Abstract
Ferroptosis is an iron-dependent cell death caused by impaired glutathione metabolism, lipid peroxidation and mitochondrial failure. Emerging evidences report a role for ferroptosis in Friedreich's Ataxia (FRDA), a neurodegenerative disease caused by the decreased expression of the mitochondrial protein frataxin. Nrf2 signalling is implicated in many molecular aspects of ferroptosis, by upstream regulating glutathione homeostasis, mitochondrial function and lipid metabolism. As Nrf2 is down-regulated in FRDA, targeting Nrf2-mediated ferroptosis in FRDA may be an attractive option to counteract neurodegeneration in such disease, thus paving the way to new therapeutic opportunities. In this study, we evaluated ferroptosis hallmarks in frataxin-silenced mouse myoblasts, in hearts of a frataxin Knockin/Knockout (KIKO) mouse model, in skin fibroblasts and blood of patients, particularly focusing on ferroptosis-driven gene expression, mitochondrial impairment and lipid peroxidation. The efficacy of Nrf2 inducers to neutralize ferroptosis has been also evaluated.
Keywords: Friedreich ataxia, Nrf2, Ferroptosis, Redox imbalance, Sulforaphane, EPI-743, Lipid peroxides, Mitochondria
1. Introduction
Ferroptosis is a pathway of iron-dependent cell death characterized by decreased glutathione peroxidase 4 activity (GPX4), increased lipid peroxidation and mitochondrial impairment [[1], [2], [3]].
Although the execution phase of ferroptosis is a direct result of lipid peroxidation, iron represents one of the main actors responsible for the accumulation of lipid peroxides, thus impacting on the cell sensitivity to ferroptosis [1,4,5].
Increased iron is detected in brain regions of patients with several neurodegenerative diseases, including motor neuron diseases, multiple sclerosis, Huntington's disease and Friedreich's ataxia (FRDA) [[6], [7], [8]]. In FRDA, in particular, the decreased expression of the protein frataxin, caused by the GAA repeat expansion in the first intron of both alleles of the FXN gene, induces mitochondrial iron accumulation, chronic oxidative stress and mitochondrial dysmorphology [[9], [10], [11], [12], [13]]. Frataxin has a critical role in iron metabolism, participating to the biosynthesis of iron-sulfur clusters, the prosthetic groups essential for the function of the respiratory chain enzymes.
Recently, Cotticelli et al. [14] and our group [15] reported evidences for ferroptotic pathway activation in cellular models of FRDA. Increased ferroptosis susceptibility has been found in patient and murine-derived fibroblasts treated with a known ferroptosis inducer (erastin or buthionine sulfoximine) [15], whereas decreased cell death has been detected by using a ferroptosis inhibitor (SRS11-92) [16].
Nrf2 regulates many genes directly or indirectly involved in modulating ferroptosis [17,18]. Thus, beside its important role in maintaining cellular redox balance, Nrf2 may be critical for protection against ferroptosis.
Nrf2 is neuroprotective in models of neurodegeneration, where it promotes ferroptosis resistance by regulating the expression of proteins fundamental for iron signalling (ferritin and ferroportin) as well as of enzymes responsible for glutathione synthesis (SLC7A11, GCLC/GLCM, and GSS), NADPH generation and lipid peroxides neutralization (GPX4) [19,20].
As to date, no cure and FDA-approved treatments for FRDA exist and Nrf2 signalling has been shown to be defective in several in vitro and in vivo disease models [15,[21], [22], [23], [24], [25]], here we explore the possibility to target Nrf2 to counteract ferroptosis in FRDA.
Several inhibitors of ferroptosis have been already described, such as lipoxygenase (LOX) inhibitors (tocopherols/tocotrienols, flavonoids), iron chelators (deferoxamine), lipophilic antioxidants, or agents depleting polyunsaturated fatty acids (PUFAs) [[26], [27], [28]]. However, directly acting on Nrf2, which operates on upstream multifaceted ferroptosis-actors, could be more effective in counteracting ferroptosis than inhibitors specifically directed towards single ferroptosis-inducing enzymes or noxious ferroptosis by-products. In particular, in this study, by using skin fibroblasts of patients with FRDA we analysed primary events characterizing ferroptosis (i.e. mitochondrial impairment, lipid peroxidation, glutathione imbalance, DNA oxidation) and evaluated the efficacy of Nrf2 inducers to neutralize ferroptosis. Before addressing patients’ cells, we evaluated ferroptosis in two mouse models of the disease: 1) a myoblasts cell line transfected with siRNAs targeting FXN mRNA, and 2) a frataxin Knockin/Knockout (KIKO) mouse model, which closely recapitulates the clinical human phenotype [29,30]. Slc7a11, the gene inhibiting the cystine-glutamate antiporter (known as system Xc- or xCT), Ncoa4 (the nuclear receptor coactivator 4) that plays an important role in ferritinophagy, Gpx4 that protects against lipid peroxidation, Sat1 that is implicated in the polyamine metabolism and Slc39a14, a non-transferrin-bound iron chaperone responsible for iron-mediated ferroptosis [[31], [32], [33]] have been measured to understand the predisposition of these models to ferroptosis. Finally, given the importance of monitoring the progression of the disease and the effectiveness of therapies, we investigated the blood of patients as a potential systemic sensor of tissue ferroptosis.
2. Materials and methods
2.1. C2C12 cultures and transfection
C2C12 myoblasts were obtained from the European Collection of Cell Cultures (Salisbury, UK), cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/ml penicillin/streptomycin and 2 mM glutamine (Lonza Sales, Basel, Switzerland), and maintained at 37 °C, 5% CO2 air. One day after plating, C2C12 cells were transfected with a pool of siRNAs directed against FXN mRNA. Cells transfected with scramble siRNAs having no homology to other mouse mRNAs were used as controls. Cells were transfected by electroporation as described previously [34]. Transfection efficiency was evaluated by co-transfecting scramble rhodamine-conjugated siRNAs. Only experiments that gave 80% transfection efficiency were considered.
2.2. Hearts from KIKO mice
Knock-in knock-out (KIKO) mice were purchased from Jackson Laboratories (#014162, Maine USA). KIKO mice are generated by animals bearing a (GAA)230 expansion repeat “knock in” targeted to the endogenous Fxn locus coupled with a Fxn targeted “knock out” mutation allele disrupting exon 4. Littermate C57BL/6 mice (WT) were used as controls. Experiments on animals were conducted in accordance with accepted standard of humane animal care after the approval by relevant local (Institutional Animal Care and Use Committee, Tor Vergata University) and national (Ministry of Health, license no. 324/2018-PR) committees. Mice were maintained at 21.0 °C and 55.0 ± 5.0% relative humidity under a 12 h/12 h light/dark cycle. Food and water were given ad libitum. WT and KIKO male mice were sacrificed at 9 months or 16 months of age.
2.3. Fibroblasts cultures from FRDA patients and treatments
Skin biopsies were taken from two clinically affected (and genetically proven) FRDA patients (one male and one female, 848/848 and 680/350 GAA repeats, respectively) and three age-matched controls (Ctrls). Fibroblasts were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 50 units/ml penicillin, 50 μg/mL streptomycin, 0.4% (v/v), at 37 °C in 5% CO2. Fibroblasts were cultured to 70% confluence and incubated for 24 h with 10 μM SFN or 1 μM EPI-743, diluted in culture medium (drugs dosing has been chosen following ref. [35]). Ferroptotic cell death was induced in patient-derived fibroblast cells using the GPX4 inhibitor RSL3 [36]. After washing, cells were lysed in Total RNA Purification Plus Kit (Norgen Biotek Corp., Torold, ON, Canada), according to the manufacturer's protocol for RNA extraction and subjected to quantitative Real-Time PCR, or lysed with RIPA buffer including DTT and protease inhibitors for Western blotting analysis. Cells were used at similar 9–11 passage numbers and were tested for mycoplasma contamination. The assays were performed in triplicates. All the participants signed an informed consent and the study was approved by the Ethics Committee of “Bambino Gesù” Children's Hospital (code 1166/2016; date of approval 08/06/2016).
