Abstract
Advances in gene discovery have identified genetic variants in the solute carrier family 6 member 1 gene as a monogenic cause of neurodevelopmental disorders, including epilepsy with myoclonic atonic seizures, autism spectrum disorder and intellectual disability. The solute carrier family 6 member 1 gene encodes for the GABA transporter protein type 1, which is responsible for the reuptake of the neurotransmitter GABA, the primary inhibitory neurotransmitter in the central nervous system, from the extracellular space. GABAergic inhibition is essential to counterbalance neuronal excitation, and when significantly disrupted, it negatively impacts brain development leading to developmental differences and seizures. Aggregation of patient variants and observed clinical manifestations expand understanding of the genotypic and phenotypic spectrum of this disorder. Here, we assess genetic and phenotypic features in 116 individuals with solute carrier family 6 member 1 variants, the vast majority of which are likely to lead to GABA transporter protein type 1 loss-of-function. The knowledge acquired will guide therapeutic decisions and the development of targeted therapies that selectively enhance transporter function and may improve symptoms. We analysed the longitudinal and cell type-specific expression of solute carrier family 6 member 1 in humans and localization of patient and control missense variants in a novel GABA transporter protein type 1 protein structure model. In this update, we discuss the progress made in understanding and treating solute carrier family 6 member 1-related disorders thus far, through the concerted efforts of clinicians, scientists and family support groups.
Keywords: SLC6A1 haploinsufficiency, SLC6A1-related disorders, GABA transporter 1, neurodevelopmental disorders, seizures
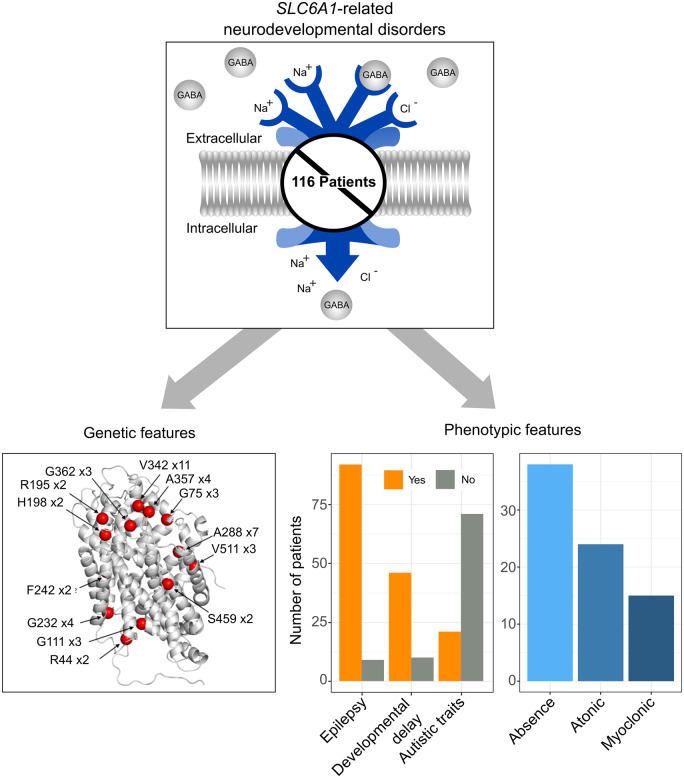
Genetic variants in the SLC6A1 gene have been identified as a monogenic cause of neurodevelopmental disorders, including epilepsy with myoclonic atonic seizures, autism spectrum disorder and intellectual disability. Here, we assess the genetic and phenotypic features observed in 116 patients with SLC6A1 variants.
Graphical Abstract
Graphical Abstract.
Introduction
Solute carrier family 6 member 1 (SLC6A1)-related disorders are emerging as a common cause of developmental and epileptic encephalopathies, since initial descriptions in 2015 (Carvill et al., 2015). SLC6A1 encodes the GABA transporter protein type 1 (GAT1), which is responsible for the reuptake of GABA into presynaptic neurons and glia (Bröer and Gether, 2012). Disruption of SLC6A1 is a prominent cause of neurodevelopmental disorders, including autism spectrum disorder, intellectual disability and seizures of varying types and severity. In the current three largest genomic screens of individuals with epilepsy (8565, 9170 and 9769 patients, respectively), SLC6A1 was listed among the top 10–20 genes with the highest number of pathogenic variants (Lindy et al., 2018; Epi25 Collaborative, 2019, p. 25; Truty et al., 2019). In the most extensive autism sequencing study to date (N = 11 986), SLC6A1 was among the top 10 genes, with the most significant variant enrichment in autism patients compared to 23 598 controls (Satterstrom et al., 2019). Recently, exome sequencing of individuals with schizophrenia found rare de novo missense variants in SLC6A1 to be associated with schizophrenia in three patients (Rees et al., 2020), extending the phenotype spectrum beyond epilepsy. Overall, the incidence of SLC6A1-related disorders is estimated to be 2.65 (90%CI: 2.38–2.86) per 100 000 births(López-Rivera et al., 2020).
