Abstract
Skin cancer is the most common cancer worldwide, with rapidly increasing incidence and consistent mortality. Skin cancer encompasses melanoma and non-melanoma skin cancer, which in turn is mainly divided into cutaneous squamous cell carcinoma and basal cell carcinoma. Small noncoding microRNAs (miRNAs) regulate protein expression after transcription and play a role in the development and progression of skin cancer. Deregulated expression of miRNAs in skin cancer is associated with cell proliferation, angiogenesis, metastasis, apoptosis, immune response, and drug resistance. Specific patterns of miRNAs in specific skin cancer types can be used as diagnostic markers. For therapeutic purposes, both miRNA and chemically modified variants thereof as well as miRNA antagonists (antagomiRs) or RNA inhibitors may be applied topically. Due to their specific physicochemical properties, physical or chemical diffusion promoters are used with varying degrees of success. There is no question by now that such preparations have a high potential for the treatment of epithelial skin tumors in particular.
Keywords: MicroRNA, Skin cancer, Therapeutic target, Topical therapy
Introduction
MicroRNAs (miRNAs) are noncoding RNAs, which post-transcriptionally regulate protein expression by binding to the 3′ untranslated region (3′UTR) of target mRNAs. Thereby, miRNAs induce translational repression or mRNA cleavage. The individual miRNA variants are subject to very different, mostly, tissue-specific expression and regulation patterns, which play a key role in a large number of physiological and pathophysiological processes. The miRNAs described so far have been named and catalogued in a public database (miRBase) in accordance with standardized criteria, with more than 12,000 variants now recorded [1, 2, 3, 4, 5, 6, 7]. miRNAs interact with transcription factors in a regulatory network and control basal cell biological processes (Fig. 1). The pattern and quantity of the miRNAs expressed in the cell nucleus are tissue-specific and allow conclusions to be drawn about the functional state of the cell, such as its division and differentiation status. Although initial data are available on miRNA patterns in skin tissue and specific skin diseases, there is a need to characterize new miRNA pathways. Since the integration of miRNAs into regulatory processes is also of vital importance in transformation processes of the cell (e.g., in oncogenesis), there is already an intensive discussion about the possibility of using miRNAs as therapeutic target structures. Tumor-specific patterns of deregulated miRNAs in frequent skin tumors have been identified, which may allow a targeted therapeutic approach [8]. This could be achieved by intracellularly targeting and substituting certain miRNAs (so-called miRNA mimetics), but also by the use of synthesized oligonucleotides as antagonists of special overexpressed miRNAs (so-called antagomiRs) or target protectors to block selective miRNA-binding sites (RNA inhibitor [RNAi]) [8]. For the development and establishment of miRNA-based therapies, potential off-target effects as well as questions of chemical stability, release, distribution, and immune stimulation have to be considered and ensured or excluded. In addition, it is known that some tissues are able to partially absorb miRNAs via endocytosis [9]. The susceptibility of miRNAs to proteases may also be relevant for their chemical stability [10]. It is also known about cells with accelerated division behavior (e.g., tumor cells) that transiently released miRNAs are as well eliminated faster, making them less suitable as therapeutic target structures [11]. Various experimental approaches have been described regarding the use of miRNA-based therapies for the treatment of skin tumors, but there have been only a few clinical investigations so far [12, 13].
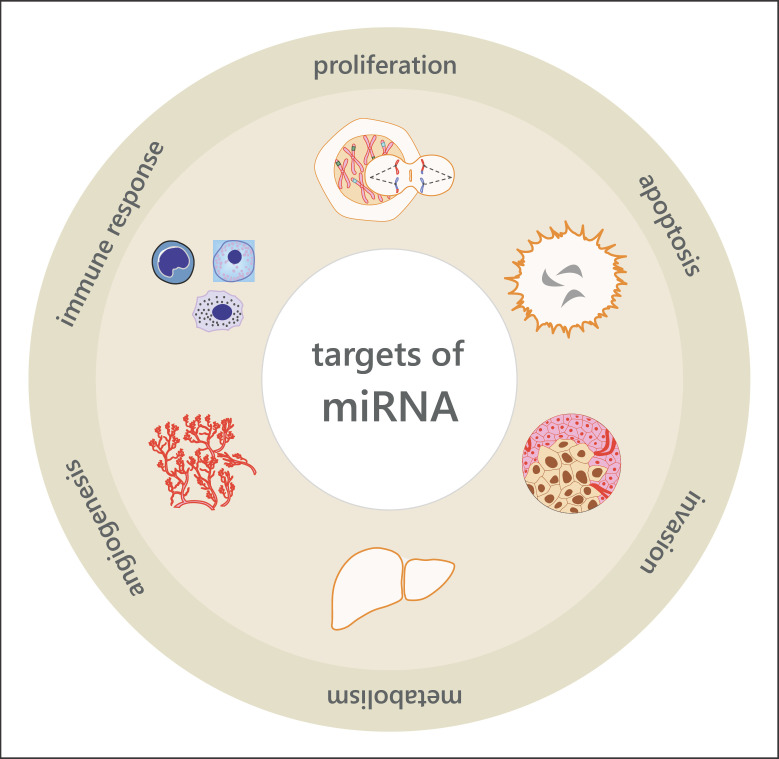
Fig. 1.
Overview of the role of miRNAs in the regulation of physiological and pathophysiological processes in cells.
miRNA Biogenesis and Function
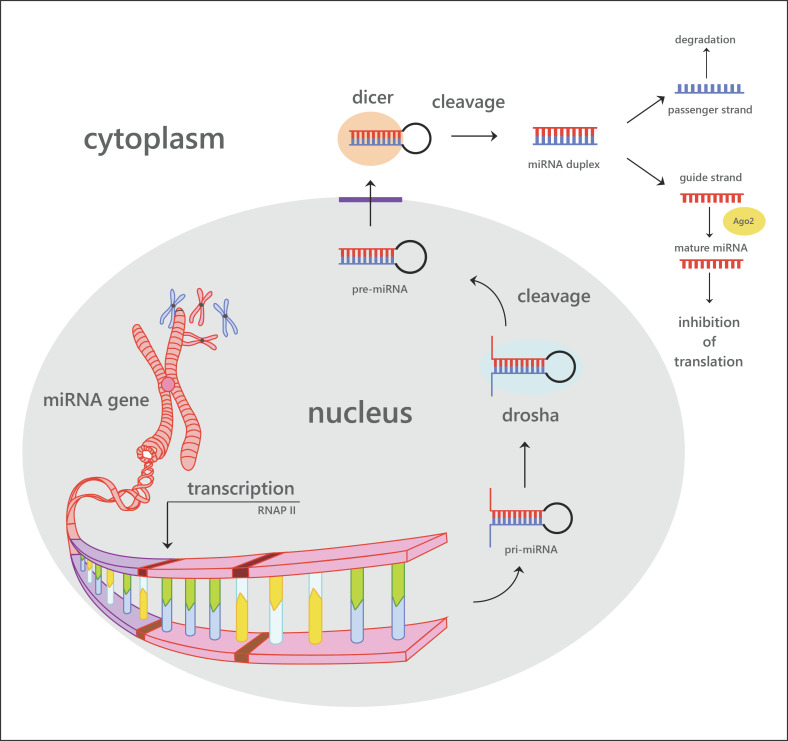
miRNAs are endogenously expressed small noncoding RNAs (20–22 nucleotides) that occur both intra- and intergenically [14]. Besides miRNAs, noncoding RNAs also include small-interfering RNAs (siRNAs), piwi-interacting RNAs, small nuclear RNAs, and small nucleolar RNAs [15, 16]. The primary miRNAs are transcribed by RNA polymerase II (Fig. 2) [17]. The resulting long (∼1 kb) transcript forms a hairpin loop, which is nuclear-cleaved by the Drosha RNase III endonuclease in a 60–70 nt precursor miRNA (pre-miRNA) [18]. Ran-GTP and the export receptor exportin-5 actively transport the pre-miRNA from the nucleus to the cytoplasm. Here, the pre-miRNA is further processed by the RNase III endonuclease Dicer, which cleaves the hairpin loop, resulting in an miRNA:miRNA duplex. One strand of the miRNA:miRNA duplex, the mature miRNA, is loaded into the RNA-induced silencing complex (RISC), while the other strand is degraded [18]. By complementarity of 2–8 residues of the miRNA, the RISC is guided to an miRNA seed sequence in the 3′UTR of target mRNAs [19]. The binding of the miRNA-RISC complex induces a translational repression or destabilization of mRNA [20, 21]. Because miRNAs do not need a perfect complementarity to the targeted 3′UTR, 1 miRNA has the potential to regulate the expression of hundreds of genes [10, 22]. miRNAs were shown to have regulatory functions in a variety of processes such as embryonic development [23], metabolism [24], cell proliferation [25], as well as disease development and progression [26].