2.4. Blood sample collection
Blood samples from patients (Table 1) were collected into EDTA Vacutainer Tubes (Becton Dickinson, Rutherford, NY, USA) and leukocytes were isolated by adding 10% dextran. After 45 min at room temperature, the upper phase was centrifuged at 2600×g (5 min) and the pellet washed with 0.9% NaCl and stored at −20 °C until the analysis. Plasma was obtained by centrifuging whole blood at 450×g for 3 min and stored at −80 °C until 4 hydroxynonenal (4-HNE) measurements. All the participants signed an informed consent and the study was approved by the Ethics Committee of “Bambino Gesù” Children's Hospital (code 1166/2016; date of approval 08/06/2016).
Table 1.
Clinical data of patients with FRDA.
| Patients | Age | Sex | Genetics | Onset | SARA | |
|---|---|---|---|---|---|---|
| New Diagnosis | BA | 13 | F | 848/682 | 5 | 25 |
| CF | 8 | M | 1000/1000 | 4 | 10.5 | |
| GV | 9 | F | 866/del 9q21.11,390 kb | 4 | 10 | |
| DE | 8 | F | 848/900 | 7 | 12 | |
| Under Idebenone | SD | 4 | M | 900/833 | 4 | N.A. |
| DBE | 12 | F | 999/999 | 5 | 24.5 | |
| CM | 26 | F | 448/848 | 4 | 36 | |
| LA | 19 | F | 915/715 | 9 | 15.5 | |
| AB | 13 | M | 766/766 | 4 | 7 | |
| DA | 18 | M | 900/700 | 9 | 25.5 | |
| BD | 9 | F | 1116/1116 | 3 | 10 | |
| AS | 58 | M | 430/1000 | 40 | 12 | |
| AF | 61 | M | 430/1000 | 40 | 9.5 | |
| AA | 33 | F | 430/900 | 23 | 12.5 | |
N.A.: uncooperative patient, measure not applicable.
2.5. Quantitative Real Time PCR (qRT-PCR)
One μg RNA samples was reverse transcribed with the SuperScript™ First-Strand Synthesis system and random hexamers as primers (Life Technologies, Carlsbad, CA, USA). The expression levels of SLC7A11, NCOA4, GPX4, SLC39A14, SAT1, FXN, NRF2, SOD2, and GCL were measured by qRT-PCR in an ABI PRISM 7500 Sequence Detection System (Life Technologies) using Power SYBR Green I dye chemistry (ThermoFisher Scientific, Walthman, MA, USA). Data were analysed using the 2−ΔΔCt method with TBP (TATA box binding protein) as housekeeping gene and shown as fold change relative to controls.
2.6. Assessment of lipid peroxidation by C11-Bodipy (581/591) fluorescent staining
Fibroblasts were incubated with 5 μM C11-BODIPY581/591 (D3861, Thermofisher Scientific, USA) for 45 min at 37 °C using a modified protocol from Refs. [37] and images were acquired with a Leica DMi8 fluorescence microscope (Leica Microsystems, Germany). Stock solutions were made by dissolving 1 mg C11-BODIPY581/591 in 50 μl dimethyl formamide (DMF).
2.7. 4-HNE plasma detection
The quantitative measurement of 4-HNE was performed in plasma of FRDA patients by a competitive ELISA kit (Lipid Peroxidation 4-HNE Assay kit, Abcam, Cambridge, UK). Samples absorbance was detected on a microplate reader (Enspire, PerkinElmer, USA) at 450 nm and quantified using a standard curve.
2.8. Immunofluorescence
Immunofluorescence staining was performed after cell fixation in 4% (v/v) formaldehyde (Sigma-Aldrich) and permeabilization with 0.1% Triton X-100 in PBS, supplemented with 1% BSA. Samples were incubated with the mouse anti-TOM20 (1:500, Santa Cruz Biotechnology, Dallas, TX, USA) primary antibody for 1 h at r.t. and with the FITC-conjugated (1:250) secondary antibody (Jackson ImmunoResearch, Cambridge, United Kingdom) for 1 h at r.t. A mouse monoclonal anti-GS-pro antibody (1:100; 101-A, Virogen, Watertown, MA, USA) was used to visualize the glutathionylated proteins. After washing three times in PBS, cells were incubated with the secondary antibody 555 Goat Anti-Mouse IgG (1:500) for 1 h at r. t. (Invitrogen/Molecular Probes Corp, CA, USA). Hoechst 33342 (Invitrogen, CA, USA) was added for 15 min, and fluorescence preserved using the Prolong Gold mounting solution (Thermo Fisher Scientific, Walthman, MA, USA). Ten fields were randomly taken for each sample using a DMI6000B inverted microscope (Leica, Germany), equipped with a Pan-Neofluar 20×/0.75 objective lens. Data are represented as percentage of positive cells/total cells (evaluated by the number of total nuclei).
2.9. Western blot analysis
Mouse hearts were homogenized (1:10 w/v) in RIPA buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 12 mM deoxycholic acid, 0.5% Nonidet P-40, and protease and phosphatase inhibitors). Fibroblasts (1 × 106) were lysed on ice with RIPA buffer, including DTT and protease inhibitors. Fourty μg proteins were subjected to SDS PAGE on 4–12% denaturing gel and probed with the following antibodies: NRF2 (1:1000, Thermo Fisher Scientific, USA), GPX4 (1:1000, Thermo Fisher Scientific, USA), Frataxin (1:500, Santa Cruz Biotechnology, USA), Hsp60 (1:10000) and vinculin (1:10000, Sigma) as loading controls. Immunoreactive bands were detected using the Lite Ablot Extend Long Lasting Chemiluminescent substrate (Euroclone, Milan, Italy). Signals derived from appropriate HRP-conjugated secondary antibodies (Bethyl Laboratories, Montgomery, TX, USA) were captured by Chemi DocTM XRS 2015 (Bio-Rad Laboratories, Hercules, CA, USA) and densitometric analysis was performed using Image Lab software (Version 5.2.1, Bio-Rad Laboratories).
2.10. γH2AX and TRF1 co-immunostaining
Cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 and blocked in PBS/BSA1%. Samples were then co-immunostained over night at 4 °C, using a rabbit telomeric protein TRF1 antibody (Santa Cruz Biotechnology, CA, USA) in combination with a mouse γH2AX antibody (Millipore, MA, USA). After washes in PBS/BSA1%, samples were incubated with the secondary antibodies (anti-mouse Alexa 546 and anti-rabbit Alexa 488, Invitrogen). Finally, slides were washed in PBS/1% BSA, counterstained with DAPI and analysed with fluorescence microscopy using an Axio-Imager M1 microscope equipped with a coupled charged device (CCD) camera. The frequency of γH2AX foci and γH2AX/TRF1 colocalization dots per cell were scored in 100 nuclei in at least two independent experiments.
2.11. Statistical analysis
Statistical analysis was performed using the Graphpad/Prism 5.0 Software (San Diego, CA, USA). Considering the small number of animals (n = 3 mice each group) and patients (n = 2 for skin biopsies, n = 4 from new diagnosis and n = 10 under Idebenone treatment), we performed statistical analysis with non-parametric Student's t-test. Analyses on each mouse and human sample were repeated in triplicate. All data are presented as mean ± standard error (SEM) or standard deviation (SD). Statistical significance was defined as *p < 0.05, **p < 0.001, ***p < 0.001 compared to healthy controls, and #p < 0.05, ##p < 0.01, ###p < 0.001 compared to untreated cells.
