Abstract
Advances in deep learning and neural networking have allowed clinicians to understand the impact that artificial intelligence (AI) could have on improving clinical outcomes and resources expenditures. In the realm of genitourinary (GU) cancers, AI has had particular success in improving the diagnosis and treatment of prostate, renal, and bladder cancers. Numerous studies have developed methods to utilize neural networks to automate prognosis prediction, treatment plan optimization, and patient education. Furthermore, many groups have explored other techniques, including digital pathology and expert 3D modeling systems. Compared to established methods, nearly all the studies showed some level of improvement and there is evidence that AI pipelines can reduce the subjectivity in the diagnosis and management of GU malignancies. However, despite the many potential benefits of utilizing AI in urologic oncology, there are some notable limitations of AI when combating real-world data sets. Thus, it is vital that more prospective studies be conducted that will allow for a better understanding of the benefits of AI to both cancer patients and urologists.
Keywords: Artificial Intelligence, prostate cancer, renal cancer, bladder cancer, clinical trials, androgen deprivation therapy
Introduction
Incidences of cancer have been on the rise globally [1]. Of these, genitourinary (GU) cancers are projected to lead to over 33,000 deaths [2]. With the increasing frequency of GU cancers, efforts have been undertaken to increase the efficacy of diagnosis and treatment. However, these efforts require further optimization and innovation as the costs of diagnosis and management are increasing concomitantly [3]. The field of urology has historically been at the forefront of innovation, such as with the early introduction of Robotic Surgery in the treatment of GU cancers [4]. Robotic surgery is among the technological advancement that incorporates Artificial Intelligence (AI).
AI is a subfield of computer science that attempts to develop a computer’s intelligence to mimic humans in the way they work and think. Machine learning is an integral part of AI which applies statistical models to machines and allows them to act independently in an intelligent way without a set of explicit commands [5,6]. Deep learning is a subclass of machine learning that makes use of artificial neural networks (ANN) which are meant to resemble the way biological nervous systems are structured and how they process information [7]. ANNs can have multi-layered collections of artificial nodes that act as neurons, which accept input, process data, and finally pass it along to other neurons. AI is a rapidly growing field that has been investigated for utilization in the diagnosis and management of GU diseases [8].
Within other fields of medicine, AI has been investigated as an aid in information management, diagnostics, and physician decision making [9-11]. According to the recent NIH Roadmap for Foundational Research on Artificial Intelligence in medical imaging, AI based algorithms will have a significant impact on the practice of clinical medical imaging within the next decade [12]. Furthermore, in diagnostics, machine learning has been successfully used to provide automatic interpretations of physiological function to help in diagnosis of diseases. For example, machine learning has been used to evaluate pulmonary function tests to determine diagnoses of obstructive lung disease [13]. AI has also been used to diagnose fractures by analyzing skeletal radiographs with an accuracy comparable to that of senior level orthopedic surgeons [14]. AI is also under investigation as a tool in treatment decision making for cardiovascular diseases by optimizing treatment algorithms [15]. In light of these advancements in other fields, AI has strong potential for diagnostic and treatment optimization within urologic oncology. Figure 1 summarizes the current applications of AI in GU cancers.
Figure 1.
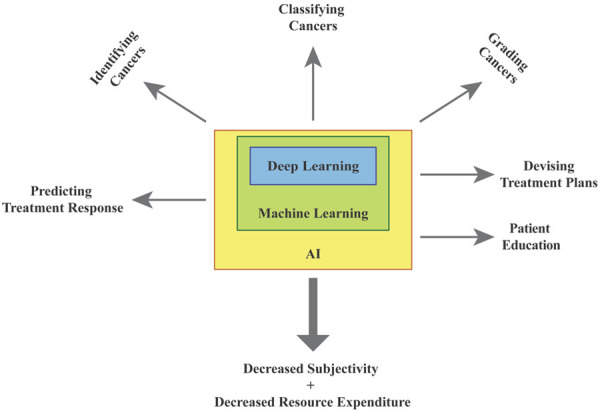
Summarizes the current applications of AI in GU cancers.
As the true power of AI has only recently been coming to light, we can predict that its use is likely to increase in GU cancer diagnosis and management. Although there are numerous examples of AI being used in the diagnosis and treatment of prostate, bladder, and renal cancer, it is only recent that AI was also used to predict the prognosis of testicular cancer [16]. Furthermore, there is limited data on the use and subsequent limitations of AI in the management and treatment of other less common urologic malignancies, including penile, urethral, and ureteral cancers. As more studies come to light, there will be more sources of prospective validation and increases in the size of the databases that will cumulatively improve overall the accuracy of AI pipelines. The current review focuses on highlighting the recent advancements and limitations of AI in urologic oncology.