Fig. 2.
Overview of the steps of biosynthesis of miRNA, their regulation, and functional integration.
miRNAs in Basal Cell Carcinoma
Basal cell carcinoma (BCC) is the most common invasive skin cancer in the world. It arises from the basal skin cell layer dividing the epidermis from the dermis. BCC development is mainly induced by UV radiation [27]. The constitutive activation of the hedgehog signaling pathway, induced by inactivating PTCH1 mutations, represents the most frequent oncogenic transformation in BCC [28, 29, 30]. Metastasis of BCC is unusual; however, metastasized disease has a poor prognosis [31]. The first study on miRNAs in BCC compared the expression pattern between nodular and infiltrating disease. In their analyses, Heffelfinger et al. [32] identified 20 miRNAs with significant differences in expression between nodular and infiltrative BCCs. Furthermore, they validated let-7g and miR-183 with an increased expression in nodular BCCs, while miR-150, miR-31, miR-146a, and miR-886-5p had a significant higher expression in invasive BCCs [32]. In a microarray-based analysis, miRNA expression was investigated in BCC lesions and adjacent non-lesional skin [33]. Sand et al. [33] identified 16 significantly upregulated and 10 significantly downregulated miRNAs in BCC lesions. In a second study, Sand and colleagues [34] showed 33 upregulated miRNAs in BCC under neoadjuvant vismodegib therapy by next-generation sequencing. Some studies investigated the functional role of miRNAs in BCC development and progression. miR-203 was shown to be downregulated in BCC lesions in a Hedgehog signaling pathway dependent manner [35]. Overexpression of miR-203 in a BCC mouse model results in the reduction of tumor growth. Additionally, it was shown that the tumor suppressor miR-203 directly targets c-JUN, resulting in a reduced proliferation and delayed G1 to S phase transition [35]. Sun and Jiang [36] demonstrated miR-451a as a BCC tumor suppressor miRNA. In their study, they showed that miR-451a expression is reduced in human and mouse BCCs. Further, they confirmed the transcription factor TBX1 as a direct target of miR-451a. The overexpression of miR-451a suppressed BCC cell growth through G1 cell cycle arrest [36]. The presented studies show that miRNA dysregulation contributes to BCC development and progression. However, the small number of functional miRNA studies in BCCs leads to an intensification of this research for a better understanding of the molecular mechanisms in BCC.
miRNAs in Cutaneous Squamous Cell Carcinoma
The cutaneous squamous cell carcinoma (cSCC) is a skin tumor, derived from epidermal keratinocytes. It is the second most common human cancer with an increasing incidence [37]. The main risk factor for cSCCs is UV radiation, potentiated by immunosuppression [37]. Most patients with primary cSCC have an excellent prognosis. However, metastatic cSCCs are still challenging. Recent studies have analyzed the role of miRNAs in cSCC development, progression, and metastasis. Sand et al. [38] first performed a comparative miRNA analysis between cSCC biopsies and adjacent healthy skin. In their analysis, they showed 13 significantly elevated and 18 downregulated miRNAs in cSCC tumors. miR-31 was shown to be the highest upregulated miRNA in cSCC compared to healthy skin [38]. In functional studies, it was shown that miR-31 overexpression induces cell motility and colony formation ability of tumor cells [39]. In addition, miR-31 was shown to directly target the tumor suppressor RhoTBT1, inducing cell proliferation and invasion in the cSCC cell line A-431 [40]. A well-studied miRNA in cSCC is the oncogenic miR-365. Studies show that miR-365 promotes cSCC proliferation and progression in vivo and in vitro [41, 42, 43, 44]. Functional analyses show that miR-365 inhibits apoptosis by directly targeting BAX [44] and forcing the cell cycle progression by NFIB repression [42, 43]. As shown in recent studies, miR-186, miR-221, miR-506, and miR-142-5p contribute to cSCC tumor growth [45, 46, 47, 48, 49]. miR-186 promotes proliferation and inhibits apoptosis by targeting RETREG1 [48] and APAF1 [46]. Gong et al. [47] reported that miR-221 promotes cell proliferation and colony growth by repressing PTEN. Furthermore, it was shown that inhibition of miR-506 induces apoptosis and decreases invasion and migration of cSCC cell lines [49]. In this study, LAMC1 and p65 were identified as direct targets of miR-506. Cancer stem cells have the capability of self-renewal, differentiation, and tumorigenesis. Therefore, they are important actors in tumorigenesis and metastasis. miR-142-5p was shown to induce cancer stem cell-like properties in cSCC cells by inhibition of PTEN [45].
Another controversially discussed miRNA in cSCC is miR-199a [50, 51, 52]. On the one hand, miR-199a has been shown to inhibit the proliferation and migration of cSCC through targeting CD44 [50]. In addition, Kim and colleagues [51] showed that miR-199a directly regulates the expression of BCAM, FZD6, WNT7a, DDR1, and PODXL, which inhibits migration capability of skin keratinocytes. On the other hand, Wang et al. [52] reported that miR-199a induces the invasion of cSCC cells by suppression of E-cadherin. Other miRNAs are more clearly described as tumor suppressors in cSCC. In a recently published research, miR-497 was reported to target FAM114A2 and SERPINE1 [53, 54], inhibiting cell growth, migration [53], cell cycle progression, as well as cSCC cell viability [54]. miR-34a and miR-181a also interfere with cSCC growth. miR-34a targets HMGB1 and inhibits cancer cell proliferation, migration, and invasion [55]. By directly targeting KRAS, miR-181a mediates MAPK signaling pathway and suppresses cSCC tumor growth in vivo. Cañueto et al. [56] reported that expression pattern of miR-203 and miR-205 may help to predict the prognosis of cSCC. They showed that miR-203 expression is associated with a positive outcome, while miR-205 correlates with a poor outcome of cSCC. Taken together, these studies show that miRNAs play an important role in the establishment, progression, and metastasis of cSCC. miRNAs act both as oncomiRs and as tumor suppressors. Additionally, miRNAs can be used as prognostic markers for the outcome of cSCC patients.
