Abstract
α-tocopherol transfer protein (TTP) was previously reported to self-aggregate into 24-meric spheres (α-TTPS) and to possess transcytotic potency across mono-layers of human umbilical vein endothelial cells (HUVECs). In this work, we describe the characterisation of a functional TTP variant with its vitamer selectivity shifted towards γ-tocopherol. The shift was obtained by introducing an alanine to leucine substitution into the substrate-binding pocket at position 156 through site directed mutagenesis. We report here the X-ray crystal structure of the γ-tocopherol specific particle (γ-TTPS) at 2.24 Å resolution. γ-TTPS features full functionality compared to its α-tocopherol specific parent including self-aggregation potency and transcytotic activity in trans-well experiments using primary HUVEC cells. The impact of the A156L mutation on TTP function is quantified in vitro by measuring the affinity towards γ-tocopherol through micro-differential scanning calorimetry and by determining its ligand-transfer activity. Finally, cell culture experiments using adherently grown HUVEC cells indicate that the protomers of γ-TTP, in contrast to α-TTP, do not counteract cytokine-mediated inflammation at a transcriptional level. Our results suggest that the A156L substitution in TTP is fully functional and has the potential to pave the way for further experiments towards the understanding of α-tocopherol homeostasis in humans.
Keywords: Nanoparticle, Vitamin E, Antioxidant, Transcytosis, Cytokine
1. Introduction
Vitamin E is a fat-soluble antioxidant that protects cell membranes from oxidative damage [1,2]. In total four tocopherol and four tocotrienol congeners are distinguished by their prefixes (α-to δ-) and recognized to feature vitamin E activity. All vitamin E iso-forms are exclusively produced by organisms performing oxygenic photosynthesis. Hence, virtually all vitamin E is initially provided through the consumption of plants and seeds [3]. The α-tocopherol isomer is the most abundant form of vitamin E in green leaves of many plants [4], whereas the γ-tocopherol isomer is mainly found in seeds [5]. Vegetable oils such as soybean oil (68%), corn oil (77%) or linseed oil (88%) contain higher amounts of γ-tocopherol than α-tocopherol.
In higher animals as well as in humans vitamin E is absorbed through the gut and transferred to the liver via lipo-proteins. Depending on the dietary habits of people the isomeric composition of the absorbed vitamin E may differ significantly. For example, it is estimated that in the US the average amount of consumed γ-tocopherol is ten-fold higher than that of α-tocopherol [6]. However, the amount of vitamin E isomers found in blood and tissues of humans remains consistent, regardless of the nutritional composition of vitamin E and thereby its bio-availability. α-tocopherol is the most abundant form of vitamin E [7] found in human tissues (80%). Studies on vitamin E metabolism indicate that in vivo levels of γ-tocopherol also depend on ω- and/or β-oxidation pathways [8].
Overall, the evolutionary determinants driving isomeric vitamin E selectivity towards α-tocopherol in mammals have not been fully understood yet. Initially, it was proposed that differences in antioxidant potency e.g. of γ-tocopherol [9] compared to α-tocopherol might be involved in driving selectivity. However, it is rather unlikely that the evolutionary preference for α-tocopherol is solely governed by the rather small differences in antioxidant activities in vitro [10]. Another rationale has emerged from the observation that the oxidation of a generic tocopherol may be associated with a relative increase of cytotoxic Michael adducts mostly in the case of isomers other than the α-form [11].
Nevertheless, current knowledge indicates that two independent processes predominantly govern vitamin E selectivity within the liver. Firstly, the metabolic processing of vitamin E [12] and second by the preferred sequestration of α-tocopherol over the other congeners by the α-tocopherol binding protein(α-TTP) [8,[13], [14], [15]], thus removing it from the metabolic degradation process.
Recent studies in our lab have provided X-ray structural and in silico mechanistic evidence that upon binding to α-tocopherol, α-TTP acquires the tendency to form spherical nanoparticles composed of 24 α-TTP units (α-TTPS) [16,17]. The findings indicate that α-TTPS may promote isomer selective delivery of α-tocopherol into barrier protected tissues in man. In addition, a functional A156L substitution has been introduced into α-TTP by site-directed mutagenesis possessing superior affinity towards γ-tocopherol over α-tocopherol (γ-TTP) [18].
In this study, we have pursued a convergent approach aiming at the structural elucidation of tetracosameric γ-TTPS nano-particles featuring customized γ-tocopherol specificity through the rational A156L modification within the binding pocket of the protomer. The comparison of γ-TTP bound to γ-tocopherol with native α-TTP bound to α-tocopherol provides quantitative biophysical evidence for the in vitro functionality of both ligand complexes. We also provide evidence for efficient transcytosis of both tetracosameric particle variants over an endothelial layer.
To complete our study, differential data of the corresponding protomeric isomer complexes on canonical inflammatory pathways in adherent cell culture is presented. It is proposed that the carrier effect of protomeric TTP attenuates the cell's inflammatory processes in dependency of the carried tocopherol itself, not the TTP construct.
2. Results
Past computational and experimental data identified the A156L mutant of α-TTP as a potential functional γ-TTP [18]. To validate this hypothesis, we first characterised structurally the γ-tocopherol-A156L mutant complex, demonstrating the occurrence of binding of γ-tocopherol in the native ligand binding site of the protein.
2.1. Crystal structure of tetracosameric γ-TTP complexed with γ-tocopherol
The A156L mutant was successfully expressed, purified and crystallised in complex with RRR-γ-tocopherol. Using crystallization conditions similar to those used for tetracosameric α-TTPS nano-spheres [16], we also found γ-TTP assembled in the same supramolecular organization. Specifically, the crystals of γ-TTP featured a I432 space group in a cubic unit cell of edge a = 168.65 Å. The X-ray diffraction pattern allowed the determination of an atomic model of γ-TTP complexed to RRR-γ-tocopherol refined to a 2.24 Å resolution (Rwork:Rfree 18.2%:21.8%, see Table A.5). The asymmetric unit consists of a single γ-TTP that includes residues 48 to 278, in complex with one γ-tocopherol molecule. The replacement at position 156 alanine with leucine was stable, see Fig. 1B. The atomic model of the tetracosameric assembly of γ-TTP was found in its oxidized state with a completely folded C-terminal helix and twelve disulfide bonds at position C80 connecting adjacent protomers over the two-fold symmetry axis, Fig. 1A.
Fig. 1.