Following the second SLC6A1 Symposium organized by the SLC6A1 Connect Foundation (https://slc6a1connect.org, 1 October 2020, date last accessed) that convened academic scientists, physicians and family advocacy organizations, we summarize the current state of research and future directions. We collected and curated the largest dataset of SLC6A1 variants to date with accompanying clinical phenotyping (N = 116). Our study represents a significant step forward to define the clinical and genotypic spectrum of SLC6A1-related disorders and ultimately will help to guide clinical management.
The SLC6A1 gene
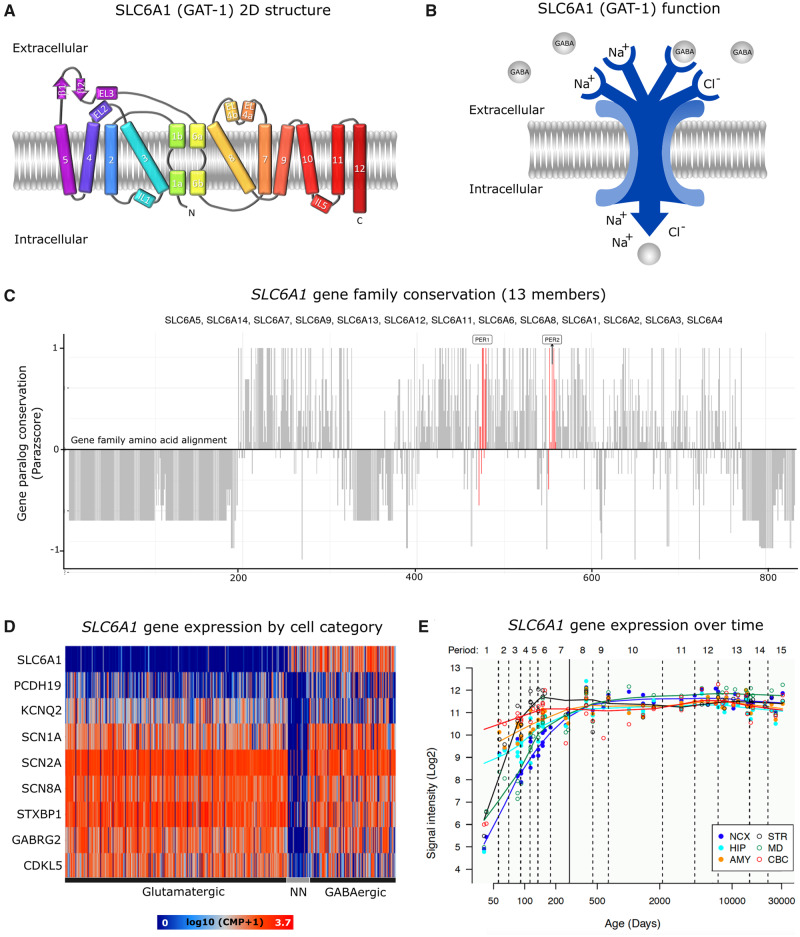
The SLC6A1 gene is located on chromosome 3 (Genome Research Consortia human assembly version 38 genomic coordinates: 3:10 992 733–11 039 248) and contains 15 exons. The encoded GAT1 protein has 12 transmembrane domains that form a single chain transporter (Fig. 1A). The primary function of GABA transporters is to lower the concentration of GABA in the extracellular space (Scimemi, 2014) (Fig. 1B). There are six major splice isoforms of human GAT1 that differ by alternative use of exons three to five. The transcript ENST00000287766 is the longest isoform of SLC6A1 and is considered canonical (Hunt et al., 2018); accordingly, most genetic variants are mapped into its sequence. The exact topology of GAT1 remains unclear due to the lack of a mammalian crystal structure. Still, homology models based on a 20–25% sequence identity to GAT1 (Yamashita et al., 2005) have allowed the identification of essential residues for substrate and sodium binding in transmembrane domains one, three, six and eight, which are necessary for the conformational transitions during the transport process (Fig. 1A and B). The SLC6A1 gene belongs to a gene family of 20 paralogues. The proteins encoded by 13 of these genes exhibit above 80% sequence identity, and six of them can transport GABA with different degrees of substrate specificity. These paralogues have been associated with a variety of neurodevelopmental disorders (Bröer and Gether, 2012). Upon linear protein sequence alignment of the 13 most paralogue-conserved gene family members, disease-associated variants reported in different family members cluster significantly together in two amino acid regions in comparison to variants from the general population. These regions are known as pathogenic variant enriched regions and any missense variant found within is 106 times more likely to be classified as pathogenic than benign (Fig. 1C) (Pérez-Palma et al., 2020). However, as in the case of many other genetic etiologies linked to neurodevelopmental disorders, disease-causing variants in SLC6A1 among affected individuals are broadly distributed along its sequence (Johannesen et al., 2018).
Figure 1.