miRNAs in Melanoma
Melanoma is the most lethal form of skin cancer with increasing incidence [57]. miRNA contribution and function in melanoma development, progression, metastasis, and drug resistance have been widely studied [58, 59, 60, 61, 62]. For this report, we have focused on prominent melanoma-associated miRNAs and the latest studies of recent years. miR-21 is an established oncomiR that is highly expressed in various cancers. In malignant melanoma, miR-21 has been shown to have significantly increased expression compared to nevi and normal melanocytes [63, 64, 65]. Furthermore, plasma miR-21 was reported to be associated with the tumor burden of melanoma patients [66]. Functional analysis showed that miR-21 targets SPRY1, PDCD4, and PTEN; regulates the ERK/NF-κB signaling pathway; and affects proliferation, migration, and apoptosis [67, 68, 69]. In addition, miR-21 increases melanoma invasiveness by targeting MMP3 [70]. The blocking of miR-21 by antisense oligonucleotides improves the sensitivity of the A375 cell line toward cisplatin [71]. miR-211 that is normally enriched in pigmented cells is reported to be downregulated in melanoma cells compared to normal melanocytes [72, 73]. However, miR-211 is regulated by MITF and was shown to regulate cell invasiveness by targeting NUAK1 [74], BRN2 [72], and RAB22A [75]. In addition, miR-211 contributes to resistance against BRAF inhibitor [76, 77], while the loss of miR-211 reduces the sensitivity of melanoma cells to cisplatin [78]. miR-203 was shown to act as tumor suppressor in several cancers [79, 80, 81, 82, 83]. Recent research reported a decreased expression of miR-203 in melanoma tissue and a link with prognosis of melanoma patients [84, 85]. Furthermore, low expression of miR-203 has been shown to inhibit migration, invasion, and proliferation of melanoma by targeting BIM1 [86] and SLUG [87]. More recent studies, however, contradict this and show an overexpression of miR-203 as a marker for an early phase of metastasis [88].
miRNAs as Therapeutic Approach
In recent decades, miRNAs have become the focus of attention as novel therapeutic targets. In vivo studies on different mouse disease models have shown the efficacy of miRNA targeting therapeutic strategies [89, 90, 91, 92, 93]. Currently, RNA molecules are already being investigated in clinical studies as therapeutic molecules mostly for systemic application. The miR-122 inhibitor miravirsen was tested as a novel therapeutic strategy for HCV infections (ClinicalTrials.gov identifier: NCT01200420) [94, 95]. In addition, patisiran, the first RNAi-based therapy, was approved by the FDA in 2018 [96, 97].
In order to therapeutically influence the intracytoplasmic concentration and function of miRNA (miRNA-based therapy), (1) defective miRNA (or slightly modified miRNA to improve chemical stability) can be substituted or (2) antagomiRs or RNAi can be used to counteract overexpression of miRNA.
One challenge for the therapeutic use of oligonucleotides to replace or inhibit miRNAs is the stability. RNAs are highly fragile for degradation by endo- and exonucleases. Chemical modifications of the oligonucleotide backbone or at the ribose were shown to increase the stability and delivery efficiency for in vivo applications. The substitution of the α-oxygen at the phosphate by a sulfur, methyl, or borano group improves the stability of the oligonucleotides [98]. Introduction of 2′-O-methyl (2′-O-methyl-(2′-O-Me) oligonucleotides) or the incorporation of a 2′,4′ methylene bridge at the ribose, to form a bicyclic oligonucleotide (locked nucleic acid oligonucleotides), protects against nucleases degradation and improves the binding ability to potential target RNAs [99, 100, 101]. The terminal introduction of cholesterol, biotin, or amino modification enhances the cellular uptake and the delivery and can increase the stability of oligonucleotides [101]. For in vivo applications, multiple modifications are used in combination to achieve the best functional efficiency and stability.
Their respective galenic formulations depend on the physicochemical properties and cannot be fixed in general terms. However, special challenges arise during the development of suitable preparations to ensure the chemical stability of the active substance within a suitable vehicle, to implement sufficient release behavior from the vehicle, to optimize intracellular bioavailability at the target cells, and to observe regulatory, patent, and commercial framework conditions. Although the use of miRNAs as active molecules in topical therapy offers significant advantages over systemic application, it also poses a number of challenges, which will be summarized below. Suitable miRNAs in sufficient and reproducible quality and quantity must be available for the application. The galenic formulation must primarily ensure the chemical and functional stability of the miRNAs, but at the same time, it should also provide a physicochemical framework for the highest possible release and diffusion rate of hydrophilic miRNAs. Depending on the indication for use, cells of the vital epidermis or the upper corium can be regarded as the galenic target compartment within the skin organ. In connection with oncological indications, it is also necessary to realize an optimized distribution behavior of the therapeutic agent intratumorally. When applied to intact epidermis, permeation of the stratum corneum is naturally necessary. Since the epicutaneous application of naked miRNA does not lead to an efficient transfection due to the high hydrophilicity and the large molecular mass, vehicle strategies for transcorneal transport are necessary. The use of preparations on severely barrier-disturbed epidermis, for example, for certain carcinomas, erosive or peracutely inflamed skin, may lead to conditions that make the use of conventional vehicles seem possible. If the stratum corneum is only functionally active to a limited extent, penetration promotion strategies must be considered. For this purpose, physical and chemical methods are available, which offer options for an intact introduction of miRNA, antagomiRs, or RNAis [102].
Physical Methods
The principle of physical penetration promotion is based on the initiation of diffusion through artificial microchannels, which are produced by physical intervention using microneedles, microjets, laser or electroporation, sonopheresis, or iontopheresis for a limited period of time. Various technical procedures and application strategies have been established and tested for the application of the respective methods. The selection of one of the possible methods depends on the molecular mass of the therapeutic agent and its physicochemical properties and vulnerability. Against this background and for practical reasons, the topical application of miRNAs in intracutaneous tumor tissue is particularly suitable for microneedles or microjets. For this purpose, commercially available systems are already established, so that the prerequisites for the application are largely given and can be adapted to the respective indication [102].
Chemical Methods
The use of chemical penetration promoters or modern vehicle strategies derived from them (so-called supramolecular aggregates) poses a significantly higher challenge. The interaction of such enhancer molecules leads to a targeted interaction with the molecular organizational structure of SC, which leads to changes in the diffusion conditions, especially in the area of the intercellular lipid matrix. Due to the hydrophilic character of miRNAs, enhancers of the polar diffusion route (i.e., alcohols, amides, surfactants, and sulfoxides) are of particular interest. For hydrophilic macromolecules, experience with microemulsion systems, liposome variations, and nanoparticles is available [102]. These colloidal vehicle systems mediate the proportional transfer of the active agent to or into the cell and realize its release in bioactive form. These very complex aggregates are complex to manufacture and very cost-intensive for practical use. The experimental application, especially in animal experiments and diffusion models, clearly shows that the epicutaneous application of miRNAs is possible [103, 104, 105]. Furthermore, the use of so-called cell penetrating peptides is propagated [106]. These are peptides that spontaneously permeate cell membranes and can also transport covalently or non-covalently bound nucleic acids or nanoparticles [107]. Data on the siRNA application of so-called “skin penetrating and cell entering” peptides, trans-activating transcription activator peptides, and poly-arginine (poly-R) peptides are available [108, 109, 110, 111]. Nanoparticles or aggregates of nanoparticles with spherical nucleic acids have also been investigated in various variations for their suitability [112]. Especially for variations with gold nanoparticles data are available [113]. Variations of liposomal structures have been validated for many years with regard to their suitability as drug delivery systems, also with regard to miRNA [114, 115, 116, 117, 118, 119]. Only through the development of so-called transfersomes (ultra-deformable liposomes) could incorporated macromolecules such as peptides, DNA vaccines, and proteins be effectively transported transmembrane or transcorneally [120, 121, 122]. However, the evidence for the suitability for the effective miRNA transport after epicutaneous application is lacking for transfersomes as well as for further developed variations, such as ethanol-rich ethosomes or transethosomes [16]. In contrast, liposome variants of mixtures of surfactant, ethanol, and cholesterol (secosomes) or of lecithin phospholipids and cationic lipids (LeciPlex) for the transport of siRNA, pre-miRNA, and anti-miRNA have already been successfully investigated [123, 124]. Due to the comparatively uncomplicated production, complexes of miRNA and chitosan were investigated with high expectations [125, 126, 127, 128]. Unfortunately, no relevant transfection efficiency was shown, so that their suitability seems questionable. Chemical modifications of miRNA, which increase the stability of the agent and lead to an improved diffusion ability, also attracted attention [129, 130]. Such modifications have also been described for RNAi (self-delivering RNAi) [131, 132].
