Abstract
Objective
To identify dysregulated metabolic pathways in amyotrophic lateral sclerosis (ALS) versus control participants through untargeted metabolomics.
Methods
Untargeted metabolomics was performed on plasma from ALS participants (n=125) around 6.8 months after diagnosis and healthy controls (n=71). Individual differential metabolites in ALS cases versus controls were assessed by Wilcoxon rank-sum tests, adjusted logistic regression and partial least squares-discriminant analysis (PLS-DA), while group lasso explored sub-pathway-level differences. Adjustment parameters included sex, age and body mass index (BMI). Metabolomics pathway enrichment analysis was performed on metabolites selected by the above methods. Finally, machine learning classification algorithms applied to group lasso-selected metabolites were evaluated for classifying case status.
Results
There were no group differences in sex, age and BMI. Significant metabolites selected were 303 by Wilcoxon, 300 by logistic regression, 295 by PLS-DA and 259 by group lasso, corresponding to 11, 13, 12 and 22 enriched sub-pathways, respectively. ‘Benzoate metabolism’, ‘ceramides’, ‘creatine metabolism’, ‘fatty acid metabolism (acyl carnitine, polyunsaturated)’ and ‘hexosylceramides’ sub-pathways were enriched by all methods, and ‘sphingomyelins’ by all but Wilcoxon, indicating these pathways significantly associate with ALS. Finally, machine learning prediction of ALS cases using group lasso-selected metabolites achieved the best performance by regularised logistic regression with elastic net regularisation, with an area under the curve of 0.98 and specificity of 83%.
Conclusion
In our analysis, ALS led to significant metabolic pathway alterations, which had correlations to known ALS pathomechanisms in the basic and clinical literature, and may represent important targets for future ALS therapeutics.
Introduction
Amyotrophic lateral sclerosis (ALS) is a progressive, fatal neurodegenerative disease of motor neurons,1 characterised by complex genetics2 and disease mechanisms,3 as well as environmental influences.4 5 Although a handful of genes are strongly linked to ALS,2 genetic causes are not known in the majority of sporadic cases. However, ALS genes affect several shared cellular processes, including proteostasis, autophagy, mitochondrial function, cytoskeletal organisation and axonal transport.2 3 Metabolic abnormalities are also implicated, such as amino acid, pyruvate and lipid metabolism.6–8 Increasing the pathological complexity is the gene-time-environment hypothesis of ALS, which states that environmental exposures superimposed on a genetic risk profile trigger metabolic abnormalities that initiate neurodegeneration.9
Metabolites ultimately reflect the coordinated influence of genetics, epigenetics and transcriptomics, as well as serving as evidence of environmental exposure through xenobiotics. Metabolites are also a reflection of dysregulated cellular processes and pathological state. For instance, a recognition that oxidative stress is an ALS hallmark, through the detection of oxidised metabolites in biosamples, led to clinical trials of the antioxidant edaravone,10 which is now US Food and Drug Administration (FDA)-approved for treating ALS. Thus, despite the genetic and clinical heterogeneity of ALS, metabolites may be a unifying feature through shared cellular processes and environmental contact. Metabolomics has emerged as a new frontier for understanding pathological mechanisms, biomarkers and evaluating environmental impact on disease.11 Metabolomics is the untargeted, system-wide and simultaneous analysis of all metabolites present in a sample. Its untargeted nature allows agnostic evaluation of metabolites to identify diagnostic or prognostic biomarker panels, understand complex pathophysiology, identify drug target candidates, or reveal potentially novel, hypothesis-generating avenues.12 In ALS, a small number of studies have identified altered metabolites in biofluids, but from metabolite datasets of around 400 metabolites or fewer.13–15 Our aim was to identify differential metabolites and enriched pathways in ALS versus control participants using a commercial untargeted metabolomics platform, which characterises an extensive number of compounds. Metabolomics analysis could lead to novel hypotheses, therapeutic targets and potential biomarkers for ALS.
Methods
Participants and biosamples
ALS and neurologically healthy control participants were enrolled at the University of Michigan (UM), as previously reported.5 Briefly, all patients seen at the UM ALS clinic over 18 years and able to communicate in English were asked to provide plasma shortly after diagnosis. Sex-matched and age-matched control subjects also provided plasma. Participant demographics were collected, for example, sex, age, height, weight, ALS disease characteristics. Blood was drawn from participants that had not been asked to fast, as it was deemed irresponsible to request this from a large number of ALS participants, who have blood drawn in conjunction with their standard clinical care. Blood samples were collected following good clinical practice into lavender EDTA tubes and stored temporarily for a maximum of 2 hours at 4°C. Tubes were then centrifuged at 2000g for 10 min at 4°C and the plasma supernatant was aliquoted into cryovials and directly transferred to the −80°C freezer for storage.
Metabolomic profiling
Plasma samples were shipped on dry ice to Metabolon (Durham, North Carolina), and stored at −80°C at their facility until they were analysed for untargeted metabolomics profiling using ultrahigh performance liquid chromatography-tandem mass spectroscopy (UPLC-MS/MS) following their published protocol.16 17 In brief, metabolites were extracted from plasma using methanol and recovery and internal standards were added to assess extraction efficiency and instrument performance, respectively. Metabolites were then analysed by reverse-phase UPLC-MS/MS in both positive and negative ion mode and hydrophilic interaction chromatography UPLC-MS/MS. A total of 1051 known metabolites (see online supplemental table S1) were identified by retention time/index, mass-to-charge ratio and chromatographic data against authenticated standards, followed by data curation to ensure correct chemical identification. Day-to-day variability was accounted for by normalising metabolite levels. This was accomplished daily by equating the metabolite median across samples for that day to one and normalising the metabolite within each sample proportionately against the median. Metabolites that were not detected in over 60% of samples were excluded from further analysis. Any missing values for remaining metabolites were imputed to the minimal observed value for each metabolite, per metabolon protocols.16 17
jnnp-2020-323611supp001.pdf (6.2MB, pdf)
Descriptive analysis
Descriptive summaries of demographic and clinical characteristics were calculated by case/control status. χ2 and Wilcoxon rank-sum tests identified significant case/control discrepancies in demographic and clinical data. Missing metabolite data counts and percentages and demographic information were tabulated. Spearman correlations determined whether medication use and exogenous drug metabolites affected other metabolite concentrations. Non-parametric Wilcoxon rank-sum test, referred to as unadjusted, evaluated non-normally distributed metabolomics data for significant case/control differences for each metabolite. Benjamini-Hochberg correction adjusted for multiple testing (see online supplemental table S2). Logistic regression models, referred to as adjusted, were constructed, by regressing each natural log-transformed and standardised metabolite against case/control status one at a time. Logistic regression models were adjusted for sex, age (as quartiles) and body mass index (BMI, as quartiles). Subjects missing sex, age and BMI were omitted. Benjamini-Hochberg correction adjusted for multiple testing (see online supplemental table S3).
Partial least squares-discriminant analysis
Partial least squares-discriminant analysis (PLS-DA) identifies metabolites that carry the greatest group-separating information, as represented by the first latent variable, which we performed using the R package mixOmics.18 Score plots illustrate differences between case versus control groups. The variable importance in projection (VIP) score of each metabolite, a weighted sum of the squared correlations between PLS-DA components and metabolites,19 contributed significantly to the separation of case versus controls for VIP >1.20
Group lasso
Group lasso regressed all metabolites against case/control status simultaneously, adjusting for sex, age and BMI (see online supplemental table S4). Group lasso selects entire sub-pathways to simultaneously account for within-sub-pathway correlation structure. The gglasso R package was used to implement group lasso with natural log-transformed and standardised metabolite data. Fivefold cross-validation was used to select the tuning parameter corresponding to a sparse model within one SE of the minimum cross-validation error. Once the tuning parameter, corresponding to the group lasso penalty was finalised, group lasso was refit to the full dataset to obtain the final model.
