Abstract
Background
Lumefantrine and mefloquine are used worldwide in artemisinin-based combination therapy (ACT) of malaria. Better understanding of drug susceptibility and resistance is needed and can be obtained from studies of genetic crosses.
Methods
Drug response phenotypes of a cross between Plasmodium falciparum lines 803 (Cambodia) and GB4 (Ghana) were obtained as half-maximal effective concentrations (EC50s) and days to recovery (DTR) after 24 h exposure to 500 nM lumefantrine. EC50s of mefloquine, halofantrine, chloroquine, and dihydroartemisinin were also determined. Quantitative trait loci (QTL) analysis and statistical tests with candidate genes were used to identify polymorphisms associated with response phenotypes.
Results
Lumefantrine EC50s averaged 5.8-fold higher for the 803 than GB4 parent, and DTR results were 3–5 and 16–18 days, respectively. In 803 × GB4 progeny, outcomes of these two lumefantrine assays showed strong inverse correlation; these phenotypes also correlated strongly with mefloquine and halofantrine EC50s. By QTL analysis, lumefantrine and mefloquine phenotypes mapped to a chromosome 5 region containing codon polymorphisms N86Y and Y184F in the P. falciparum multidrug resistance 1 protein (PfMDR1). Statistical tests of candidate genes identified correlations between inheritance of PfK13 Kelch protein polymorphism C580Y (and possibly K189T) and lumefantrine and mefloquine susceptibilities. Correlations were detected between lumefantrine and chloroquine EC50s and polymorphisms N326S and I356T in the CVIET-type P. falciparum chloroquine resistance transporter (PfCRT) common to 803 and GB4.
Conclusions
Correlations in this study suggest common mechanisms of action in lumefantrine, mefloquine, and halofantrine responses. PfK13 as well as PfMDR1 and PfCRT polymorphisms may affect access and/or action of these arylaminoalcohol drugs at locations of hemoglobin digestion and heme metabolism. In endemic regions, pressure from use of lumefantrine or mefloquine in ACTs may drive selection of PfK13 polymorphisms along with versions of PfMDR1 and PfCRT associated with lower susceptibility to these drugs.
Keywords: Malaria, Artemisinin-based combination chemotherapy, Drug resistance, Halofantrine, Quantitative trait loci analysis
Graphical abstract
Highlights
-
•
Two different lumefantrine response assays show inverse correlation in a P. falciparum cross.
-
•
Lumefantrine, mefloquine, and halofantrine phenotypes correlate and may have pathways of action in common.
-
•
Lumefantrine and mefloquine susceptibilities are linked to PfMDR1, PfCRT, and PfK13 polymorphisms.
-
•
Lumefantrine and mefloquine in ACTs may drive selection of PfMDR1, PfCRT, and PfK13 polymorphisms including PfK13 580Y.
1. Introduction
The availability of effective, safe, and affordable antimalarial drugs is increasingly threatened by the emergence and spread of drug resistant parasites (Phillips et al., 2017). Resistance to partner drugs in artemisinin-based combination therapies (ACTs) is an important case in point, as ACTs are presently recommended worldwide as first-line medicines for the treatment of malaria (World Health Organization, 2015). Artemether-lumefantrine (AL, Coartem®) was the first ACT produced under international good manufacturing practices. Since its approval by the United States Food and Drug Administration in 2009, AL has also become available as Coartem® Dispersible for pediatric use and is now the most widely used ACT for the treatment of uncomplicated Plasmodium falciparum malaria, particularly in Africa (Premji, 2009; Nzila et al., 2012). Efficacies of AL treatment in large studies have generally been better than 95% (Makanga and Krudsood, 2009; Premji, 2009), although recrudescences are reported in nonimmune adults and travelers despite appropriate lumefantrine (LUM) concentrations in the blood (Farnert et al., 2012; Sonden et al., 2017). Because LUM is a lipophilic arylaminoalcohol that has limited absorption on an empty stomach, AL is recommended to be taken with a fatty food meal to achieve adequate bioavailability (White et al., 1999; Denis et al., 2006; Premji, 2009). Additionally, the importance of adherence to the treatment regimen has been quantitatively demonstrated in outcome modeling studies (Challenger et al., 2017).
While much about the action of LUM as an antimalarial has yet to be elucidated, evidence suggests the drug can inhibit hemozoin formation in the digestive vacuole (DV) of the erythrocytic-stage malaria parasite, where the hemozoin serves as a crystalline repository for the sequestration and detoxification of ferri-protoporphyrin heme molecules released by hemoglobin digestion (Pradines et al., 1999; Combrinck et al., 2013). LUM response variations result from P. falciparum coding polymorphisms and copy number differences in the P. falciparum multidrug resistance 1 gene (pfmdr1) including: (1) codon N86Y, Y184F, and D1246Y substitutions relative to the 3D7 ‘wild-type’ sequence (Duraisingh et al., 2000; Sisowath et al., 2005; Venkatesan et al., 2014; Wurtz et al., 2014; Baraka et al., 2015); and (2) amplifications of regions of P. falciparum chromosome 5 containing the pfmdr1 gene (Price et al., 2006; Sidhu et al., 2006; Nair et al., 2007; Gadalla et al., 2011; Venkatesan et al., 2014). Selection of the pfmdr1 N86/184F/D1246 haplotype has been reported from studies of AL-treatments and recrudescences in patients in Asia and Africa (Sisowath et al., 2007; Happi et al., 2009; Baliraine and Rosenthal, 2011; Malmberg et al., 2013; Conrad et al., 2014). Likewise, selection of the wild-type K76 codon in the P. falciparum chloroquine resistance transporter gene, pfcrt, has been associated with pressure from AL treatment in a number of studies (Sisowath et al., 2009; Eyase et al., 2013; Conrad et al., 2014; Venkatesan et al., 2014; Baraka et al., 2015). Dramatic increases in the prevalence of wild-type PfCRT K76 and PfMDR1 N86 were associated with discontinuation of chloroquine (CQ) and deployment of AL in western Kenya, although AL continued to be efficacious with these changes (Achieng et al., 2015). EC50s of CQ and amodiaquine (AQ) have been found to decrease reciprocally with increases in the EC50s of LUM and mefloquine (MEF), and these reciprocal changes have been associated with opposite shifts in the prevalence of mutant vs. wild-type PfCRT and PfMDR1 polymorphisms (Humphreys et al., 2007; Eyase et al., 2013; Venkatesan et al., 2014; Sondo et al., 2016).
Experimental crosses of Plasmodium parasites are a powerful means to identify genes of drug resistance and other important malaria phenotypes. With the human malaria parasites, crosses have enabled the mapping and characterization of genes that determine in vitro responses of P. falciparum to antifolate antimalarials (Peterson et al., 1988, 1990; Wang et al., 1997); the 4-aminoquinolines CQ and AQ along with their active metabolites (Wellems et al., 1991; Fidock et al., 2000; Sá et al., 2009); the arylaminoalcohol drugs LUM, MEF, and halofantrine (HLF), as well as the endoperoxide drugs artemisinin, arteflene, artemether, dihydroartemisinin, and artesunate (Duraisingh et al., 2000). Genetic crosses have also supported the evaluations of in vivo phenotypes and genetic determinants of P. falciparum artemisinin response and of Plasmodium vivax CQ resistance in monkey models (Sá et al., 2018, 2019). Other biological investigations with P. falciparum crosses have led to the discoveries of a major parasite ligand for species-specific erythrocyte invasion (Hayton et al., 2008), family members of the channel that determines nutrient uptake by parasitized erythrocytes (Nguitragool et al., 2011), and a key molecule mediating parasite evasion of the mosquito immune system (Molina-Cruz et al., 2013).
The present study was undertaken to assess LUM parasite susceptibilities using two different drug response assays, compare these susceptibilities with those of other arylaminoalcohol drugs such as MEF and HLF, and search for genes that affect phenotypes of differential susceptibility in a cross between clonal lines of Cambodian (803) and Ghanaian (GB4) P. falciparum parasites (Sá et al., 2018). The genetic determinants of susceptibility in these two geographical regions are of particular interest, as LUM and MEF phenotypes have been reported to differ between Africa and Southeast Asia (Oduola et al., 1993; Dama et al., 2017) and parasite susceptibilities to LUM and MEF as well as HLF are known to correlate with one another (Basco et al., 1998; Pradines et al., 2006; Basco and Ringwald, 2007; Eyase et al., 2013). Here we describe findings consistent with previous reports of arylaminoalcohol drug responses mediated by PfMDR1 and PfCRT polymorphisms, provide evidence that mutations in the P. falciparum Kelch 13 protein (PfK13) associate with reduced parasite susceptibility to LUM and MEF, and present a hypothesis that LUM and MEF may drive selection of PfK13 580Y and perhaps other Kelch protein mutations in P. falciparum populations pressured by ACTs containing these drugs.