Conclusions
The therapeutic use of miRNAs, antagomiRs, and RNAis has great potential. The topical application of such biotechnological agents and the realization of a sufficient bioavailability of bioactive agents in the target cells place the highest demands on galenic development, suitable vehicle, and enhancer strategies as well as the assurance of chemical and biological stability. In addition, special experimental strategies are required to characterize developed preparations preclinically and to produce them in a standardized way. Even though initial data on the effectiveness of topical preparations in animal experiments have not yet been obtained, it is still necessary to develop special experimental strategies.
Conflict of Interest Statement
All authors declare no conflicts of interest.
Funding Sources
The authors did not receive any funding.
Author Contributions
D.G. and J.W. wrote the manuscript. C.S. commented on the manuscript.
References
- 1.Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–38. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 2.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006 Jan 1;34((Database issue)):D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008 Jan;36((Database issue)):D154–8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Griffiths-Jones S. miRBase: microRNA sequences and annotation. Curr Protoc Bioinformatics. 2010 Mar; doi: 10.1002/0471250953.bi1209s29. [DOI] [PubMed] [Google Scholar]
- 5.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011 Jan;39((Database issue)):D152–7. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014 Jan;42((Database issue)):D68–73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozomara A, Birgaoanu M, Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019 Jan 8;47((D1)):D155–D62. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sand M, Gambichler T, Sand D, Skrygan M, Altmeyer P, Bechara FG. MicroRNAs and the skin: tiny players in the body's largest organ. J Dermatol Sci. 2009 Mar;53((3)):169–75. doi: 10.1016/j.jdermsci.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Love TM, Moffett HF, Novina CD. Not miR-ly small RNAs: big potential for microRNAs in therapy. J Allergy Clin Immunol. 2008 Feb;121((2)):309–19. doi: 10.1016/j.jaci.2007.12.1167. [DOI] [PubMed] [Google Scholar]
- 10.Schirle NT, Sheu-Gruttadauria J, MacRae IJ. Structural basis for microRNA targeting. Science. 2014 Oct 31;346((6209)):608–13. doi: 10.1126/science.1258040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soifer HS, Rossi JJ, Saetrom P. MicroRNAs in disease and potential therapeutic applications. Mol Ther. 2007 Dec;15((12)):2070–9. doi: 10.1038/sj.mt.6300311. [DOI] [PubMed] [Google Scholar]
- 12.Zhang P, Chen J, Li T, Zhu YY. Use of small RNA as antiaging cosmeceuticals. J Cosmet Sci. 2013 Nov-Dec;64((6)):455–68. [PubMed] [Google Scholar]
- 13.Lawrence P, Ceccoli J. Advances in the application and impact of MicroRNAs as therapies for skin disease. BioDrugs. 2017 Oct;31((5)):423–38. doi: 10.1007/s40259-017-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004 Oct;14((10A)):1902–10. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q, Paroo Z. Biochemical principles of small RNA pathways. Annu Rev Biochem. 2010;79:295–319. doi: 10.1146/annurev.biochem.052208.151733. [DOI] [PubMed] [Google Scholar]
- 16.Ross K. Towards topical microRNA-directed therapy for epidermal disorders. J Control Release. 2018 Jan 10;269:136–47. doi: 10.1016/j.jconrel.2017.11.013. [DOI] [PubMed] [Google Scholar]
- 17.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004 Oct 13;23((20)):4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002 Sep 2;21((17)):4663–70. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009 Jan 23;136((2)):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ambros V. The functions of animal microRNAs. Nature. 2004 Sep 16;431((7006)):350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004 Jan 23;116((2)):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 22.Shivdasani RA. MicroRNAs: regulators of gene expression and cell differentiation. Blood. 2006 Dec 1;108((12)):3646–53. doi: 10.1182/blood-2006-01-030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karp X, Ambros V. Developmental biology. Encountering microRNAs in cell fate signaling. Science. 2005 Nov 25;310((5752)):1288–9. doi: 10.1126/science.1121566. [DOI] [PubMed] [Google Scholar]
- 24.Jordan SD, Krüger M, Willmes DM, Redemann N, Wunderlich FT, Brönneke HS, et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat Cell Biol. 2011 Apr;13((4)):434–46. doi: 10.1038/ncb2211. [DOI] [PubMed] [Google Scholar]
- 25.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33((4)):1290–7. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005 Jun 9;435((7043)):834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 27.Gallagher RP, Hill GB, Bajdik CD, Fincham S, Coldman AJ, McLean DI, et al. Sunlight exposure, pigmentary factors, and risk of nonmelanocytic skin cancer. I. Basal cell carcinoma. Arch Dermatol. 1995 Feb;131((2)):157–63. [PubMed] [Google Scholar]
- 28.Gailani MR, Ståhle-Bäckdahl M, Leffell DJ, Glynn M, Zaphiropoulos PG, Pressman C, et al. The role of the human homologue of Drosophila patched in sporadic basal cell carcinomas. Nat Genet. 1996 Sep;14((1)):78–81. doi: 10.1038/ng0996-78. [DOI] [PubMed] [Google Scholar]
- 29.Reifenberger J, Wolter M, Knobbe CB, Köhler B, Schönicke A, Scharwächter C, et al. Somatic mutations in the PTCH, SMOH, SUFUH and TP53 genes in sporadic basal cell carcinomas. Br J Dermatol. 2005 Jan;152((1)):43–51. doi: 10.1111/j.1365-2133.2005.06353.x. [DOI] [PubMed] [Google Scholar]
- 30.Bonilla X, Parmentier L, King B, Bezrukov F, Kaya G, Zoete V, et al. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat Genet. 2016 Apr;48((4)):398–406. doi: 10.1038/ng.3525. [DOI] [PubMed] [Google Scholar]
- 31.Walling HW, Fosko SW, Geraminejad PA, Whitaker DC, Arpey CJ. Aggressive basal cell carcinoma: presentation, pathogenesis, and management. Cancer Metastasis Rev. 2004 Aug-Dec;23((3–4)):389–402. doi: 10.1023/B:CANC.0000031775.04618.30. [DOI] [PubMed] [Google Scholar]
- 32.Heffelfinger C, Ouyang Z, Engberg A, Leffell DJ, Hanlon AM, Gordon PB, et al. Correlation of global microRNA expression with basal cell carcinoma subtype. G3 (Bethesda) 2012 Feb;2((2)):279–86. doi: 10.1534/g3.111.001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sand M, Skrygan M, Sand D, Georgas D, Hahn SA, Gambichler T, et al. Expression of microRNAs in basal cell carcinoma. Br J Dermatol. 2012 Oct;167((4)):847–55. doi: 10.1111/j.1365-2133.2012.11022.x. [DOI] [PubMed] [Google Scholar]
