Abstract
CYP2C9 gene encodes an enzyme involved in the metabolism of a wide variety of drugs which include celecoxib. This study investigated the frequencies of the alleles and genotypes of CYP2C9*1, CYP2C9*2, and CYP2C9*3 among Filipinos who underwent surgery, and to determine the association of CYP2C9 polymorphisms with post-operative pain relief via COX-2 inhibitors. Response to celecoxib was determined using the numerical rating scale (0-10) on the 24th and 48th hour of surgery. The CYP2C9 alleles were detected by real-time PCR. For CYP2C9*1 and CYP2C9*3, the allele frequencies among Filipinos were 99% and 1% respectively, which is similar with other East Asians. CYP2C9*2 alleles were not detected. The frequencies of CYP2C9*1/*1 and CYP2C9*1/*3 genotypes were 98% and 2% respectively. At 24 hours post-surgery, the average pain score was 2.57 ± 1.03, while on 48 hours post-surgery, the average pain score was 0.67 ± 0.61 among those who have the wild-type CYP2C9*1 allele. The average pain score on the 24th and 48th hour post-operatively was observed to be 2.5 ± 0.71 and 0.5 ± 0.71 respectively among two patients classified as intermediate metabolizer carrying the CYP2C9*1/*3 genotype. Low frequencies of CYP2C9 polymorphisms were observed in the present study, this pattern was similar with other Asians except Indians, and considerably lower than Caucasians. Our results suggest that CYP2C9 genotyping is not routinely needed for Filipinos but must be considered among mixed races. Consequently, a more personalized therapeutic strategy was derived from these data, resulting in good clinical outcomes and less adverse drug effects.
Keywords: Celecoxib, COX-2, CYP2C9*1, CYP2C9*2, CYP2C9*3, polymorphism, post-operative pain
Introduction
The cytochrome P450 2C9 (CYP2C9) is one of the enzymes responsible for the metabolism of a wide range of drugs such as anticoagulants, hypoglycemic agents, non-steroidal anti-inflammatory drugs (NSAIDs) and selective COX-2 inhibitors [1-4]. Polymorphisms in the CYP2C9 gene have been associated with decreased enzyme activity and alteration of celecoxib pharmacokinetic parameters. Particularly, individuals with low enzyme activity (e.g. slow metabolizers) have a high propensity for adverse drug reactions [5].
CYP2C9 gene is located at chromosome 10q23.33 which shows genetic polymorphisms. To date, more than 60 allelic variants of the CYP2C9 gene have been described [4]. CYP2C9*1 is the wild-type allele and is associated with normal enzyme activity and normal metabolism of drugs. The two most common variants are CYP2C9*2 and CYP2C9*3. The CYP2C9*2 allele is the result of a 430C>T transition in exon 3 leading to an Arg-to-Cys substitution at amino acid position 144 of the CYP2C9 molecule. On the other hand, CYP2C9*3 is the result of a 1075A>C transversion in exon 7 causing an Ile-to-Leu substitution at amino acid position 359. Studies have shown that both alleles are associated with significantly reduced enzyme activity [6-8]. In addition, carriers of the CYP2C9*3 variant are at risk for complications such as bleeding following use of warfarin. The frequencies of these two variants vary between different ethnic populations. For CYP2C9*2, the allele frequency among Caucasians ranges from 10-20%, whereas CYP2C9*2 has not been detected among East Asian populations such as Chinese, Japanese and Koreans. For CYP2C9*3, the allele frequency among East Asians varies from 1-6% [1,9].
Selective COX-2 inhibitor is a type of NSAID that has been utilized in post-operative pain management. Previous studies showed that COX-2 has some advantage over traditional or nonselective NSAID especially in certain group of patients undergoing tonsillectomy, eye and neurosurgical procedures, and other types of surgeries which contraindicate the use of traditional NSAID. Development of COX-2 and other pain relievers with less post-operative side effects made comeback investigations to effectively alleviate pain and suffering of post-operative patients and promote speedy recovery from surgery [10-12]. However, COX-2 administration despite its popularity, still requires cautious monitoring to prevent peri-operative adverse effects like traditional NSAID leading to renal injury, stomach ulcers and bleeding [13].