SLC6A1 main features. (A) Schematic representation of the 2D structure of GAT1. (B) Diagram of GAT1 GABA transport function. (C) Paralogue conservation of the SLC6A1 gene family members. Higher values denote the degree of conservation. The pathogenic enriched regions are shown in red bars. (D) Cell type-specific expression of SLC6A1 and other frequently mutated epilepsy and neurodevelopmental disorder-associated genes (http://celltypes.brain-map.org/rnaseq/human). (E) Dynamic gene expression of SLC6A1 along with entire development and adulthood in the cerebellar cortex (CBC), mediodorsal nucleus of the thalamus (MD), striatum (STR), amygdala (AMY), hippocampus (HIP) and 11 areas of the neocortex (NCX) (http://hbatlas.org).
The function of GAT-1
The SLC6A1 gene is expressed in the mammalian central nervous system, predominantly in the adult frontal cortex (Gamazon et al., 2018). Unlike other GABA transporters, GAT1 is almost exclusively expressed in GABAergic axon terminals but can also be present in astrocytes, oligodendrocytes and microglia (Minelli et al., 1995; Melone et al., 2015; Fattorini et al., 2017, 2020) (Fig. 1D). During embryonic development, GAT1 is expressed within fully functional GABAergic neurons even before glutamatergic excitatory activity, but complete development of the GABAergic system continues through adolescence (Kilb, 2012; Wu and Sun, 2015) (Fig. 1E). GABA transporters couple translocation of GABA with the dissipation of the electrochemical gradient for sodium and chloride. By moving these ions across the membrane in a fixed ratio with GABA (1 GABA: 2 Na+: 1 Cl−), GAT1 generates a stoichiometric current (Lester et al., 1994) (Fig. 1B). At rest, the driving force for sodium and chloride forces these ions to move from the extracellular space towards the cell cytoplasm, promoting rapid binding and intracellular translocation of extracellular GABA within milliseconds of its release. This fast removal system prevents GABA from activating neighboring synapses (Isaacson et al., 1993). In addition to the transport of GABA, GAT1 is also an ion channel that generates two ionic currents: (i) an inward sodium current activated by GABA binding to GAT1 and (ii) a leak current mediated by alkali ions, which occurs in the absence of GABA (Risso et al., 1996; MacAulay et al., 2002). Lastly, in the absence of GABA, GAT1 generates a sodium-dependent capacitive current (Mager et al., 1993). Through these currents, GAT1 activation can create local changes in membrane potential or a shunt for membrane resistance. In GAT1 deficient mice, tonic inhibition is increased and the decay time of evoked phasic currents mediated by GABAA receptors is prolonged, whereas the frequency, amplitude and kinetics of spontaneous GABAA postsynaptic currents is not changed (Jensen et al., 2003; Chiu et al., 2005; Bragina et al., 2008; Cope et al., 2009).
Interestingly, other works on GAT1-deficient mice show that the frequency of miniature inhibitory postsynaptic currents is decreased, an effect that is associated with increased expression of enzymes that contributes to GABA synthesis in presynaptic terminals of inhibitory neurons (i.e. GAD65/67, periaqueductal gray matter) (Bragina et al., 2008; Conti et al., 2011). Other works in the striatum indicate that GAT1 shape GABAergic transmission through pre- and post-synaptic mechanisms. Together, these findings highlight multiple molecular and cellular functions of GAT1 that demonstrate the complex etiology leading to clinical symptoms of patients with SLC6A1-related disorders. Given that GABA homeostasis is critical for brain development, it is plausible that early intervention strategies will be essential for the well-being of the patients.
SLC6A1-related disorders
In 2015, a 3p microdeletion involving only SLC6A1 and SLC6A11 was described in a patient with Doose Syndrome, a developmental epileptic encephalopathy associated with intellectual disability and early-onset epilepsy with myoclonic atonic seizures (previously myoclonic atonic epilepsy). Furthermore, likely pathogenic variants in SLC6A1 were identified in 6/160 (4%) individuals with a previously undiagnosed early-onset epilepsy with myoclonic atonic seizures (Carvill et al., 2015) and additional studies identified individual patients with autism spectrum disorder and developmental epileptic encephalopathy carrying variants in SLC6A1 (Rauch et al., 2012; Sanders et al., 2012). In the first case series, 34 patients with variants in SLC6A1 were clinically described in 2018 (Johannesen et al., 2018). Notably, nearly all genetic variants reported arose de novo and not present in the general population (Lek et al., 2016).
