). Rli47 has recently emerged as an important regulatory element in virulence and especially during stress response (Toledo-Arana et al., 2009; Archambaud et al., 2012; Mujahid et al., 2012; Marinho et al., 2019; Anast and Schmitz-Esser, 2020; Cortes et al., 2020).
Virulence
The importance of the CDGC in the pathogenicity of many enteric pathogens such as Salmonella, Escherichia, Enterococcus, and Clostridium, is well established (Garsin, 2010; Thiennimitr et al., 2011; Nuccio and Baumler, 2014; Faber et al., 2017). In Salmonella and Escherichia, the eut gene regulator EutR is essential for the development of disease because, in addition to regulating the eut genes, EutR also influences key virulence operons. Known EutR-dependent virulence factors are Salmonella genes of the pathogenicity island 2 (SPI-2), including the SPI-2 positive gene regulator, ssrB (Anderson et al., 2015); as well as E. coli virulence genes ler, qseC, stx2a, and an important type III secretion system (LEE effacement locus) (Kendall et al., 2012; Luzader et al., 2013; Gonyar and Kendall, 2014). During intestinal inflammation caused by Salmonella infection, it was found that microbiota-derived EA and PD were utilized as electron donors in respiration with tetrathionate as the required final electron acceptor (Thiennimitr et al., 2011; Faber et al., 2017). Interestingly, respiration with electron donors EA and PD promoted Salmonella expansion in the presence of commensal bacteria, but not during infection of germ-free mice. These data suggest that the utilization of EA and PD enhances Salmonella virulence in the GI tract by shifting metabolism towards respiration on commensal microbiota fermentative byproducts and electron acceptors made available through intestinal inflammation. Also, Salmonella (Nuccio and Baumler, 2014; Matthews et al., 2015) and commensal E. coli (Dogan et al., 2014) isolates not associated with GI disease often have absent or disrupted PD utilization operons, and it was found that bacteria that are associated with food poisoning more often possessed the long-form eut operon containing BMC shell component genes (like that of L. monocytogenes) (Tsoy et al., 2009).
The CDGC has recently been highlighted to be important for L. monocytogenes pathogenicity. A study that used a tiling array approach showed that the pdu and eut genes were significantly upregulated by L. monocytogenes in the GI tract and blood of mice (Toledo-Arana et al., 2009). Interestingly, a deletion mutant of the L. monocytogenes pduD gene showed attenuated virulence and faster clearing of L. monocytogenes in the GI tract, highlighting the importance of PD utilization for the fitness of L. monocytogenes during infection in the GI tract (Schardt et al., 2017). Archambaud et al. (2012) found that the expression of the CDGC was induced in the GI tract of gnotobiotic mice during orally acquired listeriosis compared to growth in broth. Remarkably, they found a much higher expression of these genes in the transcriptomic response of L. monocytogenes when Lactobacillus was present in the GI tract (Archambaud et al., 2012).
These data suggest that the utilization of the CDGC in the GI tract may increase competitive fitness over commensal bacteria (that can only metabolize fermentable carbon sources) by enabling L. monocytogenes to grow on non-fermentable carbon and nitrogen sources. Enhancing L. monocytogenes competitive fitness in the GI tract may increase the likelihood of successful invasion into the intestinal epithelium.
Once L. monocytogenes establishes in the GI tract, it internalizes into the epithelial cells (EC) of the host by hijacking host cellular machinery, and it then proceeds to replicate intracellularly (Radoshevich and Cossart, 2018). Additionally, L. monocytogenes can translocate the epithelial lining via the paracellular route after the partitioning of host cell tight junctions, mediated by the Listeria adhesion protein LAP (Drolia and Bhunia, 2019). Both pdu and eut genes were significantly induced during replication in caco-2 cells (a human EC line); however, only an insertion knockout mutant of eutB, which mediates the first step in EA metabolism, exhibited a decrease in intracellular replication. ΔeutB displayed significant attenuation of intracellular growth, second only to mutants of inlA, the gene encoding the protein responsible for internalization into ECs (Joseph et al., 2006). It has been proposed that non-specific phospholipases, such as PlcA/PlcB, cleave phosphatidylethanolamine from host cell membranes for utilization by intracellularly replicating L. monocytogenes (Joseph et al., 2006; Mellin et al., 2014; Staib and Fuchs, 2014).
The next crucial step in the systemic spread of L. monocytogenes is disseminating to peripheral organs via uptake and replication in monocytes, such as macrophages (Radoshevich and Cossart, 2018). Mellin et al. (2014) found that an eutB deletion mutant was highly attenuated in an intravenous mouse infection model and exhibited significantly lower numbers of L. monocytogenes in spleen and liver cells. Macrophages are phagocytic cells and will autonomously engulf invading bacteria within vacuoles known as phagosomes. Bacteria-containing phagosomes will then combine with lysosomes containing toxic substances such as reactive oxygen species. The fusion will form a phagolysosome that will destroy the confined bacteria. For L. monocytogenes to survive transient or extended periods within these vacuoles, it must employ a repertoire of virulence factors. The environment within these macrophagic vacuoles is acidic, and it is known that L. monocytogenes excretes Listeriolysin O (LLO) and the phospholipases PlcA/PlcB to promote neutralization and escape from host cell vacuoles (Radoshevich and Cossart, 2018). EA can freely permeate bacterial cell membranes at neutral pH. However, EA is protonated in acid conditions and requires the EA permease, EutH, to be internalized by L. monocytogenes. Anderson et al. (2018) elucidated that EutH is important for the intracellular survival and replication of S. Typhimurium and L. monocytogenes. EutH did not promote intestinal infection but instead provides a crucial advantage in the dissemination of L. monocytogenes to the liver and spleen. At 2 h post-infection of macrophages by L. monocytogenes, a deletion mutant of eutH (ΔeutH) exhibited a similar reduction in cell numbers as a mutant of the gene encoding LLO when compared with the wildtype control. The ΔeutH detrimental phenotype was not observed at 5 or 8 h post internalization; however, the LLO mutant consistently showed a high reduction of intracellular derived CFUs. This may be explained by the fact that LLO has essential functions in many aspects of intramacrophage survival (Osborne and Brumell, 2017) and that once L. monocytogenes is free of the acidic conditions of the phagosome, EutH does not have an essential role in survival (Osborne and Brumell, 2017; Anderson et al., 2018). L. monocytogenes has been shown to escape from phagosomes as early as 30 min post macrophage internalization (Tilney and Portnoy, 1989; Myers et al., 2003). However, the majority of L. monocytogenes cells remains confined to the phagosome 2–3 h post-infection (de Chastellier and Berche, 1994; Singh et al., 2008). Anderson et al. (2018) also proposed that there may be a functional link between LLO and EutH; however, a hypothesis that suggests a function was not proposed in the study. These findings highlight the importance of EutH for the survival and replication of L. monocytogenes in macrophages in response to vacuole acidification.
