Abstract
Fermented foods and particularly beer have accompanied the development of human civilization for thousands of years. Saccharomyces cerevisiae, the dominant yeast in the production of alcoholic beverages, probably co-evolved with human activity. Considering that alcoholic fermentations emerged worldwide, the number of strains used in beer production nowadays is surprisingly low. Thus, the genetic diversity is often limited. This is among others related to the switch from a household brewing style to a more artisan brewing regime during the sixteenth century and latterly the development of single yeast isolation techniques at the Carlsberg Research Laboratory in 1883, resulting in process optimizations in the brewing industry. However, due to fierce competition within the beer market and the increasing demand for novel beer styles, diversification is becoming increasingly important. Moreover, the emergence of craft brewing has influenced big breweries to rediscover yeast as a significant contributor to a beer’s aroma profile and realize that there is still room for innovation in the fermentation process. Here, we aim at giving a brief overview on how currently used S. cerevisiae brewing yeasts emerged and comment on the rationale behind replacing them with novel strains. We will present potential sources of yeasts that have not only been used in beer brewing before, including natural sources and sources linked to human activity but also an overlooked source, such as yeast culture collections. We will briefly comment on common yeast isolation techniques and finally touch on additional challenges for the brewing industry in replacing their current brewer’s yeasts.
Keywords: Saccharomyces cerevisiae, breweries, diversity, reservoirs, strain collection, domestication
Introduction
While some anthropologists believe that the discovery to process foods over an open fire as much as 1.8 million years ago was crucial for the evolution of the human species, the early history of human societies is unquestionably linked to the domestication of plants and animals (Zeder, 2006), and the subsequent development of fermented foods during the Neolithic period (Tamang et al., 2020). Fermented beverages have been an important part of civilizations for economic and health reasons for thousands of years (Campbell-Platt, 1994; Vilela, 2019), and alcoholic beverages in particular played a vital role in their traditional culture and social life (Legras et al., 2007; Dietrich et al., 2012).
Beer is one of the oldest fermented beverages known in human history. Archeological remains and pictographic evidence of cereal based beer-like brews have been found across the Fertile Crescent, the oldest dating back to the twelfth millennium BC (Katz and Maytag, 1991; Michel et al., 1992; Hornsey, 2003; Black et al., 2006; Legras et al., 2007; Sicard and Legras, 2011; Liu et al., 2018). China also has a long history of beer-like beverages (McGovern et al., 2004; Wang et al., 2016), and in Europe, Celtic tribes spread beer brewing across the continent up to 2,000 years ago (Corran, 1975; Nelson, 2005), where over time it developed into the modern beer-brewing process as we know it today (Hornsey, 2003; Unger, 2004). While many changes have been made to the fermentation process over time, the one constant factor needed for successful beer production has been an alcohol producing yeast (Lodolo et al., 2008). However, despite the vast number of different beer styles that have been developed all over the world (Protz, 1995; Glover, 1997), the number of yeasts used in the brewing industry today is surprisingly low.
Industrialization of Beer Production
For centuries beer production relied on spontaneous fermentations with complex mixtures of native/wild microbes, although yeasts belonging to the genus Saccharomyces usually dominate the later part of the process due to their strong ability to produce and tolerate ethanol (Steensels and Verstrepen, 2014). The early brewers had yet soon realized that repitching a new fermentation with a small portion of a just finished one resulted in faster and more reliable fermentations, a process commonly referred to as “backslopping” (Whittington et al., 2019). The constant reuse in man-made environments and the simultaneous reduction in contact to natural niches resulted in the domestication of brewer’s yeasts (Steensels et al., 2019) accompanied by a dramatic reduction in species involved in the brewing process. In Europe, this development was fueled during the middle ages by a switch from predominantly household fermentations to a more artisan brewing style, where taverns and monasteries became the predominant brewers. In addition, the introduction of new regulations in the sixteenth century standardized the brewing process and limited the number of ingredients used (the famous “Reinheitsgebot” from 1,516; Dornbusch, 1998; Hornsey, 2003), which improved beer quality but probably also reduced the numbers of yeasts used even further. Interestingly though, restricting the brewing period to the winter season not only resulted in a novel beer style commonly known as “lager” (vs. the traditional “ale,” Jentsch, 2007) but also in a novel brewing yeast lineage, that is, better adapted to lower fermentation temperatures (Unger, 2004; Dredge, 2019). During the industrial revolution of the nineteenth century, other groundbreaking technical discoveries were made that modernized the brewing process and allowed the year around production of “lager” beers. However, the most important discovery was made by the French scientist Louis Pasteur, who demonstrated that the splitting of malt sugar into alcohol and carbonic acid was due to the activity of a microbe, which is the brewer’s yeast (Pasteur, 1857, 1858). Inspired by these findings, Emil Christian Hansen developed techniques for isolation of single yeast cells at the Carlsberg Research Laboratory in the early 1880s, and the first pure yeast culture-based beer was brewed in 1883 (Hansen, 1888; Barnett and Lichtenthaler, 2001). From this time onward, the fast-growing beer industry began to replace spontaneous and “backslopping” based fermentations with its complex microbiomes by controlled fermentations using pure yeast starter cultures. Specifically, these were S. cerevisiae for “ale” and S. pastorianus for “lager” beers. Finally, while the number of microbreweries has increased since the late twentieth century, the global beer market is for the most part dominated today by only few internationally operating companies. In order to save costs and reduce complexity in the production processes, these big companies tend to reduce the already low number of brewing yeasts used within a company even further. Hence, human activity over the centuries resulted in a rather low number of specifically adapted yeasts used for beer brewing today (Gibson et al., 2007; Libkind et al., 2011; Steensels et al., 2014; Gallone et al., 2016).
Toward More Yeast Diversity in Beer Brewing
Despite all industrialization and modernization efforts, the brewing industry is considered a rather traditional one, and breweries are usually quite reluctant to change their brewing yeasts. However, there has been an increasing demand for novel beer styles from customers and consumers over the last one or two decades, which has been mainly driven by the emergence of the craft brewing community. Since craft breweries usually operate in much smaller volumes, they are more willing to experiment with raw materials to create more “explorative” beer styles, basically reversing the streamlining of the brewing processes previously outlined. With increasing competition in beer markets, the entire brewing industry has been forced to reconsider their approach and diversify their beverage portfolios. In that context, not only craft breweries but also the big players “rediscovered” that yeast has just as much influence on beer quality but more importantly on beer taste as the other raw materials and/or physical brewing parameters (White and Zainasheff, 2010). Experimenting with novel yeasts, they realized that there is still a hidden treasure to be found not only from a product point of view, but also from a marketing perspective since doing business “with a story” is becoming more and more popular. Selling a beer brewed with a 5,000-year-old yeast (Aouizerat et al., 2019) is a marketing consultant’s dream. Therefore, it is an intriguing idea to go out and isolate novel yeasts and use them for brewing purposes.
