Abstract
PARP inhibitors have shown remarkable efficacy in the clinical management of several BRCA-mutated tumors. This approach is based on the long-standing hypothesis that PARP inhibition will impair the repair of single stranded breaks, causing synthetic lethality in tumors with loss of high-fidelity double-strand break homologous recombination. While this is now well accepted and has been the basis of several successful clinical trials, emerging evidence strongly suggests that mutation to several additional genes involved in homologous recombination may also have predictive value for PARP inhibitors. While this notion is supported by early clinical evidence, the mutation frequencies of these and other functionally related genes are largely unknown, particularly in cancers not classically associated with homologous recombination deficiency. We therefore evaluated the mutation status of 22 genes associated with the homologous recombination DNA repair pathway or PARP inhibitor sensitivity, first in a pan-cancer cohort of 55,586 patients, followed by a more focused analysis in The Cancer Genome Atlas cohort of 12,153 patients. In both groups we observed high rates of mutations in a variety of HR-associated genes largely unexplored in the setting of PARP inhibition, many of which were associated also with poor clinical outcomes. We then extended our study to determine which mutations have a known oncogenic role, as well as similar to known oncogenic mutations that may have a similar phenotype. Finally, we explored the individual cancer histologies in which these genomic alterations are most frequent. We concluded that the rates of deleterious mutations affecting genes associated with the homologous recombination pathway may be underrepresented in a wide range of human cancers, and several of these genes warrant further and more focused investigation, particularly in the setting of PARP inhibition and HR deficiency.
Subject terms: Tumour biomarkers, Cancer, Cancer genetics
Introduction
Precision medicine has become standard of care in the management of several malignancies. This approach involves the identification of clinically actionable molecular features, typically via Next Generation Sequencing (NGS), and the subsequent implementation of a specific, targeted therapy. For example, Tyrosine kinase inhibitors such as imatinib, bosutinib, and dasatinib targeting the BCR-ABL fusion protein have improved outcomes in Philadelphia chromosome positive leukemia1, and similar approaches have made a considerable impact in breast cancer2, non-small cell lung cancer (NSCLC)3, and several other cancer types. This approach has significantly improved outcomes in a variety of tumor types, as a recent pan-cancer trial of 1144 patients determined that those harboring distinct molecular aberrations and treated with a matched targeted therapy had significant improvements in overall response rates, time-to-treatment failure, and overall survival4.
There is a large body of evidence strongly supporting the use of selective PARP inhibitors such as olaparib or talazoparib in BRCA-mutated tumors5. This approach has strong scientific rationale, as patients with BRCA mutations are thought to have homologous recombination deficiency (HRD), thereby limiting their ability to repair double stranded DNA breaks. The use of PARP inhibitors in these patients limits their ability to undergo single stranded break repair, leading to the accumulation of DNA damage and eventually cell death6,7. This approach has shown clinical efficacy in the management of BRCA-mutated breast, ovarian, pancreas, and prostate cancers8–11, and more recently glioblastoma multiforme and metastatic thymomas12,13.
While BRCA is a strong predictor for the efficacy of PARP inhibitors, homologous recombination (HR) involves a wide range of additional DNA repair genes, some of which may also have predictive value for PARP inhibitors. For instance, in metastatic prostate cancer, mutations to genes more modestly associated with the pathway such as ATM, CHEK2, and PALB2 are strongly associated with clinical responses to olaparib14,15. For example, ATM-deficient cell lines have shown increased sensitivity to PARP inhibition than their ATM-proficient counterparts in variety of cancer types16–20. Likewise, 88% of prostate cancer patients with CHECK2 mutations showed clinical responses to PARP inhibition14, with similar results observed in other studies, many including additional cancer histologies21–24. Similarly, PALB2 mutated breast cancer also appears to be highly sensitive to PARP inhibition25. This appears to extend to genes that are far up or downstream of the HR pathway, as PTEN deficient tumors have been suggested to respond to PARP inhibitors due to loss of RAD51, though this remains unclear and PTEN is not currently considered a useful predictor for PARP inhibition26–29. However, it is clear that there are several additional HR associated mutations that may also be informative when stratifying patients for PARP inhibition.
While several studies have explored the mutation frequency and predictive value of established HR associated genes such as BRCA1/2 or upstream HR-associated genes ATM, CHEK2, and PALB2, few have evaluated alterations to other functionally related HR genes. This is particularly true for genes more weakly associated with HRD, some of which are beginning to show predictive value for PARP inhibition30. Such genes include BARD1, BRIP1, FAAP20, FAN1, FANCE, FANCM, RAD51B, RAD51C, and RAD51D, all of which have been suggested to predict for PARP inhibitor sensitivity31–33, with additional context specific roles emerging for genes such as POLQ. For instance, though loss of POLQ appears to upregulate HR activity in HR-proficient cells, loss of POLQ is also seemingly central to PARP inhibitor sensitivity in the setting of topoisomerase, ATR, or FANCD2-deficiency34,35.
Hence, it is clear that stratifying patients based solely BRCA mutations will likely under predict for those who will derive clinical benefit from PARP inhibition. We therefore evaluated the mutation status of 22 genes with either established, emerging, or potential roles in either the HR repair pathway or PARP inhibitor sensitivity, first in a pan cancer analysis of over 55,000 patients compiled from several genomic databases, followed by a more focused analysis of The Cancer Genome Atlas (TCGA) cohort, which allowed for more insight into disease-specific mutation frequencies. Interestingly, we observed high rates of mutations in a variety of largely unexplored HR genes, many of which were associated with poor clinical outcomes. We then identified the individual cancer types in which these alterations are most frequent. Though many of the observed mutations are currently of unknown significance, these newly identified genomic alterations warrant further investigation, particularly in the setting of homologous recombination deficiency and PARP inhibition.
Methods
Pan-cancer genomic database analysis
Patient data was visualized using cBioportal for Cancer Genomics as described in the original references36,37, and DNA/RNA sequencing analyses and protocols can be found per the references listed above. Using this dataset, survival was assessed using the Kaplan Meier method. Subsequent genetic analyses were restricted to fully sequenced tumors and gene sequences compared to a reference population as described previously38. A complete list of studies included in this analysis is listed in the supplemental materials section.
TCGA database analysis
Provisional TCGA patient datasets were downloaded (https://tcga-data.nci.nih.gov/tcga/) and visualized using cBioportal for Cancer Genomics as described. Detailed information regarding the TCGA dataset and DNA sequencing analyses and protocols can be found on the TCGA data portal webpage listed above. Like the pan-cancer dataset, survival was assessed using the Kaplan Meier method, and subsequent genetic analyses were restricted to fully sequenced tumors also as described previously38.