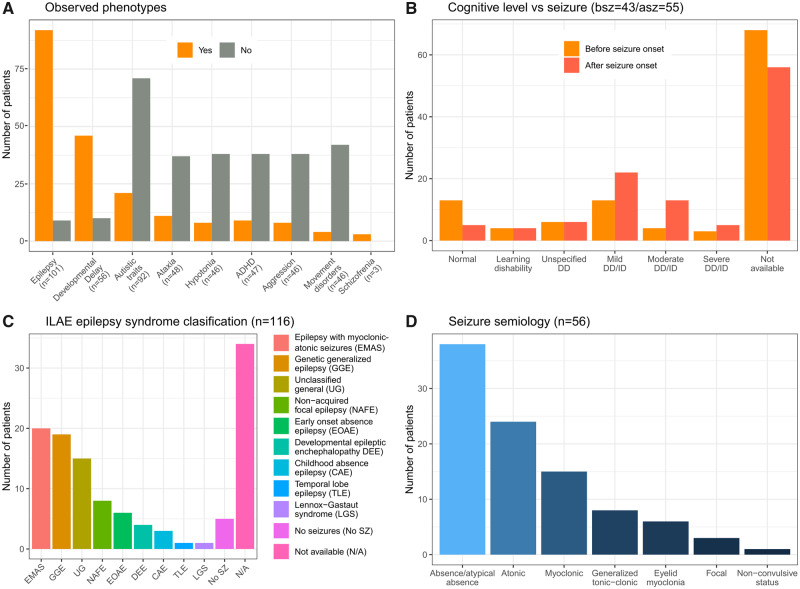
Here we present the largest collection of SLC6A1 patients, including genetic and clinical features (Supplementary Table 1). Our cohort includes 116 patients from three sources: previously reported individuals (58.6%), individual referrals to the SLC6A1 Connect Foundation (15.5%), and the Epi25 Collaborative for Large-Scale Whole Genome Sequencing in Epilepsy Collaborative Database (25.8%) (Epi25 Collaborative, 2019). Clinical and genetic data are presented in Supplementary Table 1. Data from the Epi25 Collaborative for Large-Scale Whole Genome Sequencing in Epilepsy Collaborative database were limited to genotype and International League Against Epilepsy categorization only. We describe 85 unique SLC6A1 variants. Most of patients variants observed arose de novo (40/49, 81.63%). Regarding variant type, we observed 88 missense variants, 15 protein truncating variants) that lead to a complete loss of function, seven variants in splice sites, three large deletions Copy Number Variants, two small insertions and deletions and one synonymous variant (Supplementary Table 1). In our cohort, epilepsy (92/101, 91.1%), developmental delay, and cognitive impairment (46/56, 82.1%) and autistic traits (20/92, 22.8%) were the most common clinical features (Fig. 2A). We found a similar number of males and females (55.1% female), and the mean age of seizure onset was 2.5 years (standard deviation 1.58 years). Developmental data concerning seizure-onset was available on 43 before seizure-onset and 55 after seizure-onset. Developmental delay was present in 26/43 (60.4%) before seizure-onset. After the onset of seizures, nearly all subjects demonstrate developmental delay (46/55, 83.6%), most in the mild to moderate range (35/55, 63.6%) (Fig. 2B). The most prevalent epilepsy syndrome was early-onset epilepsy with myoclonic atonic seizures (20/82, 24.3%) followed by genetic generalized epilepsy (19/82, 23.1%) and non-acquired focal epilepsy (8/82, 9.75%) (Fig. 2C). Detailed seizure semiology data was available on 56 of the 92 subjects with epilepsy; most commonly (atypical) absence seizures (38/53, 71.7%), atonic seizures (24/54, 44.44%) and myoclonic seizures (15/54, 27.77.1%) (Fig. 2D). Generalized epileptiform discharges (36/52, 69.2%) were the most commonly reported EEG abnormality among available data, especially at a frequency of 2–4 Hz (12/52, 23%). Generalized background slowing is reported in 17/52 (32.6%). Recently, an exome-wide trio sequencing study identified de novo missense variants in SLC6A1 to be associated with schizophrenia (Rees et al., 2020). No evidence of epilepsy, intellectual disability, or autism spectrum disorders was reported in the three patients described (Supplementary Table 1).
Figure 2.
Updated SLC6A1 cohort of patient’s summary statistics. We collected 116 patients from three different sources: the literature, Epi25 Collaborative for Large-Scale Whole Genome Sequencing in Epilepsy, and direct contact with the patient family foundation SLC6A1 connect. A total of 85 unique SCL6A1 variants were observed, and the clinical phenotype is expanded, and all clinical features are summarized in Supplementary Table 1. When data are available, we show (A) the phenotypes observed including additional clinical features beyond epilepsy, (B) the developmental status of patients before and after the onset of seizures, (C) the distribution of epilepsy classification (per updated International League Against Epilepsy guidelines) and (D) reported seizure semiologies.
Towards a model of SLC6A1 pathophysiology
Missense variants can cause a gain of function or loss of function; however, the expected molecular mechanism by which variants lead to SLC6A1-related disorders is loss of function or haploinsufficiency. This disease-model is supported by in vivo and in vitro experiments in both wild type and GAT1−/− mice, as well as studies on recombinant GAT1 proteins from individuals with SLC6A1 variants. However, the mechanisms by which the loss of function lead to clinical manifestations are not well understood. Recently, experimental evidence showed that seven SLC6A1 variants (five missense variants, one in-frame deletion and one nonsense variant) identified in epilepsy patients reduce GABA transport in vitro studies (Mattison et al., 2018). The residual transporter activity was found, ranging from 2% to 27% compared to the wild type.
