Abstract
Objectives
Due to the number of asymptomatic infections and limited access to high-performance antibody tests, the true prevalence and seropositivity of SARS-CoV-2 infection remains unknown. To fill this gap, the clinical performance of a point-of-care SARS-CoV-2 Rapid Antibody Assay, a chromatographic immunoassay for detecting IgM/IgG antibodies, in near patient settings was assessed.
Methods
Forty-two anti-SARS-Cov-2 positive (CoV+) and 92 anti-SARS-Cov-2 negative (CoV–) leftover samples from before December 2019 were assessed; the Elecsys® Anti-SARS-CoV-2 was used as the reference assay. Analytical specificity was tested using leftover samples collected before December 2019 from patients with common cold symptoms.
Results
The SARS-CoV-2 Rapid Antibody Test was 100.0% (95% CI 91.59–100.0) sensitive and 96.74% (95% CI 90.77–99.32) specific, with 0.00% assay failure rate. No cross-reactivity was observed against the common cold panel. Method comparison was additionally conducted by two external laboratories, using 100 CoV+ and 275 CoV– samples, also comparing whole blood versus plasma matrix. The comparison demonstrated 96.00% positive and 96.36% negative percent agreement for plasma with the Elecsys Anti-SARS-CoV-2 and 99.20% percent overall agreement between whole blood and EDTA plasma.
Conclusion
The SARS-CoV-2 Rapid Antibody Test demonstrated similar performance to the manufacturer’s data and a centralised automated immunoassay, with no cross-reactivity with common cold panels.
Keywords: SARS-CoV-2, Rapid antibody test, Past exposure
Introduction
The global COVID-19 pandemic has created an urgent and unmet clinical need to investigate reliable diagnostic tools for patients, as well as to understand the extent of exposure and spread of infection among wider populations (Centers for Disease Prevention and Control, 2020, European Centre for Disease Prevention and Control, 2020a, The World Health Organization, 2020). Acute diagnosis of the COVID-19 infection is based on identification of viral RNA via polymerase chain reaction (PCR) from swab samples, which is detectable from symptom onset for approximately 4 weeks. As is known from localised testing during outbreaks, many people who are infected with the virus do not present with any clinical symptoms; current estimates suggest around 30% of seropositive individuals are asymptomatic (Mizumoto et al., 2020, Pollan et al., 2020, Sandri et al., 2020). Those individuals carry the virus and potentially spread it to others, who may react with severe COVID-19 disease. No region in the world can perform PCR testing of every patient with common cold symptoms or who has had contact with a suspected COVID-19 patient. In addition to clinical testing of individuals with suspected COVID-19 for direct virus detection, surveillance strategies need to combine several diagnostic techniques to monitor disease kinetics in wider populations (European Centre for Disease Prevention and Control, 2020b, World Health Organization, 2020). To control the pandemic, it seems crucial to investigate who has already had an infection and developed antibodies as an immune response, and who is still vulnerable to an infection (Althoff et al., 2020, Fiore et al., 2020, MacIntyre, 2020, Sen-Crowe et al., 2020, Steinbrook, 2020).
Antibody tests are not intended to diagnose an acute COVID-19 infection; more specific diagnostic methods should be performed to obtain this (European Centre for Disease Prevention and Control, 2020a). Ongoing research into the level and duration of immunity of seropositive people will add further value to the clinical and epidemiological interpretation of positive antibody testing results. Preliminary data suggest that high-affinity antibody tests show good correlation with neutralising activity (Wu et al., 2020). Based on current evidence, immunoglobulin M (IgM) antibodies are detectable within 5 days after symptom onset and immunoglobulin G (IgG) antibodies within 5–7 days (Guo et al., 2020, Long et al., 2020, Lou et al., 2020, Okba et al., 2020, Sethuraman et al., 2020, To et al., 2020, Zhang et al., 2020, Zhao et al., 2020). Depending on the applied method, seroconversion is observed after a median of 10–13 days after symptom onset for IgM and 12–14 days for IgG, and a maximum for both is reached after 2 weeks (Amanat et al., 2020, Guo et al., 2020, Long et al., 2020, Lou et al., 2020, Okba et al., 2020, Sethuraman et al., 2020, To et al., 2020, Xiang et al., 2020, Zhang et al., 2020, Zhao et al., 2020). Individual levels and kinetics of both IgM and IgG are highly variable, which is why simultaneous detection of both is recommended.