Inclusion criteria
A literature search was performed to review recent ad-vancements in AI in the diagnosis and management of GU cancers. Publications for the literature review were identified from the PubMed®/Medline® database by searching the terms “prostate cancer”, “renal cell carcinoma”, “renal cancer”, “bladder cancer”, “artificial intelligence”, “deep learning”, “machine learning”, and “neural network” in any combination. Relevant articles written in the English language, with an emphasis on original research articles written within the last five years, were included in the literature review.
Literature review
Prostate cancer - diagnosis
The diagnosis and management of GU cancers involves parsing through a large amount of clinical data. This involves histological images, MRI imaging, biomarker screening, and more. Among the challenges in employing AI in the diagnosis of prostate cancer is developing a consistent methodology to link qualitative data from many sources to create a quantitative metric for decision making [17]. Even more challenging is the task of using AI to create a method that empowers patients to make their own decisions. Nevertheless, Auffenberg et al. recently devised a method to overcome both of these challenges. In a new model, clinical registry data was taken from over 7,500 men with prostate cancer across 45 different urological practices in the state of Michigan. This data included patient age at biopsy, pre-diagnosis prostate specific antigen (PSA) levels, Gleason scores, and more. With this, a machine learning algorithm was trained to predict primary treatment outcomes according to what other patients in similar situations had received [18]. This tool, which is now available via web and smartphone application, allows patients to easily leverage a large amount of relevant clinical data to take charge of their care. However, a disadvantage of the random forest model used in this algorithm is that it is often less interpretable than a single decision tree. In other words, it can be hard to determine what sources have the greatest impact in diagnosis leading to a black box effect which can be a disadvantage to patient usage.
Deep learning has also recently been utilized to automate Gleason grading of histological specimens from prostate biopsies. While Gleason grading is among the most important prognostic predictors for patients suffering from prostate cancer, it is time-consuming, requires experienced pathologists, and is subject to limited inter-rater reproducibility [19]. Lucas et al. recently found that convolutional neural networks that automate Gleason grading from prostatic biopsies were able to achieve a 65% concordance with an experienced GU pathologist [20]. This is similar to the inter-observer agreement between two trained pathologists described in a study by Ozkan et al. [20,21]. Similarly, Arvaniti et al. demonstrated that deep learning algorithms were successfully able to divide patients into prognostically different groups on the basis of automated Gleason grading of prostate cancer tissue microarrays in a manner comparable to trained pathologists [19]. In addition, machine learning has also been used to predict Gleason scores using multiparametric magnetic resonance (MR) images combined with texture features with a high degree of accuracy [22], thereby decreasing the need for prostatic biopsy for cancer diagnosis. Table 1A-D summarizes the findings discussed above. However, there are several roadblocks to developing the perfect AI Gleason scoring software, including that many of the trained datasets contain only a small number samples that are used to train the algorithm. Larger data sets will allow models to perform more realistically when faced with new external samples. In addition, the quality of scans, specific methods in sample collection, and human error in pathologist scoring will vary in different datasets which makes it difficult to start with a gold standard. It is hard to estimate each of these variances within a given dataset, which is a basis for probabilistic models to determine regional effects. Despite this, these studies show some promise in how AI can improve the process of Gleason scoring in prostate cancer diagnosis by saving resources, improving reliability, and reducing patient discomfort.
Table 1.
Prostate cancer review articles summary
| Legend | Name/Type of Model | No. of patients | Cancer | Summary | Data Analysis | Implications | References |
|---|---|---|---|---|---|---|---|
| 1-A | Random forest | 7543 | Prostate Cancer | A clinical registry of relevant prostate cancer data across 45 urology practices in the state of Michigan, a random forest machine learning (ML) model was developed to predict probability of receiving a given treatment based on clinical, pathologic, and demographic factors. | Accuracy AUC was 0.81 | This web- and smartphone-based platform can serve as a useful tool for patients to better understand treatment options and decisions and be able to take charge of their care. | 18. Auffenberg et al. |
| 1-B | CNN | 886 (641 for training, 245 for testing) | Prostate Cancer | A CNN model was developed to automatically assign Gleason scores to H&E stained prostate cancer tissue microarrays. | Inter-annotator agreement between the model and 2 pathologists using Cohen’s quadratic kappa statistic was kappa = 0.75 and 0.71 respectively. | Deep learning technology has the potential to assist pathologists in histopathologic grading of prostate cancer and mitigate the effects of inter-pathologist variability. | 19. Arvaniti, E., et al. |
| 1-C | CNN | 38 patients (96 biopsies) | Prostate Cancer | A CNN model was developed to detect Gleason patterns (GP) and determine grade groups (GG) from H&E stained tissue sections of 96 prostate biopsies. | The model’s ability to differentiate between malignant (GP ≥ 3) and non-malignant (GP < 3) sections reached an accuracy of 92% (F-score = 0.93). Automated GG reached a concordance of 65% with a genitourinary pathologist (kappa = 0.70). | Computer-aided automated histopathologic grading of prostate cancer has the potential to reduce both inter-pathologist variability and time required to diagnose. | 20. Lucas, M. et al. |