Interestingly, the first step in EA metabolism is the oxidation of EA to acetaldehyde and ammonia (NH3) by the EutB/EutC complex. In Salmonella, it is suggested that the EutB/EutC complex is associated with the outer surface of the BMC and injects the EutB/EutC-derived acetaldehyde into the lumen of the BMC shell (Chowdhury et al., 2014). Ammonia likely remains in the cytosol, where it can be incorporated into nitrogen metabolism. Phagolysosomes of host cells can indeed mediate nutrient capture and sequester metals such as Fe2+ and Zn2+ (Uribe-Querol and Rosales, 2017), and EA may serve as the sole nitrogen and carbon source within these vacuoles. L. monocytogenes and other bacteria, such as Salmonella, may synthesize and recycle cobalamin via ATP:Co(I)rrinoid adenosyl transferases denoted for their respective pathway: CobA (for the de novo synthesis of cobalamin), EutT, or PduO (Chowdhury et al., 2014; Costa and Escalante-Semerena, 2018; Zeng et al., 2019). Until recently, ATP:Co(I)rrinoid adenosyl transferases were described as requiring metal ions such as Fe(II) or Zn(II) for optimal activity; however, Costa and Escalante-Semerena (2018) have described a new class of EutT in L. monocytogenes which does not require metal ions for activity.
Therefore, it seems likely that EA metabolism may benefit L. monocytogenes during nutrient-limited intra-vacuolar replication. Interestingly, the impairment of protonated-EA import reduced intramacrophage cell survivability after 2 h, similar to an LLO mutant (Anderson et al., 2018). If EA metabolism only increased fitness through nutrient acquisition, it would be suspected that intracellular replication may be hindered. However, ΔeutH CFUs (determined after intramacrophage growth) were reduced by 42% when comparing 0–2 h post-infection (Anderson et al., 2018), suggesting that cell death occurred. Is it possible that the increased production of ammonia during the first step of EA metabolism may neutralize free protons that can freely permeate the cell membrane? Is it also possible that the secretion of ammonia provides localized protection against the acidic conditions of the phagosome? LLO forms pores that increase in size over time, destabilizing the pH and membrane integrity of the phagosomes. Could the time-dependent effectivity of LLO pore formation in the phagosome membrane explain why ΔeutH only exhibits a reduction of cell survivability and growth at 2 h post macrophage phagocytosis (Anderson et al., 2018)? The L. monocytogenes genome encodes a transporter that is predicted to be involved with ammonia/ammonium transport (lmo1516), and it has been shown that ammonia transport is passive and bidirectional in other bacteria (Wingreen, 2004).
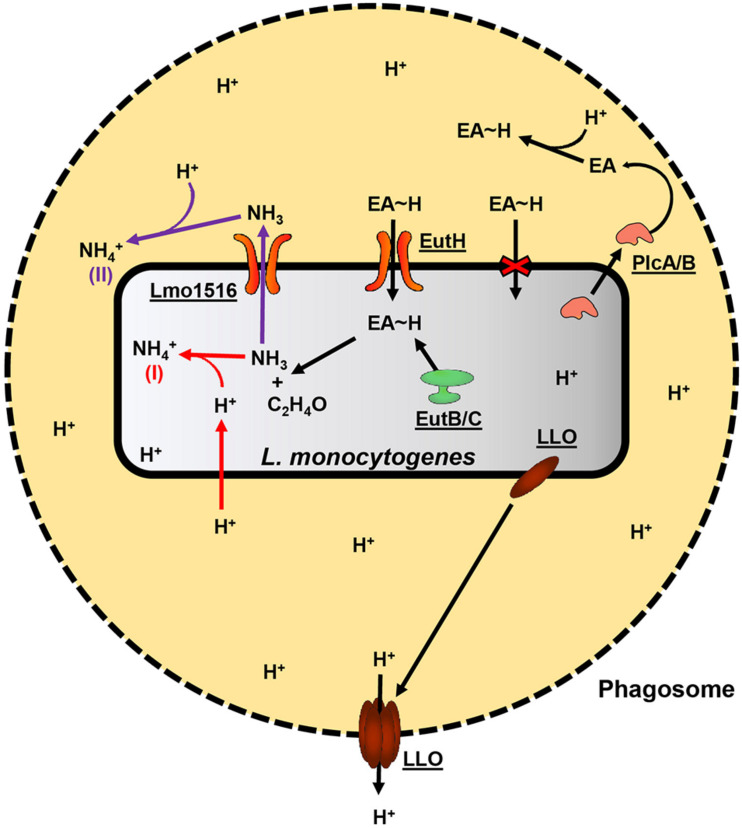
Mycobacterium tuberculosis, Helicobacter pylori, and Staphylococcus aureus utilize ammonia production for localized protection against and possibly to inhibit host defenses such as the formation of phagolysosomes and acidic conditions (Gordon et al., 1980; Ansari and Yamaoka, 2017; Zhou et al., 2019). PlcA/PlcB are supportive in the escape from host phagosomes, possibly by the degradation of the phagosome phospholipid membrane. However, PlcA/PlcB are not required for intracellular growth, unlike LLO, and the exact mechanism of how PlcA/PlcB supports escape from the phagosome is unknown (Lam et al., 2012; Radoshevich and Cossart, 2018). Therefore, we hypothesize that ammonia produced in the initial step of EA catabolism has an essential role during L. monocytogenes pathogenesis. Ammonia derived from EA metabolism could provide localized protection from the acidic conditions within the macrophage phagosome by neutralizing low pH. We further hypothesize that PlcA/PlcB contributes to the survival within macrophage phagosomes by degrading phagosome membranes. The degradation of the phagosome membrane releases EA from phosphatidylethanolamine, which L. monocytogenes can metabolize and convert to ammonia. A model of how L. monocytogenes may use EA metabolism during virulence is proposed in Figure 1. This hypothesis could be tested in future experiments by generating deletion mutants of the lmo1516 putative ammonia/ammonium transporter gene and comparing the deletion mutant with the wildtype to identify possible effects on the survival in macrophages.
