Abstract
Postprandial hyperglycemia has orchestrated untimely death among diabetic patients over the decades and regulation of α-amylase activity is now becoming a promising management option for type 2 diabetes. The present study investigated the binding interactions of three structurally diverse dichalcogenoimidodiphosphinate ligands with α-amylase to ascertain the affinity of the ligands for α-amylase using spectroscopic and molecular docking methods. The ligands were characterized using 1H and 31P NMR spectroscopy and CHN analysis. Diselenoimidodiphosphinate ligand (DY300), dithioimidodiphosphinate ligand (DY301), and thioselenoimidodiphosphinate ligand (DY302) quenched the intrinsic fluorescence intensity of α-amylase via a static quenching mechanism with bimolecular quenching constant (Kq) values in the order of x1011 M-1s-1, indicating formation of enzyme-ligand complexes. A binding stoichiometry of n≈1 was observed for α-amylase, with high binding constants (Ka). α-Amylase inhibition was as follow: Acarbose > DY301>DY300>DY302. Values of thermodynamic parameters obtained at temperatures investigated (298, 304 and 310 K) revealed spontaneous complex formation (ΔG<0) between the ligands and α-amylase; the main driving forces were hydrophobic interactions (with DY300, DY301, except DY302). UV–visible spectroscopy and Förster resonance energy transfer (FRET) affirmed change in enzyme conformation and binding occurrence. Molecular docking revealed ligands interaction with α-amylase via some key catalytic site amino acid residues (Asp197, Glu233 and Asp300). DY301 perhaps showed highest α-amylase inhibition (IC50, 268.11 ± 0.74 μM) due to its moderately high affinity and composition of two sulphide bonds unlike the others. This study might provide theoretical basis for development of novel α-amylase inhibitors from dichalcogenoimidodiphosphinate ligands for management of postprandial hyperglycemia.
Keywords: α-Amylase inhibition, Ligand-protein binding, Spectroscopy, Hyperglycemia, Anti-diabetic agents
Graphical abstract
Synopsis for the Graphical Abstract The present study investigated the binding interaction mechanisms of three structurally diverse dichalcogenoimidodiphosphinate ligands with α-amylase to ascertain the binding affinity of the ligands for α-amylase using spectroscopic techniques and molecular docking. The study provided insights for use of dichalcogenoimidodiphosphinate ligands as novel α-amylase inhibitors for management of postprandial hyperglycemia.

Highlights
-
•
Interaction of α-amylase with dichalcogenoimidodiphosphinate ligands was studied.
-
•
Spectroscopy and molecular docking explored the interaction mechanisms.
-
•
The main driving forces were hydrophobic interactions with DY300 and DY301.
-
•
The ligands quenched α-amylase fluorescence intensity by static mechanism.
-
•
Dichalcogenoimidodiphosphinate ligands inhibited α-amylase activity.
1. Introduction
Diabetes mellitus (DM) is a metabolic disorder characterized by hyperglycemia [1,2]. Hence patients suffering from DM are noted for their excessively high blood glucose concentration. Aside from hypertension and cardiovascular diseases, diabetes mellitus is another highly deleterious disease that leads to death of about 1.6 million people globally per annum [3]. DM can be classified into two types: (i) type 1 DM which is also referred to as insulin dependent diabetes mellitus (IDDM) and, (ii) non-insulin dependent diabetes mellitus (NIDDM) also known as the type 2 DM [2,4,5]. While type 1 diabetes mellitus is due to deficiency or reduced level of insulin in the body, type 2 diabetes mellitus is caused by insulin resistance (inability of insulin receptors to recognize the presence of insulin to facilitate their binding interaction) [6,7]. In a healthy non-diabetic physiological state, binding of insulin to its receptors on cells is necessary to orchestrate a series of transduction reactions that would culminate in transport of glucose from the blood into cells for glucose catabolism [7,8]. The failure of insulin-mediated glucose transport from blood to cells is what results in accumulation of glucose (hyperglycemia) in the blood of diabetic patients [9,10]. While research is still on-going to obtain a cure for diabetes mellitus, a number of approaches had been prescribed for management of the disease ranging from restriction in consumption of high carbohydrate food by patients to use of drugs for chemotherapy [9,11]. Although type 1 DM can be managed by administering insulin to a DM patient by injection, this does not work for type 2 DM patients as theirs is not due to insulin deficiency. Hence, over the years various chemotherapeutic agents such as acarbose, miglitol, glibenclamide, etc. are used in the management of type 2 DM patients [10,12]. Some of these drugs (e.g. acarbose and miglitol) are known to reduce blood glucose level in type 2 DM patients by inhibiting some key enzymes (e.g. α-amylase, α-glucosidase, sucrase, etc.) which are involved in carbohydrate metabolism. This slows down the rate of carbohydrate breakdown, especially immediately after a meal, thus preventing a sudden upsurge in blood glucose level which could be detrimental to DM patients. Alpha-amylase (E.C. 3.2.2.1) breakdowns starch via cleavage of α-1.4 glycosidic bonds in starch molecules, thereby leading to release of products such as maltose and glucose [[13], [14], [15], [16]]. As such, intake of drugs or chemical agents that have α-amylase inhibitory property is one of the significant management approaches used in suppressing or preventing postprandial hyperglycemia in type 2 DM patients [11]. The present-day challenges reported to be associated with the existing therapeutic agents such as acarbose and miglitol include excessive delay in carbohydrate digestion which results in abdominal discomfort, flatulence, etc. [10,17]. These have been linked to the extremely high inhibitory effects of these drugs on α-amylase and other carbohydrases. These side-effects have therefore opened-up a global research gap which researchers attempt to fill by synthesizing new compounds (such as novel polyphenolics, flavonoids, amides, aryl imines, quinolones, etc.), which may have moderately low yet clinically beneficial inhibitory effects on carbohydrate-degrading enzymes such as α-amylase [13,18], for the prevention of postprandial hyperglycemia without the aforementioned side-effects associated with acarbose and other drugs [12]. Although a number of investigations has been made on the α-amylase inhibitory prowess and binding interaction mechanisms of α-amylase with some synthesized compounds especially those belonging to polyphenolic and flavonoid groups [11,12,18], much has not been reported in literature regarding the amides especially the imidodiphosphinates. The imidodiphosphinates, particularly dichalcogenoimidodiphosphinate [R2P(E)NP(E)R'2]- ligands (E = O, S, Se, Te; R, R' = various aryl or alkyl groups) are a class of ligands which form complexes with metals and their metal complexes are capable of acting as (E,E) chelates which exhibit a remarkable variety of three-dimensional structures [19,20]. Due to the great coordinating versatility of these ligands, they produce both single and multinuclear metal complexes, with a variety of bonding modes [19,20]. Within the last few years, researchers have tried to investigate the likely biological impacts or pharmaceutical benefits of some of these imidodiphosphinate ligands or their metal complexes; few works have researched their possible biological effects such as potential inhibition of platelet activating factor [19], as anti-cancers [21], as well as their anti-inflammatory activity towards reumatoidis arthritis [22], based on the assumption that endogenous metals may be less toxic [19]. In spite of the attempts made so far towards understanding the biological activity of imidodiphosphinate ligands or their coordination compounds, there appears to be no report yet in literature neither regarding their potential inhibitory effects on α-amylase nor on their binding interaction mechanisms with α-amylase, a biological macromolecule (an enzyme) implicated in the regulation of postprandial blood glucose levels in type 2 diabetic patients. Before in vivo tests using animal models are carried out, it is very important to investigate using in vitro and in silico approaches the likely biological impacts of dichalcogenoimidodiphosphinate ligands on targeted biomolecules such as α-amylase [9,11,23].
In the present study, we synthesized and characterized three analogues of dichalcogenoimidodiphosphinate ligands using 1H NMR and 31P NMR spectroscopy and CHN analysis and thereafter investigated their binding interaction mechanisms with α-amylase as well as their inhibitory capacities. The three amides varied majorly in their composition of selenium and sulphur atoms (Fig. 1a). While the enzyme inhibition analysis was done using standard biochemical protocols, the binding interaction mechanisms of α-amylase with the synthesized dichalcogenoimidodiphosphinate ligands were investigated using spectroscopic techniques including UV–visible absorption spectroscopy and fluorescence spectroscopy. Thermodynamics parameters, binding constants as well as Förster resonance non-radioactive energy transfer (FRET) between α-amylase and the ligands were also determined. Molecular docking was used to probe the binding interactions between the dichalcogenoimidodiphosphinate ligands and amino acid residues present at the active site of the enzyme. To the best of our knowledge, based on existing literature, the binding interaction between α-amylase and dichalcogenoimidodiphosphinate ligands is first reported in the present study.
