Abstract
Lung adenocarcinoma (LUAD) is the most predominant subtype of lung cancers and is one of the leading causes of cancer related mortality worldwide. Despite the advancements in the field of cancer diagnostics and therapeutics, detection at an early stage using reliable biomarkers is an unmet clinical need for a plethora of cancers, including LUAD, thus attributing to poor prognosis. In view of this, to identify potential biomarkers and therapeutic candidate genes, the expression of all known human genes was screened in the publicly available ‘The Cancer Genome Atlas’ (TCGA) samples of LUAD patients which resulted in the identification of overexpressed genes. Further analysis of these genes across various patient sample datasets revealed that ZNF687, ODR4, PBXIP1, PYGO2, METTL3, PIGM and RAD1 are consistently more highly expressed in LUAD. Higher expression of these genes either alone or in combination is correlated with poor survival of LUAD patients. Hence, in this study we propose that these identified genes could serve as potential candidates as gene signatures or biomarkers for LUAD that require further investigation in large cohorts of LUAD samples.
Keywords: Bioinformatics, Cancer research, Oncology, Lung adenocarcinoma, The cancer Genome Atlas, Biomarkers, Gene expression, cBioPortal
Bioinformatics; Cancer research; Oncology; Lung adenocarcinoma, The cancer Genome Atlas, Biomarkers, Gene expression, cBioPortal.
1. Introduction
Lung adenocarcinoma (LUAD) is the most common sub-type of non-small cell lung cancer (NSCLC), which is usually detected in the periphery and upper lobes of the lungs, especially in the glandular cells of bronchial mucosa. Amongst the sub-types of NSCLC, LUAD accounts for nearly 40% of all lung cancers and its prevalence is increasing especially among non-smokers (males being at a higher risk than females) and in Asians [1]. Although the initial growth of the cancer occurs at a very early stage, the progression rate is much slower when compared to the other subtypes. However, due to poor prognosis, the overall survival rate is comparatively less than other subtypes [2]. Some studies suggest that a combined exposure to asbestos, cigarette smoke and also certain elements like chromium, nickel and arsenic increases the probability of LUAD development [3, 4]. Secreted Phospho- Protein 1 (SPP1), Ficolin 3 (FCN3), Cyclin-dependent kinase-1 (CDK1) and Mitotic Arrest Deficient 2 Like 1 (MAD2L1) have been shown to be overexpressed in LUAD and are considered as biomarkers due to their consistent expression in the patient samples analyzed from different databases [5]. Recent research suggests that the occurrence of Tumor Infiltrating Leukocytes (TILs) correlates with the prognosis of lung adenocarcinoma [6]. Given the poor prognosis of LUAD, a large number of studies are warranted towards discovering biomarkers which are highly expressed in patient samples (at various stages from very early to late) across various population demographics and occupations. The aim of this work is to determine biomarkers for LUAD, which may help in early diagnosis and thereby prolonging overall survival of patients. The RNA sequencing data from The Cancer Genome Atlas (TCGA-2014) has been used to identify highly expressed genes in LUAD [7].
2. Materials and methods
2.1. Patient datasets and gene expression analysis
For the analysis of the genes involved in LUAD, the list of 19755 human genes was downloaded from the Hugo Gene Nomenclature Committee (HGNC) (https://www.genenames.org/cgi-bin/download) and its abbreviations were retained for conducting the study [8]. To download, visualize and analyze large datasets, “cBioPortal” (http://www.cbioportal.org/) was used [9, 10]. The TCGA-2014 data set, consisting of 230 LUAD patient samples including the normal adjacent samples as reference was considered for the analysis [10]. Samples with RNA sequencing data were used to study mRNA expression, mutations and copy number variations in human genes [7]. To identify the differential mRNA expression among the samples, a minimum of two-fold higher was considered as differentially expressed. cBioPortal arrives at mRNA expression data by computing the expression of an individual gene relative to the gene's expression distribution in a reference population. The reference population includes all samples that are diploid for the gene in question (by default for mRNA), or normal samples (when specified), or all profiled samples. The returned value indicates the number of individuals deviating from the mean of expression (standard deviation) in the reference population (Z-score). This measure is useful to determine whether a gene is up- or down-regulated relative to the normal samples or all other tumor samples, and the output is provided as an oncoprint. Oncoprint is a pictorial representation of up or down regulated genes, displaying either red bars (higher expression) or blue (lower expression) in affected individuals. Each bar represents one sample. Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn/) an interactive web tool, was used for analyzing RNA sequencing expression data of 9,736 tumors and 8,587 normal samples from the TCGA and the GTEx projects employing standard statistical methods or tools [11]. Four-way analysis of variance (ANOVA), using sex, age, ethnicity and disease state (Tumor or Normal) as variables, is used for calculating the differential expression of the corresponding genes. The expression data are first Log2 (TPM+1) transformed, and the log2FC is defined as the difference between median (Tumor) and median (Normal). The Benjamini and Hochberg false discovery rate (FDR) method was used to adjust the p-value across each factor to obtain the multiple testing adjusted q-value. UALCAN (http://ualcan.path.uab.edu/index.html) is a user-friendly online web tool useful for analyzing the TCGA data for gene expression, survival and protein expression [12]. It uses Transcripts Per Million (TPM) as a measure of gene expression and provides box plots by comparing gene expression levels with corresponding normal samples in that data set. It also provides gene expression in different disease stages of LUAD. It utilizes all regular statistical methods of R and uses t-test for estimating p-value.
2.2. Patient survival plots
Kaplan-Meier curves and estimates of survival were determined using SurvExpress (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp), and the significance was calculated by log-rank test [13]. The correlation was performed between either an individual gene or a group of genes and the survival of the patients. The TCGA-2016 LUAD data consisting of 475 patients was used with death as the stratification parameter. PROGgene V2 (http://genomics.jefferson.edu/proggene/) was used to correlate the over-expression gene cluster with the survival of the patient samples across various data [14].