| 1-D | SVM | 147 | Prostate Cancer | Three ML algorithms were tested to be able to classify prostate cancer Gleason scores based on combined apparent diffusion coefficient (ADC) and T2-weighted prostate MRIs of patients with biopsy-proven prostate cancer undergoing radical prostatectomy. ML results were validated with Gleason grade patterns obtained from histopathological analysis of the excised prostates. | Distinguishing between GS 6 vs. GS ≥ 7 resulted in 93% accuracy for cancers occurring in both peripheral (PZ) and transition (TZ) zones. Distinguishing between GS 7 (3+4) vs. GS 7 (4+3) resulted in 92% accuracy for cancers occurring in both PZ and TZ. | The ability of ML algorithms to differentiate Gleason scores from multiparametric MRI images presents a unique method for non-invasive and accurate diagnosis of prostate cancer, potentially decreasing the need for prostate biopsies. | 22. Fehr, D. et al. |
| 1-E | k-nearest neighbor | 150 | Prostate Cancer | ML algorithm was developed to generate low-dose-rate brachytherapy treatment plans for patients with prostate cancer. Automatically generated plans were compared to both expert radiation therapist (RT) and brachytherapist (BT) plans. | Dosimetry and clinical quality of ML plans were found to be equivalent to RT and BT plans. Planning time was significantly reduced with the ML plan (mean 0.84 min vs. 17.88 min for RT/BT, p = 0.020). | The capability of the ML algorithm to reach equivalent clinical quality to experts in brachytherapy planning combined with its significant reduction in time and resource expenditure is a promising utilization of ML for aiding in planning treatments. | 23. Nicolae, A. et al. |
| 1-F | CNN | 418 for training | Prostate Cancer | An automatic contour propagation pipeline was created using deformable image registration (DIR) to develop online adaptive intensity-modulated proton therapy (IMPT) plans for prostate cancer. | A conservative success rate of 80% was achieved, signifying that 80% of plans generated could be used without manual correction. | IMPT is capable of delivering a localized dose of radiation to target tissue, while minimizing damage to surrounding organs. However, IMPT is more sensitive to daily anatomical changes, exacerbating treatment-associated toxicity. DIR and deep learning have the capability to create an automated plans that adapt to these daily anatomical changes and protect organs at risk (OARs). | 24. Elmahdy, M.S., et al. |
| 32 for testing |
Prostate cancer - management
AI has also made strides in influencing treatment options for prostate cancer patients. For example, Nicolae et al. used a k-nearest neighbor algorithm that allowed for the automatic generation of prostate low-dose-rate brachytherapy treatment plans in a manner that was comparable to experts. The average planning time using this approach was 0.84 minutes compared to an average of 17.88 minutes for the expert. Thus, this approach has the potential to create consistency within patients’ treatment as well as to decrease resource expenditures [23]. One limitation of the k-nearest neighbor is that although it produces a definite classification result, increased complexity in the data can create too many overlapping groups leading to an unreliable classification.
Deep learning models have also been developed to aid in Intensity-Modulated Proton Therapy (IMPT). IMPT is capable of delivering a localized dose of radiation to a target tissue while diminishing potential damage to nearby normal tissue [24,25]. Unfortunately, this method is quite sensitive to daily anatomical changes, which can exacerbate treatment-associated toxicity. Nevertheless, deep learning has also recently been used to create a pipeline which can automate contour propagation to develop treatment plans that adapt to daily anatomical changes. The algorithm was successful in treatment plans that did not need any manual correction 80% of the time [24]. A significant limitations in these studies, however, is their retrospective analysis. Due to the growing trend in surveillance, it is critical to incorporate real time effects into the models. Furthermore, the retrospective nature poses the opportunity to make decisions without controlling for confounding factors. For example, this can be seen in a study by Obermeyer et al. which found evidence of racial bias in a commonly used commercial algorithm that is used to guide health care decisions across the United States. The algorithm, which uses health care costs to predict health needs, falsely associated lower expenditures on African American patients with overall better health status in this population [26]. Despite the need for more prospective analyses, these studies exemplify how AI and precision medicine can not only save resources, but also reduce adverse effects associated with prostate cancer treatments. Table 1E, 1F summarizes the findings in prostate cancer management.
Renal cancer
Renal cancer is the most lethal GU cancer with 25% of patients having evidence of metastatic disease at initial presentation [27]. The three most common classifications of renal cancer include clear cell, papillary, and chromophobe types [28]. The proper classification is of particular importance due to differences in prognostic and therapeutic factors between classes [29], including potential responses to molecularly targeted therapies. In a recent study by Han et al., a convolutional neural network (CNN) was created to classify renal cancer into one of the three types using 3-Phase computerized tomography (CT) images that were marked with a particular region of interest. Using a training set with 135 randomly selected CT scans, a validation set of 34 biopsy-proven cases allowed for an AUC of about 0.9, regardless of the classification of the cancer. Despite the limitation that radiologists must initially mark the proposed region of interest prior to automated classification, the model has significant implications for training radiologists [30]. Furthermore, it provides a method for radiologists to confirm their findings and may reduce the need for invasive biopsies. Finally, the small sample size of 135 scans used in the model was able to accomplish fairly impressive results compared to the several hundred samples typically needed to train a CNN.
