Abstract
Double-stranded (ds)DNA, not dsRNA, has an ability to form a homologous complex with single-stranded (ss)DNA or ssRNA of homologous sequence. D-loops and homologous triplexes are homologous complexes formed with ssDNA by RecA/Rad51-family homologous-pairing proteins, and are a key intermediate of homologous (genetic/DNA) recombination. R-loop formation independent of transcription (R-loop formation in trans) was recently found to play roles in gene regulation and development of mammals and plants. In addition, the crRNA-Cas effector complex in CRISPR-Cas systems also relies on R-loop formation to recognize specific target. In homologous complex formation, ssDNA/ssRNA finds a homologous sequence in dsDNA by Watson-Crick base-pairing. crRNA-Cas effector complexes appear to actively melt dsDNA to make its bases available for annealing to crRNA. On the other hand, in D-loop formation and homologous-triplex formation, it is likely that dsDNA recognizes the homologous sequence before the melting of its double helix by using its intrinsic molecular function depending on CH2 at the 2′-position of the deoxyribose, and that the major role of RecA is the extension of ssDNA and the holding dsDNA at a position suitable for homology search. This intrinsic dsDNA function would also play a role in R-loop formation. The dependency of homologous-complex formation on 2′-CH2 of the deoxyribose would explain the absence of homologous complex formation by dsRNA, and dsDNA as sole genome molecule in all cellular organisms.
Keywords: Homologous pairing, RecA, Rad51, Homologous triplex, CH-pi interaction (CH-π interaction), CRISPR-Cas system, crRNA-Cas-effector complex, dsDNA, single-stranded DNA, ssRNA, Entropy-driven reaction, Deoxyribose
1. Introduction
It is a longstanding question why DNA, not RNA, is the sole genomic material of all cellular organisms. Possible explanations to answer this question include the chemical and biological instability of RNA, its A-form structure as its double-strand form of RNA and possible ribozyme activity. The A-form dsRNA has small grooves that prevent sequence-dependent interactions with proteins, but dsRNA has been shown to adopt also the A’ form that contains wider major groove to accommodate sequence-specific interactions with proteins [1]. Encoded sequences may induce a ribozyme activity that causes the degradation of the RNA genome, but DNA enzymes have been constructed [2]. Thus, these explanations are not qualitative differences between DNA and RNA. The ability of double-stranded (ds)DNA to form homologous complexes with single-stranded (ss)DNA or ssRNA provides a more definitive answer.
Homologous complexes are products of a reaction in which a region of a strand of dsDNA is replaced by the ssDNA or ssRNA of fully or almost fully identical sequence (homologous sequence), thus forming a hybrid duplex (less than 10 base-pairs [bps] to several kbps in length) with the complementary strand of the dsDNA (Fig. 1). Homologous complexes include D-loops [3], [4] (Fig. 1, B and E), R-loops [5] (Fig. 1, B and D) and “homologous triplexes” [6] (Figs. 1C and 2A). Homologous-complex formation with either ssDNA or ssRNA are generally observed in the cells of all living organisms. Though dsRNA is found in various cells, intermolecular homologous-complex formation with dsRNA has not thus far been reported in vivo. It is intriguing that dsRNA is generated during RNA interference but processed into ssRNA before the recognition of the complementary sequence in its target RNAs. RNA interference is a cellular system of post transcriptional gene regulation and repression of transposons (See Ref. [7], an example of review).
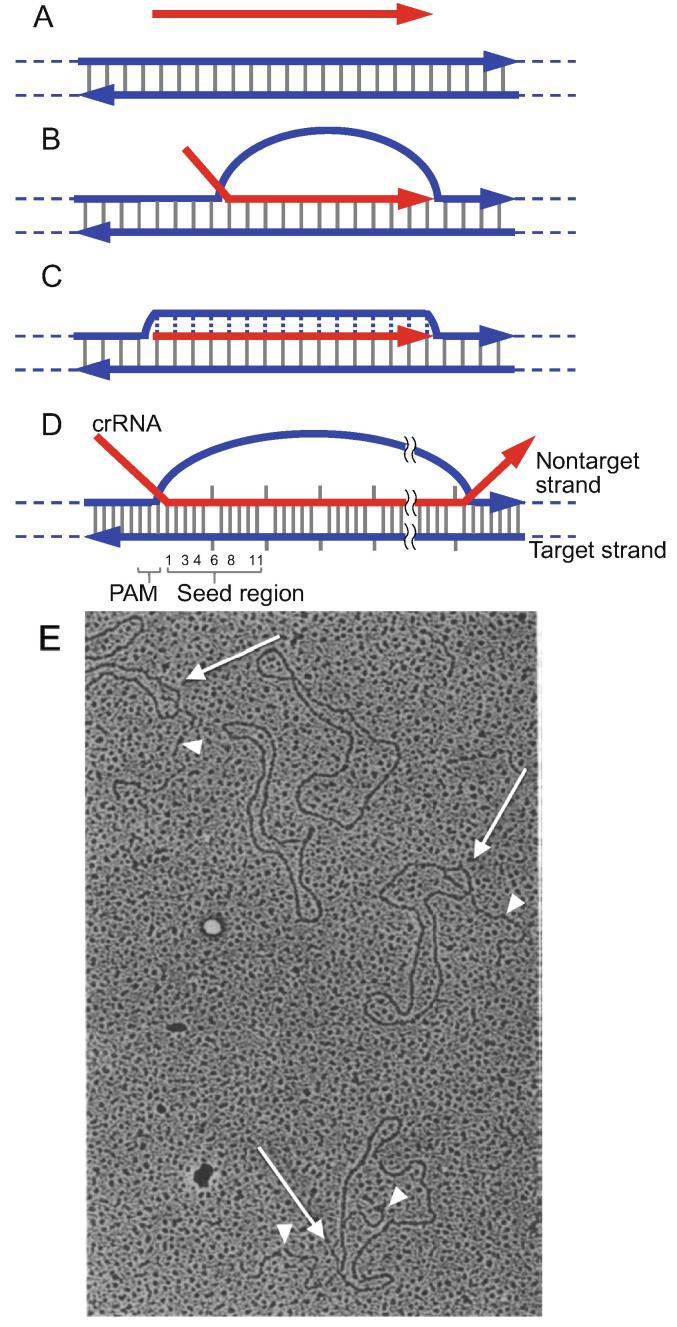
Fig. 1.
Homologous complex A. Substrates of homologous-complex formation. A pair of blue arrows represent strands of dsDNA. Vertical gray lines represent W-C base pairs. Red arrow represents a strand of homologous ssDNA or ssRNA. Arrows indicate the polarity of the strand (5′ to 3′). Broken lines flanking the arrows represent the continuity of the strands. B. D-loop/R-loop. Red arrow represents the strand derived from ssDNA for a D-loop or the strand derived from ssRNA for an R-loop. D- or R-loop length in ccc-dsDNA, measured in base pairs, is about 10 times the number of negative supercoils relaxed by the formation of the D- or R-loop. When the ssDNA exceeds this length, its terminal region(s) forms a tail(s) (See panel E). C. Homologous triplex. As in B, the red arrow represents the strand derived from ssDNA. Broken vertical blue lines represent base-pair-specific hydrogen bonds stabilizing the replaced strand in the major groove of the hybrid duplex (See Fig. 2A). D. R-loop formed by Class 1 crRNA-Cas-effector complex (Cascade, etc.). This R-loop comprises 5 segments, each of which contains a 5 bp region followed by a broken base pair (short grey vertical bars). PAM and seed regions are indicated. E. Electron micrograph of D-loops formed from negatively supercoiled ccc-dsDNA and homologous ssDNA-fragment by RecA. Arrows and arrow heads indicate D-loops and tails, respectively. This figure is reproduced from Ref. [10] and is copyrighted to the authors. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
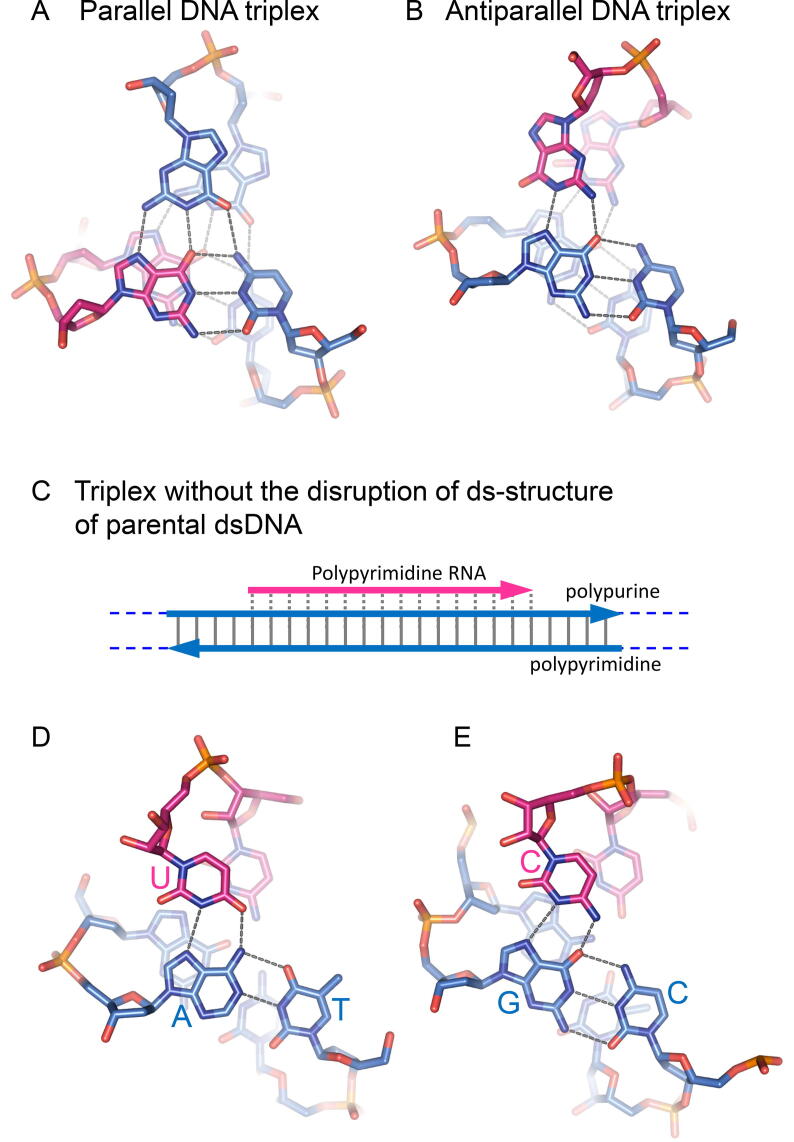
Fig. 2.
Triplexes. A. Crystal structure of a parallel DNA triplex at G·GC [32]. This parallel triplex is included in the homologous triplex formed between the invading ssDNA (pink; red strand in Fig. 1C) and the parental homologous dsDNA (sky blue; blue strand in Fig. 1C). This panel is an example of base parings in the triplex. All combinations of C·CG, A·AT and T·TA in addition to G·GC exists in the homologous triplex. Note that the pink strand forms a hybrid W-C duplex with the complementary strand of the parental dsDNA (sky blue), and has the same polarity and sequence as the replaced strand of the parental dsDNA (sky blue) during homologous-triplex formation (See Fig. 1C). B. Crystal structure of an antiparallel DNA triplex at G·GC [32]. The top strand with G (pink) and the strand with G of the parental dsDNA (sky blue) are antiparallel and paired with Hoogsteen hydrogen bonds. C. Antiparallel triplex formed from polypyrimidine ssRNA and polypurine-polypyrimidine W-C dsDNA without disruption of the double-strand structure. D. Antiparallel triplex at U in ssRNA (pink) paired with the A:T bp in dsDNA (sky blue). E. Antiparallel triplex at C in ssRNA (pink) paired with the GC W-C base pair in dsDNA (sky blue). The polypyrimidine RNA strand is parallel to the polypurine DNA strand and is antiparallel to the polypyrimidine DNA strand. Thus, this type of triplex cannot form between the transcript and the template dsDNA (See Ref. [34]). Panels A and B were generated from PDB 272D [32], and panels D and E are from PDB 1R3X [102]. All the figures of the structures in this article were prepared with PyMOL (PyMOL Molecular Graphics System, Schrödinger, LLC). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Homologous complexes with single-stranded DNA were first postulated as essential intermediates for homologous (genetic) recombination (general genetic recombination; homologous DNA recombination) [3], [8], [9], and proven to the case by the discovery that homologous-complex formation is catalyzed by either RecA [10], [11] or Rad51 [12], both of which are encoded by essential genes for homologous recombination [13], [14]. Homologous recombination is a basic genetic function in all living organisms. It is essential to cell proliferation in vertebrates [15], including humans. The double-strand structure allows dsDNA to maintain genome information in duplicate, thus enabling the accurate correction or repair of replication errors and DNA lesions by using the complementary strand as a template. When both strands of dsDNA are broken (double-strand break), homologous recombination facilitates precise repair by using dsDNA bearing the homologous sequence as a template (See Ref. [16]). Homologous recombination reorganizes genes between genomes inherited from parents, and thus, increases genetic variation, which increases the efficiency of natural selection [17]. The mechanism of homologous recombination has been reviewed numerous times (for example, see Refs. [16], [18]).
Homologous recombination of RNA genomes (RNA viruses) is rare and depends on a template-switch (or “copy-choice”) mechanism using ssRNA rather than an RNA version of a homologous-complex intermediate (See Ref. [19] for review; see also Ref. [20]). It has been reported that dsRNA is more thermally stable than dsDNA [21]. This may explain the difference between dsDNA and dsRNA in terms of their ability to form homologous complexes. The absence of any evidence of homologous-complex formation with dsRNA, even after decades of study, suggests a qualitative advantage for dsDNA over dsRNA.
Homologous (DNA) complexes with ssRNA (i.e. R-loops) have been regarded as genome-destabilizing byproducts of transcription in which the transcripts (RNA) associate with the template DNA strand [22]. Recent studies have revealed that R-loops play an important role in cellular function, as described below.
The crucial question related to homologous-complex formation is how the ssDNA or ssRNA recognizes the complementary sequence within dsDNA. A naive possibility is that the melting (disruption of double-strand structure) of the dsDNA is followed by annealing with homologous ssDNA or ssRNA to form hybrid. Studies on RecA have revealed that homologous sequences are recognized before the melting of the dsDNA, in the presence of ATP [23], [24], [25]. Unlike the annealing of complementary ssDNA or ssRNA, all bases in dsDNA that form stable Watson-Crick (W-C) base pairs are not readily available to recognize sequence complementarity with ssDNA or ssRNA.
In this minireview, we describe the biological functions of homologous-complex formation with ssDNA or ssRNA as well as the biochemical and structural features of homologous-complex formation with ssDNA or ssRNA by proteins that catalyze formation. We also discuss the possible molecular mechanisms suggested by molecular-structural analyses and recommend the use of computational simulation methods to further investigate outstanding questions.
2. D-loops, R-loops, and homologous triplexes as homologous (DNA) complexes with ssDNA or ssRNA
2.1. D-loops and R-loops
D-loops and R-loops are typical homologous complexes in which the replaced strand of dsDNA (Fig. 1A) separates from a hybrid duplex and forms a loop (Fig. 1, B and E). D-loop formation by RecA is enhanced by negative (right-hand) supercoil, which all natural covalently closed circular dsDNA (ccc-dsDNA) isolated from cells owns [26]. The D-loop or R-loop, formed with ccc-dsDNA, is stabilized by the relaxation of its negative supercoil [27]. Under conditions that denature the double helix, ccc-dsDNA carrying a D-loop or an R-loop in relaxed form is dissociated into ccc-dsDNA with negative supercoil and free ssDNA or ssRNA. A D-loop in dsDNA that has a strand break outside the site of the D-loop is spontaneously dissociated by branch migration induced by the rotational thermal movement of dsDNA at almost 104 bps/sec at 37 °C [28].
Topoisomerases change the topological state of ccc-dsDNA and its homologous complex. However, topoisomerases do not appear to directly stimulate or inhibit RecA-catalyzed homologous-complex formation. Topoisomerases are very useful when studying the mechanism of homologous-complex formation, since untwisting (often expressed by unwinding) and rewinding of the double helix during D-loop or R-loop formation can be sensitively measured as topological changes that are quantified by the use of topoisomerase and ccc-dsDNA (See Refs. [23], [29], [30], [31]).
2.2. Homologous triplex
The other type of homologous complex is the homologous triplex, which is a type of parallel triplex [32] (Figs. 1C and 2A), in which the replaced DNA strand is held by base-pair-specific hydrogen bonds (not by Hoogsteen hydrogen bonds) in the major groove of the hybrid W-C duplex that is newly formed between the invading ssDNA or ssRNA and the complementary strand of the parental dsDNA (Fig. 1C). The homologous triplex has been shown to be a homologous complex formed by RecA in vitro between ssDNA and dsDNA oligomer with hairpin at an end [6]. The homologous triplex between relaxed ccc-dsDNA with ssDNA has been shown to be formed by a protein, Mhr1, which is essential to yeast mitochondrial homologous recombination [29]. Unlike the case of the D-loop described above, when the relaxed ccc-dsDNA carrying the homologous triplex formed by Mhr1 in the presence of a topoisomerase is dissociated into ssDNA and dsDNA by high temperatures or alkaline pH after the inactivation of the proteins, the ccc-dsDNA has been demonstrated to remain relaxed without detectable topological change [29].
Note that the homologous triplex is different from the products of triplex formation between ssDNA or ssRNA and dsDNA in the absence of disruption of the W-C base pairs of the parental dsDNA. The triplex formed in the absence of disruption of the parental dsDNA does not contain the hybrid W-C duplex (Fig. 2C). Typical triplexes are formed between polypyrimidine ssRNA and polypurine-polypyrimidine dsDNA, with the third polypyrimidine RNA strand held antiparallel to the polypyrimidine strand of the dsDNA by Hoogsteen hydrogen bonds in the major groove of the dsDNA [33] (Fig. 2, D and E). The formation of this type of triplex has various sequence constraints (See Ref. [34]). Unlike triplex formation without hybrid duplex formation, homologous triplex formation requires two parallel strands with an almost identical sequence, and accepts various base-sequences (Figs. 1C and 2A).
3. Proteins that catalyze homologous-complex formation with ssDNA and their biological function
3.1. D-loop formation by RecA/Rad51-family homologous-pairing proteins in homologous recombination
The RecA homologous-pairing protein (RecA protein, RecA) is the prototype of the RecA/Rad51-family homologous-pairing proteins that catalyze D-loop formation from dsDNA and homologous ssDNA in vitro in an ATP-dependent mode [10], [11]. RecA/Rad51-family proteins include eubacterial RecA, eukaryotic orthologue Rad51 [35], archaeal RadA [36], and Dmc1 [37]. Dmc1 is the meiosis-specific paralogue of Rad51 that is essential to reductive segregation in meiosis for gamete formation in sexual reproduction of eukaryotes.
Homologous-complex formation by RecA/Rad51/Dmc1/RadA consists of two steps: homologous pairing and branch migration [38]. Homologous pairing is a quick (within a second at 37 °C) reaction that forms the core of the homologous complex, a roughly 15 bp hybrid duplex [39], [40]. Homologous pairing requires ATP but does not require its hydrolysis. Branch migration is an ATP-hydrolysis-dependent slow reaction that elongates the hybrid duplex unidirectionally by thousands of bps at the rate of approximately 4 bps per second (at 37 °C) [38]. The RecA-family proteins form a right-handed helical filament around ssDNA or dsDNA [30], [41], [42] (Fig. 3, A and B). Homologous pairing does not require filament formation, but branch migration depends on it [43].
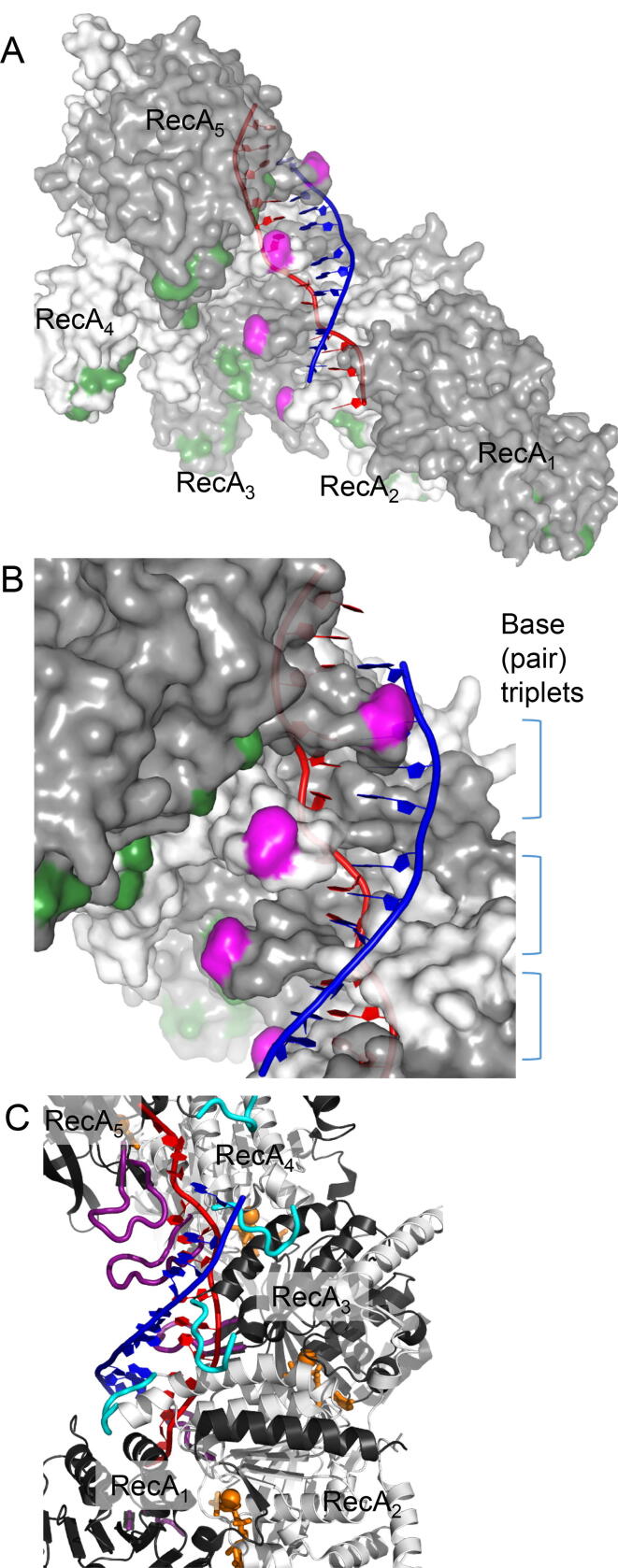
Fig. 3.
Crystal structure of active RecA-DNA filament. These panels show the 5 RecA protomers of 6 protomers per a turn of an elongated (i.e., active) RecA spiral filament containing dsDNA, which is likely to represent the hybrid duplex after D-loop formation [42] (PDB: 3CMX). The 3D structures of the RecA filament and the ssDNA in the RecA-ssDNA complex are almost identical to those of the RecA-filament and the red strand of the dsDNA in the RecA-dsDNA complex, respectively [42]. The blue strand is another strand of the dsDNA. After homologous-complex formation, the blue strand represents a strand of dsDNA paired with invading ssDNA in the hybrid duplex. A. Side view from the inside of the spiral filament. Basic amino acid residues consisting of gateway are colored green. B. Close-up of base triplets and L2 loops of which top is colored in magenta. C. ATP (analogue) molecules bound between two adjacent RecA protomers. Non-hydrolysable ATP analogues and Mg2+ ions are shown in orange. L1 and L2 loops are shown in cyan and purple, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
It is intriguing to note that the three-dimensional (3D) structure of RecA/Rad51/Dmc1/RadA’s core domain is almost identical to that of the DNA helicase domain of T7 replicative helicase-primase [44] and is also very similar to that of the DNA helicases of the same group [45]. DNA helicases are enzymes that melt dsDNA into complementary ssDNAs in an ATP-hydrolysis-dependent mode.
In eukaryotic cells, dsDNA is folded into chromatins, wherein it wraps around histone octamers and forms a beads-on-a-string structure [46]. Nucleosomes on the dsDNA prevent D-loop formation by Rad51, but Dmc1 forms D-loops preferentially in the nucleosome-depleted regions [47]. Rad54, a protein that interacts with Rad51 and is required for homologous recombination, stimulates remodeling of the chromatin structure to enable Rad51 to form D-loops [48]. A histone chaperone, Nap1, interacts with Rad54 and stimulates Rad54-mediated eviction of the linker histone H1, further stimulating D-loop formation by Rad51-Rad54 in dsDNA in higher-ordered chromatin with the linker histone H1 [49]. Thus, even in the cell, D-loop formation occurs at nucleosome-free regions of dsDNA.
3.2. Homologous complex formation by ATP-independent homologous-pairing proteins
Homologous triplexes have been identified as in vitro products formed in the absence of ATP by budding yeast mitochondrial protein Mhr1. MHR1 is an essential gene for homologous recombination, DNA replication, and DNA partitioning (segregation) into daughter cells in budding yeast mitochondria, which lack the RecA/Rad51 orthologue. In budding yeast, mitochondrial DNA is replicated through a rolling-circle mode initiated at the homologous complex formed by Mhr1 (See Ref. [50] for review, [51]).
Although major homologous recombination of genomic DNA strictly requires either RecA [13] or Rad51 [14], homologous recombination of plasmid DNA and homologous recombination of inverted repeats are detected even in the absence of active RecA and Rad51. These minor recombination events are promoted by eubacterial RecT and eukaryotic Rad52 in vivo. RecT and Rad52 catalyze homologous-complex formation in the absence of ATP in vitro [52], [53]. RecT and Rad52 belong to the same structural family of proteins [54], but Mhr1 belongs to a structurally distinct group and has a 3D structure similar to that of RecA/Rad51 [55].
3.3. TRF2 (telomere-repeat-binding factor 2) for telomere-loop formation
A telomere loop is a large lariat-like dsDNA loop of variable size (several to dozens of kbps in human cultured cells) that forms at the ends of chromosomes called telomeres. Telomere loops protect the chromosomal termini. For example, in humans, telomeres are thousands of repeats of the 5′-TTAGGG-3′/3′-AATCCC-5′ sequence. In the telomere loop, the 3′ ssDNA tail of the repeats invades an inner repeat to form a homologous complex [56]. TRF1 and TRF2 are major telomere-binding proteins that recognize the telomere-repeat sequence. In vivo, TRF2 is required for telomere-loop formation, but TRF1 is not [57]. In vitro, TRF2, but not TRF1, promotes telomere-loop formation of dsDNA with ssDNA (both of which have telomere repeat sequences) in an ATP-independent mode [31], [56], [58]. Note that the negative supercoils and telomere sequences in substrate dsDNA stimulate spontaneous formation in the absence of protein, resulting in a high background of TRF2-promoted formation in vitro [31].
4. Homologous complex formation with ssRNA
4.1. In vivo R-loop formation in trans
Various tools are available for analyzing R-loops in an entire genome at a single-base resolution. For example, DNA-RNA hybrid immunoprecipitation (RDIP or DRIP) coupled with high-throughput sequencing is an effective tool. The combination of RDIP and strand-specific sequencing of complementary DNA (RDIP-seq), or single-strand DNA ligation-based library construction after DRIP combined with next generation sequencing (ssDRIP-seq), enables us to locate strand-specific R-loops in an entire genome ([59] and see Ref. [60]). The presence of DNA-RNA hybrid duplexes is confirmed by sensitivity to RNase H, which specifically degrades RNA in DNA-RNA hybrid duplexes [61]. Studies using these new tools have shown that R-loops are abundant in the genomes of human [59] and plant (See Ref. [60]) cells, and have revealed that R-loops, rather than acting as lesions to destabilize genomes, are involved in gene regulation for various genetic and developmental functions. Most R-loops are thought to be formed co-transcriptionally (in association with transcription; see Ref. [62] for review), but it has recently been shown that some R-loops are formed independent of transcription. This type of R-loop formation is called R-loop formation in trans [59].
Studies on a long non-coding RNA (lncRNA) called APOLO (AUXIN-REGULATED PROMOTER LOOP) in Arabidopsis thaliana have shown that this lncRNA forms R-loops in trans at multiple distant-target genetic loci for lateral root development [63]. Seeking a second, independent approach to obtain experimental support, the authors of this study introduced, in addition to DRIP, a new DRIP-independent protocol called RNA isolation by DNA purification (RIDP), which uses biotinylated DNA probes to isolate the DNA of specified loci. APOLO-lncRNA transcription is activated by a phytohormone, auxin, during lateral root development. APOLO-lncRNA-mediated R-loop formation modulates chromatin loops at the target loci (at least 200 loci). R-loop formation at each target locus occurs at a pair of target sequences that are distant on the DNA but in close proximity via the chromatin loop, resulting in decrease of LHP1 (plant Polycomb Repressive Complex 1 component), decrease in repressive mark (histone methylation [H3K27me3]), opening of the chromatin loop, and activation of transcription at the target locus. The invasion of the target site by APOLO lncRNA can thus coordinate the transcription of spatially non-associated auxin-responsive genes [63]. APOLO lncRNA forms R-loops at regions containing a consensus sequence, GAAGAA(G/C), which is required for R-loop formation [63]. At present, it is not known which protein catalyzes R-loop formation with APOLO lncRNA.