3. Results
3.1. FXN silencing modulates ferroptosis markers in myoblasts
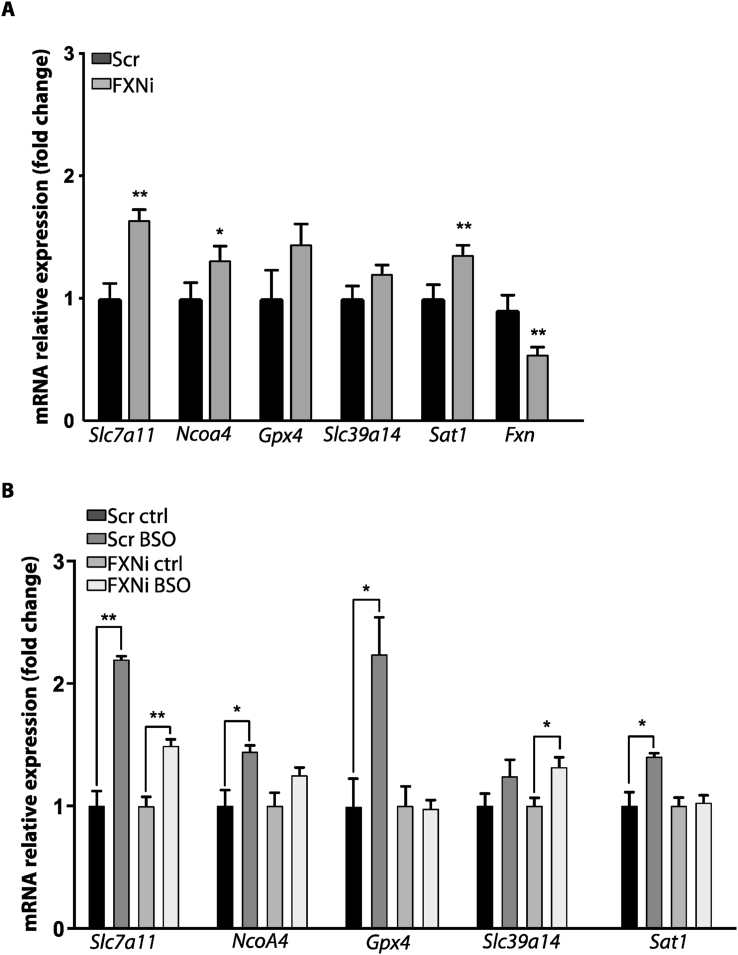
Mouse C2C12 myoblasts have been transfected with siRNAs targeting Fxn mRNA (FXNi) or with Scr siRNAs (SCR), and mRNA expression of genes involved in ferroptosis has been determined (Fig. 1). As reported in Fig. 1A, the expression of genes pertaining to ferroptosis pathways were significantly up-regulated in FXNi compared to Scr myoblasts. In particular, even though the expression of the protective gene Slc7a11 was increased (+65%), genes favouring ferroptosis such as Ncoa4 and Sat1 were up-regulated (+32% and +36%, respectively) in FXNi myoblasts. By contrast, the mRNA level of the anti-ferroptotic Gpx4 remained unchanged (Fig. 1A). Importantly, FXN down-regulation appeared to shift FXNi cells towards a pro-ferroptosis condition. Indeed, even though cell death was not observed (data not shown), upon treatment with buthionine sulfoximine (BSO), a drug able to induce iron-mediated oxidative stress [14,38], FXNi myoblasts failed to up-regulate anti-ferroptotic genes such as Slc7a11, Gpx4 and Sat1. By contrast, these genes were significantly up-regulated in Scr myoblasts. Moreover, in BSO-treated FXNi myoblasts the lack of the anti-ferroptotic response was associated with the significant induction of the intracellular iron transporter Slc39a14 (Fig. 1B). Therefore, as a consequence of frataxin deficiency, myoblasts show a dysregulation of iron homeostasis, which is not accompanied by the induction of genes counteracting ferroptosis, thus shifting cell status towards a pro-ferroptotic condition that could be exploited as marker of disease state.
Fig. 1.
qRT-PCR analysis of ferroptosis-related genes expression in mouse C2C12 myoblasts transfected with siRNAs targeting FXN mRNA (FXNi) or with Scr siRNAs (SCR). (A) Gene expression under basal conditions. B) Gene expression after treatment with the ferroptosis inducer buthionine sulfoximine (BSO). Three independent experiments were performed and analyses repeated in triplicate. Values were expressed as mean ± SD. *p < 0.05, **p < 0.01, compared with controls' group (by Student's two-tailed t-test).
3.2. Evidence of ferroptosis in the cardiac tissue of KIKO FRDA mice
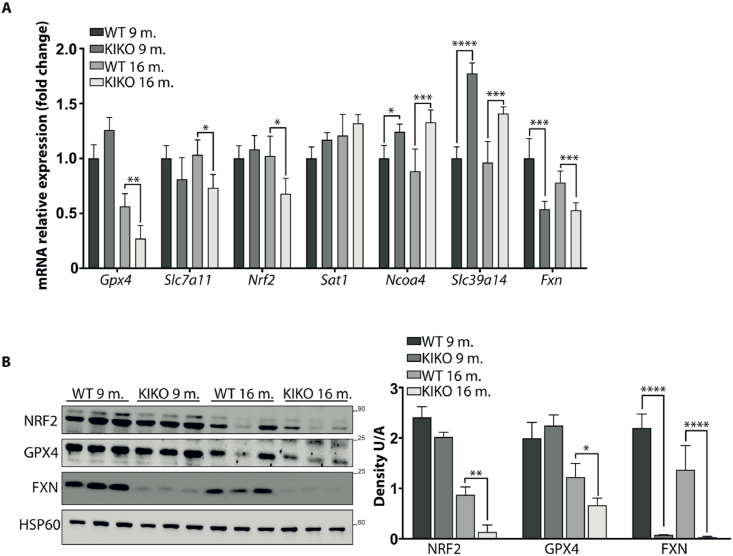
The expression of ferroptosis-related genes (Slc7a11, Ncoa4, Gpx4, Slc9a14, Sat1) has been also evaluated in hearts of KIKO mice at 9- and 16-months of age. As shown in Fig. 2A, the pro-ferroptotic Ncoa4 and Slc39a14 transcripts increased as early as 9-months, while the anti-ferroptotic Gpx4, Slc7a11 and Nrf2 mRNAs significantly decreased at 16-months, thus evidencing a trend throughout the disease progression leading to the weakening of the protective defences against cardiac ferroptosis. This reduced ability to counteract ferroptosis during aging was also confirmed by analysing NRF2 and GPX4 protein levels by Western blot (Fig. 2B). Both proteins progressively decreased in mouse hearts with age, either in WT or in KIKO mice. However, this trend was particularly pronounced in KIKO hearts, where NRF2 and GPX4 were 93% and 70% reduced at 16 months, with respect to 9 months of age. By contrast, WT hearts displayed 63% and 39% reduction of NRF2 and GPX4 in old compared to young mice.
Fig. 2.
Ferroptosis-related genes expression in hearts of WT and KIKO FRDA mice at 9- and 16-months of age. A) qRT-PCR analysis. B) Representative Western blot (right) and densitometric analysis (left) of FXN, NRF2, and GPX4 protein levels. HSP60 has been used as loading control. Experiments were performed in triplicates on n = 3 mice each group and values expressed as mean ± SD. *p < 0.05, **p < 0.01, ****p < 0.001, compared to controls' group (by Student's two-tailed t-test).