It has been shown that SLC6A1 variants also cause impaired protein trafficking. Characterization of a missense variant in SLC6A1 (G234S) associated with Lennox–Gastaut syndrome leads to reduced protein expression in both cell surface and total protein levels in heterologous and rat cortical neurons (Cai et al., 2019). The surface protein level of the mutant is about 70% of the wild type, while the GABA uptake of the mutant GAT1 is ∼30% of the wild type. This suggests the mutant GAT1 had impaired GABA uptake in addition to impaired protein trafficking (Cai et al., 2019). Similarly, a recent report on SLC6A1 (P361T) associated with epilepsy and autism indicates that the mutant GAT-1 had endoplasmic reticulum retention and enhanced degradation (Wang et al., 2020).
There is no specific animal model of SLC6A1-related disorders. Heterozygous (Het) GAT1 knockout mice are phenotypically normal despite having diminished GABA reuptake capacity (Chiu et al., 2005; Cope et al., 2009). The homozygous GAT1 knockout animals exhibit a constant tremor, abnormal gait, reduced strength, absence seizures and mobility, as well as anxious behaviors (Chiu et al., 2005; Cope et al., 2009). This model partially recapitulates features of the human disease, including mobility and cognitive impairment (Johannesen et al., 2018). Ex vivo hippocampal and thalamic recordings show that GAT1 homozygous knockout mice have an increase in GABAAR tonic inhibition (Jensen et al., 2003; Cope et al., 2009), which is known to contribute to generalized spike-wave discharge typical of absence seizure(Crunelli V et al., 2020).
Prediction of GAT1 structure and evaluation of missense variants
Truncating variants are very likely to lead to loss-of-function due to lower levels of GAT1 and subsequent haploinsufficiency. However, the molecular consequence of missense variants can range from benign to damaging, mostly determined by the effect of the variants on protein function. We explored the pathogenic variants’ positions in the GAT1 sequence in the context of the functionally critical sites and domains (Fig. 3A). To further investigate the benign variants from the general population and pathogenic variants from patients in 3D space, we generated the structure of GAT1 using the RaptorX server [Fig. 3B, based on template structure: 4XPT (Wang et al., 2015)]. We then mapped the variants’ positions on the structure (Fig. 3C and D). Differential spatial segregation of variants is notable upon 3D mapping. The majority of the patient variants are located in the helical-transmembrane segments (32 out of 88, 36.4%) and inter-helical hinges (42 out of 88, 47.8%) (Fig. 3D, left) while genome aggregation consortia variants cluster in the cytoplasmic domain (38 out of 121, 31.4%, Fig. 3C). We observed ten patient variants also present in the general population (genome aggregation consortia) (Supplementary Table 1). The pathogenicity of these variants can be challenged according to the ACMG guidelines (Richards et al., 2015). We further highlighted the amino acid positions harboring recurrent patient variants and those also found in genome aggregation consortia (Fig. 3D, right). Out of 13 recurrent variant positions, six positions form a cluster near the extracellular (top) part of the structure. All three schizophrenia-related variants’ positions are located on the outer surface of the structure, mutated residues more than 70% exposed to solvent as measured by the residue’s accessible surface area.
Figure 3.
GAT1 3D structure model and spatial distribution of patient and general population missense variants in SLC6A1. There is no experimentally solved protein structure of the eukaryotic GAT1 protein (Fig. 2A). We predicted the GAT-1 structure using a homology model from a dopamine transporter (Protein Data Bank ID: 4xp4) (Waterhouse et al., 2018) and a multi-template based serotonin transporter (Protein Data Bank IDs: 4xpt, and 6awn) using RaptorX web server (Wang et al., 2016). The resulting novel model for GAT1 exhibited high quality and confidence (P-value = 3.67e−13). (A) Linear GAT1 protein sequence. The 599-residue long sodium- and chloride-dependent GAT1 consists of 12 helical domains (light purple), 1 extracellular (light green) and 2 cytoplasmic (light orange). (B) Predicted 3D structure, domains (colored as in A), and special binging sites (colored spheres). Numbers (in red) above the reference amino acids in sequence reflect the count of patients carrying a missense variant altering that amino acid. (C) Genome aggregation consortia variants (blue spheres) variant positions mapped on 3D and (D) patient variants (orange spheres). Recurrent variants are highlighted in red (upper-left structure) as well as patient variants also found in the general population (N = 10, bottom-left structure).
Existing treatment
Available clinical data for individuals with disease-causing variants in SLC6A1 is limited, and most patients require an interdisciplinary team, including neurologists, developmental pediatricians, genetic counsellors and speech and occupational therapists for comprehensive management (Kuo et al., 2016; Katkin et al., 2017). There is insufficient data available to guide pharmacotherapy in SLC6A1-related disorders. Thus treatment is guided by existing strategies for the specific clinical epilepsy syndromes, rather than underlying genetic etiology, using broad-spectrum anti-seizure medications, including valproic acid, lamotrigine or benzodiazepines. In a prior study, 20 of 31 patients became seizure-free with anti-seizure medication, and valproic acid was the most effective drug. Lamotrigine and ethosuximide also showed success (Johannesen et al., 2018). Importantly, there was not a correlation between seizure control and cognitive outcome.
