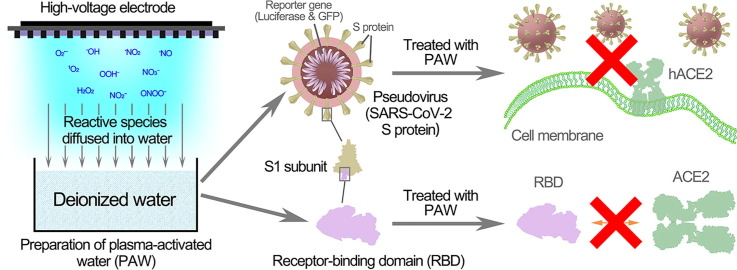
Graphical abstract
Keywords: Plasma-activated water, SARS-CoV-2, Receptor-binding domain (RBD), Pseudoviruses, Reactive species
Abstract
SARS-CoV-2 is a highly contagious virus and is causing a global pandemic. SARS-CoV-2 infection depends on the recognition of and binding to the cellular receptor human angiotensin-converting enzyme 2 (hACE2) through the receptor-binding domain (RBD) of the spike protein, and disruption of this process can effectively inhibit SARS-CoV-2 invasion. Plasma-activated water efficiently inactivates bacteria and bacteriophages by causing damage to biological macromolecules, but its effect on coronavirus has not been reported. In this study, pseudoviruses with the SARS-CoV-2 S protein were used as a model, and plasma-activated water (PAW) effectively inhibited pseudovirus infection through S protein inactivation. The RBD was used to study the molecular details, and the RBD binding activity was inactivated by plasma-activated water through the RBD modification. The short-lived reactive species in the PAW, such as ONOO−, played crucial roles in this inactivation. Plasma-activated water after room-temperature storage of 30 days remained capable of significantly reducing the RBD binding with hACE2. Together, our findings provide evidence of a potent disinfection strategy to combat the epidemic caused by SARS-CoV-2.
1. Introduction
The COVID-19 pandemic caused by the SARS-CoV-2 outbreak has resulted in approximately 20 million infections and more than 739,000 deaths as of the end of August 11, 2020 [1], [2], [3]. The pandemic has caused a severe threat to public health and safety, which urgently requires effective therapeutic and control strategies [4]. To date, no drug or vaccine for SARS-CoV-2 has been officially approved [5]. The development of effective strategies to prevent the transmission and infection of SARS-CoV-2 can alleviate the current situation to a certain extent.
The first step of most viral infections is the binding between viral particles and surface receptors of the host cell. In the human coronaviruses, the spike (S) glycoproteins embedded in its envelope surface participant in binding with the host receptors [6], [7]. The spike glycoproteins are homotrimeric and each monomer includes two subunits: globular S1 and membrane-proximal S2 [8]. The S1 subunit consists of the N-terminal domain (NTD) and the receptor-binding domain (RBD), and the RBD is responsible for recognizing and binding with the cellular receptor—human angiotensin-converting enzyme 2 (hACE2) to initiate the infection process of coronaviruses [8], [9], [10]. Thus, disruption of the interaction between the RBD and ACE2 can prevent coronavirus infection, and the RBD can be an effective target of disinfectants and drugs.
Cold atmospheric-pressure plasma (“plasma” hereafter) generates numerous reactive species, such as hydrogen peroxide (H2O2), nitrite (NO2 −), and peroxynitrite (ONOO−) as well as electrons, ions, and UV photons near room temperature, thus making plasma attractive for biomedical and environmental applications [11], [12], [13], [14], [15], [16], [17], [18]. Plasma has been widely studied in bacterial inactivation and is potent for sterilization and therapy of infectious diseases [19], [20]. Previous studies have reported that a form of plasma efficiently inactivated feline calicivirus and ɸ174 and λ bacteriophages [21], [22], [23], [24], [25]. The biological effect of plasma is dose-dependent, which is important in the application of plasma biomedicine [26], [27]. Plasma-activated water (PAW) prepared by the plasma treatment of water also efficiently inactivated T4, ɸ174, and MS2 bacteriophages and exhibited an effect similar to that of the direct plasma treatment [28]. Su et al. demonstrated that the treatment of Newcastle disease virus with PAW effectively decreased its infectivity [29]. The microbial inactivation activity of PAW is attributed to the storage of reactive species in water, which has advantages of precise control of reactive species dosage and uniformity, as well as facilitating the therapy of infectious diseases in deep tissue such as abdominal infection [30], [31], [32]. However, the inactivation effects of plasma or PAW on coronavirus have not been reported.
SARS-CoV-2 experiments are limited to performance in a biosafety level-3 laboratory, which greatly restricts research [10]. SARS-CoV-2 entry into cells is based on the binding of its S protein to ACE2 [7], [10]. The utilization of a pseudovirus system with SARS-CoV-2 S protein could alleviate this limitation [33]. A pseudovirus with SARS-CoV-2 S protein was used as a model for studying the entry mechanism and neutralization assays [10], [33].