- 34.Sand M, Bechara FG, Gambichler T, Sand D, Friedländer MR, Bromba M, et al. Next-generation sequencing of the basal cell carcinoma miRNome and a description of novel microRNA candidates under neoadjuvant vismodegib therapy: an integrative molecular and surgical case study. Ann Oncol. 2016 Feb;27((2)):332–8. doi: 10.1093/annonc/mdv551. [DOI] [PubMed] [Google Scholar]
- 35.Sonkoly E, Lovén J, Xu N, Meisgen F, Wei T, Brodin P, et al. MicroRNA-203 functions as a tumor suppressor in basal cell carcinoma. Oncogenesis. 2012 Mar 12;1:e3. doi: 10.1038/oncsis.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun H, Jiang P. MicroRNA-451a acts as tumor suppressor in cutaneous basal cell carcinoma. Mol Genet Genomic Med. 2018 Nov;6((6)):1001–9. doi: 10.1002/mgg3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alam M, Ratner D. Cutaneous squamous-cell carcinoma. N Engl J Med. 2001 Mar 29;344((13)):975–83. doi: 10.1056/NEJM200103293441306. [DOI] [PubMed] [Google Scholar]
- 38.Sand M, Skrygan M, Georgas D, Sand D, Hahn SA, Gambichler T, et al. Microarray analysis of microRNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci. 2012 Dec;68((3)):119–26. doi: 10.1016/j.jdermsci.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 39.Wang A, Landén NX, Meisgen F, Lohcharoenkal W, Ståhle M, Sonkoly E, et al. MicroRNA-31 is overexpressed in cutaneous squamous cell carcinoma and regulates cell motility and colony formation ability of tumor cells. PloS One. 2014;9((7)):e103206. doi: 10.1371/journal.pone.0103206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin N, Zhou Y, Lian X, Tu Y. MicroRNA-31 functions as an oncogenic microRNA in cutaneous squamous cell carcinoma cells by targeting RhoTBT1. Oncol Lett. 2017 Mar;13((3)):1078–82. doi: 10.3892/ol.2017.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou M, Liu W, Ma S, Cao H, Peng X, Guo L, et al. A novel onco-miR-365 induces cutaneous squamous cell carcinoma. Carcinogenesis. 2013 Jul;34((7)):1653–9. doi: 10.1093/carcin/bgt097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou M, Zhou L, Zheng L, Guo L, Wang Y, Liu H, et al. miR-365 promotes cutaneous squamous cell carcinoma (CSCC) through targeting nuclear factor I/B (NFIB) PloS One. 2014;9((6)):e100620. doi: 10.1371/journal.pone.0100620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou L, Wang Y, Ou C, Lin Z, Wang J, Liu H, et al. microRNA-365-targeted nuclear factor I/B transcriptionally represses cyclin-dependent kinase 6 and 4 to inhibit the progression of cutaneous squamous cell carcinoma. Int J Biochem Cell Biol. 2015 Aug;65:182–91. doi: 10.1016/j.biocel.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 44.Zhou L, Gao R, Wang Y, Zhou M, Ding Z. Loss of BAX by miR-365 promotes cutaneous squamous cell carcinoma progression by suppressing apoptosis. Int J Mol Sci. 2017 May 30;18((6)):1157. doi: 10.3390/ijms18061157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bai X, Zhou Y, Chen P, Yang M, Xu J. MicroRNA-142-5p induces cancer stem cell-like properties of cutaneous squamous cell carcinoma via inhibiting PTEN. J Cell Biochem. 2018 Feb;119((2)):2179–88. doi: 10.1002/jcb.26379. [DOI] [PubMed] [Google Scholar]
- 46.Tian J, Shen R, Yan Y, Deng L. miR-186 promotes tumor growth in cutaneous squamous cell carcinoma by inhibiting apoptotic protease activating factor-1. Exp Ther Med. 2018 Nov;16((5)):4010–8. doi: 10.3892/etm.2018.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gong Z-H, Zhou F, Shi C, Xiang T, Zhou C-K, Wang Q-Q, et al. miRNA-221 promotes cutaneous squamous cell carcinoma progression by targeting PTEN. Cell Mol Biol Lett. 2019;24((1)):9. doi: 10.1186/s11658-018-0131-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu X, Liu Y, Ai P, He S, Liu L, Chen C, et al. MicroRNA-186 promotes cell proliferation and inhibits cell apoptosis in cutaneous squamous cell carcinoma by targeting RETREG1. Exp Ther Med. 2019 Mar;17((3)):1930–8. doi: 10.3892/etm.2019.7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou J, Zhang Y, Han Z, Dong Z, Cao T, Wei A, et al. miR-506 contributes to malignancy of cutaneous squamous cell carcinoma via targeting of P65 and LAMC1. Cell Cycle. 2019 Feb;18((3)):333–45. doi: 10.1080/15384101.2019.1568747. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Wang SH, Zhou JD, He QY, Yin ZQ, Cao K, Luo CQ. MiR-199a inhibits the ability of proliferation and migration by regulating CD44-Ezrin signaling in cutaneous squamous cell carcinoma cells. Int J Clin Exp Pathol. 2014;7((10)):7131–41. [PMC free article] [PubMed] [Google Scholar]
- 51.Kim BK, Kim I, Yoon SK. Identification of miR-199a-5p target genes in the skin keratinocyte and their expression in cutaneous squamous cell carcinoma. J Dermatol Sci. 2015 Aug;79((2)):137–47. doi: 10.1016/j.jdermsci.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 52.Wang S, Cao KE, He Q, Yin Z, Zhou J. miR-199a-5p induces cell invasion by suppressing E-cadherin expression in cutaneous squamous cell carcinoma. Oncol Lett. 2016 Jul;12((1)):97–101. doi: 10.3892/ol.2016.4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mizrahi A, Barzilai A, Gur-Wahnon D, Ben-Dov IZ, Glassberg S, Meningher T, et al. Alterations of microRNAs throughout the malignant evolution of cutaneous squamous cell carcinoma: the role of miR-497 in epithelial to mesenchymal transition of keratinocytes. Oncogene. 2018 Jan 11;37((2)):218–30. doi: 10.1038/onc.2017.315. [DOI] [PubMed] [Google Scholar]
- 54.Wei XH, Gu XL, Zhou XT, Ma M, Lou CX. miR-497 promotes the progression of cutaneous squamous cell carcinoma through FAM114A2. Eur Rev Med Pharmacol Sci. 2018 Nov;22((21)):7348–55. doi: 10.26355/eurrev_201811_16272. [DOI] [PubMed] [Google Scholar]
- 55.Li S, Luo C, Zhou J, Zhang Y. MicroRNA-34a directly targets high-mobility group box 1 and inhibits the cancer cell proliferation, migration and invasion in cutaneous squamous cell carcinoma. Exp Ther Med. 2017 Dec;14((6)):5611–8. doi: 10.3892/etm.2017.5245. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56.Cañueto J, Cardeñoso-Álvarez E, García-Hernández JL, Galindo-Villardón P, Vicente-Galindo P, Vicente-Villardón JL, et al. MicroRNA (miR)-203 and miR-205 expression patterns identify subgroups of prognosis in cutaneous squamous cell carcinoma. Br J Dermatol. 2017 Jul;177((1)):168–78. doi: 10.1111/bjd.15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015 Mar 1;136((5)):E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 58.Alegre E, Sanmamed MF, Rodriguez C, Carranza O, Martín-Algarra S, González A. Study of circulating microRNA-125b levels in serum exosomes in advanced melanoma. Arch Pathol Lab Med. 2014 Jun;138((6)):828–32. doi: 10.5858/arpa.2013-0134-OA. [DOI] [PubMed] [Google Scholar]