The distribution of CYP2C9 gene polymorphisms have been extensively studied in many populations. However, data among Filipinos are lacking. Here, we determined the allelic and genotype frequencies of CYP2C9*1, CYP2C9*2 and CYP2C9*3 among Filipinos and compared with other ethnic populations. In addition, we investigated the association of CYP2C9 polymorphisms with post-operative pain relief via COX-2 inhibitors. The characterization of CYP2C9 polymorphisms among various ethnicity could contribute to the optimization of a wide range of drugs which include celecoxib.
Materials and methods
Study participants
A total of 99 unrelated patients were enrolled in this prospective observational study covering the period of December 2017 to October 2019. There were 32 (32%) males and 67 (68%) females. The study included patients 21 to 60 years old, with American Society of Anesthesiologists (ASA) classification 1 and 2, who underwent ENT procedures such as thyroidectomy, functional endoscopic sinus surgery, tympanoplasty, mastoidectomy, tonsillectomy and excision biopsy (Table 1). This population was chosen because these patients were already on full diet following surgery, thus they can be given oral analgesics such as celecoxib. Patients were excluded if any of the following are present: history of COX-2 inhibitor allergy; those who took COX2 inhibitors or any analgesics in the pre-operative period; patients with history of gastrointestinal toxicity to NSAIDs; patients with creatinine of less than 30 ml/minute; pregnant; and patients with ischemic heart disease.
Table 1.
Patient characteristics and type of surgery performed
| Characteristics | Frequency n (%) |
|---|---|
| Sex | |
| Male | 32 (32) |
| Female | 67 (68) |
| Age | Mean 44 |
| Surgery performed | |
| Thyroidectomy | 56 (57) |
| Functional endoscopic sinus surgery | 22 (22) |
| Tympanoplasty/mastoidectomy | 9 (9) |
| Tonsillectomy | 7 (7) |
| Excision biopsy | 5 (5) |
All patients enrolled for this study were Filipinos, ethnically and primarily Malay, Malay-Chinese, Malay-Polynesian, and those 4th generation Filipino American or Filipino-European descent. All patients signed written informed consent prior to their enrolment. This study was approved by the Institutional Ethics Review Committee of St. Luke’s Medical Center.
Genomic DNA extraction and CYP2C9 genotyping by real-time PCR
Approximately 4 mL of blood was drawn from each patient in EDTA anti-coagulated tube. Genomic DNA was isolated and purified using QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) according to manufacturer’s instructions. The DNA concentration and purity were analyzed using Nanodrop 1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Genotyping of CYP2C9*2 and CYP2C9*3 variants was carried out using ABI™ 7500 Fast Real-Time PCR System (Applied Biosystems, California, USA) based on 5’ nuclease assay (Figure 1). The two variants were genotyped in separate SNP Assays: SNP ID rs1799853 for CYP2C9*2 and SNP ID rs1057910 for CYP2C9*3 but utilized the same run method (hold at 95°C for 10 min followed by 50 cycles of 92°C for 15 min and 60°C for 1 min and 30 sec). Negative and no template controls were included for every run to ensure the quality of genotyping results. Allelic discrimination analysis of CYP2C9*2 and CYP2C9*3 variations was performed using SDS 2.3 software (Applied Biosystems, California, USA) (Figure 1).
Figure 1.