Isolation of Novel Brewing Yeast
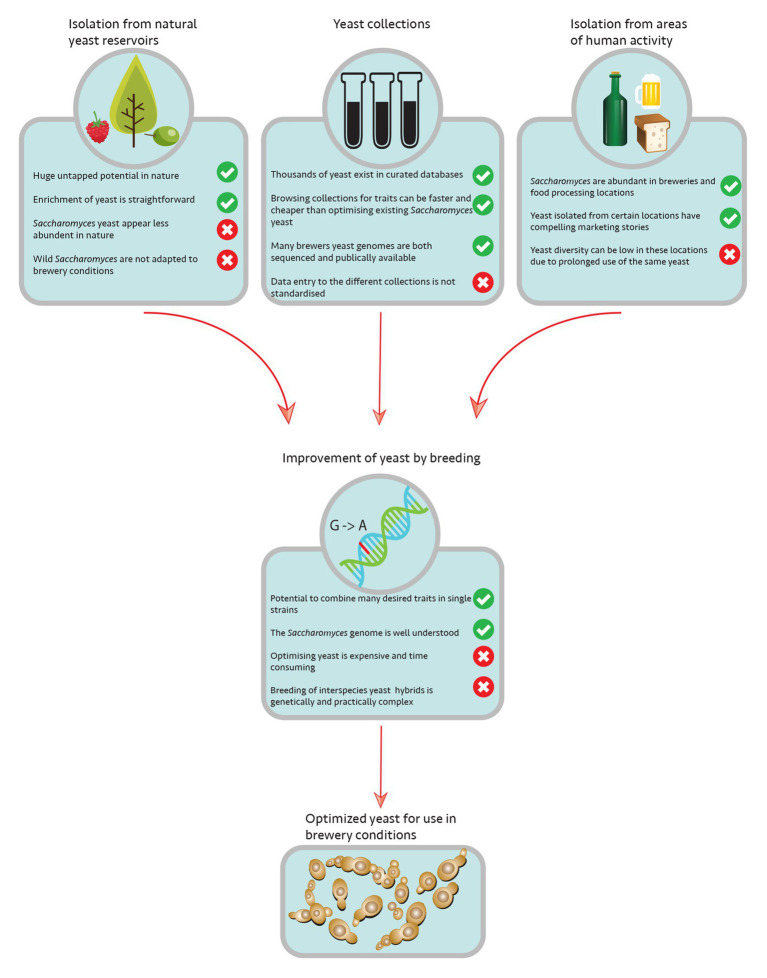
The most obvious sources for isolating yeast strains are the ones immediately linked to human S. cerevisiae-based activities (Figure 1), such as baking bread and fermenting wine or sake. Due to the high economic impact of these processes, a lot of research has been conducted on the yeasts involved in making these products (Ohya and Kashima, 2019; Parapouli et al., 2020). Several wine, sake, and baker’s yeasts have been analyzed for their beer brewing potential over the years. However, only few baker’s yeasts showed promising potential (Canonico et al., 2014; Marongiu et al., 2015; Rossi et al., 2018; Cubillos et al., 2019; Tokpohozin et al., 2019). The phylogenetic analyses of industrial S. cerevisiae strains in recent times have shown that most strains analyzed fell into a handful of clades and that were characterized by a certain production process (Gallone et al., 2016; Gonçalves et al., 2016; Kang et al., 2019). Further analyses could link these clades to genetic and phenotypic differences that most likely resulted from specifically adapting to the respective fermentation conditions, a phenomenon typical for domestication events (Driscoll et al., 2009; Purugganan and Fuller, 2009; Steensels et al., 2019). Interestingly, most baker’s yeast strains are closely related to their beer brewing counterparts, sharing several genetic and phenotypic features (Gonçalves et al., 2016), something that can be explained by the strong interconnection of baking and brewing in the early days. These analyses also confirmed a common “problem” for all these domesticated yeasts, a lack of genetic but sometimes also phenotypic variability especially within a clade. In fact, besides the fermenting microbe, these processes share quite a similar history. Except for sake fermentation, which was first mentioned in the eighth century, they emerged in the Neolithic period (Evans, 1990; McGovern et al., 1996, 1997; Cavalieri et al., 2003; Sicard and Legras, 2011). They were initially based on spontaneous fermentations, but have developed into highly engineered processes using only a small number of specialized pure yeast starter cultures since then (Heard and Fleet, 1985; Henick-Kling et al., 1998; Domizio et al., 2007; Daenen et al., 2008; Randez-Gil et al., 2013; Borneman et al., 2016). In order to avoid isolating yeast that is similar to what is already in use, one could potentially focus attention to more traditional fermentations, where indigenous yeasts may have escaped parts of the domestication processes and therefore provide a higher natural diversity. In addition, other natural substrates/raw materials than grains and grapes are sometimes used to create these traditional alcoholic beverages such as the honey-based mead, adding another level of complexity. Only in recent years have people started to look into the beer brewing potential of yeasts from traditional fermentations, such as cachaça spirits (Araújo et al., 2018), pulque, tequila (Cubillos et al., 2019), or sub-Saharan alcoholic beverages (Johansen et al., 2019). However, only the historically proven old-style beers such as Finnish sahti and the traditional Norwegian kveik beer, both using juniper instead of or in addition to hops, or the bread based Russian kvass have shown promising results so far (Dlusskaya et al., 2008; Ekberg et al., 2015; Preiss et al., 2018). There are still many traditional beverages left to explore such as traditional European low alcoholic beverages (Baschali et al., 2017) or fermented drinks produced by the Aboriginal people in Australia even before European arrival (Mangaitch, Way-a-linah, and Kambuda; Brady, 2008). Further research is needed to isolate and characterize yeast from these drinks. In addition to alcoholic beverages, other fermented foods could also serve as a target for novel yeast isolation. In 1987, Campbell-Platt reported the existence of around 3,500 different fermented foods and beverages, but there might be more than 5,000 varieties of these products being consumed in the world today (Campbell-Platt, 1987; Tamang, 2010). The role of S. cerevisiae in nonalcoholic fermentations has so far been less studied because among other things research is often complicated by the presence of other microorganisms, mainly lactic acid bacteria. Furthermore, S. cerevisiae could not only be identified in many dairy based fermentations but also in fermentations used to refine other agricultural products such as coffee or cocoa beans (Tofalo et al., 2019) that could be tested in beer fermentations. In addition, industrial bioethanol production has been reported to be a rich source of S. cerevisiae strains (Basso et al., 2008). Particularly, in Brazilian bioethanol plants, the specific operating conditions allow for the entry and growth of wild yeast strains (Amorim et al., 2011). The initial yeast starter cultures were replaced over time by indigenous strains present in sugar cane molasses that would outperform the domesticated yeasts due to their resistance to high ethanol levels and temperature fluctuations (Beato et al., 2016; Reis et al., 2017; Kechkar et al., 2019). Recently, several S. cerevisiae strains isolated from Brazilian distilleries have been investigated in high gravity beer fermentations (Christofoleti-Furlan et al., 2020), clearly showing they harbor a potential for use in the brewing industry.
Figure 1.
Overview of microbial reservoirs and strategies to introduce novel Saccharomyces diversity to breweries.