2. Materials and methods
2.1. P. falciparum parasites of the 803 × GB4 cross and in vitro cultivation
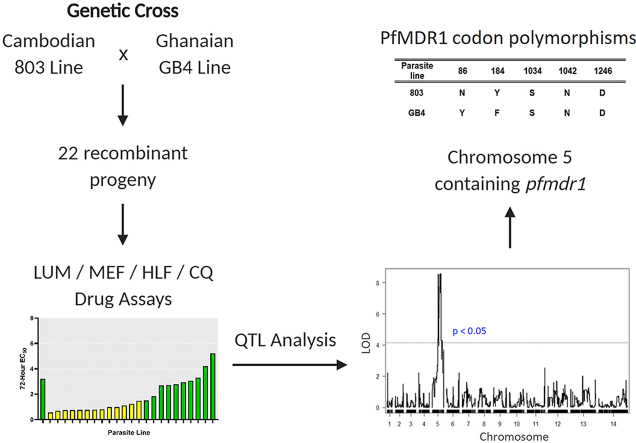
Production of the 803 × GB4 cross and recovery of the independent recombinant progeny for genetic linkage studies was as previously described (Sá et al., 2018). Briefly, gametocytes were induced from blood stage cultures in vitro, mixed, and fed to Anopheles mosquitoes. After infection of the mosquitoes and confirmation of recombinant oocysts, sporozoites were allowed to develop in the mosquito salivary glands, purified, and cryopreserved. Sporozoite populations carrying markers of both parents (sporozoites, like blood stages, are haploid) were later thawed and inoculated into a Pan troglodytes chimpanzee to produce broods of liver stage parasites and subsequent populations of recombinant blood-stage progeny. Blood samples were collected 18–40 days after the inoculation and the parasites were cultivated briefly in vitro before cloning by limiting dilution. After sorting and characterization of more than 400 parasite clones by microsatellite fingerprinting, a subset of clones representing broods of independent recombinants was identified and typed by microarray genome analysis with 3629 single nucleotide polymorphisms.
Parasite lines from the 803 × GB4 cross were maintained in 10 mL cultures with human O+ red blood cells (RBCs) (Virginia Blood Bank, VA, USA) at ~5% hematocrit supplemented with complete media (CM) composed of RPMI-1640 (Sigma-Aldrich; St. Louis, MI), 2.0% AlbuMAX™ II wt/vol (Life Technologies™; Grand Island, NY), 0.02 mg/ml gentamycin solution, and 0.21% sodium bicarbonate (KD Medical; Columbia, MD) following established culture methods (Cranmer et al., 1997). Cultures were incubated at 37 °C in a 90% nitrogen/5% oxygen/5% carbon dioxide environment. Parasitemia and stage distributions were monitored by thin blood smears methanol-fixed and stained with 20% Giemsa for 15–30 min as well as by flow cytometry (Amaratunga et al., 2014). Synchronization of parasites was performed as described (Lambros and Vanderberg, 1979) by suspension of the pellet from 10 mL of cell culture (0.5 mL packed cells) for 5–10 min in 5 mL of 5% sorbitol wt/vol, followed by a 30 mL wash with CM.
2.2. 72-Hour half-maximal effective concentration (EC50) assays
CQ, MEF, HLF, and LUM were purchased from Sigma-Aldrich (St. Louis, MO). Stock solutions were stored at 5 mM or 10 mM in dimethylsulfoxide at −20 °C. The 72 h dose response assays followed previous methods using SYBR Green I and 96-well plates (Smilkstein et al., 2004; Lane et al., 2018). Geometric mean half-maximal effective concentrations (EC50s) with confidence intervals were calculated from the response curves using non-linear regression with variable slope on Prism v.8 (GraphPad Software Inc.; La Jolla, CA). Assays were repeated for each line a minimum of three times to obtain geometric mean and confidence interval.
2.3. Growth recovery times after a 24 h 500 nM LUM exposure
Parasites were cultivated to 2% parasitemia at 5% hematocrit with at least 70% ring stage parasites, then exposed for 24 h to 500 nM LUM (Sigma-Aldrich, St. Louis, MO). These parameters were chosen in view of the blood concentrations and frequency of parasite clearances reported 24 h after treatment (Valecha et al., 2012), thus approximating physiological conditions of exposure better in vitro than EC50 assays with LUM concentrations in the low nM range. Following this exposure, the cells were washed three times with 50 mL of CM and returned to culture with daily medium changes, maintaining 5% hematocrit until they again reached 2% parasitemia. Days to recovery (DTR) were counted as the number of days for parasites in culture to reach the parasitemia recorded at the beginning of each experiment. Assays for each parasite line were repeated a minimum of three times, with the exceptions of 36H9 and 40E7 (n = 2) to obtain mean and confidence interval.
2.4. Genome-wide quantitative trait-loci analysis and statistical evaluations
Quantitative trait-loci (QTL) analysis was performed using the R/qtl package as described (Broman et al., 2003). For the 803 × GB4 parents and for each of the studied progeny, the mean DTR and geometric mean EC50 measures were provided as phenotypes, and the corresponding genotypes of each parasite were entered from the previously established DNA microarray datasets of 3629 single-nucleotide polymorphisms (SNPs) across the genome (Sá et al., 2018). QTL searches for primary loci were performed using marker regression with 10 centimorgan (cM) spacing with these SNPs used for locus inheritance. P-values were determined from 1000 permutations. Significant (p < 0.05) logarithm of odds (LOD) scores were taken to indicate genetic linkage. LOD intervals were determined from the span of the peak exceeding the calculated significance cutoff.
Spearman's rank correlations of EC50 and DTR phenotypes and Mann-Whitney U tests of candidate gene polymorphisms against these phenotypes were performed using Prism v.8.
2.5. Microsatellite genotyping of parasite lines
Genetic verification of parasite lines was performed by PCR amplification of 12 variable markers as described (Figan et al., 2018). Capillary electrophoresis was performed on an ABI 3730XL machine and analyzed by GeneMapper v4.3 software (Applied Biosystems; La Jolla, CA). Distinguishable sizes of each marker identify the 803 and GB4 parents. Each unique progeny clone (segregant) from 803 × GB4 cross is identified by its particular combination of these 12 markers.
3. Results and discussion
3.1. Drug response phenotypes of parents and progeny of the 803 × GB4 cross
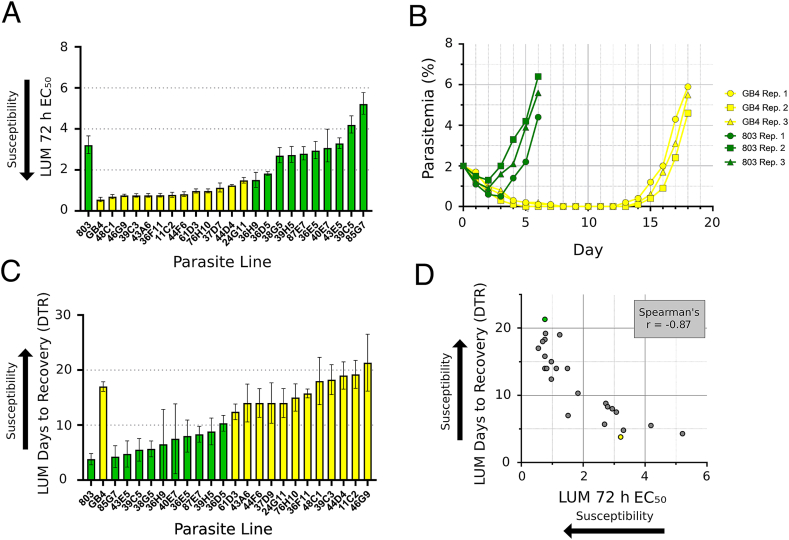
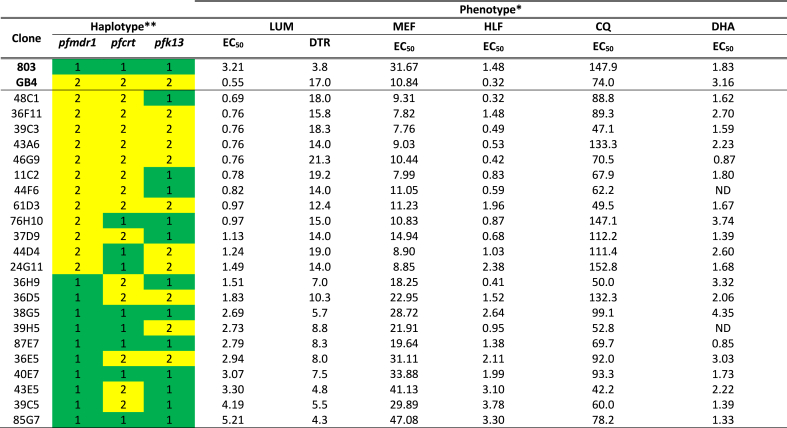
LUM drug responses of the parental clones 803 and GB4 plus 22 of the independent recombinant progeny were assessed by standard EC50 assays. The geometric mean LUM EC50 of 803 was 5.8-fold greater than GB4 (3.21 nM, 95% Confidence Interval 2.80–3.66 nM vs. 0.55 nM, 95% CI 0.46–0.67 nM, respectively; Fig. 1A, Table S1). MEF, HLF, and CQ EC50s were obtained along with those of LUM; dihydroartemisinin (DHA) responses were determined previously (Sá et al., 2018). Comparisons of the MEF and HLF responses showed that the Cambodian 803 line, as for LUM, was less susceptible than Ghanaian GB4 to these drugs: the geometric mean EC50s of 803 relative to GB4 were 2.9-fold greater with MEF and 4.6-fold greater with HLF, whereas these were 2.0-fold greater with CQ and 1.7-fold reduced with DHA (Table 1).