FIGURE 1.
Proposed model of EA oxidation and its contribution to the pathogenicity of L. monocytogenes. A black dashed perimeter indicates the membrane of the acidic phagosome. Free protons are denoted “H+,” and protein names or L. monocytogenes EGD-e locus tags are underlined. L. monocytogenes secretes LLO and PlcA/PlcB during its lifecycle within host-cell phagosomes. LLO will associate with the membrane of the phagosome and form pores, allowing protons to diffuse into the host cell cytosol, which, in turn, raises the internal pH of the phagosome. PlcA/PlcB may assist vesicle escape by degrading the host phospholipid membranes. In addition to the already present EA, degrading phosphatidylethanolamine found in host membranes would release additional EA into the vesicle environment. EA is quickly protonated (EA∼H) in the acidic conditions of the phagosome and is internalized by L. monocytogenes by the EutH transporter. The oxidation of the imported EA by the EutB/EutC complex yields acetaldehyde and ammonia. The acetaldehyde and ammonia may enter general carbon and nitrogen metabolism; however, the production of ammonia may contribute to the survival of L. monocytogenes during intracellular life in at least two mechanisms. In the first mechanism, denoted by a red “(I)” and red arrows, ammonia spontaneously reacts with a free proton (which may freely diffuse across L. monocytogenes cell membrane) within the Listeria cytosol and thereby increase intracellular pH by producing ammonium. In the second mechanism, denoted by a purple “(II)” and purple arrows, ammonia is excreted from L. monocytogenes and spontaneously reacts with protons outside the Listeria cell, yielding ammonium, and thereby providing a degree of localized protection from the acidic conditions of the phagosome.
Thus, the analysis of the CDGC in L. monocytogenes seems to indicate that both EA and PD utilization is crucial in the establishment of L. monocytogenes in the GI tract, and the presence of eut genes and EA utilization may promote the systemic spread and intracellular replication during listeriosis infections. Additionally, EA may serve as a nutrient source within the macrophage cytosol. Once L. monocytogenes escapes from the macrophage phagosome, EA is available in phospholipids derived from the destroyed phagosome membrane. However, it seems more likely that, since EA metabolism is not as energetically favorable as the utilization of other nitrogen and carbon sources, and L. monocytogenes likely shifts metabolism toward more easily metabolizable nutrient sources (Sauer et al., 2019). A possible utilization of EA by L. monocytogenes after macrophage escape would need to be verified in future experiments.
Implications for Survival in Food and Food Production Environments
Although it is well established that the CDGC may contribute to the survival of foodborne pathogens in food and food production environments (Chan et al., 2008; Fox et al., 2011; Srikumar and Fuchs, 2011; Goudeau et al., 2013; Tang et al., 2015; Kaval and Garsin, 2018; Anast and Schmitz-Esser, 2020), functional characterizations of the L. monocytogenes CDGC importance during food and food production associated stresses are far more limited than studies that assess its contribution to virulence. PD and EA are found in many food products as constituents of glycoconjugates or lipids and are used as additives of various food products and containers (Cameron et al., 1998; Tang et al., 2015). Tang et al. (2015) found that L. monocytogenes grown on cold vacuum-packed salmon significantly increased expression of the CDGC compared to growth in rich media. They proposed that L. monocytogenes induced the CDGC to utilize PD and EA found in salmon, increasing proliferation on the food matrix within the vacuum packaging (Tang et al., 2015). Carnobacterium piscicola is naturally found on salmon and can inhibit the growth of Listeria. When C. piscicola was co-cultured with L. monocytogenes, it was elucidated that C. piscicola attenuated the growth of L. monocytogenes partially by glucose depletion. During this inhibition, L. monocytogenes significantly increased the expression of the CDGC (Nilsson et al., 2005). L. monocytogenes also increased expression of the pdu gene cluster in co-culture biofilm with Bacillus subtilis but not in mono-culture biofilm (Tirumalai, 2015). Recently, a study from our laboratory that co-cultured L. monocytogenes with either a Brevibacterium or Psychrobacter food isolate found that the CDGC was highly upregulated in a modular way and was dependent on co-culture condition (Anast and Schmitz-Esser, 2020). We also observed that Rli47 was by far the most expressed gene during co-cultivation, further linking the functions of Rli47 and the CDGC (Anast and Schmitz-Esser, 2020). The authors of some recent studies (Tang et al., 2015; Tirumalai, 2015; Anast and Schmitz-Esser, 2020) concur that L. monocytogenes is most likely searching for alternative nutrients to fit into competitive niches, perhaps in a more cooperative approach.