4.2. R-loop formation by the crRNA-Cas-effector complex in CRISPR-Cas immune systems
CRISPR (clusters of regularly interspaced short palindromic repeats)-Cas (CRISPR-associated) systems are adaptive immune systems that act against invading viruses and plasmids found in prokaryotes and archaea. When DNA viruses and plasmids invade the cell, their DNA is processed into short DNA sequences, which are integrated in the form of spacer sequences in the CRISPR loci as a memory of the infection. crRNA (CRISPR RNA) is a processed transcript of CRISPR (for example, 61 nucleotides for Escherichia coli) that binds to Cas protein(s) to form a crRNA-Cas-effector complex in which the crRNA works as a guide RNA that recruits the effector complex to the homologous sequence (called a “protospacer”) of its target dsDNA by forming an R-loop. This R-loop formation results in the degradation of the foreign DNA. Two crRNA-Cas-effector complexes that have been extensively studied are: Cascade (a CRISPR-associated complex for antiviral defense) in E. coli and other eubacteria (Class 1, a multi-effector protein complex) and the effector complexes of Cas9 (Class 2, a single effector protein), which is used in gene-editing technology. The details and classification of CRISPR-Cas systems have been reviewed multiple times [64], [65], [66].
crRNA-Cas-effector complexes (containing guide RNA) first recognize a PAM (protospacer adjacent motif) sequence in target DNA by protein-DNA interaction that does not involve guide RNA. R-loops are then formed to complete target recognition, wherein the spacer sequence of the guide RNA and the complementary sequence (target strand) of the protospacer sequence form a hybrid duplex. PAM comprises a 2 to 3 bp sequence adjacent to either the 5′ end (Cascade, Fig. 1D) or the 3′ end (Cas9) of the non-target strand of the protospacer in the target dsDNA. This R-loop formation induces the cleavage of the target DNA.
5. Mechanisms of homologous-complex formation with ssDNA/ssRNA
5.1. RecA/Rad51-family ATP-dependent homologous-pairing proteins
RecA/Rad51-family proteins share, with a minor difference, a common mechanism for D-loop formation. Most research on the reaction steps of D-loop formation has focused on RecA. In D-loop formation, RecA first binds to ssDNA, even in the presence of dsDNA [67], [68], then forms a right-handed spiral filament around the DNA at a rate of 6 RecA protomers per turn [42] (Fig. 3, A and B). In the presence of ATP or of a nonhydrolyzable ATP analogue, ssDNA in the filament is extended 1.5 fold longer than B-form dsDNA with the same base sequence (active filament [69]). The 3D structure of the RecA/Rad51-DNA active filaments in the presence of a nonhydrolyzable ATP-analogue has been determined by X-ray crystallography [42] (Fig. 3, A and B) and by high-resolution cryo-electron microscopy [70], [71]. The active RecA-ssDNA filament (extended filament [42], [69]) structure shows that ssDNA binds to the primary DNA-binding site that contains the folded L1 and L2 loop regions (Fig. 3C). The L1 and L2 loops are disordered in the inactive RecA-filament (in the absence of DNA and the ATP analogue [72]). In the active RecA/Rad51-DNA complex, the extension of ssDNA is not uniform. The RecA protomer binds stoichiometrically to the ssDNA, with one protomer to three nucleotides, and pairs of folded L2 loops of all adjacent RecA protomers sandwich 3 nucleotides (nucleotide triplet) of DNA (Fig. 3C), in which the base distance is slightly larger than that of B-form DNA (3.83 to 4.66 Å vs. 3.5 Å). The base distance between triplets is further extended (7.8 Å) by the L2 loop (Fig. 3C). Cryo-electron microscopic observation has revealed the similar structure of the ssDNA and the L2 loop in the active Rad51-ssDNA filament [70].
By binding to ssDNA in the presence of ATP or a nonhydrolyzable ATP-analogue, RecA is activated for sequence-independent binding to dsDNA [67]. The RecA-ssDNA filament then slides along the dsDNA [39] to search for a homologous sequence between the ssDNA in the RecA-filament and the dsDNA. Once a homologous sequence is found, a homologous complex-core (nascent homologous complex) is formed [67], without ATP-hydrolysis [38]. The binding sites of ATP are located between the adjacent RecA protomers, and ATP-binding to the RecA-ssDNA filament modulates the spatial configuration of the adjacent RecA protomer. This spatial configuration change results in the extension of the filament associated with the extension of the bound ssDNA (compare [42] with [72]) as well as the activation of the dsDNA binding to the filament. The binding surface of dsDNA, consisting of several amino-acid residues (called the gateway), is located on the C-terminal domain of RecA and at the boundaries between adjacent RecA protomers on the extended RecA filament ([73], and refs. cited) (Fig. 3).
The smallest detectable homologous complex formed by RecA/Rad51 contains a 6–8 bp hybrid duplex [40], [74], [75], and the smallest stable homologous complex contains a 15 bp hybrid duplex [40]. Alongside the nucleotide triplets in the RecA/Rad51-ssDNA filament, the hybrid duplex is elongated from 6 - 8 bps to 15 bps in 3 bp increments, in the absence of ATP-hydrolysis [39], [40].
The nascent homologous complex could be a homologous triplex [6] (See 2.2). The hybrid duplex in the nascent complex is spontaneously elongated in association with D-loop growth by the rotational stress generated by the negative supercoils of the substrate ccc-dsDNA (See Refs. [43], [76]), or by an ATP-hydrolysis-dependent function of RecA [77].
The gateway leads the bound dsDNA to the L2 loop (Fig. 3, B and C), where the dsDNA encounters ssDNA in the RecA filament. Homologous-sequence recognition between dsDNA and ssDNA then takes place in the vicinity of the L2 loop. A recent study revealed that the active unit for ATP-dependent homologous pairing is a pair of adjacent RecA protomers and that filament formation is not essential to this process [43]. ATP-hydrolysis-dependent branch migration depends on filament formation (See Ref. [78] for review).
5.2. Role of RecA/Rad51 in homologous complex formation
The primary model for homologous-sequence recognition is the melting of dsDNA at the gateway to make the dsDNA bases available to form W-C base pairs with ssDNA. Melting induces untwisting of the double helix. Several studies have described the ssDNA and ATP or ATP-analogue-dependent untwisting of dsDNA by RecA under conditions for homologous-complex formation. Untwisting of dsDNA bound to RecA-heterologous ssDNA filament had been observed in the presence of an nonhydrolyzable analogue, supporting this melting-before-annealing model [79], but the later studies in the presence of ATP revealed that the untwisting of dsDNA bound to RecA-ssDNA filaments requires homology between dsDNA and the ssDNA in the RecA-filaments [23], [25], supporting homologous-sequence recognition in the absence of the melting of dsDNA. Since RecA, ssDNA, and dsDNA form a huge network of molecules with an irregular shape under these conditions [80], the individual RecA-DNA filaments had not been identified by either electron microscopy or crystallographic analysis.
Although the initial binding takes a longer time [81] by using the gateway [82], RecA can bind to dsDNA in the absence of ssDNA (in the presence of ATP). Electron micrographs taken under conditions for dsDNA-untwisting in the absence of ssDNA by RecA show well-ordered filaments [30], [41]. The crystal structure of RecA-dsDNA filament formed in the presence of an ATP-analogue is almost identical to that of active RecA-ssDNA filament, and dsDNA bound at the center of RecA-dsDNA filaments at the primary DNA-binding sites is untwisted but conserves base pairing. No base pair is disrupted in the filament [42]. Note that the dsDNA untwisting so far observed does not require ATP-hydrolysis, and it is induced by binding to the primary DNA-binding site in the extended RecA-filament induced by ATP-binding.
The features of dsDNA bound to the gateway or RecA-ssDNA spiral filament had not been revealed until recently. Yang et al. solved by use of cryo-electron microscope, high resolution 3D structures of active RecA-ssDNA filaments consisting of nine protomers and dsDNA bound to gateway [83]. They analyzed the complexes formed by the RecA-ssDNA and heterologous dsDNA and by RecA-ssDNA and homologous dsDNA. Based on these structures, the authors proposed a mechanistic model for D-loop formation: dsDNA bound to the gateways of the RecA-ssDNA spiral filament encounters an L2 loop, and this interaction causes the local melting of the double helix (up to 15 bps or more), followed by annealing with the ssDNA at the center [83]. This model appears to be well supported by various 3D structures revealed by their analysis, but does not explain why DNA, but not RNA, can form homologous complex with ssDNA or ssRNA. Note that Yang collected the cryo-electron microscopic images in the presence of a nonhydrolyzable ATP-analogue, the condition where the untwist of dsDNA by RecA bound to heterologous ssDNA had been observed [79] (For discussion, see above and below).
The sole model that explains why only DNA can form homologous complexes was proposed based on the unique Nuclear Magnetic Resonance (NMR) 3D structure of ssDNA oligomers bound to homologous-complex-forming proteins. The NMR analysis (transfer NOE) has revealed the unique, ATP-analogue-dependent 3D structure of extended ssDNA oligomers induced by transient binding to RecA/Rad51 (Fig. 4A). In this structure, the distance between adjacent bases is extended 1.5 fold of B-form DNA (Fig. 4B). B-form DNA is stabilized by stacking between adjacent bases. Instead of base-stacking, the extended ssDNA structure is stabilized by CH-π (CH-pi) interaction, an attractive molecular force [84], between the CH2 moiety at the 2′ position on the deoxyribose and the base of the following nucleotide [85], [86] (Fig. 4A). A similar extended ssDNA structure was induced also by all tested proteins (RecT, Mhr1, and RecO) that catalyze homologous-complex formation between dsDNA and ssDNA in an ATP-independent fashion [87]. Thus, the extended DNA structure stabilized by CH-π interaction is a general intermediate of the homologous-complex formation of dsDNA and ssDNA. Considering the variation in the 3D structure of homologous-complex forming proteins, the DNA structure stabilized by CH-π interaction in the general intermediate suggests that the homologous complex formation is an intrinsic DNA function rather than a function of specific proteins [87], [88].