Currently available rapid antibody tests require improved accuracy before being recommended by competent authorities and used by healthcare professionals in the wider population (Centers for Disease Prevention and Control, 2020, European Centre for Disease Prevention and Control, 2020). The US Food and Drug Administration (FDA) released technical requirements for antibody tests on 04 April 2020, which include a specificity of ≥95% and cross-reactivity testing for common cold and other coronaviruses (US Food and Drug Administration, 2020). There are different high-throughput Anti-SARS-CoV-2 antibody tests available; the assay selected as the reference for the current study (Elecsys Anti-SARS-CoV-2 Immunoassay) is based on electrochemiluminescence (ECLIA), using a double-antigen sandwich test principle and a recombinant protein representing the antigen for the determination of antibodies to SARS-CoV-2, namely the nucleocapsid protein (N) (Roche Diagnostics GmbH, 2020). It provides a qualitative result with a sensitivity of 100.0% (95% CI 88.10–100.0) at ≥ 14 days after PCR confirmation and a specificity of 99.81% (95% CI 99.65–99.91).
Rapid tests, also called point-of-care (PoC) tests, combine immunoassay and chromatography for a qualitative detection of antibodies. The selected test – Anti-SARS-CoV-2 Rapid Antibody Test (SD Biosensor, Chungcheongbuk-do, Republic of Korea) – is a CE marked lateral flow assay displaying a visual ‘yes/no’ answer for the selective detection of specific IgG and/or IgM antibodies to SARS-CoV-2, with two separate coloured bands for IgG and IgM (SD SD Biosensor, 2020). The manufacturer states a specificity of 98.65% and sensitivity beyond 14 days after symptom onset of 99.03%, tested on 103 PCR-confirmed CoV + and 222 Cov– samples (SD SD Biosensor, 2020). The samples had also been tested for early sensitivity between 7–14 days after symptom onset, with a result of 92.59%. SD Biosensor has performed several cross-reactivity studies involving numerous specimens, including influenza A and B.
The current study further validated and extended the manufacturers’ clinical performance and cross-reactivity data by performing a matrix and method comparison to expand the limited external data and gain additional data on the overall assay performance.
Materials and methods
The assay
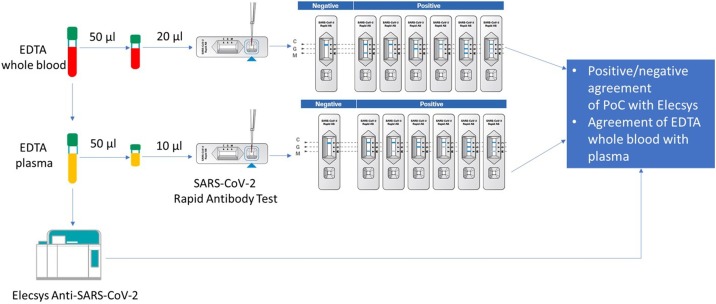
The assay (Figure 1 ) needs 10 μL human serum or plasma, or 20 μL whole venous or capillary blood sample to be filled into the pre-formed well of the test device. Three pre‑coated lines mark the ‘C’ control line, and the ‘G’ and ‘M’ test lines for IgG and IgM. The monoclonal chicken IgY antibody is coated on the ‘C’ region, and monoclonal anti‑human IgG and monoclonal anti-human IgM antibodies are coated on the ‘G’ and ‘M’ test line regions. During the test, SARS-CoV-2-specific antibodies in the sample interact with recombinant SARS‑CoV‑2 protein (nucleocapsid and spike protein) conjugated with colloidal gold particles forming antibody-antigen gold particle complexes. This complex migrates on the membrane via capillary action until the ‘M’ and ‘G’ test lines, where it will be captured by the monoclonal anti‑human IgG antibody or monoclonal anti‑human IgM antibody. A violet test line would be visible in the result window if SARS‑CoV‑2-specific antibodies were present in the sample. The intensity of the coloured test line varies depending upon the amount of SARS-CoV-2 antibodies present in the sample. Even if the colour is faint, the test result should be interpreted as a positive result. The control line is used as a procedural control and should always appear if the test procedure is properly performed and the test reagents are working. According to the instructions for use, the assay should be read between 10–15 min after addition of test materials.
Figure 1.
External method and matrix comparison – workflow.
