Abstract
The COVID-19 pandemic negatively impacted the 2019/20 WHO European Region influenza surveillance. Compared with previous 4-year averages, antigenic and genetic characterisations decreased by 17% (3,140 vs 2,601) and 24% (4,474 vs 3,403). Of subtyped influenza A viruses, 56% (26,477/47,357) were A(H1)pdm09, 44% (20,880/47,357) A(H3). Of characterised B viruses, 98% (4,585/4,679) were B/Victoria. Considerable numbers of viruses antigenically differed from northern hemisphere vaccine components. In 2020/21, maintaining influenza virological surveillance, while supporting SARS-CoV-2 surveillance is crucial.
Keywords: surveillance, influenza virus, SARS-CoV-2, virus characterization, COVID-19, pandemic, Europe
The ending of the 2019/20 influenza season in the World Health Organization (WHO) European Region coincided with the start of the first wave of the coronavirus disease (COVID-19) pandemic. This study assesses potential impacts of the pandemic on influenza surveillance and presents characteristics of influenza viruses detected in the Region in 2019/20, relative to contemporary components of influenza vaccines for the northern hemisphere (NH).
Influenza virological surveillance in Europe, influenza season 2019/20
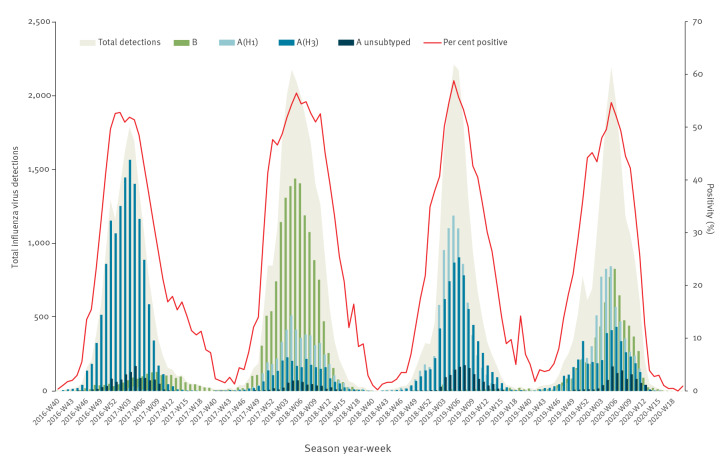
In the WHO European Region, the 2019/20 influenza season started in week 47 2019, peaked for 2 weeks, weeks 05 and 06 2020, and returned to baseline levels (< 10% positivity in sentinel samples) very rapidly in week 13 2020, following widespread public health and social measures implemented to control COVID-19 (Figure 1). Influenza type A viruses (120,493; 72.9%) dominated over type B (44,774; 27.1%). Of 47,357 subtyped influenza A viruses, 26,477 (56%) were A(H1)pdm09 and 20,880 (44%) were A(H3) viruses. The lineage of 4,679 B viruses was determined and 4,585 (98%) were B/Victoria lineage viruses [1,2].
Figure 1.
Number of positive sentinel specimens and positivity by week of reporting, week 40 2016 to week 20 2020, over four consecutive seasons, WHO European Region, 2016/17–2019/20
WHO: World Health Organization.
National Influenza Centres (NICs) in the Region collect influenza virological surveillance data, conduct genetic and antigenic characterisation of viruses and report to The European Surveillance System (TESSy) on a weekly basis. The WHO Collaborating Centres (WHO CC) in London and in Atlanta (at the Centers for Disease Control and Prevention (CDC)) provide NICs with post-infection ferret antisera or other antisera raised against egg and/or cell culture-propagated vaccine/reference viruses for antigenic characterisation or typing/subtyping using haemagglutination inhibition (HAI) assays. WHO CC London also provides a list of reference sequences for the assignment of viruses to haemagglutinin (HA) gene clades/subclades following Sanger or next generation sequencing (NGS) [3]. NICs share representative influenza-positive samples with the WHO CC for in depth antigenic and genetic analyses essential for decision-making at vaccine composition meetings (VCMs).
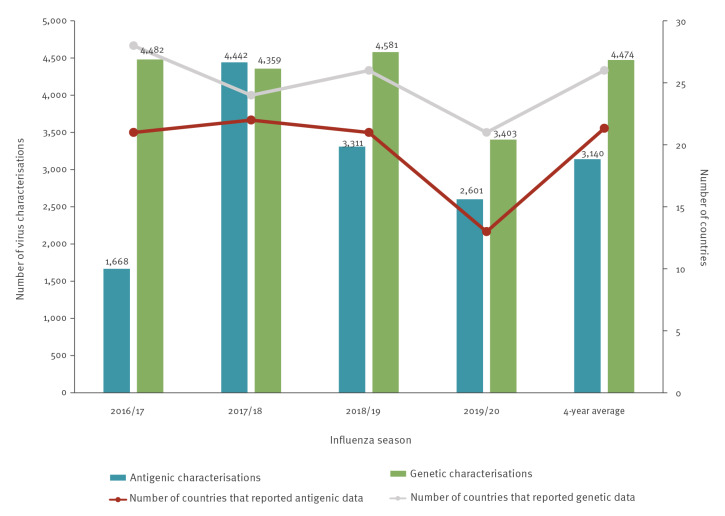
Fifty Member States of the WHO European Region reported 165,267 influenza virus detections between week 40 2019 through week 20 2020. Relative proportions of circulating influenza A(H3), A(H1)pdm09 and B/Victoria lineage viruses varied between countries [1,4]. Only 24 of the 50 countries reporting influenza detection data contributed virus characterisation data. Of all viruses detected, 2% (2,601/165,267) were antigenically and 2% (3,403/165,267) were genetically characterised ahead of the 2020 southern hemisphere (SH) VCM [1]. Virus characterisation data were used to determine the similarity of circulating viruses to the components of influenza vaccines for the 2019/20 NH influenza season and to assess implications of the COVID-19 pandemic on influenza surveillance and its output.