General view of γ-TTP (α-TTP A156L) in complex with γ-tocopherol. (A) View of the tetracosameric nano-sphere of γ-TTPS. (B) Zoom of the binding pocket with all its residues visualized; in orange are the residues highlighted that form the niche around C8 of α-tocopherol in the wild type α-TTP. The surface of the binding pocket is colored depending its hydrophobicity. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.1.1. The γ-tocopherol binding site in γ-TTP
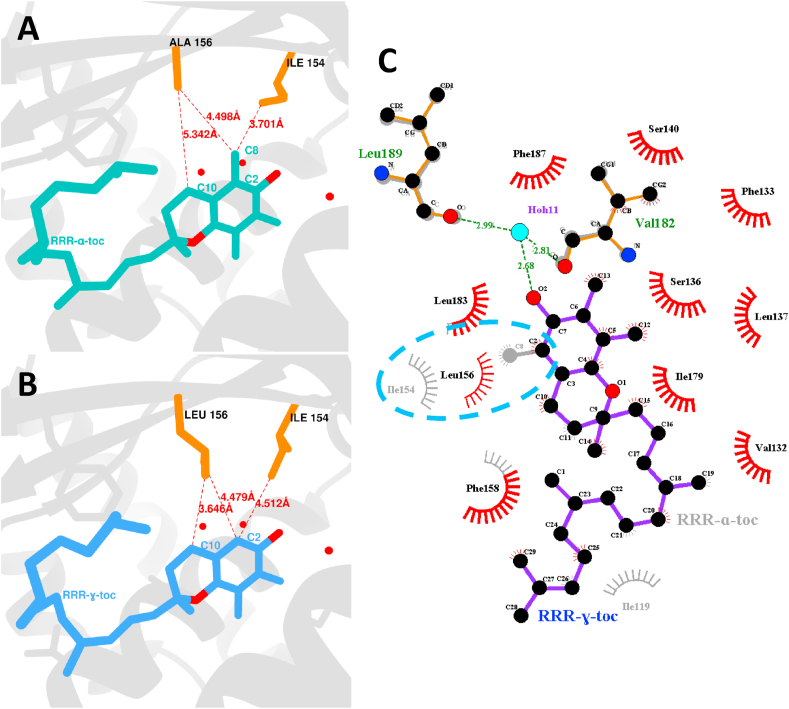
The binding pocket of γ-TTP is, with exception of position 156, virtually identical to that of α-TTP. Indeed, the same hydrophobic residues accommodate γ-tocopherol in the binding pocket. As previously reported [19] for the native α-TTP/α-tocopherol complex, several residues such as S136, S140 and three water molecules surround the chromanol moiety of γ-tocopherol. One water molecule connects the phenolic hydroxyl of the ligand to the backbone carbonyl of V182 and L189. The U-turn of the prenyl tail is similarly positioned within the binding pocket as for α-tocopherol bound to α-TTP, confirming prediction by computational modelling [18]. Fig. 2 shows a schematic comparison between the binding pocket of γ-TTP and native α-TTP, revealing minor differences between the two. Indeed, the substitution of alanine at position 156 by leucine fills the niche formerly formed by I194, V191, I154 and L183, so compensating for the void generated by the missing methyl group at C2 of chromanol moiety in γ-tocopherol. Fig. 2C evidences the new hydrophobic interaction between atom C2 of the ligand and L156 in substitution of the interaction between I154 and the methyl group of α-tocopherol. This demonstrates the compensatory effect of the newly introduced alanine 156 by re-establishing optimal distance between γ-tocopherol and the binding pocket surface. Calculations using the online tool CASTp [20] revealed a reduction of the binding pocket volume from an original value of 1034.6 Å3 in α-TTP to 993.5 Å3 in the γ-TTP mutant. Seemingly due to steric clashes, the reduction of the niche formed by residues I194, V191, I154 and L183 makes the binding of the natively preferred α-tocopherol to the γ-TTP mutant energetically unfavourable.
Fig. 2.
(A&B) Real space comparison of the binding pockets of wild type α-TTP complexed to RRR-α-tocopherol and of γ-TTP complexed to RRR-γ-tocopherol, respectively. (A) In the binding pocket of α-TTP in complex with α-tocopherol I154 is retracted, in order to create space for the niche surrounding the methyl group attached to C2 of α-tocopherol. (B) In contrast, in the binding pocket of γ-TTP bound to RRR-γ-tocopherol the void caused by the missing C8 is filled by L156 maintaining a similar distance (4.479 Å) to C2 as has A156 in the atomic model of wild type α-TTP (4.498 Å). (C) Schematic analysis (ligplot) of the binding pocket between α-TTP and γ-TTP; besides the newly introduced L156 very little discrepancies exist between both the atomic models.
2.2. Thermodynamics an kinetics of γ-tocopherol binding to γ-TTP
Quantification of the binding affinity and the exchange kinetic rates is critical to asses if the A156L mutant can effectively act as a γ-TTP in vitro and in the cell. The monomeric protomer of α-TTP shows high binding affinity to its ligand, and it has been associated with inter-membrane tocopherol transport activity, as widely reported in the literature [[21], [22], [23], [24], [25]]. On the contrary, in the tetracosameric assembly the mobile lid helix (residues 198–221), which is responsible for the ligand access to its binding site, is closed and sterically locked. This prevents TTPS from possessing vitamin E transfer activity. In fact, any physiological role of the tetracosameric assembly is likely associated to cardiovascular vitamin E transport after sequestration by monomeric units in liver cells [16]. For this reasons, the binding affinities and transfer rates between engineered γ-TTP and tocopherol were estimated using the monomeric protomers only.
2.2.1. Determination of the binding affinity
The binding affinity to γ-tocopherol is critical for the functionality of γ-TTP in a physiological context, in particular, its ability to solubilise γ-tocoherol from the liver into the systemic circulation of a model organism. For example, mild AVED mutant such as A120T, and H101Q, have similar transfer characteristics for yet markedly lower affinities to α-tocopherol [26].
The determination of the dissociation constant was done by an analytical methodology based on micro-differential scanning calorimetry (mDSC), which allows the individual estimate of the protein unfolding temperature and associated thermodynamic quantities: the melting temperature, , and the unfolding enthalpies, entropies at ( and the change in heat capacity by unfolding () for the apo and holo states (see SI for details 4.5). Related results are presented in Table 1.
Table 1.
Thermodynamic parameters from mDSC experiments for the respective samples.
| Sample |
|
|
|
|
|---|---|---|---|---|
| [K] | [kcal mol−1] | [kcal mol−1] | [kcal mol−1 K−1] | |
| APO α-TTP | 326.54 0.76 | 94.13 0.33 | 0.29 8.90 10−4 | 2.50 0.28 |
| APO γ-TTP | 327.97 0.06 | 86.56 3.56 | 0.26 0.01 | 2.36 0.31 |
| α-TTP + α-tocopherol | 341.07 0.06 | 150.75 1.90 | 0.44 0.01 | 2.16 0.25 |
| γ-TTP + γ-tocopherol | 336.53 0.12 | 149.97 5.21 | 0.45 0.02 | 2.06 0.20 |
| Tocopherols are all in RRR-stereoisomer configuration. All data are denoted as averages SEM. | ||||
The related unfolding free energies were determined using the Gibbs-Helmholtz equation (1)) using thermodynamic values from the mDSC experiment (Table 1).
| (1) |
The free energy of binding is simply obtained from a thermodynamic cycle Fig. 3, as the difference between the unfolding free energies of the apo- and holo-states:
| (2) |
Fig. 3.
Scheme of the thermodynamic cycle that emphasizes the relation between ligand binding and protein unfolding. (A) Classical thermodynamic cycle that describes a model in which the unfolding of the protein in complex does not influence the binding. Hence, in this model the ligand stays bound to the protein, despite of its structural integrity. (B) In this scheme unfolding of the protein causes the ligand to disassociate from the binding protein and the binding to the unfolded state is virtually inexistent (). (C) Schematic illustration of coupling between the unfolding equilibrium of the protein and the binding equilibrium to the ligand. Increments in thermal stability reflect the addition of binding free energy to the free energy of the protein fold ; the resulting free energy is the unfolding free energy of the protein-ligand complex.
and consequently, the dissociation constant :
| (3) |
The thermodynamic analysis in this study follows that previously reported [18]. In this instance, however, it is applied to compare an apo- and a holo-state of the associated protein. The energetic difference between apo-α-TTP and holo-α-TTP accounts for the absolute binding observed. In the previous study, mDSC was used to compare binding of and tocopherol ligands to the same protein, thus reporting the of binding for the two isoforms. The resulting exponential plot for as a function of the temperature is shown in Fig. 4. At physiological temperature (310.15 K) we calculated a dissociation constant of 14.25 nM for the native α-TTP + α-tocopherol complex, and of 53.79 nM for γ-TTP + γ-tocopherol as shown in Fig. 4B, corresponding to a difference of only 0.8 kcal mol−1 in binding free energy for the two complexes.
Fig. 4.
(A) Gibbs-Helmholtz plots of and as functions of temperature. It can be observed that the general stability of α-TTP (apoprotein and holoprotein) is quite similar to γ-TTP. However, of γ-TTP features a strikingly sharper decline than that of the wild type. The difference between both curves at any temperature corresponds to . The fact that of γ-TTP decreases much faster with increasing temperature reflects the lower overall affinity of γ-TTP for γ-tocopherol than α-TTP for α-tocopherol. (B) Arrhenius plots of the dissociation equilibrium () for α-TTP + α-tocopherol and γ-TTP + γ-tocopherol, respectively. Values for the at 24.85 ∘C (298 K) and at 36.85 ∘C (310 K) are highlighted within the plots.