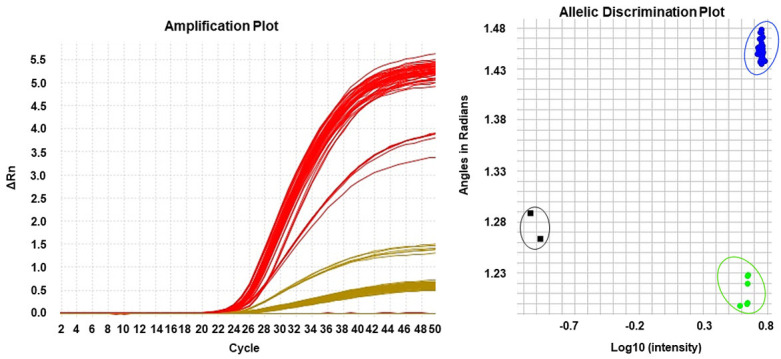
Representative amplification plot for real-time PCR using the 5’ nuclease assay. Allelic discrimination plot showing a cluster of samples categories as AA genotype, homozygous wild-type (blue); AC genotype, heterozygous (green); and no template controls (black).
Post-operative pain management
On the day of every patient’s surgery, standard ASA monitoring was performed including non-invasive blood pressure monitoring, pulse oximetry, capnography, and electrocardiogram. The patients were given 40 mg intravenous (IV) dose of parecoxib intraoperatively. Twelve hours after the IV dose, post-operative care was initiated by administration of first 200 mg oral dose of celecoxib. Likewise, the same oral dose was given within 24 to 48 hours post-operative period. Follow up was done to check the patient’s response to celecoxib and this was accomplished using the numerical pain score on the 24th and 48th hour post-operative period. Tramadol was chosen as rescue medication for standardization, it was administered in 50 mg IV dose every 8 hours as needed for pain score of greater than 4 (scale of 0 to 10) despite intake of celecoxib (Table 2).
Table 2.
Numerical rating scale for pain assessment
| Score | Interpretation | Classification of patients according to drug response | Class definition |
|---|---|---|---|
| 0 | No pain | Responder | NRS of 0-3 after being given 1 dose of celecoxib |
| 1-3 | Mild pain | Responder | NRS of 0-3 after being given 1 dose of celecoxib |
| 4-6 | Moderate pain | Intermediate | NRS of 4-6 after being given more than 1 dose of celecoxib |
| 7-10 | Severe pain | Non-responder | NRS of 7-10 or those who have been given celecoxib but still needed rescue medications |
NRS = numerical rating scale.
Statistical analysis
Data were analyzed using the SPSS software and GraphPad QuickCalcs for categorical data (GraphPad Software, California, USA). Hardy-Weinberg Equilibrium was analyzed by the chi-square test. The genotype distribution and allele frequencies were estimated using 95% confidence interval for every observed proportion. Fisher’s exact test was used to compare allele frequencies of CYP2C9*2 and CYP2C9*3 polymorphism obtained from this study to other population. A P value of less than .05 was considered statistically significant.
Results
Allele and genotype frequencies of CYP2C9*1/*2/*3 single nucleotide polymorphisms among Filipinos
The frequency of CYP2C9*1/*2/*3 alleles were 99%, 0% and 1% respectively. Ninety-eight percent (97/99) of the patients were genotyped as CYP2C9*1/CYP2C9*1 and 2% (2/99) as CYP2C9*1/CYP2C9*3 (Table 3). The observed genotype frequency distribution of CYP2C9*3 did not show a deviation from the Hardy-Weinberg equilibrium.
Table 3.
CYP2C9 genotype and allele frequency
| Genotype distribution (n = 99 patients) | ||
|
| ||
| Genotype | Frequency; n (%) | 95% CI |
|
| ||
| CYP2C9*1/CYP2C9*1 | 97 (98) | 92-100 |
| CYP2C9*1/CYP2C9*2 | 0 | 0-4 |
| CYP2C9*1/CYP2C9*3 | 2 (2) | 0-8 |
| CYP2C9*2/CYP2C9*2 | 0 | 0-4 |
| CYP2C9*2/CYP2C9*3 | 0 | 0-4 |
|
| ||
| Allele frequency (n = 198 alleles) | ||
|
| ||
| Allele | Frequency; n (%) | 95% CI |
|
| ||
| CYP2C9*1 | 196 (99) | 96-100 |
| CYP2C9*2 | 0 | 0-2 |
| CYP2C9*3 | 2 (1) | 0-4 |
None of the patients were found to be heterozygous (CT) or homozygous (TT) for CYP2C9*2 alleles. On the other hand, CYP2C9*3 genotyping showed prevalence of 98% (97/99) for the wild-type (AA) and 2% for the heterozygous (AC). No homozygous variant (CC) was identified in this study (Table 4).