Fig. 1a.
Structures of dichalcogenoimidodiphosphinate ligands (DY300, DY301 and DY302) investigated in the present study.
2. Material and methods
2.1. Material and reagents used
Lyophilized α-amylase from Bacillus subtilis (Mayer Baker, India) was used in this study. Under an atmosphere of dry nitrogen, all syntheses occurred by using a standard Schlenk line, connected to a vacuum pump. All chemicals (from Merck and Fluka) were used without any further purification(s). All solvents were anhydrous and obtained either through distillation or storage through molecular sieves. Elemental analyses were performed by the microanalysis section of the Institute of Organic Chemistry and Macromolecular Chemistry, Friedrich-Schiller-Universität, Jena. NMR measurements were done on Bruker Avance (III) 400 MHz FT-NMR spectrometer through deuterated solvent(s). 1H NMR analyses were referenced to SiMe4, while 31P NMR spectra were measured in reference to tetraoxophosphate (V) acid, H3PO4. Melting point was recorded on a Gallenkamp melting point apparatus.
2.2. Synthesis and characterization of imidodiphosphinate ligands
2.2.1. Synthesis of [iPr2P(Se)NHP(Se)iPr2] (diselenoimidodiphosphinate ligand, DY300)
This ligand DY300 was synthesized as described by Ref. [37,39,41]. In hot toluene (50 mL), a solution of chlorodiisopropylphosphine (25 g, 26 mL, 164 mmol) was added in drops to a solution of 1, 1, 1, 3, 3, 3-hexamethyldisilazane (13.20 g, 17 mL, 82 mmol) for 30 min. At 90°C, this mixture was heated and stirred for 3 h, and the reaction cooled to room temperature. Selenium powder (12.95 g, 164 mmol) was added and the mixture stirred and refluxed for 6 h. An off-yellow solution was obtained and left to cool overnight. Solution was filtered to give an off-white crude product. Dissolution in hot dichloromethane, Celite filtration (for excess selenium removal) and solvent removal under reduced pressure produced a white crystalline powder. Yield: 11.13 g (33%), M. Pt: 140–142°C. Elemental analysis: Calculated for C12H29NP2Se2: C, 35.39; H, 7.18; N, 3.44%. Found: C, 36.45; H, 7.28; N, 3.40%. 1H NMR, (δ, CDCl3, 400 MHz): δ = 1.34 (m, 24H, 8xCH(CH3)2); 2.70 (m, 4H, 4xCH(CH3)2); 3.10 (s, 1H, –NH). 31P{1H} NMR: 90.05 ppm. 1JP-Se = 751 Hz.
2.2.2. Synthesis of [iPr2P(S)NHP(S)iPr2]1 (dithioimidodiphosphinate ligand, DY301)
This ligand DY301 was synthesized as described by Ref. [37,38,40]. In hot toluene (50 mL), a solution of chlorodiisopropylphosphine (25 g, 26 mL, 164 mmol) was added in drops to a solution of 1, 1, 1, 3, 3, 3-hexamethyldisilazane (13.20 g, 17 mL, 82 mmol) for 30 min. At 90°C, mixture was refluxed with stirring for 3 h, and the reaction cooled to room temperature. Sulphur powder (5.30 g, 164 mmol) was added and the mixture stirred and refluxed for 6 h and cooled overnight. A creamy solution was obtained and filtered to give a white residue. This was washed with CS2 (2 × 10 ml) and n-hexane (25 ml). A white fluffy, free-flowing crystalline product obtained. Yield: 9.94 g (39%), M. Pt: 148–150°C. Elemental analysis: Calculated for C12H29NP2S2: C, 45.98; H, 9.33; N, 4.47; S, 20.46%. Found: C, 45.97; H, 9.50; N, 4.50; S, 20.10%. 1H NMR, (δ, CDCl3, 400 MHz): δ = 1.30 (m, 24H, 8xCH(CH3)2); 2.50 (m, 4H, 4xCH(CH3)2); 2.80 (s, 1H, –NH). 31P{1H} NMR: 90.62 ppm.
2.2.3. Synthesis of [iPr2P(S)NHP(Se)iPr2]1 (thioselenoimidodiphosphinate ligand, DY302)
This ligand DY302 was synthesized as described by Ref. [42]. In hot toluene (30 mL), a solution of chlorodiisopropylphosphine (25 g, 25.80 mL, 163.80 mmol) was added in drops to a solution of 1, 1, 1, 3, 3, 3-hexamethyldisilizane (13.20 g, 17 ml, 82 mmol) for 30 min. At 90°C, mixture was refluxed for 3 h and cooled to room temperature. Selenium powder (6.47 g, 82 mmol) was added and the solution stirred overnight. Sulphur powder (2.63 g, 82 mmol) was added and the mixture stirred for further 6 h. A white solution was obtained, filtered, air-dried and dissolved in hot dichloromethane. Hot filtration to remove excess reactant followed by solvent removal under reduced pressure produced a crystalline creamy/off-white product. Yield: 10.35 g (35%), M. Pt: 138–140°C. Elemental analysis: Calculated for C12H29NP2SSe: C, 40.00; H, 8.11; N, 3.89; S, 8.90%. Found: C, 40.22; H, 8.33; N, 3.93; S, 9.08%. 1H NMR, (δ, CDCl3, 400 MHz): δ = 1.34 (m, 24H, 8xCH(CH3)2); 2.50 (m, 2H, Se P–CH(CH3)2); 2.75 (m, 2H, S P–CH(CH3)2); 3.20 (s, 1H, -NH). 31P{1H}NMR: 93.04, 89.15 ppm. 1JP-Se = 504 Hz.
The reaction scheme for all the synthesized dichalcogenoimidodiphosphinate ligands is as shown in Fig. 1b.
Fig. 1b.
Reaction scheme for the synthesis of dichalcogenoimidodiphosphinate ligands.
2.2.4. Preparation of ligand and acarbose stock/working solutions
Each ligand was weighed (0.2 g), dissolved with 5 mL of absolute ethanol (HPLC Grade, Sigma Aldrich) and diluted to a final volume of 100 mL with distilled water. Subsequent concentrations of the ligand used for the experiment were made from the stock via serial dilution using distilled water as diluent. A stock solution of acarbose (Glucobay®), a standard anti-diabetic drug, was prepared in a similar manner to a concentration of 1000 μg/mL. Thereafter, other concentrations of the standard α-amylase inhibitor (acarbose) were prepared via serial dilutions of the stock and used as positive control.
2.3. Protein binding and interaction studies
2.3.1. UV absorption spectroscopic analysis
The binding interactions between α-amylase and each of the three dichalcogenoimidodiphosphinate ligands (DY300, DY301 and DY302) were investigated using ultraviolet (UV)-visible absorption spectroscopy. UV–visible absorption spectra were recorded on a Shimadzu UV-1650pc spectrophotometer following the methods of Ernest et al. [24] and Wang et al. [25] with some modifications. 100 μL of different concentrations (0.0000, 0.0667, 0.1333, 0.1667 and 0.2000 mg mL-1) of dichalcogenoimidodiphosphinate ligands were added to 3 mL of α-amylase solution (0.5 mg mL-1) prepared with 200 mM phosphate buffer (pH 6.9) containing 6 mM NaCl. The absorption spectra were recorded from 200 to 400 nm after 5 min using a 1 mL Quartz cuvette at 298 K. The absorption spectra of α-amylase solution read in the absence of the ligands (but with 100 μL of the buffer) under the same experimental conditions served as reference [25].