2.2.2. Thermodynamic analysis of ligand binding
mDSC experiments allowed not only for the quantification of binding free energy, but also for a qualitatively analysis of thermodynamic features of the binding event, as illustrated in Fig. 5A. In both cases, the major contribution to the binding is enthalpic, as expected for hydrophobic substrates ( = −30.79 kcal mol−1 for α-TTP + α-tocopherol and = −51.33 kcal mol−1 for γ-TTP + γ-tocopherol), and for the large network of van der Waals contacts formed between the ligand and several amino-acid residues in the binding pocket, see Fig. 5B. The entropic contribution () is unfavourable at 310 K, (23.83 kcal mol−1 for α-TTP wt + α-tocopherol and 45.28 kcal mol−1 for γ-TTP + γ-tocopherol). This is also expected in molecular binding events where there is a neat loss in the translational entropy. Interestingly, even though the resulting binding free energies at 310 K are very similar, the two complexes show clear differences in the two enthalpic and entropic components. The reasons for these differences may be related to the different volume of the binding pocket, that on the one hand increases favorable binding interactions, but one the other hand likely restricts the motion of the flexible phytyl tail of the ligand. An increased value of for γ-TTP is an indication of a stronger temperature dependency of the dissociation constant. This may affect the behaviour at body temperature in a future in vivo experiment compared to in vitro data collected a room conditions.
Fig. 5.
(A) Thermodynamic signature of the binding of α-tocopherol to α-TTP and of γ-tocopherol to γ-TTP, respectively. In both case a strong favorable ethalpic contribution () exceeds the unfavourable entropic contribution (), resulting in both cases in a negative binding free energy ; and thus in the binding of the ligand with a dissociation constant () at 36.85 ∘C (310 K) of 14.25 nM for α-TTP/α-tocopherol and of 53.79 nM for γ-TTP/γ-tocopherol, respectively. (B) Illustration of the hydrogen bond network within the binding of α-TTP. The four trapped water molecules within the binding pocket together with residues Y100, Y117, S136, S140 V182, L189 and the ligand itself make up the large hydrogen bond network responsible for the strong ethalpic contribution () to the total binding ().
2.2.3. Characterisation of kinetic rates of γ-TTP
A functional γ-TTP must show an efficient uptake/release of the ligand between its pocket and the environment. To verify that the A156L mutation did not affect dramatically such properties, we measured in vitro transfer activity and compared those to native α-TTP. Specifically, release rates were determined by monitoring the transfer of tocopherol from holo-TTP to acceptor liposomes by fluorescence resonance energy transfer (FRET), see Fig. A.8 Transfer rates in both directions were determined by a FRET assay as described in methods 4.7. Additionally, we measured release rates via an independent colorimetric assay that works without SUVs and instead provides detergent micelles (sodium-cholate) as an acceptor for tocopherols (details in supplementary information, see p. 27).
Table A.6 summarizes all the measured rates and Fig. A.9 illustrates each completed experiment. Figure A.10 illustrates and Table A.7 summarizes all results of the ABTS•+ decolorization assay.
In the colorimetric assay the observed release rate () of α-tocopherol by native α-TTP is 1.15 10−3 1.27 10−5 s−1 and of γ-tocopherol 3.36 10−3 5.99 10−5 s−1. The fact that the release rate of γ-tocpherol is approximately three-fold higher reflects the lower affinity of wild type α-TTP towards γ-trocopherol in relation to α-tocopherol. In the γ-TTP mutant, the release rate of γ-tocopherol approximates that of wild type.
With the rate constants ( and ) measured by FRET as described thoroughly in supplementary information (see p. A.1), it is possible to estimate the partition coefficient () of the ligand between the protein and the liposome membrane. quantifies the relative preference of the ligand between two specific loci, in this case it the binding pocket of TTP and its incorporation within a lipid bi-layer Table 2. The more the of γ-TTP/γ-tocopherol approximates the partition coefficient of native α-TTP/α-tocopherol the better. Importantly, γ-tocopherols partitions into γ-TTP with very similar ratio compared to that of α-tocopherol into α-TTP. This suggests that γ-TTP can have similar efficiency in the extraction of γ-tocopherol from the endosomal compartments than its native counterpart.
Table 2.
Partitions coefficients of tocopherols for wild type α-TTP and γ-TTP.
| Protein | Ligand | ||
|---|---|---|---|
| α-TTP | α-tocopherol | 2.72 | 0.0 |
| α-TTP | γ-tocopherol | 4.05 | +1.33 |
| γ-TTP | α-tocopherol | 3.08 | +0.37 |
| γ-TTP | γ-tocopherol | 2.54 | −0.18 |
2.3. Activity of γ-TTP in cell cultures
Having demonstrated that γ-TTP maintains very similar binding and exchange properties for γ-tocopherol as native α-TTP for α-tocopherol, we proceeded to investigate the properties of γ-TTP in cellular cultures in vitro.
2.3.1. Transcytosis of γ-TTP through a HUVEC monolayer
The native form of tocopherol transfer protein loaded with α-tocopherol (α-TTP) was readily transported through a tight endothelial monolayer in our HUVEC culture model as previously described [16]. The modified TTP carrying γ-tocopherol displayed similar properties in vitro as it was also transported through a HUVEC monolayer by a transcytotic mechanism (Fig. 6). All monomeric, oligomeric and tetracosahedral forms of γ-TTP were transported to a considerable extent (Fig. 6). For α-TTPs, we observed a 10.3-fold (monomeric α-TTP), 10.4-fold (intermediate α-TTP) and 18.0-fold (α-TTPS) increase in flux through the endothelial cell layer compared to that of RITC-dextran, which is a marker for unregulated paracellular flux. γ-TTPs showed a 6.1-fold (monomeric γ-TTP), 16.4-fold (intermediate γ-TTP) and 18.6-fold (γ-TTPS) increase in flux compared to that of RITC-dextran. The faster uptake for the largest TTPS constructs suggests the presence of a recognition-mediated mechanism for cell incorporation and transfer, instead of a diffusion-dominated event.
Fig. 6.
The transcytotic efficacy of tetracosameric α-TTPs and γ-TTPs (α-TTP A156L) compared to its monomer form were monitored in a transwell model system comprising confluent and maturely developed monolayers of HUVECs. Human transferrin served as the positive control. Simultaneous application of rhodamine isothiocyanate - (RITC) labeled dextran confirmed the integrity of the HUVEC cell monolayers and served to determine the paracellular flux. Measurements report a 10-fold–18-fold increase in the flux through the endothelial cell layer of both α- and γ-TTPs compared to that of RITC-dextran. Our data also show that α- and γ-TTPs cross the endothelium at a flux rate 2–6 times faster than human transferrin with increasing speed the more complex the protein assemblies.
2.3.2. Monomeric α-TTP attenuates cytokine-induced transcription of pro-inflammatory cytokines and proteins
In a second set of experiments, HUVEC cells were first grown against 1 ml TTP sample in PBS (control only PBS) and 3 ml HUVEC growth media, and then incubated with recombinant human TNF (rhTNF), rhIL1F2 and rhIFNG.
This resulted in a substantial induction of the proinflammatory cytokines IL6, CCL2, and TNF, the inflammation-associated enzyme COX2 (Fig. 7 A-D, left boxes) and the adhesion molecule ICAM1 (Fig. 7 E, left boxes) as mRNA steady state levels increased by 49-427-fold compared to untreated controls. The same test was conducted by first soaking the HUVEC cells in a 12.5 μM solution of substrate-loaded TTP proteins. In particular, we repeated independent tests using solutions of α- or γ-TTP, loaded with α-and γ-tocopherol, respectively, in either their monomeric, oligomeric, or TTPS protomeric states. Growing primary HUVECs as adherent mono-layer prevents transcytosis and thus forces a fraction of TTP proteins into the cytoplasmic environment. Strikingly, pre-treatment of HUVECs with monomeric α-TTP significantly decreased cytokine-induced mRNA levels of IL6, CCL2, TNF and ICAM1 (Fig. 7 A-C, E, central boxes). Contrary to α-TTP, monomeric γ-TTP did not counteract cytokine-mediated inflammation in HUVECs. Transcription of IL6, CCL2, TNF, COX2 and ICAM1 was not diminished by γ-TTP pre-treatment prior to pro-inflammatory cytokine application (Fig. 7 A-E, right boxes). Thus, cytokine-mediated increases in mRNA levels of selected target genes remained significantly elevated compared to the respective control following γ-TTP pre-treatment and was only reduced by α-TTP pre-treatments as depicted in Fig. 7. Initial data suggest putative pro-inflammatory activity of oligomeric TTPs per se, as oligomeric α-TTP and γ-TTP each induced considerable cytokine expression in HUVECs compared to their respective monomeric proteins. Interestingly, cells treated with α-TTPS demonstrated elevated mRNA levels of pro-inflammatory cytokines and adhesion proteins similar to those of untreated cells, indicating that TTPS constructs are incompetent for ligand release in HUVEC.