Table 4.
Frequency of CYP2C9*2 and CYP2C9*3 variants
| CYP2C9*2 genotype | n = 99 | Prevalence (%) | 95% CI |
| CC | 99 | 100 | 95-100 |
| CT | 0 | 0 | 0-4 |
| TT | 0 | 0 | 0-4 |
| Allele | Number of alleles | Frequency | 95% CI |
| C | 198 | 100 | 98-100 |
| T | 0 | 0 | 0-2 |
| CYP2C9*3 genotype | |||
| AA | 97 | 98 | 92-98 |
| AC | 2 | 2 | 0-8 |
| CC | 0 | 0 | 0-4 |
| Allele | Number of alleles | Frequency | 95% CI |
| A | 196 | 99 | 96-100 |
| C | 2 | 1 | 0-4 |
Predicted phenotype and mean pain scores at 24 and 48-hours post-surgery
Based on the CYP2C9 genotypes, patients predicted metabolizer phenotypes were used to assess and monitor the patient response to celecoxib. Homozygous wild-type was predicted to be extensive metabolizer, heterozygous variants were classified as intermediate metabolizer, while compound heterozygous and homozygous variants were poor metabolizer. In this study, the genotype-derived poor metabolizer phenotype was absent in the study population, whereas 98% (97/99) of the patients was predicted to be extensive metabolizer and 2% (2/99) to be intermediate metabolizers. At 24 hours post-surgery, the average pain score was 2.57 ± 1.03, while on 48 hours post-surgery, the average pain score was 0.67 ± 0.61 among those who have the wild-type CYP2C9*1 allele. The average pain score on the 24th and 48th hour post-operatively was observed to be 2.5 ± 0.71 and 0.5 ± 0.71 respectively among two patients classified as intermediate metabolizer carrying the CYP2C9*1/*3 genotype (Table 5).
Table 5.
Predicted phenotype and mean pain scores at 24 and 48-hours post-surgery
| Genotype | Predicted phenotype | n = 99 | Pain score (24 hours) | Pain score (48 hours) |
|---|---|---|---|---|
| Homozygous wild-type | Extensive metabolizer | 97 | 2.57 ± 1.03 | 0.67 ± 0.61 |
| CYP2C9*1/CYP2C9*1 | ||||
| Heterozygous | Intermediate metabolizer | - | - | - |
| CYP2C9*1/CYP2C9*2 | 2 | 2.5 ± 0.71 | 0.5 ± 0.71 | |
| CYP2C9*1/CYP2C9*3 | ||||
| Compound heterozygous | Poor metabolizer | - | - | - |
| CYP2C9*2/CYP2C9*3 | ||||
| Homozygous variant | Poor metabolizer | - | - | - |
| CYP2C9*2/CYP2C9*2 | ||||
| CYP2C9*3/CYP2C9*3 |
Allele frequencies of CYP2D6*2/*3 reported from various ethnic populations
Table 6 shows the CYP2C9*2 and CYP2C9*3 allele frequencies among various populations. No significant differences were found between Filipinos and other Asians, except for North Indian population. The allele frequencies of CYP2C9*2 and CYP2C9*3 were significantly different (P = <.001) between Filipinos and Caucasian populations.
Table 6.