Yeast Origin and Diversity
As just exemplified by the Brazilian bioethanol production, the isolation of wild yeast strains from natural reservoirs could be potentially very interesting, as these could bear novel features useful for biotechnological applications (Figure 1). Even though S. cerevisiae is one of the most extensively studied organisms, it has only recently evolved into a model system for population genomic, biogeographic, and ecological studies (for review, see Alsammar and Delneri, 2020). Based on its metabolism, it was initially thought that S. cerevisiae would mainly reside on the surface of fruits and could therefore be found in respective environments, such as orchards and wineries. Surprisingly, in recent years, it was shown that the yeasts present in wineries seem mainly not to originate from grapes since their presence on intact grapes is rather scarce (Mortimer and Polsinelli, 1999; Taylor et al., 2014). Instead, it seems that yeast cells are most likely transferred to these ecosystems via insect vectors that feed off damaged grapes (Stefanini et al., 2012; Buser et al., 2014; Dapporto et al., 2016). The potential of insects as a natural reservoir for wild S. cerevisiae strains is still unclear and needs further investigation (Stevic, 1962). While some researchers believe that S. cerevisiae lives a nomadic life style since it can adapt to a wide variety of growth conditions (Goddard and Greig, 2015), more recent studies indicated that certain trees, especially from the order Fagales and the surrounding soil seem to be a natural niche for the organism (Hittinger, 2013). In fact, the most primitive S. cerevisiae populations to date, which showed a significantly higher genetic variability than previously isolated strains, were isolated from remote primeval forests in China (Wang et al., 2012; Boynton and Greig, 2014; Liti, 2015; Duan et al., 2018; Peter et al., 2018; Bendixsen et al., 2020). Even though S. cerevisiae is generally considered to be a terrestrial yeast, very few strains have been isolated from marine waters in coastal areas (Zaky et al., 2014) or freshwater lakes and rivers (Sláviková and Vadkertiová, 1995, 1997). Although the strains isolated from such habitats possess distinct phenotypical characteristics (Šťovíček et al., 2010, 2014), the question of whether these sites are an additional natural niche for S. cerevisiae or whether it only reflects human activity in the area, which needs further investigation. There are two main challenges when isolating wild S. cerevisiae strains. Due to the vast geographical distribution of strains linked to human activity, special care needs to be taken in choosing the right habitat during sampling to avoid isolating strains of the “common” genotypes. In addition, the low number of isolates in general poses another challenge to this approach asking for proper isolation techniques (Beech and Davenport, 1971). In a recent report, Boynton and colleagues discussed two of the most common isolation techniques, direct plating and so-called enrichment cultures, comparing their feasibility in isolating yeasts of the genus Saccharomyces from forest samples (Boynton et al., 2019). The results confirmed the general acceptance that enrichment culturing techniques favor the isolation of rare Saccharomyces species such as S. cerevisiae. However, since each enrichment protocol may select for certain strain features, sampling methods need to be carefully adjusted to interest and sampling habitats, the development of marine yeast isolation protocols being one example (Zaky et al., 2016). Besides the technical and biological challenges, the Convention of Biological Diversity (CBD) and its supplementary agreement, the Nagoya Protocol should be followed when isolating organisms from natural reservoirs. Proper documentation during the sampling process is required to ensure the conservation and sustainable use of biological diversity under this legal framework (Boundy-Mills et al., 2016).
Yeast Culture Collections
While isolating and characterizing novel S. cerevisiae strains from the wild for industrial applications is an intriguing idea, only big breweries usually have the personnel and the technical requirements to run respective projects or are willing to outsource these tasks to scientific collaborators. Instead, most breweries will rely on a small number of commercially available yeasts that are provided by few companies specialized on brewing yeasts (Quain, 2006). However, another invaluable source of yeasts strains seems to have been overlooked in the brewing industry for a long time, such as yeast culture collections (Figure 1; Daniel and Prasad, 2010). This comes as a little surprise since one of first culture collections, though not publicly available, was established at the Carlsberg brewery shortly after Hansen had developed the pure culture isolation techniques (Hansen, 1888; Barnett, 2007). Ever since the first service culture collections were established around 1,900 (Uruburu, 2003), the number of culture collections has increased significantly, ranging from well-known, large public repositories to dozens of smaller public collections. Even though the collections are generally considered to represent only a small fraction of the microbial diversity (Boundy-Mills, 2012), they can be an invaluable source of well-characterized strains that were isolated from various natural or human activity associated niches. In addition, especially the larger collections have started to offer scientific services, which could be another option for breweries to outsource prospective projects. Boundy-Mills and colleagues recently published a detailed analysis of modern yeast culture collections listing the most important collections and illustrating the challenges and opportunities for their users (Boundy-Mills et al., 2016).
Saccharomyces Hybrids and Beyond
In this mini review, we have focused on S. cerevisae. However, it should be mentioned that interspecies hybrids between S. cerevisiae and usually another member of the Saccharomyces genus play a significant role in industrial fermentation processes including beer brewing. For example, it was found that up to 25% of the top fermenting yeasts isolated from Belgian Lambic/Trappist beer production are interspecies hybrids between S. cerevisiae and S. kudriavzevii (González et al., 2008; Peris et al., 2012, 2018). More importantly, the previously mentioned S. pastorianus “lager” yeast, responsible for 90% of the beer produced worldwide today, was suggested to be a hybrid already decades ago (for an early review, see Polaina, 2002). However, it was only in 2011 that the missing non-S. cerevisiae parent of the “lager” yeast lineage was isolated and identified as S. eubayanus (Libkind et al., 2011). Hybrids often show a better performance under certain conditions since they combine features otherwise unique to one of the parental species, e.g., the high fermentative capacity of S. cerevisiae and the cold tolerance of S. kudriavzevii or S. eubayanus, resulting in hybrids suitable for cold temperature fermentations (Mertens et al., 2015; Krogerus et al., 2017; Nikulin et al., 2018; Ortiz-Tovar et al., 2018; Baker et al., 2019). In addition, hybrids can also benefit from a less well-understood effect called heterosis or hybrid vigor, where the resulting hybrid shows a greater fitness than one would expect based on each parental strain (Lindegren et al., 1953; Hebly et al., 2015; Bernardes et al., 2017). Although hybrids have a huge potential for industrial use, they themselves are usually less suitable for performing genetic crosses since most strains will suffer from hybrid sterility, resulting in very low numbers of viable progeny with a clear mating type (Sipiczki, 2018, 2019; Ono and Greig, 2020; Sipiczki et al., 2020). While there were attempts to create novel “lager” strains by crossing meiotic offspring of “lager” yeasts already in the early 1980s (Gjermansen and Sigsgaard, 1981), it was the identification of S. eubayanus that changed the concept of creating novel “lager” yeasts significantly. Once S. eubayanus was available novel S. cerevisiae × S. eubayanus hybrids could be generated by “simple” interspecies crosses, and several reports over the last few years have clearly indicated the high potential of these novel hybrids (Krogerus et al., 2015; Mertens et al., 2015; Nikulin et al., 2018; Langdon et al., 2019). In addition, not only the increasing number of strains but also species within the Saccharomyces genus might facilitate the generation of a wider selection of intra- and inter-species hybrids with hopefully exciting novel features (Nikulin et al., 2018). Finally, as mentioned in the beginning, the beer brewing process was initiated by spontaneous fermentation for centuries, and still is for certain beer types today. A vast number of so-called nonconventional yeasts have been isolated from these mixed fermentations over the years and their impact on fermentation performance and flavor profiles has been studied (Spencer et al., 2002; Michel et al., 2016a; Varela, 2016; Gibson et al., 2017; Capece et al., 2018b; Holt et al., 2018; Canonico et al., 2019; Colomer et al., 2019; Cubillos et al., 2019). In addition to using pure yeast cultures from these nonconventional yeast species in brewing of novel beer styles or special beers (Steensels et al., 2015; Michel et al., 2016b; Capece et al., 2018a; De Francesco et al., 2018; Gutiérrez et al., 2018; Ravasio et al., 2018; Colomer et al., 2020; Iattici et al., 2020; Zdaniewicz et al., 2020), these yeasts could be used, where genetically possible, to expand the concept of novel interspecies hybrids for the brewing industry beyond the Saccharomyces genus adding even more genetic diversity to brewer’s yeasts.
Outlook
Since prehistoric times, humans have used S. cerevisiae to convert sugar containing raw materials into flavorful alcoholic beverages, such as beer (Michel et al., 1992; Piskur et al., 2006; Steensels and Verstrepen, 2014). At the same time the organism has been domesticated over the span of centuries, resulting in the evolution of highly specific strain lineages optimized for beer brewing but dramatically reduced in genetic variability (McMurrough et al., 1996; Brown et al., 2010; Steensels and Verstrepen, 2014; Gallone et al., 2016; Gonçalves et al., 2016). While it is an attractive idea to breathe new life into current brewer’s yeasts by introducing novel wild yeasts into industrial applications, their use may not be as straight forward as anticipated. Most natural isolates will lack features that are necessary for the industrial scale beer production and the genetic diversity may not always correspond to a phenotypic one (Camarasa et al., 2011). This implies that strains will need to be improved (Figure 1). However, due to consumer concerns, the use of modern genetic approaches is usually objected (Bamforth, 2006; Saerens et al., 2010). Therefore, applied researchers currently rely on classic yeast breeding techniques. Interestingly enough that these techniques were initially developed by the brewing industry, when Øjvind Winge discovered the sexual cycle of S. cerevisiae and used his knowledge to combine desirable brewing traits by crossing different strains at the Carlsberg Research Laboratory in the 1930s (Winge, 1935; Barnett, 2007). So the brewing industry could end up renewing their applied yeast research and their strain portfolio by going back to their own roots (Gibson et al., 2017; Iattici et al., 2020).