Clear cell renal cell carcinoma is typically graded according to the Fuhrman method, which relies on nuclear pleomorphic pattern analysis. Unfortunately, analysis of pathological slides can lead to inconsistencies among different pathologists [31]. However, Holdbrook et al. devised a support vector machine (SVM) that allows for automated grading of these specimens directly from histopathological whole-slide images of biopsies. SVMs use many subsets of features, known as support vectors, to make their final decisions, which makes them quick and efficient. Although this makes them ideal to use when there is a clear separation within classification groups, these models tend to do more poorly in noisy data and often do not give probability estimates in their results. However, the results of this study demonstrated a correlation between the generated automated image scores and another multigene assay based scoring system that has been known to accurately predict prognosis [31].
Lin et al. expanded on this study by developing yet another machine learning model to distinguish between high- and low-grade clear cell renal cell carcinoma using a variety of different types of CT images. This method proved the most successful, with three-phase CT images resulting in an AUC of 0.87 when compared to the classifications within a pathology database that were reconfirmed by an experienced pathologist [32]. Thus, these studies illustrate the potential of AI in determining prognostic factors in the diagnosis of renal cancer and in minimizing the risks, resources, and subjectivity associated with unnecessary biopsy collection and manual pathological analysis.
AI has also been used to optimize treatment strategies in patients suffering from renal cell carcinoma. Renal cell carcinoma may be pharmacologically treated by cytokine therapy or tyrosine kinase inhibitors [33]. However, treatment with different systemic therapies tends to lend itself to significant variability in prognosis. Thus, Buchner et al. recently developed an artificial neural network that allows for the input of multiple parameters, including but not limited to treatment type, histological parameters, body mass index (BMI), and age. Using this data, the algorithm was able to accurately predict 36 month survival in patients according to different therapy types with 91% accuracy in the validation cohort [34]. A limitation of ANNs as used in this study, however, involves its use of multiple parameters by which the network builds its recommendation. If cases develop where clinical characteristics are treated equally as important as all other features, it could lead to the overfitting and overrepresenting of certain parameters. Nevertheless, these findings demonstrates the ability of AI to develop personalized pharmacological treatment plans for renal cancer patients based on a multitude of prognostic factors and automated outcome analyses. Table 2 summarizes the advancements of AI in renal cancer management.
Table 2.
Renal cancer review articles summary
| Legend | Name/Type of Model | No. of patients | Cancer | Summary | Data Analysis | Implications | References |
|---|---|---|---|---|---|---|---|
| 2-A | CNN | 135 for training | Renal Cancer | A deep learning neural network was developed to distinguish between three major subtypes of renal cell carcinoma (clear cell, papillary, and chromophobe) using computed tomography images, with regions of interest marked by radiologists. Automated results were trained with biopsy-proven samples. | The network showed 85% accuracy with AUC of 0.9. | ML models that can classify subtypes of cancer from CT images may have an important role in reducing the need for invasive biopsies and in training radiologists to identify further signs of specific diagnoses. | 29. Han, S. et al. |
| 34 for testing | |||||||
| 2-B | SVM | 59 | Renal Cancer | An automated image classification pipeline was created to detect relevant nuclear pleomorphic patterns from histopathologic tissue of patients with clear cell renal cell carcinoma and grade these images using the Fuhrman grading scale. | The results demonstrated a correlation (R = 0.59) between the automated pipeline and an already-established multigene assay-based scoring system. | An automated pipeline that is able to grade histopathologic samples of clear cell renal cell carcinoma into high and low grades has important clinical implications in terms of treatment planning and reducing inter-pathologist variability in grading. | 30. Holdbrook, D.A. et al. |
| 2-C | naïve Bayes | 231 patients | Renal Cancer | A ML model was developed to predict the Fuhrman grade of clear cell renal cell carcinoma from single- and three-phase CT images. Automated results were confirmed with pathology-proven samples. | The model based on three-phase CT images achieved the best diagnostic performance with AUC = 0.87. | ML has the potential to minimize the risks, resources, and subjectivity associated with biopsy collection and manual pathological analysis for grading of clear cell renal cell carcinoma. | 31. Lin, F. et al. |
| (232 pathologically-proven clear cell renal cell carcinoma lesions) | |||||||
| 2-D | ANN | 175 | Renal Cancer | A ML algorithm was developed to predict 36-month survival in patients with renal cell carcinoma using a multitude of clinical prognostic data, including tumor grade, vessel invasion, and pathologic T classification, to input into the network. | The ANN achieved an accuracy of 95%. | AI has the potential to aid in developing personalized treatment plans for patients with renal cancer based on varied prognostic factors and automated ML outcome analyses. | 33. Buchner, A. et al. |
Bladder cancer