2.3. Protein expression and gene functional analysis
String database (https://string-db.org) [15] or cBioPortal was used for constructing the network diagram for shortlisted genes. Human Protein Atlas (HPA) (https://www.proteinatlas.org/) was used to obtain antibody-stained images for LUAD tissue samples for various proteins [16]. Protein expression was also obtained for the different stages of LUAD from quantitative proteomics studies of CPTAC samples using the UALCAN web tool (http://ualcan.path.uab.edu/index.html). To determine the gene expression in various stages of the disease, GEPIA or CANCERTOOL (http://web.bioinformatics.cicbiogune.es/CANCERTOOL/) was used [17]. The expression of the genes was studied in various databases [18, 19, 20, 21]. For process and functional analysis, FunRich software (version 3.1.3; 64-bit) was used [22].
3. Result and discussion
3.1. Identification of differentially-expressed genes in LUAD patient samples
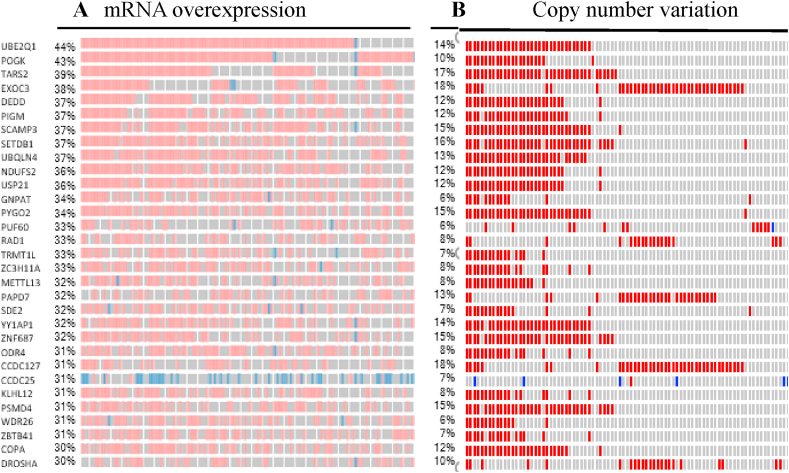
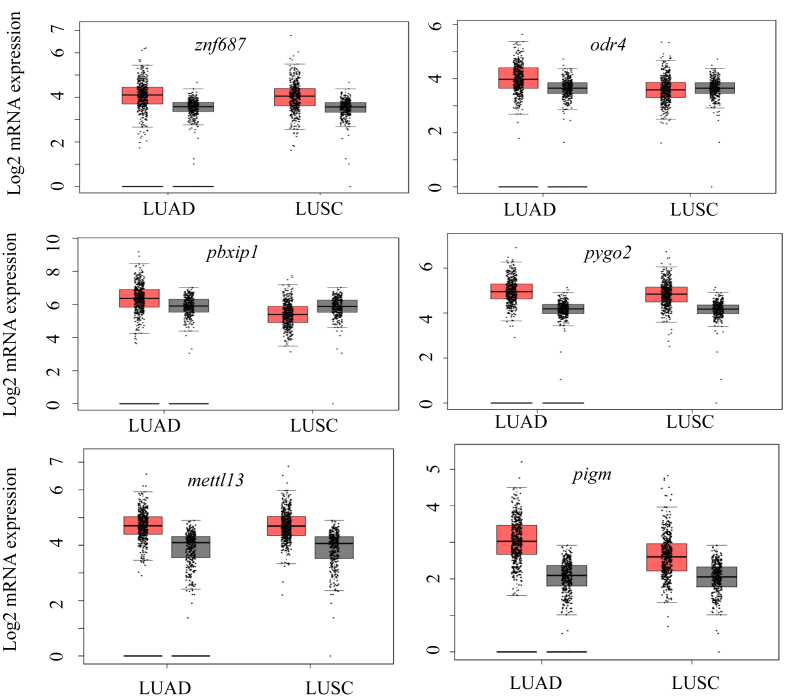
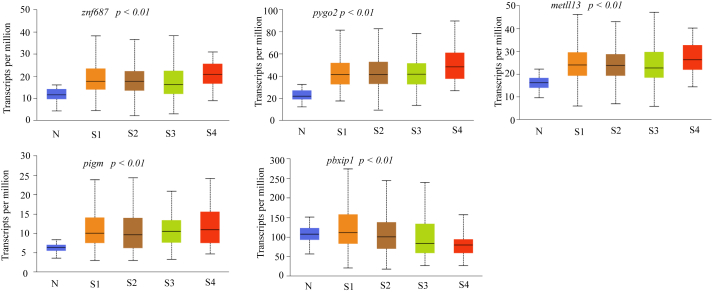
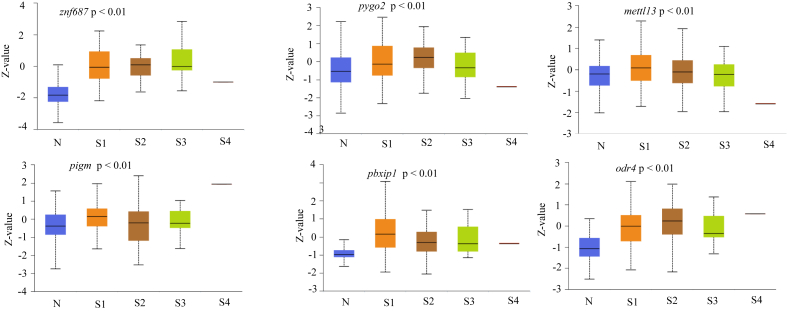
In order to determine differentially-expressed genes in LUAD patient samples without any bias, a list of all known human genes (obtained from HGNC) was screened for expression in the patient data sets of LUAD available in TCGA database. To shortlist the genes we chose a minimum of two-fold higher expression in the disease condition. We also implemented higher expression in at least 30% of the samples to obtain a manageable number of genes for further analysis. Since multiple studies have suggested that the overexpression of certain genes even in a narrow range patient data set could be considered as a biomarker, we hypothesized that overexpression of the gene in at least 30% of the patient data set could have an immense effect on diagnostic value of LUAD [23, 24, 25]. This resulted in identification of several genes that were expressed two-fold or more in LUAD (Figure 1A). Higher copy number or gene amplification is correlated to the higher expression of genes in many cancers to meet the demands of the tumor physiology and function [26, 27, 28, 29]. Our data also reveals that some of these genes show increased copy number across several patient samples in addition to higher gene expression (Figure 1B). We further validated the higher expression of these genes in large numbers of LUAD and Lung Squamosal Carcinoma (LUSC) samples. Data show that genes such as znf687, odr4, pbxip1, pygo2, mettl3, and pigm show the over-expression of mRNA in LUAD samples (Figure 2). Except odr4 and pbxip1, the afore mentioned genes also show high mRNA expression in LUSC (Figure 2). Furthermore, these genes have a higher mRNA expression in LUAD samples in different stages of the disease (Figure 3). As shown in Figure 3, higher expression of these genes is statistically significant in stage 1 LUAD with respect to normal samples, indicating that these could be valuable potential candidates for early stage detection of the disease. These genes also express higher protein levels in LUAD samples in different stages (Figure 4).