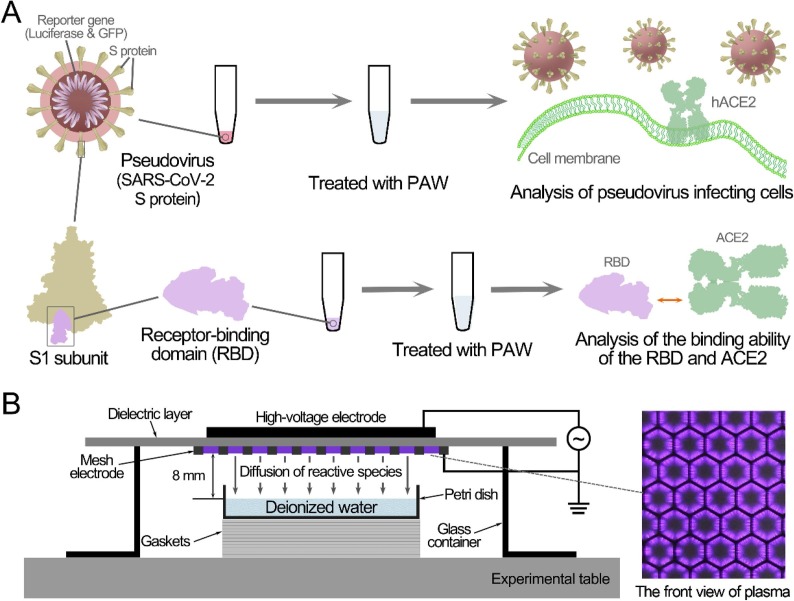
In this study, a pseudovirus and the RBD were employed as models to evaluate the inactivation of SARS-CoV-2 by PAW and the related mechanisms (Fig. 1 A). We aim to demonstrate the antiviral ability of PAW against SARS-CoV-2, thus providing a new disinfection strategy to combat the epidemic.
Fig. 1.
Schematic diagram of the experimental setup (A) and preparation of plasma-activated water (B).
2. Materials and method
2.1. Plasma device and treatments
As shown in Fig. 1B, the surface discharge plasma device consisted of a high-voltage plane electrode, a liquid-facing grounded mesh electrode, and a dielectric layer (made of polytetrafluoroethylene) sandwiched between the two electrodes. Each mesh element had a hexagonal shape, and the surface plasma was generated in the mesh elements when a sinusoidal high voltage was applied [28], [34]. Natural air was employed as the working gas and the discharge power density was about 0.25 W/cm2 at approximate 23 kHz. The peak to peak voltage and current were approximate 8.36 kV and 572.4 mA, respectively (Fig. S1). Under such conditions, the plasma had good mesh-to-mesh homogeneity. Deionized water (5 mL) in a quartz dish (70 mm × 70 mm) was located beneath the surface plasma, which was smaller than the area of the surface plasma (80 mm × 80 mm). The depth of the deionized water was kept at 1 mm, whereas the air gap between the plasma and the liquid surface was 8 mm. For PAW pretreated for different durations, the minimum pretreatment duration for effective antiviral activity was 5 min and a 10-min pretreatment led to optimal antiviral activity. Results with 5-min and 10-min pretreatment were included here.
2.2. Pseudovirus based inactivation assay
Pseudovirus incorporated with the SARS-CoV-2 S protein and the reporter gene firefly luciferase were supplied by Genscript (Nanjing, China) and COS-7 and HEK-293T cells overexpressing human ACE2 were supplied by Vitalstar (Beijing, China) and GENEWIZ (Suzhou, China), respectively. COS-7 and HEK-293T cells were cultured in the complete medium containing Dulbecco’s modified Eagle’s medium (DMEM, Thermo-Fisher), 10% (v/v) fetal bovine serum (FBS, Gibco), and 1% (v/v) penicillin–streptomycin (Beyotime) at 37 °C in 5% CO2. The pseudovirus treated with PAW-5 min and PAW-10 min and untreated pseudovirions were mixed with 4 times the volume of the complete medium. Then, the mixture was added to hACE2-overexpressing cells in a 96-well plate. After incubation for 3 h, the virus-containing medium was replaced with fresh medium and cultured for 48 h. Then, pseudovirus infections were measured using ONE-Glo™ Luciferase Assay System (Promega), and cell vitalities were measured using CellTiter-Glo® Luminescent Cell Vitality Assay (Promega).
2.3. Transmission electron microscopy (TEM)
Pseudoviruses treated with PAW-5 min and PAW-10 min were diluted 20 times, dropped onto carbon-coated grids, and incubated for 10 min at 22 °C. Then, the excess liquid was discarded, and the grids were covered with 1% uranyl acetate for 30 s at 22 °C. These stained samples were examined by using an FEI Talos F200C transmission electron microscope operating at 200 kV at × 45,000 magnification.
2.4. Surface plasmon resonance (SPR)
The biotinylated SARS-CoV-2 RBD and human ACE2 proteins were purchased from ACRO Biosystems (Beijing, China). SPR analysis was performed according to a published procedure [8]. hACE2 was immobilized to a CM5 sensorchip (GE Healthcare) at a level of ~700 response units (RUs) using a Biacore T200 (GE Healthcare) and a running buffer composed of 10 mM HEPES, 150 mM NaCl, 3 mM EDTA, and 0.05% Tween 20, pH 7.4. Serial dilutions of the SARS-CoV-2 RBD were flowed through with a concentration ranging from 125 to 1.9 nM. The SARS-CoV-2 RBD was injected at a flow rate of 30 μL/min for an association phase of 120 s and this was followed by a dissociation phase of 330 s. The resulting data were fit to a 1:1 binding model using Biacore Evaluation Software (GE Healthcare).