2.3.2. Fluorescence quenching analysis
The intrinsic fluorescence quenching spectra of α-amylase both in the absence and presence of the ligands (DY300, DY301 and DY302) were measured using Cary Eclipse Fluorescence Spectrophotometer (Agilent Technologies) following the procedures described by Lin et al. [26] and Tang et al. [27] with slight modifications. Briefly, 3 mL of α-amylase solution (0.5 mg/mL) prepared with 0.2 mol L-1 phosphate buffer (pH 6.9) containing 0.006 mol L-1 NaCl was incubated with 100 μL of the ligands (0.0000, 0.0667, 0.1333, 0.1667 and 0.2000 mg mL-1) for 5 min at three temperatures (298, 304 and 310 K). The intrinsic fluorescence intensities of the free enzyme and enzyme-ligand mixtures were measured at an excitation wavelength of 280 nm and emission wavelengths ranging from 290 - 500 nm. The values of Stern-Volmer quenching constant (Ksv), the binding constant (Ka), and the binding stoichiometry for α-amylase (n, equivalent to the number of binding sites per enzyme molecule) [28] were calculated from the Stern-Volmer equation (Eq. (1)) and the modified Stern-Volmer equation (Eq. (2)). The Fo and F denote the fluorescence intensities of α-amylase with or without any of the dichalcogenoimidodiphosphinate ligands, respectively; τ0 is the lifetime of a fluorophore in the absence of the quencher (τ0 = 10-8 s for a biomolecule) [28,29]; Kq represents the bimolecular quenching constant, which is equal to Ksv/τ0; and [Q] is the concentration of the quencher.
| (1) |
| (2) |
2.3.3. Resonance energy transfer analysis
The UV/vis absorption spectra of the ligands (DY300, DY301 and DY302) were recorded at a concentration of 0.20 mg/mL at room temperature over a wavelength range of 200–500 nm in the absence of the enzyme. UV–vis-IR (a/e) FluorTools software was used for the analysis of overlap integral of emission fluorescence and UV-absorption spectral plots.
2.3.4. Assay for inhibition of α-amylase activity
Determination of the α-amylase inhibitory property of the ligands was carried out following procedures described by Adefegha et al. [10] with slight modifications. Each ligand (DY300, DY301 or DY302) at an increasing concentration (500 μL, 66.70–200.00 μg mL-1 in 0.2 M sodium phosphate buffer, pH 6.9) was added to α-amylase (E.C 3.2.1.1) solution (500 μL, 5 mg/mL), and incubated for 10 min at 37°C. Thereafter, starch hydrolysis reaction was initiated by adding 500 μL of 2% starch solution to the reaction mixture. The reaction was terminated after incubating at 37°C for 10 min via addition of 250 μL of DNS reagent and 12% sodium potassium tartrate (250 μL). The mixture was diluted with 3 mL distilled water after incubating in a hot water bath (95–98°C) for 5 min. The absorbance values were measured at 540 nm using a UV–Visible spectrophotometer (UV-1650pc, Shimadzu, Japan) after cooling to room temperature. The reference had 500 μL of buffer solution in place of the α-amylase solution. Percent inhibition was calculated by:
| (3) |
where, Abssam is the absorbance of the sample (DY300, DY301 and DY302) and Absref is the absorbance of reference.
2.3.5. Molecular docking analysis
Molecular docking study was performed using AutoDock Tool v1.5.6. Three dimensional structure of human pancreatic α-amylase (PDB ID: 5E0F) was downloaded from the RCSB Protein Data Bank (http://www.rcsb.org/pdb). Water molecules and co-crystallised ligands were removed while polar hydrogens were added to the protein (receptor) before assigning Kollman charges. The grid box size for the receptor-ligand interaction was set to 80 x 72 × 66 Å in the x, y and z dimension with 0.375 Å spacing while the grid box site was −7.836, 10.509 and −22.951 Å (x, y and z). The structures of the ligands were sketched and optimized using ChemDraw Ultra 12.0. Polar hydrogens were added to the structures and Gasteiger charges assigned. The outputs of the docking analysis were rendered by PyMol (http://www.pymol.org/). LigPlot+ was used for the analysis of the protein-ligand interactions.
2.4. Statistical analysis
Data were analyzed and presented as mean ± standard error of mean of triplicate measurements. Graphs were plotted using Microsoft Excel and OriginPro (version 9.1).
3. Results and discussion
3.1. Inhibition effect of dichalcogenoimidodiphosphinate ligands against α-amylase
The inhibitory effects of the three structurally diverse ligands (DY300, DY301 and DY302) on the starch-hydrolyzing action of α-amylase were investigated in the present study and their relative α-amylase inhibitory capacities expressed in terms of their IC50 (the concentration of inhibitor at which 50% of enzyme catalytic activity is suppressed) [11,13]. From the results shown in Fig. 2, the IC50 values of diselenoimidodiphosphinate ligand (DY300), dithioimidodiphosphinate ligand (DY301) and thioselenoimidodiphosphinate ligand (DY302) towards α-amylase inhibition were in the order: DY302 > DY300 > DY301. This indicated that dithioimidodiphosphinate ligand (DY301) inhibited α-amylase the most while thioselenoimidodiphosphinate ligand (DY302) inhibited the starch-hydrolyzing enzyme the least. Aside from the impact of the four isopropyl groups present in each of these ligands, the observed variation in α-amylase inhibition exhibited by these ligands could be ascribed to the different compositions of atoms in the ligand, with respect to selenium and sulphur. While DY302 (with the least inhibitory effect, IC50 = 375.60 ± 1.19 μM) has one atom each of selenium and sulphur, each also attached to the phosphorus having the isopropyl groups in its moiety, DY301 (which has the highest inhibitory effect, IC50 = 268.11 ± 0.74 μM) possesses two sulphur atoms (and no selenium atom) attached to the phosphorus atoms in its moiety which are doubly bonded to each other. However, the ligand with the intermediate inhibitory effect (DY300, IC50 = 372.14 ± 0.75 μM), has two selenium atoms (and no sulphur atom) doubly bonded to the phosphorus carrying the isopropyl groups within the structure. Hence, the α-amylase inhibitory effect of the three ligands investigated in this present study increased mostly with increase in the number of sulphur atoms in the ligands and also partly with increase in selenium atoms. As confirmed from our molecular docking results (detailed discussion provided in Section 3.7), the selenium and sulphur atoms within the ligands were involved in some non-covalent interactions with some of the amino acid residues present at the catalytic site of the α-amylase. Hence, this contributed to the overall inhibitory effects of the dichalcogenoimidodiphosphinate ligands on the starch-hydrolyzing enzyme [12,13,18]. It is noteworthy to affirm that the dichalcogenoimidodiphosphinate ligands exhibited a moderate yet clinically more beneficial inhibitory prowess against α-amylase when compared to the extremely high inhibitory effect of a standard α-amylase inhibitor (acarbose, IC50 = 167.78 ± 0.93 μM) obtained under similar experimental conditions of this study (Fig. 2). This implies that the potential use of these analogues of imidodiphosphinate ligand would, perhaps, not excessively delay the breakdown of α-1,4-glycosidic bonds in starch molecules during digestion in diabetic patients as reported in literature regarding use of acarbose with which flatulence, diarrhoea, high blood pressure etc (resulting from excessive α-amylase inhibition) have been associated [10,13]. Therefore, the moderate inhibitory prowess of these dichalcogenoimidodiphosphinate ligands on α-amylase might be a good indication for their potential use as alternatives to acarbose in prevention of postprandial hyperglycemia in diabetic subjects after subsequent complementary clinical trials [9,10,23].
Fig. 2.
Inhibition of α-amylase activity (%) by structural analogues of dichalcogenoimidodiphosphinate ligands. (A) Diselenoimidodiphosphinate ligand (DY300); (B) dithioimidodiphosphinate ligand (DY301), (C) thioselenoimidodiphosphinate ligand (DY302) and (D) acarbose. The extrapolated IC50 of DY300, DY301 and DY302 for α-amylase were 372.14 ± 0.75, 268.11 ± 0.74, 375.60 ± 1.19 and 167.78 ± 0.93 μM, respectively.