Figure 1.
Co-relation between mRNA expression profile and copy number alteration (CNA) of the various genes in LUAD. (A) mRNA upregulation for genes in LUAD patient samples from TCGA 2014 (N = 230). (B) CNA status of same set of genes in the sample dataset. Blue bars indicate lower mRNA levels in A or deletion in B as compared to the normal whereas red bars indicate over expression or amplification. Each bar indicates a patient sample.
Figure 2.
Comparative expression of the genes in LUAD (T = 483, N = 347) and LUSC (T = 486, N = 338). Red and grey boxes indicate the tumor and the normal samples respectively (T = Tumor samples, N=Normal patients). The box plots were obtained from GEPIA.
Figure 3.
Expression of various genes at different stages of LUAD at mRNA level in TCGA samples. N indicates Normal sample, S1 – S4 indicates different stages of LUAD, having 59, 277, 85 and 28 samples respectively. P < 0.01 is with respect to normal versus stage1 LUAD. The box plots were obtained from UALCAN.
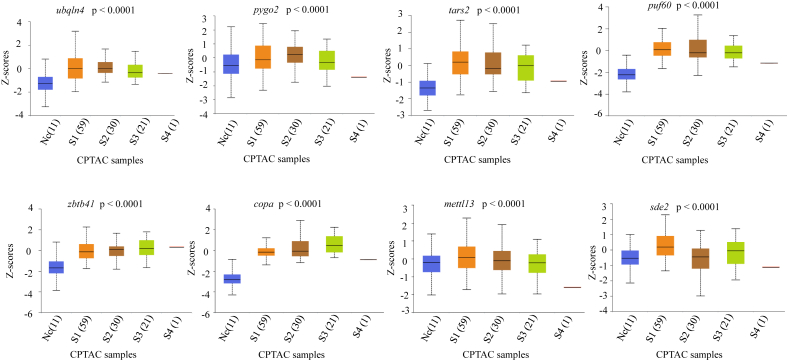
Figure 4.
Protein expression level of various genes at different stages of LUAD in CPTAC samples. N indicates Normal sample, S1 – S4 indicates different stages of LUAD having 11, 55, 30, 21, and 1 sample respectively. P-value is with respect to normal versus stage 1 LUAD for all the proteins expression. Z-values represent standard deviations from the median across samples for the given cancer type. The box plots were obtained from UALCAN.
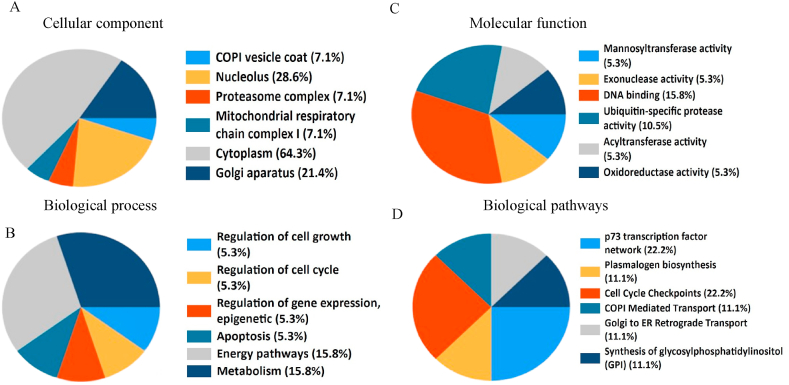
We further examined whether these genes are over-expressed in other cancers. Results indicate that znf687 showed statistically-significant higher expression in esophageal, low grade glioma, liver and thyroid cancers (Figure S1). Similarly, some of the genes, such as odr4, pbxip1, pygo2, mettl3, and pigm and rad1 also show statistically-significant higher expression in many other cancers (Figures S2-S7). To further confirm the expression of these genes at the protein level in LUAD, immunostaining data from Human Protein Atlas (HPA) was analyzed. Some of these genes show higher protein expression status in LUAD in comparison to the normal lung tissue sample (Figure 5) while others show no differential expression between normal and LUAD samples (Figures S8 and 9). Higher protein expression of these genes was further confirmed by analyzing the CPTAC samples pertaining to LUAD at different stages of the disease (Figure 6). To understand the biological relevance or functions of these genes, FunRich web tool was used. As shown in Figure 7, genes were categorized based on their expression locations, the biological process they operate, and the functional and biological pathways in which they participate. It was observed that most of them are expressed in the cytoplasm or the nucleolus (Figure 7A). Many of these genes are involved in the regulation of cell cycle, metabolism and energy pathways (Figure 7B) which are shown to have significant importance in the context of cancer [30, 31]. When it comes to the functions of these genes, many have DNA binding or exonuclease activity (Figure 7C). Many of these genes also operate in the p73 network and cell cycle checkpoints (Figure 7D).