2.5. Enzyme-linked immunosorbent assay (ELISA)
The hACE2 protein was diluted with a coating buffer (15 mM Na₂CO3, 35 mM NaHCO3, 7.7 mM NaN3, pH 9.6) to 1 μg/mL and added to the wells of 96-well plates (100 μL per well). The plates were incubated at 4 °C overnight and washed with PBS-T. Then, the wells were blocked with PBS-T containing 2% BSA at 37 °C for 1.5 h and washed. Next, 100 μL of biotinylated SARS-CoV-2 RBD (3.90625–1000 ng/mL, diluted with PBS-T containing 0.5% BSA) was added to each well and incubated at 37 °C for 1 h. The wells were washed and incubated with 100 μL of horseradish peroxidase-conjugated streptavidin (Thermo Fisher) at 37 °C for 1 h. After washing, TMB substrate (Thermo Fisher) and 2 M H2SO4 were sequentially added to the wells. Then, the optical density (OD) was measured using a microplate reader (Thermo Scientific Varioskan Flash) at 450 nm.
2.6. Mass spectrometry
The untreated RBD and RBD treated with PAW-10 min (~1 μg) were analyzed using an analytical column ( 2.0 mm × 50 mm, 5 μm) on an Ultimate 3000 RSLCnano system connected to a Q-Exactive Plus mass spectrometer (Thermo Scientific). Samples were eluted using a binary solvent system with 2% acetonitrile (ACN) and 0.1% formic acid (phase A), and 98% ACN and 0.1% formic acid (phase B). The full-scan mass spectrometry (MS) mode was operated with the following parameters: automatic gain control (AGC) target, 1e6; resolution, 17,500 FWHM; scan range, 1000–9000 m/z; and maximum injection time, 50 ms. The results were analyzed by Protein Deconvolution 2.0 SP2.
2.7. Measurement of reactive species in PAW
The concentrations of H2O2 and NO2 −/NO3 − in PAW were measured by using a hydrogen peroxide/peroxidase assay kit (AAT Bioquest) and nitrite/nitrate colorimetric assay kit (Beyotime Biotechnology), respectively. •OH and ONOO− were measured by using fluorescent probes the 3′-(p-aminophenyl) fluorescein (APF, Sigma), 3ʹ-(p-hydroxyphenyl) fluorescein (HPF, Sigma), and coumarin boronic acid pinacolate ester (CBA, Cayman) at final concentrations of 5 μM, 5 μM, and 20 μM, respectively. After incubation for 10 min, the fluorescence intensities were detected by using a microplate reader (Thermo Scientific Varioskan Flash) at the corresponding excitation and emission wavelengths (APF and HPF: 490/515 nm; CBA: 332⁄470 nm).
3. Results
3.1. Inactivation of pseudovirus with S protein by PAW
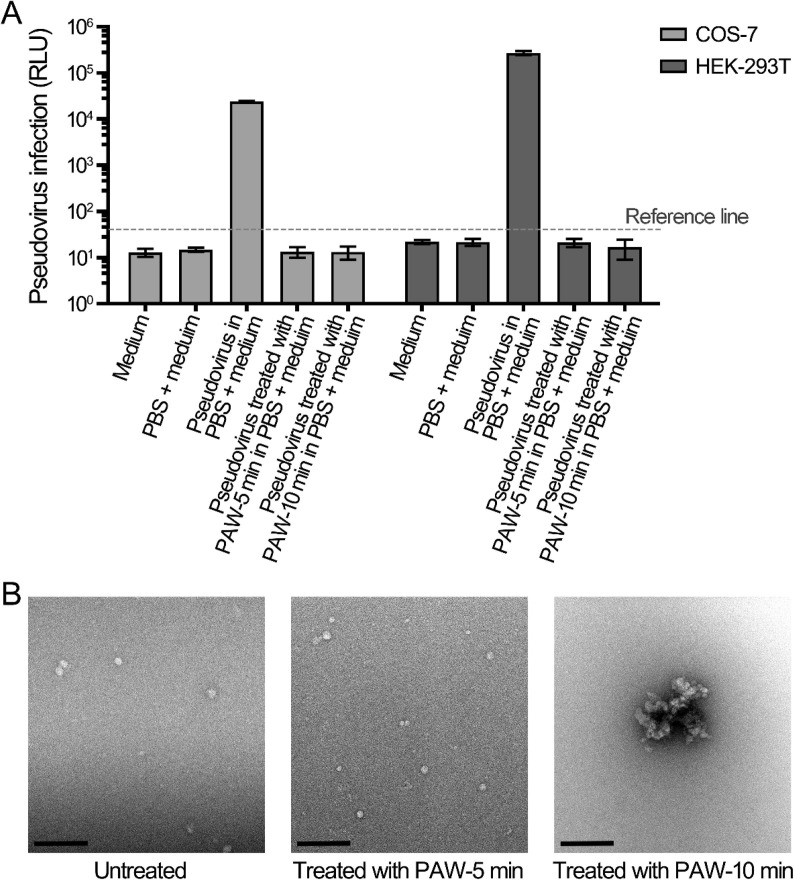
A variety of gaseous reactive species were generated by surface discharge in air and then dissolved in water to produce PAW, as shown in Fig. 1B. PAW has been frequently reported for its efficient inactivation of bacteria and viruses [28], [30]. Here, pseudovirus incorporated with SARS-CoV-2 S protein was used as a model for SARS-CoV-2, and COS-7 and HEK-293T cells overexpressing human ACE2 (hACE2-COS-7 and hACE2-HEK-293T) were used as recipient cells to evaluate the inactivation effect of PAW. Two doses of PAW, namely PAW-5 min and PAW-10 min, were selected by treating deionized water with plasma for 5 min and 10 min, respectively. The infection of pseudovirus and PAW-treated pseudovirus did not significantly affect cell viability (Fig. S2). The values of relative light unit (RLU) of hACE2-COS-7 and hACE2-HEK-293T cells infected with untreated pseudovirus were 24,071 and 271,295, respectively, whereas those of both kinds of cells with pseudovirus treated with PAW-5 min and PAW-10 min were 13.4 and 13.2 and 21.2 and 16.8, respectively, which were similar to the values of the control group without pseudovirus (Fig. 2 A). Based on the cell backgrounds, the reference line was set at the value of RLU 40 (101.6), more than 500 times lower than the RLU of untreated pseudovirus (Fig. 2A). The results indicated that pseudovirus infection was effectively inhibited by the PAW treatment at both of its selected doses.