3.3. Ferroptosis occurs in fibroblasts of FRDA patients
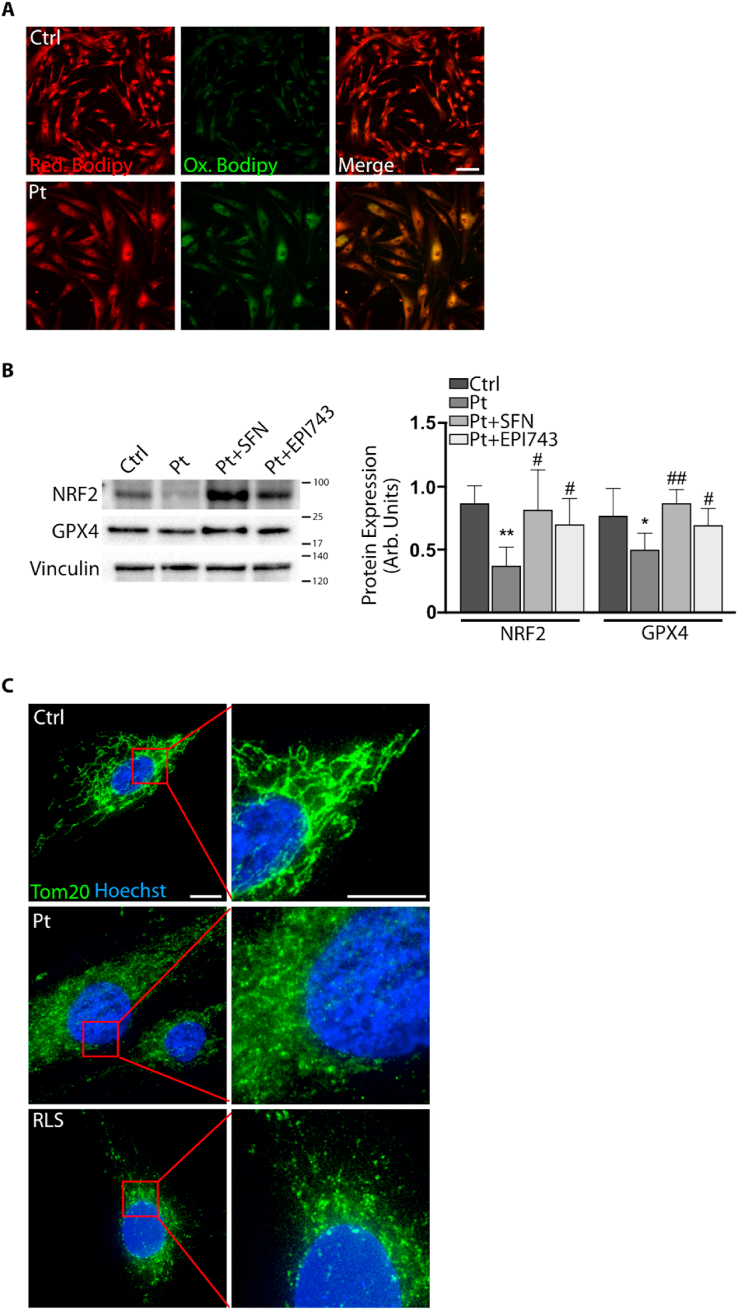
To investigate whether ferroptosis biomarkers were altered also in FRDA patients, we analysed fibroblasts obtained from skin biopsies of two FRDA patients. Using the fluorescent probe Bodipy-C11, we found an accumulation of oxidized lipids in FRDA fibroblasts compared to controls (Fig. 3A), indicating an increased extent of lipid peroxidation in patients. Consistent with these data, Western blot analysis revealed a significant decrease (35%) of Gpx4 protein in FRDA fibroblasts (Fig. 3B). Moreover, as defective mitochondrial morphology represents another sign of ferroptosis, and altered mitochondria have been reported as a consequence of GPX4 silencing [[39], [40], [41], [42]], we analysed the mitochondrial network in fibroblasts of patients (Fig. 3C). Through immunofluorescence analysis using an anti-TOM20 antibody, we evidenced small and fragmented mitochondria, closely resembling the morphological defects classified as category II/III by Neitemer [43] and Jelinek [44] among the ferroptosis-related mitochondrial categories.
Fig. 3.
Ferroptosis markers in fibroblasts of patients with FRDA. A) Lipid peroxides detected by using the fluorescent lipophilic Bodipy-C11 sensor (in green the oxidized lipids, in red the total lipids, scale bar = 100 μm). B) Representative Western blot (right) and densitometric analysis (left) of NRF2, and GPX4 protein levels in FRDA fibroblasts. Vinculin has been used as loading control. C) Immunofluorescence of mitochondrial network by using the anti-TOM20 antibody. As positive ferroptosis control, fibroblasts have been treated with 0.2 μM ferroptosis inducer RSL-3 for 24 h. Scale Bar = 25 μm. Three independent experiments were performed on fibroblasts obtained from n = 2 FRDA patients and values were expressed as mean ± SD. *p < 0.05, **p < 0.01, compared to controls' cells, and #p < 0.05, ##p < 0.01, compared to treated cells (by Student's two-tailed t-test). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.4. FRDA fibroblasts display increased protein oxidation and DNA damage
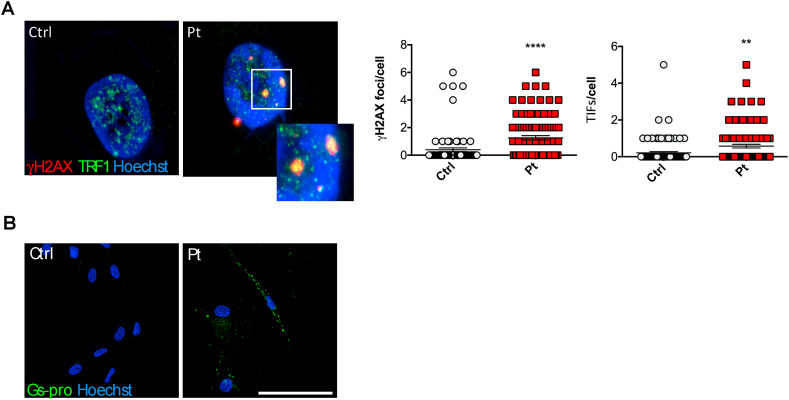
To better characterise ferroptosis-related events, we analysed the DNA damage and protein glutathionylation in patients’ fibroblasts. We firstly evaluated the presence of DNA damage and telomere dysfunction by analysing the γ-H2AX foci and the telomere dysfunction-induced foci (TIF). As shown in Fig. 4A, we found that FRDA cells display a significant higher frequency of γ-H2AX foci compared to controls (p < 0.001). Notably, as previously reported [45], FRDA cells had also a greater TIF frequencies with respect to controls (p < 0.01), suggesting that FRDA fibroblasts accumulate DNA damage at telomeres, a DNA region highly sensitive to oxidative damage [46,47]. An increase of protein oxidation has been even evidenced in FRDA cells (Fig. 4B), as visualized by immunofluorescence analysis using an antibody specifically directed to glutathionylated proteins (GSPro). Overall, these findings confirmed FRDA as a ferroptotic/oxidative disorder.
Fig. 4.
Redox imbalance in FRDA fibroblasts. A) DNA damage evaluated by the γ-H2AX foci analysis (red) and telomere dysfunction-induced foci (TRF1) (green). On the right, the quantification of γ-H2AX and TRF1 foci in FRDA patient's fibroblasts. B) Immunofluorescence of GS-pro using the anti-GS-pro antibody specifically probing glutathionylated proteins (green). Hoechst staining was performed for nuclear staining (blue). Scale Bar = 100 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.5. Rescue of ferroptosis and redox imbalance in FRDA cells by Nrf2 induction
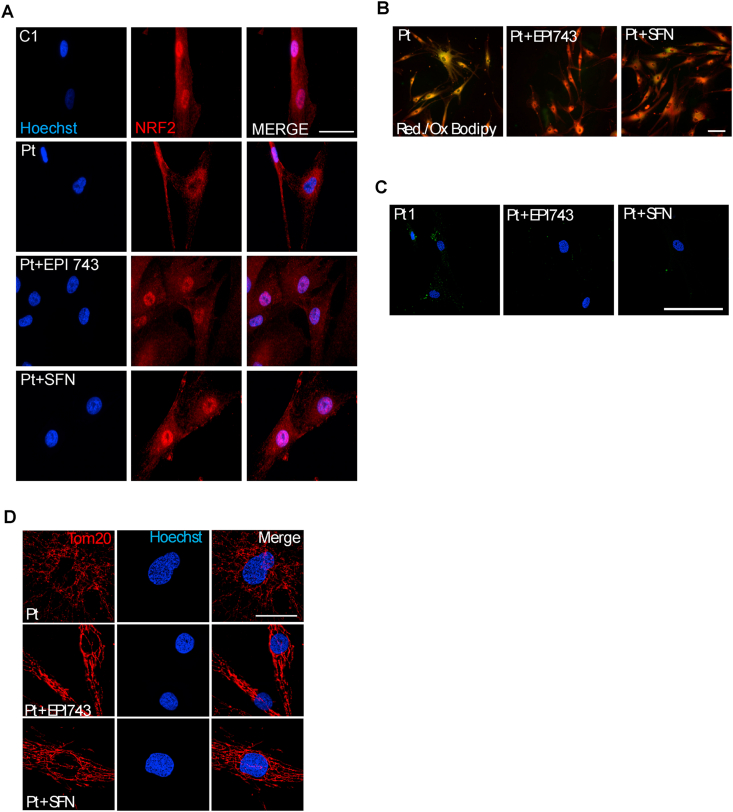
As redox imbalance can be efficiently counteracted by Nrf2 inducers [18,48] either in FRDA fibroblasts [23] and neural stem cells [49], we attempted at evaluating the effect of two Nrf2 activators on ferroptosis and oxidative stress markers in FRDA cells. FRDA fibroblasts were treated with EPI-743, which potently prevents ferroptosis in cells of patients with mitochondriopathies [37], or with SFN, a well-known Nrf2 activator enhancing mitochondrial function and biogenesis in in vivo and in vitro FRDA models [50]. In FRDA fibroblasts, both treatments promoted the nuclear translocation of Nrf2 (Fig. 5A) and the inhibition of redox imbalance and ferroptosis. Actually, we observed a consistent decrease of both lipid peroxides (Fig. 5B) and glutathionylated proteins (Fig. 5C). Even mitochondria, which in untreated FRDA fibroblasts appeared round, small and fragmented, upon such treatments displayed a tubular network, recapitulating the morphology of control cells (Fig. 5D).