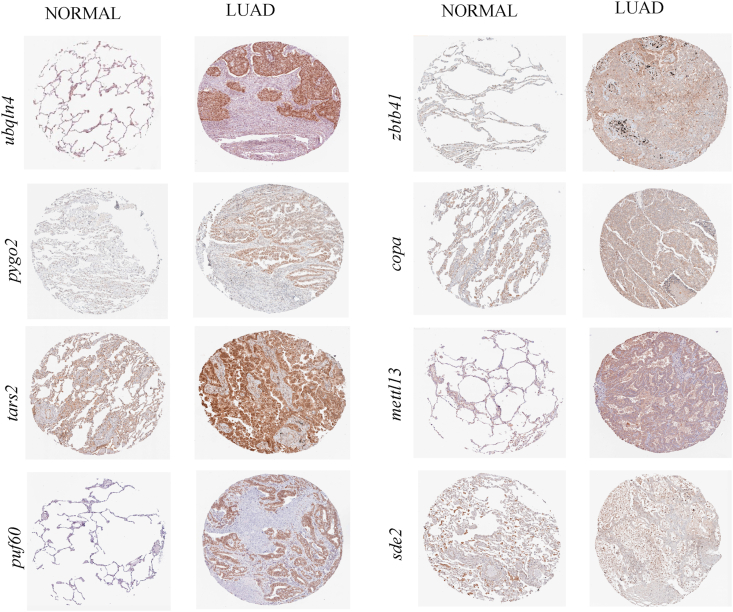
Figure 5.
Immunohistochemical staining representing the protein higher expression of the genes in various LUAD tissues. Images were obtained from human protein atlas.
Figure 6.
Protein expression in CPTAC (Clinical Proteomic Tumor Analysis Consortium) samples of LUAD. Z-values represent standard deviations from the median across samples for the given cancer type. Log2 Spectral count ratio values from CPTAC were first normalized within each sample profile, then normalized across samples. The stated p values are with respect to normal versus stage 1 and less than 0.05 unless stated otherwise. The box plots were obtained from UALCAN.
Figure 7.
Gene set enrichment analysis. A, B, C, D LUAD higher expressed genes were analyzed with Funrich software for their cellular component, biological process, molecular function and biological pathways.
3.2. Patient survival and clinical significance of higher gene expression
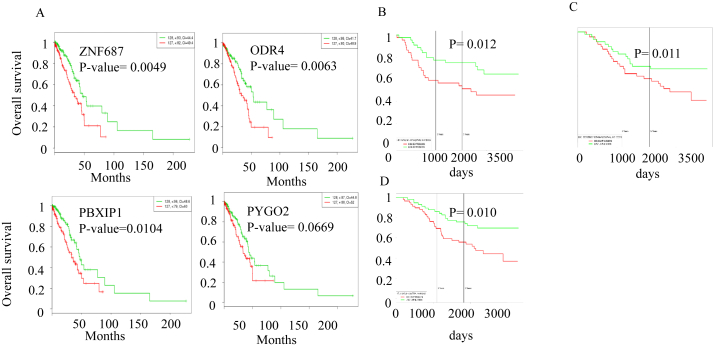
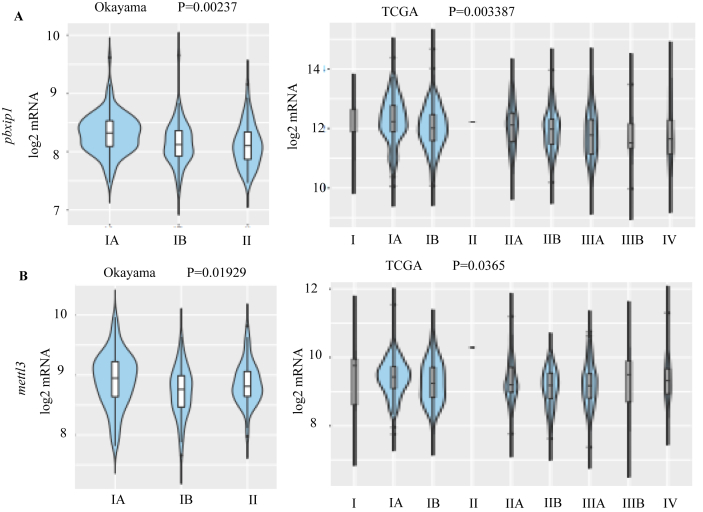
To assess the clinical significance of higher expression of these genes on patient survival, SurvExpress containing TCGA data set pertaining to LUAD was used. Results show that higher expression of znf687, odr4 and pbxip1 significantly correlate with poor survival (Figure 8A) whereas mettl3, pigm and rad1 did not show a statistically-significant outcome on patient survival. Further analysis showed that higher expression of some of these genes in combination has a statistically-significant effect on patient survival outcome (Figure 8B, C and D). It is interesting to note that genes such as dedd, scamp3, setdb1, ubqln4, ndufs2, gnpat, rad1, zc3h11a, and klhl12 are common in data sets where patient survival is significantly compromised by their higher expression as shown in Figures 8B, C and D. We also analyzed whether any of these genes show a stage-wise expression pattern in LUAD and thereby serve the utility of being stage- specific diagnostic markers. Violin plots for pbxip1 and mettl3 from various LUAD data sets show that these two genes are expressed more during stages 1 and 1A of LUAD (Figure 9A and B). Higher expression of znf687 is correlated with poor survival of patients in stage 3B of LUAD (Figure S10).