3.2. Quenching of α-amylase fluorescence intensity by dichalcogenoimidodiphosphinate analogues
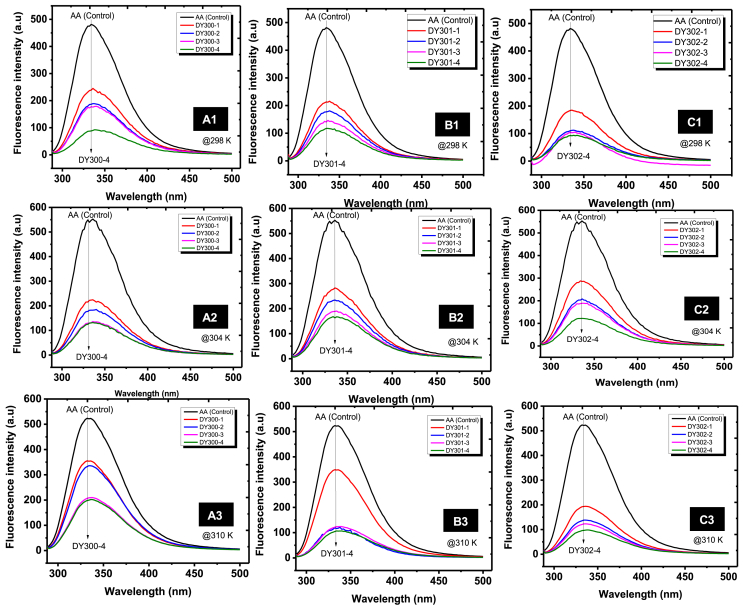
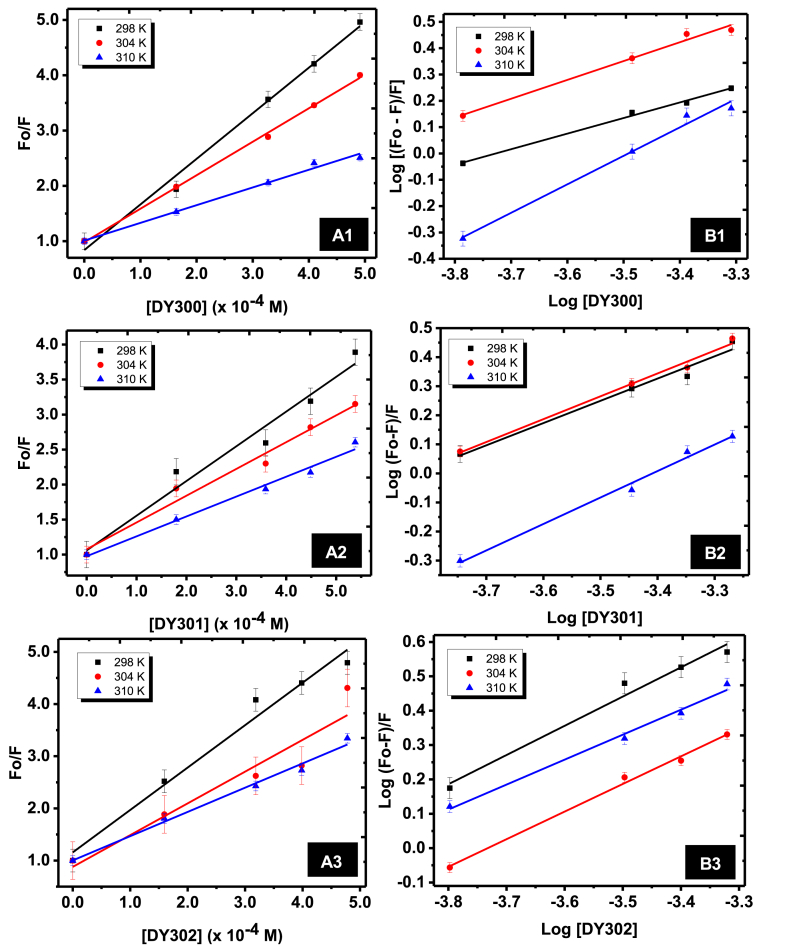
Fluorescence quenching analysis was carried out in the present study to determine the binding affinities of dichalcogenoimidodiphosphinate ligands to α-amylase [30]. Fig. 3 shows the fluorescence intensity spectra of α-amylase at various temperatures (298, 304 and 310 K) in the presence of the three ligands investigated (DY300, DY301 and DY302), respectively. Increase in the concentration of dichalcogenoimidodiphosphinate ligand at constant concentration of α-amylase resulted in decrease in the fluorescence intensity of α-amylase. At 298 K, the free enzyme exhibited a maximum fluorescence at 336 nm, which shifted towards 353, 335 and 351 nm in the presence of 0.20 mg mL-1 of DY300, DY301 and DY302, respectively. Similar alterations in α-amylase maximum fluorescence were also observed at 304 and 310 K. The observed shifts implied that the ligands exhibited potential quenching effects on α-amylase fluorescence and perhaps altered the structural architecture of the enzyme. To understand the fluorescence quenching mechanism of α-amylase by these dichalcogenoimidodiphosphinate analogues, Stern–Volmer parameters were deduced from the plots of Fo/F versus Log [Q] (Fig. 4A). As shown in Table 1, the values of the bimolecular quenching constant (Kq) obtained in this study for α-amylase interaction with the three dichalcogenoimidodiphosphinate analogues (DY300, DY301 and DY302) were in the order of x 1011 M−1 s−1, significantly higher than the maximum scattering collision quenching constant value of 2.0 × 1010 M−1 s−1. This indicated that these ligands quenched α-amylase fluorescence via a static quenching mechanism (which involves complex formation) and not by dynamic quenching [30,31]. Moreover, the observed decrease in Stern-Volmer quenching constant (Ksv) values as well as the bimolecular quenching constant (Kq) values as temperature increased was an indication that high temperature did not favour the quenching process, and the binding interactions between these ligands and α-amylase were exothermic in nature [27,30].
Fig. 3.
Fluorescence intensity of α-amylase in the absence and presence of varying concentrations of dichalcogenoimidodiphosphinate ligands under different temperature conditions (298, 304 and 310 K) at pH 6.8. (A) DY300; (B) DY301, and (C) DY302.
Fig. 4.
Stern-Volmer (A) and modified Stern-Volmer (B) plots for binding interactions between α-amylase and dichalcogenoimidodiphosphinate ligands (DY300, DY301 and DY302) at pH 6.8 under different temperature conditions (298, 304 and 310 K).
Table 1.
Stern-Volmer quenching constant (Ksv), bimolecular quenching constant (Kq), binding constant (Ka) and number of binding sites (n) involved in the interaction between α-amylase and dichalcogenoimidodiphosphinate ligands.
| Temp. (K) | Ksv (x103 M-1) | Kq (x1011 M-1 s-1) | aR2 | Ka (x103 M-1) | n | bR2 |
|---|---|---|---|---|---|---|
| DY300 | ||||||
| 298 | 7.83 ± 0.05 | 7.83 ± 0.05 | 0.9876 | 0.16 ± 0.01 | 0.59 ± 0.03 | 0.9952 |
| 304 | 6.01 ± 0.01 | 6.01 ± 0.01 | 0.9984 | 0.72 ± 0.02 | 0.72 ± 0.06 | 0.9868 |
| 310 | 3.23 ± 0.02 | 3.23 ± 0.02 | 0.9911 | 6.03 ± 0.03 | 1.09 ± 0.08 | 0.9888 |
| DY301 | ||||||
| 298 | 4.299 ± 0.05 | 4.30 ± 0.05 | 0.9668 | 0.749 ± 0.03 | 0.77 ± 0.08 | 0.9770 |
| 304 | 3.377 ± 0.03 | 3.38 ± 0.03 | 0.9749 | 0.914 ± 0.02 | 0.79 ± 0.05 | 0.9911 |
| 310 | 2.371 ± 0.02 | 2.37 ± 0.02 | 0.9801 | 1.092 ± 0.02 | 0.91 ± 0.06 | 0.9911 |
| DY302 | ||||||
| 298 | 7.358 ± 0.07 | 7.36 ± 0.07 | 0.9759 | 2.303 ± 0.03 | 0.85 ± 0.09 | 0.9793 |
| 304 | 4.967 ± 0.11 | 4.97 ± 0.11 | 0.9038 | 0.886 ± 0.02 | 0.80 ± 0.04 | 0.9946 |
| 310 | 4.009 ± 0.03 | 4.01 ± 0.03 | 0.9885 | 0.669 ± 0.02 | 0.73 ± 0.05 | 0.9910 |
R2, R-squared values for the linear regression involving the Stern-Volmer quenching constant.
R2, R-squared values for the linear regression involving the binding constant, Ka.