List of studies included in TCGA analysis
Data from each of the following studies was compiled and visualized as described above: Pan-Lung Cancer (TCGA, Nat Genet 2016), Adrenocortical Carcinoma (TCGA, Provisional), Cholangiocarcinoma (TCGA, Provisional), Bladder Urothelial Carcinoma (TCGA, Provisional), Colorectal Adenocarcinoma (TCGA, Provisional), Breast Invasive Carcinoma (TCGA, Provisional), Brain Lower Grade Glioma (TCGA, Provisional), Merged Cohort of LGG and GBM (TCGA, Cell 2016), Glioblastoma Multiforme (TCGA, Provisional), Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma (TCGA, Provisional), TCGA data for Esophagus-Stomach Cancers (TCGA, Nature 2017), Esophageal Carcinoma (TCGA, Provisional), Stomach Adenocarcinoma (TCGA, Provisional), Uveal Melanoma (TCGA, Provisional), Head and Neck Squamous Cell Carcinoma (TCGA, Provisional), Kidney Renal Clear Cell Carcinoma (TCGA, Provisional), Kidney Chromophobe (TCGA, Provisional), Kidney Renal Papillary Cell Carcinoma (TCGA, Provisional), Liver Hepatocellular Carcinoma (TCGA, Provisional), Lung Adenocarcinoma (TCGA, Provisional), Lung Squamous Cell Carcinoma (TCGA, Provisional), Lymphoid Neoplasm Diffuse Large B-cell Lymphoma (TCGA, Provisional), Acute Myeloid Leukemia (TCGA, Provisional), Ovarian Serous Cystadenocarcinoma (TCGA, Provisional), Pancreatic Adenocarcinoma (TCGA, Provisional), Pancreatic Adenocarcinoma (ICGC, Nature 2012), Mesothelioma (TCGA, Provisional), Prostate Adenocarcinoma (TCGA, Provisional), Skin Cutaneous Melanoma (TCGA, Provisional), Pheochromocytoma and Paraganglioma (TCGA, PanCancer Atlas), Sarcoma (TCGA, Provisional), Testicular Germ Cell Cancer (TCGA, Provisional), Thymoma (TCGA, Provisional), Thyroid Carcinoma (TCGA, Provisional), Uterine Carcinosarcoma (TCGA, Provisional), Uterine Corpus Endometrial Carcinoma (TCGA, Provisional).
Inclusion/exclusion criteria
All genomic analyses were restricted to fully sequenced tumors. All studies listed were included in pan-cancer survival analyses, though mutation frequencies were limited to samples with an N ≥ 25.
Statistical analysis
Data were analyzed by either student’s T test, Xi squared test, or ANOVA fit to a general linear model in Minitab express, the validity of which was tested by adherence to the normality assumption and the fitted plot of the residuals. Results were considered significant at either p or q < 0.05 unless otherwise noted.
Results
Mutations to genes associated with the homologous recombination pathway predict for poor clinical outcomes in a large pan-cancer study
To determine the frequency of pathway mutations in a large sample size, we first evaluated the mutation status of 22 key homologous recombination genes in a pooled pan-cancer cohort of 55,586 patients from 32 different cancer types (individual studies detailed in the supplemental methods). These genes include: ATM, BARD1, BRCA1, BRCA2, BRIP1, CDK12, CHEK2, DMC1, FAAP20, FAN1, FANCD2, FANCE, FANCL, FANCM, PALB2, POLQ, RAD51, RAD51B, RAD51C, RAD51D, RAD54L, and XRCC3. HR pathway mutations were common in this cohort, affecting 7117 (13.4%) of patients. ATM and BRCA2 mutations were most common, affecting 2160 (4.1%) and 1452 (2.7%) of patients respectively, followed by BRCA1 (822 or 1.5%), CDK12 (805 or 1.5%), and POLQ (634 or 1.19%). These mutation frequencies are summarized in Table 1.
Table 1.
Mutation frequencies of genes associated with the homologous recombination DNA repair pathway in a pan cancer cohort (N = 55,586).
| Gene | Observed mutations pan cancer (N = 55,586) | Mutation frequency (%) |
|---|---|---|
| ATM | 2160 | 4.056 |
| BRCA2 | 1452 | 2.727 |
| BRCA1 | 822 | 1.544 |
| CDK12 | 805 | 1.512 |
| POLQ | 634 | 1.191 |
| BRIP1 | 573 | 1.076 |
| PALB2 | 507 | 0.952 |
| FANCM | 494 | 0.928 |
| CHEK2 | 479 | 0.899 |
| BARD1 | 429 | 0.806 |
| FANCD2 | 377 | 0.708 |
| RAD54L | 235 | 0.441 |
| FAN1 | 189 | 0.355 |
| RAD51C | 166 | 0.312 |
| FANCE | 134 | 0.252 |
| RAD51B | 150 | 0.282 |
| RAD51D | 112 | 0.210 |
| RAD51 | 109 | 0.205 |
| DMC1 | 84 | 0.158 |
| FANCL | 70 | 0.131 |
| FAAP20 | 39 | 0.073 |
| XRCC3 | 34 | 0.064 |
| Any HR mutation | 7117 | 13.365 |
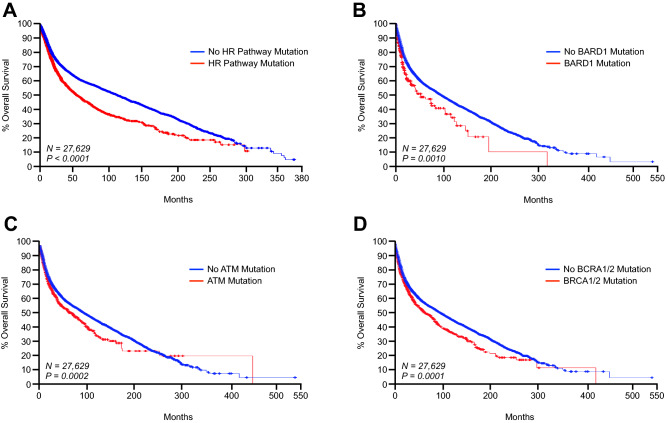
Of the initial 55,586 patients, survival data was available for 33,633 (60.5%). Of these 33,633 patients, 4472 (13.3%) had an identifiable mutation to the queried HR genes, whereas 29,161 (86.7%) did not. Additionally, patients with any HR pathway mutation had significantly poorer outcomes, with a median overall survival of 60.5 months compared to the 105.91 months in patients with no HR pathway mutation (Fig. 1, Table 2). Interestingly, several HR genes were independent predictors of poor outcomes including ATM, BARD1, BRCA2, CDK12, DMC1, FAAP20, PALB2, and POLQ, though it is important to note that it is unlikely that these patients were treated with a PARP inhibitor (Fig. 1, Table 2).
Figure 1.
Mutations to genes associated with the homologous recombination pathway predict for poor clinical outcomes in a large pan-cancer study. We determined the mutation status of ATM, BARD1, BRCA1, BRCA2, BRIP1, CDK12, CHEK2, DMC1, FAAP20, FAN1, FANCD2, FANCE, FANCL, FANCM, PALB2, POLQ, RAD51, RAD51B, RAD51C, RAD51D, RAD54L, and XRCC3 in a in a pooled pan-cancer cohort of 55,586 patients from 32 different cancer types. Of these patients, survival data was available from 27,629, which were used for subsequent analyses. Kaplan Meier plots are displayed showing overall survival from patients with or without a mutation to: (A) one or more of the genes listed above, (B) BARD1, (C) ATM, or (D) BRCA1 and/or BRCA2.