Bladder cancer is the ninth most common type of cancer worldwide and causes significant morbidity and mortality in the aging population [35]. Unfortunately, there have been no improvements in the five-year survival rates for bladder cancer in over thirty years [36]. Although urine biomarkers have been discussed in the diagnosis of bladder cancer, cystoscopy remains the gold standard [37]. However, correctly recognizing cystoscopic findings can prove difficult for many clinicians and success is contingent on the clinician’s experience. Eminaga et al. tries to take an AI approach to the problem by developing a convolutional neural network that used 479 different cases that contained 44 different urological findings. After training the algorithm, it was able to successfully identify all of the images with cancer-containing lesions [38]. This shows how AI can be used to improve accuracy and reduce variability in analyzing cystoscopic images. This study was able to overcome the daunting task for a CNN to be able to identify regions given the variable input data from different clinicians by its large sample size and numerous distinct findings used to train the algorithm. However, although cystoscopy is the standard in the diagnosis of bladder cancer, it is an invasive and expensive procedure and some efforts have been made to move to more non-invasive methods. Sokolov et al. recently devised a machine learning based method that was able to detect bladder cancer using images of cells that were extracted from urine samples. This method showed 94% diagnostic accuracy, which demonstrated a significant improvement over cystoscopy [39]. However, this model relied on calculating surface features rather than analyzing them as would be done with a neural network. In cases with complex images and noisy features, this has potential to lead to inconsistent results. Overall, despite limitations, these studies together highlight how AI can be used to improve, or perhaps even shift, the standard of care in the diagnosis of bladder cancer.
A diagnosis of bladder cancer is typically followed by staging in order to determine prognosis and potential therapeutic options. Neoadjuvant chemotherapy is usually recommended when the cancer grade is at or exceeds T2, at which point the tumor has reached the muscle in the bladder wall. Garapati et al. trained a CNN that accurately used images from CT Urography to stratify bladder cancer patients into one of two groups: equal to or above stage T2 or below stage T2 [40]. This model shows the potential of AI in not only the diagnosis of bladder cancer, but also in determining prognostically relevant information, which has major effects on treatment options.
AI has also been shown to have the potential to directly impact the treatment of bladder cancers. Although cystectomy has been considered the gold standard for treating invasive cancers, there is strong evidence that neoadjuvant chemotherapy prior to cystectomy can prolong life. Nevertheless, drug-associated toxicities along with inadequate chemotherapy responses can lead to unnecessary resource expenditures and impacts on patients’ quality of life [41]. Recently, Cha et al. devised a study that analyzed different radiomics-based predictive models using deep learning to accurately classify tumors according to their chemotherapy responses. The model used over 6,000 pre- and post-treatment paired regions of interests from CT scans and compared the results to that of two expert radiologists. The differences between the deep learning algorithms and the radiologists in predicting chemotherapy responses did not reach statistical significance [42]. Although the model is promising, the use of only two radiologists allows for the possibility that the pipeline is fitting for radiologist’s bias. As a whole, these findings demonstrates how AI can be used to monitor treatment responses, allowing clinicians to spare resources and avoid unnecessary drug-associated toxicity in patients’ whose cancers are resistant to chemotherapy. Table 3 summarizes the advancements of AI in bladder cancer management.
Table 3.
Bladder cancer review articles summary
| Legend | Name/Type of Model | No. of patients | Cancer | Summary | Data Analysis | Implications | References |
|---|---|---|---|---|---|---|---|
| 3-A | CNN | 479 patients with 44 different urologic findings | Bladder Cancer | Multiple CNN models were developed to identify various urologic findings from cystocopic images. | The Xception-based CNN achieved the highest F1 score (99.52%), and all models were correctly able to identify cancerous lesions. | This study demonstrates the potential for artificial intelligence to be used to improve accuracy in diagnosis and reduce examiner-variability in analyzing cystoscopic images. | 37. Eminaga, O. et al. |
| 3-B | Random Forests (multiple) | 43 patients without bladder cancer | Bladder Cancer | Three different machine learning methods were used to analyze atomic force microscopy (AFM) images of urine samples to non-invasively identify evidence of bladder cancer. | This method showed 94% diagnostic accuracy, which is a statistically significant improvement in diagnostic accuracy compared to current gold standard cystoscopy. | AI has the ability to aid in diagnosis of bladder cancer from non-invasive methods with high accuracy, implying a plausible shift in the standard of care in the diagnosis of bladder cancer from more invasive methods. | 38. Sokolov, I. et al. |
| 25 patients with pathologically-proven bladder cancer | |||||||
| 3-C | Multiple (NN, SVM, Random forest) | 76 patients with 84 bladder cancer lesions | Bladder Cancer | A machine learning algorithm was developed to stage bladder cancer into high (≥ pathologic stage T2) and low (< pathologic stage T2) based on CT urography images. | AUC for each of the models ranged between 0.88-0.97. | AI has the potential to accurately determine prognostically relevant information, such as stage of bladder cancer, which has major effects on treatment planning. | 39. Garapati, S.S. et al. |
| 3-D | CNN | 82 patients with 87 bladder cancers for training | Bladder Cancer | A CNN was used to develop 3 predictive models that can distinguish between bladder cancers that have fully responded to neoadjuvant chemotherapy and those that have not based on analysis of pre- and post-treatment CT images. | All 3 models performed comparably in terms of AUC to the two expert radiologists. | ML can be used to monitor treatment responses, allowing clinicians to conserve resources and avoid unnecessary drug-associated toxicity in patients whose cancers are resistant to chemotherapy. | 41. Cha, K.H. et al. |
| 41 patients with 43 bladder cancers for testing |
Discussion
The involvement of AI in healthcare has been discussed for the last two decades, but only recently have we started working towards harnessing its true potential. The use of AI in clinical decision-making takes advantage of its ability to pick out patterns in large data sets and use a training algorithm to make accurate predictions. While the volume of data makes this task extremely difficult and time consuming for humans, computers however have been shown to do it efficiently and accurately. Moreover, processes such as identifying, classifying, and staging cancerous lesions use significant resources and are open to a level of subjectivity. Automation of these processes could help conserve these resources and reduce this level of subjectivity, thus improving the efficiency and accuracy of how GU cancer patients are diagnosed and treated.
The estimated 3-year total Medicare cost of the diagnosis and treatment of prostate cancer alone in men over the age of 70 is 1.2 billion dollars [3]. This, combined with the increasing incidence of GU cancers, demonstrates the importance of conserving resources without sacrificing outcomes in these patients [2]. Incorporating AI in the diagnosis and treatment of these patients is one way this could successfully be accomplished. While there may be an initial investment in implementing the technology, the eventual cost savings could certainly be significant.
The accurate grading of tumors plays a crucial role in determining both prognosis and potential therapeutic options. Automating these processes could allow clinicians to make more accurate decisions in managing the care of these patients. Furthermore, this may help patients avoid the side effects of therapies that are not likely to be successful. Thus, by reducing subjectivity it is possible to reduce the morbidity and mortality in cancer patients while improving quality of life. Overall, as the clinical data available to physicians continues to expand, AI will likely have an increased role in automation, pattern recognition, and predictive modeling. Therefore, it is imperative to create pipelines that can serve to further the field of urologic oncology while focusing on improving patient care.
Acknowledgements
We thank all the mentors (Dr. Dipen J Parekh), collaborators (Dr. Joshua M Hare, Dr. Bonnie Blomberg and Dr. Andrew V Schally), and interns for their insights, suggestions and support during this study. Additionally, we would like to thank the Clinician Scientist Development Grant from American Cancer Society (1335-78-CSDG-19-016-01-CSM), American Urological Association Research Scholar Award and Clinical and Translational Science Institute supported by NIH (UL1TR002736), and the Sexual Medicine Society of North America (SMSNA) for HA and RR. Supported by the American Urological Association Research Scholar Award and Stanley Glaser Award to HA.
Disclosure of conflict of interest
None.
References
- 1.You W, Henneberg M. Cancer incidence increasing globally: the role of relaxed natural selection. Evol Appl. 2018;11:140–152. doi: 10.1111/eva.12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagheri MH, Ahlman MA, Lindenberg L, Turkbey B, Lin J, Cahid Civelek A, Malayeri AA, Agarwal PK, Choyke PL, Folio LR, Apolo AB. Advances in medical imaging for the diagnosis and management of common genitourinary cancers. Urol Oncol. 2017;35:473–491. doi: 10.1016/j.urolonc.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trogdon JG, Falchook AD, Basak R, Carpenter WR, Chen RC. Total Medicare costs associated with diagnosis and treatment of prostate cancer in elderly men. JAMA Oncol. 2019;5:60–66. doi: 10.1001/jamaoncol.2018.3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hemal AK, Menon M. Robotics in urology. Curr Opin Urol. 2004;14:89–93. doi: 10.1097/00042307-200403000-00007. [DOI] [PubMed] [Google Scholar]
- 5.Koza J, Bennett Iii F, Andre D, Keane M. Automated design of both the topology and sizing of analog electrical circuits using genetic programming. 1998 [Google Scholar]
- 6.Bishop CM. Pattern Recognition and Machine Learning (Information Science and Statistics) Springer-Verlag. 2006 [Google Scholar]
- 7.Aizenberg IN, Aizenberg NN, Vandewalle JP. Multi-valued and universal binary neurons: theory, learning and applications. Kluwer Academic Publishers. 2000 [Google Scholar]
- 8.Anagnostou T, Remzi M, Lykourinas M, Djavan B. Artificial neural networks for decision-making in urologic oncology. Eur Urol. 2003;43:596–603. doi: 10.1016/s0302-2838(03)00133-7. [DOI] [PubMed] [Google Scholar]
- 9.Puri M, Pathak Y, Sutariya VK, Tipparaju S, Moreno W. Artificial neural network for drug design, delivery and disposition. 2015 [Google Scholar]
- 10.Anwar SM, Majid M, Qayyum A, Awais M, Alnowami M, Khan MK. Medical image analysis using convolutional neural networks: a review. J Med Syst. 2018;42:226. doi: 10.1007/s10916-018-1088-1. [DOI] [PubMed] [Google Scholar]