Certain strains of L. monocytogenes are primarily known for their prolonged persistence in food production environments (Carpentier and Cerf, 2011; Ferreira et al., 2014). These strains often harbor genes that confer increased tolerance to stresses associated with these habitats, i.e., protection against methods used to control microbial growth in food production operations. The literature strongly suggests that the CDGC may provide survival advantages to L. monocytogenes during exposure to disinfectants, growth inhibitors, desiccation, and during cold temperature (Chan et al., 2008; Fox et al., 2011; Mattila et al., 2012; Tessema et al., 2012; Hingston et al., 2017; Suo et al., 2018; Kragh and Truelstrup Hansen, 2019; Cortes et al., 2020).
In one study, the transcriptional profiles of a persistent vs. a non-persistent L. monocytogenes strain were assessed in response to exposure to the quaternary ammonia compound disinfectant, benzethonium chloride (Fox et al., 2011). In the persistent strains, the expression of the CDGC was highly upregulated compared to the non-persistent strains. This study concluded that the utilization of this operon in response to disinfectant stress might confer a survival advantage to L. monocytogenes, therefore prolonging persistence in food production facilities (Fox et al., 2011). Many growth inhibitors, such as organic acids, are commonly added to foods to inhibit the growth of Listeria (Bucur et al., 2018). The eut two-component response regulator EutV/EutW, the EA transporter EutH and the positive regulator of PD metabolism PocR were all downregulated when L. monocytogenes was exposed to media acidified to pH five by hydrochloric, acetic, or lactic acid (Tessema et al., 2012). In contrast, another study found that pocR and eutV/eutW were highly induced during lactic acid exposure at pH 3.4, suggesting that the induction of the CDGC in response to acidic stress may be highly dependent on condition (Cortes et al., 2020). Sodium lactate is a widely used food additive that does not influence pH, enhances the organoleptic properties of meat products, and may increase the water compacity of foods (Papadopoulos et al., 1991). When L. monocytogenes was exposed to 4% (wt/wt) sodium lactate and cultivated in ready-to-eat meat products, nearly the entire CDGC (n = 66 genes) was highly downregulated (Suo et al., 2018).
Limiting or removing water from food and food production environments is a standard method of microbial control. When L. monocytogenes was desiccated for short and extended timepoints on steel plates, it was revealed that 16 CDGC genes altered in expression (11 upregulated and five downregulated). Interestingly, both eutV/eutW were downregulated in all five measured timepoints (Kragh and Truelstrup Hansen, 2019). Maintaining food at cold temperatures is another prominent method used for controlling the growth of pathogens and other spoilage organisms. The transcriptional response of L. monocytogenes was also assessed regarding different temperatures relevant to the food production environment (20 vs. 4°C). Remarkably, the authors of a recent study found that at 4°C L. monocytogenes increased expression of 17 genes of the eut pathway, while in contrast, many of the pdu genes and almost the entire operon for de novo synthesis of cobalamin was downregulated (Hingston et al., 2017). This study also observed high expression levels of aspocR, suggesting that PocR did not induce the pdu genes. The consistent induction of the eut genes and repression of the pdu genes at colder temperatures may indicate that EA may be more preferably metabolized at low temperatures than PD. This theory is supported by a previously unrecognized precedent of the importance of EA metabolism during cold temperatures in L. monocytogenes. Chan et al. (2008) created deletion mutants of L. monocytogenes sigma factors, two-component regulatory systems, and negative regulators to investigate their contribution during cold adaptation. Interestingly, the response regulator eutV deletion mutant had significantly lower cell counts than the wildtype after 12 days of incubation at 4°C (Chan et al., 2008). Later, a homolog of eutV in Enterococcus faecalis was described to be a part of the two-component response regulatory system controlling the EA metabolism genes (Del Papa and Perego, 2008; Fox et al., 2009). Noteworthy, the genomic organization of the eut gene pathway is conserved between Enterococcus faecalis and L. monocytogenes. Lastly, Mattila et al. (2012) demonstrated that during exposure of L. monocytogenes to 3 and 37°C, both eutV/eutW are positively regulated by σL, the alternative sigma factor shown to be required for efficient growth during low temperatures and organic acid exposure. Remarkably, PocR was positively influenced by σL during growth at 3°C but in contrast, was downregulated by σL during cultivation at 37°C. These transcriptomic data suggest that σL positively regulates both pdu and eut genes at colder temperatures, but represses PocR at optimal temperature.
The studies mentioned above suggest that the CDGC, especially the EA utilization module, contributes to the survival of L. monocytogenes in food and food production environments. The deletion of one eut gene leads to impaired growth at refrigeration temperatures. However, the mechanism of how EA metabolism may be necessary for the proliferation of L. monocytogenes at cold temperatures is currently unknown. It is tempting to speculate that PD and EA may be more readily utilized during exposure to food production environment-associated stress conditions than other, more efficient carbon and energy substrates such as glucose.
Conclusion and Future Directions
Although evident attenuation of virulence is observed when EA and PD metabolism genes are deleted, the molecular mechanisms of how key EA and PD metabolic genes contribute to pathogenicity remain unknown in Listeria. Although several transcriptomic studies show that the CDGC is differentially expressed during food and food production environment relevant stress conditions, functional characterization of how CDGC mechanisms may contribute to L. monocytogenes stress tolerance is, to the best of our knowledge, currently unavailable. Thus, determining the contribution of the CDGC to survival under food and food production associated stress conditions may – in the future – reveal novel countermeasures for implementation into good management practices of food production plants. Finally, at least two alternative sigma factors, a two-component regulatory system, non-coding RNAs such as Rli39, aspocR, Rli47, and Rli55, are involved in regulating the CDGC, emphasizing the complexity of the regulatory network of CDGC. Alternative sigma factors and non-coding RNAs have emerged as critical regulatory entities in both virulence and general stress response that may have global impacts on gene expression.