Table 2.
Select mutations to genes associated with the homologous recombination DNA repair pathway are associated with poor survival in a pan cancer cohort (N = 33,633).
| Gene | Median months survival without mutation | Median months survival with mutation | P value |
|---|---|---|---|
| All genes | 105.91 | 60.50 | P < 0.0001 |
| ATM | 99.97 | 71.68 | P = 1.610 × 10–4 |
| BARD1 | 98.80 | 53.77 | P = 1.34 × 10–4 |
| BRCA1 | 99.00 | 82.63 | P = 0.203 |
| BRAC2 | 99.90 | 73.16 | P = 1.577 × 10–4 |
| BRCA1/2 | 100.62 | 73.16 | P = 1.792 × 10–5 |
| BRIP1 | 98.80 | 80.00 | P = 0.139 |
| CHEK2 | 86.37 | 98.5 | P = 0.872 |
| CDK12 | 99.40 | 53.15 | P = 5.195 × 10–3 |
| DMC1 | 98.77 | 50.3 | P = 5.579 × 10–3 |
| FAAP20 | 98.70 | 37.71 | P = 0.0136 |
| FAN1 | 98.70 | 76.97 | P = 0.833 |
| FANCD2 | 98.70 | 74.00 | P = 0.606 |
| FANCE | 98.32 | – | – |
| FANCL | 98.50 | 109/00 | P = 0.885 |
| FANCM | 98.83 | 57.59 | P = 0.0567 |
| PALB2 | 98.90 | 50.72 | P = 0.0114 |
| POLQ | 99.53 | 63.50 | P = 1.141 × 10–3 |
| RAD51 | 98.50 | 72.01 | P = 0.809 |
| RAD51B | 98.370 | 156.9 | P = 0.549 |
| RAD51C | 98.70 | 42.00 | P = 0.137 |
| RAD51D | 98.70 | 75.43 | P = 0.121 |
| RAD54L | 98.50 | 77.00 | P = 0.367 |
| XRCC3 | 98.37 | 109.00 | P = 0.295 |
Mutations to genes associated with the homologous recombination pathway similarly predict poor clinical outcomes in The Cancer Genome Atlas cohorts
While these data suggest that as a whole, HR pathway mutations may have prognostic value, these results may be skewed should HR mutations be more frequently observed in more aggressive cancers. Additionally, given the relatively small sample sizes of several individualized cancer cohorts included in our pan-cancer analysis and varied methods of measuring outcomes, we next repeated the study, this time restricting our analysis to the cancer genome atlas (TCGA) datasets (N = 12,153). Though the combined TCGA dataset has a smaller combined sample size, these data represent a smaller number of cancer types typically with larger sample sizes in each. Additionally, while outcomes were not available for each for roughly half of patients in the previous pan cancer dataset, in the TCGA dataset survival data was available for nearly all patients.
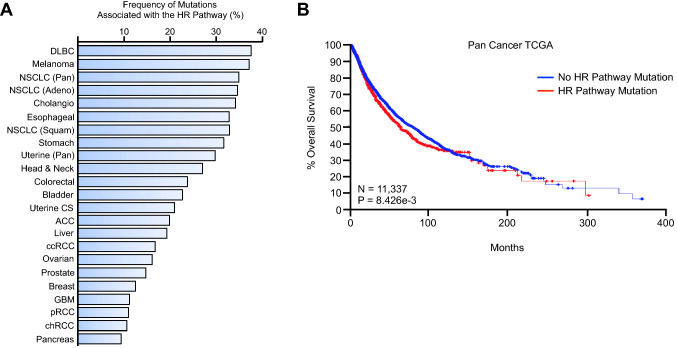
Using this new sample set, we determined the rate of mutations to the HR pathway both overall and by by cancer type (Fig. 2A). HR pathway mutations were particularly common among diffuse large B cell lymphomas and melanoma patients, with combined mutation rates of 37.5 and 37.33%, respectively (Fig. 2A). This was closely followed by lung adenocarcinoma (34.78%), cholangiocarcinoma (34.29%), pan-esophageal cancer (32.97%), squamous cell lung cancer (32.96%), pan-stomach cancer (31.65%), and pan-uterine cancer (30%) (Fig. 2A). Several other cancer types had mutation frequencies between 20 and 30%, including pan-head and neck, colorectal, uterine carcinoma, and adenoid cystic carcinoma (Fig. 2A). Once again, mutations affecting the combined gene set were associated with poor outcomes in the combined cancer cohort (Fig. 2B), with several mutations to select also independently predicting for poor outcomes (Supplementary Table S1).
Figure 2.
Mutations to genes associated with the homologous recombination pathway are frequent in several cancer histologies for which PARP inhibitors are not currently approved. (A) We determined the mutation status of ATM, BARD1, BRCA1, BRCA2, BRIP1, CDK12, CHEK2, DMC1, FAAP20, FAN1, FANCD2, FANCE, FANCL, FANCM, PALB2, POLQ, RAD51, RAD51B, RAD51C, RAD51D, RAD54L, and XRCC3 in a in The Cancer Genome Atlas (TCGA) pan-cancer cohort of 12,153 patents, and show the combined mutation frequency across all genes by percent for the most represented cancer types. DLBC: diffuse large B cell lymphoma, NSCLC: non-small cell lung cancer, ACC: adenoid cystic carcinoma, ccRCC: clear cell renal cell carcinoma, GBM: glioblastoma multiforme, pRCC: papillary renal cell carcinoma, chRCC: chromophobe renal cell carcinoma. (B) Of these 12,153 patients, survival data was available from 11,337, which were used for subsequent analyses. A Kaplan–Meier plot is displayed showing overall survival from patients with or without a mutation to one or more of the genes listed above.
Mutations to genes associated with the homologous recombination pathway are heterogeneous and frequently associate with mutations to a variety of unrelated genes
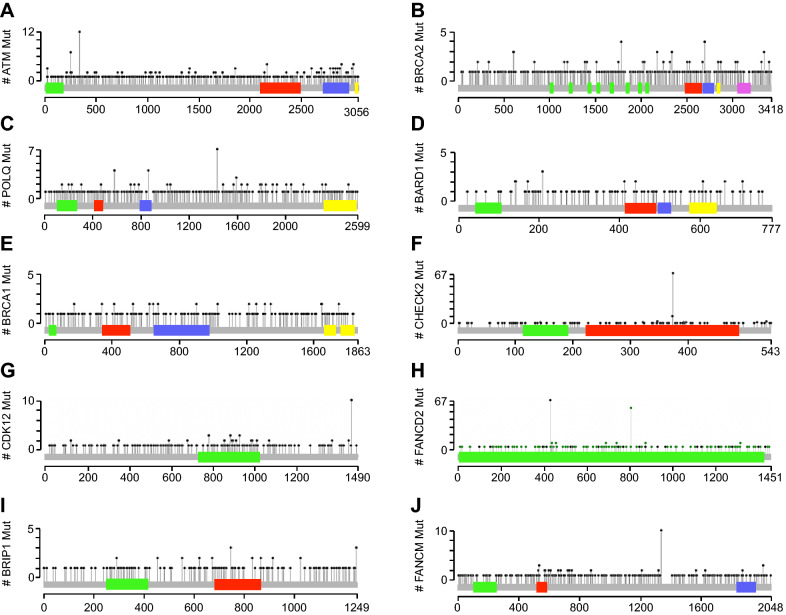
We next analyzed the frequency of mutations affecting each individual gene. Once again, ATM was the most frequently altered gene, with mutation observed in 4% of all cases (Supplementary Table S2). With respect to ATM, we observed a total of 485 mutations, 339 of which were missense, 142 truncating, and 4 in-frame mutations of unknown significance (Fig. 3A). This was followed by BARD1 (2.51%), BRCA1 (2.49%), and BRCA2 (1.98%), each with a similar distribution of mutations. While mutations to POLQ, FANCM, CHECK2, CDK12, and FANCD2 were among the most heterogeneous, these had a relatively low frequency, most effecting only one patient (Fig. 3B–J).