Fig. 7.
Primary human umbilical vein endothelial cells (HUVECs) were allowed to form a tight monolayer within 7 days of pre-culturing. At day 7 tocopherol transfer proteins (TTPs) carrying either α-tocopherol (α-TTP) or γ-tocopherol (α-TTP A156L, referred as γ-TTP) were added for 4 h followed by cytokine (TNF: 50 ng/ml, IL1F2: 1 ng/ml, IFNG: 50 ng/ml) stimulation for another 4 h mRNA levels of IL6 (A), CCL2 (B), TNF(C), COX2 (D) and ICAM1 (E) were significantly increased by cytokine stimulation (controls, left boxes) while α-TTP significantly reduced cytokine-induced mRNA expression of both IL6, CCL2, TNF and ICAM1 (central boxes). γ-TTP was not able to attenuate cytokine-mediated transcription of any pro-inflammatory cytokine or protein measured (right boxes). Boxplots display outliers, if applicable. N = 1 in triplicate. Statistical evaluation with R with assumed significance at p < 0.05. Relevant significant differences are designated with asterisks as follows: *p < 0.05, **p < 0.01, ***p < 0.001.
3. Discussion and conclusions
HUVECs originate from human umbilical veins, representing large vessel endothelium that is capable of both responding to and inducing an inflammatory environment. In this context, HUVECs have been shown to express various toll-like receptors and to secrete cytokines, chemokines and adhesion molecules including ICAM1 [27]. Moreover, nicotinamide adenine dinucleotide phosphate oxidase (Nox), which is a key player in cardiac inflammation and heart failure, is a major producer of reactive oxygen species (ROS) in vascular cells, so in HUVECs. In this setting, Nox can be readily induced by inflammatory stimuli. Given that the Nox-isoforms Nox2 and Nox4 are both expressed in endothelial cells [28], application of inflammatory cytokines is assumed to promote the formation of cellular ROS. Additionally, TNF induces a pro-inflammatory environment in human endothelial cells via COX and NOS and disrupts redox homoeostasis [29] accounting for lipid peroxidation and DNA hydroxylation [30]. Applying TNF, IL1F2 and IFNG to HUVECs induces inflammation and the release of reactive species (e.g., superoxide, hydrogen peroxide, hydroxyl radical) and chemical mediators (e.g., cytokines, chemokines, eicosanoids, complement components) thereby inducing tissue damage and redox stress [30]. Incubation of mature HUVEC monolayers with rhTNF, rhIL1F2 and rhIFNG resulted in marked induction of the transcription of the pro-inflammatory cytokines IL6, CCL2, and TNF, the enzyme COX2, which stokes inflammatory processes, and the adhesion molecule ICAM1 (Fig. 7) [27].
As reported, the in vitro antioxidant activities of the major lipophilic tocopherol isomers (α, β, γ, and δ) may depend very much on the test system used [[10], [31]]. Furthermore, tocotrienols may represent better antioxidants in membrane systems, while tocopherols are better in solution [32]. Interestingly, α-tocopherol preferentially accumulates in human tissues and serum irrespective of the genetic background with simultaneous discrimination of γ-tocopherol [[10], [33]].
As we report (Fig. 6), both monomeric and oligomeric α-TTP and γ-TTP are readily taken up by HUVECs, transported to the basolateral side and finally secreted (transcytosis). Prior to this study, this had been solely shown for α-TTP [16]. Thus, we conclude that γ-TPP not only exhibits equivalent biophysical but also physiological properties to α-TTP within in vitro cultured cells. As tocopherols are readily released intracellularly by TTP constructs, they potentially play a key role in the prevention of cellular inflammatory processes as mediated by cytokines and ROS. As described in the literature, α-tocopherol is able to reduce redox stress in vitro. Therefore, α-tocopherol, intracellularly released from α-TTP, may attenuate cytokine-mediated oxidative stress and inflammation (Fig. 7). Although γ-TTP was readily taken up by HUVECs, γ-TTP had no such effect. Therefore, inflammation was counteracted by α-TTP while γ-TTP failed to rescue the endothelium from pro-inflammatory stimuli. The transcription factor nuclear factor “kappa-light-chain-enhancer” of activated B-cells (NF-κB) orchestrates the transcription of numerous cytokines, amongst others [34]. Importantly, not only ligand-dependent induction of signal transduction (e.g. TNF) but also intracellular redox homoeostasis are centrally involved in the activation of NF-κB [34]. Particularly, hydrogen peroxide has been shown to activate NF-κB and antioxidants may block NF-κB activation [30,35,36]. Thus, α-tocopherol may have favourably impacted cellular redox status, thereby inhibiting NF-κB-dependent pro-inflammatory cytokine production.
Structural and thermodynamical characterisation of γ-TTP presented in this works were meant to verify that γ-TTP shows similar chemical, biophysical and physiological properties in vitro, e.g., vitamin release and transcytosis, than α-TTP. Thus, the beneficial effect of α-TPP with respect to attenuation of inflammatory processes seems to primarily depend on the carried tocopherol itself, not the TTP construct. This conclusion is supported by recent findings of the research group of Cook-Mills, showing a protective association for α-tocopherol but not with γ-tocopherol in asthma inception in a human cohort study [37].
In this work we have sought to provide the most comprehensive characterisation of the γ-tocopherol-specific γ-TTP mutant. This includes the solution of its crystal structure, the analysis of its transfer activity through partition coefficients (), as well as the determination of its affinity through complementary dissociation constants. These measurements collectively represent a robust quantitative in vitro characterisation of γ-TTP's functionality. The release vs. sequestration ratio of both wild type α-TTP and γ-TTP for each respective tocopherol is of highest importance for rigorous analysis of transfer activity, see Table A.6. We report that the magnitude of the related ratio for both TTP variants broadly remains in the same range. represents a powerful tool for the analysis of transfer activity, allowing for the estimation of overall transfer protein functionality. is the quotient between the respective transfer rates as described by equation (2). Using equation (2) we were able to normalize to the sequestration rate. Hence, values capture the relative increase in release or simply the normalized general release velocity. In this study the observed decrease from 2.72 to 4.05 in the when comparing wild type α-TTP - α-tocopherol with wild type α-TTP - γ-tocopherol indicates a significant loss of functionality. In counterpoint, the determined value of 2.54 for γ-TTP - γ-tocopherol indicates full functionality.
The crystal structure of γ-TTP complexed with γ-tocopherol revealed that the substitution of leucine by alanine at position 156 fomented compensatory effects that favor the binding of γ-tocopherol. Accordingly, a low discrepancy of only 0.019 Å for the van der Waals (vdW) contacts between the carbon of A156 to C8 of α-tocopherol (4.498 Å) and the δ1-carbon of L156 to C2 of γ-tocopherol (4.479 Å) was observed. Moreover, in the wild type structure I154 creates a vdW contact (3.701 Å) between its δ1-carbon and C8 of α-tocopherol, whereas in the mutant the binding occurs to position C2 of γ-tocopherol (4.515 Å). The related extension of 0.811 Å of the vdW contact proved disadvantageous and may well contribute to the lower affinity of the mutant type bound to γ-tocopherol compared to the wild type bound to α-tocopherol. The extension may partially be compensated by a shortening of 1.696 Å of the vdW contact between the δ1-carbon of L156 and C10 of γ-tocopherol. Other amino-acid residues and the phytyl chain of the ligand were barely shifted as illustrated in Fig. 2C. We conclude that the niche composed by I194, V191, I154, and L183 around C8 of α-tocopherol in the binding pocket of native α-TTP forms a network of hydrophobic interactions each of which is near its optimal van der Waals distances.