One important aspect of CDGC that should be addressed in future research is to identify whether EA catabolism may have an effect on the host immune system and commensal bacteria of the intestine. Acetate is the final end-product of EA catabolism, and it has been shown to modulate the immune response by suppressing IgA production in the GI tract (Maslowski et al., 2009; Wu et al., 2017). It is possible that EA metabolism resulting in the production of acetate by enteric pathogens disrupts the host immune response. This hypothesis has been previously proposed by others (Kaval and Garsin, 2018), and to the best of our knowledge, has yet to be experimentally verified. In addition, it may be important to test if the utilization of PD is restricted to the intestinal mucosa as a source of rhamnose or fucose. These two sugars are released from complex carbohydrates by commensal bacteria in the GI tract and may not be available in the host blood, lymphatic fluid, or during survival in monocytes and hepatocytes in concentrations or forms that are feasible for L. monocytogenes to metabolize them. Possible ways to test if the catabolism of these sugars is essential for the intestinal survival of L. monocytogenes would be to delete key genes involved in the metabolism of fucose, rhamnose, and PD and then test L. monocytogenes survival or growth in the GI tract, or during in vitro growth by using defined media with and without fucose and rhamnose.
In future studies that assess the CDGC of L. monocytogenes, experiments should be designed to observe the global effects of EA and PD metabolism by transcriptomic and non-targeted metabolomic approaches to best represent the comprehensive phenotype of L. monocytogenes in regards to the tested condition. Much of the CDGC of L. monocytogenes and its contribution to virulence and stress survival has been elucidated, but further study should be encouraged to uncover more specific mechanisms in which the CDGC enables Listeria to survive food and food production associated stress conditions, which can subsequently lead to foodborne illness.
Author Contributions
JA conceived and wrote the first draft of the manuscript and analyzed data. SS-E, TB, and JA wrote and approved the final manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. SS-E and JA were supported by the USDA National Institute of Food and Agriculture Hatch project nos. 1011114 and 1018898 and by the USDA National Institute of Food and Agriculture, Agricultural and Food Research Initiative Competitive Program, grant number: 2019-67017-29687.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.601816/full#supplementary-material
References
- Allerberger F., Wagner M. (2010). Listeriosis: a resurgent foodborne infection. Clin. Microbiol. Infect. 16 16–23. 10.1111/j.1469-0691.2009.03109.x [DOI] [PubMed] [Google Scholar]
- Anast J. M., Schmitz-Esser S. (2020). The transcriptome of Listeria monocytogenes during co-cultivation with cheese rind bacteria suggests adaptation by induction of ethanolamine and 1,2-propanediol catabolism pathway genes. PLoS One 15:e0233945. 10.1371/journal.pone.0233945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C. J., Clark D. E., Adli M., Kendall M. M. (2015). Ethanolamine signaling promotes Salmonella niche recognition and adaptation during infection. PLoS Pathog. 11:e1005278. 10.1371/journal.ppat.1005278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C. J., Satkovich J., Koseoglu V. K., Agaisse H., Kendall M. M. (2018). The ethanolamine permease EutH promotes vacuole adaptation of Salmonella enterica and Listeria monocytogenes during macrophage infection. Infect. Immun. 86:e00172-18. 10.1128/IAI.00172-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari S., Yamaoka Y. (2017). Survival of Helicobacter pylori in gastric acidic territory. Helicobacter 22:10.1111/hel.12386. 10.1111/hel.12386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambaud C., Nahori M. A., Soubigou G., Becavin C., Laval L., Lechat P., et al. (2012). Impact of Lactobacilli on orally acquired listeriosis. Proc. Natl. Acad. Sci. U.S.A. 109 16684–16689. 10.1073/pnas.1212809109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldoma L., Aguilar J. (1988). Metabolism of L-fucose and L-rhamnose in Escherichia coli: aerobic-anaerobic regulation of L-lactaldehyde dissimilation. J. Bacteriol. 170 416–421. 10.1128/jb.170.1.416-421.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell C. M., Scarlett F. A., Turner J. M. (1976). Ethanolamine catabolism by bacteria, including Escherichia coli. Biochem. Soc. Trans. 4 495–497. 10.1042/bst0040495 [DOI] [PubMed] [Google Scholar]
- Bobik T. A., Havemann G. D., Busch R. J., Williams D. S., Aldrich H. C. (1999). The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B(12)-dependent 1, 2-propanediol degradation. J. Bacteriol. 181 5967–5975. 10.1128/JB.181.19.5967-5975.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchrieser C., Rusniok C., Kunst F., Cossart P., Glaser P., Listeria C. (2003). Comparison of the genome sequences of Listeria monocytogenes and Listeria innocua: clues for evolution and pathogenicity. FEMS Immunol. Med. Microbiol. 35 207–213. 10.1016/S0928-8244(02)00448-0 [DOI] [PubMed] [Google Scholar]
- Bucur F. I., Grigore-Gurgu L., Crauwels P., Riedel C. U., Nicolau A. I. (2018). Resistance of Listeria monocytogenes to stress conditions encountered in food and food processing environments. Front. Microbiol. 9:2700. 10.3389/fmicb.2018.02700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron D. C., Altaras N. E., Hoffman M. L., Shaw A. J. (1998). Metabolic engineering of propanediol pathways. Biotechnol. Prog. 14 116–125. 10.1021/bp9701325 [DOI] [PubMed] [Google Scholar]