Metabolism pathway analysis
Pathway enrichment analysis was performed using our in-house R package richR (https://github.com/hurlab/richR/). Sub-pathways (115 in total), annotated by Metabolon and 1051 identified metabolites were used as background pathway and metabolite sets, respectively. The significant metabolites identified by unadjusted Wilcoxon, adjusted logistic regression, group lasso and PLS-DA were evaluated for over-representation in each sub-pathway by modified Fisher’s exact test. A hypergeometric test was performed for each candidate sub-pathway. Sub-pathways with a p value <0.05 were deemed significantly enriched.
Metabolites correlation network analysis
In addition to the pathway analysis relying on prior knowledge, we also applied a data-driven approach to explore the potential associations based on group lasso-selected metabolites. Highly correlated and significant metabolite pairs with a Spearman’s rank-order correlation coefficient (ρ>0.5) and a p value of <0.05 were identified. A correlated metabolite network was constructed in Cytoscape V.3.7.2, a network visualisation and analysis platform.
Classification prediction analysis using machine learning
To examine the feasibility of predicting metabolite-based ALS cases, we constructed machine learning classification models using seven widely used algorithms, generalised boosted models, linear discriminant analysis, prediction analysis for microarrays, random forest (RF), regularised logistic regression with elastic net regularisation (RLR), recursive partitioning and regression trees and support vector machine (SVM). We used the R package caret 21 to build and evaluate the performance of machine learning models using 10-fold cross-validation. Synthetic minority oversampling technique was used to balance case/control sets to account for imbalance.22 Prediction accuracy metrics for area under the curve (AUC), sensitivity (SENS) and specificity (SPEC), were calculated for each model, which were visualised through receiver operating characteristic curves generated by R package pROC.23 Metabolites were ranked and reported based on their contribution to model performance using the varImp function in the caret package.
Statistical software
All statistical and prediction analyses were performed in R statistical computing software.
Results
Participants
This study included 125 ALS and 71 control participants that did not differ significantly by sex, age, BMI, smoking status and military service history (table 1). Controls achieved higher educational levels, consistent with prior publications.24 Cases reflected a typical ALS population with a median diagnostic age of 62.2 years (IQR 52.7–68.7), an interval between symptom onset and diagnosis of 1.01 years (IQR 0.68–1.51) and onset segment of bulbar 30.4%, cervical 30.4% and lumbar 39.2%. Plasma was collected within 0.57 years of diagnosis (IQR 0.36–0.75).
Table 1.
Participant demographics
| Covariate | ALS cases (n=125) |
Controls (n=71) |
P value |
| Age at plasma collection (years)* | 63.0 (53.0–69.0) | 61.0 (53.0–65.0) | 0.362 |
| Sex | 0.972 | ||
| Female | 51 (40.8) | 28 (39.4) | |
| Male | 74 (59.2) | 43 (60.6) | |
| BMI at study entry (kg/m2)† | 25.6 (22.8–29.6) | 26.6 (23.9–30.3) | 0.116 |
| Smoking | 0.569 | ||
| Non-smoker | 68 (54.4) | 33 (46.5) | |
| Former smoker | 43 (34.4) | 26 (36.6) | |
| Current smoker | 13 (10.4) | 10 (14.1) | |
| Missing | 1 (0.8) | 2 (2.8) | |
| Family history of ALS | |||
| No | 111 (88.8) | ||
| Yes | 10 (8.0) | ||
| Missing | 4 (3.2) | ||
| Age at diagnosis (years) | 62.2 (52.7–68.7) | ||
| El Escorial criteria | |||
| Suspected | 3 (2.4) | ||
| Possible | 19 (15.2) | ||
| Probable, LS | 37 (29.6) | ||
| Probable | 42 (33.6) | ||
| Definite | 23 (18.4) | ||
| Missing | 1 (0.8) | ||
| Onset segment | |||
| Bulbar | 38 (30.4) | ||
| Cervical | 38 (30.4) | ||
| Lumbar | 49 (39.2) | ||
| Time between diagnosis and blood draw (years)* | 0.57 (0.36–0.75) | ||
| Time between symptom onset and diagnosis (years) | 1.01 (0.68–1.51) | ||
| PEG tube present (%) | 6 (4.8) |
Table of descriptive statistics for the overall participant study population. Continuous variables represented as the median (25th–75th percentile) and for categorical variables as n (%). P values correspond to Wilcoxon rank-sum tests for continuous variables and χ2 tests for categorical variables. C9orf72 status and ALS family history do not have p values because counts were too small for hypothesis testing.
*Median, 25th percentile and 75th percentile are computed using 123 cases and 69 controls (2 cases and 2 controls are missing).
†Median, 25th percentile and 75th percentile are computed using 117 cases and 61 controls (8 cases and 10 controls are missing).
ALS, amyotrophic lateral sclerosis; BMI, body mass index; GED, Graduate Equivalency Diploma; HS, high school; LS, lab supported; m, metres; NA, not available; PEG, percutaneous endoscopic gastrostomy.
Metabolite profiling
We identified 1051 known metabolites, which were evaluated by descriptive statistics by case/control status (see online supplemental table S1). Of the 1051 identified metabolites, 144 had missingness >60% and were removed from further analyses. Of the remaining 907 metabolites, 8 were known metabolites of commonly used drugs, which showed only weak or no correlations with the 899 endogenous metabolites (see online supplemental figure S1). Metabolites with negative correlations of <−0.2, which would bias the association away from the null hypothesis, did not significantly differ between cases and controls; therefore, their impact is negligible. We also separately examined riluzole, which was removed due to >60% missingness in cases and controls. Riluzole also had only weak correlations with other metabolites (data not shown). Overall, since these drug metabolites did not have strong correlations (ρ>0.5) with other metabolites, because drug metabolites had a high rate of missingness, and because typical drugs used in ALS have not been shown to influence metabolite results after drug washout,25 we did not include the drug metabolites in the analysis. This ensured that drug metabolites would not influence case/control association status. Thus, 899 metabolites were used for case/control downstream analysis.
Differential case/control metabolites
Wilcoxon rank-sum tests identified 303 significant differential metabolites (adjusted p value <0.05), which we visualised in a volcano plot (see online supplemental figure S2) and in significance plots at the super-pathway and sub-pathway levels (see online supplemental figure S3).
Adjusted differential case/control metabolites
Next, the logistic regression models, adjusted for sex, age and BMI, identified 300 metabolites presented in significance plots at the super-pathway and sub-pathway levels (see online supplemental figure S4) and as ORs for metabolites by sub-pathway (see online supplemental figure S5). There were differences among metabolites selected by unadjusted Wilcoxon rank-sum test (see online supplemental figure S3) versus logistic regression analysis, adjusted for sex, age and BMI (see online supplemental figure S4). Thirty-four metabolites identified by unadjusted Wilcoxon were no longer statistically significant after adjusting for clinical factors (see online supplemental figure S6A). On the other hand, adjusted logistic regression uniquely selected 31 metabolites, including glutamate-related metabolites, suggesting that sex, age and BMI may significantly affect metabolite relationships between cases and controls.