Future treatment
There is a clear unmet medical need for improved treatment options for SLC6A1-related disorder. GAT1 is a potential candidate for therapeutic development based on the following observations: (i) it has a known biological function based on in vitro and in vivo studies (Jensen et al., 2003; Scimemi, 2014), (ii) there are known antiepileptic drugs such as valproic acid, tiagabine and vigabatrin that can modulate GABA concentrations (Schousboe et al., 2014) and (iii) the transporters’ relatively small size (599 aa) makes SLC6A1 a plausible candidate for viral-mediated gene therapy using adeno-associated virus vectors (Chamberlain et al., 2016). Alternatively, antisense oligonucleotides therapy can be used to specifically increase productive SLC6A1 mRNA and consequently restore levels of GAT1 protein. Antisense oligonucleotides strategies targeting non-productive splicing events have shown promising results in animal models of other haploinsufficiency neurodevelopmental disorders like Dravet syndrome (Han et al., 2020; Lim et al., 2020).
Restoration of the GAT1 transporter function may provide therapeutic benefit, but two challenges arise. First, is the need to understand the regulation of GAT1 expression and activity in a developmental context, and second, the reversibility of symptoms is unknown. Patient-derived induced pluripotent stem cell experiments (i.e. in vitro neuronal cultures and organoids) allow researchers to probe for the functional effects of SLC6A1 variants on neuronal excitability, developmental progression and network behavior. Importantly, this can be done in human neurons, under physiological expression levels of the transporter and under each patient’s unique genetic background. Transgenic rodent models provide the opportunity to assay neurons in the context of the brain to capture non-autonomous, circuit-level and behavioral effects. Understanding how alterations in GAT1 function affect both developing and mature neuronal integration, as well as network function, is critical to enable effective interventions.
Towards precision therapeutics
To develop treatments for patients with SLC6A1-related disorders it is critical to define the full phenotypic spectrum of the disease. Observational studies are needed to characterize the natural course of the disease and to identify appropriate end-points for use in future interventional trials. This cross-sectional review of available clinical data is a critical first step. Based on the expanded cohort described here, epilepsy and neurodevelopmental disabilities appear to be the core features of SLC6A1-related disorders. However, additional assessments of gait and mobility or movement disorders may also be of interest.
Further progress will require collaboration between clinicians, scientists and family organizations such as the SLC6A1-Connect Foundation (https://slc6a1connect.org). Significant limitations of a retrospective dataset include the lack of uniformity of presented data and risk that the same patient may be presented in multiple de-identified datasets. Collaborations between crucial stakeholders will lead to the development of disease-specific tools for collecting and analysing patient data, including greater data consistency, longitudinal phenotyping and standardized evaluation of medication use and response. More extensive population studies may also elucidate relationships between SLC6A1 variants and milder phenotypes. Furthermore, the use of EEG and imaging biomarkers can be explored. EEG may prove to be a valuable biomarker, given the high prevalence of abnormal electrical activity in clinical reports, however little is known about the EEG signature in individuals with SLC6A1-related disorder without seizures. Finally, standardized neuropsychological assessments to characterize developmental disabilities are needed, including patient-centred outcome measures that are jointly developed with substantial input from patient advocates and family organizations.
A key challenge in the pursuit of precision therapeutics is the identification of eligible individuals. While access to genetic testing is improving, it remains a significant barrier in some countries. Identification of early phenotypic features that should prompt appropriate evaluation will support early diagnosis. Alongside deep phenotyping, functional analysis of variants will be necessary to assess genotype-phenotype correlations. Finally, the implementation of high-throughput analysis of the functional impact of variants on channel function would accelerate drug development considerably.
Conclusion
SLC6A1-related disorders are neurodevelopmental disorders caused by aberrant GABA neurotransmission secondary to impaired functioning of GAT1. Based on a review of 116 individuals with SLC6A1-related disorder, developmental delay, epilepsy, autism and motor dysfunction, including stereotypies and ataxia, are the most common clinical features. Data from the literature and our analysis on GAT1 structure support loss-of-function as the primary disease-associated molecular pathology. A comprehensive translational research programme needs to be developed (i) to better understand the underlying pathophysiology, (ii) to develop targeted therapies for SLC6A1-related disorder and (iii) to define the full clinical spectrum of the disease.
Supplementary material
Supplementary material is available at Brain Communications online.
Supplementary Material
Acknowledgements
The authors acknowledge the collaborative efforts to the families affected by SLC6A1-related disorders who are supporting these efforts, especially the SLC6A1 Connect Foundation. In addition, we acknowledge the efforts of clinicians and genetic counsellors supporting patients and their families.
Competing interests
The authors report no competing interests.