- 59.Zhang J, Lu L, Xiong Y, Qin W, Zhang Y, Qian Y, et al. MLK3 promotes melanoma proliferation and invasion and is a target of microRNA-125b. Clin Exp Dermatol. 2014 Apr;39((3)):376–84. doi: 10.1111/ced.12286. [DOI] [PubMed] [Google Scholar]
- 60.Xu D, Chen X, He Q, Luo C. MicroRNA-9 suppresses the growth, migration, and invasion of malignant melanoma cells via targeting NRP1. Onco Targets Ther. 2016;9:7047–57. doi: 10.2147/OTT.S107235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J, Na S, Liu C, Pan S, Cai J, Qiu J. MicroRNA-125b suppresses the epithelial-mesenchymal transition and cell invasion by targeting ITGA9 in melanoma. Tumour Biol. 2016 May;37((5)):5941–9. doi: 10.1007/s13277-015-4409-8. [DOI] [PubMed] [Google Scholar]
- 62.Bu P, Luo C, He Q, Yang P, Li X, Xu D. MicroRNA-9 inhibits the proliferation and migration of malignant melanoma cells via targeting sirituin 1. Exp Ther Med. 2017 Aug;14((2)):931–8. doi: 10.3892/etm.2017.4595. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Jiang L, Lv X, Li J, Li J, Li X, Li W, et al. The status of microRNA-21 expression and its clinical significance in human cutaneous malignant melanoma. Acta Histochem. 2012 Oct;114((6)):582–8. doi: 10.1016/j.acthis.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 64.Satzger I, Mattern A, Kuettler U, Weinspach D, Niebuhr M, Kapp A, et al. microRNA-21 is upregulated in malignant melanoma and influences apoptosis of melanocytic cells. Exp Dermatol. 2012 Jul;21((7)):509–14. doi: 10.1111/j.1600-0625.2012.01510.x. [DOI] [PubMed] [Google Scholar]
- 65.Wandler A, Riber-Hansen R, Hager H, Hamilton-Dutoit SJ, Schmidt H, Nielsen BS, et al. Quantification of microRNA-21 and microRNA-125b in melanoma tissue. Melanoma Res. 2017 Oct;27((5)):417–28. doi: 10.1097/CMR.0000000000000374. [DOI] [PubMed] [Google Scholar]
- 66.Saldanha G, Potter L, Shendge P, Osborne J, Nicholson S, Yii N, et al. Plasma microRNA-21 is associated with tumor burden in cutaneous melanoma. J Invest Dermatol. 2013 May;133((5)):1381–4. doi: 10.1038/jid.2012.477. [DOI] [PubMed] [Google Scholar]
- 67.Yang CH, Yue J, Pfeffer SR, Handorf CR, Pfeffer LM. MicroRNA miR-21 regulates the metastatic behavior of B16 melanoma cells. J Biol Chem. 2011 Nov 11;286((45)):39172–8. doi: 10.1074/jbc.M111.285098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiao J, Fan Y, Zhang Y. Expression and clinicopathological significance of microRNA-21 and programmed cell death 4 in malignant melanoma. J Int Med Res. 2015 Oct;43((5)):672–8. doi: 10.1177/0300060515583707. [DOI] [PubMed] [Google Scholar]
- 69.Mao XH, Chen M, Wang Y, Cui PG, Liu SB, Xu ZY. MicroRNA-21 regulates the ERK/NF-κB signaling pathway to affect the proliferation, migration, and apoptosis of human melanoma A375 cells by targeting SPRY1, PDCD4, and PTEN. Mol Carcinog. 2017 Mar;56((3)):886–94. doi: 10.1002/mc.22542. [DOI] [PubMed] [Google Scholar]
- 70.Martin del Campo SE, Latchana N, Levine KM, Grignol VP, Fairchild ET, Jaime-Ramirez AC, et al. MiR-21 enhances melanoma invasiveness via inhibition of tissue inhibitor of metalloproteinases 3 expression: in vivo effects of MiR-21 inhibitor. PLoS One. 2015;10((1)):e0115919. doi: 10.1371/journal.pone.0115919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang HL, Si LB, Zeng A, Long F, Qi Z, Zhao R, et al. MicroRNA-21 antisense oligonucleotide improves the sensitivity of A375 human melanoma cell to cisplatin: an in vitro study. J Cell Biochem. 2018 Apr;119((4)):3129–41. doi: 10.1002/jcb.26455. [DOI] [PubMed] [Google Scholar]
- 72.Boyle GM, Woods SL, Bonazzi VF, Stark MS, Hacker E, Aoude LG, et al. Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigment Cell Melanoma Res. 2011 Jun;24((3)):525–37. doi: 10.1111/j.1755-148X.2011.00849.x. [DOI] [PubMed] [Google Scholar]
- 73.Babapoor S, Horwich M, Wu R, Levinson S, Gandhi M, Makkar H, et al. microRNA in situ hybridization for miR-211 detection as an ancillary test in melanoma diagnosis. Mod Pathol. 2016 May;29((5)):461–75. doi: 10.1038/modpathol.2016.44. [DOI] [PubMed] [Google Scholar]
- 74.Bell RE, Khaled M, Netanely D, Schubert S, Golan T, Buxbaum A, et al. Transcription factor/microRNA axis blocks melanoma invasion program by miR-211 targeting NUAK1. J Invest Dermatol. 2014 Feb;134((2)):441–51. doi: 10.1038/jid.2013.340. [DOI] [PubMed] [Google Scholar]
- 75.Yu H, Yang W. MiR-211 is epigenetically regulated by DNMT1 mediated methylation and inhibits EMT of melanoma cells by targeting RAB22A. Biochem Biophys Res Commun. 2016 Aug 5;476((4)):400–5. doi: 10.1016/j.bbrc.2016.05.133. [DOI] [PubMed] [Google Scholar]
- 76.Díaz-Martínez M, Benito-Jardón L, Alonso L, Koetz-Ploch L, Hernando E, Teixidó J. miR-204-5p and miR-211-5p contribute to BRAF inhibitor resistance in melanoma. Cancer Res. 2018 Feb 15;78((4)):1017–30. doi: 10.1158/0008-5472.CAN-17-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sahoo A, Sahoo SK, Joshi P, Lee B, Perera RJ. MicroRNA-211 loss promotes metabolic vulnerability and BRAF inhibitor sensitivity in melanoma. J Invest Dermatol. 2019 Jan;139((1)):167–76. doi: 10.1016/j.jid.2018.06.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li N, Liu Y, Pang H, Lee D, Zhou Y, Xiao Z. Methylation-mediated silencing of MicroRNA-211 decreases the sensitivity of melanoma cells to cisplatin. Med Sci Monitor: Int Med J Exp Clin Res. 2019 Mar 1;25:1590–9. doi: 10.12659/MSM.911862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou P, Jiang N, Zhang GX, Sun Q. MiR-203 inhibits tumor invasion and metastasis in gastric cancer by ATM. Acta Biochim Biophys Sin. 2016 Aug;48((8)):696–703. doi: 10.1093/abbs/gmw063. [DOI] [PubMed] [Google Scholar]