- 11.Hamet P, Tremblay J. Artificial intelligence in medicine. Metabolism. 2017;69S:S36–S40. doi: 10.1016/j.metabol.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Langlotz CP, Allen B, Erickson BJ, Kalpathy-Cramer J, Bigelow K, Cook TS, Flanders AE, Lungren MP, Mendelson DS, Rudie JD, Wang G, Kandarpa K. A roadmap for foundational research on artificial intelligence in medical imaging: from the 2018 NIH/RSNA/ACR/the academy workshop. Radiology. 2019;291:781–791. doi: 10.1148/radiol.2019190613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das N, Topalovic M, Janssens W. Artificial intelligence in diagnosis of obstructive lung disease: current status and future potential. Curr Opin Pulm Med. 2018;24:117–123. doi: 10.1097/MCP.0000000000000459. [DOI] [PubMed] [Google Scholar]
- 14.Olczak J, Fahlberg N, Maki A, Razavian AS, Jilert A, Stark A, Sköldenberg O, Gordon M. Artificial intelligence for analyzing orthopedic trauma radiographs. Acta Orthop. 2017;88:581–586. doi: 10.1080/17453674.2017.1344459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krittanawong C, Zhang H, Wang Z, Aydar M, Kitai T. Artificial intelligence in precision cardiovascular medicine. J Am Coll Cardiol. 2017;69:2657–2664. doi: 10.1016/j.jacc.2017.03.571. [DOI] [PubMed] [Google Scholar]
- 16.Linder N, Taylor JC, Colling R, Pell R, Alveyn E, Joseph J, Protheroe A, Lundin M, Lundin J, Verrill C. Deep learning for detecting tumour-infiltrating lymphocytes in testicular germ cell tumours. J Clin Pathol. 2019;72:157–164. doi: 10.1136/jclinpath-2018-205328. [DOI] [PubMed] [Google Scholar]
- 17.Albertsen PC. Patient Decision-making: where are we going? Eur Urol. 2019;75:908–909. doi: 10.1016/j.eururo.2018.10.024. [DOI] [PubMed] [Google Scholar]
- 18.Auffenberg GB, Ghani KR, Ramani S, Usoro E, Denton B, Rogers C, Stockton B, Miller DC, Singh K Michigan Urological Surgery Improvement Collaborative. askMUSIC: leveraging a clinical registry to develop a new machine learning model to inform patients of prostate cancer treatments chosen by similar men. Eur Urol. 2019;75:901–907. doi: 10.1016/j.eururo.2018.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arvaniti E, Fricker KS, Moret M, Rupp N, Hermanns T, Fankhauser C, Wey N, Wild PJ, Rüschoff JH, Claassen M. Claassen, automated gleason grading of prostate cancer tissue microarrays via deep learning. Sci Rep. 2018;8:12054. doi: 10.1038/s41598-018-30535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lucas M, Jansen I, Savci-Heijink CD, Meijer SL, de Boer OJ, van Leeuwen TG, de Bruin DM, Marquering HA. Deep learning for automatic Gleason pattern classification for grade group determination of prostate biopsies. Virchows Arch. 2019;475:77–83. doi: 10.1007/s00428-019-02577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozkan TA, Eruyar AT, Cebeci OO, Memik O, Ozcan L, Kuskonmaz I. Interobserver variability in Gleason histological grading of prostate cancer. Scand J Urol. 2016;50:420–424. doi: 10.1080/21681805.2016.1206619. [DOI] [PubMed] [Google Scholar]
- 22.Fehr D, Veeraraghavan H, Wibmer A, Gondo T, Matsumoto K, Vargas HA, Sala E, Hricak H, Deasy JO. Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc Natl Acad Sci U S A. 2015;112:E6265–6273. doi: 10.1073/pnas.1505935112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicolae A, Morton G, Chung H, Loblaw A, Jain S, Mitchell D, Lu L, Helou J, Al-Hanaqta M, Heath E, Ravi A. Evaluation of a machine-learning algorithm for treatment planning in prostate low-dose-rate brachytherapy. Int J Radiat Oncol Biol Phys. 2017;97:822–829. doi: 10.1016/j.ijrobp.2016.11.036. [DOI] [PubMed] [Google Scholar]
- 24.Elmahdy MS, Jagt T, Zinkstok RT, Qiao Y, Shahzad R, Sokooti H, Yousefi S, Incrocci L, Marijnen CAM, Hoogeman M, Staring M. Robust contour propagation using deep learning and image registration for online adaptive proton therapy of prostate cancer. Med Phys. 2019;46:3329–3343. doi: 10.1002/mp.13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kooy HM, Grassberger C. Intensity modulated proton therapy. Br J Radiol. 2015;88:20150195. doi: 10.1259/bjr.20150195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Obermeyer Z, Powers B, Vogeli C, Mullainathan S. Dissecting racial bias in an algorithm used to manage the health of populations. Science. 2019;366:447–453. doi: 10.1126/science.aax2342. [DOI] [PubMed] [Google Scholar]