Figure 3.
Location of the most frequently represented mutations in genes associated with the homologous recombination pathway in The Cancer Genome Atlas cohort. Lolipop plots displaying the most common mutations to (A) ATM (B) BRCA2 (C) POLQ (D) BARD1 (E) BRCA1 (F) CHEK2 (G) CDK12 (H) FANCD2 (I) BRIP1 (J) FANCM.
We subsequently analyzed the entirety of the mutations identified in this study using the OncoKB precision oncology knowledge base39. This approach predicts for mutations most likely to alter protein function, as well as compares these mutations to those reported in the literature to provide additional insight into which are likely oncogenic, neutral mutations, or of unknown significance. While the majority of mutations identified in this study have yet to be uncharacterized, a sizeable fraction was analogous to those reported previously to have a role in PARP inhibitor sensitivity and/or HRD and likely to have oncogenic function, though this requires further exploration (Supplementary Table S3). Interestingly, several HR-associated mutations often co-occurred in the same patients, suggesting patients with select HR-associated mutations are likely to incur additional HR-associated mutations (Supplementary Table S4). Additionally, patients with HR-associated mutations also harbored mutations to several non-HR genes with higher frequency than those without HR associated mutations (Supplementary Table S5), several of which were also independent predictors of poor clinical outcomes (Supplementary Table S6).
Mutations to genes associated with the homologous recombination pathway are frequent in several cancer histologies for which PARP inhibitors are not currently approved
In order to identify genes that may be the most useful in determining the status of the HR pathway in select cancer types, we next determined the mutation frequency of these 22 genes in the eight cancers with the highest rate of HR mutation. As mentioned previously, HR mutations were observed most frequently in diffuse large B cell lymphoma, affecting roughly 38% of patients, though this may be inflated given the small sample size of the study (N = 47). In diffuse large B cell lymphoma patients, ATM mutations were the most frequently represented, affecting nearly 15% of patients, followed by POLQ which was mutated in 10.6% (Supplementary Table S7). Other mutations were less common, but again the relevance of these findings are limited due to the small sample size, and warrant exploration in a larger cohort.
Also as mentioned, HR mutations were also common in cutaneous melanomas (37.5%), though this represented data from 288 patients. In this group, BRCA mutations were observed in 11.5% of patients, though mutations to ATM, BRIP1, FANCM, and other genes were also common (Table 3). This was paralleled by both lung adenocarcinoma (N = 660) and squamous lung cancers (N = 484), which had an overall BRCA mutation frequency of 7.7% and 10.7%, respectively (Table 4). These cancers also had high rates of ATM, POLQ, FANCM mutations, as well as those to several other genes (Table 5). ATM, BRCA, CHECK2, and CDK12 mutations were frequent in cholangiocarcinoma (Supplementary Table S8, N = 34) with similar results in uterine carcinoma (Supplementary Table S9, N = 57), though the significance of these results is limited by small sample sizes.
Table 3.
Mutation frequencies of genes associated with the homologous recombination DNA repair pathway in the TCGA Cutaneous Melanoma cohort (N = 288).
| Gene | Observed mutations melanoma (N = 288) | Mutation frequency (%) |
|---|---|---|
| ATM | 16 | 5.556 |
| BARD1 | 1 | 0.347 |
| BRCA1 | 15 | 5.208 |
| BRAC2 | 20 | 6.9444 |
| BRCA1/2 | 33 | 11.458 |
| BRIP1 | 15 | 5.208 |
| CHEK2 | 7 | 2.4305 |
| CDK12 | 4 | 1.388 |
| DMC1 | 11 | 3.819 |
| FAAP20 | 2 | 0.694 |
| FAN1 | 2 | 0.694 |
| FANCD2 | 13 | 4.514 |
| FANCE | 2 | 0.694 |
| FANCL | – | – |
| FANCM | 17 | 5.903 |
| PALB2 | 2 | 0.694 |
| POLQ | 7 | 2.431 |
| RAD51 | 1 | 0.347 |
| RAD51B | 2 | 0.694 |
| RAD51C | 4 | 1.389 |
| RAD51D | – | – |
| RAD54L | – | – |
| XRCC3 | – | – |
| Any HR mutation | 108 | 37.500 |
Table 4.
Mutation frequencies of genes associated with the homologous recombination DNA repair pathway in the TCGA Pan Lung Cancer cohort (N = 1144).
| Gene | Observed mutations lung adenocarcinoma (N = 660) | Mutation frequency (%) | Observed mutations lung squamous cell carcinoma (N = 484) | Mutation frequency (%) |
|---|---|---|---|---|
| ATM | 59 | 8.939 | 28 | 5.785 |
| BARD1 | 15 | 2.273 | 7 | 1.446 |
| BRCA1 | 24 | 3.636 | 24 | 4.959 |
| BRAC2 | 30 | 4.545 | 30 | 6.198 |
| BRCA1/2 | 51 | 7.727 | 52 | 10.744 |
| BRIP1 | 23 | 3.485 | 5 | 1.033 |
| CHEK2 | 13 | 1.967 | 9 | 1.860 |
| CDK12 | 22 | 3.333 | 15 | 3.099 |
| DMC1 | 1 | 0.152 | 0 | – |
| FAAP20 | 0 | – | 1 | 0.207 |
| FAN1 | 11 | 1.667 | 4 | 0.826 |
| FANCD2 | 7 | 1.061 | 7 | 1.446 |
| FANCE | 5 | 0.758 | 1 | 0.207 |
| FANCL | 3 | 0.455 | 7 | 1.446 |
| FANCM | 44 | 6.667 | 20 | 4.132 |
| PALB2 | 13 | 1.970 | 13 | 2.686 |
| POLQ | 42 | 6.364 | 37 | 7.645 |
| RAD51 | 1 | 0.152 | 3 | 0.620 |
| RAD51B | 3 | 0.455 | 6 | 1.240 |
| RAD51C | 6 | 0.910 | 5 | 1.033 |
| RAD51D | 3 | 0.455 | 4 | 0.826 |
| RAD54L | 8 | 1.212 | 3 | 0.620 |
| XRCC3 | 0 | – | 2 | 0.413 |
| Any HR mutation | 235 | 35.606 | 166 | 34.298 |
Table 5.