Fig. 2.
Inactivation of pseudovirus by plasma-activated water. (A) Infection of untreated and PAW-treated pseudovirus on COS-7 and HEK-293T cells. The pseudoviruses treated with PAW-5 min and PAW-10 min, and untreated pseudoviruses in PBS were mixed with 4 volumes of medium. The mixture as added to COS-7 and HEK-293T cells for infection. (B) Transmission electron microscopy (TEM) analysis of pseudoviruses treated with PAW. The pseudoviruses treated with PAW-5 min and PAW-10 min, and untreated pseudovirus were negative-stained and examined by using TEM. The bars represent 200 nm.
To determine the mechanism of the inactivation effect on pseudoviruses, morphological changes in pseudoviruses treated with PAW were investigated using transmission electron microscopy (TEM). The untreated pseudovirus exhibited a spherical structure, with a diameter of 20–40 nm (Fig. 2B). The pseudovirus treated with PAW-5 min exhibited little morphological changes, while that treated with PAW-10 min aggregated and formed large complexes (Fig. 2B). The morphological study suggested that the effects of PAW on pseudoviruses can be divided into two modes: one was to inactivate the virus without causing morphological changes, and the other was to inactivate the virus and cause the virus aggregation. The above results suggested that the reactive species in PAW inactivated the S proteins of the pseudoviruses and further caused interaction between the S proteins of adjacent pseudoviruses, leading to pseudovirus aggregation.
3.2. Effect of PAW on the SARS-CoV-2 RBD
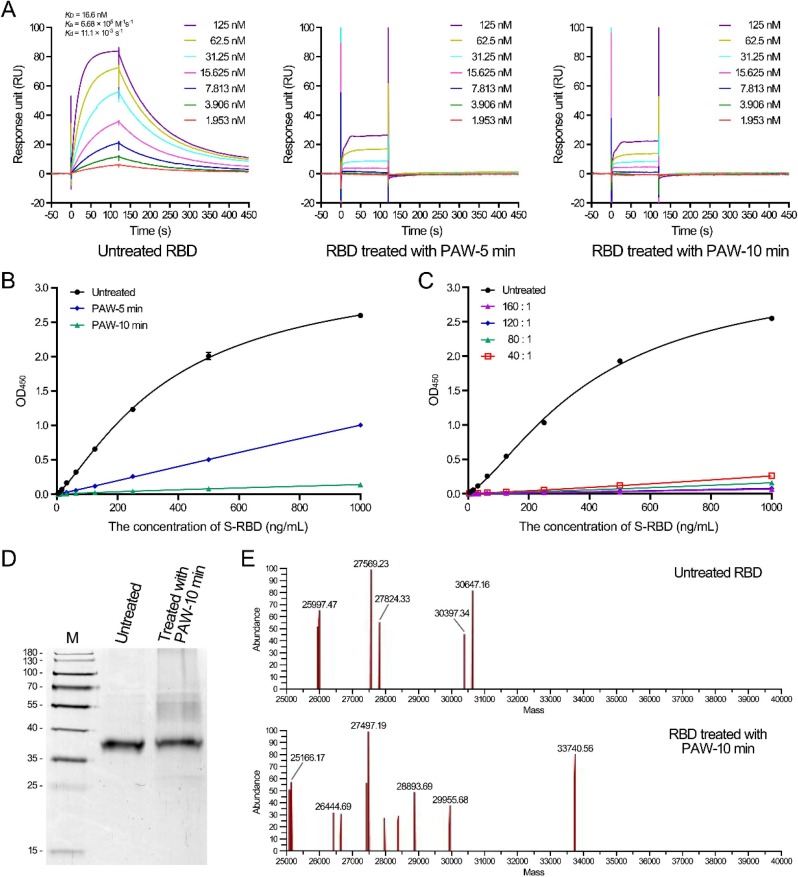
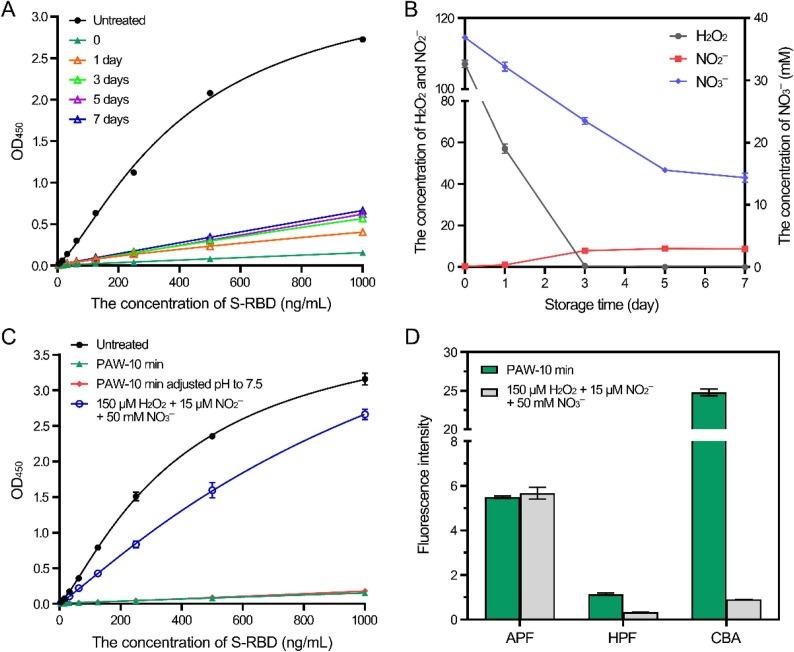
The RBD domain is located at the top side of the S protein and is the key component in the first step of virus entry. To further understand the mechanism of PAW on the S protein, the RBD was employed as a representative and the effects of PAW on the RBD were investigated. First, the binding affinity of the RBD to hACE2 was measured and the K d value between untreated RBD and hACE2 was 16.6 nM, consistent with 4.4–44.2 nM, as previously reported [6], [35], [36]. However, the RBD treated with PAW-5 min or PAW-10 min exhibited little binding activity to hACE2 (Fig. 3 A). The result was further verified by enzyme-linked immunosorbent assay (ELISA). The concentration for 50% of the maximal effect (EC50) of the untreated RBD was approximately 370 ng/mL, whereas that of the RBD treated with PAW-5 min or PAW-10 min was at least 270 times higher, as calculated by a fitted curve (Fig. 3B). Changing the volume ratios of PAW to the RBD had little effect on the inactivation effects (Fig. 3C). These results suggested that PAW effectively damaged the RBD protein to prevent its binding with hACE2.
Fig. 3.