- 27.Rossi SH, Klatte T, Usher-Smith J, Stewart GD. Epidemiology and screening for renal cancer. World J Urol. 2018;36:1341–1353. doi: 10.1007/s00345-018-2286-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jonasch E, Gao J, Rathmell WK. Renal cell carcinoma. BMJ. 2014;349:g4797. doi: 10.1136/bmj.g4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muglia VF, Prando A. Renal cell carcinoma: histological classification and correlation with imaging findings. Radiol Bras. 2015;48:166–174. doi: 10.1590/0100-3984.2013.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han S, Hwang SI, Lee HJ. The classification of renal cancer in 3-phase CT images using a deep learning method. J Digit Imaging. 2019;32:638–643. doi: 10.1007/s10278-019-00230-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holdbrook DA, Huber RG, Marzinek JK, Stubbusch A, Schmidtchen A, Bond PJ. Automated renal cancer grading using nuclear pleomorphic patterns. Pharmacol Res. 2019;147:104372. doi: 10.1016/j.phrs.2019.104372. [DOI] [PubMed] [Google Scholar]
- 32.Lin F, Cui EM, Lei Y, Luo LP. CT-based machine learning model to predict the Fuhrman nuclear grade of clear cell renal cell carcinoma. Abdom Radiol (NY) 2019;44:2528–2534. doi: 10.1007/s00261-019-01992-7. [DOI] [PubMed] [Google Scholar]
- 33.Dreicer R. Tyrosine kinase inhibitors compared with cytokine therapy for metastatic renal cell carcinoma: overview of recent clinical trials differentiating clinical response and adverse effects. Clin Genitourin Cancer. 2006;5(Suppl 1):S19–23. doi: 10.3816/cgc.2006.s.003. [DOI] [PubMed] [Google Scholar]
- 34.Buchner A, Kendlbacher M, Nuhn P, Tüllmann C, Haseke N, Stief CG, Staehler M. Outcome assessment of patients with metastatic renal cell carcinoma under systemic therapy using artificial neural networks. Clin Genitourin Cancer. 2012;10:37–42. doi: 10.1016/j.clgc.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 35.Malats N, Real FX. Epidemiology of bladder cancer. Hematol Oncol Clin North Am. 2015;29:177–189. vii. doi: 10.1016/j.hoc.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Berdik C. Unlocking bladder cancer. Nature. 2017;551:S34. doi: 10.1038/551S34a. [DOI] [PubMed] [Google Scholar]
- 37.Maas M, Bedke J, Stenzl A, Todenhöfer T. Can urinary biomarkers replace cystoscopy? World J Urol. 2019;37:1741–1749. doi: 10.1007/s00345-018-2505-2. [DOI] [PubMed] [Google Scholar]
- 38.Eminaga O, Eminaga N, Semjonow A, Breil B. Diagnostic classification of cystoscopic images using deep convolutional neural networks. JCO Clin Cancer Inform. 2018:1–8. doi: 10.1200/CCI.17.00126. [DOI] [PubMed] [Google Scholar]
- 39.Sokolov I, Dokukin ME, Kalaparthi V, Miljkovic M, Wang A, Seigne JD, Grivas P, Demidenko E. Noninvasive diagnostic imaging using machine-learning analysis of nanoresolution images of cell surfaces: detection of bladder cancer. Proc Natl Acad Sci U S A. 2018;115:12920–12925. doi: 10.1073/pnas.1816459115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garapati SS, Hadjiiski L, Cha KH, Chan HP, Caoili EM, Cohan RH, Weizer A, Alva A, Paramagul C, Wei J, Zhou C. Urinary bladder cancer staging in CT urography using machine learning. Med Phys. 2017;44:5814–5823. doi: 10.1002/mp.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buttigliero C, Tucci M, Vignani F, Scagliotti GV, Di Maio M. Molecular biomarkers to predict response to neoadjuvant chemotherapy for bladder cancer. Cancer Treat Rev. 2017;54:1–9. doi: 10.1016/j.ctrv.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 42.Cha KH, Hadjiiski L, Chan HP, Weizer AZ, Alva A, Cohan RH, Caoili EM, Paramagul C, Samala RK. Bladder cancer treatment response assessment in CT using radiomics with deep-learning. Sci Rep. 2017;7:8738. doi: 10.1038/s41598-017-09315-w. [DOI] [PMC free article] [PubMed] [Google Scholar]