Mutation frequencies of genes associated with the homologous recombination DNA repair pathway in the TCGA Esophageal cohort (N = 185).
| Gene | Observed mutations esophageal adenocarcinoma (N = 89) | Mutation frequency (%) | Observed mutations esophageal squamous cell carcinoma (N = 96) | Mutation frequency (%) |
|---|---|---|---|---|
| ATM | 11 | 12.360 | 10 | 10.417 |
| BARD1 | 3 | 3.371 | 2 | 2.083 |
| BRCA1 | 3 | 3.371 | 3 | 3.125 |
| BRAC2 | 5 | 5.618 | 4 | 4.167 |
| BRCA1/2 | 7 | 7.865 | 7 | 7.292 |
| BRIP1 | 3 | 3.371 | 2 | 2.083 |
| CHEK2 | 3 | 3.371 | 2 | 2.083 |
| CDK12 | 3 | 3.371 | 2 | 2.083 |
| DMC1 | 2 | 2.247 | 0 | – |
| FAAP20 | 0 | – | 0 | – |
| FAN1 | 1 | 1.124 | 0 | – |
| FANCD2 | 5 | 5.618 | 1 | 1.042 |
| FANCE | 1 | 1.124 | 0 | – |
| FANCL | 0 | – | 0 | – |
| FANCM | 8 | 8.989 | 3 | 3.125 |
| PALB2 | 1 | 1.124 | 0 | – |
| POLQ | 4 | 4.494 | 4 | 4.167 |
| RAD51 | 3 | 3.371 | 0 | – |
| RAD51B | 1 | 1.124 | 0 | – |
| RAD51C | 0 | – | 0 | – |
| RAD51D | 0 | – | 0 | – |
| RAD54L | 1 | 1.124 | 0 | – |
| XRCC3 | 0 | – | 0 | – |
| Any HR mutation | 36 | 40.449 | 25 | 26.042 |
In esophageal cancers, HR mutations were common to both adenocarcinoma (N = 89) and squamous (N = 96) cancers, though they were more frequent to the former (Table 5). While ATM, BRCA, and POLQ mutations were similarly prevalent in both cancer types, adenocarcinoma patients had a high frequency to mutations effecting FANCM (8.9%) and FANCD2 (5.6%), comprising a majority of the difference between the two cancers in overall HR mutation rate (Table 5).
In stomach cancer, mutation rates also varied extensively depending on cancer subtype. For instance, in the four subtypes represented in the TCGA stomach adenocarcinoma cohort, HR mutations were most common in mucinous adenocarcinoma by percent at 41%, though this represents a very small sample size of only 21 patients (Table 6). In tubular (N = 61), diffuse adenocarcinomas (N = 70), and non-specified carcinomas (N = 228), rates were 37.7%, 21.4%, and 32.5% respectively (Table 6). However, the relative distribution of HR mutation among subtypes were varied, though all subtypes had relatively high rates of ATM, BRCA, and POLQ mutations, with FANCM mutations common to mucinous and non-specified carcinomas (Table 6). Finally, we evaluated squamous cancers of the head and neck (N = 512), which had an overall HR mutation frequency of 27.1%. This group had little in the way of ATM mutations (2.9%), though we observed comparatively high rates of BRCA, CHECK and POLQ mutations (Table 7).
Table 6.
Mutation frequencies of genes associated with the homologous recombination DNA repair pathway in the TCGA Stomach adenocarcinoma cohort (N = 395).
| Gene | Observed mutations mucinous adenocarcinoma (N = 21) | Observed mutations tubular adenocarcinoma (N = 61) | Observed mutations diffuse adenocarcinoma (N = 70) | Observed mutations non-specified adenocarcinoma (N = 288) |
|---|---|---|---|---|
| ATM | 4 (19.05%) | 5 (8.20%) | 3 (4.29%) | 26 (11.40%) |
| BARD1 | 0 (0%) | 2 (3.28%) | 1 (1.43%) | 10 (4.39%) |
| BRCA1 | 1 (4.76%) | 3 (4.92%) | 2 (2.86%) | 9 (3.95%) |
| BRAC2 | 3 (4.29%) | 5 (8.20%) | 5 (7.14%) | 24 (10.53%) |
| BRCA1/2 | 4 (19.05%) | 6 (9.84%) | 6 (8.57%) | 30 (13.16%) |
| BRIP1 | 2 (9.52%) | 2 (3.28%) | 0 (0%) | 2 (0.88%) |
| CHEK2 | 1 (4.76%) | 2 (3.28%) | 0 (0%) | 6 (2.63%) |
| CDK12 | 0 (0%) | 5 (8.20%) | 2 (2.86%) | 12 (5.26%) |
| DMC1 | 1 (4.76%) | 0 (0%) | 0 (0%) | 7 (3.07%) |
| FAAP20 | 1 (4.76%) | 0 (0%) | 1 (1.43%) | 1 (0.44%) |
| FAN1 | 0 (0%) | 3 (4.92%) | 0 (0%) | 8 (3.51%) |
| FANCD2 | 2 (9.52%) | 2 (3.28%) | 2 (2.86%) | 9 (3.95%) |
| FANCE | 0 (0%) | 0 (0%) | 1 (1.43%) | 6 (2.63%) |
| FANCL | 1 (4.76%) | 0 (0%) | 1 (1.43%) | 3 (1.32%) |
| FANCM | 2 (9.52%) | 2 (3.28%) | 5 (7.14%) | 20 (8.77%) |
| PALB2 | 1 (4.76%) | 1 (1.64%) | 0 (0%) | 9 (3.95%) |
| POLQ | 4 (19.05%) | 2 (3.28%) | 1 (1.43%) | 24 (10.53%) |
| RAD51 | 0 (0%) | 0 (0%) | 0 (0%) | 1 (0.44%) |
| RAD51B | 0 (0%) | 0 (0%) | 0 (0%) | 2 (0.88%) |
| RAD51C | 0 (0%) | 0 (0%) | 0 (0%) | 2 (0.88%) |
| RAD51D | 1 (4.76%) | 2 (3.28%) | 0 (0%) | 1 (0.44%) |
| RAD54L | 0 (0%) | 0 (0%) | 0 (0%) | 4 (1.75%) |
| XRCC3 | 0 (0%) | 1 (1.64%) | 0 (0%) | 3 (1.32%) |
| Any HR mutation | 9 (42.68%) | 23 (37.70%) | 15 (21.43%) | 74 (32.46%) |
Table 7.