Author Contributions
KL and VS were the main contributors to the abstract, main text, and reference list. RF provided the Figure. MK and JF were involved in the initial planning of the manuscript and provided helpful comments throughout the preparation of the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
All authors of this article are employed by the company Carlsberg A/S, Copenhagen, Denmark. VS is funded through the “Beer Fingerprinting Project” grant no. 7045-00014A of the Innovation Fund Denmark.
Acknowledgments
We thank Claes Gjermansen and Erik Lund for their helpful comments on the manuscript. Figure 1 was created using design elements from Freepik.com.
References
- Alsammar H., Delneri D. (2020). An update on the diversity, ecology and biogeography of the Saccharomyces genus. FEMS Yeast Res. 20:foaa013. 10.1093/femsyr/foaa013, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim H. V., Lopes M. L., de Castro Oliveira J. V., Buckeridge M. S., Goldman G. H. (2011). Scientific challenges of bioethanol production in Brazil. Appl. Microbiol. Biotechnol. 91, 1267–1275. 10.1007/s00253-011-3437-6, PMID: [DOI] [PubMed] [Google Scholar]
- Aouizerat T., Gutman I., Paz Y., Maeir A. M., Gadot Y., Gelman D., et al. (2019). Isolation and characterization of live yeast cells from ancient vessels as a tool in bio-archaeology. mBio 10, e00388–e00419. 10.1128/mBio.00388-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo T. M., Souza M. T., Diniz R. H. S., Yamakawa C. K., Soares L. B., Lenczak J. L., et al. (2018). Cachaça yeast strains: alternative starters to produce beer and bioethanol. Antonie Van Leeuwenhoek 111, 1749–1766. 10.1007/s10482-018-1063-3, PMID: [DOI] [PubMed] [Google Scholar]
- Baker E. P., Peris D., Moriarty R. V., Li X. C., Fay J. C., Hittinger C. T. (2019). Mitochondrial DNA and temperature tolerance in lager yeasts. Sci. Adv. 5:eaav1869. 10.1126/sciadv.aav1869, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamforth C. (2006). Brewing: New technologies. Cambridge: Woodhead Publishing Limited. [Google Scholar]
- Barnett J. A. (2007). A history of research on yeasts 10: foundations of yeast genetics. Yeast 24, 799–845. 10.1002/yea.1513, PMID: [DOI] [PubMed] [Google Scholar]
- Barnett J. A., Lichtenthaler F. W. (2001). A history of research on yeasts 3: Emil Fischer, Eduard Buchner and their contemporaries, 1880-1900. Yeast 18, 363–388. , PMID: [DOI] [PubMed] [Google Scholar]
- Baschali A., Tsakalidou E., Kyriacou A., Karavasiloglou N., Matalas A. -L. (2017). Traditional low-alcoholic and non-alcoholic fermented beverages consumed in European countries: a neglected food group. Nutr. Res. Rev. 30, 1–24. 10.1017/S0954422416000202, PMID: [DOI] [PubMed] [Google Scholar]
- Basso L. C., Amorim H. V. D., Oliveira A. J. D., Lopes M. L. (2008). Yeast selection for fuel ethanol production in Brazil. FEMS Yeast Res. 8, 1155–1163. 10.1111/j.1567-1364.2008.00428.x, PMID: [DOI] [PubMed] [Google Scholar]
- Beato F. B., Bergdahl B., Rosa C. A., Forster J., Gombert A. K. (2016). Physiology of Saccharomyces cerevisiae strains isolated from Brazilian biomes: new insights into biodiversity and industrial applications. FEMS Yeast Res. 16:fow076. 10.1093/femsyr/fow076, PMID: [DOI] [PubMed] [Google Scholar]
- Beech F. W., Davenport R. R. (1971). “Chapter V isolation, purification and maintenance of yeasts” in Methods in microbiology. ed. Booth C. (London: Academic Press; ), 153–182. [Google Scholar]
- Bendixsen D. P., Gettle N., Gilchrist C., Zhang Z., Stelkens R. (2020). Tracking the origin of baker’s yeast: Seven new long-read genomes of highly divergent, wild Saccharomyces cerevisiae strains from East Asia. BioRxiv [Preprint]. Available at: https://www.biorxiv.org/content/10.1101/2020.06.03.131607v1.full (Accessed June 20, 2020).
- Bernardes J. P., Stelkens R. B., Greig D. (2017). Heterosis in hybrids within and between yeast species. J. Evol. Biol. 30, 538–548. 10.1111/jeb.13023, PMID: [DOI] [PubMed] [Google Scholar]
- Black J. A., Cunningham G., Robson E., Zólyomi G. (2006). The literature of ancient Sumer. Oxford: Oxford University Press. [Google Scholar]
- Borneman A. R., Forgan A. H., Kolouchova R., Fraser J. A., Schmidt S. A. (2016). Whole genome comparison reveals high levels of inbreeding and strain redundancy across the spectrum of commercial wine strains of Saccharomyces cerevisiae. G3 (Bethesda) 6, 957–971. 10.1534/g3.115.025692, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boundy-Mills K. (2012). Yeast culture collections of the world: meeting the needs of industrial researchers. J. Ind. Microbiol. Biotechnol. 39, 673–680. 10.1007/s10295-011-1078-5, PMID: [DOI] [PubMed] [Google Scholar]
- Boundy-Mills K. L., Glantschnig E., Roberts I. N., Yurkov A., Casaregola S., Daniel H. -M., et al. (2016). Yeast culture collections in the twenty-first century: new opportunities and challenges. Yeast 33, 243–260. 10.1002/yea.3171, PMID: [DOI] [PubMed] [Google Scholar]
- Boynton P. J., Greig D. (2014). The ecology and evolution of non-domesticated Saccharomyces species. Yeast 31, 449–462. 10.1002/yea.3040, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boynton P. J., Kowallik V., Landermann D., Stukenbrock E. H. (2019). Quantifying the efficiency and biases of forest Saccharomyces sampling strategies. Yeast 36, 657–668. 10.1002/yea.3435, PMID: [DOI] [PubMed] [Google Scholar]
- Brady M. (2008). “First taste of alcohol” in First taste: How indigenous Australians learned about grog. ed. MacKenzie M. (Deakin, A.C.T: Alcohol Education and Rehabilitation Foundation; ). [Google Scholar]
- Brown C. A., Murray A. W., Verstrepen K. J. (2010). Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr. Biol. 20, 895–903. 10.1016/j.cub.2010.04.027, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buser C. C., Newcomb R. D., Gaskett A. C., Goddard M. R. (2014). Niche construction initiates the evolution of mutualistic interactions. Ecol. Lett. 17, 1257–1264. 10.1111/ele.12331, PMID: [DOI] [PubMed] [Google Scholar]
- Camarasa C., Sanchez I., Brial P., Bigey F., Dequin S. (2011). Phenotypic landscape of Saccharomyces cerevisiae during wine fermentation: evidence for origin-dependent metabolic traits. PLoS One 6:e25147. 10.1371/journal.pone.0025147, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell-Platt G. (1987). Fermented foods of the world: Dictionary and guide. London: Butterworth. [Google Scholar]
- Campbell-Platt G. (1994). Fermented foods — a world perspective. Food Res. Int. 27, 253–257. 10.1016/0963-9969(94)90093-0 [DOI] [Google Scholar]
- Canonico L., Comitini F., Ciani M. (2014). Dominance and influence of selected Saccharomyces cerevisiae strains on the analytical profile of craft beer refermentation. J. Inst. Brew. 120, 262–267. 10.1002/jib.133 [DOI] [Google Scholar]