- 80.Zheng XB, Chen XB, Xu LL, Zhang M, Feng L, Yi PS, et al. miR-203 inhibits augmented proliferation and metastasis of hepatocellular carcinoma residual in the promoted regenerating liver. Cancer Sci. 2017 Mar;108((3)):338–46. doi: 10.1111/cas.13167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shen J, Zhang J, Xiao M, Yang J, Zhang N. MiR-203 suppresses bladder cancer cell growth and targets the Twist1. Oncol Res. 2018 Sep 14;26((8)):1155–65. doi: 10.3727/096504017X15041934685237. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 82.Wang B, Li X, Zhao G, Yan H, Dong P, Watari H, et al. miR-203 inhibits ovarian tumor metastasis by targeting BIRC5 and attenuating the TGFβ pathway. J Exp Clin Cancer Res. 2018 Sep 21;37((1)):235. doi: 10.1186/s13046-018-0906-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xia Y, Wang Y, Wang Q, Ghaffar M, Wang Y, Sheng W, et al. Increased miR-203-3p and reduced miR-21-5p synergistically inhibit proliferation, migration, and invasion in esophageal cancer cells. Anticancer Drugs. 2019 Jan;30((1)):38–45. doi: 10.1097/CAD.0000000000000687. [DOI] [PubMed] [Google Scholar]
- 84.Bu P, Yang P. MicroRNA-203 inhibits malignant melanoma cell migration by targeting versican. Exp Ther Med. 2014 Jul;8((1)):309–15. doi: 10.3892/etm.2014.1708. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 85.Wang K, Zhang ZW. Expression of miR-203 is decreased and associated with the prognosis of melanoma patients. Int J Clin Exp Pathol. 2015;8((10)):13249–54. [PMC free article] [PubMed] [Google Scholar]
- 86.Chang X, Sun Y, Han S, Zhu W, Zhang H, Lian S. MiR-203 inhibits melanoma invasive and proliferative abilities by targeting the polycomb group gene BMI1. Biochem Biophys Res Commun. 2015 Jan 2;456((1)):361–6. doi: 10.1016/j.bbrc.2014.11.087. [DOI] [PubMed] [Google Scholar]
- 87.Lohcharoenkal W, Das Mahapatra K, Pasquali L, Crudden C, Kular L, Akkaya Ulum YZ, et al. Genome-wide screen for microRNAs reveals a role for miR-203 in melanoma metastasis. J Invest Dermatol. 2018 Apr;138((4)):882–92. doi: 10.1016/j.jid.2017.09.049. [DOI] [PubMed] [Google Scholar]
- 88.Sahranavardfard P, Firouzi J, Azimi M, Khosravani P, Heydari R, Emami Razavi A, et al. MicroRNA-203 reinforces stemness properties in melanoma and augments tumorigenesis in vivo. J Cell Physiol. 2019 Nov;234((11)):20193–205. doi: 10.1002/jcp.28619. [DOI] [PubMed] [Google Scholar]
- 89.Corsten MF, Miranda R, Kasmieh R, Krichevsky AM, Weissleder R, Shah K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007 Oct 1;67((19)):8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- 90.Tivnan A, Tracey L, Buckley PG, Alcock LC, Davidoff AM, Stallings RL. MicroRNA-34a is a potent tumor suppressor molecule in vivo in neuroblastoma. BMC Cancer. 2011 Jan 25;11:33. doi: 10.1186/1471-2407-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Babar IA, Cheng CJ, Booth CJ, Liang X, Weidhaas JB, Saltzman WM, et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci U S A. 2012 Jun 26;109((26)):E1695–704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.He XX, Chang Y, Meng FY, Wang MY, Xie QH, Tang F, et al. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene. 2012 Jul 12;31((28)):3357–69. doi: 10.1038/onc.2011.500. [DOI] [PubMed] [Google Scholar]
- 93.Qian H, Yang C, Yang Y. MicroRNA-26a inhibits the growth and invasiveness of malignant melanoma and directly targets on MITF gene. Cell Death Discov. 2017;3:17028. doi: 10.1038/cddiscovery.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013 May 2;368((18)):1685–94. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 95.Ottosen S, Parsley TB, Yang L, Zeh K, van Doorn LJ, van der Veer E, et al. In vitro antiviral activity and preclinical and clinical resistance profile of miravirsen, a novel anti-hepatitis C virus therapeutic targeting the human factor miR-122. Antimicrob Agents Chemother. 2015 Jan;59((1)):599–608. doi: 10.1128/AAC.04220-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hoy SM. Patisiran: first global approval. Drugs. 2018 Oct;78((15)):1625–31. doi: 10.1007/s40265-018-0983-6. [DOI] [PubMed] [Google Scholar]
- 97.Kristen AV, Ajroud-Driss S, Conceição I, Gorevic P, Kyriakides T, Obici L. Patisiran, an RNAi therapeutic for the treatment of hereditary transthyretin-mediated amyloidosis. Neurodegener Dis Manag. 2019 Feb;9((1)):5–23. doi: 10.2217/nmt-2018-0033. [DOI] [PubMed] [Google Scholar]
- 98.Sharma VK, Watts JK. Oligonucleotide therapeutics: chemistry, delivery and clinical progress. Future Med Chem. 2015;7((16)):2221–42. doi: 10.4155/fmc.15.144. [DOI] [PubMed] [Google Scholar]
- 99.Chabot S, Orio J, Castanier R, Bellard E, Nielsen SJ, Golzio M, et al. LNA-based oligonucleotide electrotransfer for miRNA inhibition. Mol Ther. 2012 Aug;20((8)):1590–8. doi: 10.1038/mt.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bernardo BC, Ooi JY, Lin RC, McMullen JR. miRNA therapeutics: a new class of drugs with potential therapeutic applications in the heart. Future Med Chem. 2015;7((13)):1771–92. doi: 10.4155/fmc.15.107. [DOI] [PubMed] [Google Scholar]
- 101.Ni S, Yao H, Wang L, Lu J, Jiang F, Lu A, et al. Chemical modifications of nucleic acid aptamers for therapeutic purposes. Int J Mol Sci. 2017 Aug 2;18((8)) doi: 10.3390/ijms18081683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Münch S, Wohlrab J, Neubert RHH. Dermal and transdermal delivery of pharmaceutically relevant macromolecules. Eur J Pharm Biopharm. 2017 Oct;119:235–42. doi: 10.1016/j.ejpb.2017.06.019. [DOI] [PubMed] [Google Scholar]
- 103.Goebel AS, Schmaus G, Neubert RH, Wohlrab J. Dermal peptide delivery using enhancer molecules and colloidal carrier systems: part I: carnosine. Skin Pharmacol Physiol. 2012;25((6)):281–7. doi: 10.1159/000341085. [DOI] [PubMed] [Google Scholar]
- 104.Neubert RHH, Sommer E, Schölzel M, Tuchscherer B, Mrestani Y, Wohlrab J. Dermal peptide delivery using enhancer moleculs and colloidal carrier systems. Part II: Tetrapeptide PKEK. Eur J Pharm Biopharm. 2018 Mar;124:28–33. doi: 10.1016/j.ejpb.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 105.Sommer E, Neubert RHH, Mentel M, Tuchscherer B, Mrestani Y, Wohlrab J. Dermal peptide delivery using enhancer molecules and colloidal carrier systems. Part III: Tetrapeptide GEKG. Eur J Pharm Sci. 2018 Nov 1;124:137–44. doi: 10.1016/j.ejps.2018.08.034. [DOI] [PubMed] [Google Scholar]
- 106.Liu Q, Lin Z, Liu Y, Du J, Lin H, Wang J. Delivery of miRNA-29b using R9-LK15, a novel cell-penetrating peptide, promotes osteogenic differentiation of bone mesenchymal stem cells. Biomed Res Int. 2019;2019:3032158. doi: 10.1155/2019/3032158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Abes R, Arzumanov AA, Moulton HM, Abes S, Ivanova GD, Iversen PL, et al. Cell-penetrating-peptide-based delivery of oligonucleotides: an overview. Biochem Soc Trans. 2007 Aug;35((Pt 4)):775–9. doi: 10.1042/BST0350775. [DOI] [PubMed] [Google Scholar]