Glossary
- GAT1 =
GABA transporter protein type 1
- SLC6A1 =
solute carrier family 6 member 1
References
- Bragina L, Marchionni I, Omrani A, Cozzi A, Pellegrini‐Giampietro DE, Cherubini E, et al. GAT-1 regulates both tonic and phasic GABAA receptor-mediated inhibition in the cerebral cortex. J Neurochem 2008; 105: 1781–93. [DOI] [PubMed] [Google Scholar]
- Bröer S, Gether U. The solute carrier 6 family of transporters. Br J Pharmacol 2012; 167: 256–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai K, Wang J, Eissman J, Wang J, Nwosu G, Shen W, et al. A missense mutation in SLC6A1 associated with Lennox–Gastaut syndrome impairs GABA transporter 1 protein trafficking and function. Exp Neurol 2019; 320: 112973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvill GL, Mcmahon JM, Schneider A, Zemel M, Myers CT, Saykally J, et al. Mutations in the GABA transporter SLC6A1 cause epilepsy with myoclonic-atonic seizures. Am J Hum Genet 2015; 96: 808–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain K, Riyad JM, Weber T. Expressing transgenes that exceed the packaging capacity of adeno-associated virus capsids. Hum Gene Ther Methods 2016; 27: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C-S, Brickley S, Jensen K, Southwell A, Mckinney S, Cull-Candy S, et al. GABA transporter deficiency causes tremor, ataxia, nervousness, and increased GABA-induced tonic conductance in cerebellum. J Neurosci 2005; 25: 3234–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti F, Melone M, Fattorini G, Bragina L, Ciappelloni S, A role for GAT-1 in presynaptic GABA Homeostasis? [Internet]. Front Cell Neurosci 2011; 5: 2. https://www.frontiersin.org/articles/10.3389/fncel.2011.00002/full [DOI] [PMC free article] [PubMed]
- Cope DW, Di Giovanni G, Fyson SJ, Orbán G, Errington AC, Lőrincz ML, et al. Enhanced tonic GABAA inhibition in typical absence epilepsy. Nat Med 2009; 15: 1392–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crunelli V, Lőrincz ML, Mccafferty C, Lambert RC, Leresche N, Di Giovanni G, et al. Clinical and experimental insight in pathophysiology, comorbidity and therapy of absence seizures. Brain 2020; 143: 2341–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epi25 Collaborative. Ultra-rare genetic variation in the epilepsies: a whole-exome sequencing study Of 17,606 individuals. Am J Hum Genet 2019; 105: 267–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattorini G, Catalano M, Melone M, Serpe C, Bassi S, Limatola C, et al. Microglial expression of GAT-1 in the cerebral cortex. Glia 2020; 68: 646–55. [DOI] [PubMed] [Google Scholar]
- Fattorini G, Melone M, Sánchez‐Gómez MV, Arellano RO, Bassi S, Matute C, et al. GAT-1 mediated GABA uptake in rat oligodendrocytes. Glia 2017; 65: 514–22. [DOI] [PubMed] [Google Scholar]
- Gamazon ER, Segrè AV, van de Bunt M, Wen X, Xi HS, Hormozdiari F, et al. Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat Genet 2018; 50: 956–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Chen C, Christiansen A, Ji S, Lin Q, Anumonwo C, et al. Antisense oligonucleotides increase Scn1a expression and reduce seizures and SUDEP incidence in a mouse model of Dravet syndrome. Sci Transl Med 2020; 12: 558. [DOI] [PubMed] [Google Scholar]
- Hunt SE, Mclaren W, Gil L, Thormann A, Schuilenburg H, Sheppard D, et al. Ensembl variation resources [Internet]. Database (Oxford) 2018; 2018: 1–12. https://academic.oup.com/database/article/doi/10.1093/database/bay119/5255129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson JS, Solis JM, Nicoll RA. Local and diffuse synaptic actions of GABA in the hippocampus. Neuron 1993; 10: 165–75. [DOI] [PubMed] [Google Scholar]
- Jensen K, Chiu C-S, Sokolova I, Lester HA, Mody I. GABA transporter-1 (GAT1)-deficient mice: differential tonic activation of GABAA Versus GABAB receptors in the hippocampus. J Neurophysiol 2003; 90: 2690–701. [DOI] [PubMed] [Google Scholar]
- Johannesen KM, Gardella E, Linnankivi T, Courage C, De Saint Martin A, Lehesjoki A-E, et al. Defining the phenotypic spectrum of SLC6A1 mutations. Epilepsia 2018; 59: 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katkin JP, Kressly SJ, Edwards AR, Perrin JM, Kraft CA, Richerson JE, et al. Guiding principles for team-based pediatric care. Pediatrics 2017; 140: E20171489. [DOI] [PubMed] [Google Scholar]
- Kilb W. Development of the GABAergic system from birth to adolescence. Neuroscientist 2012; 18: 613–30. [DOI] [PubMed] [Google Scholar]
- Kuo DZ, Houtrow AJ, Disabilities C, On CW, Recognition and management of medical complexity [Internet]. Pediatrics 2016; 138: 6. https://pediatrics.aappublications.org/content/138/6/e20163021 [DOI] [PubMed]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester HA, Mager S, Quick MW, Corey JL. Permeation properties of neurotransmitter transporters. Annu Rev Pharmacol Toxicol 1994; 34: 219–49. [DOI] [PubMed] [Google Scholar]