Influenza virus characterisation in the WHO European Region in light of the COVID-19 pandemic
The spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) occurred in March 2020, relatively late in the course of the influenza season in the Region, and the total number of influenza virus detections was comparable to previous seasons. However, the COVID-19 pandemic adversely affected the generation and reporting of virus characterisation data.
Compared with the previous 4-year averages, a lower number of countries contributed antigenic and genetic data in 2019/20 (13 and 21 vs 21 and 26, respectively) (Figure 2), and the number of antigenic and genetic characterisations decreased by 17% (2,601 vs 3,140) and 24% (3,403 vs 4,474) respectively (Figure 2). The most pronounced decrease was observed in the number of countries reporting antigenic characterisations, possibly reflecting reduced access to laboratory resources and equipment, biosafety concerns or pressure on human resources. Notably, virus characterisation reports effectively stopped in March 2020, and few influenza viruses were detected thereafter, whereas in previous years positive samples were collected and viruses characterised throughout the year.
Figure 2.
Number of countries reporting influenza virus characterisation data and number of influenza virus characterisations by season, WHO European Region, 2016/17–2019/20
WHO: World Health Organization.
Genetic and antigenic analysis of circulating influenza viruses, 2019/20
For specimens collected from week 40 2019 to 20 2020, genetic characterisation data of 3,403 viruses were reported to TESSy by 21 countries and antigenic characterisation data of 2,601 viruses by 13 countries. Table 1 and Table 2 provide the full list of numbers of viruses in each antigenic group and genetic clade, reporting category by week of sample collection.
Table 1. Antigenic characteristics of influenza viruses as reported to TESSy by week of sampling, WHO European Region, week 40 2019–week 20 2020 (n = 2,601 viruses characterised in 13 countries).
| Year and week |
Influenza B | Influenza A | TOTAL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yamagataa | Victoriab | H3 | H1 | |||||||||
| B/Phuket/3073/2013-likec | No category | B/Colorado/06/2017-liked | B/Washington/02/2019-likee | A/Switzerland/8060/2017-like | No category | A/Singapore/INF-16-0019/2016-like | A/South Australia/34/2019-likef | A/Kansas/14/2017-likeg | No category | A/Brisbane/02/2018-likeh | ||
| 2019 | ||||||||||||
| 40 | 0 | 0 | 1 | 3 | 0 | 0 | 1 | 0 | 2 | 0 | 2 | 9 |
| 41 | 0 | 0 | 3 | 0 | 0 | 1 | 0 | 0 | 3 | 0 | 1 | 8 |
| 42 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 6 | 0 | 3 | 10 |
| 43 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 9 | 0 | 3 | 15 |
| 44 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 13 | 0 | 4 | 20 |
| 45 | 0 | 0 | 0 | 5 | 1 | 1 | 1 | 0 | 13 | 0 | 11 | 32 |
| 46 | 0 | 0 | 4 | 11 | 2 | 3 | 1 | 1 | 10 | 1 | 8 | 41 |
| 47 | 0 | 0 | 2 | 12 | 0 | 2 | 0 | 1 | 64 | 0 | 8 | 89 |
| 48 | 0 | 0 | 6 | 13 | 1 | 0 | 1 | 1 | 66 | 3 | 10 | 101 |
| 49 | 0 | 1 | 6 | 9 | 0 | 2 | 1 | 1 | 90 | 7 | 15 | 132 |
| 50 | 0 | 0 | 12 | 22 | 4 | 6 | 1 | 0 | 75 | 6 | 37 | 163 |
| 51 | 0 | 0 | 25 | 27 | 0 | 4 | 2 | 0 | 116 | 11 | 35 | 220 |
| 52 | 1 | 0 | 10 | 9 | 1 | 4 | 6 | 0 | 12 | 5 | 22 | 70 |
| 2020 | ||||||||||||
| 1 | 0 | 0 | 2 | 7 | 2 | 2 | 1 | 1 | 2 | 10 | 34 | 61 |
| 2 | 0 | 0 | 8 | 13 | 0 | 3 | 0 | 1 | 17 | 12 | 51 | 105 |
| 3 | 0 | 0 | 23 | 51 | 0 | 0 | 2 | 0 | 24 | 14 | 72 | 186 |
| 4 | 0 | 0 | 21 | 20 | 0 | 0 | 5 | 0 | 36 | 3 | 105 | 190 |
| 5 | 1 | 0 | 29 | 25 | 0 | 0 | 11 | 0 | 63 | 17 | 94 | 240 |
| 6 | 0 | 0 | 26 | 23 | 0 | 0 | 10 | 0 | 53 | 18 | 75 | 205 |
| 7 | 0 | 0 | 16 | 24 | 0 | 0 | 23 | 0 | 39 | 24 | 63 | 189 |
| 8 | 0 | 0 | 28 | 8 | 0 | 0 | 12 | 0 | 26 | 14 | 63 | 151 |
| 9 | 0 | 0 | 24 | 6 | 0 | 0 | 5 | 0 | 50 | 7 | 69 | 161 |
| 10 | 0 | 0 | 15 | 3 | 0 | 0 | 1 | 0 | 33 | 11 | 36 | 99 |
| 11 | 0 | 0 | 6 | 5 | 0 | 0 | 2 | 0 | 16 | 5 | 22 | 56 |
| 12 | 0 | 0 | 8 | 1 | 0 | 1 | 2 | 0 | 5 | 3 | 10 | 30 |
| 13 | 0 | 0 | 5 | 0 | 0 | 0 | 1 | 0 | 4 | 2 | 5 | 17 |
| 14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 2 | 1 | 280 | 300 | 11 | 31 | 89 | 8 | 847 | 173 | 859 | 2,601 |
TESSy: The European Surveillance System; WHO: World Health Organization.