Study design
A performance analysis was conducted at Roche Diagnostics (Penzberg, Mannheim, Germany) using 42 Elecsys Anti-SARS-CoV-2 confirmed CoV + and 92 leftover samples from healthy donors, collected before December 2019, and additionally confirmed by the Elecsys Anti-SARS-CoV-2 Cov– (56 lithium heparin plasma, 36 EDTA plasma). Cross-reactivity testing was conducted with 18 samples from individuals expressing signs and symptoms of a common cold (i.e. sore throat, cough, fever) collected before December 2019. Additional matrix equivalence and readout time analysis captured 159 Elecsys referenced Cov– samples, consisting of 55 heparin plasma, 55 EDTA plasma and 49 serum samples. Additionally, an independent comparison was performed with matched EDTA plasma and whole blood samples from a total of 375 anonymised leftover samples at two external testing sites. In total, 100 samples tested positive for SARS-CoV-2 antibodies by the Elecsys anti-SARS-CoV2 immunoassay and 275 negative samples (25 CoV + and 75 Cov– subjects at MVZLM Ruhr GmbH, Essen and 75 CoV + and 200 Cov– subjects at MVZ Labor Dr. Limbach & Kollegen GbR Heidelberg). No information on PCR result or time of sample collection related to symptom onset was available. All samples were analysed with the Anti-SARS-CoV-2 Rapid Antibody Assay and results were directly compared with the EDTA plasma sample from the Elecsys Anti-SARS-CoV-2 Assay (see Figure 1 for workflow). All investigations were performed according to a single determination and the manufacturer’s instructions for use.
Statistical analyses
Point estimates and 95% CI values were calculated for sensitivity and specificity. To determine positive and negative percent agreement (PPA and NPA), the Elecsys Anti-SARS-CoV-2 result was used as a comparator: »Non-reactive« (cut-off index [COI] <1.0) was a negative result and »Reactive« (COI ≥ 1.0) a positive result. The rapid test was considered positive in cases when either IgG or IgM showed a coloured line, even if the line was faint. The Clopper–Pearson exact method was used for the calculation of the two-sided 95% confidence intervals (CIs).
Data availability
Qualified researchers may request access to individual patient level data through the clinical study data request platform (https://vivli.org/). Further details on Roche's criteria for eligible studies are available here: https://vivli.org/members/ourmembers/. For further details on Roche's Global Policy on the Sharing of Clinical Information and how to request access to related clinical study documents, see here: https://www.roche.com/research_and_development/who_we_are_how_we_work/clinical_trials/our_commitment_to_data_sharing.htm.
Results
Clinical performance evaluation
The assay presented a qualitative visual test result without the need for a readout instrument like other rapid antibody devices on the market. Some lines were quite faint, and according to manufacturer’s instruction they were interpreted as a positive result. Handling was easy with one sample-transfer step and three drops of buffer dropped into the well after the blood sample; the results read at 10–15 min. Everything needed to conduct the test was included into the test package except for quality control material, lancets and a transferring pipette for 10 μL. All test cassettes were correctly assembled, opening the foil pouch was easy without danger of destroying the desiccant and no membranes were scratched.
Forty-two left-over samples with prior Elecsys-confirmed positive SARS-CoV-2 antibody detection were included in the sensitivity analysis. The overall sensitivity was 100.0% (95% CI 91.59–100.0) (Table 1). Ninety-two samples from healthy donors before December 2019 presented an overall specificity of 96.74% (95% CI 90.77–99.32) with no difference between EDTA and lithium-heparin plasma: 97.22% (95% CI: 85.47–99.93) and 96.43% (95% CI 87.69–99.56) (Table 1a, Table 1b) .
Table 1a.
Performance data of the manufacturer, internal clinical performance test, matrix equivalence test, and external method comparison.
| SD Biosensor data**, % (95% CI) |
Internal test, % (95% CI) |
Matrix evaluation, % (95% CI) | External comparison % (95% CI) |
|
|---|---|---|---|---|
| Samples tested, n | 195+/222– | 42+/92– | 159– | 100+/275– |
| Reference method | PCR | Elecsys® Anti-SARS-CoV-2 Antibody Assay | Elecsys® Anti-SARS-CoV-2 Antibody Assay | NA |
| Specificity versus reference method |
98.65 (96.10–99.72) |
96.74 (90.77–99.32) |
96.23 (91.97–98.60) |
96.00*** (90.07–98.90) |
| Sensitivity versus reference method | NA | |||
| 7–14 days* | 92.59 (82.11-97.94) | |||
| 14 days* | 99.03(94.71-99.98) | |||
| All time periods | 100 (91.59-100.00) | 96.36*** (93.41-98.24) |
NA, not applicable; +/–, positive/negative for SARS-CoV-2 determined by respective reference method *Post symptom onset.