Fig. 5.
Rescue of ferroptosis and redox imbalance in FRDA fibroblasts after NRF2 induction by EPI-743 and SFN. A) Representative images of immunofluorescence of FRDA fibroblasts (Pt) probed with the anti-NRF2 antibody, indicating the nuclear translocation in patient's cells after treatment with NRF2 inducers. Scale Bar = 100 μm. B) Representative images of BODIPY-C11 fluorescent staining of patient's fibroblasts showing a decrease of lipid peroxides (green) after NRF2 induction (in red the total lipids, in yellow the coincident staining). Scale Bar = 100 μm. C) Decrease of GS-Pro immunofluorescent signal in patient's fibroblasts after EPI-743 and SFN treatments. Scale Bar = 100 μm. D) Immunofluorescence of mitochondrial network by using the anti-TOM20 antibody. Scale Bar = 50 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.6. Blood as sensor of ferroptosis in FRDA
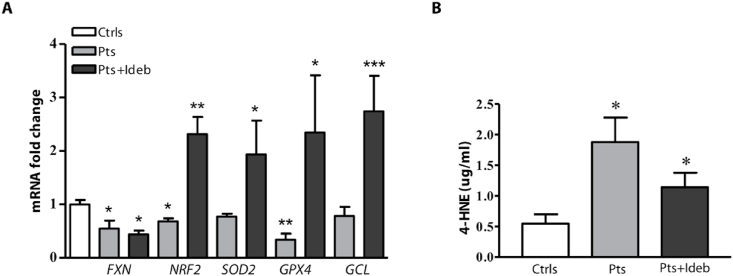
In order to evaluate whether blood samples could be used to monitor systemic ferroptosis, we analysed the expression of some ferroptosis-related genes (FXN, NRF2, SOD2, GPX4, GCL) in leukocytes of 4 newly diagnosed FRDA patients and in 10 FRDA patients who underwent Idebenone therapy (see Table 1 for clinical data). As shown in Fig. 6, all these genes were down-regulated in blood of patients with respect to healthy subjects, and Idebenone treatment led to a significant increase of the expression both of NRF2 (2.3-fold) and its down-stream genes (1.9-fold SOD2, 2.3-fold GPX4, and 2.7-fold GCL) (Fig. 6A). We also measured plasma levels of the ferroptosis marker 4-HNE, the major by-product of lipid peroxidation [[51], [52], [53]], and we found a 3–fold increase in patients with FRDA (1.88 ± 0.4 vs 0.55 ± 0.15 μg/ml, p < 0.05), compared to healthy subjects (Fig. 6B). Notably, the plasma 4-HNE content decreased in patients treated with Idebenone.
Fig. 6.
Ferroptosis markers in blood of patients with FRDA. (A) Real-time PCR analysis of frataxin (FXN), NRF2 and its target genes (NRF2, SOD2, GPX4, GCL) performed on leukocytes isolated from blood of n = 4 newly diagnosed patients and n = 10 patients undergoing Idebenone therapy. (B) 4-HNE plasma levels in patients with and without Idebenone therapy. Analyses were performed in triplicate on n = 4 newly diagnosed patients and on n = 10 patients under Idebenone. Values represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 respect to healthy subjects (by Student's two tailed t-test).
4. Discussion
In this study, we attempted at understanding whether ferroptosis, an iron-dependent regulated form of cell death caused by iron dysmetabolism and accumulation of lipid peroxides, may underlie the pathogenesis of FRDA. Moreover, we evaluated whether the induction of Nrf2 could prevent ferroptosis in FRDA and whether the analysis of ferroptosis markers in blood samples could represent a valuable tool to assess systemic ferroptosis in FRDA patients.
FRDA is an autosomal recessive inherited neurodegenerative disorder, characterised by progressive spinocerebellar ataxia, cardiomyopathy and type 2 diabetes [54]. It is caused by a homozygous GAA repeat expansion mutation within intron 1 of the FXN gene. The GAA expansion mutation reduces the expression of frataxin, a ubiquitous mitochondrial protein involved in iron-sulfur cluster (ISC) and heme biosynthesis. The frataxin deficiency inhibits mitochondrial respiration and promotes production of reactive oxygen species (ROS), causing mitochondrial dysfunction, oxidative stress and mitochondrial iron accumulation [55,56].
Recently, ferroptosis has been suggested to occur in FRDA [14,15] and increased sensitivity to erastin and BSO, two known ferroptosis inducers, together with a protective effect of two ferroptosis inhibitors (SRS11-92 and Fer-1) have been described in FRDA cellular models [14,15], thus paving the way to ferroptosis as a new pathogenic mechanism in FRDA.
Ferroptosis has a crucial role in neurodegeneration, and mounting evidence indicate ferroptosis as the main driver of neurological cell death in several neurodegenerative diseases [57]. As the brain is particularly susceptible to the oxidative damage for its high levels of oxidizable PUFAs, high rates of ROS production and low levels of endogenous antioxidants, the targeting of ferroptosis may represent a valid therapeutic strategy to counteract neurodegeneration, also considering the reversible nature of the process [[58], [59], [60]]. In light of this, anti-ferroptosis strategies are emerging in order to minimize neuronal cell death [61], and inhibitors of ferroptosis have been tested in models of Parkinson's, Alzheimer's and Huntington's diseases as well as in traumatic and haemorrhagic brain injury [[62], [63], [64], [65], [66], [67]].
Multiple actors are involved in triggering ferroptosis, with the execution phase as a direct result of LOXs-catalysed lipid peroxidation [5,40,[68], [69], [70]]. All the machinery involved in iron import, export, storage and turnover are responsible for the lipid peroxides accumulation, and the decreased expression of GPX4, a protective enzyme able to detoxify lipid peroxides, also strongly contributes to increase cell susceptibility to ferroptosis [1,4,71].
To elucidate if ferroptosis occurs in FRDA, we analysed three models of the disease, i.e. myoblasts silenced for the Fxn gene, cardiac tissues isolated from KIKO mice, and fibroblasts of patients. Our findings show that both Fxn-silenced myoblasts and KIKO hearts show alteration of the expression of some ferroptosis-related genes and exhibit a reduced adaptation to iron-mediated oxidative stress. Interestingly, a reduced expression of Nrf2 and Gpx4 protein amounts was evidenced in old KIKO mice, when compared to age-matched controls, indicating a progressive weakening of the protective anti-ferroptosis response throughout the disease progression. Notably, the decrease of Nrf2 protein levels are more pronounced with respect to the mRNA levels in KIKO hearts. The same trend was observed in fibroblasts of FRDA patients [35]. This may depend on the increased protein degradation, as the activity of this transcription factor is mainly modulated by post-translational events involving the ubiquitin/proteasome system. In line with this hypothesis, SFN treatment, which is known to block NRF2 degradation, is able to restore NRF2 levels of FRDA fibroblasts to those of normal people. Based on this evidence we can hypothesize that protein degradation is the principal mechanism involved in NRF2 decrease and the down-regulation of mRNA levels participates in this process at minor extent.
Signs of ferroptosis were detectable also in human fibroblasts obtained from skin biopsies of two patients, where a significant increase of lipid peroxidation and a decrease of GPX4 expression have been found. As additional indicators of ferroptosis, we further analysed the FRDA mitochondrial morphology and, according to the classification by Neitemer [43] and Jelinek [44], defective mitochondria closely resembling the category II/III have been detected in fibroblasts of patients. They consisted of dysmorphological short mitochondria still organized in long tubules and distributed throughout the cytosol.