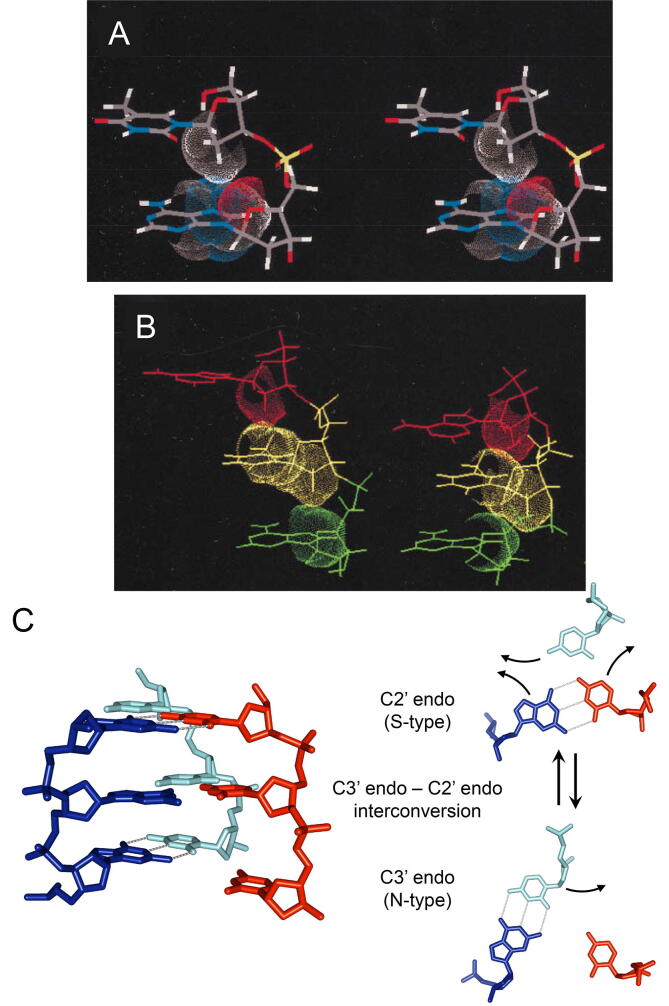
Fig. 4.
Extended DNA structure induced by binding to RecA/Rad51 (ATP-dependent) and ATP-independent homologous-pairing proteins, and a model for homology recognition by RecA/Rad51. A. Stereo view of extended ssDNA structure stabilized by CH-π interaction, an attractive molecular force [84], upon binding to RecA and Rad51 in the presence of an ATP-analogue [85], [86] and to ATP-independent homologous-pairing proteins [87]. Van der Waals spheres of a 2′-methylene moiety in a 5′ side residue (thymine) and of the base in the 3′ side residue (adenine) are indicated, and the contact of these surfaces represent CH-π interaction. B. Side views of extended DNA by binding to RecA (left) and B-form DNA (right) for a comparison. Van der Waals spheres of adjacent residues are indicated. In the extended DNA (left), in stead of base-base stacking, CH-π interaction stabilizes the distance between the planes of the adjacent bases (or axial rise per base) at nearly 5 Å, approximately 1.5 fold larger than that (3.5 Å) of B-form DNA (right), in which the structure is stabilized by base-base stacking. C. Model for homology search and W-C base-pairing between dsDNA extended by CH-π interaction and ssDNA (extended by binding to RecA). The 2D model (on the right) is the view from the bottom of the 3D model (on the left). Bases of dsDNA (dark and light blue, bottom) are randomly flipped out by the interconversion of sugar puckers from the C3′ endo to the C2′ endo (middle). An ssDNA (red) approaches the minor groove of the dsDNA (bottom). Only if the flipped base (dark blue) is complementary to the base of the ssDNA, the base-pair switch occurs (top) and the replace base (light blue) is in the major groove of the hybrid base pair that is newly formed. When this base-pair switch takes place between complementary sequences of dsDNA and ssDNA, homologous recognition and homologous-complex-core formation are accomplished simultaneously, and the replaced strand is left in the major groove of the newly formed hybrid duplex (See Fig. 2A). Panels A and B were reproduced with minor modification from Ref. [85], with permission (Copyright 1997, National Academy of Sciences, U.S.A.). Panel C is the reproduction of the Fig. 4 in Ref. [86], with permission (Copyright 1998, National Academy of Sciences, U.S.A.), generated using the coordinates of PDB code 1I1V (Model Archive DOI: https://doi.org//10.2210/pdb1I1V/pdb [86]). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Models of extended dsDNA stabilized by CH-π interaction were constructed [86]. In normal dsDNA, base pairs randomly opens and the mean lifetime of W-C base pairs at 35 °C is in the range of milliseconds [89]. In the extended dsDNA, dsDNA bases in base pairs opened can be flipped out spontaneously by the interconversion of sugar puckers, and can thus interact with the bases of nearby ssDNA. This interaction explains the mechanism for homologous-sequence recognition between dsDNA and ssDNA without the melting of the double helix [86], [88] (Fig. 4C). The CH-π interaction between a sugar and a base is unique to DNA and does not exist in RNA, and thus, the interaction explains the unique ability of dsDNA to form homologous complexes. In Fig. 4C, both strands of dsDNA are assumed to be elongated, but when the CH-π interaction occurs in one of the two strands, it results in DNA bending [90]. Thus, this model works with bent dsDNA. Lipfert el al. found that the characteristic transition rate of RNA during plectonemic buckling is two orders of magnitude slower than that of dsDNA [91]. This finding suggests that dsDNA has a high enough flexibility to allow sharp local bending of dsDNA (on a scale of approximately 5 nm in length). The two orders of magnitude difference between dsDNA and dsRNA also explain the absence of homologous-complex formation by dsRNA. The cryo-electron microscopic study published by Yang shows a 3D structure that appears RecA-ssDNA filament that forms complex with a heterologous bent dsDNA [83].
The absence of active melting of dsDNA by RecA in nascent homologous-complex formation is supported by kinetic analysis of homologous-complex formation by RecA [92]. Using oligo DNA labeled with fluorescent dyes, Xiao et al found that nascent homologous-complex formation from dsDNA and extended ssDNA on an active RecA-ssDNA filament was driven entropically rather than enthalpically [92]. Their results support the conclusion that RecA-catalyzed homologous-complex formation relies on dynamic intrinsic DNA functions [88], [92].
5.3. Uncatalyzed homologous-complex formation
A recent magnetic-tweezers experiment revealed that the mechanical stretching of ssDNA induces homologous-triplex formation with homologous dsDNA, not with heterologous dsDNA, at room temperature in the absence of any protein. The ssDNA in the homologous complex was shown to be sensitive to a dsDNA-specific endonuclease, indicating that ssDNA forms a hybrid duplex with the complementary strand of dsDNA [93]. This finding shows that the active melting of dsDNA is not required for homologous-complex formation, and supports a view that homologous complex formation is an intrinsic DNA function.
Regarding the uncatalyzed homologous-triplex formation by mechanically stretched ssDNA and dsDNA described above [93], we can speculate that RecA/Rad51 plays a critical role in homologous-complex formation via the extension of ssDNA and the holding of dsDNA at a location suitable for homology-search interaction with the extended ssDNA. The homologous sequence in the extended ssDNA may be recognized by dsDNA through base-flipping enabled by transient CH-π interaction-dependent dsDNA extension [86], [88] and/or bending [90].
5.4. Role played by the crRNA-Cas-effector complex in R-loop formation
Single-molecule analysis using rotor-bead tracking (RBT) or an equivalent technique has shown that R-loops are formed within 100 ms by Cas9 [94]. R-loop formation by Cascade is faster than by Cas9, and the R-loops formed by Cascade are more stable than those formed by Cas9 [95]. R-loop formation involves transient discrete intermediates, including initial DNA-RNA hybridization within the seed region (11 bps; Figs. 1D and 5A) of the protospacer sequence adjacent to the PAM sequence of the target DNA [94].
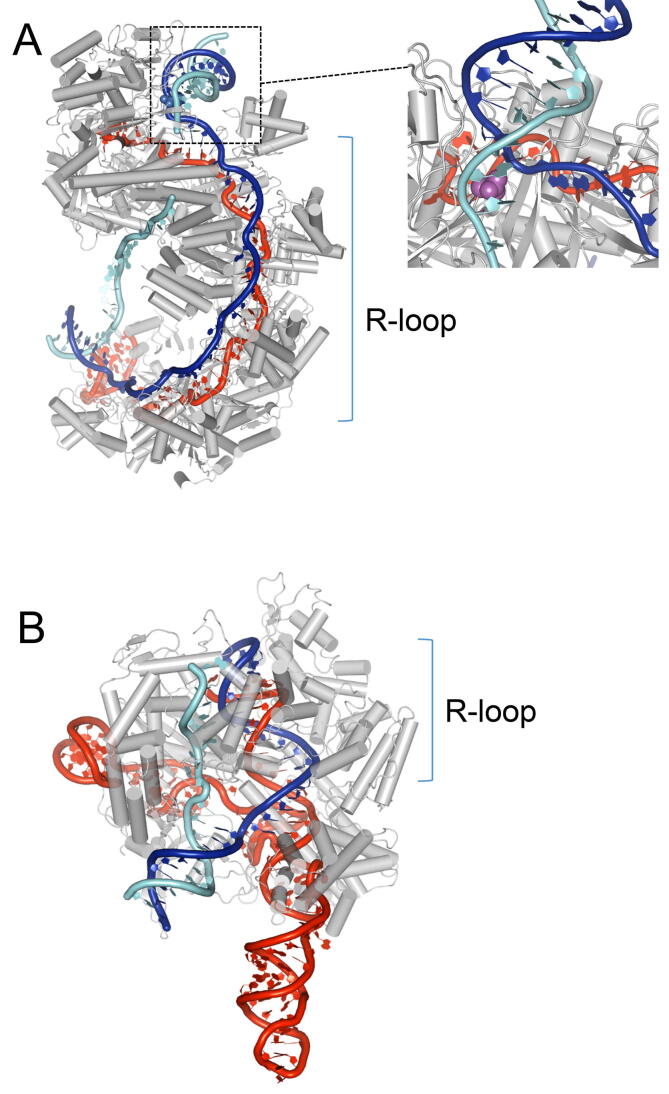
Fig. 5.