**according to Anti-SARS-CoV-2 Rapid Antibody Assay Method sheet.
***positive/negative percent agreement with Elecsys Anti-SARS-CoV-2 in EDTA plasma, data in italics as generated by comparative analysis, not clinical performance analysis.
Table 1b.
Internal comparison Rapid Test to Elecsys Anti-SARS-CoV-2.
| Elecsys positive | Elecsys negative | |
|---|---|---|
| Rapid AB positive | 42 | 3 |
| Rapid AB negative | 0 | 89 |
An additional matrix evaluation and readout time analysis was performed with 159 SARS-CoV-2 negative samples to confirm equal results throughout the pre-defined readout time window. At readout times ≤10 min, specificity was slightly higher versus at 15 min (Table 2); however, detectable signals were ‘weaker’ or less well defined at ≤10 min compared with 15 min readout time. No relevant performance differences were noticed between serum, heparin and EDTA plasma (Table 2).
Table 2.
Matrix evaluation and readout time analysis.
| Read-out time |
|||||
|---|---|---|---|---|---|
| 8 min | 10 min | 12 min | 15 min | ||
| Total | Number of samples tested (negative) | 159 | 159 | 159 | 159 |
| False positive | 4 | 5 | 6 | 6 | |
| Specificity % (95% CI) | 97.48 (93.68–99.31) | 96.86 (92.81–98.97) | 96.23 (91.97–98.60) | 96.23 (91.97–98.60) | |
| Serum | Number of samples tested (negative) | 49 | 49 | 49 | 49 |
| False positive | 1 | 2 | 2 | 2 | |
| Specificity % (95% CI) | 97.96 (89.15–99.95) | 95.92 (86.02–99.50) | 95.92 (86.02–99.50) | 95.92 (86.02–99.50) | |
| Heparin plasma | Number of samples tested (negative) | 55 | 55 | 55 | 55 |
| False positive | 1 | 1 | 2 | 2 | |
| Specificity % (95% CI) | 98.18 (90.28–99.95) | 98.18 (90.28–99.95) | 96.36 (87.47–99.56) | 96.36 (87.47–99.56) | |
| EDTA plasma | Number of samples tested (negative) | 55 | 55 | 55 | 55 |
| False positive | 2 | 2 | 2 | 2 | |
| Specificity % (95% CI) | 96.36 (87.47–99.56) | 96.36 (87.47–99.56) | 96.36 (87.47–99.56) | 96.36 (87.47–99.56) | |
Cross-reactivity was tested in 18 samples from a common cold panel, collected before December 2019 without further information on the exact pathogens of the specimens. For all 18 measurements, a coloured control line was obtained, indicating that the test worked properly. No colour appeared on the test lines for IgG and IgM, thus leading to a negative result and resulting in a specificity of 100.0% (95% CI 81.47–100.0%). The results demonstrated that the test is not reacting with antibodies directed against related pathogens from common cold infections.
External method and matrix comparison
The total results for both testing methods (plasma/whole blood samples) were 96.00% (95% CI 90.07–98.90)/94.00% (95% CI 87.40–97.77) for positive percent agreement rate and 96.36% (95% CI 93.41–98.24)/96.00% (95% CI 92.96–97.99) for negative percent agreement rate (Table 3 ). The overall percent agreement rate between whole blood and EDTA plasma was 99.20% (95% CI 97.68–99.83) and the positive/negative percent agreement rates for whole blood versus EDTA plasma were 98.11% (95% CI 93.35–99.77)/99.63 (95% CI 97.95–99.99). Ten EDTA samples and 11 whole blood samples were detected as antibody-positive by the Anti-SARS-CoV-2 Rapid Antibody Assay but antibody-negative by the Elecsys assay (see Table 4a for the respectively detected immunoglobulin classes and the respective signal intensity on the 11 rapid tests). Ninety-six plasma and 94 whole blood samples out of 100 Elecsys-positive samples were detected positive by the rapid test (see Table 4b for the respective immunoglobulin classes and signal intensity of the six rapid test results). The matrix comparison for the Anti-SARS-CoV-2 Rapid Antibody Assay found 106 plasma samples with an antibody-positive test result, including two samples that had negative tests results with whole blood. One of 269 negative test results on the rapid test with plasma displayed a positive result with whole blood.