a Within influenza B Yamagata lineage, no viruses were reported as not belonging to a pre-defined antigenic category.
b For influenza B Victoria lineage, no viruses were reported as being B/Brisbane/60/2008-like.
c Vaccine component in quadrivalent both northern (2019/20 season) and southern (2020 season) hemispheres.
d Vaccine component for use in northern hemisphere 2019–2020 season.
e Vaccine component for the southern hemisphere 2020 season.
f Vaccine component for the southern hemisphere 2020 season.
g Vaccine component for the northern hemisphere 2019–2020 season.
h Vaccine component for both northern (2019–2020 season) and southern (2020 season) hemispheres.
Table 2. Genetic characteristics of influenza viruses as reported to TESSy by week of sampling, WHO European Region, week 40 2019–week 20 2020 (n = 3,403 viruses characterised in 21 countries).
| Year and week |
Influenza B | Influenza A | TOTAL | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yamagataa | Victoriaa | H1a | H3a | ||||||||||||||
| Subgroup not listed |
3b
_ B/ Phuket/3073/2013 |
Subgroup not listed |
1A(Δ162-164)Β_ B/ Washington /02/2019c |
No clade |
1Α(Δ162-163) _ B/ Colorado/06/2017d |
1A(Δ162-164) _ B/ Hong Kong/269/2017 |
Subgroup not listed |
6Β.1Α7 _ A/ Slovenia/1489/2019 |
6Β.1Α1 _ A/ Brisbane/02/2018e |
6Β.1Α5Α _ A/ Norway/3433 /2018 |
6Β.1Α5Β _ A/ Switzerland/3330/2018 |
3C.2a1b +T131K- B _ A/ South Australia/34/19f |
3C.3a _ A/ Kansas/14/2017g |
3C.2a1b +T135K _ A/ La Rioja/2202/2018 |
3C.2a1b +T135K-B _ A/ Hong Kong/2675/19 |
||
| 2019 | |||||||||||||||||
| 40 | 0 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 1 | 4 | 7 | 1 | 2 | 23 |
| 41 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 6 | 0 | 10 | 5 | 1 | 1 | 25 |
| 42 | 0 | 0 | 0 | 4 | 0 | 1 | 0 | 0 | 0 | 0 | 9 | 0 | 9 | 2 | 1 | 1 | 27 |
| 43 | 0 | 1 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 1 | 13 | 4 | 2 | 4 | 36 |
| 44 | 0 | 1 | 0 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 17 | 3 | 12 | 13 | 0 | 7 | 62 |
| 45 | 0 | 0 | 0 | 13 | 0 | 0 | 0 | 1 | 0 | 0 | 12 | 8 | 10 | 19 | 2 | 4 | 69 |
| 46 | 0 | 0 | 0 | 17 | 0 | 1 | 0 | 1 | 0 | 0 | 27 | 6 | 5 | 17 | 2 | 3 | 79 |
| 47 | 0 | 2 | 1 | 32 | 0 | 0 | 0 | 0 | 1 | 1 | 23 | 2 | 24 | 22 | 3 | 9 | 120 |
| 48 | 0 | 7 | 1 | 50 | 1 | 1 | 0 | 1 | 0 | 1 | 34 | 1 | 20 | 25 | 2 | 10 | 154 |
| 49 | 1 | 2 | 2 | 41 | 0 | 2 | 1 | 0 | 1 | 3 | 40 | 2 | 20 | 46 | 2 | 5 | 168 |
| 50 | 0 | 3 | 3 | 63 | 0 | 0 | 0 | 4 | 2 | 0 | 55 | 2 | 15 | 41 | 3 | 4 | 195 |
| 51 | 1 | 2 | 0 | 60 | 0 | 7 | 0 | 1 | 2 | 4 | 51 | 1 | 21 | 60 | 11 | 8 | 229 |
| 52 | 0 | 1 | 1 | 47 | 0 | 3 | 2 | 1 | 1 | 2 | 56 | 2 | 26 | 53 | 6 | 7 | 208 |
| 2020 | |||||||||||||||||
| 1 | 0 | 3 | 2 | 47 | 0 | 1 | 0 | 5 | 1 | 7 | 95 | 3 | 31 | 37 | 3 | 6 | 241 |
| 2 | 0 | 3 | 6 | 48 | 0 | 0 | 0 | 4 | 6 | 1 | 123 | 4 | 27 | 39 | 5 | 5 | 271 |
| 3 | 0 | 1 | 2 | 92 | 0 | 2 | 0 | 0 | 5 | 2 | 170 | 6 | 25 | 63 | 7 | 7 | 382 |
| 4 | 0 | 0 | 4 | 41 | 0 | 0 | 0 | 0 | 3 | 0 | 98 | 3 | 16 | 35 | 11 | 3 | 214 |
| 5 | 0 | 1 | 11 | 40 | 0 | 1 | 1 | 0 | 0 | 0 | 92 | 2 | 17 | 46 | 6 | 3 | 220 |
| 6 | 0 | 0 | 5 | 41 | 0 | 0 | 1 | 1 | 0 | 3 | 55 | 3 | 20 | 47 | 4 | 4 | 184 |
| 7 | 0 | 1 | 2 | 41 | 0 | 1 | 0 | 0 | 1 | 0 | 48 | 2 | 23 | 31 | 5 | 0 | 155 |
| 8 | 0 | 0 | 0 | 21 | 0 | 0 | 0 | 0 | 0 | 0 | 26 | 0 | 5 | 18 | 0 | 0 | 70 |
| 9 | 0 | 0 | 0 | 19 | 0 | 0 | 0 | 1 | 0 | 2 | 21 | 0 | 4 | 15 | 2 | 1 | 65 |