Molecular structure of R-loop formation by crRNA-Cas-effector complexes. crRNA, target DNA strand and non-target DNA strand are shown in red, dark blue and light blue, respectively. A. Cryo-electron microscopic 3D structures of crRNA Cascade (Class 1) that contains a full R-loop formed between the crRNA and target dsDNA (PDB code 5U0A [99]). The wedge inserted at the point of strand separation of parental dsDNA is represented by magenta spheres. B. Crystal structure of crRNA Cas9 (Class 2) complex that contains an R-loop of the crRNA and target dsDNA (PDB code 5F9R [101]). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The need for high-resolution imaging of the 3D structures of crRNA-Cas-effector complexes with or without target dsDNA has been met by X-ray crystallography (Cascade [Class 1, Type I] [96], [97], RNA-silencing Cmr complex [Class 1, Type III] [98]), cryo-electron microscopy (Cascade [99], [100]), and by both techniques together (Cas9 [Class 2, Type II] [101]). These techniques all reveal differences in R-loop structure between class 1 (Cascade and RNA-silencing Cmr complex) crRNA-Cas-effector complexes and class 2 (Cas9) crRNA-Cas-effector complexes, as well as common unique 3D structures that point to the recognition mechanism of the protospacer sequence in the target dsDNA.
The 3 bp PAM sequence of target DNA is recognized by interaction with a subunit of Cascade. The PAM-adjacent protospacer region of the dsDNA is then bent approximately 90° and the two strands are separated. At this point, a wedge is inserted to disrupt the first base pair and to help flip out the first several bases from both DNA strands [99], [100] (Fig. 5A). Thus, PAM recognition leads to the bending of dsDNA, resulting in the spontaneous melting of the dsDNA, which is captured by the crRNA to form the seed bubble. The seed bubble is elongated along a crRNA guide in 6 base increments to form a full R-loop [97], [99].
In the R-loops formed by the Class 1 Cas-effector complex (Cascade and RNA silencing Cmr complex), the crRNA hybrid duplex and the target strand of the protospacer region of the target DNA are not a canonical double helix, but rather an extensively untwisted ladder-like structure. The R-loop consists of 5 segments, each including a 5 bp region followed by a disrupted base pair [97], [99], [100] (Figs. 1D and 5A). Every sixth unpaired nucleotide of crRNA binds tightly to one of the 6 well-organized Cas7 protein subunits of the cascade along the crRNA [96]. The unfolded crRNA structure is not changed before or after R-loop formation [100].
Upon recognition of a PAM sequence in dsDNA, Cas9 (Class 2, Type II) also bends dsDNA, providing enough stress to melt the dsDNA and stimulate R-loop formation with crRNA, though the bend is only about 30° [99], [101] (Fig. 5B). Cas9 has no wedge structure, showing that the wedge found in Cascade is not a general structure required to melt dsDNA. In dsDNA bound to Cas9, the non-target strand of the protospacer dsDNA adjacent to a PAM sequence is held separate from the crRNA by extensive interaction with the protein [101] (Fig. 5B). The R-loop formed by Cas9 is a continuous 20 bp A-form DNA-RNA-hybrid duplex [101], in contrast to the segmented ladder-like structure formed by Cascade.
6. Summary and perspectives
It has been suggested as described above that, in R-loop formation by crRNA-Cas-effector complexes, proteins plays an active role in dsDNA melting in the seed region for hybrid-duplex formation with crRNA.
In homologous-complex formation by RecA, on the other hand, the interaction between DNA and the protein is crucial for the extension of ssDNA. D-loop formation by the extended RecA-ssDNA filament and dsDNA is entropy-driven rather than enthalpy-driven, suggesting that homology recognition occurs without melting of the double helix by RecA activity [92], as is the case with uncatalyzed homologous-complex formation of dsDNA and mechanically stretched ssDNA [93]. Thus, once the ssDNA is sufficiently extended by RecA, spontaneous interaction with the dsDNA transiently and weakly captured by the protein is sufficient to initiate hybrid-duplex formation. Local bending of dsDNA upon encountering ssDNA could contribute to the initiation of hybrid-duplex formation. The model contains CH-π interaction between deoxy-sugar and base (Fig. 4) explains homologous-complex formation in the absence of preceding dsDNA melting and the DNA-specific ability of homologous-complex formation.
This raises the intriguing question of whether recognition of homologous sequence between ssDNA/ssRNA and dsDNA without the preceding melting of dsDNA double helix is a general principle for homologous complex formation. This possibility has not been excluded as a mechanism for R-loop formation by crRNA-Cas-effector complexes.
Currently available analytical techniques are not sufficient to test and evaluate the various hypotheses for homology recognition and homologous-complex formation between ssDNA and dsDNA before the melting of the double helix. Such hypotheses include the contribution of base-flipping by bending and/or by transient extension of dsDNA, the role played by CH-π interaction, and the contribution of RecA/Rad51. Current techniques are limited to X-ray crystallography, high resolution cryo-electron microscopy, NMR analyses and single molecule analysis, and these techniques do not have sufficient time- or spatial-resolution, or direct information about the order of events and time elapsed in a reaction sequence. On the other hand, molecular dynamic simulation by computational means has enormous potential as a more effective way to evaluate these hypotheses. It should be possible to simulate homology recognition on a larger time scale, since the rate of spontaneous base-flipping depends on the rate of thermal W-C base pair opening and the mean lifetime of W-C base pairs at 35 °C is in the range of milliseconds [89]. This will require significant development in the field of computer simulation well beyond existing methods.
References
- 1.Tanaka Y., Fujii S., Hiroaki H., Sakata T., Tanaka T., Uesugi S. A'-form RNA double helix in the single crystal structure of r(UGAGCUUCGGCUC) Nucl Acids Res. 1999;27:949–955. doi: 10.1093/nar/27.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Y., Breaker R.R. Deoxyribozymes: new players in the ancient game of biocatalysis. Curr Opin Struct Biol. 1999;9(3):315–323. doi: 10.1016/S0959-440X(99)80042-6. [DOI] [PubMed] [Google Scholar]
- 3.Holloman W.K., Wiegand R., Hoessli C., Radding C.M. Uptake of homologous single-stranded fragments by superhelical DNA: a possible mechanism for initiation of genetic recombination. Proc Natl Acad Sci. 1975;72(6):2394–2398. doi: 10.1073/pnas.72.6.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu L.F., Wang J.C. On the degree of unwinding of the DNA helix by ethidium. Biochim Biophys Acta. 1975;395(4):405–412. doi: 10.1016/0005-2787(75)90064-7. [DOI] [PubMed] [Google Scholar]
- 5.Thomas M., White R.L., Davis R.W. Hybridization of RNA to double-stranded DNA: formation of R-loops. Proc Natl Acad Sci. 1976;73(7):2294–2298. doi: 10.1073/pnas.73.7.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rao B.J., Chiu S.K., Radding C.M. Homologous recognition and triplex formation promoted by RecA protein between duplex oligonucleotides and single-stranded DNA. J Mol Biol. 1993;229:328–343. doi: 10.1006/jmbi.1993.1038. [DOI] [PubMed] [Google Scholar]
- 7.Gutbrod M.J., Martienssen R.A. Conserved chromosomal functions of RNA interference. Nat Rev Genet. 2020;21(5):311–331. doi: 10.1038/s41576-019-0203-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitehouse H.L.K. A theory of crossing-over by means of hybrid deoxyribonucleic acid. Nature. 1963;199(4898):1034–1040. doi: 10.1038/1991034a0. [DOI] [PubMed] [Google Scholar]
- 9.Holliday R. A mechanism for gene conversion in fungi. Genetic Res Camb. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 10.Shibata T., DasGupta C., Cunningham R.P., Radding C.M. Purified Escherichia coli recA protein catalyzes homologous pairing of superhelical DNA and single-stranded fragments. Proc Natl Acad Sci. 1979;76(4):1638–1642. doi: 10.1073/pnas.76.4.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McEntee K., Weinstock G.M., Lehman I.R. Initiation of general recombination catalyzed in vitro by the recA protein of Escherichia coli. Proc Natl Acad Sci. 1979;76(6):2615–2619. doi: 10.1073/pnas.76.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265(5176):1241–1243. doi: 10.1126/science:8066464. [DOI] [PubMed] [Google Scholar]
- 13.Clark A.J., Margulies A.D. Isolation and characterization of recombination-deficient mutants of Escherichia coli K12. Proc Natl Acad Sci. 1965;53(2):451–459. doi: 10.1073/pnas.53.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakai S., Matsumoto S. Two types of radiation-sensitive mutants in yeast. Mutat Res. 1967;4:129–136. doi: 10.1016/0027-5107(67)90064-4. [DOI] [PubMed] [Google Scholar]
- 15.Sonoda E., Sasaki M.S., Buerstedde J.-M., Bezzubova O., Shinohara A., Ogawa H. Rad51 deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szostak J.W., Orr-Weaver T.L., Rothstein R.J., Stahl F.W. The double-strand-break repair model for recombination. Cell. 1983;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 17.Goddard M.R., Godfray H.C.J., Burt A. Sex increases the efficacy of natural selection in experimental yeast populations. Nature. 2005;434(7033):636–640. doi: 10.1038/nature03405. [DOI] [PubMed] [Google Scholar]
- 18.San Filippo J., Sung P., Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77(1):229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 19.Simon-Loriere E., Holmes E.C. Why do RNA viruses recombine? Nat Rev Microbiol. 2011;9(8):617–626. doi: 10.1038/nrmicro2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiao X., Qiao J., Mindich L. Anin vitrosystem for the investigation of heterologous RNA recombination. Virology. 1997;227(1):103–110. doi: 10.1006/viro.1996.8311. [DOI] [PubMed] [Google Scholar]