PLS-DA differential case/control metabolites
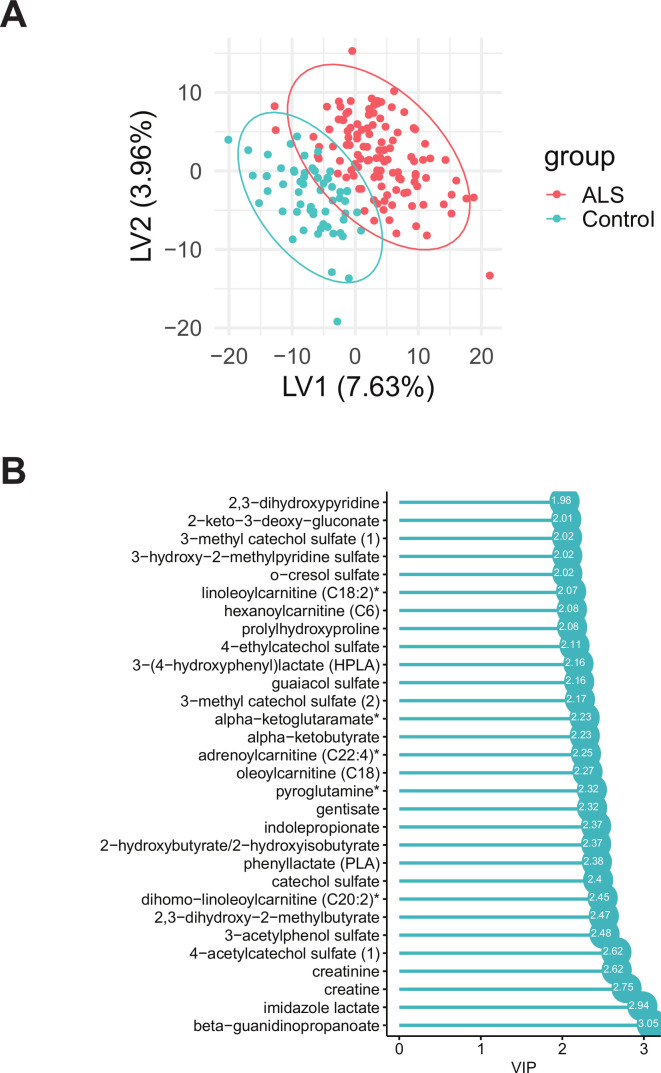
PLS-DA identified 295 metabolites with VIP >1 that separated cases from controls (figure 1A). The top 30 metabolites, with highest VIP and greatest contribution to case/control separation, are presented in a VIP score plot (figure 1B). Top metabolites (sub-pathway) with the highest VIP included beta-guanidinopropanoate (xenobiotics), imidazole lactate (histidine metabolism), creatine (creatine metabolism), creatinine (creatine metabolism) and 4-acetylcatechol sulfate (xenobiotics).
Figure 1.
Partial least squares-discriminant analysis (PLS-DA) analysis of amyotrophic lateral sclerosis (ALS) cases versus controls. (A) PLS-DA score plot of ALS cases (red) versus controls (blue); each dot represents an individual subject. (B) The variable importance in projection (VIP) score plot of the top 30 PLS-DA metabolites, which most significantly separate cases from controls. A total of 295 metabolites had VIP >1. Asterisks denote compounds that have not been confirmed against a standard, but whose identity the analytical platform is confident in.
Group lasso differential case/control metabolites
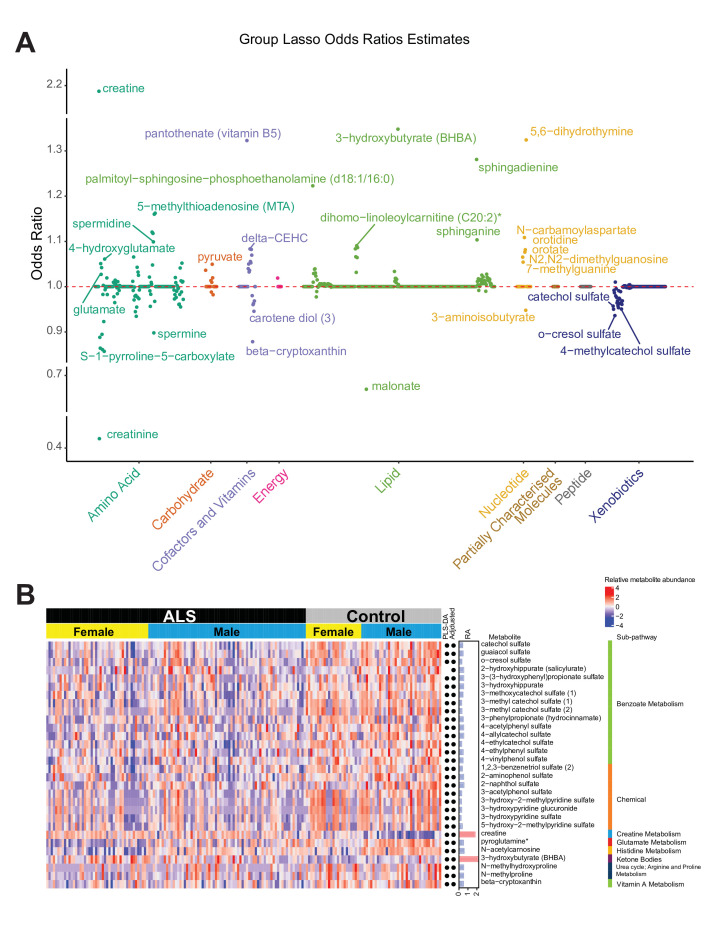
Sub-pathways shared by metabolites provide biologically meaningful groupings and are critical for a deeper understanding of disease pathomechanisms. Group lasso incorporates additional group information (ie, sub-pathways) when determining significant differences between cases versus controls. Sex-adjusted, age-adjusted and BMI-adjusted group lasso identified 259 differential metabolites (ORs≠1) across 32 sub-pathways (see online supplemental figure S7). Heatmaps illustrate the 30 metabolites with the highest differences in relative abundance between case and control groups (figure 2) and also for all metabolites (see online supplemental figure S8). Group lasso-selected metabolites did not overlap fully with the other models (see online supplemental figure S9). Whereas, unadjusted Wilcoxon, adjusted logistic regression and PLS-DA only uniquely identified 19, 7 and 3 metabolites, respectively, group lasso uniquely selected 123 metabolites. These model differences are further highlighted by enriched pathways, as discussed later, and demonstrate the group lasso effect of considering sub-pathways for discriminating cases from controls.
Figure 2.
Group lasso of amyotrophic lateral sclerosis (ALS) cases versus controls. (A) The ORs of each metabolite within its respective super-pathway. The dashed red line represents an OR of 1. All OR values are available in online supplemental table S4. (B) A heatmap of relative levels for the 30 metabolites with the highest differences in relative abundance in cases versus controls. Metabolites selected by partial least squares-discriminant analysis and adjusted models are marked by a black dot next to the differences in relative abundance (RA) column. Pink bars, OR >1, RA >0 (over-represented metabolites); blue bars, OR <1, RA <0 (under-represented metabolites). A heatmap of all metabolites is available in online supplemental figure S8. Asterisks denote compounds that have not been confirmed against a standard, but whose identity the analytical platform is confident in.