Once established ferroptosis in our models and given that Nrf2 modulates almost all genes implicated in ferroptosis, we wondered if the Nrf2 activation may directly alleviate the ferroptotic stress in FRDA. Indeed, Nrf2 is down-regulated in the disease [[21], [22], [23],25,35] and it is inducible by several drugs, some of those already tested in FRDA fibroblasts [23] and in neural stem cells from KIKO mice [49]. Thus, in order to provide some insights for novel therapies, we decided to evaluate the effect of EPI-743, which potently prevents ferroptosis in cells of patients with mitochondriopathies [37], and of SFN, which enhances mitochondrial function and biogenesis in vivo and in vitro models [50], on ferroptosis markers in FRDA fibroblasts. We found that the drug-mediated Nrf2 induction plays a protective action on ferroptosis hallmarks, as shown by the decreased lipid peroxidation and increased GPX4 expression upon EPI743- and SFN-treatment of FRDA cells. Also, the mitochondrial network appeared less fragmented after such treatments, as they displayed a morphology resembling that of controls. Finally, we explored blood as a potential sensor of systemic ferroptosis by measuring gene expression and lipid peroxidation in isolated FRDA leukocytes and plasma. In patients, the ferroptosis-related genes appeared altered and a significant accumulation of lipid peroxides was observed. Of note, 4-HNE content dropped to control values after antioxidant therapy.
In conclusion, it is important to point out that, despite the recognized role of Nrf2 in FRDA, the link among frataxin deficiency, ISC assembly defects and Nrf2 signalling pathway remains currently unclear in this disease. What is known is that frataxin acts as an iron chaperone in the first steps of ISC biogenesis and Nrf2, besides ensuring an efficient antioxidant response, is also implicated in the regulation of iron metabolism, contributing to maintain the iron bioavailability needed for ISC assembly [20]. Nevertheless, the exact correlation between frataxin-mediated ISC biosynthesis and Nrf2 signalling remains at the moment unknown, even representing the major challenge in order to decipher the pathogenic mechanism underlying FRDA.
5. Conclusions
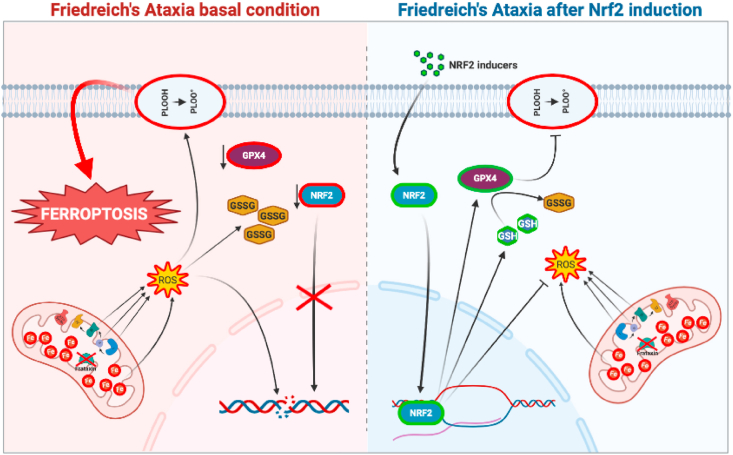
In order to understand if ferroptosis underlies the pathogenic mechanism in FRDA, we used several models of the disease and found alteration of ferroptosis-related genes that was not buffered by the induction of an effective anti-ferroptosis protective system. A rescue of ferroptosis and redox imbalance was obtained in FRDA fibroblasts upon treatment with NRF2 inducers (i.e. EPI-743 and SFN). By analysing blood samples from patients treated or not with the antioxidant Idebenone, we also demonstrated that blood reflects tissue ferroptotic signatures and may be a useful tool to monitor the disease progression and therapy efficacy. Finally, our findings highlight a close correlation between Nrf2 and ferroptosis in FRDA, and support Nrf2 activation as a promising line of therapy to prevent the ferroptosis-induced neurodegeneration (see Fig. 7).
Fig. 7.
Representative model summarizing the main findings of this study.
In FRDA, frataxin deficiency causes a decrease of NRF2, GPX4 and GSH levels, with a consequent increase of lipid peroxides. An altered mitochondrial morphology, signs of protein oxidation and glutathionylation and DNA damage are also evidenced, overall suggesting a condition of ferroptosis (left). Upon NRF2 induction (right), the GPX4 expression and GSH biosynthesis increase, thus reducing lipid peroxides and DNA/protein oxidation. Also the mitochondrial morphology appears recovered, indicating a protective effect of the NRF2 induction against ferroptosis.
Author contributions
Designed, supervised experiments and finalized the manuscript: Piergiorgio La Rosa, Sara Petrillo, Katia Aquilano, Enrico Bertini and Fiorella Piemonte.
Performed experiments: Piergiorgio La Rosa, Sara Petrillo, Riccardo Tuchi, Francesco Berardinelli.
Human samples: Gessica Vasco, Tommaso Schirinzi and Enrico Bertini.
Interpreted data and wrote manuscript: Piergiorgio La Rosa, Katia Aquilano, Enrico Bertini and Fiorella Piemonte.
Critical revision of the manuscript for important intellectual content: Daniele Lettieri Barbato, Maria Teresa Fiorenza, Gessica Vasco, Tommaso Schirinzi, Enrico Bertini.
Funding
This work was supported by Friedreich's Ataxia Research Alliance (General Research Grant 2017–2018) and National Ataxia Foundation (Seed Money Grant 2016) to K.A.
Declaration of competing interest
The authors declare no conflicts of interest.
List of abbreviations
- BODIPY
Boron dipyrromethene
- BSO
Buthionine sulfoximine
- TOM 20
Mitochondrial import receptor subunit 20
- FRDA
Friedreich's Ataxia
- FXN
Frataxin
- GCLC
Glutamate cysteine ligase catalytic subunit
- GPX4
Glutathione peroxidase 4
- GS-pro
Glutathionylated proteins
- KIKO
Knock-in knock-out
- LOX
Lipoxygenase
- PUFA
Polyunsatured fatty acids
- qRT-PCR
quantitative Real Time PCR
- ROS
Reactive oxygen species
- WT
Wild type
- RSL3
(1S,3R)-RSL3
- SFN
Sulforaphane
- TIF
Telomere dysfunction-induced foci
- γ-H2AX
phosphorylated form of H2A histone family member X
References
- 1.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., Patel D.N., Bauer A.J., Cantley A.M., Yang W.S. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stockwell B.R., Friedmann Angeli J.P., Bayir H., Bush A.I., Conrad M., Dixon S.J., Fulda S., Gascón S., Hatzios S.K., Kagan V.E. Ferroptosis: a regulated cell death Nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–285. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galluzzi L., Vitale I., Aaronson S.A., Abrams J.M., Adam D., Agostinis P., Alnemri E.S., Altucci L., Amelio I., Andrews D.W. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao M., Monian P., Quadri N., Ramasamy R., Jiang X. Glutaminolysis and transferrin regulate ferroptosis. Mol. Cell. 2015;59:298–308. doi: 10.1016/j.molcel.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang W.S., Kim K.J., Gaschler M.M., Patel M., Shchepinov M.S., Stockwell B.R. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E4966–E4975. doi: 10.1073/pnas.1603244113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwan J.Y., Jeong S.Y., Van Gelderen P., Deng H.X., Quezado M.M., Danielian L.E., Butman J.A., Chen L., Bayat E., Russell J. Iron accumulation in deep cortical layers accounts for MRI signal abnormalities in ALS: correlating 7 tesla MRI and pathology. PloS One. 2012;7 doi: 10.1371/journal.pone.0035241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li K., Reichmann H. Role of iron in neurodegenerative diseases. J. Neural. Transm. 2016;123:389–399. doi: 10.1007/s00702-016-1508-7. [DOI] [PubMed] [Google Scholar]
- 8.Sheykhansari S., Kozielski K., Bill J., Sitti M., Gemmati D., Zamboni P., Singh A.V. Redox metals homeostasis in multiple sclerosis and amyotrophic lateral sclerosis: a review. Cell Death Dis. 2018;9:348. doi: 10.1038/s41419-018-0379-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delatycki M.B., Camakaris J., Brooks H., Evans-Whipp T., Thorburn D.R., Williamson R., Forrest S.M. Direct evidence that mitochondrial iron accumulation occurs in Friedreich ataxia. Ann. Neurol. 1999;45:673–675. [PubMed] [Google Scholar]
- 10.Wong A., Yang J., Cavadini P., Gellera C., Lonnerdal B., Taroni F., Cortopassi G. The Friedreich's ataxia mutation confers cellular sensitivity to oxidant stress which is rescued by chelators of iron and calcium and inhibitors of apoptosis. Hum. Mol. Genet. 1999;8:425–430. doi: 10.1093/hmg/8.3.425. [DOI] [PubMed] [Google Scholar]