Comparison of CYP2C9*2 and CYP2C9*3 allele frequencies from various ethnic populations
| Population | n | Number of alleles | CYP2C9*2 frequency (P value) | CYP2C9*3 frequency (P value) | Reference |
|---|---|---|---|---|---|
| Asian | |||||
| Filipino | 99 | 198 | 0 | 1.0 | Present study |
| Indian | 89 | 178 | 4.5 (.0023) | 10.1 (<.001) | [9] |
| Malay | 183 | 366 | 1.0 (NS) | 3.0 (NS) | [18] |
| Japanese | 140 | 280 | 0 (NS) | 1.8 (NS) | [20] |
| Korean | 574 | 1148 | 0 (NS) | 1.1 (NS) | [21] |
| Chinese | 394 | 788 | 0.1 (NS) | 3.6 (NS) | [22] |
| African | |||||
| Ethiopian | 150 | 300 | 4.3 (.0023) | 2.3 (NS) | [19] |
| Caucasian | |||||
| Romanian | 332 | 664 | 11.3 (<.001) | 9.3 (<.001) | [8] |
| Italian | 157 | 314 | 11.0 (<.001) | 9.0 (<.001) | [19] |
| French | 151 | 302 | 15.0 (<.001) | 8.0 (<.001) | [22] |
| British | 100 | 200 | 12.5 (<.001) | 8.5 (<.001) | [25] |
| American | 100 | 200 | 8.0 (<.001) | 6.0 (.01) | [26] |
| Swedish | 430 | 860 | 10.7 (<.001) | 7.4 (<.001) | [27] |
P value was calculated using Fisher’s exact test; NS = not significant.
Discussion
The frequency of CYP2C9*1, CYP2C9*2 and CYP2C9*3 have been reported in various ethnic populations worldwide [14-17]. The frequency of the wild-type allele CYP2C9*1 and the common CYP2C9*3 variant among Han Chinese were 94.48% and 2.94% respectively [5]. The frequency of the wild-type allele among Koreans was reported to be 0.934 [1]. In another study, the frequency of CYP2C9*3 among Malays was 3% [18]. In contrast, the allele frequencies of CYP2C9*1 (80%), CYP2C9*2 (11%) and CYP2C9*3 (9%) among Italians were similar with other Caucasian populations [19]. The results of the present study showed a frequency of 99% for CYP2C9*1 and 1% for CYP2C9*3 among Filipinos. Our findings are comparable to the frequencies of wild-type CYP2C9*1 and CYP2C9*3 variant allele among other East Asian populations. In this study, no CYP2C9*2 variant was detected. This finding agreed with previous studies among East Asian countries like Japan, China, Korea, and Malaysia; suggestive that CYP2C9*2 is rare or almost absent in this region [20-22]. Meanwhile, 2% of the patients were predicted as intermediate metabolizers for acquiring heterozygous CYP2C9*3; this variant has been reported to have reduced enzymatic activity. To the best of our knowledge, this is the first report of CYP2C9 allele frequency among Filipinos, which was also primarily used to assess the predicted metabolizer phenotypes of post-operative patients taking COX-2 inhibitors.
Patients with CYP2C9*1/CYP2C9*3 genotype have responded well to celecoxib with the 24-hour mean pain score of 2.5 ± 0.71 and 48-hour mean pain score of 0.5 ± 0.71, even lower than those with homozygous CYP2C9*1. However, considerations in the treatment strategies should be made since they were predicted as intermediate metabolizers compared to those participants which can rapidly eliminate the drug after delivery of therapeutic effect. Fortunately, celecoxib and other COX-2 inhibitors have a broad therapeutic window which may be favorable for these patients against unwanted adverse drug reactions. In 2016, Kim and colleagues reported that the plasma concentration of celecoxib and its final metabolite, celecoxib carboxylic acid was determined to correlate the effects of CYP2C9 polymorphisms in drug clearance. This provided further information on how each phenotypic group differs in clearing celecoxib once it has delivered its therapeutic pain relief [23]. However, this study cannot confirm previous findings that the plasma concentration of celecoxib is correlated to the effects of CYP2C9 polymorphisms. This serves as a limitation of this study.
The CYP2C9 polymorphisms are relevant in terms of predicting the efficacy and adverse effects of NSAIDS, hypoglycemic agents and anticoagulants belonging to the class of vitamin K epoxide reductase inhibitors. Among the patients enrolled, there was no reported side effect encountered secondary to COX-2 inhibitor throughout the study period.