Treatment of the SARS-CoV-2 RBD with plasma-activated water inhibited the interaction between the RBD and hACE2. (A) Analysis of the binding between untreated and PAW-treated RBDs to hACE2 by surface plasmon resonance assay. Binding curves of immobilized hACE2 with untreated RBD (left), RBD treated with PAW-5 min (middle), and RBD treated with PAW-10 min (right). The results of untreated RBD binding were fit to a 1:1 binding model. (B) Analysis of the binding between untreated and PAW-treated RBDs to hACE2 by ELISA. The binding curves of untreated and PAW-treated RBDs to the immobilized hACE2 were analyzed. (C) Treatment of the RBD with PAW-10 min at different volume ratios. The binding curves of untreated and PAW-treated RBDs to the immobilized hACE2 were analyzed by ELISA. (D) SDS-polyacrylamide gel electrophoresis of untreated and PAW-treated RBDs. The untreated RBD and RBD treated with PAW-10 min (~400 ng) were subjected to electrophoresis in a 12% SDS-polyacrylamide gel. The gel was visualized by silver staining. (E) Mass spectrum of untreated and PAW-treated RBDs. The untreated RBD and RBD treated with PAW-10 min were analyzed by LC-MS.
PAW contains various reactive species, and therefore we hypothesized that the inactivation effect of PAW was caused by the reaction of reactive species with RBD proteins. To test this, the changes in the RBD protein were analyzed by SDS-PAGE and mass spectrometry (Fig. 3D and 3E). The theoretical molecular weight of the untreated RBD is 28.7 kDa, and the protein migrated at the position of 38 kDa on SDS-PAGE (Fig. 3D). After treatment with PAW-10 min, the amount of the RBD protein at 38 kDa decreased to approximately 69.7% and migrated more slowly, with a weak band appearing at ~ 28 kDa and smeared bands appearing at positions with molecular weights between 45 and 65 kDa (Fig. 3D; Fig. S3). The mass spectrum also exhibited the mass changes in the RBD protein after PAW treatment (Fig. 3E).
3.3. Reactive species in PAW and their relevance to the inactivation effect
To investigate the chemical mechanisms, the inactivation effects and reactive species of PAW were compared after PAW storage for different times. PAW stored at room temperature (approximately 25 °C) for 7 days exhibited a slightly weaker effect on the RBD than that of the freshly prepared PAW, probably due to the decay of reactive species during storage (Fig. 4 A). According to the literature, H2O2, NO2 −, and NO3 − are the main long-lived species in PAW, and their concentrations in the freshly prepared PAW were 107.10 μM, 0.34 μM, and 36.89 mM, respectively. During storage for 7 days, the concentration of H2O2 decreased rapidly to the detection limit, and the concentration of NO3 − gradually decreased by ~ 2.5-fold, whereas that of NO2 – slightly increased from 0.3 to 8.8 μM (Fig. 4B). The pH value of the freshly prepared PAW was approximately 1.8 and slightly increased during PAW storage.