- Canonico L., Galli E., Ciani E., Comitini F., Ciani M. (2019). Exploitation of three non-conventional yeast species in the brewing process. Microorganisms 7:11. 10.3390/microorganisms7010011, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capece A., Romaniello R., Pietrafesa A., Siesto G., Pietrafesa R., Zambuto M., et al. (2018a). Use of Saccharomyces cerevisiae var. boulardii in co-fermentations with S. cerevisiae for the production of craft beers with potential healthy value-added. Int. J. Food Microbiol. 284, 22–30. 10.1016/j.ijfoodmicro.2018.06.028 [DOI] [PubMed] [Google Scholar]
- Capece A., Romaniello R., Siesto G., Romano P. (2018b). Conventional and non-conventional yeasts in beer production. Fermentation 4:38. 10.3390/fermentation4020038 [DOI] [Google Scholar]
- Cavalieri D., McGovern P. E., Hartl D. L., Mortimer R., Polsinelli M. (2003). Evidence for S. cerevisiae fermentation in ancient wine. J. Mol. Evol. 57, S226–S232. 10.1007/s00239-003-0031-2, PMID: [DOI] [PubMed] [Google Scholar]
- Christofoleti-Furlan R. M., Portugal C. B., Varize C. S., Muynarsk E. S. M., Alcarde A. R., Basso L. C. (2020). Unraveling Brazilian bioethanol yeasts as novel starters for high-gravity brewing. Food Res. Int. 135:109282. 10.1016/j.foodres.2020.109282, PMID: [DOI] [PubMed] [Google Scholar]
- Colomer M. S., Chailyan A., Fennessy R. T., Olsson K. F., Johnsen L., Solodovnikova N., et al. (2020). Assessing population diversity of Brettanomyces yeast species and identification of strains for brewing applications. Front. Microbiol. 11:637. 10.3389/fmicb.2020.00637, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomer M. S., Funch B., Forster J. (2019). The raise of Brettanomyces yeast species for beer production. Curr. Opin. Biotechnol. 56, 30–35. 10.1016/j.copbio.2018.07.009, PMID: [DOI] [PubMed] [Google Scholar]
- Corran H. S. (1975). A history of brewing. Newton Abbot: David & Charles. [Google Scholar]
- Cubillos F. A., Gibson B., Grijalva-Vallejos N., Krogerus K., Nikulin J. (2019). Bioprospecting for brewers: exploiting natural diversity for naturally diverse beers. Yeast 36, 383–398. 10.1002/yea.3380, PMID: [DOI] [PubMed] [Google Scholar]
- Daenen L., Sterckx F., Delvaux F. R., Verachtert H., Derdelinckx G. (2008). Evaluation of the glycoside hydrolase activity of a Brettanomyces strain on glycosides from sour cherry (Prunus cerasus L.) used in the production of special fruit beers. FEMS Yeast Res. 8, 1103–1114. 10.1111/j.1567-1364.2008.00421.x, PMID: [DOI] [PubMed] [Google Scholar]
- Daniel H. -M., Prasad G. S. (2010). The role of culture collections as an interface between providers and users: the example of yeasts. Res. Microbiol. 161, 488–496. 10.1016/j.resmic.2010.01.011, PMID: [DOI] [PubMed] [Google Scholar]
- Dapporto L., Stefanini I., Rivero D., Polsinelli M., Capretti P., De Marchi P., et al. (2016). Social wasp intestines host the local phenotypic variability of Saccharomyces cerevisiae strains. Yeast 33, 277–287. 10.1002/yea.3173, PMID: [DOI] [PubMed] [Google Scholar]
- De Francesco G., Sannino C., Sileoni V., Marconi O., Filippucci S., Tasselli G., et al. (2018). Mrakia gelida in brewing process: an innovative production of low alcohol beer using a psychrophilic yeast strain. Food Microbiol. 76, 354–362. 10.1016/j.fm.2018.06.018, PMID: [DOI] [PubMed] [Google Scholar]
- Dietrich O., Heun M., Notroff J., Schmidt K., Zarnkow M. (2012). The role of cult and feasting in the emergence of Neolithic communities. New evidence from Göbekli Tepe, South-Eastern Turkey. Antiquity 86, 674–695. 10.1017/S0003598X00047840 [DOI] [Google Scholar]
- Dlusskaya E., Jänsch A., Schwab C., Gänzle M. G. (2008). Microbial and chemical analysis of a kvass fermentation. Eur. Food Res. Technol. 227, 261–266. 10.1007/s00217-007-0719-4 [DOI] [Google Scholar]
- Domizio P., Lencioni L., Ciani M., Di Blasi S., Pontremolesi C., Sabatelli M. P. (2007). Spontaneous and inoculated yeast populations dynamics and their effect on organoleptic characters of Vinsanto wine under different process conditions. Int. J. Food Microbiol. 115, 281–289. 10.1016/j.ijfoodmicro.2006.10.052, PMID: [DOI] [PubMed] [Google Scholar]
- Dornbusch H. D. (1998). Prost!: The story of German beer. Boulder: Brewers Publications. [Google Scholar]
- Dredge M. (2019). A brief history of lager: 500 years of the world’s favourite beer. London: Kyle Books. [Google Scholar]
- Driscoll C. A., Macdonald D. W., O’Brien S. J. (2009). From wild animals to domestic pets, an evolutionary view of domestication. Proc. Natl. Acad. Sci. 106, 9971–9978. 10.1073/pnas.0901586106, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S. -F., Han P. -J., Wang Q. -M., Liu W. -Q., Shi J. -Y., Li K., et al. (2018). The origin and adaptive evolution of domesticated populations of yeast from Far East Asia. Nat. Commun. 9:2690. 10.1038/s41467-018-05106-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekberg J., Gibson B., Joensuu J. J., Krogerus K., Magalhães F., Mikkelson A., et al. (2015). Physicochemical characterization of sahti, an ‘ancient’ beer style indigenous to Finland. J. Inst. Brew. 121, 464–473. 10.1002/jib.246 [DOI] [Google Scholar]
- Evans L. H. (1990). “Yeast strains for baking” in Yeast technology. eds. Spencer J. F. T., Spencer D. M. (Berlin: Springer-Verlag; ), 13–45. [Google Scholar]
- Gallone B., Steensels J., Prahl T., Soriaga L., Saels V., Herrera-Malaver B., et al. (2016). Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell 166, 1397.e16–1410.e16. 10.1016/j.cell.2016.08.020, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson B., Geertman J. -M. A., Hittinger C. T., Krogerus K., Libkind D., Louis E. J., et al. (2017). New yeasts-new brews: modern approaches to brewing yeast design and development. FEMS Yeast Res. 17. 10.1093/femsyr/fox038, PMID: [DOI] [PubMed] [Google Scholar]
- Gibson B. R., Lawrence S. J., Leclaire J. P. R., Powell C. D., Smart K. A. (2007). Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiol. Rev. 31, 535–569. 10.1111/j.1574-6976.2007.00076.x, PMID: [DOI] [PubMed] [Google Scholar]
- Gjermansen C., Sigsgaard P. (1981). Construction of a hybrid brewing strain of Saccharomyces carlsbergensis by mating of meiotic segregants. Carlsb. Res. Commun. 46, 1–11. 10.1007/BF02906193 [DOI] [Google Scholar]
- Glover B. (1997). The world encyclopedia of beer. New York: Lorenz Books. [Google Scholar]
- Goddard M. R., Greig D. (2015). Saccharomyces cerevisiae: a nomadic yeast with no niche? FEMS Yeast Res. 15:fov009. 10.1093/femsyr/fov009, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves M., Pontes A., Almeida P., Barbosa R., Serra M., Libkind D., et al. (2016). Distinct domestication trajectories in top-fermenting beer yeasts and wine yeasts. Curr. Biol. 26, 2750–2761. 10.1016/j.cub.2016.08.040, PMID: [DOI] [PubMed] [Google Scholar]