Fig. 1.
Lumefantrine response phenotypes of parents and progeny from the 803 × GB4 cross. Parasites indicated in green carry the 803 pfmdr1 locus encoding N86 and Y184, whereas parasites indicated in yellow carry the GB4 pfmdr1 locus encoding 86Y and 184F. A Results from 72 h half-maximal effective concentration (EC50) of growth inhibition assays for 22 progeny and parental lines using SYBR green, a sensitive fluorescence indicator of double-stranded DNA levels (Smilkstein et al., 2004). B Parasitemia of parental lines 803 and GB4 after a 24 h 500 nM LUM exposure with three biological replicates. C Days to 2% parasitemia recovery (DTR) after a 24 h 500 nM LUM exposure for 22 progeny and parental lines. D Scatter plot between the EC50 and DTR values of the individual parents and progeny. 803 is indicated in green and GB4 is indicated in yellow. Data from individual phenotype determinations are provided in Table S1 and Table S2. EC50 means and confidence intervals were calculated from three biological replicates for all parasites except 36H9 and 40E7 (n = 2). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
** Parental 803 and GB4 haplotypes are indicated by ‘1’ (green) and ‘2’ (yellow), respectively.
* UM DTR values are taken from Tables S1 and S2 of this report. DHA EC50s are from supplementary Table S1 of Sá et al. (2018), except for subsequently determined mean ± standard errors in the mean (SEM) values of 1.39±0.04 (37D9; n=37), 3.32±0.41 (36H9; n=3), 2.22±0.30 (43E5; n=3), 2.60±0.47 (44D4; n=3) published here. See the cited Tables for confidence intervals and SEMs. EC50 values are in nanomolar; DTR values in days. ND = not determined.
Table 1.
Summary of antimalarial drug phenotypes and pfmdr1, pfcrt, and pfk13 haplotypes in the 803 × GB4 cross.
Geometric mean LUM EC50s of the individual 803 × GB4 parents and progeny were 0.55–5.21 nM (Table 1), a range that overlapped with those of 0.19–12.5 nM and 1.4–3.4 nM reported by Nsobya et al. (2010) and Dama et al. (2017) but was lower than the ranges from other studies by Basco et al. (1998) (95% CI 10.4–13.6 nM), Pradines et al. (2006) (5.7–8.2 nM), Mwai et al. (2009) (Interquartile Range 29–96 nM), Eyase et al. (2013) (IQR of 9.5–52.4 nM in 2011), and Wurtz et al. (2014) (95% CI 18.8–26.9 nM). Assay details such as timings of drug exposure, compositions of culture media, incubation conditions employed, and the in vitro status and synchrony of parasite populations are among potential reasons for the differences in these ranges. We note that the higher range EC50 values remain well below bloodstream levels achieved after a standard dose of LUM, although exposure times of 72 h approach the 3–6 day half-life of the drug in vivo (White et al., 1999; Valecha et al., 2012). In the absence of accepted standards for LUM-resistant P. falciparum parasites, much remains to be resolved about the LUM levels indicative of drug sensitivity, tolerance, and resistance in these susceptibility assays. Reference parasites with benchmark phenotypes of LUM resistance have yet to be established either as culture-adapted lines from clinical treatment failures or as selected lines after LUM pressure in vitro.
To study the LUM responses of parasites after exposures closer to those of drug treatment in vitro (Valecha et al., 2012), we developed an assay that employs 500 nM LUM exposure (~100x EC50) for 24 h. Outcomes of this assay were assessed by measuring the days to recovery (DTR) to starting parasitemia. Three to five days were required for the drug-exposed 803 parasites to recover in these assays, whereas the drug-exposed GB4 parasites required 16–18 days (Fig. 1B, Table S2). Interestingly, no dormant forms were observed after exposure of the parasites to 500 nM LUM for 24 h, consistent with the report of Chavchich et al. (2016); further, in separate assessments, we were unable to find evidence for any reduced LUM susceptibility to the drug by the parasites that repopulated the culture after surviving 500 nM LUM exposure.
Twenty-two recombinant progeny from the 803 × GB4 cross were assessed by the LUM EC50 and DTR assays. In both of these assays, the progeny phenotypes ranged between and above those of the 803 and GB4 parental lines (Fig. 1C; Table 1), and an inverse relationship was evident between the individual parasite responses in the two assays (Fig. 1D). The two assays yielded a strong negative coefficient of correlation by Spearman's rank calculation (r = −0.87, 95% CI: −0.95 to −0.72, p < 0.001; Table 2).
Table 2.
Spearman and Mann-Whitney U statistics on drug phenotypes and candidate gene polymorphisms in the 803 × GB4 cross.
| Comparison | p-value (two-tailed) | Significant? | ||
|---|---|---|---|---|
| Spearman correlation | r coefficient | 95% confidence interval | ||
| PfMDR1 vs. PfCRT haplotype | 0.32 | −0.10 to 0.65 | 0.12 | no |
| PfMDR1 vs. PfK13 haplotype | 0.34 | −0.08 to 0.66 | 0.10 | no |
| LUM EC50vs. LUM DTR | −0.87 | −0.95 to −0.72 | <.001 | yes *** |
| LUM EC50vs. MEF EC50 | 0.85 | 0.67 to 0.93 | <.001 | yes *** |
| LUM EC50vs. HLF EC50 | 0.79 | 0.56 to 0.91 | <.001 | yes *** |
| LUM EC50vs. CQ EC50 | 0.00 | −0.42 to 0.41 | 0.99 | no |
| LUM EC50vs. DHA EC50 | −0.12 | −0.52 to 0.33 | 0.61 | no |
| LUM DTR vs. MEF EC50 | −0.89 | −0.95 to −0.75 | <.001 | yes *** |
| LUM DTR vs. HLF EC50 | −0.67 | −0.85 to −0.35 | <.001 | yes *** |
| LUM DTR vs. CQ EC50 | 0.03 | −0.39 to 0.44 | 0.88 | no |
| LUM DTR vs. DHA EC50 | −0.05 | −0.47 to 0.39 | 0.82 | no |
| |
||||
|
Mann-WhitneyUTest |
Medianphenotype of803haplotype |
Medianphenotype ofGB4haplotype |
||
| PfMDR1 haplotype vs. LUM DTR | 7.0 | 15.8 | <.001 | yes *** |
| PfMDR1 haplotype vs. LUM EC50 | 2.94 | 0.78 | <.001 | yes *** |
| PfMDR1 haplotype vs. MEF EC50 | 29.89 | 9.31 | <.001 | yes *** |
| PfMDR1 haplotype vs. HLF EC50 | 1.99 | 0.68 | 0.005 | yes ** |
| PfMDR1 haplotype vs. CQ EC50 | 78.2 | 88.8 | 0.61 | no |
| PfMDR1 haplotype vs. DHA EC50 |
1.95 |
1.74 |
0.89 |
no |
| PfCRT haplotype vs. LUM DTR | 8.3 | 14.0 | 0.14 | no |
| PfCRT haplotype vs. LUM EC50 | 2.73 | 0.82 | 0.02 | yes * |
| PfCRT haplotype vs. MEF EC50 | 21.91 | 11.05 | 0.22 | no |
| PfCRT haplotype vs. HLF EC50 | 1.48 | 0.68 | 0.08 | no |
| PfCRT haplotype vs. CQ EC50 | 99.1 | 70.5 | 0.048 | yes * |
| PfCRT haplotype vs. DHA EC50 |
1.78 |
1.93 |
0.91 |
no |
| PfK13 haplotype vs. LUM DTR | 7.5 | 14.0 | 0.03 | yes * |
| PfK13 haplotype vs. LUM EC50 | 2.69 | 0.97 | 0.06 | trend |
| PfK13 haplotype vs. MEF EC50 | 19.64 | 10.44 | 0.04 | yes * |
| PfK13 haplotype vs. HLF EC50 | 1.38 | 1.03 | 0.52 | no |
| PfK13 haplotype vs. CQ EC50 | 78.2 | 89.3 | 0.78 | no |
| PfK13 haplotype vs. DHA EC50 | 1.77 | 2.15 | 0.62 | no |
Analysis of results from the 803 × GB4 parents and progeny showed that the EC50s of LUM were positively and strongly correlated with those of MEF and HLF (r = 0.85, 95% CI: 0.67 to 0.93, p < 0.001; and r = 0.79, 95% CI: 0.56 to 0.91, p < 0.001, respectively; Table 2), consistent with such correlations in a number of previous studies (Basco et al., 1998; Pradines et al., 2006; Basco and Ringwald, 2007; Eyase et al., 2013). This analysis found no correlation between the EC50s of LUM and CQ (r = 0.00, 95% CI: −0.42 to 0.41, p = 0.99), in agreement with the results of several reports (Basco et al., 1998; Pradines et al., 1999; Nsobya et al., 2010) but not others that identified a moderate inverse LUM-CQ correlation (Mwai et al., 2009; Eyase et al., 2013). Finally, our analysis of the EC50s of LUM and those of DHA (determined by Sá et al. (2018)) showed that these drug responses were uncorrelated (r = −0.12, 95% CI: −0.52 to 0.33, p = 0.61; Table 2); this result agrees with previous findings of no LUM-DHA correlation by Nsobya et al. (2010) and Amambua-Ngwa et al. (2017), although it differs from the weak positive LUM-DHA correlations reported from some other studies (Pradines et al., 2006; Basco and Ringwald, 2007; Mwai et al., 2009).
3.2. Polymorphisms identified by quantitative trait loci (QTL) analysis and correlation tests with genes known to affect drug responses
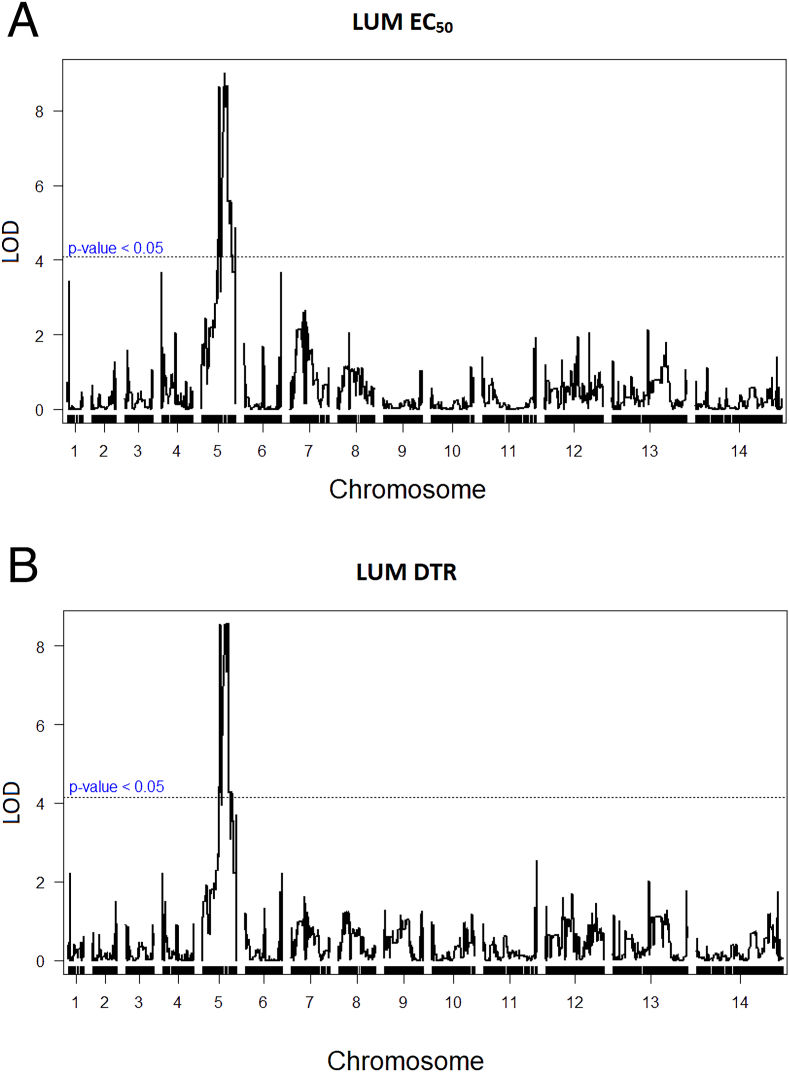
QTL primary scans of LUM EC50 and DTR phenotypes identified a major peak on Chromosome 5 with maximal LOD scores of 8.94 (p < 0.005) and 8.55 (p < 0.005) respectively (Fig. 2A and B). This peak spans a ~500 kilobase region between the locations of SNPs 702,055 and 1,171,094 of Chromosome 5, with the maximum likelihood location at 966,290. Searches of the PlasmoDB database identified 117 annotated genes within these regions, notably including pfmdr1 at location 957,890 to 962,149 (PF3D7_0523,000; Table S3), a candidate gene previously associated to LUM response variations ex vivo, in vivo, and in vitro (Duraisingh et al., 2000; Nzila et al., 2012; Venkatesan et al., 2014; Veiga et al., 2016).
Fig. 2.
Quantitative trait loci (QTL) analysis of lumefantrine phenotypes in the 803 × GB4 cross. A QTL plot from LUM 72 h half-maximal effective concentration (EC50) determinations. Maximal logarithm of odds (LOD) score is at location 966,290 on Chromosome 5 (p < 0.005). B QTL plot from the parasitemia Days to Recovery (DTR) LUM data, with a maximal LOD score at location 966,290 on Chromosome 5 (p < 0.005). Dashed lines in the figure panels mark the 0.05 significance threshold.
QTL analysis of the MEF EC50 phenotypes likewise identified a primary peak at the pfmdr1 locus (Fig. S1A; p < 0.005). However, scans of the HLF and CQ EC50 phenotypes identified no peaks of statistical significance (Figs. S1B and C; p = 0.34; and p = 0.82 for HLF and CQ, respectively). The absence of a strong pfcrt signal with the CQ EC50 values is consistent with the fact the 803 and GB4 parents both have ‘CVIET’ pfcrt alleles carrying the 76T codon determinant of CQ resistance (Table 3) that can be modulated by the effects of other genes, including pfmdr1 (Sá et al., 2009; Dhingra et al., 2019).
Table 3.
Polymorphisms in the PfMDR1, PfCRT, and PfK13 sequences of P. falciparum lines 3D7 (reference line), 803, and GB4.
| PfMDR1 |
PfCRT |
PfK13 |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Parasite line | 86 | 184 | 1034 | 1042 | 1246 | copyno.a | 72 | 74 | 75 | 76 | 220 | 271 | 326 | 356 | 371 | 189 | 580 |
| 3D7 | N | Y | S | N | D | 1 | C | M | N | K | A | Q | N | I | R | K | C |
| 803 | N | Y | S | N | D | 1 | C | I | E | T | S | E | S | T | I | K | Y |
| GB4 | Y | F | S | N | D | 1 | C | I | E | T | S | E | N | I | I | T | C |
Details of copy number analysis are provided in Table S4.
Using the reported 803 and GB4 genome sequences (Garimella et al., 2020) and copy number analysis, we checked for presence pfmdr1 polymorphisms including the codon variations that are known to occur in other P. falciparum lines at positions 86, 184, 1034, 1042, and 1246 (Foote et al., 1990). A pfmdr1 copy number of 1 was obtained for both GB4 and 803, the same as for the control 3D7 line (Table S4). Relative to the pfmdr1 canonical 3D7 sequence (‘wild-type’), mutant codons 86Y and 184F were identified in the GB4 sequence, whereas all five codons in the 803 sequence matched those of the 3D7 reference sequence (Table 3). The PfMDR1 haplotypes in the 803 × GB4 cross thus differ only at two sites in the 803 (N86/Y184) and GB4 (86Y/184F) parents; accordingly, the results of this study are silent on other combinations of PfMDR1 polymorphisms. While further investigations will be needed to clarify the drug response phenotypes of these other combinations, the linkage of PfMDR1 N86 to the less sensitive LUM and MEF phenotypes of 803 relative to GB4 is consistent with previous reports associating N86 with reduced susceptibility to these drugs (Duraisingh et al., 2000; Sisowath et al., 2005; Dokomajilar et al., 2006; Mwai et al., 2009; Lekana-Douki et al., 2011; Li et al., 2014; Wurtz et al., 2014). The effect of the PfMDR1 Y184F polymorphism on LUM response in association with N86 is less clear: PfMDR1 N86 and Y184 were selected in reinfections after AL treatment in studies in Burkina Faso (Zongo et al., 2007; Somé et al., 2010), but PfMDR1 N86 and 184F were more prevalent in new infections after AL treatment in a Zanzibar cohort (Sisowath et al., 2007). In coastal Kenya, a slight increase of PfMDR1 184F in association with N86 and D1246 was attributed to AL pressure between 2006 and 2013 (Okombo et al., 2014); however, PfMDR1 184F was not associated with LUM or MEF responses in a study in Senegal (Wurtz et al., 2014) and, in Western Kenya, prevalence of Y184 allele was found to be increased in parasite populations post-AL treatment compared to populations before treatment (Achieng et al., 2015).
Although QTL analysis of the drug susceptibility phenotypes in the 803 × GB4 cross identified no loci in addition to pfmdr1 by scans with genome microarrays, we note that mutations in candidate genes including pfcrt codons N326S and I356T and pfk13 codon C580Y (all of which occur in the 803 parent; Table 3) have been associated with multigenic backgrounds that mediate parasite fitness and susceptibility to a variety of antimalarials (Dhingra et al., 2019). In previous studies of the 803 × GB4 cross, we showed that the PfK13 polymorphism C580Y is linked to survival rates of dihydroartemisinin (DHA)-exposed rings but that it does not determine EC50 levels with this drug (Sá et al., 2018). Tests for correlation of the pfmdr1 haplotype with those of pfcrt or pfk13 alleles did not yield significance at the p < 0.05 level (Spearman rank coefficients r = 0.32, 95% CI: −0.10 to 0.65, p = 0.12; and r = 0.34, 95% CI: −0.08 to 0.66, p = 0.10; Table 2). However, the correlation tests suggested an association of the mutant 326S and 356T codons in the 803 parent's CVIET-type PfCRT allele with greater CQ EC50s in the 803 × GB4 progeny (p = 0.048; Table 2), consistent with the findings of Dhingra et al. (2019) for these same codon changes in the CVIET allele of the Dd2 parasite. LUM response EC50s also suggested a possible influence of PfCRT mutations 326S and 356T in the cross (p = 0.02), but this influence did not reach significance in the DTR assays (Table 2). The well-established ability of PfCRT wild-type K76 to support the selection of LUM-tolerant parasite populations under AL pressure and to reduce the LUM susceptibility of laboratory-transformed parasites (Dhingra et al., 2019) is not observable in the 803 × GB4 progeny, as all have a CVIET-type background from one parent or the other, i.e. all carry PfCRT 76T.
Analysis with the PfK13 haplotypes also showed significant associations with the LUM DTR and MEF EC50 response phenotypes (p = 0.03 and p = 0.04, respectively). These remarkable results for the two aminoarylalcohols did not extend to HLF, nor was there any significant association between PfK13 haplotype and the CQ or DHA EC50 phenotypes (Table 2). Consistent with these results, LUM and DHA responses have been found to be uncorrelated or only weakly correlated with each other in a number of previous studies (Section 3.1); likewise, only weak positive correlations have been reported for MEF – DHA (Pradines et al., 2006; Basco and Ringwald, 2007).
3.3. Evidence for multiple loci and common mechanisms in the drug response phenotypes modulated by PfMDR1, PfCRT, and PfK13
The strong agreement between results of the LUM EC50 and DTR assays suggests that these different phenotypes of drug response involve common pathways and mechanisms. Further, the progeny phenotypes from both of these assays did not resolve into clear groups clustered about one parent or the other: instead, these phenotypes occurred in inversely correlated distributions spanning the phenotypes of the parents, with some of the progeny presenting beyond the bound of the upper parental phenotype in each assay (Fig. 1A, C). Broad distributions of this nature suggest the presence of multiple loci affecting drug susceptibility and parasite growth. The possibility of such loci is consistent with effects of PfK13 and PfCRT in addition to PfMDR1 on LUM and MEF susceptibilities. As with multigenically-determined phenotypes in other genetic systems (Bloom et al., 2013), larger numbers of independent recombinant progeny and new resources of P. falciparum crosses from humanized mice (Vaughan et al., 2015; Vendrely et al., 2020) complemented by modern tools of ‘forward’ and ‘reverse’ genetics (de Koning-Ward et al., 2015) will help to identify and characterize such loci.
Little is known about the mechanisms of LUM action or of potential parasite resistance to the drug (Premji, 2009). The well-established linkage of PfMDR1 and PfCRT polymorphisms to LUM susceptibility along with the location of PfMDR1 (Cowman et al., 1991) and PfCRT (Fidock et al., 2000; Cooper et al., 2002) at the DV is consistent with proposals that LUM, like CQ, promotes heme toxicity by forming complexes that inhibit hemozoin formation (Pradines et al., 1999; Combrinck et al., 2013). The evidence in this study that PfK13 polymorphisms correlate with LUM and MEF response phenotypes is likewise suggestive of effects on heme metabolism and heme-dependent pathways in the parasite. Recent studies have shown that PfK13 can mediate hemoglobin uptake (Yang et al., 2019; Birnbaum et al., 2020) and that disruption of DegP2 in Toxoplasma gondii and of a DegP2 homolog in P. falciparum point to mitochondrial porphyrin levels and heme metabolism as a target of DHA (Harding et al., 2020). Considering the likelihood of effects on other drugs that target heme in this scenario, various polymorphisms of PfMDR1, PfCRT, and PfK13 may alter access and action of LUM and MEF at their heme target, thus modulating toxicity.
In light of the above observations, we hypothesize that the use of LUM and MEF in malaria endemic regions may drive selection of PfK13 as well as PfMDR1 and PfCRT polymorphisms. Indeed, a study of treatment failures following MEF-containing ACT found greater chances of recrudescence with P. falciparum strains containing PfK13 propeller mutations and multiple copies of PfMDR1 (Phyo et al., 2016). Results presented here suggest that MEF may have contributed to selection of the PfK13 propeller mutations and their association with treatment failures.
4. Conclusions
LUM is a vital partner in what is perhaps the leading ACT worldwide for the treatment of uncomplicated P. falciparum malaria, and MEF continues to be widely used in ACTs by a number of countries in South American and Southeast Asia, including Cambodia (World Health Organization, 2019). In this study, the results from two different drug response assays with LUM, a 72 h EC50 growth inhibition assay and a DTR parasitemia recovery assay following 24 h drug exposure, showed strong inverse correlation in the parents and 22 independent recombinant progeny of a cross between Cambodian (803) and Ghanaian (GB4) lines of P. falciparum. Strong correlations were also found between LUM, MEF, and HLF susceptibilities in the 803 × GB4 cross. By QTL analysis and Mann-Whitney U tests with candidate genes known to be involved in heme metabolism, LUM, MEF, and HLF phenotypes were associated with inheritance of PfMDR1, confirming previous findings that the wild-type N86 residue confers decreased susceptibility to these drugs. Statistical tests likewise identified moderately reduced CQ susceptibility from mutations N326S and I356T in the 803 version of the CVIET-type PfCRT carried by both parents; however, the well-known reduction of LUM susceptibility from wild-type PfCRT K76 could not be assessed as all parents and progeny contained PfCRT 76T. Finally, statistical tests of PfK13 Kelch protein polymorphisms identified significant reductions of LUM and MEF susceptibility with inheritance of the PfK13 580Y mutation in the 803 parent. In endemic regions where LUM and MEF are used in ACTs, pressure on parasite populations from these partner drugs may boost the prevalence of PfK13 580Y and other Kelch polymorphisms involved in drug susceptibility.
Ethics approval and consent to participate
Not applicable.
Availability of data and materials
All data generated or analyzed in this study are provided in the main text of this article or as supplementary information in supporting files.
Authors’ contributions
STW, KDL, NBG, and TEW designed the study and wrote the manuscript. STW, KDL, NBG, AL, and JMS collected data, STW, KDL, JM, RLC, RSR, JMS, and TEW contributed resources and analyzed results. All authors read, revised, and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no competing interests
Acknowledgements
The authors thank Christine E. Figan for her assistance in microsatellite genotyping, Kiran V. Garimella for providing PacBio DNA sequences for the parental lines, and Michael P. Fay for discussion of statistical correlation tests. This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Disease, National Institutes of Health.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2020.10.009.