Fig. 4.
Reactive species in PAW. (A) Inactivation effects of PAW after storage. PAW-10 min was stored for the indicated times, and the inactivation effects on the SARS-CoV-2 RBD were examined. (B) Long-lived species during storage. The H2O2 and NO2−/NO3− in PAW-10 min stored for the indicated times were measured. (C) Comparison of the inactivation effects of PAW and of the mixed solution of H2O2, NO2−, and NO3−. The inactivation effects of PAW-10 min, PAW-10 min with the pH adjusted to 7.5, and the mixed solution of H2O2 (150 μM), NO2− (15 μM), and NO3− (50 mM) on the SARS-CoV-2 RBD were analyzed. (D) Short-lived species in PAW and the mixed solution of H2O2, NO2−, and NO3−. The fluorescence intensities of the three probes in PAW-10 min and the H2O2 and NO2−/NO3− mixture were measured.
To determine whether long-lived species played a key role, a mixed solution of 150 μM H2O2, 15 μM NO2 −, and 50 mM NO3 − was prepared, with their concentrations higher than those in the PAW, but this solution exhibited only slight inhibition effects on the RBD. The pH value of PAW adjusted to pH 7.0–7.5 using 1 M NaOH did not affect the inhibition effect (Fig. 4C). Further, ozone water and ultraviolet-treated water exhibited little inactivation effect on RBD, while the direct treatment of ultraviolet exhibited a slight inactivation effect (Fig. S4). Therefore, long-lived species do not appear to have played a major role in this process. Because of the limitations of current detection probes for short-lived reactive species, only a few short-lived species can be detected in PAW. For this study, 3′-(p-aminophenyl) fluorescein (APF) and 3ʹ-(p-hydroxyphenyl) fluorescein (HPF) were used as probes for •OH and ONOO−, and coumarin boronic acid pinacolate ester (CBA) was used as a probe for ONOO−. The measurement results were used to compare the short-lived species in the PAW and the mixed solution of H2O2, NO2 −, and NO3 −. The fluorescence intensities of HPF and CBA in PAW were much higher than those in the mixed solution, whereas those of APF in both were similar (Fig. 4D). These results suggested that several short-lived reactive species were present in PAW, including ONOO−, which probably played crucial roles in this inactivation process. Furthermore, the inactivation effect of PAW was also compared with that of medical hydrogen peroxide (3% H2O2). Although both inactivated the RBD, the inactivation effect of PAW was much stronger than that of medical hydrogen peroxide (Fig. S5).
4. Discussion
The ongoing public health crisis caused by SARS-CoV-2 has become a global pandemic. Disinfection of viruses in the environment can effectively prevent their transmission, and an efficient inactivation strategy needs to be established. Based on the pseudovirus model, PAW inactivated pseudoviruses and inhibited infection, which reflected the inactivation ability of PAW on SARS-CoV-2. The strong infectivity of SARS-CoV-2 is due to the high affinity to hACE2, which mainly relies on the RBD of the S protein [6], [36]. The entry receptors hACE2 in cells are indispensable, and the infection of the pseudovirus to the cells without overexpression of hACE2 was hardly detectable [37], [38]. Therefore, the RBD is considered to be an important target for neutralizing antibodies to block virus infection and even to control the pandemic [39], [40]. PAW efficiently inactivated the pseudovirus incorporated with S protein and the RBD of the S protein to disrupt binding to hACE2, thus preventing the viral entry into cells.
The short-lived species generated in PAW, such as ONOO– and O2 •–, selectively react with the amino acids in proteins. Based on previous studies, the tyrosine and tryptophan in protein are highly sensitive to ONOO– and formed 3-nitrotyrosine, 6-nitrotryptophan, and dityrosine, which caused protein aggregation and fragmentation [41]. Oxidation modification or nitration modification caused slight changes in the molecular weights of proteins, therefore the band of PAW-treated RBD in ~38 kDa was modestly greater than that of the untreated RBD protein. The results of mass spectrometry further demonstrated that the molecular weights of RBD proteins were all changed, suggesting that the reactive species in PAW non-specifically reacted and modified the RBD proteins. The treatment of RBD by PAW also caused aggregation (45–65 kDa) and fragmentation (~28 kDa). Therefore, we suggest that the reactive species in PAW reacted with the amino acids of RBD and induced oxidative modification, aggregation, and fragmentation with this resulting in the inactivation of RBD. The reaction of the reactive species in PAW is nonspecific and thus PAW inevitably affects other domains of S protein, which can synergistically enhance the inactivation of S protein. Based on the images of transmission electron microscopy, the treatment of PAW did not disrupt the envelopes of pseudoviruses. Therefore, these support our conclusion that the effect of PAW was on part of S protein outside the envelope.