Table 3.
External comparison to Elecsys Anti-SARS-CoV-2 per matrix and immunoglobulin class.
| Number of results (Whole blood) |
Number of results (EDTA Plasma) |
|||||||
|---|---|---|---|---|---|---|---|---|
| IgM | IgG | IgM+IgG | Total | IgM | IgG | IgM+IgG | Total | |
| SARS-CoV-2 positive (confirmed by Elecsys) | 1 | 57 | 36 | 94 | 0 | 54 | 42 | 96 |
| SARS-CoV-2 negative (confirmed by Elecsys) | 0 | 0 | 0 | 264 | 0 | 0 | 0 | 265 |
| False positive results (compared with Elecsys) | 4 | 5 | 2 | 11 | 3 | 5 | 2 | 10 |
| False negative results (compared with Elecsys) | 0 | 0 | 0 | 6 | 0 | 0 | 0 | 4 |
| OPA (95% CI) | 95.47% (92.84–97.34) | 96.27% (93.82–97.94) | ||||||
| PPA (95% CI) | 94.00% (87.40–97.77) | 96.00% (90.07–98.90) | ||||||
| NPA (95% CI) | 96.00% (92.96–97.99) | 96.36% (93.41–98.24) | ||||||
OPA, overall percentage agreement; PPA, positive percentage agreement; NPA, negative percentage agreement.
Table 4a.
Method comparison: Discrepant positive samples - detected signal intensity and immunoglobulin class.
| Sample number | Rapid AB results – whole blood |
Rapid AB results – EDTA plasma |
Elecsys results (EDTA plasma) |
|||
|---|---|---|---|---|---|---|
| IgG | IgM | IgG | IgM | Result | COI | |
| 1 | – | X (weak) | – | X (weak) | Non-reactive | 0.089 |
| 2 | – | X (weak) | – | X (weak) | Non-reactive | 0.119 |
| 3 | – | X (weak) | – | X (weak) | Non-reactive | 0.089 |
| 4 | X (weak) | – | X (weak) | – | Non-reactive | 0.079 |
| 5 | – | X (weak) | – | – | Non-reactive | 0.069 |
| 6 | X (weak) | X (weak) | X (weak) | X (weak) | Non-reactive | 0.229 |
| 7 | X | X | – | Non-reactive | 0.146 | |
| 8 | X (weak) | – | X (weak) | – | Non-reactive | 0.221 |
| 9 | X (weak) | – | X (weak) | – | Non-reactive | 0.269 |
| 10 | X | X | X | X | Non-reactive | 0.189 |
| 11 | X | X | – | Non-reactive | 0.378 | |
AB, antibody; COI, cut-off index.
Table 4b.
Method comparison: Discrepant negative samples - detected signal intensity and immunoglobulin class.
| Sample number | Rapid AB results – whole blood |
Rapid AB results – EDTA plasma |
Elecsys results (EDTA plasma) |
|||
|---|---|---|---|---|---|---|
| IgG | IgM | IgG | IgM | Result | COI | |
| 1 | – | – | – | – | Reactive | 1.21 |
| 2 | – | – | – | – | Reactive | 1.99 |
| 3 | – | – | X (weak) | – | Reactive | 39.54 |
| 4 | – | – | – | – | Reactive | 2.32 |
| 5 | – | – | – | – | Reactive | 2.34 |
| 6 | – | – | X (weak) | – | Reactive | 16.7 |
AB, antibody.
Discussion
The COVID-19 pandemic has created an urgent need for antibody testing of large populations, to determine seroprevalence and potential immunity, which will be of more importance if more conclusive scientific data on correlation between these factors become available (Centers for Disease Prevention and Control, 2020, European Centre for Disease Prevention and Control, 2020a, Ozcurumez et al., 2020, Theel et al., 2020, Zhang et al., 2020). The anticipated need for high testing capacities in PoC settings outside of large hospitals calls for the development and validation of high-performance rapid antibody tests as reliable diagnostic instruments, in addition to the centralised antibody tests that are available. The evaluated Anti-SARS-CoV-2 Rapid Antibody Test complies with the acceptance criteria, defined by the FDA EUA on 04 April 2020, for SARS-CoV-2 antibody test developments (US Food and Drug Administration, 2020).