- González S. S., Barrio E., Querol A. (2008). Molecular characterization of new natural hybrids of Saccharomyces cerevisiae and S. kudriavzevii in brewing. Appl. Environ. Microbiol. 74, 2314–2320. 10.1128/AEM.01867-07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez A., Boekhout T., Gojkovic Z., Katz M. (2018). Evaluation of non-Saccharomyces yeasts in the fermentation of wine, beer and cider for the development of new beverages. J. Inst. Brew. 124, 389–402. 10.1002/jib.512 [DOI] [Google Scholar]
- Hansen E. C. (1888). Undersogelser over alkoholgjaersvampenes fysiologi og morfologi. VII. Meddelelser Carlsberg Lab. 2, 220–256. [Google Scholar]
- Heard G. M., Fleet G. H. (1985). Growth of natural yeast flora during the fermentation of inoculated wines. Appl. Environ. Microbiol. 50, 727–728. 10.1128/AEM.50.3.727-728.1985, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebly M., Brickwedde A., Bolat I., Driessen M. R. M., de Hulster E. A. F., van den Broek M., et al. (2015). S. cerevisiae × S. eubayanus interspecific hybrid, the best of both worlds and beyond. FEMS Yeast Res. 15:fov005. 10.1093/femsyr/fov005, PMID: [DOI] [PubMed] [Google Scholar]
- Henick-Kling T., Edinger W., Daniel P., Monk P. (1998). Selective effects of sulfur dioxide and yeast starter culture addition on indigenous yeast populations and sensory characteristics of wine. J. Appl. Microbiol. 84, 865–876. 10.1046/j.1365-2672.1998.00423.x [DOI] [Google Scholar]
- Hittinger C. T. (2013). Saccharomyces diversity and evolution: a budding model genus. Trends Genet. 29, 309–317. 10.1016/j.tig.2013.01.002, PMID: [DOI] [PubMed] [Google Scholar]
- Holt S., Mukherjee V., Lievens B., Verstrepen K. J., Thevelein J. M. (2018). Bioflavoring by non-conventional yeasts in sequential beer fermentations. Food Microbiol. 72, 55–66. 10.1016/j.fm.2017.11.008, PMID: [DOI] [PubMed] [Google Scholar]
- Hornsey I. S. (2003). A history of beer and brewing. Cambridge: Royal Society of Chemistry. [Google Scholar]
- Iattici F., Catallo M., Solieri L. (2020). Designing new yeasts for craft brewing: when natural biodiversity meets biotechnology. Beverages 6:3. 10.3390/beverages6010003 [DOI] [Google Scholar]
- Jentsch M. (2007). Top-fermented beer specialities in focus. Brauwelt Int. 5, 332–334. [Google Scholar]
- Johansen P. G., Owusu-Kwarteng J., Parkouda C., Padonou S. W., Jespersen L. (2019). Occurrence and importance of yeasts in indigenous fermented food and beverages produced in sub-Saharan Africa. Front. Microbiol. 10:1789. 10.3389/fmicb.2019.01789, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K., Bergdahl B., Machado D., Dato L., Han T. -L., Li J., et al. (2019). Linking genetic, metabolic, and phenotypic diversity among Saccharomyces cerevisiae strains using multi-omics associations. Gigascience 8:giz015. 10.1093/gigascience/giz015, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz S., Maytag F. (1991). Brewing an ancient beer. Archaeology 44, 22–23. [Google Scholar]
- Kechkar M., Sayed W., Cabrol A., Aziza M., Ahmed Zaid T., Amrane A., et al. (2019). Isolation and identification of yeast strains from sugarcane molasses, dates and figs for ethanol production under conditions simulating algal hydrolysate. Braz. J. Chem. Eng. 36, 157–169. 10.1590/0104-6632.20190361s20180114 [DOI] [Google Scholar]
- Krogerus K., Magalhães F., Vidgren V., Gibson B. (2015). New lager yeast strains generated by interspecific hybridization. J. Ind. Microbiol. Biotechnol. 42, 769–778. 10.1007/s10295-015-1597-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogerus K., Seppänen-Laakso T., Castillo S., Gibson B. (2017). Inheritance of brewing-relevant phenotypes in constructed Saccharomyces cerevisiae × Saccharomyces eubayanus hybrids. Microb. Cell Factories 16:66. 10.1186/s12934-017-0679-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon Q. K., Peris D., Baker E. P., Opulente D. A., Nguyen H. -V., Bond U., et al. (2019). Fermentation innovation through complex hybridization of wild and domesticated yeasts. Nat. Ecol. Evol. 3, 1576–1586. 10.1038/s41559-019-0998-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legras J. -L., Merdinoglu D., Cornuet J. -M., Karst F. (2007). Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol. Ecol. 16, 2091–2102. 10.1111/j.1365-294X.2007.03266.x, PMID: [DOI] [PubMed] [Google Scholar]
- Libkind D., Hittinger C. T., Valério E., Gonçalves C., Dover J., Johnston M., et al. (2011). Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc. Natl. Acad. Sci. U. S. A. 108, 14539–14544. 10.1073/pnas.1105430108, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindegren C. C., Braham J. E., Calle J. D. D. (1953). Heterosis in Saccharomyces. Nature 172, 800–802. 10.1038/172800a0, PMID: [DOI] [PubMed] [Google Scholar]
- Liti G. (2015). The fascinating and secret wild life of the budding yeast S. cerevisiae. elife 4:e05835. 10.7554/eLife.05835, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang J., Rosenberg D., Zhao H., Lengyel G., Nadel D. (2018). Fermented beverage and food storage in 13,000 y-old stone mortars at Raqefet cave, Israel: investigating Natufian ritual feasting. J. Archaeol. Sci. Rep. 21, 783–793. 10.1016/j.jasrep.2018.08.008 [DOI] [Google Scholar]
- Lodolo E. J., Kock J. L. F., Axcell B. C., Brooks M. (2008). The yeast Saccharomyces cerevisiae– the main character in beer brewing. FEMS Yeast Res. 8, 1018–1036. 10.1111/j.1567-1364.2008.00433.x, PMID: [DOI] [PubMed] [Google Scholar]
- Marongiu A., Zara G., Legras J. -L., Del Caro A., Mascia I., Fadda C., et al. (2015). Novel starters for old processes: use of Saccharomyces cerevisiae strains isolated from artisanal sourdough for craft beer production at a brewery scale. J. Ind. Microbiol. Biotechnol. 42, 85–92. 10.1007/s10295-014-1525-1, PMID: [DOI] [PubMed] [Google Scholar]
- McGovern P. E., Glusker D. L., Exner L. J., Voigt M. M. (1996). Neolithic resinated wine. Nature 381, 480–481. 10.1038/381480a0 [DOI] [Google Scholar]
- McGovern P. E., Hartung U., Badler V. R., Glusker D. L., Exner L. J. (1997). The beginnings of winemaking and viniculture in the ancient near east and Egypt. Expedition 39, 3–21. [Google Scholar]
- McGovern P. E., Zhang J., Tang J., Zhang Z., Hall G. R., Moreau R. A., et al. (2004). Fermented beverages of pre- and proto-historic China. Proc. Natl. Acad. Sci. U. S. A. 101, 17593–17598. 10.1073/pnas.0407921102, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurrough I., Madigan D., Donnelly D., Hurley J., Doyle A. -M., Hennigan G., et al. (1996). Control of ferulic acid and 4-vinyl guaiacol in brewing. J. Inst. Brew. 102, 327–332. 10.1002/j.2050-0416.1996.tb00918.x [DOI] [Google Scholar]