3.3. Binding constants and number of binding site(s) on α-amylase
For a protein-ligand binding interaction that involved complex formation, determination of the binding constant (Ka) and the number of binding site(s) (n) was achieved using the modified Stern-Volmer’s plot of log[(F0–F)/F] versus log[Q] (Fig. 4B). The results of Ka and n for the interactions between the dichalcogenoimidodiphosphinate analogues with α-amylase at three different temperatures are as shown in Table 1. Values of the binding or association constant (Ka) are used in estimating the affinity or degree of association between a protein and a ligand, with a higher Ka value indicating a closer association (stronger affinity) between the fluorophore and quencher [27,28,30]. In the present study, the Ka values of α-amylase for the three ligands at 298 K were in the order DY300<DY301<DY302. This suggested that the affinity between thioselenoimidodiphosphinate ligand (DY302) and α-amylase was the highest at that temperature [30], whereas that between diselenoimidodiphosphinate ligand (DY300) and α-amylase was the lowest. It is, however, noteworthy to mention that values of binding constants (Ka) measured at the investigated temperatures were all in the order of x103 M-1, which indicated a moderately strong affinity between the enzyme and all the ligands. The values of the binding stoichiometry (n) between α-amylase and the three dichalcogenoimidodiphosphinate ligands used in this study, which are equivalent to the number of binding sites per enzyme molecule [28], were approximately equal to 1 at the three temperatures investigated. This showed that the dichalcogenoimidodiphosphinate ligands interacted with α-amylase molecule via a single binding site [25,32]. In addition, the value of the binding stoichiometry (n ≈ 1) also indicated that the biological macromolecule (α-amylase) interacted with each ligand in a ratio of 1:1 [25,32].
3.4. Conformational change in α-amylase
One of the effective techniques for monitoring the changes in protein conformation during binding with ligand is absorption spectroscopy [32,33]. In the present study, the absorption spectra of α-amylase in the presence of increasing concentrations of each of the dichalcogenoimidodiphosphinate ligands investigated (DY300, DY301 and DY302) is as shown in Fig. 5. Generally, absorption peak at ∼207 nm is ascribed to the π → π∗ transition of carbonyl group in peptide bond [32], whereas absorption peak around 278 nm is assigned to π → π∗ transition of aromatic groups (for example tyrosine, phenylalanine and tryptophan residues in proteins) [32,34]. Our results in Fig. 5 showed that the absorption peaks of α-amylase at ∼207 nm and around 278 nm increased regularly in the presence of the ligands. This implied that each of the three ligands combined with α-amylase and altered its conformation thereby increasing the aromatic group π → π ∗ transition probabilities. Furthermore, this also signified the formation of complexes between the dichalcogenoimidodiphosphinate ligands and α-amylase; which is in agreement with results of the fluorescence spectroscopy reported in this study that the ligands quenched the intrinsic fluorescence of α-amylase via a static mechanism.
Fig. 5.
UV–visible spectra of α-amylase in the absence and presence of varying concentrations of the dichalcogenoimidodiphosphinate ligands at pH 6.8 and 298 K; (A) DY300; (B) DY301, and (C) DY302.
3.5. Binding distances and non-radioactive energy transfer
One notable mechanism used in describing transfer of energy from a donor molecule to an acceptor molecule through nonradioactive dipole–dipole coupling is the Förster resonance energy transfer (FRET) which is widely used in estimating the spatial distances between the donor (protein) and the acceptor (interacting ligand) [34]. Generally, an overlap in the emission spectrum of the donor (α-amylase) with the absorption spectrum of the acceptor (the dichalcogenoimidodiphosphinate ligand) results in non-radioactive transfer of energy from the donor (tryptophan residues in α-amylase) to the acceptor [35]. Following the Förster non-radioactive energy transfer theory, the distance between donor and acceptor could therefore be estimated. The energy transfer efficiency (E) can be calculated as stated in equation (4) [34,35]:
| (4) |
where F and Fo are the fluorescence intensities of α-amylase in the presence and absence of acceptor, respectively; Ro represents the critical distance when the energy transfer efficiency is 50% while r denotes the average distance between the donor and acceptor. Ro can be estimated as indicated in equation (5) [34,35]:
| (5) |
where k2 denotes spatial orientation factor of the dipole with k2 = 2/3 for random orientation in fluid solution; N denotes the average refractive index of the medium (N = 1.288 for α-amylase solution in the present case). The fluorescence quantum yield (Φ or Φx) of α-amylase can be measured by comparing the fluorescence intensity of α-amylase solution (0.5 mg/mL) and a standard solution (bovine serum albumin, BSA) under identical condition. Equation (6) can be used to calculate the quantum yield of α-amylase as follow [26]:
| (6) |
Where Ax and Astd denote the absorbances of α-amylase and BSA, respectively, at the excitation wavelength of BSA while Fx and Fstd represent the fluorescence intensities of α-amylase and BSA, respectively. Φstd is the fluorescence quantum yield of the standard, which is 0.118 for BSA [34] while Φx is the fluorescence quantum yield of α-amylase (Φ). Based on Eq. (6), the value of Φx was calculated to be 0.0292. The spectral overlap integral [J(λ)] between the emission spectrum of the donor and the absorption spectrum of the acceptors can be determined based on equation (7) [35]:
| (7) |
where F(λ) is the normalized fluorescence intensity of the donor at the wavelength (λ) and ε(λ) is the extinction coefficient of the acceptor at the wavelength.
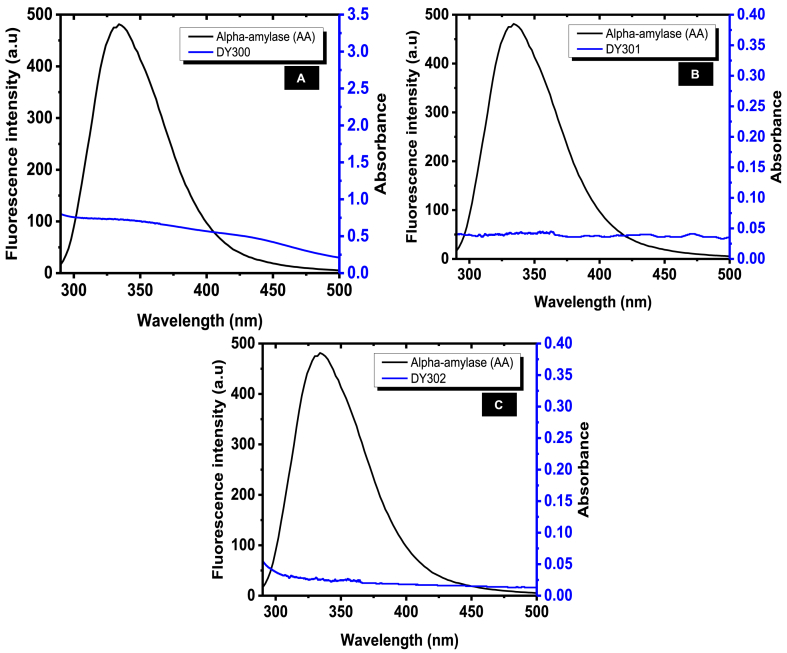
The overlap of the fluorescence emission spectrum of α-amylase and the absorption spectra of the three dichalcogenoimidodiphosphinate ligands are as shown in Fig. 6. The calculated values of J, E, Ro and r for the enzyme-ligand complexes at 298 K are as shown in Table 2. Noticeably, the binding distances (r in nm) between α-amylase and each of the ligands was r < 8 nm and 0.5Ro < r < 1.5Ro. This indicated that the fluorescence quenching of α-amylase by these dichalcogenoimidodiphosphinate ligands was also a non-radiation energy transfer process [26,35]. In addition, since the value of r > Ro it indicated further, and affirmed other results of the present study, that the fluorescence quenching of α-amylase induced with DY300, DY301, and DY302 occurred via static quenching mechanism [26].
Fig. 6.
The overlap between emission spectrum of α-amylase (0.5 mg mL-1) and absorption spectrum of (A) DY300, (B) DY301, and (C) DY302 (0.2 mg mL-1) at room temperature.
Table 2.