Figure 8.
Clinical significance of the higher expressed gene on the patient survival in LUAD (N = 475). (A) Genes that are significantly expressed in LUAD. (B) Prognostic and Predictive Gene Signature for Adjuvant Chemotherapy in Resected Non-Small Cell Lung Cancer for genes dedd, scamp3, setdb1, ubqln4, ndufs2, usp21, gnpat, puf60, rad1, zc3h11a, mettl13, papd7, yy1ap1, ccdc25, klhl1 (C) Effect on the survival of the diseased individuals by the overexpressed genes in Expression Profile-Defined Classification of Lung Adenocarcinoma data set. for genes pogk, dedd, pigm, scamp3, setdb1, ndufs2, usp21, gnpat, pygo2, rad1, klhl12, psmd4, wdr26, copa (D) Effect on the survival of individuals by the overexpressed genes such as scamp3 setdb1, ubqln, ndufs2, usp21, gnpat, pygo2, rad1, zc3h11a, yy1ap1, znf687, ccdc127, ccdc25, klhl12 in Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis dataset. Green line and red line indicates survival status of the patient samples in which genes are under expressed and over expressed respectively compared to the normal sample. The survival plots were obtained from PROGgeneV2.
Figure 9.
mRNA expression level of a) pbxip1 and b) mettl13 in the different stages of LUAD in different datasets - OKAYAMA (N = 246) and TCGA (N = 514). A p-value of 0.05 or less is considered as significant. Violin plots were obtained from CANCERTOOL.
Since, pbxip1 and znf687 showed common results across several databases, we wanted to identify their interacting partners and verify whether they could be used as signatures for LUAD (Figure S11). Also, gene expression profile from TCGA LUAD 2014 data revealed that some of the genes that are networked to pbxip1 and znf687 show higher mRNA expression in the LUAD (Figure S12). Furthermore, higher expression of pbxip1 and znf687 network genes also have statistically-significant poor survival among the affected patients (Figures S13-S14). It has been reported that overexpression of znf687 enhances tumorigenicity in hepatocellular carcinoma [32].
4. Discussion
In our study, we identified several over-expressed genes in LUAD and validated them with various bioinformatic tools in LUAD patient samples. It would be interesting to know how many of these genes are already known or identified as potential candidate biomarkers. From the shortlisted genes as indicated in Figure 1, genes such as odr4, pbxip1, mettl3, pigm and rad1 are identified as being upregulated in LUAD in this study and have not been reported as markers for LUAD. Interestingly, all these genes show higher expression at the level of mRNA and protein. Genes such as setdb1 and ndufs2 are shown to be upregulated in lung cancer [33, 34], whereas other genes are known to be up/downregulated or are employed as biomarkers for many other cancer types as indicated in the supplementary tables. Nevertheless, all of these genes show higher expression in LUAD patient samples either at the level of mRNA/or protein or both. Early detection and diagnosis of cancers is of tremendous importance in making appropriate decisions for treatment and for better prognosis. Unfortunately, it is still an unmet clinical need that has to be addressed. In this study, genes have been identified which have a statistically significant higher expression pattern in LUAD (stagewise) with respect to normal samples.
There are several approaches/mathematical models that are being employed for determining biomarkers for several diseases. One among them is the Dynamical Network Biomarkers (DNB) and as per this approach, each individual may not have exactly the same DNB even for the same disease, i.e. the markers could vary from person to person [35]. Recent studies have reported potential biomarkers using, single cell RNA sequencing studies in samples either derived from xenografts or patient samples for many cancers and reported potential biomarkers [36]. However, there is no correlation or common overlapping genes as biomarkers between our study and DNB studies pertaining to lung cancer due to the difference in approaches. Identification of reliable biomarkers with high sensitivity and specificity is highly desirable for better management of many cancers [37]. Although there are several research reports on biomarkers for LUAD available, these reports have limited application potential. It would be interesting to test if any of the genes that show higher expression in our study are potential candidates to be used as reliable biomarkers either alone or in combinations and as gene signatures for LUAD. Since a significant proportion of the patient's samples shows higher expression of these genes, we believe that further validation of these genes in a large cohort of patient samples is warranted for developing a suitable and dependable biomarker for LUAD.
Declarations
Author contribution statement
Deepak Sayeeram: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Teesta V Katte: Performed the experiments; Wrote the paper.
Saloni Bhatia, Anushree Jai Kumar, Avinesh Kumar, Jayashree G, Rachana D.S, Avinash Arvind Rasalkar: Performed the experiments.
Nalla Reddy Harsha Vardhan: Performed the experiments; Analyzed and interpreted the data.
Rajya Lakshmi Malempati: Analyzed and interpreted the data; Wrote the paper.