Moreover, in the atomic model of γ-TTP + γ-tocopherol we observe that the mutation A156L partially reestablishes this optimal distance network. However, we noted that the average distance to γ-tocopherol was prolonged and therefore off the optimal distance. Therefore, the A156L γ-TTP mutation tolerates the binding of γ-tocopherol but may not necessarily increase the dissociation constant () of wild type TTP for this ligand. In previous experimental work, we calculated in silico and derived the relative binding free energy () between RRR-γ-tocopherol and RRR-α-tocopherol bound to α-TTP A156L. There we observed that γ-TTP binds preferentially RRR-γ-tocopherol by a calculated of −1.192.28 kcal mol−1, and an experimental of −3.423.04 kcal mol−1 [18]. Here we have reapplied that approach leading to the determination of the absolute binding free energy of γ-TTP to γ-tocopherol. mDSC was applied because, in contrast to punctual isothermal calorimetry, it offers relatively precise monitoring of the dissociation constant over a range of temperatures. Both methods do however allow for the determination of the contributing enthalpic and entropic terms to the binding free energy . We determined a for α-tocopherol bound to wild type α-TTP of 14.25 nM at 36.85∘C, which is comparable to previously reported values of 25 nM [38] and 9.8 nM [24], respectively. The discrepancies arise from the fact that it may be difficult to precisely determine the working temperature in classical assays, for example we observed a of 25 nm at approximately 40.35∘C (313.5 k) and of 9.8 nM at 34.35∘C (307.5 K). For γ-TTP and γ-tocopherol we determined a of 53.79 nM at 36.85∘C (310 K), which is 3.8 times lower than the reference value for α-TTP and α-tocopherol. However, this value still remains within the acknowledged functional range. We conclude that the observed preference for γ-tocopherol and the impressive small of 0.18 between γ-TTP/γ-tocopherol and α-TTP/α-tocopherol both indicate that γ-TTP may act as a functional substitute for wild type α-TTP. Since both α-TTP and γ-TTP exhibit equivalent biophysical but also physiological properties within in vitro cultured cells, the beneficial effect of α-TPP for physiology seems to primarily depend on the carried tocopherol itself, not the TTP construct. This conclusion is fully supported by recent findings of the research group of Cook-Mills, showing a protective association for α-tocopherol but not with γ-tocopherol in asthma inception in a human cohort study [37].
Building on these findings, a natural future step will be testing our conceptual framework in an in vivo context by expanding current research towards the use of a γ-TTP knock-in mouse model that may exhibit modifications in phenotype and thereby provide clues about how the homeostasis of tocopherols is associated with their physiologic functions.
4. Material and methods
4.1. Expression and purification
TTP Proteins were expressed in Escherichia coli strain BL21(DE3) and purified essentially as previously described [16,18].
4.2. Preparation of TTP ligand-complexes
Protein-ligand complex formation was induced by dialysing freshly prepared apo-TTP in the presence of detergent solubilized tocopherol. In brief, a droplet of 1 mg of α- or γ-tocopherol was overlaid with 40.9 mg of solid sodium cholate and subsequently suspended in 1 ml of elution buffer (20 mM Tris, 100 mM NaCl, 150 mM imidazole, pH 8.0). The suspension was bath sonicated until all material had dissolved to a clear solution. Apo-α-TTP (11 ml at 2.5 mg/ml) was complemented with the tocopherol-sodium cholate solution at 9:1 (v/v) ratio and transferred into a CelluSep T3 dialysis tubular membrane with an MWCO range of 12–14 kDa (Membranes Filtration Products. TX, USA). Dialysis was performed in two steps against 3 l buffer (20 mM Tris, 100 mM NaCl, pH 8.0) each for 6 h at 4∘C. The dialysate (12 ml) was filtered through a Millex GP 0.22 μm filter (EMD Milipore, MA, USA), supplemented with Triton X-100 at a final concentration of 0.01% (v/v), reduced to 2 ml and separated by preparative gel filtration chromatography (GFC). Fractions corresponding to the size of the monomeric TTP ligand-complex were pooled and concentrated to 20 mg/ml using Vivaspin concentrators (MWCO 10 kDa; Satorius, Göttingen, Germany) and directly used for crystallization. Fractions corresponding to the size of α- or γ-TTPS nano-spheres were pooled and concentrated using Vivaspin concentrators (MWCO 30 kDa) to 10 mg/ml re-purified by analytical GFC.
4.3. Crystallization and structure determination of γ-TTP
Crystals were grown by either hanging or sitting-drop vapor diffusion using reservoir solutions ranging from 10 to 15% PEG-4000, 100–175 mM ammonium sulfate in 100 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes) sodium pH 7.5 at 18∘C. Freshly prepared monomeric γ-TTP ligand-complex was used in a concentration range between 12 and 22 mg/ml. Highest quality crystals of fully reduced γ-TTPS were observed within two weeks at drop ratios of protein over reservoir ranging between 3/1 and 2/1 (v/v). Crystals had cubic shape with edge length ranging between 20 and 80 μm. Isomorphous crystals of fully oxidized γ-TTPS were collected after two months. All crystals were flash frozen in nitrogen after adding glycerol in two steps to a final concentration of 20% (v/v). Diffraction data were collected at the Swiss Light Source (SLS) synchrotron beamline X06DA (PSI Villigen) at 100 K, employing a Dectris Pilatus 2 M CCD detector (DECTRIS Ltd., Baden, Switzerland). All data were indexed, integrated and scaled with XDS [39]. Phaser-MR was used for calculating the initial phases with the truncated structure model (residues 47–275) of monomeric α-TTP (PDB ID: 1OIP) as search structure. The atomic model γ-TTPS was refined by iterative cycles of manual model building using COOT [40] and restrained refinements using the Phenix program suite [41]. Coordinates and structure factors of the structure has been deposited in the RCSB Protein Data Bank with ID code 6ZPD.
4.4. Size exclusion chromatography
Preparative and analytical GFC of monomeric TTP and tetracosameric TTPS was performed on HiLoad 16/60 Supersose 75 prep grade and on Superose 6 10/300 columns respectively (GE Healthcare, Little Chalfont, UK), both attached to an ÄKTA Purifier chromatography system (GE Healthcare, Little Chalfont, UK). Runs were performed in SEC buffer (10 mM Tris, 100 mM NaCl, pH 8.0) at flow rates ranging from 0.5 (analytical) to 1.5 ml/min (preparative) at 6 ∘C. Both SEC columns were calibrated using commercially available protein calibration kits (GE Healthcare, Little Chalfont, UK).
4.5. Micro-differential scanning calorimetry
Micro-differential scanning calorimetry (mDSC) was performed on a TA Instruments Nano DSC calorimeter running Nano DSCRun Software v4.2.4 (TA Instruments, DE, USA). Each DSC experiment required 300 μl of protein sample as well as the same volume of reference buffer (20 mM TRIS-HCL, 40 mM NaCl, pH 7.4). Protein samples were measured in duplicate and the concentration of protein samples was determined by amino acid analysis in order to gather exact knowledge of protein quantities applied to the DSC experiments. Protein concentration ranged from 0.3846 to 0.5713 mg ml−1. Each sample was centrifuged at 13,200 rpm for 10 min at 4 ∘C and degassed for 5 min prior to DSC experiments. Reference buffer was used to generate the baselines that were subtracted from the sample to generate the DSC thermograms, which were analysed. Data were collected under 3 bar pressure from 20 to 80 ∘C using a 2 K min−1 temperature increment with a 10-min equilibration period prior to the scans. Raw data processing and visualization was performed with NanoAnalyze Software v3.1.2 (TA Instruments, DE, USA). The calorimeric enthalpy of unfolding () was calculated by integrating the Cp versus T curve using a sigmoidal baseline with 1st order fit for the initial as well as for the final baseline. The difference in the heat capacity of unfolding was determined by quantifying the difference of heat capacity between the pre-translational inflexion point before the endothermic peak and the post-translational inflexion point after the endothermic peak.