| 10 | 0 | 0 | 0 | 25 | 0 | 0 | 0 | 2 | 0 | 2 | 30 | 0 | 7 | 18 | 1 | 0 | 85 |
| 11 | 0 | 0 | 0 | 27 | 0 | 1 | 0 | 0 | 0 | 0 | 14 | 0 | 11 | 12 | 1 | 0 | 66 |
| 12 | 0 | 0 | 0 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 0 | 7 | 3 | 0 | 0 | 33 |
| 13 | 0 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 4 | 1 | 0 | 0 | 18 |
| 14 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| 15 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 16 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 18 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 2 | 28 | 40 | 819 | 1 | 22 | 5 | 22 | 23 | 28 | 1,121 | 52 | 386 | 679 | 81 | 94 | 3,403 |
TESSy: The European Surveillance System; WHO: World Health Organization.
a For influenza B viruses of the Yamagata lineage, no viruses were reported as not belonging to a predefined genetic clade (‘no clade’), while for influenza B viruses of the Victoria lineage, no viruses of clade 1A without amino-acid deletions, represented by the B/Brisbane/60/2008, were detected. For influenza A(H1) no viruses were reported as ‘no clade’ and no viruses were detected as being part of subclade 6B.1A6, represented by A/Ireland/84630/2018. For influenza A(H3) no viruses were reported with ‘no clade’ and no viruses had a ‘subgroup not listed’.
b Vaccine component in quadrivalent both northern (2019/20 season) and southern (2020 season) hemispheres.
c Vaccine component for the southern hemisphere 2020 season.
d Vaccine component for use in northern hemisphere 2019/20 season.
e Vaccine component for both northern (2019/20 season) and southern (2020 season) hemispheres.
f Vaccine component for the southern hemisphere 2020 season.
g Vaccine component for the northern hemisphere 2019/20 season.
Among A(H1)pdm09 viruses, of the 1,246 that were genetically characterised, 1,121 (90%) belonged to the 6B.1A5A group, moreover, of the 1,032 antigenically characterised, the majority (n = 859; 83%) were similar to the A/Brisbane/02/2018 vaccine virus. However, 173 A(H1)pdm09 viruses were not attributed to any predefined antigenic category, indicative of possible antigenic drift; of these viruses, genetic information was reported for only 48, nine of which had the HA1 N156K amino-acid substitution in antigenic site Sa. Overall, 16% (168/1,049) of A(H1)pdm09 viruses with genetic sequence information were 6B.1A5A-156K.
Of the 1,240 genetically characterised A(H3) viruses, the majority (n = 679; 55%) belonged to clade 3C.3a and were antigenically similar to the NH 2019/20 vaccine virus A/Kansas/14/2017. The remainder belonged to subclade 3C.2a1b and were antigenically distinct [5]. Of the 986 antigenically characterised viruses, most (n = 847, 86%) were characterised as A/Kansas/14/2017-like. The high proportion of viruses antigenically characterised as clade 3C.3a viruses probably reflects issues with characterisation of subclade 3C.2a1b viruses by HAI; 3C.2a1b viruses do not agglutinate red blood cells well and therefore were less tested with HAI [3,6,7].
Of 917 genetically characterised type B viruses, the B/Victoria-lineage accounted for 887 (97%), with 819 (92%) of these belonging to clade 1A(Δ162–164-B) and being antigenically distinct from the clade 1A(Δ162–163) vaccine virus B/Colorado/06/2017. Only 30/917 (3%) of type B viruses were assigned to the B/Yamagata-lineage, and 28 of these were assigned to clade 3, remaining antigenically similar to the B/Phuket/3073/2013 vaccine virus.