Divijendra Natha Reddy S: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
Divijendra Natha Reddy S was supported by a grant ECR/2016-001685 from Department of Science and Technology, Govt. of India.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Supplementary Figures
References
- 1.Travis W.D., Brambilla E., Noguchi M., Nicholson A.G., Geisinger K.R., Yatabe Y., Beer D.G., Powell C.A., Riely G.J., Van Schil P.E., Garg K., Austin J.H., Asamura H., Rusch V.W., Hirsch F.R., Scagliotti G., Mitsudomi T., Huber R.M., Ishikawa Y., Jett J., Sanchez-Cespedes M., Sculier J.P., Takahashi T., Tsuboi M., Vansteenkiste J., Wistuba I., Yang P.C., Aberle D., Brambilla C., Flieder D., Franklin W., Gazdar A., Gould M., Hasleton P., Henderson D., Johnson B., Johnson D., Kerr K., Kuriyama K., Lee J.S., Miller V.A., Petersen I., Roggli V., Rosell R., Saijo N., Thunnissen E., Tsao M., Yankelewitz D. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Classification of Lung Adenocarcinoma. J. Thorac. Oncol. 2011;6:244–285. doi: 10.1097/JTO.0b013e318206a221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu Z., Gu X., Zhong R., Zhong H. Tumor-infiltrating CD45RO+ memory cells correlate with favorable prognosis in patients with lung adenocarcinoma. J. Thorac. Dis. 2018;10:2089–2099. doi: 10.21037/jtd.2018.03.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mollo F., Pira E., Piolatto G., Bellis D., Burlo P., Andreozzi A., Bontempi S., Negri E. Lung adenocarcinoma and indicators of asbestos exposure. Int. J. Canc. 1995;60:289–293. doi: 10.1002/ijc.2910600302. [DOI] [PubMed] [Google Scholar]
- 4.Kuo C.Y., Wong R.H., Lin J.Y., Lai J.C., Lee H. Accumulation of chromium and nickel metals in lung tumors from lung cancer patients in Taiwan. J. Toxicol. Environ. Health Part A Curr. Issues. 2006;69:1337–1344. doi: 10.1080/15287390500360398. [DOI] [PubMed] [Google Scholar]
- 5.Shi Y.-X., Zhu T., Zou T., Zhuo W., Chen Y.-X., Huang M.-S., Zheng W., Wang C.-J., Li X., Mao X.-Y., Zhang W., Zhou H.-H., Yin J.-Y., Liu Z.-Q. Prognostic and predictive values of CDK1 and MAD2L1 in lung adenocarcinoma. Oncotarget. 2016;7:85235–85243. doi: 10.18632/oncotarget.13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu X., Li Y.Q., Li Q.G., Ma Y.L., Peng J.J., Cai S.J. ITGAE defines CD8+ tumor-infiltrating lymphocytes predicting a better prognostic survival in colorectal cancer. EBioMedicine. 2018;35:178–188. doi: 10.1016/j.ebiom.2018.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collisson E.A., Campbell J.D., Brooks A.N., Berger A.H., Lee W., Chmielecki J., Beer D.G., Cope L., Creighton C.J., Danilova L., Ding L., Getz G., Hammerman P.S., Neil Hayes D., Hernandez B., Herman J.G., Heymach J.V., Jurisica I., Kucherlapati R., Kwiatkowski D., Ladanyi M., Robertson G., Schultz N., Shen R., Sinha R., Sougnez C., Tsao M.-S., Travis W.D., Weinstein J.N., Wigle D.A., Wilkerson M.D., Chu A., Cherniack A.D., Hadjipanayis A., Rosenberg M., Weisenberger D.J., Laird P.W., Radenbaugh A., Ma S., Stuart J.M., Averett Byers L., Baylin S.B., Govindan R., Meyerson M., Rosenberg M., Gabriel S.B., Cibulskis K., Sougnez C., Kim J., Stewart C., Lichtenstein L., Lander E.S., Lawrence M.S., Getz G., Kandoth C., Fulton R., Fulton L.L., McLellan M.D., Wilson R.K., Ye K., Fronick C.C., Maher C.A., Miller C.A., Wendl M.C., Cabanski C., Ding L., Mardis E., Govindan R., Creighton C.J., Wheeler D., Balasundaram M., Butterfield Y.S.N., Carlsen R., Chu A., Chuah E., Dhalla N., Guin R., Hirst C., Lee D., Li H.I., Mayo M., Moore R.A., Mungall A.J., Schein J.E., Sipahimalani P., Tam A., Varhol R., Gordon Robertson A., Wye N., Thiessen N., Holt R.A., Jones S.J.M., Marra M.A., Campbell J.D., Brooks A.N., Chmielecki J., Imielinski M., Onofrio R.C., Hodis E., Zack T., Sougnez C., Helman E., Sekhar Pedamallu C., Mesirov J., Cherniack A.D., Saksena G., Schumacher S.E., Carter S.L., Hernandez B., Garraway L., Beroukhim R., Gabriel S.B., Getz G., Meyerson M., Hadjipanayis A., Lee S., Mahadeshwar H.S., Pantazi A., Protopopov A., Ren X., Seth S., Song X., Tang J., Yang L., Zhang J., Chen P.-C., Parfenov M., Wei Xu A., Santoso N., Chin L., Park P.J., Kucherlapati R., Hoadley K.A., Todd Auman J., Meng S., Shi Y., Buda E., Waring S., Veluvolu U., Tan D., Mieczkowski P.A., Jones C.D., Simons J.V., Soloway M.G., Bodenheimer T., Jefferys S.R., Roach J., Hoyle A.P., Wu J., Balu S., Singh D., Prins J.F., Marron J.S., Parker J.S., Neil Hayes D., Perou C.M., Liu J., Cope L., Danilova L., Weisenberger D.J., Maglinte D.T., Lai P.H., Bootwalla M.S., Van Den Berg D.J., Triche T., Jr., Baylin S.B., Laird P.W., Rosenberg M., Chin L., Zhang J., Cho J., DiCara D., Heiman D., Lin P., Mallard W., Voet D., Zhang H., Zou L., Noble M.S., Lawrence M.S., Saksena G., Gehlenborg N., Thorvaldsdottir H., Mesirov J., Nazaire M.