List of abbreviations
- 803
Cambodian P. falciparum parent of the 803 × GB4 cross
- ACT
artemisinin-based combination therapy
- AL
artemether-lumefantrine
- CI
confidence interval
- CQ
chloroquine
- DTR
days to recover starting parasitemia
- DV
digestive vacuole
- EC50
half maximal effective concentration to inhibit parasite growth
- GB4
Ghanaian P. falciparum parent of the 803 × GB4 cross
- HLF
halofantrine
- LOD
logarithm of odds
- LUM
lumefantrine
- MEF
mefloquine
- QTL
quantitative trait loci
- RBC
red blood cell
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Achieng A.O., Muiruri P., Ingasia L.A., Opot B.H., Juma D.W., Yeda R., Ngalah B.S., Ogutu B.R., Andagalu B., Akala H.M., Kamau E. Temporal trends in prevalence of Plasmodium falciparum molecular markers selected for by artemether-lumefantrine treatment in pre-ACT and post-ACT parasites in western Kenya. Int. J. Parasitol. Drugs Drug Resist. 2015;5:92–99. doi: 10.1016/j.ijpddr.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amambua-Ngwa A., Okebe J., Mbye H., Ceesay S., El-Fatouri F., Joof F., Nyang H., Janha R., Affara M., Ahmad A., Kolly O., Nwakanma D., D’Alessandro U. Sustained ex vivo susceptibility of Plasmodium falciparum to artemisinin derivatives but increasing tolerance to artemisinin combination therapy partner quinolines in the Gambia. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.00759-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaratunga C., Neal A.T., Fairhurst R.M. Flow cytometry-based analysis of artemisinin-resistant Plasmodium falciparum in the ring-stage survival assay. Antimicrob. Agents Chemother. 2014;58:4938–4940. doi: 10.1128/AAC.02902-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliraine F.N., Rosenthal P.J. Prolonged selection of pfmdr1 polymorphisms after treatment of falciparum malaria with artemether-lumefantrine in Uganda. J. Infect. Dis. 2011;204:1120–1124. doi: 10.1093/infdis/jir486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraka V., Tinto H., Valea I., Fitzhenry R., Delgado-Ratto C., Mbonye M.K., Van Overmeir C., Rosanas-Urgell A., Van Geertruyden J.P., D’Alessandro U., Erhart A. In vivo selection of Plasmodium falciparum pfcrt and pfmdr1 variants by artemether-lumefantrine and dihydroartemisinin-piperaquine in Burkina Faso. Antimicrob. Agents Chemother. 2015;59:734–737. doi: 10.1128/AAC.03647-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basco L.K., Bickii J., Ringwald P. In vitro activity of lumefantrine (benflumetol) against clinical isolates of Plasmodium falciparum in Yaoundé, Cameroon. Antimicrob. Agents Chemother. 1998;42:2347–2351. doi: 10.1128/aac.42.9.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basco L.K., Ringwald P. Molecular epidemiology of malaria in Cameroon. XXIV. Trends of in vitro antimalarial drug responses in Yaounde, Cameroon. Am. J. Trop. Med. Hyg. 2007;76:20–26. [PubMed] [Google Scholar]
- Birnbaum J., Scharf S., Schmidt S., Jonscher E., Hoeijmakers W.A.M., Flemming S., Toenhake C.G., Schmitt M., Sabitzki R., Bergmann B., Frohlke U., Mesen-Ramirez P., Blancke Soares A., Herrmann H., Bartfai R., Spielmann T. A Kelch13-defined endocytosis pathway mediates artemisinin resistance in malaria parasites. Science. 2020;367:51–59. doi: 10.1126/science.aax4735. [DOI] [PubMed] [Google Scholar]
- Bloom J.S., Ehrenreich I.M., Loo W.T., Lite T.L., Kruglyak L. Finding the sources of missing heritability in a yeast cross. Nature. 2013;494:234–237. doi: 10.1038/nature11867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman K.W., Wu H., Sen S., Churchill G.A. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Challenger J.D., Bruxvoort K., Ghani A.C., Okell L.C. Assessing the impact of imperfect adherence to artemether-lumefantrine on malaria treatment outcomes using within-host modelling. Nat. Commun. 2017;8:1373. doi: 10.1038/s41467-017-01352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavchich M., Van Breda K., Rowcliffe K., Diagana T.T., Edstein M.D. The spiroindolone KAE609 does not induce dormant ring stages in Plasmodium falciparum parasites. Antimicrob. Agents Chemother. 2016;60:5167–5174. doi: 10.1128/AAC.02838-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combrinck J.M., Mabotha T.E., Ncokazi K.K., Ambele M.A., Taylor D., Smith P.J., Hoppe H.C., Egan T.J. Insights into the role of heme in the mechanism of action of antimalarials. ACS Chem. Biol. 2013;8:133–137. doi: 10.1021/cb300454t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M.D., LeClair N., Arinaitwe E., Wanzira H., Kakuru A., Bigira V., Muhindo M., Kamya M.R., Tappero J.W., Greenhouse B., Dorsey G., Rosenthal P.J. Comparative impacts over 5 years of artemisinin-based combination therapies on Plasmodium falciparum polymorphisms that modulate drug sensitivity in Ugandan children. J. Infect. Dis. 2014;210:344–353. doi: 10.1093/infdis/jiu141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper R.A., Ferdig M.T., Su X.-z., Ursos L.M.B., Mu J.B., Nomura T., Fujioka H., Fidock D.A., Roepe P.D., Wellems T.E. Alternative mutations at position 76 of the vacuolar transmembrane protein PfCRT are associated with chloroquine resistance and unique stereospecific quinine and quinidine responses in Plasmodium falciparum. Mol. Pharmacol. 2002;61:35–42. doi: 10.1124/mol.61.1.35. [DOI] [PubMed] [Google Scholar]
- Cowman A.F., Karcz S., Galatis D., Culvenor J.G. A P-glycoprotein homologue of Plasmodium falciparum is localized on the digestive vacuole. J. Cell Biol. 1991;113:1033–1042. doi: 10.1083/jcb.113.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranmer S.L., Magowan C., Liang J., Coppel R.L., Cooke B.M. An alternative to serum for cultivation of Plasmodium falciparum in vitro. Trans. R. Soc. Trop. Med. Hyg. 1997;91:363–365. doi: 10.1016/s0035-9203(97)90110-3. [DOI] [PubMed] [Google Scholar]
- Dama S., Niangaly H., Ouattara A., Sagara I., Sissoko S., Traore O.B., Bamadio A., Dara N., Djimde M., Alhousseini M.L., Goita S., Maiga H., Dara A., Doumbo O.K., Djimde A.A. Reduced ex vivo susceptibility of Plasmodium falciparum after oral artemether-lumefantrine treatment in Mali. Malar. J. 2017;16:59. doi: 10.1186/s12936-017-1700-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning-Ward T.F., Gilson P.R., Crabb B.S. Advances in molecular genetic systems in malaria. Nat. Rev. Microbiol. 2015;13:373–387. doi: 10.1038/nrmicro3450. [DOI] [PubMed] [Google Scholar]
- Denis M.B., Tsuyuoka R., Lim P., Lindegardh N., Yi P., Top S.N., Socheat D., Fandeur T., Annerberg A., Christophel E.M., Ringwald P. Efficacy of artemether-lumefantrine for the treatment of uncomplicated falciparum malaria in northwest Cambodia. Trop. Med. Int. Health. 2006;11:1800–1807. doi: 10.1111/j.1365-3156.2006.01739.x. [DOI] [PubMed] [Google Scholar]
- Dhingra S.K., Gabryszewski S.J., Small-Saunders J.L., Yeo T., Henrich P.P., Mok S., Fidock D.A. Global spread of mutant PfCRT and its pleiotropic impact on Plasmodium falciparum multidrug resistance and fitness. mBio. 2019;10 doi: 10.1128/mBio.02731-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokomajilar C., Nsobya S.L., Greenhouse B., Rosenthal P.J., Dorsey G. Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob. Agents Chemother. 2006;50:1893–1895. doi: 10.1128/AAC.50.5.1893-1895.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duraisingh M.T., Roper C., Walliker D., Warhurst D.C. Increased sensitivity to the antimalarials mefloquine and artemisinin is conferred by mutations in the pfmdr1 gene of Plasmodium falciparum. Mol. Microbiol. 2000;36:955–961. doi: 10.1046/j.1365-2958.2000.01914.x. [DOI] [PubMed] [Google Scholar]
- Eyase F.L., Akala H.M., Ingasia L., Cheruiyot A., Omondi A., Okudo C., Juma D., Yeda R., Andagalu B., Wanja E., Kamau E., Schnabel D., Bulimo W., Waters N.C., Walsh D.S., Johnson J.D. The role of Pfmdr1 and Pfcrt in changing chloroquine, amodiaquine, mefloquine and lumefantrine susceptibility in western-Kenya P. falciparum samples during 2008-2011. PloS One. 2013;8 doi: 10.1371/journal.pone.0064299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnert A., Ursing J., Tolfvenstam T., Rono J., Karlsson L., Sparrelid E., Lindegardh N. Artemether-lumefantrine treatment failure despite adequate lumefantrine day 7 concentration in a traveller with Plasmodium falciparum malaria after returning from Tanzania. Malar. J. 2012;11:176. doi: 10.1186/1475-2875-11-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidock D.A., Nomura T., Talley A.K., Cooper R.A., Dzekunov S.M., Ferdig M.T., Ursos L.M., Sidhu A.B., Naudé B., Deitsch K.W., Su X.Z., Wootton J.C., Roepe P.D., Wellems T.E. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol. Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figan C.E., Sa J.M., Mu J., Melendez-Muniz V.A., Liu C.H., Wellems T.E. A set of microsatellite markers to differentiate Plasmodium falciparum progeny of four genetic crosses. Malar. J. 2018;17:60. doi: 10.1186/s12936-018-2210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote S.J., Kyle D.E., Martin R.K., Oduola A.M., Forsyth K., Kemp D.J., Cowman A.F. Several alleles of the multidrug-resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature. 1990;345:255–258. doi: 10.1038/345255a0. [DOI] [PubMed] [Google Scholar]