Mutation frequencies of genes associated with the homologous recombination DNA repair pathway in the TCGA Head & Neck squamous cell carcinoma cohort (N = 512).
| Gene | Observed mutations head and neck squamous cell carcinoma (N = 512) | Mutation frequency (%) |
|---|---|---|
| ATM | 15 | 2.930 |
| BARD1 | 9 | 1.758 |
| BRCA1 | 11 | 2.148 |
| BRAC2 | 23 | 4.492 |
| BRCA1/2 | 34 | 6.641 |
| BRIP1 | 13 | 2.539 |
| CHEK2 | 21 | 4.102 |
| CDK12 | 7 | 1.367 |
| DMC1 | 1 | 0.195 |
| FAAP20 | – | – |
| FAN1 | 5 | 0.977 |
| FANCD2 | 10 | 1.953 |
| FANCE | – | – |
| FANCL | 4 | 0.781 |
| FANCM | 10 | 1.953 |
| PALB2 | 7 | 1.367 |
| POLQ | 22 | 4.2975 |
| RAD51 | 1 | 0.195 |
| RAD51B | 1 | 0.195 |
| RAD51C | 3 | 0.586 |
| RAD51D | 2 | 0.391 |
| RAD54L | 5 | 0.977 |
| XRCC3 | – | – |
| Any HR mutation | 139 | 27.148 |
Discussion
The efficacy of PARP inhibitors in BRCA-mutated tumors stems largely from the known roles for PARP in mediating single stranded break repair40,41. Thus, initial trials were based on the hypothesis that inhibiting the repair of single stranded breaks will cause synthetic lethality in tumors with loss of high-fidelity double-strand break homologous recombination40. As discussed, this approach has shown tremendous efficacy in several BRCA-mutant cancers, including those of the breast, ovary, prostate, colon, thymus, and pancreas10,13,42. Olaparib became the first FDA-approved PARP inhibitor based on results from Study 19, a randomized, placebo-controlled trial in ovarian cancer showing an improvement in both progression-free and overall survival43.
Additionally, olaparib was soon approved for BRCA-mutated breast cancer following the phase III OlympiAD trial, which showed improvements in both response rate and progression-free survival when compared to standard therapy44. Subsequently, PARP inhibitors have shown efficacy in the second line, and olaparib, rucaparib and niraparib have now been approved as maintenance therapy for HR deficient ovarian cancer patients following platinum-based chemotherapy45–47. However, while PARP inhibitors have no doubt improved clinical outcomes in BRCA-mutated tumors, there is mounting biologic evidence that other molecular subsets may also derive clinical benefit from PARP inhibitors30. These include patients with genomic alterations in ATM, BARD1, BRIP1, CHEK2, FAAP20, FAN1, FANCE, FANCM, PALB2, POLQ, RAD51B, RAD51C, and RAD51D31–33. While mutations of these and other HR genes are certainly less established indicators of HRD, those affecting ATM and PALB2 have already been shown to associate with responsiveness to PARP inhibition14.
Thus, when evaluating a pan-cancer cohort, we found that by expanding our search to include several HR genes beyond those most frequently associated with PARP inhibitors, there may be several additional patient groups who also have genetic loss of HRD and may therefore also respond to PARP inhibition. This is consistent to results observed in a similar study, which also found that expanding criteria identifies a larger group of patients who potentially harbor defects to the HR pathway48. In our study, when restricting our analysis to BRCA-mutated tumors, we found that only ~ 4% of patients are represented. When including ATM-mutated tumors, this number more than doubles to 8.36%. However, when including the other genes in our panel, as many as 13.36% of patients are now represented. While we cannot conclusively state that the entirety of these patients are in fact HR deficient and would derive clinical benefit from PARP inhibition, as mutations to BARD1, CDK12, DMC1, PALB2, and POLQ seem to predict for poor outcomes in this cohort, their predictive value for PARP inhibition is not established and warrants continued exploration.
This is particularly true for the many cancer histologies identified in this study for which PARP inhibitors are not widely used or FDA approved. For instance, though limited by a small sample size, we found that nearly 40% of diffuse large B cell lymphoma patients harbor mutations to genes associated with the HR pathway, though BRCA mutations were only observed in 6.38%. Though early evidence supports the addition of the PARP inhibitor veliparib to bendamustine and rituximab in B-cell lymphomas49, the role for PARP inhibitors in diffuse large B-cell lymphoma is still under investigation. Still, recent evidence points to additional predictive criteria expanding beyond BRCA mutations, with less-studied HR-associated genes such as LMO2 appearing to predict for sensitivity to PARP inhibition50.
As discussed, we also identified a high frequency in HR mutations in cutaneous melanoma patients. Murine models have supported a pro-metastatic role for PARP-1, and PARP inhibition is showing early promise in combination with radiotherapy in murine models of uveal melanoma 51,52. However, like with diffuse large B cell lymphoma, clinical data is rather limited. A 2013 phase II study suggests that the PARP inhibitor rucaparib cooperates with temozolomide in metastatic melanoma53, though there are a relatively small number of subsequent clinical studies, likely as BRCA1/2 mutations are not typically considered a cause of malignant melanoma54. However, in the TCGA cohort examined in our study, we found that BRCA mutations are represented in as many as 11.5% of cutaneous melanoma patients, with many patients also harboring mutations to ATM, BRIP1, CHECK2, DMC1, FANCD2, FANCM, and POLQ. As 37.5% of this patient cohort had at least one mutation affecting the HR pathway, the use of these and other mutations warrant consideration when exploring PARP inhibitors in subsequent clinical trials.
Using this expanded gene panel, we found that HRD in lung, bile duct, esophageal, stomach, uterine, and head and neck cancers may also be underreported. This may be of clinical significance, as PARP inhibitors are showing early promise in several of these cancer histologies, particularly when combined with chemotherapy or radiation55–64. However, we must note that an inherent limitation of our study is though we identified several mutations in HR associated genes, relatively few have been characterized, particularly with respect to either HRD or PARP inhibition. Additionally, as our data is largely dependent on sequencing from formalin fixed paraffin embedded tissues, these rates of mutation may be inflated due to technical artifacts. Hence, it is not clear how many patients identified in this study will in fact have HRD or would benefit from PARP inhibition. Additionally, clinical response to PARP inhibition is not solely driven by HRD, involving several other factors including replication, oxidative, and ER stress65–69.
Therefore, specific alterations to these and other genes warrant further investigation prior to any being proposed as a reliable surrogate for HRD, particularly in the setting of other cellular processes. Further, should PARP inhibitors be combined with other DNA-damaging agents such as chemo or radiotherapy, a patient’s HRD status may become less relevant, as early evidence suggests that such approaches may have efficacy in multiple TP53 mutated but HR-intact tumor types70. However, improving the selection criteria for PARP inhibition in monotherapy or without additional DNA-damaging agents will require careful evaluation of these and potentially other HR associated genes in hopes of identifying the patients who will most benefit from this approach.
Supplementary information
Acknowledgements
This work was supported by NIH R01 CA216410 from the National Cancer Institute and Veteran Administration Awards BX002703 and BX004855 to A. Rana, and by NIH F30CA236031 from the National Cancer Institute and UIC Award for Graduate Research to D.R Principe.
Abbreviations
- PARP
Poly ADP ribose polymerase
- HR
Homologous recombination
- NGS
Next generation sequencing
- NSCLC
Non-small cell lung cancer
- TCGA
The Cancer Genome Atlas
Author contributions
D.P., M.N., and R.K. performed patient genomic analyses. D.P. designed the study, assimilated data into tables/figures, and drafted the manuscript. A.R. provided oversight with laboratory personnel, edited the manuscript, and provided funding support.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Daniel R. Principe and Matthew Narbutis.
Supplementary information
is available for this paper at 10.1038/s41598-020-76975-6.