The reactive species in PAW are dissolved from gaseous reactive species that are produced by the high-voltage breakdown of air. The mixture of long-lived H2O2, NO2 –, and NO3 –, ozone water, and ultraviolet-treated water exhibited little inactivation effects on RBD, suggesting that short-lived species played an important role in this inactivation process in our experiments. In contrast to disinfectants derived from inorganic or organic chemicals, PAW treatment does not result in residual chemical contamination. In the SARS-CoV-2 epidemic, the most commonly used disinfectants include medical alcohol (70–75% ethanol) and chlorine-containing disinfectants. Alcohol has potential safety risks, while chlorine-containing disinfectants are irritant to the human body. In contrast, PAW does not exhibit these two harmful aspects.
As a new disinfectant, the storage time to maintain the inactivation effect of PAW must be taken into account. After storage for 30 days, PAW-10 min was still capable of strong inactivation effects, stronger than those of medical hydrogen peroxide and similar to those of PAW-5 min (Fig. S6). The long-term effectiveness of PAW provides a basis for the development of a disinfectant.
In conclusion, PAW prevented the infection of pseudovirus with SARS-CoV-2 S protein through mechanisms including inactivation of the RBD of the S protein and prevention of RBD binding with hACE2. PAW can be stored for 30 days at least and is of low cost and with no residue, thus demonstrating its clear potential as a new disinfectant. Further studies on SARS-CoV-2 are needed to explore a novel strategy for disinfection to combat the COVID-19 epidemic.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank Dr. Huimin Tong at Instrument Analysis Center of Xi’an Jiaotong University for the assistance with the analysis of transmission electron microscopy. This work was supported by National Natural Science Foundation of China (52041702, 51977174 and 51722705), Fundamental Research Funds for the Central Universities, and Doctoral Fund of Ministry of Education of China (2018T111062).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cej.2020.127742.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K., Washington State V.C.I.T., Case F. Novel coronavirus in the United States. N. Engl. J. Med. 2019;382(2020):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiang Y.T., Li W., Zhang Q., Jin Y., Rao W.W., Zeng L.N., Lok G.K.I., Chow I.H.I., Cheung T., Hall B.J. Timely research papers about COVID-19 in China. Lancet. 2020;395:684–685. doi: 10.1016/S0140-6736(20)30375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y.Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181:223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang D., Comish P., Kang R. The hallmarks of COVID-19 disease. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H., Zhou Y., Zhang M., Wang H., Zhao Q., Liu J. Updated approaches against SARS-CoV-2. Antimicrob. Agents Chemother. 2020;64:e00483–20. doi: 10.1128/AAC.00483-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., Wang Q., Zhou H., Yan J., Qi J. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 9.Li F., Li W.H., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 10.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bekeschus S., Clemen R., Niessner F., Sagwal S.K., Freund E., Schmidt A. Medical gas plasma jet technology targets murine melanoma in an immunogenic fashion. Adv. Sci. 2020;7:1903438. doi: 10.1002/advs.201903438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colagar A.H., Memariani H., Sohbatzadeh F., Omran A.V. Nonthermal atmospheric argon plasma jet effects on Escherichia coli biomacromolecules. Appl. Biochem. Biotechnol. 2013;171:1617–1629. doi: 10.1007/s12010-013-0430-9. [DOI] [PubMed] [Google Scholar]
- 13.Kvam E., Davis B., Mondello F., Garner A.L. Nonthermal atmospheric plasma rapidly disinfects multidrug-resistant microbes by inducing cell surface damage. Antimicrob. Agents Chemother. 2012;56:2028–2036. doi: 10.1128/AAC.05642-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin A., Gorbanev Y., De Backer J., Van Loenhout J., Van Boxem W., Lemiere F., Cos P., Dewilde S., Smits E., Bogaerts A. Non-thermal plasma as a unique delivery system of short-lived reactive oxygen and nitrogen species for immunogenic cell death in melanoma cells. Adv. Sci. 2019;6:1802062. doi: 10.1002/advs.201802062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magureanu M., Mandache N.B., Parvulescu V.I. Degradation of pharmaceutical compounds in water by non-thermal plasma treatment. Water Res. 2015;81:124–136. doi: 10.1016/j.watres.2015.05.037. [DOI] [PubMed] [Google Scholar]
- 16.Moreau M., Orange N., Feuilloley M.G.J. Non-thermal plasma technologies: New tools for bio-decontamination. Biotechnol. Adv. 2008;26:610–617. doi: 10.1016/j.biotechadv.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Assadi A.A., Bouzaza A., Wolbert D. Comparative study between laboratory and large pilot scales for VOC's removal from gas streams in continuous flow surface discharge plasma. Chem. Eng. Res. Des. 2016;106:308–314. [Google Scholar]
- 18.Assadi A.A., Bouzaza A., Wolbert D. Study of synergetic effect by surface discharge plasma/TiO2 combination for indoor air treatment: Sequential and continuous configurations at pilot scale. J. Photoch. Photobio. A. 2015;310:148–154. [Google Scholar]