- Carpentier B., Cerf O. (2011). Review–persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food Microbiol. 145 1–8. 10.1016/j.ijfoodmicro.2011.01.005 [DOI] [PubMed] [Google Scholar]
- Chan Y. C., Hu Y., Chaturongakul S., Files K. D., Bowen B. M., Boor K. J., et al. (2008). Contributions of two-component regulatory systems, alternative sigma factors, and negative regulators to Listeria monocytogenes cold adaptation and cold growth. J. Food Prot. 71 420–425. 10.4315/0362-028x-71.2.420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiara M., Caruso M., D’Erchia A. M., Manzari C., Fraccalvieri R., Goffredo E., et al. (2015). Comparative genomics of Listeria sensu lato: genus-wide differences in evolutionary dynamics and the progressive gain of complex, potentially pathogenicity-related traits through lateral gene transfer. Genome Biol. Evol. 7 2154–2172. 10.1093/gbe/evv131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury C., Sinha S., Chun S., Yeates T. O., Bobik T. A. (2014). Diverse bacterial microcompartment organelles. Microbiol. Mol. Biol. Rev. 78 438–468. 10.1128/MMBR.00009-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes B. W., Naditz A. L., Anast J. M., Schmitz-Esser S. (2020). Transcriptome sequencing of Listeria monocytogenes reveals major gene expression changes in response to lactic acid stress exposure but a less pronounced response to oxidative stress. Front. Microbiol. 10:3110. 10.3389/fmicb.2019.03110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa F. G., Escalante-Semerena J. C. (2018). A new class of EutT ATP:Co(I)rrinoid adenosyltransferases found in Listeria monocytogenes and other Firmicutes does not require a metal ion for activity. Biochemistry 57 5076–5087. 10.1021/acs.biochem.8b00715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Chastellier C., Berche P. (1994). Fate of Listeria monocytogenes in murine macrophages: evidence for simultaneous killing and survival of intracellular bacteria. Infect. Immun. 62 543–553. 10.1128/IAI.62.2.543-553.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Papa M. F., Perego M. (2008). Ethanolamine activates a sensor histidine kinase regulating its utilization in Enterococcus faecalis. J. Bacteriol. 190 7147–7156. 10.1128/JB.00952-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan B., Suzuki H., Herlekar D., Sartor R. B., Campbell B. J., Roberts C. L., et al. (2014). Inflammation-associated adherent-invasive Escherichia coli are enriched in pathways for use of propanediol and iron and M-cell translocation. Inflamm. Bowel Dis. 20 1919–1932. 10.1097/mib.0000000000000183 [DOI] [PubMed] [Google Scholar]
- Drolia R., Bhunia A. K. (2019). Crossing the intestinal barrier via Listeria adhesion protein and internalin A. Trends Microbiol. 27 408–425. 10.1016/j.tim.2018.12.007 [DOI] [PubMed] [Google Scholar]
- Faber F., Thiennimitr P., Spiga L., Byndloss M. X., Litvak Y., Lawhon S., et al. (2017). Respiration of microbiota-derived 1,2-propanediol drives Salmonella expansion during Colitis. PLoS Pathog. 13:e1006129. 10.1371/journal.ppat.1006129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira V., Wiedmann M., Teixeira P., Stasiewicz M. J. (2014). Listeria monocytogenes persistence in food-associated environments: epidemiology, strain characteristics, and implications for public health. J. Food Prot. 77 150–170. 10.4315/0362-028X.JFP-13-150 [DOI] [PubMed] [Google Scholar]
- Fox E. M., Leonard N., Jordan K. (2011). Physiological and transcriptional characterization of persistent and nonpersistent Listeria monocytogenes isolates. Appl. Environ. Microbiol. 77 6559–6569. 10.1128/AEM.05529-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K. A., Ramesh A., Stearns J. E., Bourgogne A., Reyes-Jara A., Winkler W. C., et al. (2009). Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc. Natl. Acad. Sci. U.S.A. 106 4435–4440. 10.1073/pnas.0812194106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsin D. A. (2010). Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat. Rev. Microbiol. 8 290–295. 10.1038/nrmicro2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonyar L. A., Kendall M. M. (2014). Ethanolamine and choline promote expression of putative and characterized fimbriae in enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 82 193–201. 10.1128/IAI.00980-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon A. H., Hart P. D., Young M. R. (1980). Ammonia inhibits phagosome-lysosome fusion in macrophages. Nature 286 79–80. 10.1038/286079a0 [DOI] [PubMed] [Google Scholar]
- Goudeau D. M., Parker C. T., Zhou Y., Sela S., Kroupitski Y., Brandl M. T. (2013). The Salmonella transcriptome in lettuce and cilantro soft rot reveals a niche overlap with the animal host intestine. Appl. Environ. Microbiol. 79 250–262. 10.1128/AEM.02290-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmans S., Bont J. A. M. (1986). Acetol monooxygenase from Mycobacterium Py1 cleaves acetol into acetate and formaldehyde. FEMS Microbiol. Lett. 36 155–158. 10.1111/j.1574-6968.1986.tb01686.x [DOI] [Google Scholar]
- Hingston P., Chen J., Allen K., Truelstrup Hansen L., Wang S. (2017). Strand specific RNA-sequencing and membrane lipid profiling reveals growth phase-dependent cold stress response mechanisms in Listeria monocytogenes. PLoS One 12:e0180123. 10.1371/journal.pone.0180123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B., Przybilla K., Stuhler C., Schauer K., Slaghuis J., Fuchs T. M., et al. (2006). Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J. Bacteriol. 188 556–568. 10.1128/JB.188.2.556-568.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaval K. G., Garsin D. A. (2018). Ethanolamine utilization in bacteria. mBio 9:e00066-18. 10.1128/mBio.00066-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai K., Fujita M., Nakao M. (1974). Lipid components of two different regions of an intestinal epithelial cell membrane of mouse. Biochim. Biophys. Acta 369 222–233. 10.1016/0005-2760(74)90253-7 [DOI] [PubMed] [Google Scholar]