The contribution to the enthalpy and entropy at the temperature is described by equations (4), (5)).
| (4) |
| (5) |
4.6. Preparation of SUVs
Acceptor small unilamellar liposome vesicles (SUVs) were prepared by mixing stocks of phosphatidylcholine (Sigma-Aldrich, MO, USA), dansyl-DHPE (Avanti Polar Lipids, AL, USA), BHT (Sigma-Aldrich, MO, USA) and tocopherol (DSM, Kaiseraugst, Switzerland) in chloroform as described in Table 3. Donor SUVs that were used to determine the return transfer (sequestration of tocopherol by apo-α-TTP), additionally contained a 25% mole fraction of tocopherol which resulted in a strong FRET signal as shown in Fig. A.8 B. The mixtures were dried under a stream of nitrogen, then 2 ml of HBS buffer (100 mM NaCl, 20 mM HEPES/NaOH pH 7.5 and 0.02% sodium azide) were added and the mixture was incubated for 15 min at room temperature. The mixture was subsequently vortexed vigorously in order to resuspend all the phospholipids (PLs), the result was a cloudy uniform suspension. The PLs suspension was then sonicated in a bath sonicator twice for 5 min and afterwards ten freeze-thaw cycles were performed until the suspension got almost transparent, however, a slight opalescence was still noticeable.
Table 3.
SUV composition overview.
| Lipids | Ratios | Type |
|---|---|---|
| (60) PC/dansyl-DHPE/BHT | 98.5/1/0.5 | Acceptor SUV |
| (60) PC/dansyl-DHPE/BHT/TOCs | 78.5/1/0.5/20 | Donor SUV |
4.7. Transfer rate measurements
The transfer rates of vitamin E were determined by monitoring the time dependent change in the FRET signal of a sample upon mixing holo-TTP with acceptor-SUVs and apo-TTP with donor-SUVs, respectively. In order to reliably monitor the kinetics an injector was used to mix the solutions of both reactants and the time dependent FRET signal was measured at every second with an Infinite M1000 Pro fluorescence plate reader (TECAN, Männedorf, Switzerland). The excitation was set at 292 nm (10 nm slit) and the emission at 525 nm (20 nm) slit, the gain was kept manually in every measurement at 100. Kinetics measurements were performed always at 37 ∘C (310.15 K) so that physiological conditions were simulated. The fluorescence signal was normalized to the starting fluorescent intensity of tocopherol bound to TTP or of tocopherol bound to SUVs as 100%, respectively. It results that the rate law of the transfer for both directions is of second-order, which explains the reason for dimensionless of the partition coefficient (). However, in this case we obtained pseudo first-order transfer rates for both reactions by fitting the fluorescence data to the sum of a single exponential process and a linear term (representing chromophore bleaching). Equation (6) was used to quantify the sequestration rate () of tocopherol from SUVs to apo-α-TTP, and equation (7) was used to quantify the release rate () of tocopherol from holo-α-TTP to SUVs.
Single exponential equation for and linear term for bleaching
| (6) |
Single exponential equation for and linear term for bleaching
| (7) |
4.7.1. Quantifying the release of tocopherol with sodium cholate micelles
ABTS was dissolved in water (7 mM) and incubated overnight in presence of 2.45 mM potassium persulfate (final concentration), in order to form ABTS•+ radicals. Vitamin E bound to TTP (end concentration 6.4 μM) is not accessible to the ABTS•+ radical (end concentration 8 μM) and hence no decolorization was observed without the presence of negatively charged acceptor micelles. Decolorization was observed at 734 nm with an Evolution Array UV/VIS (Thermo Fisher Scientific, MA, USA) upon addition of sodium cholate micelles to an end concentration of 17 mM. Since the quenching reaction of the radical is immediate one can deduce that the decolorization kinetics refers to the release kinetics of vitamin E from holo-α-TTP. Because the concentration of sodium cholate is in great excess (2656.25-fold) compared to holo-α-TTP one can use a pseudo-first-order approximation for the calculation of release rates. Thus, equation (8) shows the reaction mechanism for the release of tocopherol in this assay and equation (9) the resulting rate equation.
| (8) |
The rate equation (9) is transformed as illustrated above from a second order reaction to pseudo-first-order reaction. Due to the simplification of the reaction equation (8), it is easy to integrate and linearize the rate equation for as shown by equation (10).
| (9) |
| (10) |
4.8. Cell culture
Primary human umbilical vein endothelial cells (HUVECs) were isolated from fresh umbilical cords with the help of trypsin/EDTA as described previously [16]. HUVECs were maintained in Endothelial Cell Growth Medium (Promocell, Heidelberg, Germany) comprising 100 U/ml penicillin and 100 μg/ml streptomycin (PAN Biotech, Aidenbach, Germany) with gelatin pre-coating (Sigma-Aldrich, Darmstadt, Germany). For subculturing, cells were detached with Trypsin/EDTA (0.04%/0.03%) using the DetachKit for primary human cells from Promocell. To comparatively determine transcytosis of the tocopherol transfer proteins (TTPs) that carry α-tocopherol (α-TTP) and γ-tocopherol (α-TTP A156L, referred as γ-TTP), HUVECs were cultured in a transwell system as described before [16]. In brief, cells were allowed to form a tight monolayer within 7 days of culture while medium was changed every other day. For transcytosis measurements medium comprising RITC labeled dextran (70 kDa; marker for paracellular flux) and either 0.4 mg/ml of FITC labeled monomeric, oligomeric or spherical α-TTP or γ-TTP, respectively, was applied to the apical chamber. Transferrin served as the positive control. Transport was monitored by sampling 100 μl of basolateral medium at consecutive time points (15, 30, 45, 60, 120, 180, and 240 min) after addition of TTPs to the apical chamber. Basolateral aliquots were subsequently analysed for fluorescence with a Tecan infinite200 microplate reader (Tecan Deutschland GmbH, Crailsheim, Germany) at an excitation wavelength of 485 nm and an emission wavelength of 535 nm (FITC) followed by measurements at an excitation wavelength of 535 nm and an emission wavelength of 590 nm (RITC). For the determination of mRNA levels of selected pro-inflammatory cytokines and proteins, cells were seeded in gelatin pre-coated standard 6-well multiwell plates (Sarstedt, Nuembrecht, Germany). Cells were allowed to form a tight monolayer within 7 days of culture while medium was changed every other day. Then monomeric TTPs loaded with either α-tocopherol (α-TTP) or γ-tocopherol (γ-TTP; 0.4 mg/ml, each) were applied for 4 h, respectively. Ca and Mg free phosphate buffered saline (PBS; PAN Biotech) was used for controls. Then cell were treated with a cytokine mix consisting of human recombinant (rh) tumor necrosis factor alpha (rhTNF, 50 ng/ml), interleukin-1 beta (rhIL1F2, 1 ng/ml) and interferon-gamma (rhIFNG, 50 ng/ml; all ImmunoTools GmbH, Friesoythe, Germany) for another 4 h. Cells were washed with PBS, covered with TriFast reagent (peqlab, Erlangen, Germany) and stored at 80 ∘C until RNA isolation.
4.8.1. RT-qPCR analysis
Total RNA was extracted with TriFast reagent according to the manufacturer's instructions. RNA concentration was determined via NanoDrop measurements (NanoDrop2000c; ThermoScientific, Waltham, MA/USA). RT-qPCR was performed using a SensiFast SYBR No-ROX One-Step Kit (Bioline, London, UK) on a Rotor-Gene 6000 real-time PCR cycler (Corbett/Qiagen). DNA primer were purchased from Eurofins Genomics (Ebersberg, Germany). Relative mRNA concentrations were calculated using the respective standard curves. Target gene expression (Table 4) was normalized to the expression of the housekeeping genes glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and hypoxanthine-phospho-ribosyl-transferase 1 (HPRT1).