A similar study evaluated the relationship between polymorphisms in CYP2C9 and the pharmacokinetics of celecoxib. The study demonstrated that individuals carrying CYP2C9*1/CYP2C9*3 and CYP2C9*3/CYP2C9*3 had lower clearance than those with the wild-type allele CYP2C9*1/CYP2C9*1. In addition, the half-life was noted to be 2.7-fold higher in patients with CYP2C9*3/CYP2C9*3 than in those with the wild-type but not in those with CYP2C9*1/CYP2C9*3 [24]. The findings of this study suggest that those who carry the CYP2C9*3/CYP2C9*3 have poor metabolism of celecoxib thus, the dose of celecoxib be decreased in CYP2C9*3 carriers.
Moreover, clinical case reports have associated genotypes expressing the CYP2C9*2 and CYP2C9*3 alleles with significant reductions in both metabolism and daily dose requirements of selected CYP2C9 substrates. Individuals expressing these variant genotypes appear to be significantly more susceptible to adverse effects with the narrow therapeutic index agent warfarin and phenytoin, particularly during the initiation of therapy [6].
Genotyping of CYP2C9 is expected to have a role in predicting drug clearance and implementing individualized pharmacotherapy. Prospective clinical studies with large samples are needed to establish gene-dose and gene-effect relationships for CYP2C9 [28,29]. Thus, future pharmacogenetic studies may be done to investigate other variants that may significantly influence celecoxib dosing among Filipinos.
The outcome of this study suggests that routine CYP2C9 genotyping need not be performed among Filipinos but must be considered among mixed race patients most especially those with Caucasian lineage. This is to ensure that every patient will be treated with personalized care and prevent the occurrence of unnecessary adverse drug reactions.
Acknowledgements
This work was supported by St. Luke’s Medical Center through the Research and Biotechnology Group (Project No. 18-002) with the collaboration of the Department of Anesthesiology and Pain Management Center of Luke’s Medical Center-Quezon City.
Disclosure of conflict of interest
None.
References
- 1.Bae JW, Kim HK, Kim JH, Yang SI, Kim MJ, Jang CG, Park YS, Lee SY. Allele and genotype frequencies of CYP2C9 in a Korean population. Br J Clin Pharmacol. 2005;60:418–422. doi: 10.1111/j.1365-2125.2005.02448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae JW, Kim JH, Choi CI, Kim MJ, Kim HJ, Byun SA, Chang YS, Jang CG, Park YS, Lee SY. Effect of CYP2C9*3 allele on the pharmacokinetics of naproxen in Korean subjects. Arch Pharm Res. 2009;32:269–273. doi: 10.1007/s12272-009-1232-z. [DOI] [PubMed] [Google Scholar]
- 3.Carbonell N, Verstuyft C, Massard J, Letierce A, Cellier C, Deforges L, Saliba F, Delchier JC, Becquemont L. CYP2C9*3 loss-of-function allele is associated with acute upper gastrointestinal bleeding related to the use of NSAIDs other than aspirin. Clin Pharmacol Ther. 2010;87:693–698. doi: 10.1038/clpt.2010.33. [DOI] [PubMed] [Google Scholar]
- 4.Pratt VM, Cavallari LH, Del Tredici AL, Hachad H, Ji Y, Moyer AM, Scott SA, Whirl-Carrillo M, Weck KE. Recommendations for clinical CYP2C9 genotyping allele selection: a joint recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn. 2019;21:746–755. doi: 10.1016/j.jmoldx.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai DP, Xu RA, Hu LM, Wang SH, Geng PW, Yang JF, Yang LP, Qian JC, Wang ZS, Zhu GH, Zhang XH, Ge RS, Hu GX, Cai JP. CYP2C9 polymorphism analysis in Han Chinese populations: building the largest allele frequency database. Pharmacogenomics J. 2014;14:85–92. doi: 10.1038/tpj.2013.2. [DOI] [PubMed] [Google Scholar]