- Lim KH, Han Z, Jeon HY, Kach J, Jing E, Weyn-Vanhentenryck S, et al. Antisense oligonucleotide modulation of non-productive alternative splicing upregulates gene expression. Nat Commun 2020; 11: 3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindy AS, Stosser MB, Butler E, Downtain-Pickersgill C, Shanmugham A, Retterer K, et al. Diagnostic outcomes for genetic testing of 70 genes in 8565 patients with epilepsy and neurodevelopmental disorders. Epilepsia 2018; 59: 1062–71. [DOI] [PubMed] [Google Scholar]
- López-Rivera JA, Pérez-Palma E, Symonds J, Lindy AS, Mcknight DA, Leu C, et al. A catalogue of new incidence estimates of monogenic neurodevelopmental disorders caused by de novo variants [Internet]. Brain 2020; 143: 4. https://academic.oup.com/brain/advance-article/doi/10.1093/brain/awaa051/5803191 [DOI] [PMC free article] [PubMed]
- MacAulay N, Zeuthen T, Gether U. Conformational basis for the Li(+)-induced leak current in the rat gamma-aminobutyric acid (GABA) transporter-1. J Physiol (Lond) 2002; 544: 447–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager S, Naeve J, Quick M, Labarca C, Davidson N, Lester HA. Steady states, charge movements, and rates for a cloned GABA transporter expressed in Xenopus oocytes. Neuron 1993; 10: 177–88. [DOI] [PubMed] [Google Scholar]
- Mattison KA, Butler KM, Inglis GAS, Dayan O, Boussidan H, Bhambhani V, et al. SLC6A1 variants identified in epilepsy patients reduce γ-aminobutyric acid transport. Epilepsia 2018; 59: e135. [DOI] [PubMed] [Google Scholar]
- Melone M, Ciappelloni S, Conti F. A quantitative analysis of cellular and synaptic localization of GAT-1 and GAT-3 in rat neocortex. Brain Struct Funct 2015; 220: 885–97. [DOI] [PubMed] [Google Scholar]
- Minelli A, Brecha NC, Karschin C, DeBiasi S, Conti F. GAT-1, a high-affinity GABA plasma membrane transporter, is localized to neurons and astroglia in the cerebral cortex. J Neurosci 1995; 15: 7734–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Palma E, May P, Iqbal S, Niestroj L-M, Du J, Heyne HO, et al. Identification of pathogenic variant enriched regions across genes and gene families. Genome Res 2020; 30: 62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet 2012; 380: 1674–82. [DOI] [PubMed] [Google Scholar]
- Rees E, Han J, Morgan J, Carrera N, Escott-Price V, Pocklington AJ, et al. De novo mutations identified by exome sequencing implicate rare missense variants in SLC6A1 in schizophrenia. Nat Neurosci 2020; 23: 179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17: 405–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso S, DeFelice LJ, Blakely RD. Sodium-dependent GABA-induced currents in GAT1-transfected HeLa cells. J Physiol 1996; 490: 691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 2012; 485: 237–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterstrom FK, Walters RK, Singh T, Wigdor EM, Lescai F, Demontis D, et al. Autism spectrum disorder and attention deficit hyperactivity disorder have a similar burden of rare protein-truncating variants. Nat Neurosci 2019; 22: 1961–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schousboe A, Madsen KK, Barker-Haliski ML, White HS. The GABA synapse as a target for antiepileptic drugs: a historical overview focused on GABA transporters. Neurochem Res 2014; 39: 1980–7. [DOI] [PubMed] [Google Scholar]
- Scimemi A, Structure, function, and plasticity of GABA transporters [Internet]. Front Cell Neurosci 2014; 8: 161. https://www.frontiersin.org/articles/10.3389/fncel.2014.00161/full [DOI] [PMC free article] [PubMed]
- Truty R, Patil N, Sankar R, Sullivan J, Millichap J, Carvill G, et al. Possible precision medicine implications from genetic testing using combined detection of sequence and intragenic copy number variants in a large cohort with childhood epilepsy. Epilepsia Open 2019; 4: 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Poliquin S, Mermer F, Eissman J, Delpire E, Wang J, et al. Endoplasmic reticulum retention and degradation of a mutation in SLC6A1 associated with epilepsy and autism. Mol Brain 2020; 13: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Penmatsa A, Gouaux E. Neurotransmitter and psychostimulant recognition by the dopamine transporter. Nature 2015; 521: 322–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Li W, Liu S, Xu J. RaptorX-Property: a web server for protein structure property prediction. Nucleic Acids Res 2016; 44: W430–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 2018; 46: W296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Sun D. GABA receptors in brain development, function, and injury. Metab Brain Dis 2015; 30: 367–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl-dependent neurotransmitter transporters. Nature 2005; 437: 215–23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.