The current internal clinical performance evaluation confirmed the manufacturers’ reported clinical sensitivity of the test with an uncharacterised cohort of antibody-positive samples from a population with unknown time from symptom onset, which may reflect the typical clinical scenario for test indication. The external comparison confirmed high agreement of both the Elecsys assay and the Anti-SARS-CoV-2 Rapid Antibody Test, also with no confirmed PCR result and time of sampling from symptom onset unknown. For both the internal and external evaluation, samples could be mixed with some collected early after symptom onset. Both tests claimed <100% sensitivity <14 days after symptom onset. The Elecsys Anti-SARS-CoV-2 Antibody Test measures high-affinity IgG/IgM antibodies directed at the nucleocapsid antigen, with an excellent sensitivity of 100.0% (95% CI 88.1–100.0) at ≥ 14 days after PCR confirmation (Roche Diagnostics GmbH, 2020). The rapid test has been developed to detect both nucleocapsid-associated and spike protein-associated antibodies to increase test accuracy compared with centralised serological assays, which typically perform better than rapid tests (SD Biosensor, 2020). A limitation of this study was that the samples were not differentiated according to time of collection after symptom onset and were not PCR-tested upfront. On the other hand, PCR may already have become negative at the time of sampling and not every PCR-positive patient develops seroconversion; as such, comparisons with PCR have limited accuracy for performance comparison of a rapid versus centralised antibody assay. The comparative specificity reported in this paper is within a similar range to those reported for other rapid antibody tests; however, most other rapid tests granted Emergency Use Authorization were validated by comparison with PCR-positive samples only, which has limitations as explained above (The US Food and Drug Administration, 2020).
At the time of writing this manuscript, few studies had demonstrated the performance of rapid antibody tests external to the manufacturers (Batra et al., 2020, Jaaskelainen et al., 2020, Minteer et al., 2020, Naranbhai et al., 2020, Pallett et al., 2020). The intention of this study was to add real-world clinical evidence for a rapid antibody test. For SARS-CoV-2 tests, high specificity is a priority, particularly in low-prevalence settings; as such it would be interesting to further evaluate the interpretation of the low positive samples at the rapid test, particularly for samples that were IgM positive only, to determine if these reflect non-specific binding or true early detection. Further assessment with serial samples taken from COVID-19 patients from early infection phase onwards will help to better understand this.
The investigated Anti-SARS-CoV-2 Rapid Antibody Test provided readily evaluable results regardless of sample type (whole blood or EDTA plasma) and independent of any readout time between 10–15 min, as stated in the instructions for use. Those positive practical aspects could enable potential use outside medical environments. The assay is currently intended for professional use in laboratory and PoC environments as an aid in identifying individuals with an adaptive immune response to SARS-CoV-2, indicating prior infection. In these studies, the SARS-CoV-2 Rapid Antibody Test demonstrated excellent clinical performance without cross-reactivity to common cold samples and results comparable with data of an automated high-performance immunoassay (Elecsys).
The current data confirm and extend the manufacturers’ performance data and add further details on matrix and method comparisons. Whilst further assessments in the future will be valuable, this study supports the use of the SARS-CoV-2 Rapid Antibody Test as a reliable diagnostic instrument for SARS-CoV-2 IgM and IgG antibody detection in near-patient settings with potential extended usability outside medical environments.
Conflicts of interest/financial disclosures
Eloisa Lopez-Calle, Tanja Schneider, Eva Urlaub, Johannes Hayer, are all employees of Roche Diagnostics. Claudia Zemmrich works as a freelance contractor for Roche Diagnostics.
Ethical statement
The study was conducted in accordance with applicable regulations, including relevant European Union directives and regulations, and the principles of the Declaration of Helsinki. All samples were anonymised leftover specimens. For the samples tested at Roche Diagnostics, a statement was obtained from the Ethics Committee of the Landesärztekammer Bayern confirming that there are no objections against the transfer and the coherent use of the anonymised leftover samples. For the samples tested in MVZLM Ruhr GmbH (Essen) and at MVZ Labor Dr. Limbach & Kollegen GbR (Heidelberg) no ethics committee vote is required in accordance with MPG (Medizinproduktegesetz Deutschland).