- 6.Wang B, Wang J, Huang SQ, Su HH, Zhou SF. Genetic polymorphism of the human cytochrome P450 2C9 gene and its clinical significance. Curr Drug Metab. 2009;10:781–834. doi: 10.2174/138920009789895480. [DOI] [PubMed] [Google Scholar]
- 7.Dorado P, Sosa-Macias MG, Penas-Lledo EM, Alanis-Banuelos RE, Wong ML, Licinio J, Lares-Asseff I, Llerena A. CYP2C9 allele frequency differences between populations of mexican-mestizo, mexican-tepehuano, and spaniards. Pharmacogenomics J. 2011;11:108–112. doi: 10.1038/tpj.2010.29. [DOI] [PubMed] [Google Scholar]
- 8.Buzoianu AD, Trifa AP, Muresanu DF, Crisan S. Analysis of CYP2C9*2, CYP2C9*3 and VKORC1 1639 G>A polymorphisms in a population from South-Eastern Europe. J Cell Mol Med. 2012;16:2919–2924. doi: 10.1111/j.1582-4934.2012.01606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhary N, Kabra M, Gulati S, Gupta YK, Pandey RM, Bhatia BD. Frequencies of CYP2C9 polymorphisms in north Indian population and their association with drug levels in children on phenytoin monotherapy. BMC Pediatr. 2016;16:66. doi: 10.1186/s12887-016-0603-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilron I, Milne B, Hong M. Cyclooxygenase-2 inhibitors in postoperative pain management: current evidence and future directions. Anesthesiology. 2003;99:1198–1208. doi: 10.1097/00000542-200311000-00029. [DOI] [PubMed] [Google Scholar]
- 11.Pinheiro SP, Gates MA, De Vivo I, Rosner BA, Tworoger SS, Titus-Ernstoff L, Hankinson SE, Cramer DW. Interaction between use of non-steroidal anti-inflammatory drugs and selected genetic polymorphisms in ovarian cancer risk. Int J Mol Epidemiol Genet. 2010;1:320–331. [PMC free article] [PubMed] [Google Scholar]
- 12.Rollason V, Samer CF, Daali Y, Desmeules JA. Prediction by pharmacogenetics of safety and efficacy of non-steroidal anti-inflammatory drugs: a review. Curr Drug Metab. 2014;15:326–343. doi: 10.2174/1389200215666140202214454. [DOI] [PubMed] [Google Scholar]
- 13.Malhi H, Atac B, Daly AK, Gupta S. Warfarin and celecoxib interaction in the setting of cytochrome P450 (CYP2C9) polymorphism with bleeding complication. Postgrad Med J. 2004;80:107–109. doi: 10.1136/pmj.2003.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee CR, Goldstein JA, Pieper JA. Cytochrome P450 2C9 polymorphisms: a comprehensive review of the in-vitro and human data. Pharmacogenetics. 2002;12:251–263. doi: 10.1097/00008571-200204000-00010. [DOI] [PubMed] [Google Scholar]
- 15.Vogl S, Lutz RW, Schonfelder G, Lutz WK. CYP2C9 genotype vs. metabolic phenotype for individual drug dosing: a correlation analysis using flurbiprofen as probe drug. PLoS One. 2015;10:e0120403. doi: 10.1371/journal.pone.0120403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qayyum A, Najmi MH, Mansoor Q, Irfan M, Naveed AK, Hanif A, Kazmi AR, Ismail M. Frequency of common VKORC1 polymorphisms and their impact on warfarin dose requirement in Pakistani population. Clin Appl Thromb Hemost. 2018;24:323–329. doi: 10.1177/1076029616680478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marjani A, Gharanjik AM. Genetic polymorphism of CYP2C9 among Sistani ethnic group in Gorgan. Indian J Clin Biochem. 2018;33:208–213. doi: 10.1007/s12291-017-0660-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosdi RA, Mohd Yusoff N, Ismail R, Soo Choon T, Saleem M, Musa N, Yusoff S. High allele frequency of CYP2C9*3 (rs1057910) in a Negrito’s subtribe population in Malaysia; Aboriginal people of Jahai. Ann Hum Biol. 2016;43:445–450. doi: 10.3109/03014460.2015.1068372. [DOI] [PubMed] [Google Scholar]