Parameters related to non-radioactive energy transfer for α-amylase- dichalcogenoimidodiphosphinate complexes.
| Complex | J (cm3 M-1) | E (%) | Ro (nm) | r (nm) |
|---|---|---|---|---|
| α-Amylase-DY300 | 2.114 × 10-17 | 45.25 | 3.31 | 3.42 |
| α-Amylase-DY301 | 9.979 × 10-19 | 25.46 | 1.99 | 2.38 |
| α-Amylase-DY302 | 6.612 × 10-19 | 36.88 | 1.86 | 2.03 |
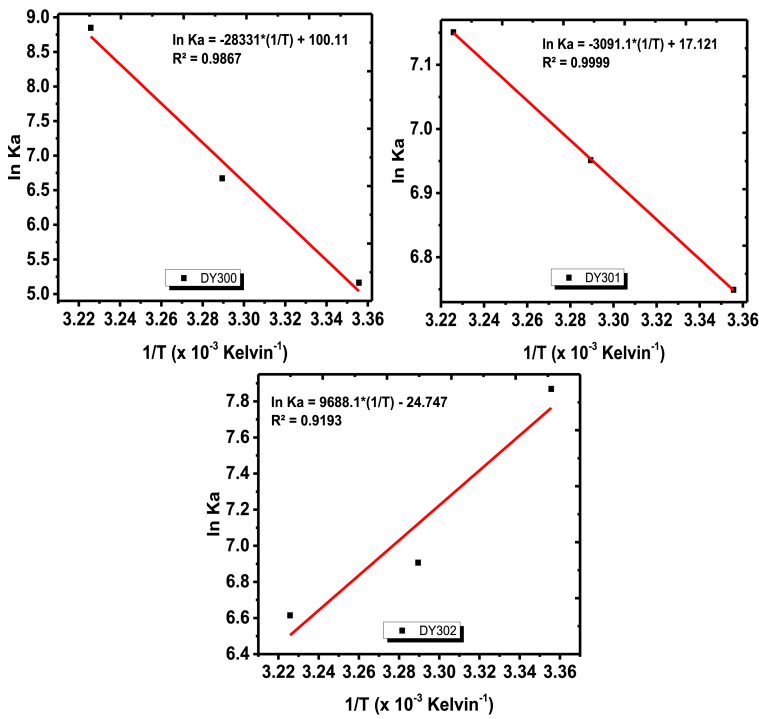
3.6. Thermodynamic parameters and nature of the binding forces
The major molecular driving forces for the binding interaction between α-amylase and the dichalcogenoimidodiphosphinate ligands were characterized by analyzing the thermodynamic parameters. The binding of a small molecule to biological macromolecules generally involves the combination of two or more interaction forces such as hydrophobic interaction, van der Waals force, electrostatic force, and hydrogen bond interaction [25,32]. The specific kind of binding interacting forces can be predicted from values of the thermodynamic parameters [change in enthalpy (ΔH), change in Gibb’s free energy (ΔG), and change in entropy (ΔS)] obtained from Van’t Hoff’s plot (Fig. 7) and the Gibbs-Helmoholtz equation (ΔG = ΔH - TΔS). The thermodynamics criteria used for the interpretation are as follows [25,36]:
-
(a)
ΔH>0 and ΔS>0 denote presence of hydrophobic bonds;
-
(b)
ΔH<0 and ΔS<0 represent hydrogen bond and/or van der Waals forces; and,
-
(c)
ΔH≈0 and ΔS>0 indicate an electrostatic force.
Fig. 7.
Van’t Hoff plots for the determination of thermodynamic parameters at pH 6.8.
The results obtained (Table 3) in this study suggested that the binding processes between α-amylase and each of the three ligands took place spontaneously since ΔG<0 at different temperatures [27]. The values of ΔH and ΔS were positive for the interaction between α-amylase and two of the ligands [diselenoimidodiphosphinate ligand (DY300) and dithioimidodiphosphinate ligand (DY301)]. This revealed that the predominant driving forces involved in their interaction processes with α-amylase were hydrophobic interactions. However, negative values of ΔH and ΔS were obtained for the interaction between α-amylase and thioselenoimidodiphosphinate ligand (DY302), which suggested that hydrogen bond and van der Waals forces were the main driving forces between the enzyme and DY302 [25,32].
Table 3.
Thermodynamic parameters of the interaction between the dichalcogenoimidodiphosphinate ligands and α-amylase at pH 6.8 under different temperatures.
| Temp. (K) | ΔH (kJ/mol) | ΔG (kJ/mol) | ΔS (kJ/mol/K) |
| DY300 | |||
| 298 | 231.35 | −12.31 | 0.818 |
| 304 | −17.22 | ||
| 310 | −22.12 | ||
| DY301 | |||
| 298 | 24.12 | −16.41 | 0.136 |
| 304 | −17.22 | ||
| 310 | −18.04 | ||
| DY302 | |||
| 298 | −79.37 | −18.92 | −0.203 |
| 304 | −17.70 | ||
| 310 | −16.48 | ||
3.7. Molecular docking
The dichalcogenoimidodiphosphinate ligands (DY300, DY301 and DY302) were also analyzed by molecular docking to further probe into the nature of their interactions with amylase protein, especially their involvement with catalytic amino acid residues [18,23]. The ligands docked with binding energies (ΔGb) of −5.5 kcal/mol for DY300 and DY301, while DY302 docked with a better binding energy of −5.7 kcal/mol. As shown in Fig. 8 and Table 4, the ligands (DY300, DY301 and DY302) docked into the active site of human pancreatic α-amylase, interacted with three key amino acid residues (Asp197, Glu233, and Asp300) involved in catalysis within the catalytic pocket [30], surrounded with other amino acid residues such as Trp58, Trp59, Tyr62, Gln63, His101, Thr163, Leu165, and Ala198 in slightly different combinations within 4 Å. The results of molecular docking revealed that none of the ligands bound to the active site of α-amylase via electrostatic interaction. However, the ligands were stabilized by non-bonded interactions to mostly non-polar amino acids in the binding pocket of α-amylase as shown in Fig. 8 and Table 4. The molecular docking results corroborated the fluorescence-based thermodynamics parameters previously reported in this study which showed that the main driving forces for the interaction between α-amylase and DY300/DY301 ligands were hydrophobic interactions, while van der Waals forces (in combination with hydrogen bond) were responsible for DY302. It is very likely that the additional driving force (hydrogen bond) indicated for DY302 ligand’s interaction with α-amylase by the fluorescence-based thermodynamics parameters, but not seen during the docking process, was one that was weakly formed when the ligand interacted with the enzyme in the aqueous (buffer) medium hence the in silico analysis was unable to reveal it. Additionally, the varying binding affinities (Table 4) of the dichalcogenoimidodiphosphinate ligands for α-amylase might be due to dipoles created in each ligand, since they differ only in their compositions of sulphur and selenium. Besides the isopropyl side group atoms, the selenium and sulphur atoms within the ligands were also involved in some non-covalent interactions with some of the amino acid residues present at the catalytic site of α-amylase [Leu165 interacted with selenium (Se) from DY300; Thr163 interacted with sulphur (S) from DY301 and DY302]. All these contributed to the observed overall inhibitory effects of the dichalcogenoimidodiphosphinate ligands on the starch-hydrolyzing enzyme.
Fig. 8.
Surface representation of α-amylase (PDB ID:5E0F) showing the ligands at the protein active site (coloured magenta). The enzyme-ligand complexes [DY300 (A1-A2), DY301 (B1–B2), and DY302 (C1–C2)] were stabilized mostly by non-covalent forces (indicated with dotted red lines), which emanated from the active site amino acid residues indicated in the inserted figures (A2, B2 and C2). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 4.
Molecular docking binding affinities of dichalcogenoimidodiphosphinate ligands with human pancreatic α-amylase.
| Compound | Binding affinity (kcal/mol) | Non-bonded interacting amino acid residues at the active site |
|---|---|---|
| DY300 | −5.5 | Trp58, Trp59, Leu165, Gln63, Tyr62, Glu233, Ala198, Asp197, Asp300, His101 |
| DY301 | −5.5 | Trp59, Leu165, Gln63, Tyr62, Asp197, Glu233, Asp300, Thr163 |
| DY302 | −5.7 | Trp59, Tyr62, Asp197, Asp300, Thr163, Leu165, Glu233 |
4. Conclusion
In this study, a comparative investigation of the binding interaction mechanisms of α-amylase with three structurally diverse dichalcogenoimidodiphosphinate ligands [diselenoimidodiphosphinate ligand (DY300), dithioimidodiphosphinate ligand (DY301) and thioselenoimidodiphosphinate ligand (DY302)] was reported for the first time. The compounds were synthesized and characterized using 1H NMR spectroscopy and CHN analysis. All the samples exhibited inhibition potential against α-amylase activity in vitro, with DY301 showing the highest inhibitory effect (IC50, 268.11 ± 0.74 μM) and DY302 the least (IC50, 375.60 ± 1.19 μM). The binding interaction studies revealed a binding stoichiometry (n) of approximately one for α-amylase, which indicated that the imidodiphosphinate ligands possibly interacted with α-amylase on a ratio of 1:1 or via a single binding site. α-Amylase intrinsic fluorescence intensity was quenched by the three ligands through static mechanisms, with bimolecular quenching constant (Kq) values in the order of x1011 M-1s-1. This suggested the formation of non-fluorescent complexes between the enzyme and the ligands. At the temperatures investigated, the values of the thermodynamic parameters showed that the binding processes between the ligands and α-amylase were spontaneous (ΔG<0); the main driving forces were hydrophobic interactions (for the interactions between α-amylase and DY300 or DY301), and a combination of hydrogen bonds and van der Waals forces (for the interaction between α-amylase and DY302). Ultraviolet–visible (UV–vis) spectroscopy indicated an interaction-induced conformational change in α-amylase structure. Förster resonance energy transfer (FRET) results revealed a non-radioactive transfer of resonance energy between the enzyme (donor) and ligands (acceptor), which adduced the feasibility of the binding interaction. The molecular docking studies affirmed the interaction of the ligands with α-amylase at its catalytic site via a number of amino acid residues such as Asp197, Glu233 and Asp300 within 4 Å. The theoretical basis provided by the present study for the inhibitory potential and binding interaction mechanisms between α-amylase and the dichalcogenoimidodiphosphinate ligands (DY300, DY301, and DY302) may find application in the development of alternative α-amylase inhibitors for the management of postprandial hyperglycemia and also provide empirical clues for further understanding of the mechanism of action and pharmacokinetics.