- Mertens S., Steensels J., Saels V., De Rouck G., Aerts G., Verstrepen K. J. (2015). A large set of newly created interspecific Saccharomyces hybrids increases aromatic diversity in lager beers. Appl. Environ. Microbiol. 81, 8202–8214. 10.1128/AEM.02464-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel M., Kopecká J., Meier-Dörnberg T., Zarnkow M., Jacob F., Hutzler M. (2016a). Screening for new brewing yeasts in the non-Saccharomyces sector with Torulaspora delbrueckii as model. Yeast 33, 129–144. 10.1002/yea.3146, PMID: [DOI] [PubMed] [Google Scholar]
- Michel R. H., McGovern P. E., Badler V. R. (1992). Chemical evidence for ancient beer. Nature 360:24. 10.1038/360024b0 [DOI] [Google Scholar]
- Michel M., Meier-Dörnberg T., Jacob F., Methner F. -J., Wagner R. S., Hutzler M. (2016b). Review: pure non-Saccharomyces starter cultures for beer fermentation with a focus on secondary metabolites and practical applications. J. Inst. Brew. 122, 569–587. 10.1002/jib.381 [DOI] [Google Scholar]
- Mortimer R., Polsinelli M. (1999). On the origins of wine yeast. Res. Microbiol. 150, 199–204. 10.1016/S0923-2508(99)80036-9, PMID: [DOI] [PubMed] [Google Scholar]
- Nelson M. (2005). The barbarian’s beverage: A history of beer in ancient Europe. Abingdon: Routledge. [Google Scholar]
- Nikulin J., Krogerus K., Gibson B. (2018). Alternative Saccharomyces interspecies hybrid combinations and their potential for low-temperature wort fermentation. Yeast 35, 113–127. 10.1002/yea.3246, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohya Y., Kashima M. (2019). History, lineage and phenotypic differentiation of sake yeast. Biosci. Biotechnol. Biochem. 83, 1442–1448. 10.1080/09168451.2018.1564620, PMID: [DOI] [PubMed] [Google Scholar]
- Ono J., Greig D. (2020). A Saccharomyces paradox: chromosomes from different species are incompatible because of anti-recombination, not because of differences in number or arrangement. Curr. Genet. 66, 469–474. 10.1007/s00294-019-01038-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Tovar G., Pérez-Torrado R., Adam A. C., Barrio E., Querol A. (2018). A comparison of the performance of natural hybrids Saccharomyces cerevisiae × Saccharomyces kudriavzevii at low temperatures reveals the crucial role of their S. kudriavzevii genomic contribution. Int. J. Food Microbiol. 274, 12–19. 10.1016/j.ijfoodmicro.2018.03.002, PMID: [DOI] [PubMed] [Google Scholar]
- Parapouli M., Vasileiadis A., Afendra A. -S., Hatziloukas E. (2020). Saccharomyces cerevisiae and its industrial applications. AIMS Microbiol. 6, 1–31. 10.3934/microbiol.2020001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasteur L. (1857). Mémoire sur la fermentation alcoolique. CR Chim. 45, 1032–1036. [Google Scholar]
- Pasteur L. (1858). Nouveaux faits concernant l’histoire de la fermentation alcoolique. CR Chim. 47, 1011–1013. [Google Scholar]
- Peris D., Lopes C. A., Belloch C., Querol A., Barrio E. (2012). Comparative genomics among Saccharomyces cerevisiae × Saccharomyces kudriavzevii natural hybrid strains isolated from wine and beer reveals different origins. BMC Genomics 13:407. 10.1186/1471-2164-13-407, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris D., Pérez-Torrado R., Hittinger C. T., Barrio E., Querol A. (2018). On the origins and industrial applications of Saccharomyces cerevisiae × Saccharomyces kudriavzevii hybrids. Yeast 35, 51–69. 10.1002/yea.3283, PMID: [DOI] [PubMed] [Google Scholar]
- Peter J., De Chiara M., Friedrich A., Yue J. -X., Pflieger D., Bergström A., et al. (2018). Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556, 339–344. 10.1038/s41586-018-0030-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskur J., Rozpedowska E., Polakova S., Merico A., Compagno C. (2006). How did Saccharomyces evolve to become a good brewer? Trends Genet. 22, 183–186. 10.1016/j.tig.2006.02.002, PMID: [DOI] [PubMed] [Google Scholar]
- Polaina J. (2002). “Brewer’s yeast: genetics and biotechnology” in Applied mycology and biotechnology agriculture and food production. eds. Khachatourians G. G., Arora D. K. (Amsterdam: Elsevier; ), 1–17. [Google Scholar]
- Preiss R., Tyrawa C., Krogerus K., Garshol L. M., van der Merwe G. (2018). Traditional Norwegian kveik are a genetically distinct group of domesticated Saccharomyces cerevisiae brewing yeasts. Front. Microbiol. 9:2137. 10.3389/fmicb.2018.02137, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protz R. (1995). Ultimate encyclopedia of beer: The definitive guide to the world’s great brews. 1st Edn. New York: Smithmark Publishers. [Google Scholar]
- Purugganan M. D., Fuller D. Q. (2009). The nature of selection during plant domestication. Nature 457, 843–848. 10.1038/nature07895, PMID: [DOI] [PubMed] [Google Scholar]
- Quain D. E. (2006). “8 - yeast supply and propagation in brewing” in Brewing. ed. Bamforth C. W. (Cambridge: Woodhead Publishing; ), 167–182. [Google Scholar]
- Randez-Gil F., Córcoles-Sáez I., Prieto J. A. (2013). Genetic and phenotypic characteristics of baker’s yeast: relevance to baking. Annu. Rev. Food Sci. Technol. 4, 191–214. 10.1146/annurev-food-030212-182609, PMID: [DOI] [PubMed] [Google Scholar]
- Ravasio D., Carlin S., Boekhout T., Groenewald M., Vrhovsek U., Walther A., et al. (2018). Adding flavor to beverages with non-conventional yeasts. Fermentation 4:15. 10.3390/fermentation4010015 [DOI] [Google Scholar]
- Reis V. R., Antonangelo A. T. B. F., Bassi A. P. G., Colombi D., Ceccato-Antonini S. R. (2017). Bioethanol strains of Saccharomyces cerevisiae characterised by microsatellite and stress resistance. Braz. J. Microbiol. 48, 268–274. 10.1016/j.bjm.2016.09.017, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi S., Turchetti B., Sileoni V., Marconi O., Perretti G. (2018). Evaluation of Saccharomyces cerevisiae strains isolated from non-brewing environments in beer production. J. Inst. Brew. 124, 381–388. 10.1002/jib.503 [DOI] [Google Scholar]
- Saerens S. M. G., Duong C. T., Nevoigt E. (2010). Genetic improvement of brewer’s yeast: current state, perspectives and limits. Appl. Microbiol. Biotechnol. 86, 1195–1212. 10.1007/s00253-010-2486-6, PMID: [DOI] [PubMed] [Google Scholar]
- Sicard D., Legras J. -L. (2011). Bread, beer and wine: yeast domestication in the Saccharomyces sensu stricto complex. CR Biol. 334, 229–236. 10.1016/j.crvi.2010.12.016, PMID: [DOI] [PubMed] [Google Scholar]
- Sipiczki M. (2018). Interspecies hybridisation and genome chimerisation in Saccharomyces: combining of gene pools of species and its biotechnological perspectives. Front. Microbiol. 9:3071. 10.3389/fmicb.2018.03071, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M. (2019). Yeast two- and three-species hybrids and high-sugar fermentation. Microb. Biotechnol. 12, 1101–1108. 10.1111/1751-7915.13390, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M., Antunovics Z., Szabo A. (2020). MAT heterozygosity and the second sterility barrier in the reproductive isolation of Saccharomyces species. Curr. Genet. 66, 957–969. 10.1007/s00294-020-01080-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sláviková E., Vadkertiová R. (1995). Yeasts and yeast-like organisms isolated from fish-pond waters. Acta Microbiol. Pol. 44, 181–189. PMID: [PubMed] [Google Scholar]