- 19.Scordo MG, Aklillu E, Yasar U, Dahl ML, Spina E, Ingelman-Sundberg M. Genetic polymorphism of cytochrome P450 2C9 in a Caucasian and a black African population. Br J Clin Pharmacol. 2001;52:447–450. doi: 10.1046/j.0306-5251.2001.01460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura M, Ieiri I, Mamiya K, Urae A, Higuchi S. Genetic polymorphism of cytochrome P450s, CYP2C19, and CYP2C9 in a Japanese population. Ther Drug Monit. 1998;20:243–247. doi: 10.1097/00007691-199806000-00001. [DOI] [PubMed] [Google Scholar]
- 21.Yoon YR, Shon JH, Kim MK, Lim YC, Lee HR, Park JY, Cha IJ, Shin JG. Frequency of cytochrome P450 2C9 mutant alleles in a Korean population. Br J Clin Pharmacol. 2001;51:277–280. doi: 10.1046/j.1365-2125.2001.00340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang JQ, Morin S, Verstuyft C, Fan LA, Zhang Y, Xu CD, Barbu V, Funck-Brentano C, Jaillon P, Becquemont L. Frequency of cytochrome P450 2C9 allelic variants in the Chinese and French populations. Fundam Clin Pharmacol. 2003;17:373–376. doi: 10.1046/j.1472-8206.2003.00148.x. [DOI] [PubMed] [Google Scholar]
- 23.Kim SH, Kim DH, Byeon JY, Kim YH, Kim DH, Lim HJ, Lee CM, Whang SS, Choi CI, Bae JW, Lee YJ, Jang CG, Lee SY. Effects of CYP2C9 genetic polymorphisms on the pharmacokinetics of celecoxib and its carboxylic acid metabolite. Arch Pharm Res. 2017;40:382–390. doi: 10.1007/s12272-016-0861-2. [DOI] [PubMed] [Google Scholar]
- 24.Prieto-Perez R, Ochoa D, Cabaleiro T, Roman M, Sanchez-Rojas SD, Talegon M, Abad-Santos F. Evaluation of the relationship between polymorphisms in CYP2C8 and CYP2C9 and the pharmacokinetics of celecoxib. J Clin Pharmacol. 2013;53:1261–1267. doi: 10.1002/jcph.169. [DOI] [PubMed] [Google Scholar]
- 25.Stubbins MJ, Harries LW, Smith G, Tarbit MH, Wolf CR. Genetic analysis of the human cytochrome P450 CYP2C9 locus. Pharmacogenetics. 1996;6:429–439. doi: 10.1097/00008571-199610000-00007. [DOI] [PubMed] [Google Scholar]
- 26.Sullivan-Klose TH, Ghanayem BI, Bell DA, Zhang ZY, Kaminsky LS, Shenfield GM, Miners JO, Birkett DJ, Goldstein JA. The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics. 1996;6:341–349. doi: 10.1097/00008571-199608000-00007. [DOI] [PubMed] [Google Scholar]
- 27.Yasar U, Eliasson E, Dahl ML, Johansson I, Ingelman-Sundberg M, Sjoqvist F. Validation of methods for CYP2C9 genotyping: frequencies of mutant alleles in a Swedish population. Biochem Biophys Res Commun. 1999;254:628–631. doi: 10.1006/bbrc.1998.9992. [DOI] [PubMed] [Google Scholar]
- 28.Moyer AM, Vitek CR, Giri J, Caraballo PJ. Challenges in ordering and interpreting pharmacogenetic tests in clinical practice. Am J Med. 2017;130:1342–1344. doi: 10.1016/j.amjmed.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 29.Daly AK, Rettie AE, Fowler DM, Miners JO. Pharmacogenomics of CYP2C9: functional and clinical considerations. J Pers Med. 2018;8:1. doi: 10.3390/jpm8010001. [DOI] [PMC free article] [PubMed] [Google Scholar]