References
- 1.An X, et al. BCR-ABL tyrosine kinase inhibitors in the treatment of Philadelphia chromosome positive chronic myeloid leukemia: A review. Leuk. Res. 2010;34:1255–1268. doi: 10.1016/j.leukres.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 2.Maximiano S, Magalhaes P, Guerreiro MP, Morgado M. Trastuzumab in the treatment of breast cancer. BioDrugs. 2016;30:75–86. doi: 10.1007/s40259-016-0162-9. [DOI] [PubMed] [Google Scholar]
- 3.Spigel DR. PARP inhibitors in lung cancer. J. Thorac. Oncol. 2012;7:S392–393. doi: 10.1097/JTO.0b013e31826df1eb. [DOI] [PubMed] [Google Scholar]
- 4.Tsimberidou AM, et al. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res. 2012;18:6373–6383. doi: 10.1158/1078-0432.CCR-12-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sisay M, Edessa D. PARP inhibitors as potential therapeutic agents for various cancers: Focus on niraparib and its first global approval for maintenance therapy of gynecologic cancers. Gynecol. Oncol. Res. Pract. 2017;4:18. doi: 10.1186/s40661-017-0055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohn EC, Lee JM, Ivy SP. The HRD decision-which PARP inhibitor to use for whom and when. Clin. Cancer Res. 2017;23:7155–7157. doi: 10.1158/1078-0432.CCR-17-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balasubramaniam S, et al. FDA approval summary: Rucaparib for the treatment of patients with deleterious BRCA mutation-associated advanced ovarian cancer. Clin. Cancer Res. 2017;23:7165–7170. doi: 10.1158/1078-0432.CCR-17-1337. [DOI] [PubMed] [Google Scholar]
- 8.Bochum S, Berger S, Martens UM. Olaparib. Recent Results Cancer Res. 2018;211:217–233. doi: 10.1007/978-3-319-91442-8_15. [DOI] [PubMed] [Google Scholar]
- 9.da Cunha Colombo Bonadio, R. R., Fogace, R. N., Miranda, V. C. & Diz, M. Homologous recombination deficiency in ovarian cancer: A review of its epidemiology and management. Clinics (Sao Paulo) 2018;73:450. doi: 10.6061/clinics/2018/e450s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golan T, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N. Engl. J. Med. 2019;381:317–327. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramakrishnan Geethakumari P, Schiewer MJ, Knudsen KE, Kelly WK. PARP inhibitors in prostate cancer. Curr. Treat. Opt. Oncol. 2017;18:37. doi: 10.1007/s11864-017-0480-2. [DOI] [PubMed] [Google Scholar]
- 12.Lesueur P, et al. Phase I/IIa study of concomitant radiotherapy with olaparib and temozolomide in unresectable or partially resectable glioblastoma: OLA-TMZ-RTE-01 trial protocol. BMC Cancer. 2019;19:198. doi: 10.1186/s12885-019-5413-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Principe DR, Kamath SD, Munshi HG, Mohindra NA. Metastatic thymoma harboring a deleterious BRCA2 mutation derives durable clinical benefit from olaparib. Oncologist. 2019 doi: 10.1634/theoncologist.2019-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mateo J, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domchek SM. Reversion mutations with clinical use of PARP inhibitors: Many genes, many versions. Cancer Discov. 2017;7:937–939. doi: 10.1158/2159-8290.CD-17-0734. [DOI] [PubMed] [Google Scholar]
- 16.Jette NR, et al. ATM-deficient cancers provide new opportunities for precision oncology. Cancers (Basel) 2020 doi: 10.3390/cancers12030687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C, Jette N, Moussienko D, Bebb DG, Lees-Miller SP. ATM-deficient colorectal cancer cells are sensitive to the PARP inhibitor olaparib. Transl. Oncol. 2017;10:190–196. doi: 10.1016/j.tranon.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weston VJ, et al. The PARP inhibitor olaparib induces significant killing of ATM-deficient lymphoid tumor cells in vitro and in vivo. Blood. 2010;116:4578–4587. doi: 10.1182/blood-2010-01-265769. [DOI] [PubMed] [Google Scholar]
- 19.Williamson CT, et al. Enhanced cytotoxicity of PARP inhibition in mantle cell lymphoma harbouring mutations in both ATM and p53. EMBO Mol. Med. 2012;4:515–527. doi: 10.1002/emmm.201200229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williamson CT, et al. ATM deficiency sensitizes mantle cell lymphoma cells to poly(ADP-ribose) polymerase-1 inhibitors. Mol. Cancer Ther. 2010;9:347–357. doi: 10.1158/1535-7163.MCT-09-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuemmel S, et al. Olaparib for metastatic breast cancer in a patient with a germline PALB2 variant. NPJ Breast Cancer. 2020;6:31. doi: 10.1038/s41523-020-00174-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith MA, et al. Initial testing (stage 1) of the PARP inhibitor BMN 673 by the pediatric preclinical testing program: PALB2 mutation predicts exceptional in vivo response to BMN 673. Pediatr. Blood Cancer. 2015;62:91–98. doi: 10.1002/pbc.25201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horak P, et al. Response to olaparib in a PALB2 germline mutated prostate cancer and genetic events associated with resistance. Cold Spring Harb. Mol. Case Stud. 2019 doi: 10.1101/mcs.a003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mateo J, et al. Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP-B): A multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 2020;21:162–174. doi: 10.1016/S1470-2045(19)30684-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buisson R, et al. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol. 2010;17:1247–1254. doi: 10.1038/nsmb.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyasaka A, et al. Anti-tumor activity of olaparib, a poly (ADP-ribose) polymerase (PARP) inhibitor, in cultured endometrial carcinoma cells. BMC Cancer. 2014;14:179. doi: 10.1186/1471-2407-14-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraser M, et al. PTEN deletion in prostate cancer cells does not associate with loss of RAD51 function: Implications for radiotherapy and chemotherapy. Clin. Cancer Res. 2012;18:1015–1027. doi: 10.1158/1078-0432.CCR-11-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dedes KJ, et al. PTEN deficiency in endometrioid endometrial adenocarcinomas predicts sensitivity to PARP inhibitors. Sci. Transl. Med. 2010;2:53ra75. doi: 10.1126/scitranslmed.3001538. [DOI] [PubMed] [Google Scholar]
- 29.Mendes-Pereira AM, et al. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol. Med. 2009;1:315–322. doi: 10.1002/emmm.200900041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pilie PG, Gay CM, Byers LA, O'Connor MJ, Yap TA. PARP inhibitors: Extending benefit beyond BRCA-mutant cancers. Clin. Cancer Res. 2019;25:3759–3771. doi: 10.1158/1078-0432.CCR-18-0968. [DOI] [PubMed] [Google Scholar]
- 31.Hoppe MM, Sundar R, Tan DSP, Jeyasekharan AD. Biomarkers for homologous recombination deficiency in cancer. J. Natl. Cancer Inst. 2018;110:704–713. doi: 10.1093/jnci/djy085. [DOI] [PubMed] [Google Scholar]
- 32.Riaz N, et al. Pan-cancer analysis of bi-allelic alterations in homologous recombination DNA repair genes. Nat. Commun. 2017;8:857. doi: 10.1038/s41467-017-00921-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Research, N Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ceccaldi R, et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature. 2015;518:258–262. doi: 10.1038/nature14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z, et al. DNA polymerase theta (POLQ) is important for repair of DNA double-strand breaks caused by fork collapse. J. Biol. Chem. 2019;294:3909–3919. doi: 10.1074/jbc.RA118.005188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cerami E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Principe DR, et al. Long-term gemcitabine treatment reshapes the pancreatic tumor microenvironment and sensitizes murine carcinoma to combination immunotherapy. Cancer Res. 2020 doi: 10.1158/0008-5472.CAN-19-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chakravarty D, et al. OncoKB: A precision oncology knowledge base. JCO Precis. Oncol. 2017 doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.del Rivero J, Kohn EC. PARP inhibitors: The cornerstone of DNA repair-targeted therapies. Oncology (Williston Park) 2017;31:265–273. [PubMed] [Google Scholar]
- 41.Ray Chaudhuri A, Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017;18:610–621. doi: 10.1038/nrm.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashworth A, Lord CJ. Synthetic lethal therapies for cancer: what's next after PARP inhibitors? Nat. Rev. Clin. Oncol. 2018;15:564–576. doi: 10.1038/s41571-018-0055-6. [DOI] [PubMed] [Google Scholar]
- 43.Ledermann J, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N. Engl. J. Med. 2012;366:1382–1392. doi: 10.1056/NEJMoa1105535. [DOI] [PubMed] [Google Scholar]
- 44.Robson M, et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N Engl J Med. 2017;377:523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 45.Moore K, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N. Engl. J. Med. 2018;379:2495–2505. doi: 10.1056/NEJMoa1810858. [DOI] [PubMed] [Google Scholar]
- 46.Coleman RL, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390:1949–1961. doi: 10.1016/S0140-6736(17)32440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mirza MR, et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N. Engl. J. Med. 2016;375:2154–2164. doi: 10.1056/NEJMoa1611310. [DOI] [PubMed] [Google Scholar]
- 48.Heeke AL, et al. Prevalence of homologous recombination-related gene mutations across multiple cancer types. JCO Precis. Oncol. 2018 doi: 10.1200/PO.17.00286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soumerai JD, et al. The PARP inhibitor veliparib can be safely added to bendamustine and rituximab and has preliminary evidence of activity in B-cell lymphoma. Clin. Cancer Res. 2017;23:4119–4126. doi: 10.1158/1078-0432.CCR-16-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parvin S, et al. LMO2 confers synthetic lethality to PARP inhibition in DLBCL. Cancer Cell. 2019;36:237–249. doi: 10.1016/j.ccell.2019.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodriguez MI, et al. PARP-1 regulates metastatic melanoma through modulation of vimentin-induced malignant transformation. PLoS Genet. 2013;9:e1003531. doi: 10.1371/journal.pgen.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Koning L, et al. PARP inhibition increases the response to chemotherapy in uveal melanoma. Cancers (Basel) 2019 doi: 10.3390/cancers11060751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Plummer R, et al. A phase II study of the potent PARP inhibitor, Rucaparib (PF-01367338, AG014699), with temozolomide in patients with metastatic melanoma demonstrating evidence of chemopotentiation. Cancer Chemother. Pharmacol. 2013;71:1191–1199. doi: 10.1007/s00280-013-2113-1. [DOI] [PubMed] [Google Scholar]
- 54.Debniak T, et al. BRCA1/2 mutations are not a common cause of malignant melanoma in the Polish population. PLoS ONE. 2018;13:e0204768. doi: 10.1371/journal.pone.0204768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bian X, et al. PTEN deficiency sensitizes endometrioid endometrial cancer to compound PARP-PI3K inhibition but not PARP inhibition as monotherapy. Oncogene. 2018;37:341–351. doi: 10.1038/onc.2017.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dinkic C, et al. PARP inhibition sensitizes endometrial cancer cells to paclitaxel-induced apoptosis. Oncol. Lett. 2017;13:2847–2851. doi: 10.3892/ol.2017.5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kageyama SI, et al. PARP inhibitor olaparib sensitizes esophageal carcinoma cells to fractionated proton irradiation. J. Radiat. Res. 2020 doi: 10.1093/jrr/rrz088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kubota E, et al. Low ATM protein expression and depletion of p53 correlates with olaparib sensitivity in gastric cancer cell lines. Cell Cycle. 2014;13:2129–2137. doi: 10.4161/cc.29212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fehling SC, Miller AL, Garcia PL, Vance RB, Yoon KJ. The combination of BET and PARP inhibitors is synergistic in models of cholangiocarcinoma. Cancer Lett. 2020;468:48–58. doi: 10.1016/j.canlet.2019.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mao Y, et al. PARP inhibitor olaparib sensitizes cholangiocarcinoma cells to radiation. Cancer Med. 2018;7:1285–1296. doi: 10.1002/cam4.1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sen T, Gay CM, Byers LA. Targeting DNA damage repair in small cell lung cancer and the biomarker landscape. Transl. Lung Cancer Res. 2018;7:50–68. doi: 10.21037/tlcr.2018.02.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang Y, et al. PARP inhibitors synergize with gemcitabine by potentiating DNA damage in non-small-cell lung cancer. Int. J. Cancer. 2019;144:1092–1103. doi: 10.1002/ijc.31770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Farago AF, et al. Combination olaparib and temozolomide in relapsed small-cell lung cancer. Cancer Discov. 2019;9:1372–1387. doi: 10.1158/2159-8290.CD-19-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jelinek MJ, et al. A phase I/II trial adding poly(ADP-ribose) polymerase (PARP) inhibitor veliparib to induction carboplatin-paclitaxel (Carbo-Tax) in patients with head and neck squamous cell carcinoma (HNSCC) Alliance A091101. J. Clin. Oncol. 2018;36:6031–6031. doi: 10.1200/JCO.2018.36.15_suppl.6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H, Zhang S, Song L, Qu M, Zou Z. Synergistic lethality between PARP-trapping and alantolactone-induced oxidative DNA damage in homologous recombination-proficient cancer cells. Oncogene. 2020;39:2905–2920. doi: 10.1038/s41388-020-1191-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.King D, et al. MYCN expression induces replication stress and sensitivity to PARP inhibition in neuroblastoma. Oncotarget. 2020;11:2141–2159. doi: 10.18632/oncotarget.27329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maya-Mendoza A, et al. High speed of fork progression induces DNA replication stress and genomic instability. Nature. 2018;559:279–284. doi: 10.1038/s41586-018-0261-5. [DOI] [PubMed] [Google Scholar]
- 68.Hou D, et al. Increased oxidative stress mediates the antitumor effect of PARP inhibition in ovarian cancer. Redox Biol. 2018;17:99–111. doi: 10.1016/j.redox.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26:417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sizemore ST, et al. Synthetic lethality of PARP inhibition and ionizing radiation is p53-dependent. Mol. Cancer Res. 2018;16:1092–1102. doi: 10.1158/1541-7786.MCR-18-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.