Ethical statement
An ethical approval was not needed for this study, as data are not identifiable back to the patients from whom they originated.
Discussion
Based on the data, influenza activity in the European Region appears to have ended abruptly in week 13 2020, earlier than previous seasons [1,8,9]. Responses to the COVID-19 pandemic, e.g. changes in access to and utilisation of healthcare and SARS-CoV-2 non-pharmaceutical control measures, such as school closures and social distancing, likely impeded continued surveillance and spread of influenza. This resulted in few influenza viruses being detected after week 13 2020 and, overall, fewer viruses being characterised, despite the obvious efforts from the laboratories under high pressure and overwhelming work load. Redirection of laboratory testing capacities to SARS-CoV-2, with shortages of laboratory supplies and human resources, could also explain the reduced level of influenza virus characterisation.
During the 2019/20 influenza season, the vast majority of influenza A(H3) viruses fell in genetic clade 3C.3a and subclade 3C.2a1b. The 3C.3a viruses were antigenically similar to the recommended 2019/20 NH vaccine virus, while 3C.2a1b viruses were antigenically distinct [5,6]. Most A(H1)pdm09 viruses fell in clade 6B.1A5A (90%) with the majority being antigenically similar to the vaccine virus. However, antigenically distinct viruses with HA1 N156K amino acid substitution were detected. Numbers of viruses in the 6B.1A5A-156K group increased rapidly in many countries simultaneously worldwide, notably in some of those that had A(H1)pdm09 epidemics, resulting in a change of the A(H1)pdm09 vaccine component for the SH 2021 influenza season [5,9]. Of the B/Victoria lineage viruses, accounting for 98% of type B viruses, the vast majority belonged to the Δ162–164-B triple deletion subgroup and were antigenically distinct from the vaccine virus.
Despite circulation of viruses antigenically distinct from vaccine components (i.e. A(H3) subclade 3C.2a1b, B/Victoria-lineage Δ162–164-B and A(H1)pdm09 6B.1A5A-156K viruses) during 2019/20, a moderately good overall level of vaccine effectiveness was observed, notably for type B and A(H1)pdm09 viruses [10]. The issue of poor recognition of circulating A(H3) viruses by immune responses to egg-propagated vaccine virus remained [11].
Data from the 2020 SH influenza season show that circulation of influenza viruses was extremely limited in the SH winter and also elsewhere for the NH inter-seasonal period [5,9,12]. Similar low levels of influenza might be expected in the WHO European Region in the 2020/21 season, if COVID-19-related public health measures are implemented. However, co-circulation of both influenza and SARS-CoV-2 viruses is possible, and should warrant resource-related and operational prioritisation efforts to ensure that continued evidence-based decisions can be made at WHO influenza VCMs. In either scenario, NICs will be challenged to ensure collection of representative specimens for influenza virus detection and subsequent virus characterisations with laboratory capacities being divided between influenza and SARS-CoV-2 surveillance [13]. The European Centre for Disease Prevention and Control (ECDC) and WHO Regional Office for Europe have issued joint interim guidance on what approaches should be used to maintain influenza surveillance during the winter period with the ongoing COVID-19 pandemic [13].
In terms of way forward, NICs play crucial roles in surveillance of seasonal influenza and zoonotic events and are responsible for arranging the essential shipments of representative specimens to the WHO CC to ensure there are sufficient data for making VCM recommendations. With increasing number of avian influenza outbreaks and continued evolution of influenza viruses in swine, there is also need for maintained vigilance in public health laboratories to ensure detection of zoonotic events for pandemic preparedness purposes [14,15]. In the 2020/21 season, efforts are needed to ensure maintenance of influenza surveillance, but also to support COVID-19 surveillance to understand SARS-CoV-2 transmission and inform national responses to the pandemic.
Acknowledgements
We express our gratitude to the TESSy data management team, especially Marius Valentin Valcu, for technical support. We would like to thank Andrew Amato for reviewing the manuscript, as well as the reviewers of the ‘Summary of influenza virus antigenic and genetic characterizations in Europe: data reported by National Influenza Centres to The European Surveillance System for weeks 40/2019 to week 20/2020’, for their valuable comments. We acknowledge all the members of the European region influenza surveillance network for their work on influenza surveillance data collection. We gratefully acknowledge the authors of the HA sequences retrieved from GISAID and used in this study. We would also like to acknowledge the physicians and nurses of sentinel network sites and intensive care units for their contribution in providing respiratory specimens. We would also like to thank the WHO CC for reference and research on influenza, Crick Worldwide Influenza Centre, The Francis Crick Institute, London, United Kingdom for their continuous tireless support to the WHO European Region influenza network.
Specific country acknowledgements
United Kingdom (England): We would like to acknowledge all staff of the Respiratory Virus Unit, National Infection Service, Public Health England
Finland: We would like to thank physicians and nurses of sentinel network sites and intensive care units for their contribution in providing respiratory specimens.