- Kendall M. M., Gruber C. C., Parker C. T., Sperandio V. (2012). Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. mBio 3:e00050-12. 10.1128/mBio.00050-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfeld C. A., Aussignargues C., Zarzycki J., Cai F., Sutter M. (2018). Bacterial microcompartments. Nat. Rev. Microbiol. 16 277–290. 10.1038/nrmicro.2018.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keto-Timonen R., Tolvanen R., Lundén J., Korkeala H. (2007). An 8-Year Surveillance of the diversity and persistence of Listeria monocytogenes in a chilled food processing plant analyzed by amplified fragment length polymorphism. J. Food Prot. 70 1866–1873. 10.4315/0362-028x-70.8.1866 [DOI] [PubMed] [Google Scholar]
- Kragh M. L., Truelstrup Hansen L. (2019). Initial transcriptomic response and adaption of Listeria monocytogenes to desiccation on food grade stainless steel. Front. Microbiol. 10:3132. 10.3389/fmicb.2019.03132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutzner E., Kern T., Felsl A., Eisenreich W., Fuchs T. M. (2016). Isotopologue profiling of the listerial N-metabolism. Mol. Microbiol. 100 315–327. 10.1111/mmi.13318 [DOI] [PubMed] [Google Scholar]
- Lam G. Y., Czuczman M. A., Higgins D. E., Brumell J. H. (2012). Interactions of Listeria monocytogenes with the autophagy system of host cells. Adv. Immunol. 113 7–18. 10.1016/B978-0-12-394590-7.00008-7 [DOI] [PubMed] [Google Scholar]
- Liu Y., Orsi R. H., Boor K. J., Wiedmann M., Guariglia-Oropeza V. (2017). Home alone: elimination of all but one alternative Sigma factor in Listeria monocytogenes allows prediction of new roles for sigma(B). Front. Microbiol. 8:1910. 10.3389/fmicb.2017.01910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzader D. H., Clark D. E., Gonyar L. A., Kendall M. M. (2013). EutR is a direct regulator of genes that contribute to metabolism and virulence in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 195 4947–4953. 10.1128/JB.00937-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinho C. M., Dos Santos P. T., Kallipolitis B. H., Johansson J., Ignatov D., Guerreiro D. N., et al. (2019). The sigma(B)-dependent regulatory sRNA Rli47 represses isoleucine biosynthesis in Listeria monocytogenes through a direct interaction with the ilvA transcript. RNA Biol. 16 1424–1437. 10.1080/15476286.2019.1632776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslowski K. M., Vieira A. T., Ng A., Kranich J., Sierro F., Yu D., et al. (2009). Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461 1282–1286. 10.1038/nature08530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews T. D., Schmieder R., Silva G. G. Z., Busch J., Cassman N., Dutilh B. E., et al. (2015). Genomic comparison of the closely-related Salmonella enterica serovars Enteritidis, Dublin and Gallinarum. PLoS One 10:e0126883. 10.1371/journal.pone.0126883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila M., Somervuo P., Rattei T., Korkeala H., Stephan R., Tasara T. (2012). Phenotypic and transcriptomic analyses of Sigma L-dependent characteristics in Listeria monocytogenes EGD-e. Food Microbiol. 32 152–164. 10.1016/j.fm.2012.05.005 [DOI] [PubMed] [Google Scholar]
- Mellin J. R., Koutero M., Dar D., Nahori M. A., Sorek R., Cossart P. (2014). Riboswitches. Sequestration of a two-component response regulator by a riboswitch-regulated noncoding RNA. Science 345 940–943. 10.1126/science.1255083 [DOI] [PubMed] [Google Scholar]
- Mellin J. R., Tiensuu T., Becavin C., Gouin E., Johansson J., Cossart P. (2013). A riboswitch-regulated antisense RNA in Listeria monocytogenes. Proc. Natl. Acad. Sci. U.S.A. 110 13132–13137. 10.1073/pnas.1304795110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujahid S., Bergholz T. M., Oliver H. F., Boor K. J., Wiedmann M. (2012). Exploration of the role of the non-coding RNA SbrE in L. monocytogenes stress response. Int. J. Mol. Sci. 14 378–393. 10.3390/ijms14010378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A., Rychli K., Zaiser A., Wieser C., Wagner M., Schmitz-Esser S. (2014). The Listeria monocytogenes transposon Tn6188 provides increased tolerance to various quaternary ammonium compounds and ethidium bromide. FEMS Microbiol. Lett. 361 166–173. 10.1111/1574-6968.12626 [DOI] [PubMed] [Google Scholar]
- Myers J. T., Tsang A. W., Swanson J. A. (2003). Localized reactive oxygen and nitrogen intermediates inhibit escape of Listeria monocytogenes from vacuoles in activated macrophages. J. Immunol. 171 5447–5453. 10.4049/jimmunol.171.10.5447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandedkar A. K. N. (1974). Report on the utilization of ethanolamine-1-14C by Mycobacterium 607. Biochem. Med. 11 67–70. 10.1016/0006-2944(74)90096-9 [DOI] [PubMed] [Google Scholar]
- Nilsson L., Hansen T. B., Garrido P., Buchrieser C., Glaser P., Knochel S., et al. (2005). Growth inhibition of Listeria monocytogenes by a nonbacteriocinogenic Carnobacterium piscicola. J. Appl. Microbiol. 98 172–183. 10.1111/j.1365-2672.2004.02438.x [DOI] [PubMed] [Google Scholar]
- Nuccio S. P., Baumler A. J. (2014). Comparative analysis of Salmonella genomes identifies a metabolic network for escalating growth in the inflamed gut. mBio 5:e00929-14. 10.1128/mBio.00929-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne S. E., Brumell J. H. (2017). Listeriolysin O: from bazooka to Swiss army knife. Philos. Trans. R. Soc. B 372:20160222. 10.1098/rstb.2016.0222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos L. S., Miller R. K., Ringer L. J., Cross H. R. (1991). Sodium lactate effect on sensory characteristics, cooked meat color and chemical composition. J. Food Sci. 56 621–626. 10.1111/j.1365-2621.1991.tb05343.x [DOI] [Google Scholar]