Author statement
O.J.A., T.T.O., F.O.A., A.A.A. and S.O.A. conceptualized the research; all authors contributed to the conduct of the experimental study, result analysis and discussion; all authors who are O.J.A., T.T.O., F.O.A., C.A.O., A.A.A., O.F.D., A.S.E., S.O.A. and M.O.O. contributed towards the preparation of the manuscript; proofread, edited and approved the final version.
Funding
Financial support was provided by Elsevier Foundation, with grant number: BBREPAEAx0nHYATS_BG.
Declaration of competing interest
The authors declare no conflict of interests.
Acknowledgements
The technical supports of Mr T.A. Adebayo and other laboratory personnel are highly appreciated. The authors are also grateful to Seeding Laboratories, USA, for donation of some of the equipment used in this study to the Centre for Chemical and Biochemical Research (CCBR), Redeemer’s University, Ede, Nigeria via Prof. E.I. Unuabonah and other colleagues.
References
- 1.Cunha W., Arantes G., Ferreira D., Lucarini R., Silva M., Furtado N., da Silva Filho A., Crotti A., Araújo A. Hypoglicemic effect of Leandra lacunosa in normal and alloxan-induced diabetic rats. Fitoterapia. 2008;79(5):356–360. doi: 10.1016/j.fitote.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Oboh G., Ademiluyi A.O., Agunloye O.M., Ademosun A.O., Ogunsakin B.G. Inhibitory effect of garlic, purple onion, and white onion on key enzymes linked with type 2 diabetes and hypertension. J. Diet. Suppl. 2019;16(1):105–118. doi: 10.1080/19390211.2018.1438553. [DOI] [PubMed] [Google Scholar]
- 3.Who . 2020. World Health Organisation Report on Diabetes in 2020.https://www.who.int/health-topics/diabetes#tab=tab_1 [Google Scholar]
- 4.Anigboro A.A., Avwioroko O.J., Tonukari N.J. Brillantasia patula aqueous leaf extract averts hyperglycermia, lipid peroxidation, and alterations in hematological parameters in alloxan-induced diabetic rats. Int. J. Biomed. Sci. 2018;6(2):43–51. [Google Scholar]
- 5.Anigboro A., Avwioroko O., Ohwokevwo O., Nzor J. Phytochemical constituents, antidiabetic and ameliorative effects of Polyalthia longifiola leaf extract in alloxan-induced diabetic rats. J. Appl. Sci. Environ. Manag. 2018;22(6):993–998. [Google Scholar]
- 6.Aziz A.u.R., Farid S., Qin K., Wang H., Liu B. Regulation of insulin resistance and glucose metabolism by interaction of PIM kinases and insulin receptor substrates. Arch. Physiol. Biochem. 2020;126(2):129–138. doi: 10.1080/13813455.2018.1498903. [DOI] [PubMed] [Google Scholar]
- 7.Atanu F., Avwioroko O., Momoh S. Anti-diabetic effect of combined treatment with Aloe vera gel and Metformin on alloxan-induced diabetic rats. J. Ayurveda Holist. Med. 2018;4(1):1–5. [Google Scholar]
- 8.Atanu F., Avwioroko O., Ilesanmi O., Oguche M. Comparative study of the effects of Annona muricata and Tapinanthus globiferus extracts on biochemical indices of diabetic rats. Phcog. J. 2019;11(6) [Google Scholar]
- 9.Rocha S., Sousa A., Ribeiro D., Correia C.M., Silva V.L., Santos C.M., Silva A.M., Araújo A.N., Fernandes E., Freitas M. A study towards drug discovery for the management of type 2 diabetes mellitus through inhibition of the carbohydrate-hydrolyzing enzymes α-amylase and α-glucosidase by chalcone derivatives. Food Function. 2019;10(9):5510–5520. doi: 10.1039/c9fo01298b. [DOI] [PubMed] [Google Scholar]
- 10.Adefegha S.A., Oboh G., Omojokun O.S., Jimoh T.O., Oyeleye S.I. In vitro antioxidant activities of African birch (Anogeissus leiocarpus) leaf and its effect on the α-amylase and α-glucosidase inhibitory properties of acarbose. J. Taibah .Univ. Med. Sci. 2016;11(3):236–242. [Google Scholar]
- 11.Ashraf J., Mughal E.U., Sadiq A., Naeem N., Muhammad S.A., Qousain T., Zafar M.N., Khan B.A., Anees M. Design and synthesis of new flavonols as dual ɑ-amylase and ɑ-glucosidase inhibitors: structure-activity relationship, drug-likeness, in vitro and in silico studies. J. Mol. Struct. 2020:128458. [Google Scholar]
- 12.Aispuro-Pérez A., López-Ávalos J., García-Páez F., Montes-Avila J., Picos-Corrales L.A., Ochoa-Terán A., Bastidas P., Montaño S., Calderón-Zamora L., Osuna-Martínez U. Synthesis and molecular docking studies of imines as α-glucosidase and α-amylase inhibitors. Bioorg. Chem. 2020;94:103491. doi: 10.1016/j.bioorg.2019.103491. [DOI] [PubMed] [Google Scholar]
- 13.Magaji U., Sacan O., Yanardag R. Alpha amylase, alpha glucosidase and glycation inhibitory activity of Moringa oleifera extracts. South Afr. J. Bot. 2020;128:225–230. [Google Scholar]
- 14.Avwioroko O.J., Tonukari N.J., Asagba S.O. Biochemical characterization of crude α-amylase of Aspergillus spp. associated with the spoilage of cassava (Manihot esculenta) tubers and processed products in Nigeria. Adv. Biochem. 2015;3(1):15–23. [Google Scholar]
- 15.Avwioroko O.J., Anigboro A.A., Unachukwu N.N., Tonukari N.J. Isolation, identification and in silico analysis of alpha-amylase gene of Aspergillus Niger strain CSA35 obtained from cassava undergoing spoilage. Biochem. Biophys. Rep. 2018;14:35–42. doi: 10.1016/j.bbrep.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avwioroko O.J., Anigboro A.A., Ejoh A.S., Atanu F.O., Okeke M.A., Tonukari N.J. Characterization of α-amylases isolated from Cyperus esculentus seeds (tigernut): biochemical features, kinetics and thermal inactivation thermodynamics. Biocatal.Agric. Biotechnol. 2019;21:101298. [Google Scholar]
- 17.Oboh G., Ademosun A.O. Shaddock peels (Citrus maxima) phenolic extracts inhibit α-amylase, α-glucosidase and angiotensin I-converting enzyme activities: a nutraceutical approach to diabetes management. Diabetes Metabolic Syndrome: Clin. Res. Rev. 2011;5(3):148–152. doi: 10.1016/j.dsx.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 18.Ganesan M., Raja K.K., Narasimhan K., Murugesan S., Kumar B.K. Design, synthesis, α-amylase inhibition and in silico docking study of novel quinoline bearing proline derivatives. J. Mol. Struct. 2020;1208:127873. [Google Scholar]
- 19.Tsoupras A.B., Roulia M., Ferentinos E., Stamatopoulos I., Demopoulos C.A., Kyritsis P. 2010. Structurally Diverse Metal Coordination Compounds, Bearing Imidodiphosphinate and Diphosphinoamine Ligands, as Potential Inhibitors of the Platelet Activating Factor, Bioinorganic Chemistry Applications 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oyetunde T., Afzaal M., Vincent M.A., O'Brien P. The deposition of cadmium selenide and cadmium phosphide thin films from cadmium thioselenoimidodiphosphinate by AACVD and the formation of an aromatic species. Dalton Trans. 2019;48(4):1436–1442. doi: 10.1039/c8dt04357d. [DOI] [PubMed] [Google Scholar]