-D., Robinson J., Getz G., Lee W., Arman Aksoy B., Ciriello G., Taylor B.S., Dresdner G., Gao J., Gross B., Seshan V.E., Ladanyi M., Reva B., Sinha R., Onur Sumer S., Weinhold N., Schultz N., Shen R., Sander C., Ng S., Ma S., Zhu J., Radenbaugh A., Stuart J.M., Benz C.C., Yau C., Haussler D., Spellman P.T., Wilkerson M.D., Parker J.S., Hoadley K.A., Kimes P.K., Neil Hayes D., Perou C.M., Broom B.M., Wang J., Lu Y., Kwok Shing Ng P., Diao L., Averett Byers L., Liu W., Heymach J.V., Amos C.I., Weinstein J.N., Akbani R., Mills G.B., Curley E., Paulauskis J., Lau K., Morris S., Shelton T., Mallery D., Gardner J., Penny R., Saller C., Tarvin K., Richards W.G., Cerfolio R., Bryant A., Raymond D.P., Pennell N.A., Farver C., Czerwinski C., Huelsenbeck-Dill L., Iacocca M., Petrelli N., Rabeno B., Brown J., Bauer T., Dolzhanskiy O., Potapova O., Rotin D., Voronina O., Nemirovich-Danchenko E., Fedosenko K.V., Gal A., Behera M., Ramalingam S.S., Sica G., Flieder D., Boyd J., Weaver J., Kohl B., Huy Quoc Thinh D., Sandusky G., Juhl H., Duhig E., Illei P., Gabrielson E., Shin J., Lee B., Rogers K., Trusty D., Brock M.V., Williamson C., Burks E., Rieger-Christ K., Holway A., Sullivan T., Wigle D.A., Asiedu M.K., Kosari F., Travis W.D., Rekhtman N., Zakowski M., Rusch V.W., Zippile P., Suh J., Pass H., Goparaju C., Owusu-Sarpong Y., Bartlett J.M.S., Kodeeswaran S., Parfitt J., Sekhon H., Albert M., Eckman J., Myers J.B., Cheney R., Morrison C., Gaudioso C., Borgia J.A., Bonomi P., Pool M., Liptay M.J., Moiseenko F., Zaytseva I., Dienemann H., Meister M., Schnabel P.A., Muley T.R., Peifer M., Gomez-Fernandez C., Herbert L., Egea S., Huang M., Thorne L.B., Boice L., Hill Salazar A., Funkhouser W.K., Kimryn Rathmell W., Dhir R., Yousem S.A., Dacic S., Schneider F., Siegfried J.M., Hajek R., Watson M.A., McDonald S., Meyers B., Clarke B., Yang I.A., Fong K.M., Hunter L., Windsor M., Bowman R.V., Peters S., Letovanec I., Khan K.Z., Jensen M.A., Snyder E.E., Srinivasan D., Kahn A.B., Baboud J., Pot D.A., Mills Shaw K.R., Sheth M., Davidsen T., Demchok J.A., Yang L., Wang Z., Tarnuzzer R., Claude Zenklusen J., Ozenberger B.A., Sofia H.J., Travis W.D., Cheney R., Clarke B., Dacic S., Duhig E., Funkhouser W.K., Illei P., Farver C., Rekhtman N., Sica G., Suh J., Tsao M.-S. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruford E., Lush M.J., Wright M.W., Sneddon T.P., Povey S., Birney E. The HGNC database in 2008: a resource for the human genome. Nucleic Acids Res. 2008;36:445–448. doi: 10.1093/nar/gkm881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., Antipin Y., Reva B., Goldberg A.P., Sander C., Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Canc. Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jianjiong Gao E.L., Aksoy Bülent Arman, Dogrusoz Ugur, Dresdner Gideon, Gross Benjamin, Onur Sumer S., Sun Yichao, Jacobsen Anders, Sinha Rileen, Ethan Cerami N.S., Sander Chris. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:1–34. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandrashekar D.S., Bashel B., Balasubramanya S.A.H., Creighton C.J., Ponce-Rodriguez I., Chakravarthi B.V.S.K., Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia (United States) 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aguirre-Gamboa R., Gomez-Rueda H., Martínez-Ledesma E., Martínez-Torteya A., Chacolla-Huaringa R., Rodriguez-Barrientos A., Tamez-Peña J.G., Treviño V. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PloS One. 2013;8:1–9. doi: 10.1371/journal.pone.0074250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goswami C.P., Nakshatri H. 2014. PROGgeneV2: Enhancements on the Existing Database; pp. 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., Kuhn M., Bork P., Jensen L.J., Von Mering C. STRING v10: protein – protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C.A.-K., Odeberg J., Djureinovic D., Takanen J.O., Hober S., Alm T., Edqvist P.-H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J.M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., Ponten F. Tissue-based map of the human proteome. Science (80-.) 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 17.Cortazar A.R., Torrano V., Martín-martín N., Caro-maldonado A., Camacho L., Hermanova I., Guruceaga E., Lorenzo-martín L.F., Quesada V. CANCERTOOL: a visualization and representation interface to exploit cancer datasets. Canc. Res. 2018;78:6320–6329. doi: 10.1158/0008-5472.CAN-18-1669. [DOI] [PubMed] [Google Scholar]
- 18.Wilkerson M.D., Yin X., Walter V., Zhao N., Cabanski C.R., Hayward M.C., Miller C.R., Socinski M.A., Parsons A.M., Thorne L.B., Haithcock B.E., Veeramachaneni N.K., Funkhouser W.K., Randell S.H., Bernard P.S., Perou C.M., Hayes D.N. Differential pathogenesis of lung adenocarcinoma subtypes involving sequence mutations, copy number, chromosomal instability, and methylation. PloS One. 2012;7 doi: 10.1371/journal.pone.0036530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chitale D., Gong Y., Taylor B.S., Broderick S., Brennan C., Somwar R., Golas B., Wang L., Motoi N., Szoke J., Reinersman J.M., Major J., Sander C., Seshan V.E., Zakowski M.F., Rusch V., Pao W., Gerald W., Ladanyi M. An integrated genomic analysis of lung cancer reveals loss of DUSP4 in EGFR -mutant tumors. Oncogene. 2009;28:2773–2783. doi: 10.1038/onc.2009.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okayama H., Kohno T., Ishii Y., Shimada Y., Shiraishi K., Iwakawa R. Identification of genes upregulated in ALK -positive and EGFR/KRAS/ALK -negative lung. Adenocarcinomas. 