- Radoshevich L., Cossart P. (2018). Listeria monocytogenes: towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 16 32–46. 10.1038/nrmicro.2017.126 [DOI] [PubMed] [Google Scholar]
- Rodionov D. A., Vitreschak A. G., Mironov A. A., Gelfand M. S. (2003). Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 278 41148–41159. 10.1074/jbc.M305837200 [DOI] [PubMed] [Google Scholar]
- Sampson E. M., Bobik T. A. (2008). Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J. Bacteriol. 190 2966–2971. 10.1128/JB.01925-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer J.-D., Herskovits A. A., O’Riordan M. X. D. (2019). Metabolism of the Gram-positive bacterial pathogen Listeria monocytogenes. Microbiol. Spect. 7. 10.1128/microbiolspec.GPP3-0066-2019 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E., Hoekstra R. M., Angulo F. J., Tauxe R. V., Widdowson M. A., Roy S. L., et al. (2011). Foodborne illness acquired in the United States–major pathogens. Emerg. Infect. Dis. 17 7–15. 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardt J., Jones G., Muller-Herbst S., Schauer K., D’Orazio S. E. F., Fuchs T. M. (2017). Comparison between Listeria sensu stricto and Listeria sensu lato strains identifies novel determinants involved in infection. Sci. Rep. 7:17821. 10.1038/s41598-017-17570-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shively J. M., Ball F., Brown D. H., Saunders R. E. (1973). Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science 182 584–586. 10.1126/science.182.4112.584 [DOI] [PubMed] [Google Scholar]
- Singh R., Jamieson A., Cresswell P. (2008). GILT is a critical host factor for Listeria monocytogenes infection. Nature 455 1244–1247. 10.1038/nature07344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikumar S., Fuchs T. M. (2011). Ethanolamine utilization contributes to proliferation of Salmonella enterica serovar Typhimurium in food and in nematodes. Appl. Environ. Microbiol. 77 281–290. 10.1128/AEM.01403-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staib L., Fuchs T. M. (2014). From food to cell: nutrient exploitation strategies of enteropathogens. Microbiology 160(Pt 6), 1020–1039. 10.1099/mic.0.078105-0 [DOI] [PubMed] [Google Scholar]
- Suo Y., Gao S., Baranzoni G. M., Xie Y., Liu Y. (2018). Comparative transcriptome RNA-Seq analysis of Listeria monocytogenes with sodium lactate adaptation. Food Control 91 193–201. 10.1016/j.foodcont.2018.03.044 [DOI] [Google Scholar]
- Tang S., Orsi R. H., den Bakker H. C., Wiedmann M., Boor K. J., Bergholz T. M. (2015). Transcriptomic analysis of the adaptation of Listeria monocytogenes to growth on vacuum-packed cold smoked salmon. Appl. Environ. Microbiol. 81 6812–6824. 10.1128/AEM.01752-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessema G. T., Moretro T., Snipen L., Heir E., Holck A., Naterstad K., et al. (2012). Microarray-based transcriptome of Listeria monocytogenes adapted to sublethal concentrations of acetic acid, lactic acid, and hydrochloric acid. Can. J. Microbiol. 58 1112–1123. 10.1139/w2012-091 [DOI] [PubMed] [Google Scholar]
- Thiennimitr P., Winter S. E., Winter M. G., Xavier M. N., Tolstikov V., Huseby D. L., et al. (2011). Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. U.S.A. 108 17480–17485. 10.1073/pnas.1107857108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney L. G., Portnoy D. A. (1989). Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J. Cell Biol. 109(4 Pt 1), 1597–1608. 10.1083/jcb.109.4.1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirumalai P. S. (2015). Metabolic gene expression shift by Listeria monocytogenes in coculture biofilms. Can. J. Microbiol. 61 327–334. 10.1139/cjm-2014-0704 [DOI] [PubMed] [Google Scholar]
- Toledo-Arana A., Dussurget O., Nikitas G., Sesto N., Guet-Revillet H., Balestrino D., et al. (2009). The Listeria transcriptional landscape from saprophytism to virulence. Nature 459 950–956. 10.1038/nature08080 [DOI] [PubMed] [Google Scholar]
- Tsoy O., Ravcheev D., Mushegian A. (2009). Comparative genomics of ethanolamine utilization. J. Bacteriol. 191 7157–7164. 10.1128/JB.00838-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe-Querol E., Rosales C. (2017). Control of phagocytosis by microbial pathogens. Front. Immunol. 8:1368. 10.3389/fimmu.2017.01368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingreen N. S. (2004). Ammonia transport. EcoSal Plus 1. 10.1128/ecosalplus.3.3.2.1 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Wu W., Sun M., Chen F., Cao A. T., Liu H., Zhao Y., et al. (2017). Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol. 10 946–956. 10.1038/mi.2016.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J., Murrieta C. M., Rule D. C., Miller K. W. (2008). Exogenous or L-rhamnose-derived 1,2-propanediol is metabolized via a pduD-dependent pathway in Listeria innocua. Appl. Environ. Microbiol. 74 7073–7079. 10.1128/AEM.01074-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z., Smid E. J., Boeren S., Notebaart R. A., Abee T. (2019). Bacterial microcompartment-dependent 1,2-propanediol utilization stimulates anaerobic growth of Listeria monocytogenes EGDe. Front. Microbiol. 10:2660. 10.3389/fmicb.2019.02660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Bhinderwala F., Lehman M. K., Thomas V. C., Chaudhari S. S., Yamada K. J., et al. (2019). Urease is an essential component of the acid response network of Staphylococcus aureus and is required for a persistent murine kidney infection. PLoS Pathog. 15:e1007538. 10.1371/journal.ppat.1007538 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.