Pathway enrichment in case/control
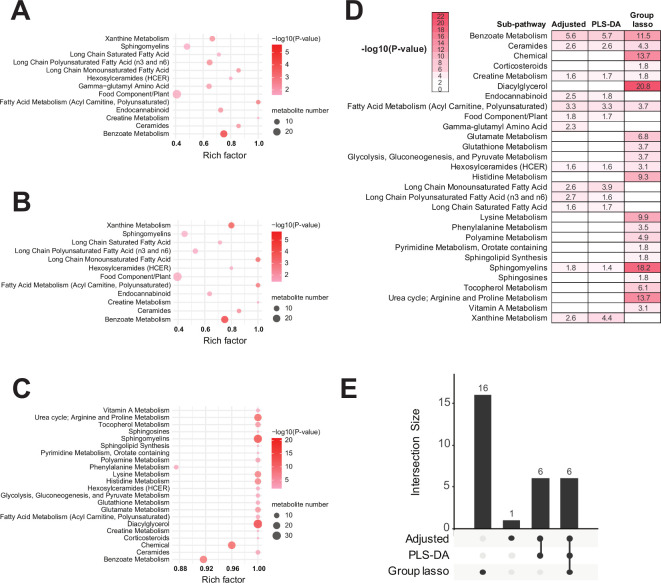
Metabolomics pathway enrichment analysis identified significantly over-represented sub-pathways among the differential metabolites. There were 11 sub-pathways enriched by unadjusted Wilcoxon-selected metabolites, 13 sub-pathways by adjusted logistic regression, 12 sub-pathways by PLS-DA and 22 sub-pathways by group lasso (see online supplemental figures S6C–E, figure 3). Unadjusted Wilcoxon, compared with adjusted logistic regression, enriched ‘tyrosine metabolism’ but not ‘sphingomyelins’, ‘gamma-glutamyl amino acid’ and ‘endocannabinoid’, indicating that sex, age and BMI adjustment may be important for interpreting ALS metabolomics data. Thus, we focused on the adjusted logistic regression, PLS-DA and group lasso models, which shared ‘benzoate metabolism’, ‘ceramides’, ‘creatine metabolism’, ‘fatty acid metabolism (acyl carnitine, polyunsaturated)’, ‘hexosylceramides’ and ‘sphingomyelins’ sub-pathways, as represented by heatmap and upset overlap plot (figure 3D, E, respectively). These pathways significantly associate with ALS. Group lasso also uniquely identified 16 sub-pathways, including, among the most significant, ‘diacylglycerol’, ‘chemical’, ‘urea cycle; arginine and proline metabolism’, ‘lysine metabolism’, ‘histidine metabolism’ and ‘glutamate metabolism’. Since these metabolic pathways are significant to ALS (see the Discussion section), this enrichment analysis suggests group lasso is an informative analytical technique for this metabolomics dataset.
Figure 3.
Pathway enrichment of adjusted logistic regression-selected, partial least squares-discriminant analysis (PLS-DA)-selected and group lasso-selected metabolites. Significantly enriched sub-pathways from metabolites selected by (A) adjusted logistic regression, (B) PLS-DA and (C) group lasso models illustrated in dot plots. Each significantly selected sub-pathway is represented by a circle characterised by three parameters. (1) The circle size represents how many metabolites were selected in the sub-pathway (see legend in grey to right of the plot for relative sizes). (2) The circle shading from light pink to red indicates the selected sub-pathway significance level according to –log10 (p value) (see legend to the right of the plot for relative colour gradient). Sub-pathways were considered significantly enriched if they met the threshold p value <0.05, which is equivalent to –log10 (p value) >1.3. (3) The circle position along the rich factor axis specifies the proportion of selected metabolites from the sub-pathway against all sub-pathway metabolites. (D) Heatmap of all significantly enriched sub-pathways and their represented in colour gradient and value according to –log10(p value). Any cell with a –log10(p value) indicates the sub-pathway was significantly enriched in metabolites selected by the corresponding models; p value <0.05 is equivalent to –log10 (p value) >1.3. (E) Upset plot of enriched pathway overlap across the three models.
Metabolite correlation analysis in case/control
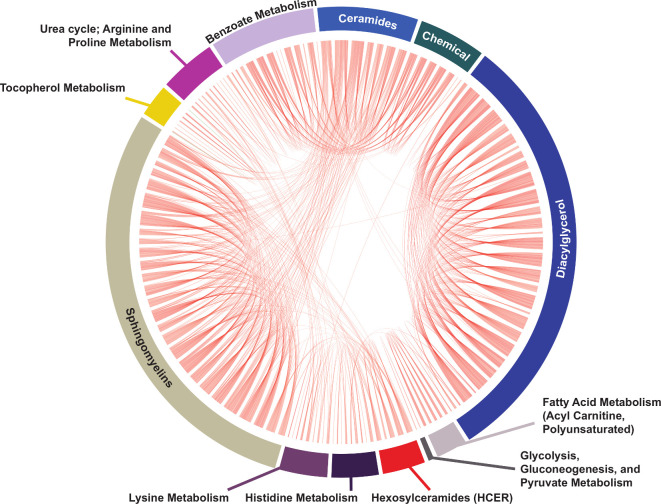
We performed a data-driven correlation analysis of the group lasso-selected metabolites to visualise interconnections between significant metabolites and their respective sub-pathways (figure 4 and online supplemental figure S10). Most interconnections arose between ‘ceramides’, ‘hexosylceramides’, ‘sphingomyelins’ and ‘diacylglycerol’ sub-pathways. Further, ‘diacylglycerol’ metabolites correlated with metabolites within ‘histidine metabolism’, ‘fatty acid metabolism (acyl carnitine, polyunsaturated)’ and ‘glutathione metabolism’ sub-pathways. Additionally, ‘sphingolipid synthesis’ metabolites correlated with ‘sphingosines’ and ‘polyamine metabolism’ metabolites. ‘Lysine metabolism’ had multiple interconnections to ‘glutamate’, ‘histidine’, ‘creatine’, ‘urea cycle; arginine and proline’ and ‘guanine containing purine’ metabolism.
Figure 4.
Metabolite correlation analysis of group lasso-selected metabolites. Correlation analysis depicted in a simplified chord diagram format of 259 significant group lasso-selected metabolites (p value <0.05) represented by chords that connect intra-sub-pathway or inter-sub-pathway significantly correlated metabolites (Spearman correlation coefficient ρ>0.5). Each sub-pathway is depicted by an annotated arc of the circle sized proportionately to the number of selected metabolites is contains. Chord correlations are colour coded by the correlation sign (red, positive, the vast majority of correlations; green, negative). Metabolite names are omitted for clarity; some sub-pathways are missed for technical reasons. The full correlation analysis, depicted in correlation network format and including all metabolite names and involved sub-pathways, is shown in online supplemental figure S10.
Classification models for predicting ALS cases
To assess the predictive power of the 259-group lasso-selected metabolites, we built and validated seven different machine learning algorithms, whose performance was evaluated by SENS, SPEC and AUC (figure 5 and online supplemental figure S11). RLR performed the best, with an AUC of 0.98. All seven models incorporated creatine, creatinine and imidazole lactate into their classification algorithms, a finding consistent with their higher absolute ORs in the case/control association models. Thus, group lasso selected metabolites were sufficient to differentiate cases and controls.
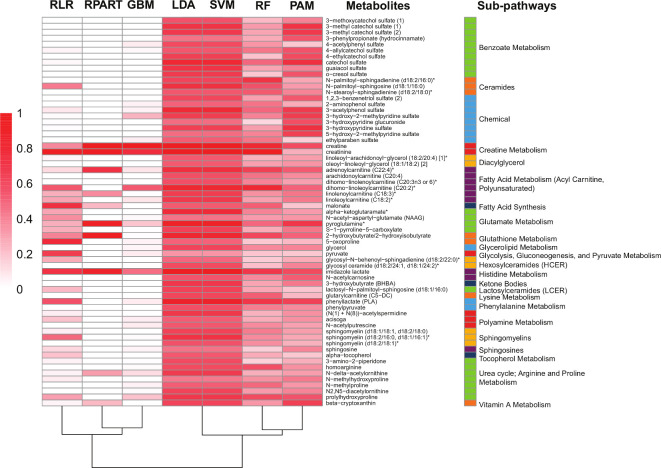
Figure 5.
Machine learning (ML) classification of group lasso-selected metabolites. Heatmap representation of metabolite importance score from the different ML models. Scores are scaled from 0 (not important) to 1 (very important) to the model’s performance. Asterisks denote compounds that have not been confirmed against a standard, but whose identity the analytical platform is confident in. ML operating characteristics are provided in online supplemental figure S11. GBM, generalised boosted models; LDA, linear discriminant analysis; PAM, prediction analysis for microarrays; RF, random forest; RLR, regularised logistic regression with elastic net regularisation; RPART, recursive partitioning and regression trees; SVM, support vector machine.
Discussion
Metabolomic profiles reflect the cumulative effect of both endogenous physiological processes and exogenous influences. Herein, we harnessed the power of advanced untargeted metabolomic analyses to gain insight into ALS disease mechanisms to identify new therapeutic opportunities. We analysed 899 metabolites after excluding metabolites with high missingness and common drug metabolites. We also omitted riluzole from our final analyses, due to its high degree of missingness and lack of strong correlations to other metabolites, as seen previously.25 Three separate approaches identified differential metabolites in ALS, 303 by unadjusted Wilcoxon, 300 by adjusted logistic regression, 295 by PLS-DA, of which 261 overlapped. Independent of the modelling approach, we found shared, critical and significant differential metabolites and sub-pathways in ALS, yielding a distinct metabolomic signature that identified potential ALS biomarkers and drug targets. These findings represent the greatest number of differential metabolites reported in ALS patients versus controls, likely due to our large dataset of 899 metabolites. Among other studies with a rich dataset,13 one identified 404 metabolites, of which 31 associated with ALS risk,14 and another identified 367 metabolites, of which 62 were significantly altered in ALS.15
PLS-DA, commonly used to identify metabolites that significantly separate cases from controls, identified creatine, creatinine and beta-guanidinopropanoate, a creatine analogue, as differential in ALS versus controls. Creatine, which is synthesised in the liver or ingested as a dietary supplement, is transported to muscle, where it recycles ATP for muscle contraction, before eventual conversion to creatinine. Previous targeted analysis shows these pathways are dysregulated in ALS,26 a relationship that persists in our untargeted analyses, where creatine metabolism tops the list of dysregulated metabolites and pathways. Clinical studies found higher creatine and lower creatinine in ALS plasma15 and cerebrospinal fluid27; plasma creatinine correlated with Revised ALS Functional Rating Scale (ALSFRS-R) scores.15 Yet, other groups did not identify changes in creatine-to-creatinine ratios in serum or erebrospinal fluid (CSF) from ALS patients versus controls.7 28 These discrepancies likely arise from ALS stage, since creatinine or changes in its levels correlate with disease severity.15 26 Dietary creatine supplementation failed to improve ALSFRS-R in a multicentre clinical trial,29 suggesting these metabolites are muscle dysfunction biomarkers and not therapeutic targets.
Besides PLS-DA, we also analysed our metabolomics data using clinically guided sex-adjusted, age-adjusted and BMI-adjusted logistic regression and group lasso models. Many metabolites exist within more than one network, which are in turn interconnected. Within these interconnected networks, some metabolites may be over-represented, whereas others may be underrepresented in these pathways, depending on disease states. In the current study, we reported individual metabolites, for example, creatine, creatinine, beta-guanidinopropanoate, but mostly focused our analyses on discovering significant pathway changes in ALS. Among the most significantly enriched pathways, ‘sphingomyelins’, ‘ceramides’, ‘benzoate metabolism’ and ‘fatty acid metabolism’ were shared among all three PLS-DA, logistic regression, group lasso, while group lasso also identified ‘diacylglycerol’, ‘chemical’, ‘urea cycle’ and ‘histamine’, ‘lysine’ and ‘glutamate’ metabolism, among others. Among pathways that were selected by logistic regression and PLS-DA but not by group lasso, ‘xanthine metabolism’ was the most significant, followed by ‘long chain monounsaturated fatty acid’. Overall, however, we suggest group lasso may be a useful approach, since our analyses show that accounting for sub-pathway architecture in complex diseases, such as ALS, may reveal additional significant insight into disease pathogenesis.
Using group lasso-selected metabolites, several prominent sub-pathways emerged that were highly significant in ALS, especially ‘diacylglycerol’ and ‘sphingomyelins’. Sphingolipids are bioactive signalling molecules in apoptosis, autophagy and inflammation,30 which are all ALS hallmarks.31 Altered sphingolipids included sphingomyelins, sphingosines, ceramides and hexosylceramides, which have been previously reported in ALS participant plasma,14 15 25 CSF,8 and spinal cord,32 33 and ALS mouse models.32–35 Targeting sphingolipid metabolism with sphingolipid inhibitors is at the forefront of cancer therapeutics,36 and has been advocated as a possible treatment for Alzheimer’s, Parkinson’s and Huntington’s disease.37 38 Our study provides evidence that sphingolipid metabolism may similarly be targeted in ALS as a potential therapeutic opportunity.
Group lasso also selected sub-pathway metabolites in arginine, proline,6 15 27 lysine,6 histidine6 28 39 and polyamine metabolism,6 as seen in ALS human and animal models. Polyamines, including spermidine, have pro-autophagy, immunomodulatory and neuroprotective properties,40 all dysregulated in ALS pathology.31 41 Spermidine rescues motor dysfunction in TDP-43 proteinopathy mice,42 and arginine, a source for spermidine biosynthesis, lengthens survival in mutant SOD1 ALS mice.43 Dietary spermidine is currently being investigated in a phase IIb trial as a treatment for cognitive decline,44 and represents another unique and untested opportunity for ALS therapeutics.
Oxidative stress and biomarkers are also pervasive in ALS,45 and this was observed in our study through enriched sub-pathways ‘glutathione metabolism’ and ‘tocopherol metabolism’. Glutathione is the most abundant intracellular thiol antioxidant that scavenges reactive oxygen species (ROS) and is also involved in xenobiotic elimination.46 Glutathione deficiencies, as occurs in ALS patients47 and culture models,48 would lead to a diminished capacity to clear ROS and elevated oxidative stress.45 Other metabolic studies also found differences in glutathione metabolism49 50 and other antioxidants, such as tocopherol15 and ascorbate,7 27 in ALS patients and model systems. Unfortunately, antioxidant therapies for ALS,51 with the exception of a moderate effect from edaravone, have failed to date, suggesting that dysregulated pathways may be a consequence of disease progression, rather than a cause.
Higher premorbid BMI is associated with slower ALS progression.52 As anticipated, we identified significant changes in sub-pathways involved in energy metabolism, a BMI marker, including ‘fatty acid metabolism (acyl carnitines, polyunsaturated)’,14 ‘glycolysis, gluconeogenesis and pyruvate metabolism’,7 25 and generally altered lipid metabolism.8 15 35 Interestingly, C9orf72 expansions, the most prevalent ALS genetic mutation, may interfere with lipid metabolism through coactivator-associated arginine methyltransferase 1, leading to lipid hypermetabolism.53 Another lipid pathway, ‘diacylglycerol’, was the most significant sub-pathway selected by group lasso. One other study reported diacylglycerols associated negatively with pre-symptomatic ALS versus healthy participants.14 Diacylglycerols are bioactive signalling lipids with roles in cytoskeleton, neuronal development, inflammation, immune cell signalling and apoptosis.54 Although these cellular processes are well-known in ALS pathology,31 41 the contribution specifically of diacylglycerols to ALS pathogenesis is unstudied and unknown. Of interest, diacylglycerol kinases, a family of enzymes that convert diacylglycerols to phosphatidic acids, is another cancer therapeutic target,55 highlighting the clinical importance of these bioactive signalling diacylglycerols, and promoting them to the list of potential new mechanism-based ALS targets.
We4 5 and others9 have shown that xenobiotic exposure from the environment may contribute to ALS. In our correlation analysis, ‘benzoate metabolism’ and ‘chemical’, both from the xenobiotic super-pathway, consisted of many metabolites containing cresol and catechol groups, which are common pesticide moieties. An integrated metabolomic and genomic study also found two xenobiotics that correlated with ALS,50 as have other studies.15 25 Single nucleotide variants of flavin-containing monooxygenases, oxidative enzymes of xenobiotics, pesticides and drugs, are more prevalent in female ALS participants,56 also underscoring that detoxification mechanisms in ALS patients may be impaired and contribute to disease onset and/or progression.
Other than genetics, and only for patients harbouring a known ALS mutation, there is currently no molecular diagnostic ALS test. We applied machine learning to the 259-group lasso-selected metabolites using seven different algorithms to determine whether these metabolites could differentiate cases from controls, and potentially serve as a diagnostic tool. RLR performed the best overall, with an AUC of 0.98% and 83% specificity, but SVM was the most sensitive (97%), suggesting machine learning guided metabolomics may be able to differentiate ALS from healthy participants. Creatine, creatinine and imidazole lactate were incorporated by all machine learning algorithms, again underscoring creatine metabolism as a possible ALS biomarker. However, our analysis was performed on plasma from individuals who had already developed ALS. It would be more useful if a diagnostic tool could identify ALS early, when treatment may be more effective. Bjornevik et al found RF and SVM could not predict whether an individual would develop ALS later in life based on an early, pre-symptomatic metabolic profile.14 Their study findings did not change when they restricted their analyses to blood draws taken within 1 (n=51) or 2 years (n=72) of ALS diagnosis. The authors suggested that since ALS is so rapidly progressing, it may not be preceded by a long preclinical stage, or that the preclinical stage may not be characterised by metabolic changes, which may rather possibly be caused by the disease. Therefore, the temporal shifts in ALS metabolic profiles prior to symptom onset remain uncertain. Since we only included ALS participants in our analysis, we cannot evaluate whether we would be able to differentiate them from individuals with a different neurological disease. Indeed, Lawton et al found that RF correctly predicted ALS versus a neurological mimic only 63% of the time, but correctly predicted ALS versus controls more often at 77%.15 Future ALS metabolomics studies as a diagnostic tool will need to analyse biosamples as early as possible in disease course and compare them to metabolic profiles from patients with similar diseases.
Our study has several strengths, foremost is the sample size; this is one of the largest untargeted metabolomics study of ALS patients versus controls, which also benefited from a large number of well-characterised metabolites. Furthermore, untargeted metabolomics evaluated aberrant pathways agnostically. By using group lasso, we adjusted for covariates, modelled all metabolites jointly and incorporated biologically meaningful groupings by sub-pathway membership. Our study does have limitations; because it was untargeted, we could not measure every metabolite in each pathway. Additionally, while our patient sample size was large, the number of metabolites surpassed sample size. We were also unable to completely control for heterogeneity, since ALS participants differed in disease stage, segment onset and genetic background, which may all impact metabolic profiles. Additionally, our study was cross-sectional, so we were unable to assess temporal changes in metabolites to highlight earlier versus later events in disease development. This also contributes towards differentiating metabolic processes that are integral to the primary disease process versus metabolic processes that may occur as downstream consequences of disease pathogenesis. Plasma analysis also reflects the systemic effect of disease development, and does not evaluate shifts in the local metabolomics milieu in certain diseased tissues. Moreover, since we only collected samples from diagnosed ALS patients, we could not determine whether there may be differences in pre-symptomatic ALS metabolite profiles that could be used to predict disease development. Finally, our study did not include plasma samples from patients with other neurological diseases, so we are unable to determine how specific the identified metabolite profiles were to ALS patients.
In summary, this untargeted metabolomics study found evidence of pathway abnormalities in ALS, both among previously established pathways (eg, energy homoeostasis, amino acid metabolism) and novel or emerging pathways (eg, diacylglycerols, benzoate metabolism), which may lead to promising new therapeutic targets for disease modification (figure 6). Targeted mechanism-based therapies are needed in ALS, and metabolomics studies can help address these research needs. Moreover, longitudinal and mechanistic studies will be able to reveal pathways preceding neurodegeneration, which will also aid in the drug discovery process.
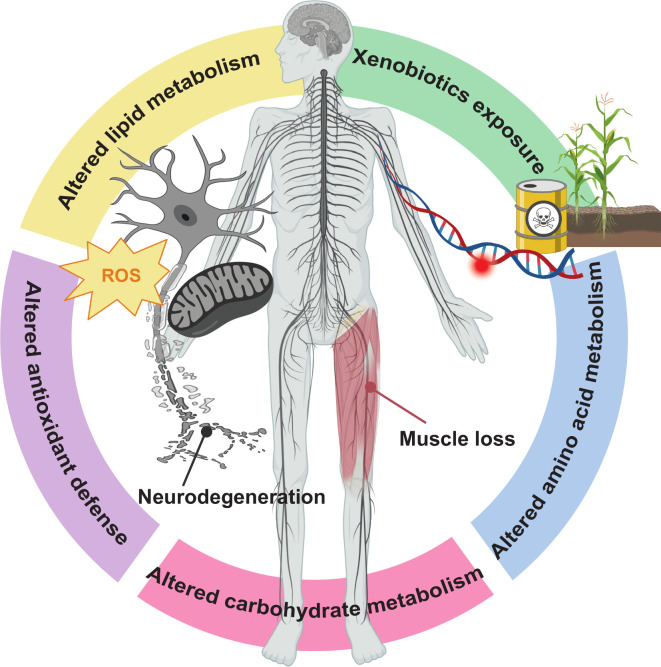
Figure 6.
Overview of metabolomics-driven pathway discovery and validation in amyotrophic lateral sclerosis (ALS). Our metabolomics analysis identified emerging pathways, which could lead to novel avenues of investigation, for example, xenobiotics and diacylglycerols. Xenobiotic exposure, for example, chemical and benzoate metabolism sub-pathways, could interact with genetic risk to increase the chance of developing ALS. The diacylglycerol (DAG) sub-pathway, belonging to the lipid metabolism super-pathway, includes signalling lipid DAG molecules with diverse biological roles, such cytoskeletal, neuronal development, inflammation, immune cell signalling and apoptosis. Although these are all aspects of ALS pathology, the DAG contribution specifically to ALS pathogenesis is not known. We also identified previously known pathways, validating our approach, for example, energy dyshomoeostasis, through lipid and carbohydrate metabolism, altered antioxidant defense and amino acid metabolism. Dysregulated lipid and carbohydrate metabolism and loss of antioxidant defense against reactive oxygen species (ROS), for instance involving mitochondria, could lead to neurodegeneration. Altered amino acid metabolism, for instance through the creatine metabolism sub-pathway, may be a consequence of muscle loss.
Acknowledgments
We are indebted to the study participants that provided samples. We thank Crystal Pacut, Stacey Jacoby, PhD, Blake Swihart, Jayna Duell, RN, Daniel Burger and Amanda Williams.
Footnotes
Contributors: SAG contributed to the study design and conceptualisation, funding acquisition, major role in data acquisition, data interpretation and manuscript writing for intellectual content. JB contributed to data analysis and manuscript revising for intellectual content. KG contributed to data analysis and manuscript revising for intellectual content. FMA contributed to data analysis and manuscript revising for intellectual content. AP contributed to data management, data interpretation and presentation of data for the manuscript. SK contributed to data analysis and manuscript revising for intellectual content. MGS contributed to data interpretation and manuscript writing and revising for intellectual content. JH contributed to data analysis and manuscript writing for intellectual content. EF contributed to the study design and conceptualisation, funding acquisition, major role in data acquisition, data interpretation, and manuscript writing and revising for intellectual content.
Funding: National ALS Registry/CDC/ATSDR (1R01TS000289); National ALS Registry/CDC/ATSDR CDCP-DHHS-US (CDC/ATSDR 200-2013-56856); NIEHS K23ES027221; NIEHS R01ES030049; NeuroNetwork for Emerging Therapies, University of Michigan; National Center for Advancing Translational Sciences at the National Institutes of Health (UL1TR002240).
Competing interests: SAG has provided medical advisory for Biogen and ITF Pharma and served on a DSMB. JB, KG, FMA, AP, SK, MGS and JH report no disclosures relevant to the manuscript. EF sits on an advisory board for Novartis.
Patient consent for publication: Not required.
Ethics approval: All participants provided verbal and written informed consent for this study, which had institutional review board approval at the University of Michigan (HUM00028826).
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available upon reasonable request. Anonymised data will be shared by request from any qualified investigator.
References
- 1. Goutman SA. Diagnosis and clinical management of amyotrophic lateral sclerosis and other motor neuron disorders. Continuum 2017;23:1332–59. 10.1212/CON.0000000000000535 [DOI] [PubMed] [Google Scholar]
- 2. Goutman SA, Chen KS, Paez-Colasante X, et al. . Emerging understanding of the genotype-phenotype relationship in amyotrophic lateral sclerosis. Handb Clin Neurol 2018;148:603–23. 10.1016/B978-0-444-64076-5.00039-9 [DOI] [PubMed] [Google Scholar]
- 3. Chia R, Chiò A, Traynor BJ. Novel genes associated with amyotrophic lateral sclerosis: diagnostic and clinical implications. Lancet Neurol 2018;17:94–102. 10.1016/S1474-4422(17)30401-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goutman SA, Boss J, Patterson A, et al. . High plasma concentrations of organic pollutants negatively impact survival in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry 2019;90:907–12. 10.1136/jnnp-2018-319785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Su F-C, Goutman SA, Chernyak S, et al. . Association of environmental toxins with amyotrophic lateral sclerosis. JAMA Neurol 2016;73:803–11. 10.1001/jamaneurol.2016.0594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Patin F, Corcia P, Vourc'h P, et al. . Omics to explore amyotrophic lateral sclerosis evolution: the central role of arginine and proline metabolism. Mol Neurobiol 2017;54:5361–74. 10.1007/s12035-016-0078-x [DOI] [PubMed] [Google Scholar]
- 7. Blasco H, Corcia P, Moreau C, et al. . 1H-NMR-based metabolomic profiling of CSF in early amyotrophic lateral sclerosis. PLoS One 2010;5:e13223. 10.1371/journal.pone.0013223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blasco H, Veyrat-Durebex C, Bocca C, et al. . Lipidomics reveals cerebrospinal-fluid signatures of ALS. Sci Rep 2017;7:17652. 10.1038/s41598-017-17389-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Al-Chalabi A, Hardiman O. The epidemiology of ALS: a conspiracy of genes, environment and time. Nat Rev Neurol 2013;9:617–28. 10.1038/nrneurol.2013.203 [DOI] [PubMed] [Google Scholar]
- 10. Yoshino H, Kimura A. Investigation of the therapeutic effects of edaravone, a free radical scavenger, on amyotrophic lateral sclerosis (phase II study). Amyotroph Lateral Scler 2006;7:241–5. 10.1080/17482960600881870 [DOI] [PubMed] [Google Scholar]
- 11. Athersuch T. Metabolome analyses in exposome studies: profiling methods for a vast chemical space. Arch Biochem Biophys 2016;589:177–86. 10.1016/j.abb.2015.10.007 [DOI] [PubMed] [Google Scholar]
- 12. Blasco H, Patin F, Madji Hounoum B, et al. . Metabolomics in amyotrophic lateral sclerosis: how far can it take us? Eur J Neurol 2016;23:447–54. 10.1111/ene.12956 [DOI] [PubMed] [Google Scholar]
- 13. Rozen S, Cudkowicz ME, Bogdanov M, et al. . Metabolomic analysis and signatures in motor neuron disease. Metabolomics 2005;1:101–8. 10.1007/s11306-005-4810-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bjornevik K, Zhang Z, O'Reilly Éilis J, et al. . Prediagnostic plasma metabolomics and the risk of amyotrophic lateral sclerosis. Neurology 2019;92:e2089–100. 10.1212/WNL.0000000000007401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lawton KA, Brown MV, Alexander D, et al. . Plasma metabolomic biomarker panel to distinguish patients with amyotrophic lateral sclerosis from disease mimics. Amyotroph Lateral Scler Frontotemporal Degener 2014;15:362–70. 10.3109/21678421.2014.908311 [DOI] [PubMed] [Google Scholar]
- 16. Dehaven CD, Evans AM, Dai H, et al. . Organization of GC/MS and LC/MS metabolomics data into chemical libraries. J Cheminform 2010;2:9. 10.1186/1758-2946-2-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Evans A, Bridgewater B, Liu Q, et al. . High resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics 2014;4. [Google Scholar]
- 18. Rohart F, Gautier B, Singh A, et al. . mixOmics: An R package for 'omics feature selection and multiple data integration. PLoS Comput Biol 2017;13:e1005752. 10.1371/journal.pcbi.1005752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Galindo-Prieto B, Eriksson L, Trygg J. Variable influence on projection (VIP) for orthogonal projections to latent structures (OPLS). J Chemom 2014;28:623–32. 10.1002/cem.2627 [DOI] [Google Scholar]
- 20. Cho H-W, Kim SB, Jeong MK, et al. . Discovery of metabolite features for the modelling and analysis of high-resolution NMR spectra. Int J Data Min Bioinform 2008;2:176–92. 10.1504/ijdmb.2008.019097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuhn M. Building predictive models in R using the CARET package 2008;28:26. [Google Scholar]
- 22. Chawla NV, Bowyer KW, Hall LO, et al. . SMOTE: synthetic minority over-sampling technique. J Artif Int Res 2002;16:321–57. [Google Scholar]
- 23. Robin X, Turck N, Hainard A, et al. . pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:77. 10.1186/1471-2105-12-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bandres-Ciga S, Noyce AJ, Hemani G, et al. . Shared polygenic risk and causal inferences in amyotrophic lateral sclerosis. Ann Neurol 2019;85:470–81. 10.1002/ana.25431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lawton KA, Cudkowicz ME, Brown MV, et al. . Biochemical alterations associated with ALS. Amyotroph Lateral Scler 2012;13:110–8. 10.3109/17482968.2011.619197 [DOI] [PubMed] [Google Scholar]
- 26. Ito D, Hashizume A, Hijikata Y, et al. . Elevated serum creatine kinase in the early stage of sporadic amyotrophic lateral sclerosis. J Neurol 2019;266:2952–61. 10.1007/s00415-019-09507-6 [DOI] [PubMed] [Google Scholar]
- 27. Wuolikainen A, Jonsson P, Ahnlund M, et al. . Multi-platform mass spectrometry analysis of the CSF and plasma metabolomes of rigorously matched amyotrophic lateral sclerosis, Parkinson's disease and control subjects. Mol Biosyst 2016;12:1287–98. 10.1039/c5mb00711a [DOI] [PubMed] [Google Scholar]
- 28. Kumar A, Bala L, Kalita J, et al. . Metabolomic analysis of serum by (1) H NMR spectroscopy in amyotrophic lateral sclerosis. Clinica Chimica Acta 2010;411:563–7. 10.1016/j.cca.2010.01.016 [DOI] [PubMed] [Google Scholar]
- 29. Shefner JM, Cudkowicz ME, Schoenfeld D, et al. . A clinical trial of creatine in ALS. Neurology 2004;63:1656–61. [DOI] [PubMed] [Google Scholar]
- 30. Hannun YA, Obeid LM. Sphingolipids and their metabolism in physiology and disease. Nat Rev Mol Cell Biol 2018;19:175–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang ML, Chiang S, Kalinowski DS, et al. . The role of the antioxidant response in mitochondrial dysfunction in degenerative diseases: cross-talk between antioxidant defense, autophagy, and apoptosis. Oxid Med Cell Longev 2019;2019:6392763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cutler RG, Pedersen WA, Camandola S, et al. . Evidence that accumulation of ceramides and cholesterol esters mediates oxidative stress-induced death of motor neurons in amyotrophic lateral sclerosis. Ann Neurol 2002;52:448–57. 10.1002/ana.10312 [DOI] [PubMed] [Google Scholar]
- 33. Dodge JC, Treleaven CM, Pacheco J, et al. . Glycosphingolipids are modulators of disease pathogenesis in amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A 2015;112:8100–5. 10.1073/pnas.1508767112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Henriques A, Croixmarie V, Bouscary A, et al. . Sphingolipid metabolism is dysregulated at transcriptomic and metabolic levels in the spinal cord of an animal model of amyotrophic lateral sclerosis. Front Mol Neurosci 2017;10:433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chaves-Filho AB, Pinto IFD, Dantas LS, et al. . Alterations in lipid metabolism of spinal cord linked to amyotrophic lateral sclerosis. Sci Rep 2019;9:11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shaw J, Costa-Pinheiro P, Patterson L, et al. . Novel sphingolipid-based cancer therapeutics in the personalized medicine era. Adv Cancer Res 2018;140:327–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Angelopoulou E, Piperi C. Beneficial effects of fingolimod in Alzheimer's disease: molecular mechanisms and therapeutic potential. Neuromolecular Med 2019;21:227–38. [DOI] [PubMed] [Google Scholar]
- 38. Di Pardo A, Maglione V. Sphingolipid metabolism: a new therapeutic opportunity for brain degenerative disorders. Front Neurosci 2018;12:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Blasco H, Nadal-Desbarats L, Pradat P-F, et al. . Biomarkers in amyotrophic lateral sclerosis: combining metabolomic and clinical parameters to define disease progression. Eur J Neurol 2016;23:346–53. 10.1111/ene.12851 [DOI] [PubMed] [Google Scholar]
- 40. Madeo F, Eisenberg T, Pietrocola F, et al. . Spermidine in health and disease. Science 2018;359. [DOI] [PubMed] [Google Scholar]
- 41. Murdock BJ, Bender DE, Segal BM, et al. . The dual roles of immunity in ALS: injury overrides protection. Neurobiol Dis 2015;77:1–12. 10.1016/j.nbd.2015.02.017 [DOI] [PubMed] [Google Scholar]
- 42. Wang IF, Guo BS, Liu YC, et al. . Autophagy activators rescue and alleviate pathogenesis of a mouse model with proteinopathies of the TAR DNA-binding protein 43. Proc Natl Acad Sci U S A 2012;109:15024–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee J, Ryu H, Kowall NW. Motor neuronal protection by L-arginine prolongs survival of mutant SOD1 (G93A) ALS mice. Biochem Biophys Res Commun 2009;384:524–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wirth M, Schwarz C, Benson G, et al. . Effects of spermidine supplementation on cognition and biomarkers in older adults with subjective cognitive decline (SmartAge)-study protocol for a randomized controlled trial. Alzheimers Res Ther 2019;11:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. D'Amico E, Factor-Litvak P, Santella RM, et al. . Clinical perspective on oxidative stress in sporadic amyotrophic lateral sclerosis. Free Radic Biol Med 2013;65:509–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Forman HJ, Zhang H, Rinna A. Glutathione: overview of its protective roles, measurement, and biosynthesis. Mol Aspects Med 2009;30:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Weiduschat N, Mao X, Hupf J, et al. . Motor cortex glutathione deficit in ALS measured in vivo with the J-editing technique. Neurosci Lett 2014;570:102–7. [DOI] [PubMed] [Google Scholar]
- 48. D'Alessandro G, Calcagno E, Tartari S, et al. . Glutamate and glutathione interplay in a motor neuronal model of amyotrophic lateral sclerosis reveals altered energy metabolism. Neurobiol Dis 2011;43:346–55. [DOI] [PubMed] [Google Scholar]
- 49. Veyrat-Durebex C, Corcia P, Piver E, et al. . Disruption of TCA cycle and glutamate metabolism identified by metabolomics in an in vitro model of amyotrophic lateral sclerosis. Mol Neurobiol 2016;53:6910–24. [DOI] [PubMed] [Google Scholar]
- 50. Yang L, Lv X, Du H, et al. . Causal effects of serum metabolites on amyotrophic lateral sclerosis: a Mendelian randomization study. Prog Neuropsychopharmacol Biol Psychiatry 2020;97:109771. [DOI] [PubMed] [Google Scholar]
- 51. Desnuelle C, Dib M, Garrel C, et al. . A double-blind, placebo-controlled randomized clinical trial of alpha-tocopherol (vitamin E) in the treatment of amyotrophic lateral sclerosis. ALS riluzole-tocopherol Study Group. Amyotroph Lateral Scler Other Motor Neuron Disord 2001;2:9–18. [DOI] [PubMed] [Google Scholar]
- 52. Dardiotis E, Siokas V, Sokratous M, et al. . Body mass index and survival from amyotrophic lateral sclerosis: a meta-analysis. Neurol Clin Pract 2018;8:437–44. 10.1212/CPJ.0000000000000521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu Y, Wang T, YJ J, et al. . A C9orf72-CARM1 axis regulates lipid metabolism under glucose starvation-induced nutrient stress. Genes Dev 2018;32:1380–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Toker A. The biology and biochemistry of diacylglycerol signalling. Meeting on molecular advances in diacylglycerol signalling. EMBO Rep 2005;6:310–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Purow B. Molecular pathways: targeting diacylglycerol kinase alpha in cancer. Clin Cancer Res 2015;21:5008–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cereda C, Gabanti E, Corato M, et al. . Increased incidence of FMO1 gene single nucleotide polymorphisms in sporadic amyotrophic lateral sclerosis. Amyotroph Lateral Scler 2006;7:227–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jnnp-2020-323611supp001.pdf (6.2MB, pdf)