Table 4.
Primer sequences used for mRNA quantification in HUVECs following treatment with the α-TTP and γ-TTP monomers in presence or absence of a cytokine mix.
| Target primer | Forward primer | Reverse primer | Annealing Temp. (∘C) |
|---|---|---|---|
| CCL2 | CAGCCAGATGCAATCAATGC | GCACTGAGATCTTCCTATTGGTGAA | 59 |
| COX2 | CATGGGGTGGACTTAAATCAT | TCTTTGACTGTGGGAGGATACA | 57 |
| GAPDH | CAATGACCCCTTCATTGACC | GATCTCGCTCCTGGAAGATG | 58 |
| HPRT1 | TGGCGTCGTGATTAGTGATG | CTCGAGCAAGACGTTCAGTC | 60 |
| ICAM1 | CTGGCGGTTATAGAGGTACG | GTGACCGTGAATGTGCTCTC | 59 |
| IL6 | TGACAAACAAATTCGGTACATCCT | TCTGCCAGTGCCTCTTTGCT | 58 |
| TNF | GGTTTGCTACAACATGGGCTACA | CCCCAGGGACCTCTCTCTA | 59 |
CCL2: C-C motif chemokine ligand 2; COX2: PTGS2 prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase); GAPDH: Glyceraldehyde-3-phosphatebdehydrogenase; HPRT1: Hypoxanthine-phosphoribosyl-transferase 1; ICAM1: intercellular adhesion molecule 1; IL6: Interleukin 6; TNF: TNF tumor necrosis factor (TNF superfamily, member 2); TTP: tocopherol transfer protein.
4.8.2. Statistical analysis
Statistical analyses on cell culture data were performed using the statistical software R version 3.6.0 [42]. The data evaluation started with the definition of an appropriate statistical model based on generalized least squares taking heteroscedasticity in consideration [43]. The model included comparisons concerning presence or absence of cytokines and treatment with the respective type of TTP versus control as well as their interaction term as fixed factors. The donor of HUVECs was regarded as random factor. The residuals were assumed to be normally distributed and to be heteroscedastic with respect to the different levels of cytokines and type of proteins. These assumptions are based on a graphical residual analysis. Based on this model, a Pseudo R2 was calculated [44] and an analysis of variances (ANOVA) was conducted, followed by multiple contrast tests [45,46] in order to compare the several levels of the influence factors as described above, respectively.
Declaration of competing interest
The authors declare no competing financial interests or personal relationships that could have influenced the work reported in this paper.
Acknowledgements
This work was supported by the Swiss National Science Foundation through Grant n. 31003A_ 130497 and 31003A_ 156419 (A.S.), and by the Research Council of Norway through the CoE Hylleraas Centre for Quantum Molecular Sciences (Grant n. 262695) (M.C.). We are grateful to Petra Schneider at the Swiss Federal Institute of Technology in Zurich (ETHZ) for her support in mDSC data collection. We would also like to thank Dr. Thomas Netscher (DSM, Basel) for the generous gift of the RRR-γ-tocopherol. Special thanks to John Culity, MD (NanoRetinal, Inc.) for proofreading this manuscript and bringing it to linguistic perfection.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101773.
Author contributions statement
Walter Aeschimann and Achim Stocker conceived the experiments, Walter Aeschimann and Stephan Kammer conducted the experiments, Achim Stocker, Michele Cascella, Walter Aeschimann, and Petra Schneider analysed the results. Stefanie Staats and Gerald Rimbach designed and conducted the cell culture experiments and analysed PCR data. All authors reviewed the manuscript.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Ingold K., Webb A., Witter D., Burton G., Metcalfe T., Muller D. Vitamin e remains the major lipid-soluble, chain-breaking antioxidant in human plasma even in individuals suffering severe vitamin e deficiency. Arch. Biochem. Biophys. 1987;259(1):224–225. doi: 10.1016/0003-9861(87)90489-9. [DOI] [PubMed] [Google Scholar]
- 2.Howard A.C., McNeil A.K., McNeil P.L. Promotion of plasma membrane repair by vitamin e. Nat. Commun. 2011;2:597. doi: 10.1038/ncomms1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Traber M.G. Mechanisms for the prevention of vitamin e excess. J. Lipid Res. 2013;54(9):2295–2306. doi: 10.1194/jlr.R032946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sattler S.E., Cahoon E.B., Coughlan S.J., DellaPenna D. Characterization of tocopherol cyclases from higher plants and cyanobacteria. evolutionary implications for tocopherol synthesis and function. Plant Physiol. 2003;132(4):2184–2195. doi: 10.1104/pp.103.024257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grusak M.A., DellaPenna D. Improving the nutrient composition of plants to enhance human nutrition and health 1. Annu. Rev. Plant Biol. 1999;50(1):133–161. doi: 10.1146/annurev.arplant.50.1.133. [DOI] [PubMed] [Google Scholar]
- 6.Traber M.G. Vitamin e regulatory mechanisms. Annu. Rev. Nutr. 2007;27:347–362. doi: 10.1146/annurev.nutr.27.061406.093819. [DOI] [PubMed] [Google Scholar]
- 7.Kayden H.J., Traber M.G. Absorption, lipoprotein transport, and regulation of plasma concentrations of vitamin e in humans. J. Lipid Res. 1993;34 343–343. [PubMed] [Google Scholar]
- 8.Grebenstein N., Schumacher M., Graeve L., Frank J. α-tocopherol transfer protein is not required for the discrimination against γ-tocopherol in vivo but protects it from side-chain degradation in vitro. Mol. Nutr. Food Res. 2014;58(5):1052–1060. doi: 10.1002/mnfr.201300756. [DOI] [PubMed] [Google Scholar]
- 9.Burton G.W., Joyce A., Ingold K.U. Is vitamin e the only lipid-soluble, chain-breaking antioxidant in human blood plasma and erythrocyte membranes? Arch. Biochem. Biophys. 1983;221(1):281–290. doi: 10.1016/0003-9861(83)90145-5. [DOI] [PubMed] [Google Scholar]
- 10.Müller L., Theile K., Böhm V. In vitro antioxidant activity of tocopherols and tocotrienols and comparison of vitamin e concentration and lipophilic antioxidant capacity in human plasma. Mol. Nutr. Food Res. 2010;54(5):731–742. doi: 10.1002/mnfr.200900399. [DOI] [PubMed] [Google Scholar]
- 11.Wang X., Thomas B., Sachdeva R., Arterburn L., Frye L., Hatcher P.G., Cornwell D.G., Ma J. Mechanism of arylating quinone toxicity involving michael adduct formation and induction of endoplasmic reticulum stress. Proc. Natl. Acad. Sci. U.S.A. 2006;103(10):3604–3609. doi: 10.1073/pnas.0510962103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brigelius-Flohe R., Traber M.G. Vitamin e: function and metabolism. Faseb. J. 1999;13(10):1145–1155. [PubMed] [Google Scholar]
- 13.Traber M., Sokol R., Burton G., Ingold K., Papas A., Huffaker J., Kayden H. Impaired ability of patients with familial isolated vitamin e deficiency to incorporate alpha-tocopherol into lipoproteins secreted by the liver. J. Clin. Invest. 1990;85(2):397. doi: 10.1172/JCI114452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Traber M.G., Arai H. Molecular mechanisms of vitamin e transport. Annu. Rev. Nutr. 1999;19(1):343–355. doi: 10.1146/annurev.nutr.19.1.343. [DOI] [PubMed] [Google Scholar]
- 15.Yokota T., Igarashi K., Uchihara T., Jishage K.-i., Tomita H., Inaba A., Li Y., Arita M., Suzuki H., Mizusawa H. Delayed-onset ataxia in mice lacking α-tocopherol transfer protein: model for neuronal degeneration caused by chronic oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 2001;98(26):15185–15190. doi: 10.1073/pnas.261456098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aeschimann W., Staats S., Kammer S., Olieric N., Jeckelmann J.-M., Fotiadis D., Netscher T., Rimbach G., Cascella M., Stocker A. Self-assembled α-tocopherol transfer protein nanoparticles promote vitamin e delivery across an endothelial barrier. Sci. Rep. 2017;7:4970. doi: 10.1038/s41598-017-05148-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peltzer R.M., Kolli H.B., Stocker A., Cascella M. Self-assembly of α-tocopherol transfer protein nanoparticles – a patchy-protein model. J. Phys. Chem. B. 2018;122:7066–7072. doi: 10.1021/acs.jpcb.8b05936. [DOI] [PubMed] [Google Scholar]