Czechia: Alexander Nagy, NRL for Influenza Centrum Epidemiology and Microbiology of National Institute of Public Health
Germany: Barbara Biere, Marianne Wedde
Greece: Maria Christoforidi
Italy: Angela Di Martino, Marzia Facchini, Giuseppina Di Mario, Antonino Bella (Istituto Superiore di Sanità, Rome), Caterina Rizzo (Bambino Gesù Children’s Hospital, Rome), the Laboratory Network for Influenza (InfluNet)
Ukraine: Larysa Radchenko, Nataliia Teteriuk, Liudmyla Leibenko, Olha Holubka
Spain: Jose María Navarro (Hospital Virgen de las Nieves, Granada), Ana Martínez-Sapiña (Hospital Miguel Servet, Zaragoza), Santiago Melón (Hospital Central de Asturias, Oviedo), Jordi Reina (Hospital Son Espases, Palma de Mallorca), Carmen Pérez (Hospital Dr Negrín, Las Palmas de Gran Canarias), Mónica Gozalo (Hospital Marqués de Valdecilla, Santander), José López-Barba (Hospital Universitario de Ceuta), Guadalupe Rodríguez (Hospital San Pedro de Alcántara, Cáceres), Sonia Pérez-Castro (Complejo Hospitalario Meixoeiro, Vigo), Miriam Blasco (Hospital San Pedro, Logroño), Antonio Moreno (Hospital Virgen de la Arrixaca, Murcia), Ana Navascués (Complejo Hospitalario de Navarra, Pamplona), Gustavo Cilla (Hospital Donostia, San Sebastián).
Sweden: Åsa Wiman, Public Health Agency of Sweden
Switzerland: We would like to thank the medical practitioners of the swiss sentinel network for their contribution in providing nasopharyngal specimens.
Portugal: We would like to acknowledge to all the members of the Médicos Sentinela network, Emergency rooms and obtstetrics network and to the National Laboratory network for influenza diagnosis (Hospitals). We would like to acknowledge all the colleagues of the laboratory and epidemiology Portuguese influenza team.
Romania: Odette Popovici and Rodica Popescu from National Institute Of Public Health. We are very grateful to all our colleagues from the Romanian local public health departments for sending specimens of suspected influenza cases.
WHO European Region Influenza Surveillance Network affiliations of authors
Austria: Monika Redlberger-Fritz, monika.redlberger@meduniwien.ac.at, National Influenza Centre for Austria, Center for Virology, Medical University Vienna, Judith Aberle, judith.aberle@meduniwien.ac.at, National Influenza Centre for Austria, Center for Virology, Medical University Vienna
Denmark: Ramona Trebbien, RATR@ssi.dk, National Influenza Center, Statens Serum Institut, Denmark, Jakob Nybo Nissen, JANN@ssi.dk, National Influenza Center, Statens Serum Institut, Denmark.
Finland: Niina Ikonen, Finnish Institute for Health and Welfare, Department of Health Security, niina.ikonen@thl.fi, Anu Haveri, Finnish Institute for Health and Welfare, Department of Health Security, anu.haveri@thl.fi
Germany: Ralf Dürrwald DuerrwaldR@rki.de
Greece: Georgia Gioula ggioula@auth.gr, National Influenza Centre for Northern Greece, Maria Exindari, mexidari@auth.gr National Influenza Centre for Northern Greece, Athanasios Kossyvakis, akossivakis@pasteur.gr, Hellenic Pasteur Institute, National Influenza Reference Laboratory of Southern Greece, Athens, Andreas Mentis, mentis@pasteur.gr, Hellenic Pasteur Institute, National Influenza Reference Laboratory of Southern Greece, Athens
Ireland: Linda Dunford, linda.dunford@ucd.ie, National Virus Reference Laboratory, University College Dublin, Ireland, Lisa Domegan, lisa.domegan@hse.ie, Health Service Executive, Health Protection Surveillance Centre, Dublin, Ireland.
Italy: Maria Rita Castrucci, mariarita.castrucci@iss.it, Istituto Superiore di Sanità, Department of Infectious Diseases, Rome, Italy, Simona Puzelli, simona.puzelli@iss.it, Istituto Superiore di Sanità, Department of Infectious Diseases, Rome, Italy
Latvia: Natalija Zamjatina, National Influenza Centre for Latvia, natalija.zamjatina@aslimnica.lv, Gatis Pakarna, National Influenza Centre for Latvia, gatis.pakarna@aslimnica.lv
Lithuania: Algirdas Griskevičius, algirdas.griskevicius@nvspl.lt, National Public Health Surveillance Laboratory, Asta Skrickiene, Centre for Communicable Diseases and AIDS, asta.skrickiene@ulac.lt
Luxembourg: Guillaume Fournier Guillaume.Fournier@lns.etat.lu, National Health Laboratory, Luxembourg, Joel Mossong Joel.Mossong@lns.etat.lu, National Health Laboratory, Luxembourg
Netherlands: Adam Meijer, adam.meijer@rivm.nl, National Institute for Public Health and the Environment, Bilthoven location of the Dutch National Influenza Centre, Bilthoven, The Netherlands, Ron Fouchier, r.fouchier@erasmusmc.nl, Erasmus University Medical Center, Rotterdam location of the Dutch National Influenza Centre, Rotterdam, The Netherlands
Norway: Karoline Bragstad, Karoline.Bragstad@fhi.no, Olav Hungnes, olav.hungnes@fhi.no, Norwegian Institute of Public Health
Portugal: Raquel Guiomar, Raquel.guiomar@insa.min-saude.pt, National Influenza Reference Laboratory, Infectious Diseases Department, National Institute of Health Dr Ricardo Jorge, Lisbon, Portugal, Pedro Pechirra, Pedro.Pechirra@insa.min-saude.pt, National Influenza Reference Laboratory, Infectious Diseases Department, National Institute of Health Dr Ricardo Jorge, Lisbon, Portugal
Romania: Mihaela Lazar, lazar.mihaela@cantacuzino.ro, National Influenza Center, “Cantacuzino” National Military-Medical Institute for Research and Development, Bucharest, Romania, Cherciu Carmen Maria, cherciu.maria@cantacuzino.ro National Influenza Center, “Cantacuzino” National Military-Medical Institute for Research and Development, Bucharest, Romania
The Russian Federation: Andrey Komissarov, a.b.komissarov@gmail.com, NIC, FSBI “Research Institute of Influenza named after A.A. Smorodintsev” Ministry of Health of the Russian Federation, St.-Petersburg, Daria Danilenko, Smorodintsev Research Institute of Influenza, daria.baibus@gmail.com. Elena Burtseva, NIC, FSBI “N.F. Gamaleya NRCEM” Ministry of Public Health of the Russian Federation, Moscow, the Russian Federation, elena-burtseva@yandex.ru
Slovakia: Elena Tichá, elena.ticha@uvzsr.sk, National Influenza Centre for Slovakia Edita Staronova, edita.staronova@uvzsr.sk, National Influenza Centre for Slovakia
Slovenia: Katarina Prosenc, katarina.prosenc@nlzoh.si, National Laboratory for Health, environment and food, Nataša Berginc, natasa.berginc@nlzoh.si, National Laboratory for Health, environment and food
Spain: Francisco Pozo, pacopozo@isciii.es, NIC-Madrid, Instituto de Salud Carlos III, Majadahonda Inmaculada Casas, icasas@isciii.es, NIC-Madrid, Instituto de Salud Carlos III, Majadahonda
Sweden: Mia Brytting, mia.brytting@folkhalsomyndigheten.se, Public Health Agency of Sweden
Switzerland: Ana Rita Gonçalves, AnaRita.GoncalvesCabecinhas@hcuge.ch, National Reference Centre for Influenza, Laboratory of Virology, Geneva University Hospitals, Geneva
Ukraine: Iryna Demchyshyna, irad@i.ua, National Influenza Centre for Ukraine
United Kingdom: Angie Lackenby, Angie.Lackenby@phe.gov.uk, National Infection Service, Public Health England, United Kingdom, Catherine Thompson, Catherine.Thompson@phe.gov.uk, National Infection Service, Public Health England, United Kingdom, Rory N Gunson, Rory.Gunson@ggc.scot.nhs.uk, West of Scotland Specialist Virology Centre, Scotland, Samantha J Shepherd, Samantha.Shepherd@ggc.scot.nhs.uk, West of Scotland Specialist Virology Centre, Scotland, Catherine Moore, catherine.moore2@wales.nhs.uk, Public Health Wales Specialist Virology Centre, Wales, Simon Cottrell, Simon.Cottrell@wales.nhs.uk Public Health Wales Communicable Disease Surveillance Centre and Vaccine Programme
European Centre for Disease Prevention and Control: Angeliki Melidou Angeliki.melidou@ecdc.europa.eu, Cornelia Adlhoch cornelia.adlhoch@ecdc.europa.eu, Oksana Martinuka oksanamartinyk@gmail.com, Pasi Penttinen Pasi.penttinen@ecdc.europa.eu, Lucia Pastore Celentano lucia.pastore.celentano@ecdc.europa.eu
WHO Regional Office for Europe: Dmitriy Pereyaslov pereyaslovd@who.int, Miriam Sneiderman sneidermanm@who.int, James Fielding James.Fielding@vidrl.org.au, Richard Pebody pebodyr@who.int
WHO Collaborating Centre for reference and research on influenza, Crick Worldwide Influenza Centre, The Francis Crick Institute, London, United Kingdom: John McCauley John.McCauley@crick.ac.uk, Rodney Daniels Rod.Daniels@crick.ac.uk
Conflict of interest: None declared.
Authors’ contributions: AM: Conceptualization, Methodology, Validation, Data curation, Formal Analysis, Visualisation, Writing original draft, Review and Editing, final approval.
DP: Methodology, Formal analysis, Writing, Review and Editing, final approval.
OH, KP, EA: Methodology, Validation, Data curation, Formal Analysis, Writing, Review and Editing.
CA, JF, MS, OM: Methodology, Data curation, Formal analysis, Writing, Review and Editing, final approval.
RP, LPC: Methodology, Validation, Data curation, Formal analysis, Writing, Review and Editing, final approval.
WHO European Region influenza surveillance network: Investigation, Resources, Data curation, Writing, Review and Editing, final approval.