- Gadalla N.B., Adam I., Elzaki S.E., Bashir S., Mukhtar I., Oguike M., Gadalla A., Mansour F., Warhurst D., El-Sayed B.B., Sutherland C.J. Increased pfmdr1 copy number and sequence polymorphisms in Plasmodium falciparum isolates from Sudanese malaria patients treated with artemether-lumefantrine. Antimicrob. Agents Chemother. 2011;55:5408–5411. doi: 10.1128/AAC.05102-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garimella K.V., Iqbal Z., Krause M.A., Campino S., Kekre M., Drury E., Kwiatkowski D., Sá J.M., Wellems T.E., McVean G. Detection of simple and complex de novo mutations with multiple reference sequences. Genome Res. 2020;30:1154–1169. doi: 10.1101/gr.255505.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happi C.T., Gbotosho G.O., Folarin O.A., Sowunmi A., Hudson T., O’Neil M., Milhous W., Wirth D.F., Oduola A.M. Selection of Plasmodium falciparum multidrug resistance gene 1 alleles in asexual stages and gametocytes by artemether-lumefantrine in Nigerian children with uncomplicated falciparum malaria. Antimicrob. Agents Chemother. 2009;53:888–895. doi: 10.1128/AAC.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding C.R., Sidik S.M., Petrova B., Gnädig N.F., Okombo J., Herneisen A.L., Ward K.E., Markus B.M., Boydston E.A., Fidock D.A., Lourido S. Genetic screens reveal a central role for heme metabolism in artemisinin susceptibility. Nat. Commun. 2020;11:4813. doi: 10.1038/s41467-020-18624-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayton K., Gaur D., Liu A., Takahashi J., Henschen B., Singh S., Lambert L., Furuya T., Bouttenot R., Doll M., Nawaz F., Mu J., Jiang L., Miller L.H., Wellems T.E. Erythrocyte binding protein PfRH5 polymorphisms determine species-specific pathways of Plasmodium falciparum invasion. Cell Host Microbe. 2008;4:40–51. doi: 10.1016/j.chom.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys G.S., Merinopoulos I., Ahmed J., Whitty C.J., Mutabingwa T.K., Sutherland C.J., Hallett R.L. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob. Agents Chemother. 2007;51:991–997. doi: 10.1128/AAC.00875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambros C., Vanderberg J.P. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 1979;65:418–420. [PubMed] [Google Scholar]
- Lane K.D., Mu J., Lu J., Windle S.T., Liu A., Sun P.D., Wellems T.E. Selection of Plasmodium falciparum cytochrome B mutants by putative PfNDH2 inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2018;115:6285–6290. doi: 10.1073/pnas.1804492115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekana-Douki J.B., Dinzouna Boutamba S.D., Zatra R., Zang Edou S.E., Ekomy H., Bisvigou U., Toure-Ndouo F.S. Increased prevalence of the Plasmodium falciparum Pfmdr1 86N genotype among field isolates from Franceville, Gabon after replacement of chloroquine by artemether-lumefantrine and artesunate-mefloquine. Infect. Genet. Evol. 2011;11:512–517. doi: 10.1016/j.meegid.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Li J., Chen J., Xie D., Monte-Nguba S.M., Eyi J.U., Matesa R.A., Obono M.M., Ehapo C.S., Yang L., Lu D., Yang H., Yang H.T., Lin M. High prevalence of pfmdr1 N86Y and Y184F mutations in Plasmodium falciparum isolates from Bioko Island, Equatorial Guinea. Pathog. Glob. Health. 2014;108:339–343. doi: 10.1179/2047773214Y.0000000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makanga M., Krudsood S. The clinical efficacy of artemether/lumefantrine (Coartem) Malar. J. 2009;8(Suppl. 1):S5. doi: 10.1186/1475-2875-8-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmberg M., Ferreira P.E., Tarning J., Ursing J., Ngasala B., Björkman A., Mårtensson A., Gil J.P. Plasmodium falciparum drug resistance phenotype as assessed by patient antimalarial drug levels and its association with pfmdr1 polymorphisms. J. Infect. Dis. 2013;207:842–847. doi: 10.1093/infdis/jis747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina-Cruz A., Garver L.S., Alabaster A., Bangiolo L., Haile A., Winikor J., Ortega C., van Schaijk B.C., Sauerwein R.W., Taylor-Salmon E., Barillas-Mury C. The human malaria parasite Pfs47 gene mediates evasion of the mosquito immune system. Science. 2013;340:984–987. doi: 10.1126/science.1235264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwai L., Kiara S.M., Abdirahman A., Pole L., Rippert A., Diriye A., Bull P., Marsh K., Borrmann S., Nzila A. In vitro activities of piperaquine, lumefantrine, and dihydroartemisinin in Kenyan Plasmodium falciparum isolates and polymorphisms in pfcrt and pfmdr1. Antimicrob. Agents Chemother. 2009;53:5069–5073. doi: 10.1128/AAC.00638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S., Nash D., Sudimack D., Jaidee A., Barends M., Uhlemann A.C., Krishna S., Nosten F., Anderson T.J. Recurrent gene amplification and soft selective sweeps during evolution of multidrug resistance in malaria parasites. Mol. Biol. Evol. 2007;24:562–573. doi: 10.1093/molbev/msl185. [DOI] [PubMed] [Google Scholar]
- Nguitragool W., Bokhari A.A., Pillai A.D., Rayavara K., Sharma P., Turpin B., Aravind L., Desai S.A. Malaria parasite clag3 genes determine channel-mediated nutrient uptake by infected red blood cells. Cell. 2011;145:665–677. doi: 10.1016/j.cell.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nsobya S.L., Kiggundu M., Nanyunja S., Joloba M., Greenhouse B., Rosenthal P.J. In vitro sensitivities of Plasmodium falciparum to different antimalarial drugs in Uganda. Antimicrob. Agents Chemother. 2010;54:1200–1206. doi: 10.1128/AAC.01412-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nzila A., Okombo J., Ohuma E., Al-Thukair A. Update on the in vivo tolerance and in vitro reduced susceptibility to the antimalarial lumefantrine. J. Antimicrob. Chemother. 2012;67:2309–2315. doi: 10.1093/jac/dks252. [DOI] [PubMed] [Google Scholar]
- Oduola A.M., Omitowoju G.O., Gerena L., Kyle D.E., Milhous W.K., Sowunmi A., Salako L.A. Reversal of mefloquine resistance with penfluridol in isolates of Plasmodium falciparum from south-west Nigeria. Trans. R. Soc. Trop. Med. Hyg. 1993;87:81–83. doi: 10.1016/0035-9203(93)90434-r. [DOI] [PubMed] [Google Scholar]
- Okombo J., Kamau A.W., Marsh K., Sutherland C.J., Ochola-Oyier L.I. Temporal trends in prevalence of Plasmodium falciparum drug resistance alleles over two decades of changing antimalarial policy in coastal Kenya. Int J Parasitol Drugs Drug Resist. 2014;4:152–163. doi: 10.1016/j.ijpddr.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson D.S., Milhous W.K., Wellems T.E. Molecular basis of differential resistance to cycloguanil and pyrimethamine in Plasmodium falciparum malaria. Proc. Natl. Acad. Sci. U. S. A. 1990;87:3018–3022. doi: 10.1073/pnas.87.8.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson D.S., Walliker D., Wellems T.E. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc. Natl. Acad. Sci. U. S. A. 1988;85:9114–9118. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M.A., Burrows J.N., Manyando C., van Huijsduijnen R.H., Van Voorhis W.C., Wells T.N.C. Malaria. Nat. Rev. Dis. Primers. 2017;3 doi: 10.1038/nrdp.2017.50. [DOI] [PubMed] [Google Scholar]
- Phyo A.P., Ashley E.A., Anderson T.J., Bozdech Z., Carrara V.I., Sriprawat K., Nair S., White M.M., Dziekan J., Ling C., Proux S., Konghahong K., Jeeyapant A., Woodrow C.J., Imwong M., McGready R., Lwin K.M., Day N.P., White N.J., Nosten F. Declining efficacy of artemisinin combination therapy against P. falciparum malaria on the Thai-Myanmar border (2003-2013): the role of parasite genetic factors. Clin. Infect. Dis. 2016;63:784–791. doi: 10.1093/cid/ciw388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradines B., Hovette P., Fusai T., Atanda H.L., Baret E., Cheval P., Mosnier J., Callec A., Cren J., Amalvict R., Gardair J.P., Rogier C. Prevalence of in vitro resistance to eleven standard or new antimalarial drugs among Plasmodium falciparum isolates from Pointe-Noire, Republic of the Congo. J. Clin. Microbiol. 2006;44:2404–2408. doi: 10.1128/JCM.00623-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradines B., Tall A., Fusai T., Spiegel A., Hienne R., Rogier C., Trape J.F., Le Bras J., Parzy D. In vitro activities of benflumetol against 158 Senegalese isolates of Plasmodium falciparum in comparison with those of standard antimalarial drugs. Antimicrob. Agents Chemother. 1999;43:418–420. doi: 10.1128/aac.43.2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premji Z.G. Coartem: the journey to the clinic. Malar. J. 2009;8(Suppl. 1):S3. doi: 10.1186/1475-2875-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price R.N., Uhlemann A.C., van Vugt M., Brockman A., Hutagalung R., Nair S., Nash D., Singhasivanon P., Anderson T.J., Krishna S., White N.J., Nosten F. Molecular and pharmacological determinants of the therapeutic response to artemether-lumefantrine in multidrug-resistant Plasmodium falciparum malaria. Clin. Infect. Dis. 2006;42:1570–1577. doi: 10.1086/503423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sá J.M., Kaslow S.R., Krause M.A., Melendez-Muniz V.A., Salzman R.E., Kite W.A., Zhang M., Moraes Barros R.R., Mu J., Han P.K., Mershon J.P., Figan C.E., Caleon R.L., Rahman R.S., Gibson T.J., Amaratunga C., Nishiguchi E.P., Breglio K.F., Engels T.M., Velmurugan S., Ricklefs S., Straimer J., Gnadig N.F., Deng B., Liu A., Diouf A., Miura K., Tullo G.S., Eastman R.T., Chakravarty S., James E.R., Udenze K., Li S., Sturdevant D.E., Gwadz R.W., Porcella S.F., Long C.A., Fidock D.A., Thomas M.L., Fay M.P., Sim B.K.L., Hoffman S.L., Adams J.H., Fairhurst R.M., Su X.Z., Wellems T.E. Artemisinin resistance phenotypes and K13 inheritance in a Plasmodium falciparum cross and Aotus model. Proc. Natl. Acad. Sci. U. S. A. 2018;115:12513–12518. doi: 10.1073/pnas.1813386115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sá J.M., Kaslow S.R., Moraes Barros R.R., Brazeau N.F., Parobek C.M., Tao D., Salzman R.E., Gibson T.J., Velmurugan S., Krause M.A., Melendez-Muniz V., Kite W.A., Han P.K., Eastman R.T., Kim A., Kessler E.G., Abebe Y., James E.R., Chakravarty S., Orr-Gonzalez S., Lambert L.E., Engels T., Thomas M.L., Fasinu P.S., Serre D., Gwadz R.W., Walker L., DeConti D.K., Mu J., Bailey J.A., Sim B.K.L., Hoffman S.L., Fay M.P., Dinglasan R.R., Juliano J.J., Wellems T.E. Plasmodium vivax chloroquine resistance links to pvcrt transcription in a genetic cross. Nat. Commun. 2019;10:4300. doi: 10.1038/s41467-019-12256-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sá J.M., Twu O., Hayton K., Reyes S., Fay M.P., Ringwald P., Wellems T.E. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18883–18889. doi: 10.1073/pnas.0911317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu A.B., Uhlemann A.C., Valderramos S.G., Valderramos J.C., Krishna S., Fidock D.A. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J. Infect. Dis. 2006;194:528–535. doi: 10.1086/507115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisowath C., Ferreira P.E., Bustamante L.Y., Dahlström S., Mårtensson A., Björkman A., Krishna S., Gil J.P. The role of pfmdr1 in Plasmodium falciparum tolerance to artemether-lumefantrine in Africa. Trop. Med. Int. Health. 2007;12:736–742. doi: 10.1111/j.1365-3156.2007.01843.x. [DOI] [PubMed] [Google Scholar]
- Sisowath C., Petersen I., Veiga M.I., Mårtensson A., Premji Z., Björkman A., Fidock D.A., Gil J.P. In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether-lumefantrine in Africa. J. Infect. Dis. 2009;199:750–757. doi: 10.1086/596738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisowath C., Stromberg J., Martensson A., Msellem M., Obondo C., Bjorkman A., Gil J.P. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem) J. Infect. Dis. 2005;191:1014–1017. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]
- Smilkstein M., Sriwilaijaroen N., Kelly J.X., Wilairat P., Riscoe M. Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob. Agents Chemother. 2004;48:1803–1806. doi: 10.1128/AAC.48.5.1803-1806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somé A.F., Séré Y.Y., Dokomajilar C., Zongo I., Rouamba N., Greenhouse B., Ouédraogo J.B., Rosenthal P.J. Selection of known Plasmodium falciparum resistance-mediating polymorphisms by artemether-lumefantrine and amodiaquine-sulfadoxine-pyrimethamine but not dihydroartemisinin-piperaquine in Burkina Faso. Antimicrob. Agents Chemother. 2010;54:1949–1954. doi: 10.1128/AAC.01413-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonden K., Wyss K., Jovel I., Vieira da Silva A., Pohanka A., Asghar M., Homann M.V., Gustafsson L.L., Hellgren U., Farnert A. High rate of treatment failures in nonimmune travelers treated with artemether-lumefantrine for uncomplicated Plasmodium falciparum malaria in Sweden: retrospective comparative analysis of effectiveness and case series. Clin. Infect. Dis. 2017;64:199–206. doi: 10.1093/cid/ciw710. [DOI] [PubMed] [Google Scholar]
- Sondo P., Derra K., Diallo Nakanabo S., Tarnagda Z., Kazienga A., Zampa O., Valéa I., Sorgho H., Owusu-Dabo E., Ouédraogo J.B., Guiguemdé T.R., Tinto H. Artesunate-amodiaquine and artemether-lumefantrine therapies and selection of pfcrt and pfmdr1 alleles in Nanoro, Burkina Faso. PloS One. 2016;11 doi: 10.1371/journal.pone.0151565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valecha N., Mohanty S., Srivastava P., Sharma S., Tyagi P., Bergqvist Y., Ringwald P. Efficacy of artemether-lumefantrine in area of high malaria endemicity in India and its correlation with blood concentration of lumefantrine. Am. J. Trop. Med. Hyg. 2012;86:395–397. doi: 10.4269/ajtmh.2012.11-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan A.M., Pinapati R.S., Cheeseman I.H., Camargo N., Fishbaugher M., Checkley L.A., Nair S., Hutyra C.A., Nosten F.H., Anderson T.J., Ferdig M.T., Kappe S.H. Plasmodium falciparum genetic crosses in a humanized mouse model. Nat. Methods. 2015;12:631–633. doi: 10.1038/nmeth.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga M.I., Dhingra S.K., Henrich P.P., Straimer J., Gnadig N., Uhlemann A.C., Martin R.E., Lehane A.M., Fidock D.A. Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat. Commun. 2016;7:11553. doi: 10.1038/ncomms11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendrely K.M., Kumar S., Li X., Vaughan A.M. Humanized mice and the rebirth of malaria genetic crosses. Trends Parasitol. 2020;36:850–863. doi: 10.1016/j.pt.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan M., Gadalla N.B., Stepniewska K., Dahal P., Nsanzabana C., Moriera C., Price R.N., Mårtensson A., Rosenthal P.J., Dorsey G., Sutherland C.J., Guérin P., Davis T.M.E., Ménard D., Adam I., Ademowo G., Arze C., Baliraine F.N., Berens-Riha N., Björkman A., Borrmann S., Checchi F., Desai M., Dhorda M., Djimdé A.A., El-Sayed B.B., Eshetu T., Eyase F., Falade C., Faucher J.F., Fröberg G., Grivoyannis A., Hamour S., Houzé S., Johnson J., Kamugisha E., Kariuki S., Kiechel J.R., Kironde F., Kofoed P.E., LeBras J., Malmberg M., Mwai L., Ngasala B., Nosten F., Nsobya S.L., Nzila A., Oguike M., Otienoburu S.D., Ogutu B., Ouédraogo J.B., Piola P., Rombo L., Schramm B., Somé A.F., Thwing J., Ursing J., Wong R.P.M., Zeynudin A., Zongo I., Plowe C.V., Sibley C.H., Asaq Molecular Marker Study G. Polymorphisms in Plasmodium falciparum chloroquine resistance transporter and multidrug resistance 1 genes: parasite risk factors that affect treatment outcomes for P. falciparum malaria after artemether-lumefantrine and artesunate-amodiaquine. Am. J. Trop. Med. Hyg. 2014;91:833–843. doi: 10.4269/ajtmh.14-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Read M., Sims P.F., Hyde J.E. Sulfadoxine resistance in the human malaria parasite Plasmodium falciparum is determined by mutations in dihydropteroate synthetase and an additional factor associated with folate utilization. Mol. Microbiol. 1997;23:979–986. doi: 10.1046/j.1365-2958.1997.2821646.x. [DOI] [PubMed] [Google Scholar]
- Wellems T.E., Walker-Jonah A., Panton L.J. Genetic mapping of the chloroquine-resistance locus on Plasmodium falciparum chromosome 7. Proc. Natl. Acad. Sci. U. S. A. 1991;88:3382–3386. doi: 10.1073/pnas.88.8.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White N.J., van Vugt M., Ezzet F. Clinical pharmacokinetics and pharmacodynamics and pharmacodynamics of artemether-lumefantrine. Clin. Pharmacokinet. 1999;37:105–125. doi: 10.2165/00003088-199937020-00002. [DOI] [PubMed] [Google Scholar]
- World Health Organization . third ed. World Health Organization; Geneva: 2015. Guidelines for the Treatment of Malaria. [Google Scholar]
- World Health Organization . World Health Organization; Geneva, Switzerland: 2019. World Malaria Report 2019. [Google Scholar]
- Wurtz N., Fall B., Pascual A., Fall M., Baret E., Camara C., Nakoulima A., Diatta B., Fall K.B., Mbaye P.S., Dieme Y., Bercion R., Wade B., Pradines B. Role of pfmdr1 in in vitro Plasmodium falciparum susceptibility to chloroquine, quinine, monodesethylamodiaquine, mefloquine, lumefantrine, and dihydroartemisinin. Antimicrob. Agents Chemother. 2014;58:7032–7040. doi: 10.1128/AAC.03494-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Yeoh L.M., Tutor M.V., Dixon M.W., McMillan P.J., Xie S.C., Bridgford J.L., Gillett D.L., Duffy M.F., Ralph S.A., McConville M.J., Tilley L., Cobbold S.A. Decreased K13 abundance reduces hemoglobin catabolism and proteotoxic stress, underpinning artemisinin resistance. Cell Rep. 2019;29:2917–2928. doi: 10.1016/j.celrep.2019.10.095. [DOI] [PubMed] [Google Scholar]
- Zongo I., Dorsey G., Rouamba N., Tinto H., Dokomajilar C., Guiguemde R.T., Rosenthal P.J., Ouedraogo J.B. Artemether-lumefantrine versus amodiaquine plus sulfadoxine-pyrimethamine for uncomplicated falciparum malaria in Burkina Faso: a randomised non-inferiority trial. Lancet. 2007;369:491–498. doi: 10.1016/S0140-6736(07)60236-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed in this study are provided in the main text of this article or as supplementary information in supporting files.