- 21.Timerbaev A.R. Advances in developing tris (8-quinolinolato) gallium (III) as an anticancer drug: critical appraisal and prospects. Metall. 2009;1(3):193–198. doi: 10.1039/b902861g. [DOI] [PubMed] [Google Scholar]
- 22.Zhang C.X., Lippard S.J. New metal complexes as potential therapeutics. Curr. Opin. Chem. Biol. 2003;7(4):481–489. doi: 10.1016/s1367-5931(03)00081-4. [DOI] [PubMed] [Google Scholar]
- 23.Chen J.-g., Wu S.-f., Zhang Q.-f., Yin Z.-p., Zhang L. α-Glucosidase inhibitory effect of anthocyanins from Cinnamomum camphora fruit: inhibition kinetics and mechanistic insights through in vitro and in silico studies. Int. J. Biol. Macromol. 2020;143:696–703. doi: 10.1016/j.ijbiomac.2019.09.091. [DOI] [PubMed] [Google Scholar]
- 24.Ernest V., Nirmala M.J., Gajalakshmi S., Mukherjee A., Chandrasekaran N. Biophysical investigation of α-Amylase conjugated silver nanoparticles proves structural changes besides increasing its enzyme activity. J. Bionanoscience. 2013;7(3):271–275. [Google Scholar]
- 25.Wang B.-L., Pan D.-Q., Zhou K.-L., Lou Y.-Y., Shi J.-H. Multi-spectroscopic approaches and molecular simulation research of the intermolecular interaction between the angiotensin-converting enzyme inhibitor (ACE inhibitor) benazepril and bovine serum albumin (BSA) Spectrochim. Acta Mol. Biomol. Spectrosc. 2019;212:15–24. doi: 10.1016/j.saa.2018.12.040. [DOI] [PubMed] [Google Scholar]
- 26.Lin M.-Z., Chai W.-M., Zheng Y.-L., Huang Q., Ou-Yang C. Inhibitory kinetics and mechanism of rifampicin on α-glucosidase: insights from spectroscopic and molecular docking analyses. Int. J. Biol. Macromol. 2019;122:1244–1252. doi: 10.1016/j.ijbiomac.2018.09.077. [DOI] [PubMed] [Google Scholar]
- 27.Tang H., Ma F., Zhao D., Xue Z. Exploring the effect of salvianolic acid C on α-glucosidase: inhibition kinetics, interaction mechanism and molecular modelling methods. Process Biochem. 2019;78:178–188. [Google Scholar]
- 28.Wang M., Shi J., Wang L., Hu Y., Ye X., Liu D., Chen J. Inhibitory kinetics and mechanism of flavonoids from lotus (Nelumbo nucifera Gaertn.) leaf against pancreatic α-amylase. Int. J. Biol. Macromol. 2018;120:2589–2596. doi: 10.1016/j.ijbiomac.2018.09.035. [DOI] [PubMed] [Google Scholar]
- 29.Lu Q., Chen C., Zhao S., Ge F., Liu D. Investigation of the interaction between gallic Acid and α-amylase by spectroscopy. Int. J. Food Prop. 2016;19(11):2481–2494. [Google Scholar]
- 30.Zheng Y., Tian J., Yang W., Chen S., Liu D., Fang H., Zhang H., Ye X. Inhibition mechanism of ferulic acid against α-amylase and α-glucosidase. Food Chem. 2020;317:126346. doi: 10.1016/j.foodchem.2020.126346. [DOI] [PubMed] [Google Scholar]
- 31.He T., Liang Q., Luo T., Wang Y., Luo G. Study on interactions of phenolic acid-like drug candidates with bovine serum albumin by capillary electrophoresis and fluorescence spectroscopy. J. Solut. Chem. 2010;39(11):1653–1664. [Google Scholar]
- 32.Wang B.-L., Pan D.-Q., Kou S.-B., Lin Z.-Y., Shi J.-H. Exploring the binding interaction between bovine serum albumin and perindopril as well as influence of metal ions using multi-spectroscopic, molecular docking and DFT calculation. Chem. Phys. 2020;530:110641. [Google Scholar]
- 33.Kumari M., Maurya J.K., Singh U.K., Khan A.B., Ali M., Singh P., Patel R. Spectroscopic and docking studies on the interaction between pyrrolidinium based ionic liquid and bovine serum albumin. Spectrochim. Acta Mol. Biomol. Spectrosc. 2014;124:349–356. doi: 10.1016/j.saa.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Shen G.-F., Liu T.-T., Wang Q., Jiang M., Shi J.-H. Spectroscopic and molecular docking studies of binding interaction of gefitinib, lapatinib and sunitinib with bovine serum albumin (BSA) J. Photochem. Photobiol. B Biol. 2015;153:380–390. doi: 10.1016/j.jphotobiol.2015.10.023. [DOI] [PubMed] [Google Scholar]
- 35.Shi J.H., Wang Q., Pan D.Q., Liu T.T., Jiang M. Characterization of interactions of simvastatin, pravastatin, fluvastatin, and pitavastatin with bovine serum albumin: multiple spectroscopic and molecular docking. J. Biomol. Struct. Dyn. 2017;35(7):1529–1546. doi: 10.1080/07391102.2016.1188416. [DOI] [PubMed] [Google Scholar]
- 36.Ross P.D., Subramanian S. Thermodynamics of protein association reactions: forces contributing to stability. Biochemistry. 1981;20(11):3096–3102. doi: 10.1021/bi00514a017. [DOI] [PubMed] [Google Scholar]
- 37.Boadi N.O., McNaughter P.D., Helliwell M., Malik M.A., Awudza J.A., O’Brien P. The deposition of PbS and PbSe thin films from lead dichalcogenoimidophosphinates by AACVD. Inorg. Chim. Acta. 2016;453:439–442. [Google Scholar]
- 38.Oyetunde D., Afzaal M., Vincent M.A., Hillier I.H., O’Brien P. Cadmium sulfide and cadmium phosphide thin films from a single cadmium compound. Inorg. Chem. 2011;50:2052–2054. doi: 10.1021/ic102309r. [DOI] [PubMed] [Google Scholar]
- 39.Levesanos N., Liyanage W.P., Ferentinos E., Raptopoulos G., Paraskevopoulou P., Sanakis Y., Choudhury A., Stavropoulos P., Nath M., Kyritsis P. Investigating the structural, spectroscopic, and electrochemical properties of [Fe {(EPiPr2) 2N} 2](E= S, Se) and the formation of iron selenides by chemical vapor deposition. Eur. J. Inorg. Chem. 2016;2016(34):5332–5339. [Google Scholar]
- 40.Ritch J.S., Chivers T., Eisler D.J., Tuononen H.M. Experimental and theoretical investigations of structural isomers of dichalcogenoimidodiphosphinate dimers: dichalcogenides or spirocyclic contact ion pairs? Chemistry–A European Journal. 2007;13(16):4643–4653. doi: 10.1002/chem.200700001. [DOI] [PubMed] [Google Scholar]
- 41.Oyetunde T., Afzaal M., Vincent M.A., O’Brien P. Aerosol-assisted CVD of cadmium diselenoimidodiphosphinate and formation of a new iPr2N2P3+ ion supported by combined DFT and mass spectrometric studies. Dalton Trans. 2016;45:18603–18609. doi: 10.1039/c6dt03186b. [DOI] [PubMed] [Google Scholar]
- 42.Oyetunde T., Afzaal M., Vincent M.A., O’Brien P. The deposition of cadmium selenide and cadmium phosphide thin films from cadmium thioselenoimidodiphosphinate by AACVD and the formation of an aromatic species. Dalton Trans. 2019;48:1436–1442. doi: 10.1039/c8dt04357d. [DOI] [PubMed] [Google Scholar]