- 21.Roberts R., Crothers D. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science. 1992;258(5087):1463–1466. doi: 10.1126/science:1279808. [DOI] [PubMed] [Google Scholar]
- 22.Allison DF, Wang GG. R-loops: formation, function, and relevance to cell stress. Cell Stress 3; 2019: 38–46. [DOI] [PMC free article] [PubMed]
- 23.Ohtani T., Shibata T., Iwabuchi M.-A., Watabe H.-O., Iino T., Ando T. ATP-dependent unwinding of double helix in closed circular DNA by RecA protein of E. coli. Nature. 1982;299(5878):86–89. doi: 10.1038/299086a0. [DOI] [PubMed] [Google Scholar]
- 24.West S.C., Cassuto E., Howard-Flanders P. Homologous pairing can occur before DNA strand separation in general genetic recombination. Nature. 1981;290(5801):29–33. doi: 10.1038/290029a0. [DOI] [PubMed] [Google Scholar]
- 25.Wu A.M., Bianchi M., DasGupta C., Radding C.M. Unwinding associated with synapsis of DNA molecules by recA protein. Proc Natl Acad Sci. 1983;80(5):1256–1260. doi: 10.1073/pnas.80.5.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shibata T., DasGupta C., Cunningham R.P., Williams J.G.K., Osber L., Radding C.M. Homologous pairing in genetic recombination. The pairing reaction catalyzed by Escherichia coli recA protein. J Biol Chem. 1981;256:7565–7572. [PubMed] [Google Scholar]
- 27.Wiegand R.C., Beattie K.L., Holloman W.K., Radding C.M. Uptake of homologous single-stranded fragments by superhelical DNA. III. The product and its enzymic conversion to a recombinant molecule. J Mol Biol. 1977;116:805–824. doi: 10.1016/0022-2836(77)90272-8. [DOI] [PubMed] [Google Scholar]
- 28.Radding C.M., Beattie K.L., Holloman W.K., Wiegand R.C. Uptake of homologous single-stranded fragments by superhelical DNA. IV. Branch migration. J Mol Biol. 1977;116:825–839. doi: 10.1016/0022-2836(77)90273-x. [DOI] [PubMed] [Google Scholar]
- 29.Ling F., Yoshida M., Shibata T. Heteroduplex joint formation free of net topological change by Mhr1, a mitochondrial recombinase. J Biol Chem. 2009;284(14):9341–9353. doi: 10.1074/jbc.M900023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stasiak A., Di Capua E. The helicity of DNA in complexes with RecA protein. Nature. 1982;299(5879):185–186. doi: 10.1038/299185a0. [DOI] [PubMed] [Google Scholar]
- 31.Amiard S., Doudeau M., Pinte S., Poulet A., Lenain C., Faivre-Moskalenko C., Angelov D., Hug N., Vindigni A., Bouvet P., Paoletti J., Gilson E., Giraud-Panis M.-J. A topological mechanism for TRF2-enhanced strand invasion. Nat Struct Mol Biol. 2007;14(2):147–154. doi: 10.1038/nsmb1192. [DOI] [PubMed] [Google Scholar]
- 32.Vlieghe D., Vanmeervelt L., Dautant A., Gallois B., Precigoux G., Kennard O. Parallel and antiparallel (G.GC)2 triple helix fragments in a crystal structure. Science. 1996;273:1702–1705. doi: 10.1126/science.273.5282.1702. [DOI] [PubMed] [Google Scholar]
- 33.Buske F.A., Mattick J.S., Bailey T.L. Potential in vivo roles of nucleic acid triple-helices. RNA Biol. 2011;8(3):427–439. doi: 10.4161/rna.8.3.14999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Z., Giles K.E., Felsenfeld G. DNA.RNA triple helix formation can function as a cis-acting regulatory mechanism at the human beta-globin locus. Proc Natl Acad Sci USA. 2019;116:6130–6139. doi: 10.1073/pnas.1900107116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shinohara A., Ogawa H., Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69(3):457–470. doi: 10.1016/0092-8674(92)90447-K. [DOI] [PubMed] [Google Scholar]
- 36.Seitz E.M., Brockman J.P., Sandler S.J., Clark A.J., Kowalczykowski S.C. RadA protein is an archaeal RecA protein homolog that catalyzes DNA strand exchange. Genes Dev. 1998;12:1248–1253. doi: 10.1101/gad.12.9.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bishop D.K., Park D., Xu L., Kleckner N. DMC1: A meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69(3):439–456. doi: 10.1016/0092-8674(92)90446-J. [DOI] [PubMed] [Google Scholar]
- 38.Cox M.M., Lehman I.R. recA protein of Escherichia coli promotes branch migration, a kinetically distinct phase of DNA strand exchange. Proc Natl Acad Sci. 1981;78(6):3433–3437. doi: 10.1073/pnas.78.6.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ragunathan K., Joo C., Ha T. Real-time observation of strand exchange reaction with high spatiotemporal resolution. Structure. 2011;19(8):1064–1073. doi: 10.1016/j.str.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi Z., Redding S.y., Lee J., Gibb B., Kwon Y., Niu H., Gaines W., Sung P., Greene E. DNA sequence alignment by microhomology sampling during homologous recombination. Cell. 2015;160(5):856–869. doi: 10.1016/j.cell.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dunn K., Chrysogelos S., Griffith J. Electron microscopic visualization of RecA-DNA filaments: evidence for a cyclic extension of duplex DNA. Cell. 1982;28(4):757–765. doi: 10.1016/0092-8674(82)90055-1. [DOI] [PubMed] [Google Scholar]
- 42.Chen Z., Yang H., Pavletich N.P. Mechanism of homologous recombination from the RecA–ssDNA/dsDNA structures. Nature. 2008;453(7194):489–494. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- 43.Yamaguchi Y, Suzuki-Nagata K, Shibata T, Mikawa T. Nonfilament-forming RecA dimer catalyzes homologous joint formation. Nucl Acids Res 46; 2018: 10855–10869. [DOI] [PMC free article] [PubMed]
- 44.Sawaya M.R., Guo S., Tabor S., Richardson C.C., Ellenberger T. Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7. Cell. 1999;99(2):167–177. doi: 10.1016/S0092-8674(00)81648-7. [DOI] [PubMed] [Google Scholar]
- 45.Yu X., Egelman E.H. The RecA hexamer is a structural homologue of ring helicases. Nat Struct Mol Biol. 1997;4(2):101–104. doi: 10.1038/nsb0297-101. [DOI] [PubMed] [Google Scholar]
- 46.Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi W., Takaku M., Machida S., Tachiwana H., Maehara K., Ohkawa Y., Kurumizaka H. Chromatin architecture may dictate the target site for DMC1, but not for RAD51, during homologous pairing. Sci Rep. 2016;6(1) doi: 10.1038/srep24228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alexeev A., Mazin A., Kowalczykowski S.C. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51–ssDNA nucleoprotein filament. Nat Struct Mol Biol. 2003;10(3):182–186. doi: 10.1038/nsb901. [DOI] [PubMed] [Google Scholar]
- 49.Machida S., Takaku M., Ikura M., Sun J., Suzuki H., Kobayashi W., Kinomura A., Osakabe A., Tachiwana H., Horikoshi Y., Fukuto A., Matsuda R., Ura K., Tashiro S., Ikura T., Kurumizaka H. Nap1 stimulates homologous recombination by RAD51 and RAD54 in higher-ordered chromatin containing histone H1. Sci Rep. 2015;4(1) doi: 10.1038/srep04863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ling F, Mikawa T, Shibata T. Enlightenment of yeast mitochondrial homoplasmy: diversified roles of gene conversion. Genes 2; 2011: 169–190. [DOI] [PMC free article] [PubMed]
- 51.Prasai K, Robinson LC, Scott RS, Tatchell K, Harrison L. Evidence for double-strand break mediated mitochondrial DNA replication in Saccharomyces cerevisiae. Nucl Acids Res; 2017. [DOI] [PMC free article] [PubMed]
- 52.Noirot P., Kolodner R.D. DNA strand invasion promoted by Escherichia coli RecT protein. J Biol Chem. 1998;273(20):12274–12280. doi: 10.1074/jbc.273.20.12274. [DOI] [PubMed] [Google Scholar]
- 53.Kagawa W., Kurumizaka H., Ishitani R., Fukai S., Nureki O., Shibata T., Yokoyama S. Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. Mol Cell. 2002;10(2):359–371. doi: 10.1016/S1097-2765(02)00587-7. [DOI] [PubMed] [Google Scholar]
- 54.Lopes A, Amarir-Bouhram J, Faure G, Petit MA, Guerois R. Detection of novel recombinases in bacteriophage genomes unveils Rad52, Rad51 and Gp2.5 remote homologs. Nucl Acids Res 38; 2010: 3952–3962. [DOI] [PMC free article] [PubMed]
- 55.Masuda T, Ling F, Shibata T, Mikawa T. Analysis of DNA-binding sites on Mhr1, a yeast mitochondrial ATP-independent homologous pairing protein. FEBS J 277; 2010: 1440–1452. [DOI] [PubMed]
- 56.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell 97; 1999: 503–514. [DOI] [PubMed]
- 57.Doksani Y., Wu J., de Lange T., Zhuang X. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013;155(2):345–356. doi: 10.1016/j.cell.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stansel R.M., de Lange T., Griffith J.D. T-loop assembly in vitro involves binding of TRF2 near the 3' telomeric overhang. EMBO J. 2001;20:5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nadel J., Athanasiadou R., Lemetre C., Wijetunga N.A., Ó Broin P., Sato H., Zhang Z., Jeddeloh J., Montagna C., Golden A., Seoighe C., Greally J.M. RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships. Epigenet Chromatin. 2015;8(1) doi: 10.1186/s13072-015-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu W., Li K., Li S., Hou Q., Zhang Y., Liu K., Sun Q. The R-loop atlas of Arabidopsis development and responses to environmental stimuli. Plant Cell. 2020;32(4):888–903. doi: 10.1105/tpc.19.00802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berkower I., Leis J., Hurwitz J. Isolation and characterization of an endonuclease from Escherichia coli specific for ribonucleic acid in ribonucleic acid-deoxyribonucleic acid hybrid structures. J Biol Chem. 1973;248:5914–5921. [PubMed] [Google Scholar]
- 62.Niehrs C., Luke B. Regulatory R-loops as facilitators of gene expression and genome stability. Nat Rev Mol Cell Biol. 2020;21(3):167–178. doi: 10.1038/s41580-019-0206-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ariel F., Lucero L., Christ A., Mammarella M.F., Jegu T., Veluchamy A., Mariappan K., Latrasse D., Blein T., Liu C., Benhamed M., Crespi M. R-loop mediated trans action of the APOLO long noncoding RNA. Mol Cell. 2020;77(5):1055–1065.e4. doi: 10.1016/j.molcel.2019.12.015. [DOI] [PubMed] [Google Scholar]
- 64.Mohanraju P., Makarova K.S., Zetsche B., Zhang F., Koonin E.V., van der Oost J. Diverse evolutionary roots and mechanistic variations of the CRISPR-cas systems. Science. 2016;353(6299):aad5147. doi: 10.1126/science:aad5147. [DOI] [PubMed] [Google Scholar]
- 65.Jiang F., Doudna J.A. CRISPR–Cas9 structures and mechanisms. Annu Rev Biophys. 2017;46(1):505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 66.Hille F., Richter H., Wong S.P., Bratovič M., Ressel S., Charpentier E. The biology of CRISPR-cas: backward and forward. Cell. 2018;172(6):1239–1259. doi: 10.1016/j.cell.2017.11.032. [DOI] [PubMed] [Google Scholar]
- 67.Shibata T., Cunningham R.P., DasGupta C., Radding C.M. Homologous pairing in genetic recombination: complexes of recA protein and DNA. Proc Natl Acad Sci. 1979;76(10):5100–5104. doi: 10.1073/pnas.76.10.5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shinohara T, Ikawa S, Iwasaki W, Hiraki T, Hikima T, Mikawa T, Arai N, Kamiya N, Shibata T. Loop L1 governs the DNA-binding specificity and order for RecA-catalyzed reactions in homologous recombination and DNA repair. Nucl Acids Res 43; 2015: 973–986. [DOI] [PMC free article] [PubMed]
- 69.Flory J., Tsang S.S., Muniyappa K. Isolation and visualization of active presynaptic filaments of recA protein and single-stranded DNA. Proc Natl Acad Sci. 1984;81(22):7026–7030. doi: 10.1073/pnas.81.22.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Short J.M., Liu Y., Chen S., Soni N., Madhusudhan M.S., Shivji M.K. High-resolution structure of the presynaptic RAD51 filament on single-stranded DNA by electron cryo-microscopy. Nucleic Acids Res. 2016;44:9017–9030. doi: 10.1093/nar/gkw783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu J., Zhao L., Xu Y., Zhao W., Sung P., Wang H.-W. Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange. Nat Struct Mol Biol. 2017;24(1):40–46. doi: 10.1038/nsmb.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Story R.M., Weber I.T., Steitz T.A. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355(6358):318–325. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 73.Kurumizaka H., Ikawa S., Sarai A., Shibata T. The mutant RecA proteins, RecAR243Q and RecAK245N, exhibit defective DNA binding in homologous pairing. Arch Biochem Biophys. 1999;365(1):83–91. doi: 10.1006/abbi.1999.1166. [DOI] [PubMed] [Google Scholar]
- 74.Hsieh P., Camerini-Otero C.S., Camerini-Otero R.D. The synapsis event in the homologous pairing of DNAs: RecA recognizes and pairs less than one helical repeat of DNA. Proc Natl Acad Sci. 1992;89(14):6492–6496. doi: 10.1073/pnas.89.14.6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ragunathan K, Liu C, Ha T. RecA filament sliding on DNA facilitates homology search. eLife 1; 2012: e00067. [DOI] [PMC free article] [PubMed]
- 76.Beattie K.L., Wiegand R.C., Radding C.M. Uptake of homologous single-stranded fragments by superhelical DNA. II. Characterization of the reaction. J Mol Biol. 1977;116:783–803. doi: 10.1016/0022-2836(77)90271-6. [DOI] [PubMed] [Google Scholar]
- 77.Burnett B., Rao B.J., Jwang B., Reddy G., Radding C.M. Resolution of the three-stranded recombination intermediate made by RecA protein – an essential role of ATP hydrolysis. J Mol Biol. 1994;238:540–554. doi: 10.1006/jmbi.1994.1313. [DOI] [PubMed] [Google Scholar]
- 78.Radding C.M. Helical interactions in homologous pairing and strand exchange driven by recA protein. J Biol Chem. 1991;266:5355–5358. [PubMed] [Google Scholar]
- 79.Cunningham R.P., Shibata T., DasGupta C., Radding C.M. Single strands induce recA protein to unwind duplex DNA for homologous pairing. Nature. 1979;281(5728):191–195. doi: 10.1038/281191a0. [DOI] [PubMed] [Google Scholar]
- 80.Gonda D.K., Radding C.M. The mechanism of the search for homology promoted by recA protein. Facilitated diffusion within nucleoprotein networks. J Biol Chem. 1986;261:13087–13096. [PubMed] [Google Scholar]
- 81.Pugh B.F., Cox M.M. General mechanism for recA protein binding to duplex DNA. J Mol Biol. 1988;203:479–493. doi: 10.1016/0022-2836(88)90014-9. [DOI] [PubMed] [Google Scholar]
- 82.Kurumizaka H, Aihara H, Ikawa S, Shibata T. Specific defects in double-stranded DNA unwinding and homologous pairing of a mutant RecA protein. FEBS Lett 477; 2000: 129–134. [DOI] [PubMed]
- 83.Yang H., Zhou C., Dhar A., Pavletich N.P. Mechanism of strand exchange from RecA–DNA synaptic and D-loop structures. Nature. 2020;586(7831):801–806. doi: 10.1038/s41586-020-2820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nishio M. The CH/pi hydrogen bond in chemistry. Conformation, supramolecules, optical resolution and interactions involving carbohydrates. Phys Chem Chem Phys. 2011;13:13873–13900. doi: 10.1039/c1cp20404a. [DOI] [PubMed] [Google Scholar]
- 85.Nishinaka T., Ito Y., Yokoyama S., Shibata T. An extended DNA structure through deoxyribose-base stacking induced by RecA protein. Proc Natl Acad Sci. 1997;94(13):6623–6628. doi: 10.1073/pnas.94.13.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nishinaka T., Shinohara A., Ito Y., Yokoyama S., Shibata T. Base pair switching by interconversion of sugar puckers in DNA extended by proteins of RecA-family: a model for homology search in homologous genetic recombination. Proc Natl Acad Sci. 1998;95(19):11071–11076. doi: 10.1073/pnas.95.19.11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Masuda T., Ito Y., Terada T., Shibata T., Mikawa T. A non-canonical DNA structure enables homologous recombination in various genetic systems. J Biol Chem. 2009;284(44):30230–30239. doi: 10.1074/jbc.M109.043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shibata T., Nishinaka T., Mikawa T., Aihara H., Kurumizaka H., Yokoyama S., Ito Y. Homologous genetic recombination as an intrinsic dynamic property of a DNA structure induced by RecA/Rad51-family proteins: a possible advantage of DNA over RNA as genomic material. Proc Natl Acad Sci. 2001;98(15):8425–8432. doi: 10.1073/pnas.111005198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Leroy J.L., Kochoyan M., Huynh Dinh T., Gueron M. Characterization of base-pair opening in deoxynucleotide duplexes using catalyzed exchange of the imino proton. J Mol Biol. 1988;200:223–238. doi: 10.1016/0022-2836(88)90236-7. [DOI] [PubMed] [Google Scholar]
- 90.Umezawa Y., Nishio M. CH/pi interactions in the crystal structure of TATA-box binding protein/DNA complexes. Bioorg Med Chem. 2000;8:2643–2650. doi: 10.1016/s0968-0896(00)00197-8. [DOI] [PubMed] [Google Scholar]
- 91.Lipfert J., Skinner G.M., Keegstra J.M., Hensgens T., Jager T., Dulin D., Kober M., Yu Z., Donkers S.P., Chou F.-C., Das R., Dekker N.H. Double-stranded RNA under force and torque: Similarities to and striking differences from double-stranded DNA. Proc Natl Acad Sci. 2014;111(43):15408–15413. doi: 10.1073/pnas.1407197111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xiao J., Lee A.M., Singleton S.F. Direct evaluation of a kinetic model for RecA-mediated DNA-strand exchange: the importance of nucleic acid dynamics and entropy during homologous genetic recombination. ChemBioChem. 2006;7(8):1265–1278. doi: 10.1002/cbic.200600038. [DOI] [PubMed] [Google Scholar]
- 93.Chen J, Tang Q, Guo S, Lu C, Le S, Yan J. Parallel triplex structure formed between stretched single-stranded DNA and homologous duplex DNA. Nucl Acids Res 45; 2017: 10032–10041. [DOI] [PMC free article] [PubMed]
- 94.Ivanov I.E., Wright A.V., Cofsky J.C., Aris K.D.P., Doudna J.A., Bryant Z. Cas9 interrogates DNA in discrete steps modulated by mismatches and supercoiling. Proc Natl Acad Sci USA. 2020;117(11):5853–5860. doi: 10.1073/pnas.1913445117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Szczelkun M.D., Tikhomirova M.S., Sinkunas T., Gasiunas G., Karvelis T., Pschera P., Siksnys V., Seidel R. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci USA. 2014;111(27):9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jackson R.N., Golden S.M., van Erp P.B.G., Carter J., Westra E.R., Brouns S.J.J., van der Oost J., Terwilliger T.C., Read R.J., Wiedenheft B. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014;345(6203):1473–1479. doi: 10.1126/science:1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mulepati S., Heroux A., Bailey S. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science. 2014;345(6203):1479–1484. doi: 10.1126/science:1256996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Osawa T., Inanaga H., Sato C., Numata T. Crystal structure of the CRISPR-Cas RNA silencing Cmr complex bound to a target analog. Mol Cell. 2015;58(3):418–430. doi: 10.1016/j.molcel.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 99.Xiao Y., Luo M., Hayes R.P., Kim J., Ng S., Ding F., Liao M., Ke A. Structure basis for directional R-loop formation and substrate handover mechanisms in Type I CRISPR-cas system. Cell. 2017;170(1):48–60.e11. doi: 10.1016/j.cell.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Halpin-Healy T.S., Klompe S.E., Sternberg S.H., Fernández I.S. Structural basis of DNA targeting by a transposon-encoded CRISPR–cas system. Nature. 2020;577(7789):271–274. doi: 10.1038/s41586-019-1849-0. [DOI] [PubMed] [Google Scholar]
- 101.Jiang F., Taylor D.W., Chen J.S., Kornfeld J.E., Zhou K., Thompson A.J., Nogales E., Doudna J.A. Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science. 2016;351(6275):867–871. doi: 10.1126/science:aad8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gotfredsen C.H., Schultze P., Feigon J. Solution structure of an intramolecular pyrimidine−purine−pyrimidine triplex containing an RNA third strand. J Am Chem Soc. 1998;120(18):4281–4289. doi: 10.1021/ja973221m. [DOI] [Google Scholar]