- 11.Rötig A., de Lonlay P., Chretien D., Foury F., Koenig M., Sidi D., Munnich A., Rustin P. Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat. Genet. 1997;17:215–217. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- 12.Lodi R., Hart P.E., Rajagopalan B., Taylor D.J., Crilley J.G., Bradley J.L., Blamire A.M., Manners D., Styles P., Schapira A.H. Antioxidant treatment improves in vivo cardiac and skeletal muscle bioenergetics in patients with Friedreich's ataxia. Ann. Neurol. 2001;49:590–596. [PubMed] [Google Scholar]
- 13.Armstrong J.S., Khdour O., Hecht S.M. Does oxidative stress contribute to the pathology of Friedreich's ataxia? A radical question. Faseb. J. 2010;24:2152–2163. doi: 10.1096/fj.09-143222. [DOI] [PubMed] [Google Scholar]
- 14.Cotticelli M.G., Xia S., Lin D., Lee T., Terrab L., Wipf P., Huryn D.M., Wilson R.B. Ferroptosis as a novel therapeutic target for Friedreich's ataxia. J. Pharmacol. Exp. Therapeut. 2019;369:47–54. doi: 10.1124/jpet.118.252759. [DOI] [PubMed] [Google Scholar]
- 15.Turchi R., Tortolici F., Guidobaldi G., Iacovelli F., Falconi M., Rufini S., Faraonio R., Casagrande V., Federici M., De Angelis L. Frataxin deficiency induces lipid accumulation and affects thermogenesis in brown adipose tissue. Cell Death Dis. 2020;11:51. doi: 10.1038/s41419-020-2253-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotticelli M.G., Crabbe A.M., Wilson R.B., Shchepinov M.S. Insights into the role of oxidative stress in the pathology of Friedreich ataxia using peroxidation resistant polyunsaturated fatty acids. Redox Biol. 2013;1:398–404. doi: 10.1016/j.redox.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dodson M., Castro-Portuguez R., Zhang D.D. NRF2 plays a critical role in mitigating lipid peroxidation and ferroptosis. Redox Biol. 2019;23:101107. doi: 10.1016/j.redox.2019.101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.La Rosa P., Bertini E.S., Piemonte F. The NRF2 signaling network defines clinical biomarkers and therapeutic opportunity in Friedreich's ataxia. Int. J. Mol. Sci. 2020:21. doi: 10.3390/ijms21030916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun X., Ou Z., Chen R., Niu X., Chen D., Kang R., Tang D. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology. 2016;63:173–184. doi: 10.1002/hep.28251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerins M.J., Ooi A. The roles of NRF2 in modulating cellular iron homeostasis. Antioxidants Redox Signal. 2018;29:1756–1773. doi: 10.1089/ars.2017.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shan Y., Schoenfeld R.A., Hayashi G., Napoli E., Akiyama T., Iodi Carstens M., Carstens E.E., Pook M.A., Cortopassi G.A. Frataxin deficiency leads to defects in expression of antioxidants and Nrf2 expression in dorsal root ganglia of the Friedreich's ataxia YG8R mouse model. Antioxidants Redox Signal. 2013;19:1481–1493. doi: 10.1089/ars.2012.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D'Oria V., Petrini S., Travaglini L., Priori C., Piermarini E., Petrillo S., Carletti B., Bertini E., Piemonte F. Frataxin deficiency leads to reduced expression and impaired translocation of NF-E2-related factor (Nrf2) in cultured motor neurons. Int. J. Mol. Sci. 2013;14:7853–7865. doi: 10.3390/ijms14047853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petrillo S., Piermarini E., Pastore A., Vasco G., Schirinzi T., Carrozzo R., Bertini E., Piemonte F. Nrf2-Inducers counteract neurodegeneration in frataxin-silenced motor neurons: disclosing new therapeutic targets for Friedreich's ataxia. Int. J. Mol. Sci. 2017;18 doi: 10.3390/ijms18102173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paupe V., Dassa E.P., Goncalves S., Auchère F., Lönn M., Holmgren A., Rustin P. Impaired nuclear Nrf2 translocation undermines the oxidative stress response in Friedreich ataxia. PloS One. 2009;4:e4253. doi: 10.1371/journal.pone.0004253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.La Rosa P., Petrillo S., Bertini E.S., Piemonte F. Oxidative stress in DNA repeat expansion disorders: a focus on NRF2 signaling involvement. Biomolecules. 2020;10 doi: 10.3390/biom10050702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy T.H., Miyamoto M., Sastre A., Schnaar R.L., Coyle J.T. Glutamate toxicity in a neuronal cell line involves inhibition of cystine transport leading to oxidative stress. Neuron. 1989;2:1547–1558. doi: 10.1016/0896-6273(89)90043-3. [DOI] [PubMed] [Google Scholar]
- 27.Khanna S., Roy S., Ryu H., Bahadduri P., Swaan P.W., Ratan R.R., Sen C.K. Molecular basis of vitamin E action: tocotrienol modulates 12-lipoxygenase, a key mediator of glutamate-induced neurodegeneration. J. Biol. Chem. 2003;278:43508–43515. doi: 10.1074/jbc.M307075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie Y., Song X., Sun X., Huang J., Zhong M., Lotze M.T., Zeh H.J.R., Kang R., Tang D. Identification of baicalein as a ferroptosis inhibitor by natural product library screening. Biochem. Biophys. Res. Commun. 2016;473:775–780. doi: 10.1016/j.bbrc.2016.03.052. [DOI] [PubMed] [Google Scholar]
- 29.Miranda C.J., Santos M.M., Ohshima K., Smith J., Li L., Bunting M., Cossée M., Koenig M., Sequeiros J., Kaplan J. Frataxin knockin mouse. FEBS Lett. 2002;512:291–297. doi: 10.1016/s0014-5793(02)02251-2. [DOI] [PubMed] [Google Scholar]
- 30.McMackin M.Z., Henderson C.K., Cortopassi G.A. Neurobehavioral deficits in the KIKO mouse model of Friedreich's ataxia. Behav. Brain Res. 2017;316:183–188. doi: 10.1016/j.bbr.2016.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liuzzi J.P., Aydemir F., Nam H., Knutson M.D., Cousins R.J. Zip14 (Slc39a14) mediates non-transferrin-bound iron uptake into cells. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13612–13617. doi: 10.1073/pnas.0606424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aydemir T.B., Cousins R.J. The multiple faces of the metal transporter ZIP14 (SLC39A14) J. Nutr. 2018;148:174–184. doi: 10.1093/jn/nxx041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoyanovsky D.A., Tyurina Y.Y., Shrivastava I., Bahar I., Tyurin V.A., Protchenko O., Jadhav S., Bolevich S.B., Kozlov A.V., Vladimirov Y.A. Iron catalysis of lipid peroxidation in ferroptosis: regulated enzymatic or random free radical reaction? Free Radic. Biol. Med. 2019;133:153–161. doi: 10.1016/j.freeradbiomed.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aquilano K., Baldelli S., La Barbera L., Lettieri Barbato D., Tatulli G., Ciriolo M.R. Adipose triglyceride lipase decrement affects skeletal muscle homeostasis during aging through FAs-PPARα-PGC-1α antioxidant response. Oncotarget. 2016;7:23019–23032. doi: 10.18632/oncotarget.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrillo S., D'Amico J., La Rosa P., Bertini E.S., Piemonte F. Targeting NRF2 for the treatment of Friedreich's ataxia: a comparison among drugs. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20205211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang W.S., Stockwell B.R. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem. Biol. 2008;15:234–245. doi: 10.1016/j.chembiol.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kahn-Kirby A.H., Amagata A., Maeder C.I., Mei J.J., Sideris S., Kosaka Y., Hinman A., Malone S.A., Bruegger J.J., Wang L. Targeting ferroptosis: a novel therapeutic strategy for the treatment of mitochondrial disease-related epilepsy. PloS One. 2019;14 doi: 10.1371/journal.pone.0214250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lippmann J., Petri K., Fulda S., Liese J. Redox modulation and induction of ferroptosis as a new therapeutic strategy in hepatocellular carcinoma. Translational Oncology. 2020;13:100785. doi: 10.1016/j.tranon.2020.100785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedmann Angeli J.P., Schneider M., Proneth B., Tyurina Y.Y., Tyurin V.A., Hammond V.J., Herbach N., Aichler M., Walch A., Eggenhofer E. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 2014;16:1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doll S., Proneth B., Tyurina Y.Y., Panzilius E., Kobayashi S., Ingold I., Irmler M., Beckers J., Aichler M., Walch A. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 2017;13:91–98. doi: 10.1038/nchembio.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maiorino M., Conrad M., Ursini F. GPx4, lipid peroxidation, and cell death: discoveries, rediscoveries, and open issues. Antioxidants Redox Signal. 2018;29:61–74. doi: 10.1089/ars.2017.7115. [DOI] [PubMed] [Google Scholar]