- 19.Maisch T., Shimizu T., Li Y.F., Heinlin J., Karrer S., Morfill G., Zimmermann J.L. Decolonisation of MRSA, S. aureus and E. coli by cold-atmospheric plasma using a porcine skin model in vitro. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0034610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S.M., Kim J.I. Decomposition of biological macromolecules by plasma generated with helium and oxygen. J. Microbiol. 2006;44:466–471. [PubMed] [Google Scholar]
- 21.Tanaka Y., Yasuda H., Kurita H., Takashima K., Mizuno A. Analysis of the inactivation mechanism of bacteriophage phi X174 by atmospheric pressure discharge plasma. IEEE Trans. Ind. Appl. 2014;50:1397–1401. [Google Scholar]
- 22.Yasuda H., Hashimoto M., Rahman M.M., Takashima K., Mizuno A. States of biological components in bacteria and bacteriophages during inactivation by atmospheric dielectric barrier discharges. Plasma Process. Polym. 2008;5:615–621. [Google Scholar]
- 23.Yasuda H., Miura T., Kurita H., Takashima K., Mizuno A. Biological evaluation of DNA damage in bacteriophages inactivated by atmospheric pressure cold plasma. Plasma Process. Polym. 2010;7:301–308. [Google Scholar]
- 24.Wu Y., Liang Y.D., Wei K., Li W., Yao M.S., Zhang J., Grinshpun S.A. MS2 virus inactivation by atmospheric-pressure cold plasma using different gas carriers and power levels. Appl. Environ. Microbiol. 2015;81:996–1002. doi: 10.1128/AEM.03322-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aboubakr H.A., Williams P., Gangal U., Youssef M.M., El-Sohaimy S.A.A., Bruggeman P.J., Goyal S.M. Virucidal effect of cold atmospheric gaseous plasma on feline calicivirus, a surrogate for human norovirus. Appl. Environ. Microbiol. 2015;81:3612–3622. doi: 10.1128/AEM.00054-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng H., Xu J.X., Li X., Liu D.W., Lu X.P. On the dose of plasma medicine: Equivalent total oxidation potential (ETOP) Phys. Plasmas. 2020;27 [Google Scholar]
- 27.Dai X., Zhang Z., Zhang J., Ostrikov K. Dosing: The key to precision plasma oncology. Plasma Process. Polym. 2020;10:1900178. [Google Scholar]
- 28.Guo L., Xu R., Gou L., Liu Z., Zhao Y., Liu D., Zhang L., Chen H., Kong M.G. Mechanism of virus inactivation by cold atmospheric-pressure plasma and plasma-activated water. Appl. Environ. Microbiol. 2018;84:e00726–18. doi: 10.1128/AEM.00726-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su X., Tian Y., Zhou H., Li Y., Zhang Z., Jiang B., Yang B., Zhang J., Fang J. Inactivation efficacy of non-thermal plasma activated solutions against Newcastle disease virus. Appl. Environ. Microbiol. 2018;84:e02836–17. doi: 10.1128/AEM.02836-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Z., Guo L., Liu D., Rong M., Chen H., Kong M.G. Chemical kinetics and reactive species in normal saline activated by a surface air discharge. Plasma Process. Polym. 2017;14:1600113. [Google Scholar]
- 31.Zhang J., Qu K., Zhang X., Wang B., Wang W., Bi J., Zhang S., Li Z., Kong M.G., Liu D., Liu C. Discharge plasma-activated saline protects against abdominal sepsis by promoting bacterial clearance. Shock. 2019;52:92–101. doi: 10.1097/SHK.0000000000001232. [DOI] [PubMed] [Google Scholar]
- 32.Zhou R.W., Zhou R.S., Wang P.Y., Xian Y.B., Mai-Prochnow A., Lu X.P., Cullen P.J., Ostrikov K., Bazaka K. Plasma-activated water: generation, origin of reactive species and biological applications. J. Phys. D Appl. Phys. 2020;53 [Google Scholar]
- 33.Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M., Lu Q., Li X., Sun Q., Liu J., Fan C., Huang W., Xu M., Wang Y. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020;9:680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo L., Zhao Y., Liu D., Liu Z., Chen C., Xu R., Tian M., Wang X., Chen H., Kong M.G. Cold atmospheric-pressure plasma induces DNA-protein crosslinks through protein oxidation. Free Radic. Res. 2018;7:783–798. doi: 10.1080/10715762.2018.1471476. [DOI] [PubMed] [Google Scholar]
- 35.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai X., Zhang X., Ostrikov K., Abrahamyan L. Host receptors: the key to establishing cells with broad viral tropism for vaccine production. Crit. Rev. Microbiol. 2020;46:147–168. doi: 10.1080/1040841X.2020.1735992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong H.L., Wu Y.T., Cao J.L., Yang R., Liu Y.X., Ma J., Qiao X.Y., Yao X.Y., Zhang B.H., Zhang Y.L., Hou W.H., Shi Y., Xu J.J., Zhang L., Wang S.J., Fu B.R., Yang T., Ge S.X., Zhang J., Yuan Q., Huang B.Y., Li Z.Y., Zhang T.Y., Xia N.S. Robust neutralization assay based on SARS-CoV-2 S-protein-bearing vesicular stomatitis virus (VSV) pseudovirus and ACE2-overexpressing BHK21 cells. Emerg. Microbes Infect. 2020;9:2105–2113. doi: 10.1080/22221751.2020.1815589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang S.B., Hillyer C., Du L.Y. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan M., Wu N.C., Zhu X.Y., Lee C.C.D., So R.T.Y., Lv H.B., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Degendorfer G., Chuang C.Y., Kawasaki H., Hammer A., Malle E., Yamakura F., Davies M.J. Peroxynitrite-mediated oxidation of plasma fibronectin. Free Radic. Biol. Med. 2016;97:602–615. doi: 10.1016/j.freeradbiomed.2016.06.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.