Acknowledgements
The authors would like to acknowledge the contributions of Rabea Held, Regina Draude, Marcus Pollok (Roche Diagnostics GmbH, Mannheim, Germany) for testing, data acquisition in the laboratory and preliminary data analysis. In addition, thanks to Imola Szebenyi, Oscar Gutierrez-Sanz, Miriam Hübner, Ludwig Pollich, Nicolai Bluthardt, Andreas Aristow (Roche Diagnostics GmbH, Penzberg, Germany) for the measurements of the clinical and cross-reactivity samples as part of the study conducted at Roche Diagnostics. Thanks to John Burden (Roche Diagnostics International Ltd. Rotkreuz, Switzerland), Sven Groesgen (Roche Diagnostics GmbH, Mannheim, Germany) and Christine Jung (Roche Diagnostics GmbH, Penzberg, Germany) for planning, set-up and analysis of the external evaluation. The authors would also like to acknowledge Matthias Metz and Daria Glukhova for conducting the measurements with the Elecsys SARS-CoV-2 method as part of the study at Roche Diagnostics.
References
- Althoff K.N., Coburn S.B., Nash D. Contact tracing: Essential to the public health response and our understanding of the epidemiology of COVID-19. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanat F., Stadlbauer D., Strohmeier S., Nguyen T., Chromikova V., McMahon M., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. medRxiv. 2020 doi: 10.1038/s41591-020-0913-5. 2020.03.17.20037713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra R., Olivieri L.G., Rubin D., Vallari A., Pearce S., Olivo A., et al. A comparative evaluation between the Abbott PanbioTM COVID-19 IgG/IgM rapid test device and Abbott ArchitectTM SARS CoV-2 IgG assay. J Clin Virol. 2020;132 doi: 10.1016/j.jcv.2020.104645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Prevention and Control . 2020. Interim Guidelines for COVID-19 Antibody Testing. Available from: https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antibody-tests-guidelines.html [accessed June 2020] [Google Scholar]
- European Centre for Disease Prevention and Control . 2020. Diagnostic testing and screening for SARS-CoV-2. Available from: https://www.ecdc.europa.eu/en/covid-19/latest-evidence/diagnostic-testing [accessed November 2020] [Google Scholar]
- European Centre for Disease Prevention and Control . 2020. Transmission of COVID-19. Available from: https://www.ecdc.europa.eu/en/covid-19/latest-evidence/transmission [accessed July 2020] [Google Scholar]
- Fiore V.G., DeFelice N., Glicksberg B.S., Perl O., Shuster A., Kulkarni K., et al. Containment of future waves of COVID-19: simulating the impact of different policies and testing capacities for contact tracing, testing, and isolation. medRxiv. 2020 doi: 10.1371/journal.pone.0247614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Ren L., Yang S., Xiao M., Chang Yang F., et al. Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID-19) Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaaskelainen A.J., Kuivanen S., Kekalainen E., Ahava M.J., Loginov R., Kallio-Kokko H., et al. Performance of six SARS-CoV-2 immunoassays in comparison with microneutralisation. J Clin Virol. 2020;129 doi: 10.1016/j.jcv.2020.104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.X., Deng H.J., Chen J., Hu J., Liu B.Z., Liao P., et al. Antibody responses to SARS-CoV-2 in COVID-19 patients: the perspective application of serological tests in clinical practice. medRxiv. 2020 2020.03.18.20038018. [Google Scholar]
- Lou B., Li T., Zheng S., Su Y., Li Z., Liu W., et al. Serology characteristics of SARS-CoV-2 infection since the exposure and post symptoms onset. medRxiv. 2020 doi: 10.1183/13993003.00763-2020. 2020.03.23.20041707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntyre C.R. Case isolation, contact tracing, and physical distancing are pillars of COVID-19 pandemic control, not optional choices. Lancet Infect Dis. 2020 doi: 10.1016/S1473-3099(20)30512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minteer C., Casanovas-Massana A., Li T., McDonald D., Wang L., Pan S.H., et al. Multi-site Validation of a SARS-CoV-2 IgG/IgM Rapid Antibody Detection Kit. medRxiv. 2020 2020.05.25.20112227. [Google Scholar]
- Mizumoto K., Kagaya K., Zarebski A., Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID-19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro Surveill. 2020;25(10) doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranbhai V., Chang C.C., Beltran W.F.G., Miller T.E., Astudillo M.G., Villalba J.A., et al. High seroprevalence of anti-SARS-CoV-2 antibodies in Chelsea, Massachusetts. J Infect Dis. 2020 doi: 10.1093/infdis/jiaa579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okba N., Müller A., Li W., Wang C., GeurtsvanKessel C., Corman V., et al. Severe Acute Respiratory Syndrome Coronavirus 2−Specific Antibody Responses in Coronavirus Disease 2019 Patients. Emerg Infect Dis J. 2020;26(7) doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcurumez M.K., Ambrosch A., Frey O., Haselmann V., Holdenrieder S., Kiehntopf M., et al. SARS-CoV-2 antibody testing-questions to be asked. J Allergy Clin Immunol. 2020;146(1):35–43. doi: 10.1016/j.jaci.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallett S.J.C., Rayment M., Patel A., Fitzgerald-Smith S.A.M., Denny S.J., Charani E., et al. Point-of-care serological assays for delayed SARS-CoV-2 case identification among health-care workers in the UK: a prospective multicentre cohort study. Lancet Resp Med. 2020;8(9):885–894. doi: 10.1016/S2213-2600(20)30315-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollan M., Perez-Gomez B., Pastor-Barriuso R., Oteo J., Hernan M.A., Perez-Olmeda M., et al. Prevalence of SARS-CoV-2 in Spain (ENE-COVID): a nationwide, population-based seroepidemiological study. Lancet. 2020 doi: 10.1016/S0140-6736(20)31483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche Diagnostics GmbH . 2020. Elecsys Anti-SARS-CoV-2. [Google Scholar]
- Sandri M.T., Azzolini E., Torri V., Carloni S., Tedeschi M., Castoldi M., et al. IgG serology in health care and administrative staff populations from 7 hospital representative of different exposures to SARS-CoV-2 in Lombardy, Italy. medRxiv. 2020 2020.05.24.20111245. [Google Scholar]
- SD Biosensor . SD Biosensor; 2020. SARS-CoV-2 Rapid Antibody Test. [Google Scholar]
- Sen-Crowe B., McKenney M., Elkbuli A. COVID-19 laboratory testing issues and capacities as we transition to surveillance testing and contact tracing. Am J Emerg Med. 2020 doi: 10.1016/j.ajem.2020.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethuraman N., Jeremiah S.S., Ryo A. Interpreting Diagnostic Tests for SARS-CoV-2. JAMA. 2020 doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- Steinbrook R. Contact Tracing, Testing, and Control of COVID-19-Learning From Taiwan. JAMA Intern Med. 2020 doi: 10.1001/jamainternmed.2020.2072. [DOI] [PubMed] [Google Scholar]
- The US Food and Drug Administration . 2020. EUA Authorized Serology Test Performance. Available from: https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/eua-authorized-serology-test-performance [accessed 03 November 2020] [Google Scholar]
- The World Health Organization . 2020. Immunity Passports in the Context of COVID-19. Available from: https://www.who.int/news-room/commentaries/detail/immunity-passports-in-the-context-of-covid-19 [accessed July 2020] [Google Scholar]
- Theel E.S., Slev P., Wheeler S., Couturier M.R., Wong S.J., Kadkhoda K. The Role of Antibody Testing for SARS-CoV-2: Is There One? J Clin Microbiol. 2020 doi: 10.1128/JCM.00797-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K.W., Tsang O.T.Y., Leung W.S., Tam A.R., Wu T.C., Lung D.C., et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US Food and Drug Administration . FDA; 2020. Policy for Coronavirus Disease-2019 Tests During the Public Health Emergency (Revised) [Google Scholar]
- World Health Organization . 2020. Surveillance strategies for COVID-19 human infection: interim guidance. Available from: https://www.who.int/publications/i/item/surveillance-strategies-for-covid-19-human-infection [accessed July 2020] [Google Scholar]
- Wu F., Wang A., Liu M., Wang Q., Chen J., Xia S., et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv. 2020 2020.03.30.20047365. [Google Scholar]
- Xiang F., Wang X., He X., Peng Z., Yang B., Zhang J., et al. Antibody Detection and Dynamic Characteristics in Patients with COVID-19. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Liu J., Li N., Liu Y., Ye R., Qin X., et al. Serological detection of 2019-nCoV respond to the epidemic: A useful complement to nucleic acid testing. medRxiv. 2020 doi: 10.1016/j.intimp.2020.106861. 2020.03.04.20030916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Qualified researchers may request access to individual patient level data through the clinical study data request platform (https://vivli.org/). Further details on Roche's criteria for eligible studies are available here: https://vivli.org/members/ourmembers/. For further details on Roche's Global Policy on the Sharing of Clinical Information and how to request access to related clinical study documents, see here: https://www.roche.com/research_and_development/who_we_are_how_we_work/clinical_trials/our_commitment_to_data_sharing.htm.