- 42.Ingold I., Berndt C., Schmitt S., Doll S., Poschmann G., Buday K., Roveri A., Peng X., Porto Freitas F., Seibt T. Selenium utilization by GPX4 is required to prevent hydroperoxide-induced ferroptosis. Cell. 2018;172:409–422. doi: 10.1016/j.cell.2017.11.048. e421. [DOI] [PubMed] [Google Scholar]
- 43.Neitemeier S., Jelinek A., Laino V., Hoffmann L., Eisenbach I., Eying R., Ganjam G.K., Dolga A.M., Oppermann S., Culmsee C. BID links ferroptosis to mitochondrial cell death pathways. Redox Biol. 2017;12:558–570. doi: 10.1016/j.redox.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jelinek A., Heyder L., Daude M., Plessner M., Krippner S., Grosse R., Diederich W.E., Culmsee C. Mitochondrial rescue prevents glutathione peroxidase-dependent ferroptosis. Free Radic. Biol. Med. 2018;117:45–57. doi: 10.1016/j.freeradbiomed.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 45.Anjomani Virmouni S., Al-Mahdawi S., Sandi C., Yasaei H., Giunti P., Slijepcevic P., Pook M.A. Identification of telomere dysfunction in Friedreich ataxia. Mol. Neurodegener. 2015;10:22. doi: 10.1186/s13024-015-0019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Zglinicki T. Role of oxidative stress in telomere length regulation and replicative senescence. Ann. N. Y. Acad. Sci. 2000;908:99–110. doi: 10.1111/j.1749-6632.2000.tb06639.x. [DOI] [PubMed] [Google Scholar]
- 47.Coluzzi E., Leone S., Sgura A. Oxidative stress induces telomere dysfunction and senescence by replication Fork arrest. Cells. 2019;8 doi: 10.3390/cells8010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kajarabille N., Latunde-Dada G.O. Programmed cell-death by ferroptosis: antioxidants as mitigators. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20194968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.La Rosa P., Russo M., D'Amico J., Petrillo S., Aquilano K., Lettieri-Barbato D., Turchi R., Bertini E.S., Piemonte F. Nrf2 induction Re-establishes a proper neuronal differentiation program in Friedreich's ataxia neural stem cells. Front. Cell. Neurosci. 2019;13:356. doi: 10.3389/fncel.2019.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lei P., Tian S., Teng C., Huang L., Liu X., Wang J., Zhang Y., Li B., Shan Y. Sulforaphane improves lipid metabolism by enhancing mitochondrial function and biogenesis in vivo and in vitro. Mol. Nutr. Food Res. 2019;63 doi: 10.1002/mnfr.201800795. [DOI] [PubMed] [Google Scholar]
- 51.Grune T., Siems W., Kowalewski J., Zollner H., Esterbauer H. Identification of metabolic pathways of the lipid peroxidation product 4-hydroxynonenal by enterocytes of rat small intestine. Biochem. Int. 1991;25:963–971. [PubMed] [Google Scholar]
- 52.Ayala A., Muñoz M.F., Argüelles S. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid Med Cell Longev. 2014;2014:360438. doi: 10.1155/2014/360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Niki E. Biomarkers of lipid peroxidation in clinical material. Biochim. Biophys. Acta. 2014;1840:809–817. doi: 10.1016/j.bbagen.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 54.Delatycki M.B., Bidichandani S.I. Friedreich ataxia- pathogenesis and implications for therapies. Neurobiol. Dis. 2019;132:104606. doi: 10.1016/j.nbd.2019.104606. [DOI] [PubMed] [Google Scholar]
- 55.Li K. Iron pathophysiology in Friedreich's ataxia. Adv. Exp. Med. Biol. 2019;1173:125–143. doi: 10.1007/978-981-13-9589-5_7. [DOI] [PubMed] [Google Scholar]
- 56.Purroy R., Medina-Carbonero M., Ros J., Tamarit J. Frataxin-deficient cardiomyocytes present an altered thiol-redox state which targets actin and pyruvate dehydrogenase. Redox Biol. 2020;32:101520. doi: 10.1016/j.redox.2020.101520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirschhorn T., Stockwell B.R. The development of the concept of ferroptosis. Free Radic. Biol. Med. 2019;133:130–143. doi: 10.1016/j.freeradbiomed.2018.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gandhi S., Abramov A.Y. Mechanism of oxidative stress in neurodegeneration. Oxid Med Cell Longev. 2012;2012:428010. doi: 10.1155/2012/428010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peña-Bautista C., Vento M., Baquero M., Cháfer-Pericás C. Lipid peroxidation in neurodegeneration. Clin. Chim. Acta. 2019;497:178–188. doi: 10.1016/j.cca.2019.07.037. [DOI] [PubMed] [Google Scholar]
- 60.Sfera A., Bullock K., Price A., Inderias L., Osorio C. Ferrosenescence: the iron age of neurodegeneration? Mech. Ageing Dev. 2018;174:63–75. doi: 10.1016/j.mad.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 61.Tang H.M., Tang H.L. Cell recovery by reversal of ferroptosis. Biol Open. 2019;8 doi: 10.1242/bio.043182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen L., Hambright W.S., Na R., Ran Q. Ablation of the ferroptosis inhibitor glutathione peroxidase 4 in neurons results in rapid motor neuron degeneration and paralysis. J. Biol. Chem. 2015;290:28097–28106. doi: 10.1074/jbc.M115.680090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Do Van B., Gouel F., Jonneaux A., Timmerman K., Gelé P., Pétrault M., Bastide M., Laloux C., Moreau C., Bordet R. Ferroptosis, a newly characterized form of cell death in Parkinson's disease that is regulated by PKC. Neurobiol. Dis. 2016;94:169–178. doi: 10.1016/j.nbd.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 64.Hambright W.S., Fonseca R.S., Chen L., Na R., Ran Q. Ablation of ferroptosis regulator glutathione peroxidase 4 in forebrain neurons promotes cognitive impairment and neurodegeneration. Redox Biol. 2017;12:8–17. doi: 10.1016/j.redox.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Q., Han X., Lan X., Gao Y., Wan J., Durham F., Cheng T., Yang J., Wang Z., Jiang C. Inhibition of neuronal ferroptosis protects hemorrhagic brain. JCI Insight. 2017;2 doi: 10.1172/jci.insight.90777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Skouta R., Dixon S.J., Wang J., Dunn D.E., Orman M., Shimada K., Rosenberg P.A., Lo D.C., Weinberg J.M., Linkermann A. Ferrostatins inhibit oxidative lipid damage and cell death in diverse disease models. J. Am. Chem. Soc. 2014;136:4551–4556. doi: 10.1021/ja411006a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zille M., Karuppagounder S.S., Chen Y., Gough P.J., Bertin J., Finger J., Milner T.A., Jonas E.A., Ratan R.R. Neuronal death after hemorrhagic stroke in vitro and in vivo shares features of ferroptosis and necroptosis. Stroke. 2017;48:1033–1043. doi: 10.1161/strokeaha.116.015609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuhn H., Banthiya S., van Leyen K. Mammalian lipoxygenases and their biological relevance. Biochim. Biophys. Acta. 2015;1851:308–330. doi: 10.1016/j.bbalip.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kagan V.E., Mao G., Qu F., Angeli J.P., Doll S., Croix C.S., Dar H.H., Liu B., Tyurin V.A., Ritov V.B. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 2017;13:81–90. doi: 10.1038/nchembio.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seiler A., Schneider M., Förster H., Roth S., Wirth E.K., Culmsee C., Plesnila N., Kremmer E., Rådmark O., Wurst W. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. Cell Metabol. 2008;8:237–248. doi: 10.1016/j.cmet.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 71.Yang W.S., Stockwell B.R. Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 2016;26:165–176. doi: 10.1016/j.tcb.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]