- 18.Helbling R.E., Aeschimann W., Simona F., Stocker A., Cascella M. Engineering tocopherol selectivity in α-ttp: a combined in vitro/in silico study. PLoS One. 2012;7 doi: 10.1371/journal.pone.0049195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meier R., Tomizaki T., Schulze-Briese C., Baumann U., Stocker A. The molecular basis of vitamin e retention: structure of human α-tocopherol transfer protein. J. Mol. Biol. 2003;331(3):725–734. doi: 10.1016/s0022-2836(03)00724-1. [DOI] [PubMed] [Google Scholar]
- 20.Dundas J., Ouyang Z., Tseng J., Binkowski A., Turpaz Y., Liang J. Castp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34(suppl 2):W116–s. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mowri H., Nakagawa Y., Inoue K., Nojima S. Enhancement of the transfer of alpha-tocopherol between liposomes and mitochondria by rat-liver protein(s) Eur. J. Biochem. 1981;117:537–542. doi: 10.1111/j.1432-1033.1981.tb06370.x. [DOI] [PubMed] [Google Scholar]
- 22.Verdon C.P., Blumberg J.B. An assay for the alpha-tocopherol binding protein mediated transfer of vitamin e between membranes. Anal. Biochem. 1988;169:109–120. doi: 10.1016/0003-2697(88)90261-8. [DOI] [PubMed] [Google Scholar]
- 23.Arita M., Sato Y., Miyata A., Tanabe T., Takahashi E., Kayden H.J., Arai H., Inoue K. Human alpha-tocopherol transfer protein: cdna cloning, expression and chromosomal localization. Biochem. J. 1995;306(2):437–443. doi: 10.1042/bj3060437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morley S., Cecchini M., Zhang W., Virgulti A., Noy N., Atkinson J., Manor D. Mechanisms of ligand transfer by the hepatic tocopherol transfer protein. J. Biol. Chem. 2008;283(26):17797–17804. doi: 10.1074/jbc.M800121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang W.X., Frahm G., Morley S., Manor D., Atkinson J. Effect of bilayer phospholipid composition and curvature on ligand transfer by the α-tocopherol transfer protein. Lipids. 2009;44(7):631–641. doi: 10.1007/s11745-009-3310-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morley S., Panagabko C., Shineman D., Mani B., Stocker A., Atkinson J., Manor D. Molecular determinants of heritable vitamin e deficiency. Biochemistry. 2004;43(14):4143–4149. doi: 10.1021/bi0363073. [DOI] [PubMed] [Google Scholar]
- 27.Khakpour S., Wilhelmsen K., Hellman J. Vascular endothelial cell toll-like receptor pathways in sepsis. Innate Immun. 2015;21(8):827–846. doi: 10.1177/1753425915606525. [DOI] [PubMed] [Google Scholar]
- 28.Voigt A., Rahnefeld A., Kloetzel P., Krüger E. Cytokine-induced oxidative stress in cardiac inflammation and heart failure–how the ubiquitin proteasome system targets this vicious cycle. Front. Physiol. 2013;4:42. doi: 10.3389/fphys.2013.00042. https://www.frontiersin.org/article/10.3389/fphys.2013.00042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X., Andresen1 B.T., Hill M., Zhang J., Booth F., Zhang C. Role of reactive oxygen species in tumor necrosis factor-alpha induced endothelial dysfunction. Curr. Hypertens. Rev. 2008;4(20559453):245–255. doi: 10.2174/157340208786241336. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2886300/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biswas S.K. Does the interdependence between oxidative stress and inflammation explain the antioxidant paradox? Oxid. Med. Cell. Longev. 2016:1–9. doi: 10.1155/2016/5698931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Traber M.G., Atkinson J. Vitamin e, antioxidant and nothing more, Free Rad. Biol. Med. 2007;43(17561088):4–15. doi: 10.1016/j.freeradbiomed.2007.03.024. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2040110/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frank J., Chin X.W.D., Schrader C., Eckert G.P., Rimbach G. Do tocotrienols have potential as neuroprotective dietary factors? Ageing Res. Rev. 2012;11:163–180. doi: 10.1016/j.arr.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 33.Schwedhelm E., Maas R., Troost R., Böger R.H. Clinical pharmacokinetics of antioxidants and their impact on systemic oxidative stress. Clin. Pharmacokinet. 2003;42(5):437–459. doi: 10.2165/00003088-200342050-00003. [DOI] [PubMed] [Google Scholar]
- 34.Liu T., Zhang L., Joo D., Sun S.-C. Nf-κb signaling in inflammation. Signal Transduct. Targeted. 2017;2(29158945):17023. doi: 10.1038/sigtrans.2017.23. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5661633/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson M.T., Staal F.J., Gitler C., Herzenberg L.A., Herzenberg L.A. Separation of oxidant-initiated and redox-regulated steps in the nf-kappa b signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 1994;91(7526398):11527–11531. doi: 10.1073/pnas.91.24.11527. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC45264/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flohé L., Brigelius-Flohé R., Saliou C., Traber M.G., Packer L. Redox regulation of nf-kappa b activation. Free Radic. Biol. Med. 1997;22(6):1115–1126. doi: 10.1016/s0891-5849(96)00501-1. http://www.sciencedirect.com/science/article/pii/S0891584996005011 [DOI] [PubMed] [Google Scholar]
- 37.Cook-Mills J., Gebretsadik T., Abdala-Valencia H., Green J., Larkin E.K., Dupont W.D., Shu X.O., Gross M., Bai C., Gao Y.-T., Hartman T.J., Rosas-Salazar C., Hartert T. Interaction of vitamin e isoforms on asthma and allergic airway disease. Thorax. 2016;71(10):954–956. doi: 10.1136/thoraxjnl-2016-208494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panagabko C., Morley S., Hernandez M., Cassolato P., Gordon H., Parsons R., Manor D., Atkinson J. Ligand specificity in the CRAL-TRIO protein family. Biochemistry. 2003;42(21):6467–6474. doi: 10.1021/bi034086v. http://www.ncbi.nlm.nih.gov/pubmed/12767229 [DOI] [PubMed] [Google Scholar]
- 39.Kabsch W. Xds. Acta Crystallogr. D. 2010;66(2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of coot. Acta Crystallogr. D. 2010;66(4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W. Phenix: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D. 2010;66(2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R. C. Team . R Foundation for Statistical Computing; Vienna, Austria: 2019. R: A Language And Environment for Statistical Computing.https://www.R-project.org/ [Google Scholar]
- 43.Carroll R.J., Ruppert D. vol. 30. CRC Press; 1988. (Transformation and Weighting in Regression). [Google Scholar]
- 44.Nakagawa S., Schielzeth H. A general and simple method for obtaining r2 from generalized linear mixed-effects models. Methods Ecol. Evol. 2013;4(2):133–142. doi: 10.1111/j.2041-210x.2012.00261.x. [DOI] [Google Scholar]
- 45.Richardson A. Multiple comparisons using r by frank bretz, torsten hothorn, peter westfall. Int. Stat. Rev. 2011;79(2) doi: 10.1111/j.1751-5823.2011.00149_23.x. [DOI] [Google Scholar]
- 46.Schaarschmidt F., Vaas L. Analysis of trials with complex treatment structure using multiple contrast tests. Hortscience. 2009;44(1):188–195. https://journals.ashs.org/hortsci/view/journals/hortsci/44/1/article-p188.xml [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.