- 108.Jamison J, Gilloteaux J, Nassiri MR, Tsai CC, Summers J. Antiviral activity of magnesium and magnesium/poly r(A-U) combinations against two RNA viruses. Nucleosides Nucleotides. 1999 Jun-Jul;18((6–7)):1221–2. doi: 10.1080/07328319908044668. [DOI] [PubMed] [Google Scholar]
- 109.Uchida T, Kanazawa T, Kawai M, Takashima Y, Okada H. Therapeutic effects on atopic dermatitis by anti-RelA short interfering RNA combined with functional peptides Tat and AT1002. J Pharmacol Exp Ther. 2011 Aug;338((2)):443–50. doi: 10.1124/jpet.111.180042. [DOI] [PubMed] [Google Scholar]
- 110.Uchida T, Kanazawa T, Takashima Y, Okada H. Development of an efficient transdermal delivery system of small interfering RNA using functional peptides, Tat and AT-1002. Chem Pharm Bull. 2011;59((2)):196–201. doi: 10.1248/cpb.59.196. [DOI] [PubMed] [Google Scholar]
- 111.Chen M, Zakrewsky M, Gupta V, Anselmo AC, Slee DH, Muraski JA, et al. Topical delivery of siRNA into skin using SPACE-peptide carriers. J Control Release. 2014 Apr 10;179:33–41. doi: 10.1016/j.jconrel.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cutler JI, Auyeung E, Mirkin CA. Spherical nucleic acids. J Am Chem Soc. 2012 Jan 25;134((3)):1376–91. doi: 10.1021/ja209351u. [DOI] [PubMed] [Google Scholar]
- 113.Ding L, Li W, Sun Q, He Y, Su B. Gold nanoparticles confined in vertically aligned silica nanochannels and their electrocatalytic activity toward ascorbic acid. Chemistry. 2014 Sep 26;20((40)):12777–80. doi: 10.1002/chem.201403426. [DOI] [PubMed] [Google Scholar]
- 114.Arias JL. Liposomes in drug delivery: a patent review (2007–present) Expert Opin Ther Pat. 2013 Nov;23((11)):1399–414. doi: 10.1517/13543776.2013.828035. [DOI] [PubMed] [Google Scholar]
- 115.Schwendener RA. Liposomes as vaccine delivery systems: a review of the recent advances. Ther Adv Vaccines. 2014 Nov;2((6)):159–82. doi: 10.1177/2051013614541440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Paliwal SR, Paliwal R, Vyas SP. A review of mechanistic insight and application of pH-sensitive liposomes in drug delivery. Drug Deliv. 2015 May;22((3)):231–42. doi: 10.3109/10717544.2014.882469. [DOI] [PubMed] [Google Scholar]
- 117.Merhautová J, Vychytilová-Faltejsková P, Demlová R, Slabý O. Systemic administration of miRNA mimics by liposomal delivery system in animal model of colorectal carcinoma. Physiol Res. 2016 Dec 21;65((Suppl 4)):S481–S88. doi: 10.33549/physiolres.933546. [DOI] [PubMed] [Google Scholar]
- 118.Yan Y, Li XQ, Duan JL, Bao CJ, Cui YN, Su ZB, et al. Nanosized functional miRNA liposomes and application in the treatment of TNBC by silencing Slug gene. Int J Nanomedicine. 2019;14:3645–67. doi: 10.2147/IJN.S207837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jiang L, Wang H, Chen S. Aptamer (AS1411)-conjugated liposome for enhanced therapeutic efficacy of miRNA-29b in ovarian cancer. J Nanosci Nanotechnol. 2020 Apr 1;20((4)):2025–31. doi: 10.1166/jnn.2020.17301. [DOI] [PubMed] [Google Scholar]
- 120.Xu J, Ding Y, Yang Y. Enhancement of mucosal and cellular immune response in mice by vaccination with respiratory syncytial virus DNA encapsulated with transfersome. Viral Immunol. 2008 Dec;21((4)):483–9. doi: 10.1089/vim.2008.0044. [DOI] [PubMed] [Google Scholar]
- 121.Jiang T, Wang T, Li T, Ma Y, Shen S, He B, et al. Enhanced transdermal drug delivery by transfersome-embedded oligopeptide hydrogel for topical chemotherapy of melanoma. ACS Nano. 2018 Oct 23;12((10)):9693–701. doi: 10.1021/acsnano.8b03800. [DOI] [PubMed] [Google Scholar]
- 122.Azimi M, Khodabandeh M, Deezagi A, Rahimi F. Impact of the transfersome delivered human growth hormone on the dermal fibroblast cells. Curr Pharm Biotechnol. 2019 Aug 9;20((14)):1194–202. doi: 10.2174/1389201020666190809120333. [DOI] [PubMed] [Google Scholar]
- 123.Jin Y, Wang S, Tong L, Du L. Rational design of didodecyldimethylammonium bromide-based nanoassemblies for gene delivery. Colloids Surf B Biointerfaces. 2015 Feb 1;126:257–64. doi: 10.1016/j.colsurfb.2014.12.032. [DOI] [PubMed] [Google Scholar]
- 124.Desmet E, Bracke S, Forier K, Taevernier L, Stuart MC, De Spiegeleer B, et al. An elastic liposomal formulation for RNAi-based topical treatment of skin disorders: Proof-of-concept in the treatment of psoriasis. Int J Pharm. 2016 Mar 16;500((1–2)):268–74. doi: 10.1016/j.ijpharm.2016.01.042. [DOI] [PubMed] [Google Scholar]
- 125.Gaur S, Wen Y, Song JH, Parikh NU, Mangala LS, Blessing AM, et al. Chitosan nanoparticle-mediated delivery of miRNA-34a decreases prostate tumor growth in the bone and its expression induces non-canonical autophagy. Oncotarget. 2015 Oct 6;6((30)):29161–77. doi: 10.18632/oncotarget.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Denizli M, Aslan B, Mangala LS, Jiang D, Rodriguez-Aguayo C, Lopez-Berestein G, et al. Chitosan nanoparticles for miRNA delivery. Methods Mol Biol. 2017;1632:219–30. doi: 10.1007/978-1-4939-7138-1_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tu L, Wang M, Zhao WY, Zhang ZZ, Tang DF, Zhang YQ, et al. miRNA-218-loaded carboxymethyl chitosan: tocopherol nanoparticle to suppress the proliferation of gastrointestinal stromal tumor growth. Mater Sci Eng C Mater Biol Appl. 2017 Mar 1;72:177–84. doi: 10.1016/j.msec.2016.10.052. [DOI] [PubMed] [Google Scholar]
- 128.Zhang X, Li K, Xing R, Liu S, Chen X, Yang H, et al. miRNA and mRNA expression profiles reveal insight into chitosan-mediated regulation of plant growth. J Agric Food Chem. 2018 Apr 18;66((15)):3810–22. doi: 10.1021/acs.jafc.7b06081. [DOI] [PubMed] [Google Scholar]
- 129.Hickerson RP, Flores MA, Leake D, Lara MF, Contag CH, Leachman SA, et al. Use of self-delivery siRNAs to inhibit gene expression in an organotypic pachyonychia congenita model. J Invest Dermatol. 2011 May;131((5)):1037–44. doi: 10.1038/jid.2010.426. [DOI] [PubMed] [Google Scholar]
- 130.Chen JS, Wu DT. Application of intronic microRNA agents in cosmetics. Methods Mol Biol. 2013;936:325–41. doi: 10.1007/978-1-62703-083-0_25. [DOI] [PubMed] [Google Scholar]
- 131.Freeley M, Derrick E, Dempsey E, Hoff A, Davies A, Leake D, et al. RNAi screening with self-delivering, synthetic siRNAs for identification of genes that regulate primary human T cell migration. J Biomol Screen. 2015 Sep;20((8)):943–56. doi: 10.1177/1087057115588288. [DOI] [PubMed] [Google Scholar]
- 132.Ligtenberg MA, Pico de Coaña Y, Shmushkovich T, Yoshimoto Y, Truxova I, Yang Y, et al. Self-delivering RNAi targeting PD-1 improves tumor-specific T cell functionality for adoptive cell therapy of malignant melanoma. Mol Ther. 2018 Jun 6;26((6)):1482–93. doi: 10.1016/j.ymthe.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]