- Sláviková E., Vadkertiová R. (1997). Seasonal occurrence of yeasts and yeast-like organisms in the river Danube. Antonie Van Leeuwenhoek 72, 77–80. 10.1023/A:1000287005253, PMID: [DOI] [PubMed] [Google Scholar]
- Spencer J. F. T., Ragout de Spencer A. L., Laluce C. (2002). Non-conventional yeasts. Appl. Microbiol. Biotechnol. 58, 147–156. 10.1007/s00253-001-0834-2, PMID: [DOI] [PubMed] [Google Scholar]
- Steensels J., Daenen L., Malcorps P., Derdelinckx G., Verachtert H., Verstrepen K. J. (2015). Brettanomyces yeasts--from spoilage organisms to valuable contributors to industrial fermentations. Int. J. Food Microbiol. 206, 24–38. 10.1016/j.ijfoodmicro.2015.04.005, PMID: [DOI] [PubMed] [Google Scholar]
- Steensels J., Gallone B., Voordeckers K., Verstrepen K. J. (2019). Domestication of industrial microbes. Curr. Biol. 29, R381–R393. 10.1016/j.cub.2019.04.025, PMID: [DOI] [PubMed] [Google Scholar]
- Steensels J., Snoek T., Meersman E., Picca Nicolino M., Voordeckers K., Verstrepen K. J. (2014). Improving industrial yeast strains: exploiting natural and artificial diversity. FEMS Microbiol. Rev. 38, 947–995. 10.1111/1574-6976.12073, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensels J., Verstrepen K. J. (2014). Taming wild yeast: potential of conventional and nonconventional yeasts in industrial fermentations. Annu. Rev. Microbiol. 68, 61–80. 10.1146/annurev-micro-091213-113025, PMID: [DOI] [PubMed] [Google Scholar]
- Stefanini I., Dapporto L., Legras J. -L., Calabretta A., Di Paola M., De Filippo C., et al. (2012). Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proc. Natl. Acad. Sci. U. S. A. 109, 13398–13403. 10.1073/pnas.1208362109, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevic S. (1962). The significance of bees (Apis sp.) and wasps (Vespa sp.) as carriers of yeast for the micoflora of grapes and the quality of wine. Arkh. Poljjoprivredne Nauke 50, 80–92. [Google Scholar]
- Šťovíček V., Váchová L., Begany M., Wilkinson D., Palková Z. (2014). Global changes in gene expression associated with phenotypic switching of wild yeast. BMC Genomics 15:136. 10.1186/1471-2164-15-136, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šťovíček V., Váchová L., Kuthan M., Palková Z. (2010). General factors important for the formation of structured biofilm-like yeast colonies. Fungal Genet. Biol. 47, 1012–1022. 10.1016/j.fgb.2010.08.005 [DOI] [PubMed] [Google Scholar]
- Tamang J. (2010). “Diversity of fermented foods” in Fermented foods and beverages of the world. eds. Tamang J., Kailasapathy K. (New York: CRC Press, Taylor and Francis Group; ), 41–84. [Google Scholar]
- Tamang J. P., Cotter P. D., Endo A., Han N. S., Kort R., Liu S. Q., et al. (2020). Fermented foods in a global age: east meets west. Compr. Rev. Food Sci. Food Saf. 19, 184–217. 10.1111/1541-4337.12520 [DOI] [PubMed] [Google Scholar]
- Taylor M. W., Tsai P., Anfang N., Ross H. A., Goddard M. R. (2014). Pyrosequencing reveals regional differences in fruit-associated fungal communities. Environ. Microbiol. 16, 2848–2858. 10.1111/1462-2920.12456, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tofalo R., Fusco V., Böhnlein C., Kabisch J., Logrieco A. F., Habermann D., et al. (2019). The life and times of yeasts in traditional food fermentations. Crit. Rev. Food Sci. Nutr. 60, 3103–3132. 10.1080/10408398.2019.1677553, PMID: [DOI] [PubMed] [Google Scholar]
- Tokpohozin S. E., Fischer S., Becker T. (2019). Selection of a new Saccharomyces yeast to enhance relevant sorghum beer aroma components, higher alcohols and esters. Food Microbiol. 83, 181–186. 10.1016/j.fm.2019.05.014, PMID: [DOI] [PubMed] [Google Scholar]
- Unger R. W. (2004). Beer in the middle ages and the renaissance. Philadelphia: University of Pennsylvania Press. [Google Scholar]
- Uruburu F. (2003). History and services of culture collections. Int. Microbiol. Off. J. Span. Soc. Microbiol. 6, 101–103. 10.1007/s10123-003-0115-2, PMID: [DOI] [PubMed] [Google Scholar]
- Varela C. (2016). The impact of non-Saccharomyces yeasts in the production of alcoholic beverages. Appl. Microbiol. Biotechnol. 100, 9861–9874. 10.1007/s00253-016-7941-6, PMID: [DOI] [PubMed] [Google Scholar]
- Vilela A. (2019). The importance of yeasts on fermentation quality and human health-promoting compounds. Fermentation 5:46. 10.3390/fermentation5020046 [DOI] [Google Scholar]
- Wang J., Liu L., Ball T., Yu L., Li Y., Xing F. (2016). Revealing a 5,000-y-old beer recipe in China. Proc. Natl. Acad. Sci. U. S. A. 113, 6444–6448. 10.1073/pnas.1601465113, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. -M., Liu W. -Q., Liti G., Wang S. -A., Bai F. -Y. (2012). Surprisingly diverged populations of Saccharomyces cerevisiae in natural environments remote from human activity. Mol. Ecol. 21, 5404–5417. 10.1111/j.1365-294X.2012.05732.x, PMID: [DOI] [PubMed] [Google Scholar]
- White C., Zainasheff J. (2010). Yeast: The practical guide to beer fermentation. Boulder: Brewers Publications. [Google Scholar]
- Whittington H. D., Dagher S. F., Bruno-Bárcena J. M. (2019). “Production and conservation of starter cultures: from ‘backslopping’ to controlled fermentations” in How fermented foods feed a healthy gut microbiota: A nutrition continuum. eds. Azcarate-Peril M. A., Arnold R. R., Bruno-Bárcena J. M. (Cham: Springer International Publishing; ), 125–138. [Google Scholar]
- Winge Ö. (1935). On haplophase and diplophase in some Saccharomycetes. Compt. Rend. Trav. Lab. Carlsberg, Ser. Physiol. 21, 77–112. [Google Scholar]
- Zaky A. S., Greetham D., Louis E. J., Tucker G. A., Du C. (2016). A new isolation and evaluation method for marine-derived yeast spp. with potential applications in industrial biotechnology. J. Microbiol. Biotechnol. 26, 1891–1907. 10.4014/jmb.1605.05074, PMID: [DOI] [PubMed] [Google Scholar]
- Zaky A. S., Tucker G. A., Daw Z. Y., Du C. (2014). Marine yeast isolation and industrial application. FEMS Yeast Res. 14, 813–825. 10.1111/1567-1364.12158, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdaniewicz M., Satora P., Pater A., Bogacz S. (2020). Low lactic acid-producing strain of Lachancea thermotolerans as a new starter for beer production. Biomol. Ther. 10:256. 10.3390/biom10020256, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeder M. A. (2006). Central questions in the domestication of plants and animals. Evol. Anthropol. Issues News Rev. 15, 105–117. 10.1002/evan.20101 [DOI] [Google Scholar]