References
- 1.European Centre for Disease Prevention and Control (ECDC) and World Health Organization. (WHO). FluNewsEurope. Joint ECDC-WHO/Europe weekly influenza update. Stockholm and Geneva: WHO and ECDC. [Accessed Oct 2020]. Available from: https://flunewseurope.org/
- 2. Mook P, Meerhoff T, Olsen SJ, Snacken R, Adlhoch C, Pereyaslov D, et al. Collective of the WHO European Region, European Influenza Surveillance Network Alternating patterns of seasonal influenza activity in the WHO European Region following the 2009 pandemic, 2010-2018. Influenza Other Respir Viruses. 2020;14(2):150-61. 10.1111/irv.12703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.European Centre for Disease Prevention and Control (ECDC) and WHO Regional Office for Europe. (WHO/Europe). European external quality assessment programme for influenza virus 2018. Stockholm and Copenhagen: ECDC and WHO/Europe; 2019. Available from: https://www.ecdc.europa.eu/sites/portal/files/documents/influenza-virus-external-quality-assessment-2018.pdf
- 4.European Centre for Disease Prevention and Control (ECDC). Influenza virus characterisation - Summary Europe, July 2020. Stockholm: ECDC; 2020. Available from: https://www.ecdc.europa.eu/en/publications-data/influenza-virus-characterisation-summary-europe-july-2020
- 5.Worldwide Influenza Centre (WIC), World Health Organisation Collaborating Centre for Reference and Research on Influenza. Report prepared for the WHO annual consultation on the composition of influenza vaccine for the Southern Hemisphere 2021. London: WIC; Sep 2020. Available from: https://www.crick.ac.uk/sites/default/files/2020-10/Crick_SH2021%20report_Full%20seasonal.pdf
- 6.Woldwide Influenza Centre (WIC), World Health Organization Collaborating Centre for Reference and Research on Influenza. Report prepared for the WHO annual consultation on the composition of influenza vaccine for the Northern Hemisphere 2019-2020. London: WIC; Feb 2019. Available from: https://www.crick.ac.uk/sites/default/files/2019-04/Crick%20VCMFeb2019%20report_toPost.pdf
- 7.European Centre for Disease Prevention and Control (ECDC). Influenza virus characterisation, Summary Europe, May 2019. Stockholm: ECDC; 2019. Available from: https://ecdc.europa.eu/sites/portal/files/documents/influenza-characterisation-report-May-2019.pdf
- 8.European Centre for Disease Prevention and Control (ECDC). Seasonal influenza 2019-2020. In: ECDC. Annual epidemiological report for 2019. Stockholm: ECDC; 2020. Available from: https://www.ecdc.europa.eu/sites/default/files/documents/AER_for_2019_influenza-seasonal.pdf
- 9.World Health Organization (WHO). Recommended composition of influenza virus vaccines for use in the 2021 southern hemisphere influenza season. Geneva: WHO; Sep 2020. Available from: https://www.who.int/influenza/vaccines/virus/recommendations/202009_recommendation.pdf?ua=1
- 10. Rose A, Kissling E, Emborg HD, Larrauri A, McMenamin J, Pozo F, et al. European IVE group Interim 2019/20 influenza vaccine effectiveness: six European studies, September 2019 to January 2020. Euro Surveill. 2020;25(10):2000153. 10.2807/1560-7917.ES.2020.25.10.2000153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woldwide Influenza Centre (WIC), World Health Organization Collaborating Centre for Reference and Research on Influenza. Report prepared for the WHO annual consultation on the composition of influenza vaccine for the Northern Hemisphere 2020-2021. London: WIC; Feb 2020. Available from: https://www.crick.ac.uk/sites/default/files/2020-03/Crick%20WIC%20Report%20Feb%202020.pdf
- 12. Olsen SJ, Azziz-Baumgartner E, Budd AP, Brammer L, Sullivan S, Pineda RF, et al. Decreased Influenza Activity During the COVID-19 Pandemic - United States, Australia, Chile, and South Africa, 2020. MMWR Morb Mortal Wkly Rep. 2020;69(37):1305-9. 10.15585/mmwr.mm6937a6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.European Centre for Disease Prevention and Control (ECDC) and WHO Regional Office for Europe (WHO/Euro). Operational considerations for influenza surveillance in the WHO European Region during COVID-19: interim guidance 2020. Stockholm and Copenhagen: ECDC and WHO/Euro; Oct 2020. Available from: https://www.ecdc.europa.eu/en/publications-data/operational-considerations-influenza-surveillance-european-region-during-covid-19#no-link
- 14.World Health Organisation Western Pacific (WHO Western Pacific). Avian Influenza Weekly Update Number 760. Manila: WHO Western Pacific; Nov 2020. Available from: https://www.who.int/docs/default-source/wpro---documents/emergency/surveillance/avian-influenza/ai-20200925.pdf?sfvrsn=30d65594_76
- 15.European Food Safety Authority (EFSA) and European Centre for Disease Prevention and Control. (ECDC), European Union Reference Laboratory for Avian Influenza (EURL). Avian influenza overview May – August 2020. Parma and Stockholm and Legnaro: EFSA and ECDC and EURL, 2020: Available from: https://www.ecdc.europa.eu/sites/default/files/documents/Avian-influenza-overview-May%E2%80%93August-2020.pdf