2012;72:100–112. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 21.Shedden K., Taylor J.M.G., Enkemann S.A., Tsao M., Yeatman T.J., Gerald W.L., Eschrich S., Jurisica I., Giordano T.J., Misek D.E., Chang A.C., Zhu C.Q., Strumpf D., Hanash S., Shepherd F.A., Ding K., Seymour L., Naoki K., Pennell N., Weir B., Verhaak R., Ladd-acosta C., Golub T., Gruidl M., Sharma A., Szoke J., Zakowski M., Rusch V., Kris M., Viale A., Motoi N., Travis W., Conley B., Seshan V.E., Meyerson M., Kuick R., Dobbin K.K., Lively T. Gene expression – based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study, Nat. Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pathan M., Keerthikumar S., Chisanga D., Alessandro R., Ang C. SHORT COMMUNICATION A novel community driven software for functional enrichment analysis of extracellular vesicles data. J. Extracell. Vesicles. 2017;6 doi: 10.1080/20013078.2017.1321455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weng C.Y., Hu X.Y., Wang Y.J. Integrated analysis of gene expression, alteration and clinical significance of carcinoembryonic antigen-related cell adhesion molecule 1 in cancer. 3 Biotech. 2020;10:1–19. doi: 10.1007/s13205-020-2122-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin S., Cao C., Meng Y., Wu P., Gao P., Zhi W., Peng T., Wu P., Gui L. Comprehensive analysis of the value of RAB family genes in prognosis of breast invasive carcinoma. Biosci. Rep. 2020;40:1–14. doi: 10.1042/BSR20201103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saha S.K., Islam S.M.R., Kwak K.S., Rahman M.S., Cho S.G. PROM1 and PROM2 expression differentially modulates clinical prognosis of cancer: a multiomics analysis. Canc. Gene Ther. 2020;27:147–167. doi: 10.1038/s41417-019-0109-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang L., Wang Y.Z., Zhu H.H., Chang Y., Di Li L., Chen W.M., Long L.Y., Zhang Y.H., Liu Y.R., Lu J., Qin Y.Z. PRAME gene copy number variation is related to its expression in multiple myeloma. DNA Cell Biol. 2017;36:1099–1107. doi: 10.1089/dna.2017.3951. [DOI] [PubMed] [Google Scholar]
- 27.Huang Y.S., Bin Liu W., Han F., Yang J.T., Hao X.L., Chen H.Q., Jiang X., Yin L., Ao L., Cui Z.H., Cao J., Liu J.Y. Copy number variations and expression of MPDZ are prognostic biomarkers for clear cell renal cell carcinoma. Oncotarget. 2017;8:78713–78725. doi: 10.18632/oncotarget.20220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuzyk A., Booth S., Righolt C., Mathur S., Gartner J., Mai S. MYCN overexpression is associated with unbalanced copy number gain, altered nuclear location, and overexpression of chromosome arm 17q genes in neuroblastoma tumors and cell lines. Gene Chromosome Canc. 2015;54 doi: 10.1002/gcc.22273. [DOI] [PubMed] [Google Scholar]
- 29.Shao X., Lv N., Liao J., Long J., Xue R., Ai N., Xu D., Fan X. Copy number variation is highly correlated with differential gene expression: a pan-cancer study. BMC Med. Genet. 2019;20:1–14. doi: 10.1186/s12881-019-0909-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao Z., Dai Z., Locasale J.W. Metabolic landscape of the tumor microenvironment at single cell resolution. Nat. Commun. 2019;10:1–12. doi: 10.1038/s41467-019-11738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 32.Zhang T., Huang Y., Liu W., Meng W., Zhao H., Yang Q., Gu S.-J., Xiao C.-C., Jia C.-C., Zhang B., Zou Y., Li H.-P., Fu B.-S. Overexpression of zinc finger protein 687 enhances tumorigenic capability and promotes recurrence of hepatocellular carcinoma. Oncogenesis. 2017;6:e363. doi: 10.1038/oncsis.2017.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu L., Ye F., Li Y.-Y., Zhan Y.-Z., Liu Y., Yan H.-M., Fang Y., Xie Y.-W., Zhang F.-J., Chen L.-H., Ding Y., Chen K.-L. Histone methyltransferase SETDB1 promotes colorectal cancer proliferation through the STAT1-CCND1/CDK6 axis. Carcinogenesis. 2019 doi: 10.1093/carcin/bgz131. [DOI] [PubMed] [Google Scholar]
- 34.Liu L., Qi L., Knifley T., Piecoro D.W., Rychahou P., Liu J., Mitov M.I., Martin J., Wang C., Wu J., Weiss H.L., Butterfield D.A., Evers B.M., O’Connor K.L., Chen M. S100A4 alters metabolism and promotes invasion of lung cancer cells by up-regulating mitochondrial complex i protein NDUFS2. J. Biol. Chem. 2019;294:7516–7527. doi: 10.1074/jbc.RA118.004365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L., Liu R., Liu Z.P., Li M., Aihara K. Detecting early-warning signals for sudden deterioration of complex diseases by dynamical network biomarkers. Sci. Rep. 2012;2:18–20. doi: 10.1038/srep00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma K.Y., Schonnesen A.A., Brock A., Van Den Berg C., Eckhardt S.G., Liu Z., Jiang N. Single-cell RNA sequencing of lung adenocarcinoma reveals heterogeneity of immune response-related genes. JCI Insight. 2019;4 doi: 10.1172/jci.insight.121387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cagle P.T., Allen T.C., Dacic S., Beasley M.B., Borczuk A.C., Chirieac L.R., Laucirica R., Ro J.Y., Kerr K.M. Revolution in lung cancer: new challenges for the surgical pathologist. Arch. Pathol. Lab Med. 2011;135:110–116. doi: 10.5858/2010-0567-RA.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures