Abstract
Introduction
Although the heritability of sporadic Alzheimer’s disease (AD) is estimated to be 60–80%, addressing the genetic contribution to AD risk still remains elusive. More specifically, it remains unclear whether genetic variants are able to affect neurodegenerative brain features that can be addressed by in vivo imaging techniques.
Methods
Targeted sequencing analysis of the coding and UTR regions of 132 AD susceptibility genes was performed. Neuroimaging data using 11C-Pittsburgh Compound B positron emission tomography (PET), 18F-fluorodeoxyglucose PET, and MRI that are available from the KBASE (Korean Brain Aging Study for Early Diagnosis and Prediction of Alzheimer’s disease) cohort were acquired. A total of 557 participants consisted of 336 cognitively normal (CN) adults, 137 mild cognitive impairment (MCI), and 84 AD dementia (ADD) groups.
Results
We called 5391 high-quality single nucleotide variants (SNVs) on AD susceptibility genes and selected significant associations between variants and five in vivo AD pathologies: (1) amyloid β (Aβ) deposition, (2) AD-signature region cerebral glucose metabolism (AD-Cm), (3) posterior cingulate cortex (PCC) cerebral glucose metabolism (PCC-Cm), (4) AD-signature region cortical thickness (AD-Ct), and (5) hippocampal volume (Hv). The association analysis for common variants (allele frequency (AF) > 0.05) yielded several novel loci associated with Aβ deposition (PIWIL1-rs10848087), AD-Cm (NME8-rs2722372 and PSEN2-rs75733498), AD-Ct (PSEN1-rs7523) and, Hv (CASS4-rs3746625). Meanwhile, in a gene-based analysis for rare variants (AF < 0.05), cases carrying rare variants in LPL, FERMT2, NFAT5, DSG2, and ITPR1 displayed associations with the neuroimaging features. Exploratory voxel-based brain morphometry between the variant carriers and non-carriers was performed subsequently. Finally, we document a strong association of previously reported APOE variants with the in vivo AD pathologies and demonstrate that the variants exert a causal effect on AD susceptibility via neuroimaging features.
Conclusions
This study provides novel associations of genetic factors to Aβ accumulation and AD-related neurodegeneration to influence AD susceptibility.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13195-020-00722-2.
Keywords: Alzheimer’s disease, Targeted panel sequencing, Genetic association, Neuroimaging, In vivo AD pathologies, PET, MRI
Background
Alzheimer’s disease (AD), the most common cause of dementia, is a neurodegenerative disease with high heritability estimated to be 60–80% [1, 2]. Previous genetic studies have elucidated causative rare variants contributing to familial AD that commonly occurs before the age of 65 (early onset). These variants are found in genes related to amyloid beta (Aβ) synthesis, such as APP (amyloid precursor protein), PSEN1 (presenilin 1), and PSEN2 (presenilin 2) [3]. However, as early-onset familial AD accounts for < 5% of AD cases [2, 3], unraveling the complex genetic contributions to sporadic AD cases, which represents the majority of AD cases that occur after age 65 (late onset), is important. To date, apolipoprotein E (APOE) ε4 allele (APOE4) is the strongest genetic factor of sporadic AD [4], which confers a 3- to 15-fold increased risk of AD [5]. Recent large-scale genome-wide association studies (GWAS) have identified novel common variants of AD in loci involved in amyloid synthesis, nervous system development, synaptic transmission, and inflammation pathways [4, 6–9]. Nevertheless, APOE4 and several other common variants identified by previous GWAS explain a modest fraction of AD heritability [4], and addressing the remainder still remains as a challenge.
Most of the previous GWAS relied on phenotypes defined by clinical diagnosis of AD dementia (ADD) [8] and not by AD pathology or its surrogate biomarkers. Therefore, it remains unclear whether previously reported genes exert associations through modulating in vivo AD pathologies. Also, applying next-generation sequencing (NGS) technologies increases the possibility of detecting rare variants with large effect on diseases, which may help to explain the missing heritability [2, 10]. Although recent efforts to aggressively integrate genome-wide effects of common and rare variants has substantially improved our understanding of AD genetics [11, 12], how individual variants function to confer such effects still requires further investigation. A number of previous studies investigated genetic association with in vivo AD pathologies using AD biomarkers, but none has utilized multiple in vivo AD pathology features and integrated them with common and rare genetic variants [13–18].
Although genome-wide approaches enable unbiased screening of novel variants, we chose to perform a targeted sequencing (TS) approach as it provided the following advantages. First, overarching hypothesis of the study assumes that there are quantifiable contributions of imaging biomarkers on the known genetic signals for LOAD, thereby aiming to provide clues of variant functions. Therefore, we decided to focus our analyses on the selected 132 genes with previous associations with LOAD. Second, compared to genome-wide approaches, the number of data points in TS is substantially smaller and allows reduced statistical penalty. Lastly, TS reduces sequencing cost and allows easier data handling.
Based on an imaging genetics approach, this work aimed to identify common and rare genetic variants closely associated with cerebral Aβ deposition and AD-type neurodegeneration by conducting a TS analysis of 132 AD-related genes in 557 deep-phenotyped participants with multi-modal brain imaging information including [11C] Pittsburgh Compound B (PiB)-positron emission tomography (PET), [18F] fluorodeoxyglucose (FDG)-PET, and magnetic resonance imaging (MRI). Our multivariable analysis approach provides a more accurate model for AD susceptibility by comparing with biomarkers for in vivo AD pathology.
Materials and methods
Participants
We evaluated 557 participants who were recruited by the Korean Brain Aging Study for the Early Diagnosis and Prediction of Alzheimer’s disease (KBASE), a prospective cohort study initiated in 2014 (Supplementary Table S1) [13]. Participants consisted of 274 cognitively normal (CN) older adults (CN-old, age ≥ 55), 62 CN young and middle-aged adult (CN-ym, age < 55), 137 individuals with mild cognitive impairment (MCI), and 84 patients with ADD. Details on the recruitment and inclusion/exclusion criteria are described in the Supplementary Methods.
Evaluation of in vivo AD pathologies
All participants underwent comprehensive clinical and neuropsychological evaluations and multi-modal brain imaging (including [11C]PiB-PET, [18F]FDG-PET, and MRI). Blood was sampled for DNA extraction. APOE genotyping was conducted as previously described [18]. More detailed information on the standardized assessment used in the KBASE cohort is described in a previous report [13]. For surrogate markers of in vivo cerebral Aβ deposition, we quantified the standardized uptake value ratio (SUVR) of global [11C] PiB retention level in the cortical region-of-interests (ROIs). [18F]FDG-PET was used to measure cerebral glucose metabolism in the AD-signature regions (AD-Cm) and posterior cingulate cortex (PCC-cm), which can capture early regional metabolic and functional deficits related to AD processes [14, 15]. T1 MRI was used to evaluate cortical thickness in the AD-signature regions (AD-Ct) [19] and intracranial volume-adjusted hippocampal volume (Hv) [16] based on previous studies. Details of acquisition and preprocessing of [11C]PiB-PET, [18F]FDG-PET, and MRI and the definition of each AD imaging biomarker are provided in the Supplementary Methods and Supplementary Table S2.
AD gene panel, sequencing, and variant calling
The 132 genes were selected according to one of the following criteria: (1) genes from previous AD GWAS (n = 32) [6–9], (2) genes with mutations reported to be causative for familial AD in the OMIM database (n = 19), (3) genes with biological relevance in AD in the KEGG pathway (hsa05010) (n = 53), or (4) manually selected genes with potential relevance in AD (n = 28) (Supplementary Table S3). The AD gene panel was designed using the Ion AmpliSeq Designer (http://ampliseq.com) and contained 5049 amplicons with sizes ranging from 125 to 275 bp. The target regions included UTR and coding exons with 10 bases of padding sequences, totaling 1.46 Mb and covering 96% of the targeted intervals. AD-targeted panel sequencing was performed with an Ion Proton sequencer (Supplementary Methods, Supplementary Fig. S1 and Supplementary Table S4). The reads were aligned to hg19 and SNVs were called using Torrent Suite v3.4.2. Variants with < 20× coverage, located in homopolymer repeats, or covered < 90% of the samples were excluded. After applying the variant filtering criteria established in our pilot experiments (Supplementary Methods and Supplementary Fig. S1), 5391 variants remained.
Association tests
Both continuous and categorical variables indicating in vivo AD pathologies were normalized and used as dependent variables to identify common variants (allele frequency (AF) > 0.05) associated with bioimaging markers (Supplementary Methods and Supplementary Table S2). Age, sex, and the number of APOE4 alleles were used as covariates. Significant associations were defined at an uncorrected P < 0.001, following the previous studies [20–22], and the Benjamini and Hochberg method was used for multiple test correction for APOE variant association tests (Supplementary Table S5). Among the rare variants (AF < 0.05), loss-of-function (LoF) variants and missense variants with strong evolutionary conservation were selected for gene-based association analyses. R (version 3.5.1) was used to conduct Fisher’s exact test with the five binominal neuroimaging traits (Supplementary Table S2) and cognitive impairment, and rare variant enrichment across each gene was adjusted by length. For APOE variants, variants with AF > 0.1 were selected and tested for association, after adjusting for age and sex. For conditional analysis, a quantitative imaging trait was used as a covariate in an association test of APOE variants and AD susceptibility.
Voxel-based analysis of multi-modal brain imaging
To identify regional PiB uptake differences, Statistical Parametric Mapping 12 (SPM12; http://www.fil.ion.ucl.ac.uk/spm) was used for the exploratory voxel-based analysis of [11C]PiB-PET. Similarly, voxel-based analyses of [18F]FDG-PET and T1 MRI were performed to visualize the differences of regional cerebral glucose metabolism or gray matter (GM) density by the presence of variants associated with AD-Cm, PCC-Cm, AD-Ct, or Hv. Additional details of the voxel-wise analysis of common variants and genes with rare variants are provided in the Supplementary Methods.
Results
Establishment of the AD panel, quality assessment, and variant calling strategy
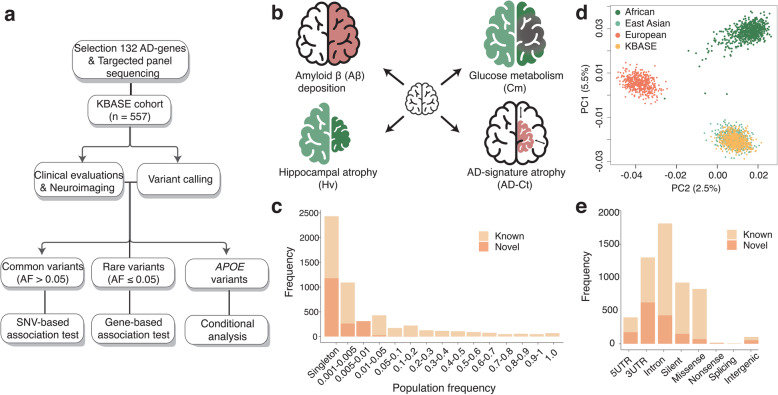
The overall study design and documented in vivo pathology biomarkers are represented in Fig. 1a, b. The clinical and demographic characteristics of the 557 participants are described in Supplementary Table S1. The mean age of the participants is 67.4 years, and 57.8% are women. The frequency of the APOE4 allele in each diagnostic group indicated a strong disease association (i.e., 9.3% and 35.7% for CN-old and dementia groups, respectively). The mean coverage depth of the AD target sequencing runs was 436.8×, and 97.8% of the targeted bases were covered more than 20×, ensuring high variant sensitivity (Supplementary Table S4).
Fig. 1.
Study design and variant profile of the cohort. a Flowchart of the study design. b Phenotyping strategies of in vivo AD pathology. c Principal component analysis (PCA) of the KBASE cohort with individuals from the 1000 Genomes Project individuals across different populations. d Distribution of variants in the KBASE cohort by population frequency. e Distribution of variants by genetic regions
We developed an in-house SNV calling method for ion proton sequencing data. Prior to data production, we optimized the variant calling pipeline through a series of pilot experiments and demonstrated 92.4% sensitivity and 93.5% specificity (Supplementary Fig. S1). To further validate the quality of the 5391 called variants, principal component analysis (PCA) of the KBASE cohort was performed with individuals from the 1000 Genomes Project, which showed that our cohort co-clustered with the East Asian group (Fig. 1c). A strong correlation of AFs between our cohort and 1000 Genomes East Asian was observed (Pearson’s r = 0.99 [95% CI, 0.989–0.991]). The majority of the variants (79.2%) are rare (AF < 0.05), and 45.2% were singletons (seen once in our cohort; Fig. 1d). The majority of the variants (88.9%) with an AF > 0.001 are already listed in the 1000 Genomes, The Genome Aggregation Database (gnomAD), and the Korean Variant Archive (KOVA) [23–25]. The variants are composed of 31.5% UTR, 1.9% intergenic, 33.7% intronic, and 32.9% coding region locations (Fig. 1e). Each individual carried an average of 369.0 heterozygous variants, 295.4 homozygous variants, and 4.4 singletons. The mean ratio of missense-to-silent variants was 0.44, and the mean ratio of transition-to-transversion variants was 2.83 per individual, comparable to other large-scale genome data sets [26].
Common variant association test
Tests that evaluated the association between common variants and Aβ deposition, AD-Cm, PCC-Cm, AD-Ct, and Hv revealed seven loci with strong associations, of which six are novel (Table 1). The CN-ym group was excluded from the test, as this group was entirely negative for the in vivo AD imaging biomarkers regardless of genotype.
Table 1.
List of common variants that significantly associated with brain imaging features (P < 1.0 × 10−3)
| AD imaging biomarker | Data type | Chr:position (hg19) | dbSNP ID | Genea | PFb | Previously reported | P |
|---|---|---|---|---|---|---|---|
| I. Cerebral amyloid-β accumulation measured by PiB-PET | |||||||
| Aβ deposition | Bin.c | chr3:39138840 | rs3732377 | GORASP1 | 0.237 | Novel | 9.32 × 10−4 |
| chr3:39139776 | rs1109643 | GORASP1 | 0.161 | Novel | 9.79 × 10−4 | ||
| chr3:39149277 | rs28362644 | GORASP1 | 0.162 | Novel | 7.02 × 10−4 | ||
| chr11:47345916 | rs2290149 | MADD | 0.082 | Novel | 2.02 × 10−4 | ||
| Quant.d | chr12:130839165 | rs10848087 | PIWIL1 | 0.105 | Novel | 5.05 × 10−4 | |
| II. Glucose metabolism levels measured by FDG-PET | |||||||
| AD-Cm | Bin. | chr1:227077809 | rs75733498 | PSEN2 | 0.083 | Novel | 1.75 × 10−4 |
| Quant. | chr7:37890267 | rs2722372 | NME8 | 0.191 | Novel | 7.63 × 10−4 | |
| PCC-Cm | Quant. | chr7:37890267 | rs2722372 | NME8 | 0.191 | Novel | 5.71 × 10−4 |
| III. Cortical thickness measured by MRI | |||||||
| AD-Ct | Bin. | chr14:73686944 | rs7523 | PSEN1 | 0.161 | Novel | 1.74 × 10−5 |
| Quant. | chr12:130839165 | rs10848087 | PIWIL1 | 0.105 | Novel | 2.94 × 10−4 | |
| IV. Hippocampal volume reduction measured by MRI | |||||||
| Hv | Quant. | chr20:55033476 | rs3746623 | CASS4 | 0.945 | Novel | 1.73 × 10−4 |
| chr20:55033647 | rs3746625 | CASS4 | 0.946 | Novel | 1.73 × 10−4 | ||
| chr20:55033713 | rs3746626 | CASS4 | 0.946 | Novel | 1.73 × 10−4 | ||
| chr20:55033856 | rs4811697 | CASS4 | 0.946 | Known [27] | 3.42 × 10−4 | ||
| Both | chr12:130839165 | rs10848087 | PIWIL1 | 0.105 | Novel | 4.24 × 10−4 | |
aThe most closely located gene from each variant
bPopulation frequency in the KBASE cohort
cCategorical variable trait transformed from normalized neuroimaging data by each criterion
dQuantitative normalized neuroimaging variable trait
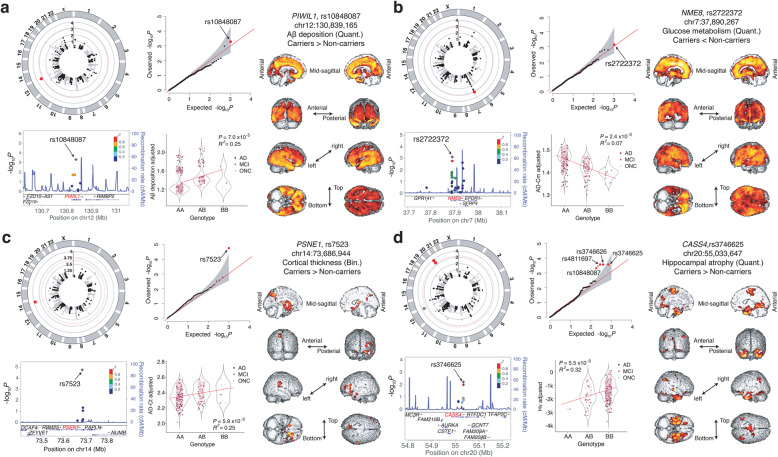
Variants in GORASP1 (rs28362644; odds ratio [OR] 0.46 [0.29–0.72]; P = 7.0 × 10−4), MADD (rs2290149; OR 2.74 [1.61–4.68]; P = 2.0 × 10−4), and PIWIL1 (rs10848087; regression coefficient [β] 0.14 [0.06–0.22]; P = 5.1 × 10−4) were significantly associated with Aβ deposition (Table 1; Fig. 2a; Supplementary Fig. S2a). Comparison with voxel-based imaging data (P < 0.01, k = 1497) revealed that carriers of MADD-rs2290149 displayed greater Aβ deposition mainly in the bilateral, lateral, and medial frontal cortices; cingulate; precuneus; and lateral temporal and inferior parietal regions. PIWIL1-rs10848087 non-carriers showed diffuse Aβ accumulation in the bilateral cerebral cortices including the frontal, temporal, parietal, occipital lobes and basal ganglia (Fig. 2a). Three SNVs near GORASP1 were identified, located in the same linkage disequilibrium (LD) block (R2 = 0.98; D′ = 0.99), and the most robust SNP, rs28362644, was used for imaging analysis (Table 1). GORASP1-rs28362644 carriers showed less Aβ deposition in the posterior hippocampal and parahippocampal regions, as well as retrosplenial and precuneus regions at a more lenient threshold compared to non-carriers (P < 0.05, k = 5058; Supplementary Fig. S2a).
Fig. 2.
Common variants that are significantly associated with neuroimaging features. For each signal, a circular Manhattan plot, quantile-quantile (Q-Q) plot, regional plot, regression plot with adjusted trait values, and voxel-based clustering analysis result are shown. a rs10848087 in PIWIL1 with cerebral Aβ deposition in global brain regions. b rs2722372 in NME8 with AD-Cm. c rs7523 in PSEN1 with AD-Ct. d rs4811697 in CASS4 with Hv
Two novel significant associations with AD-Cm were observed: PSEN2-rs75733498 (OR 0.39 [0.23–0.63]; P = 1.8 × 10−4) and NME8-rs2722372 (β − 0.04 [− 0.06 to − 0.018]; P = 7.6 × 10−4) (Table 1; Fig. 2b; Supplementary Fig. S2b). Subsequent voxel-based FDG-PET analysis revealed that PSEN2-rs75733498 non-carriers had reduced glucose metabolism levels in the bilateral fronto-temporo-parietal cortices (P < 0.01, k = 1497; Fig. 2b). On the other hand, NME8-rs2722372 carriers displayed diffuse cerebral glucose hypometabolism in the bilateral fronto-temporo-parietal cortices and subcortical structures, such as the thalamus and basal ganglia (Fig. 2b). An association between AD-Cm and NME8-rs2722372 in the PCC was also observed.
PSEN1-rs7523 (OR 0.39 [0.25–0.60]; P = 1.7 × 10−5) and PIWIL1-rs10848087 (β − 0.07 [− 0.11 to − 0.03]; P = 2.9 × 10−4) were associated with AD-Ct (Table 1; Fig. 2c; Supplementary Fig. S2c). Exploratory voxel-based analysis of T1 MRI demonstrated that PSEN1-rs7523 non-carriers showed reduced cortical thicknesses as compared to carriers in the inferior frontal, orbitofrontal, and basal ganglia (P = 2.5 × 10−5, k = 1497; Fig. 2d).
Finally, CASS4-rs3746625 (β 0.54 [0.26–0.82]; P = 1.7 × 10−4) and PIWIL1-rs10848087 (β − 0.40 [− 0.61 to − 0.19]; P = 2.3 × 10−4) strongly correlated with Hv (Table 1; Fig. 2d; Supplementary Fig. S2c). There are also three other significant SNVs in CASS4 that are located in the same LD block with rs3746625 (R2 = 1.00; D′ = 1.00). Brain images showed that GM atrophy was reduced in rs3746625 carriers than non-carriers, particularly in the bilateral medial temporal regions including the hippocampi (P < 0.01, k = 1497; Fig. 2d). To elucidate the functional implication of the CASS4 and PIWIL1 variants in the brain, ChIP-seq data from the ENCODE were utilized [28]. Notably, H3K27me3 signals, which mark heterochromatin, encompass the variants specifically in the hippocampus, but not in other parts of the brain or other organs, consistent with the results of voxel-based analysis of MRI (Supplementary Fig. S3).
The cumulative effect of common variants associated with the imaging biomarkers was evaluated by correlating AD risk score, defined as the weighted sum of the effect sizes of common variants associated with imaging features and the proportion of individuals with cognitive impairment (i.e., MCI or ADD; Supplementary Fig. S4). Common SNVs associated with each imaging biomarker (P < 0.05) explained 27–58% of cognitive impairment and 76% when used in combination.
Effect of APOE variants
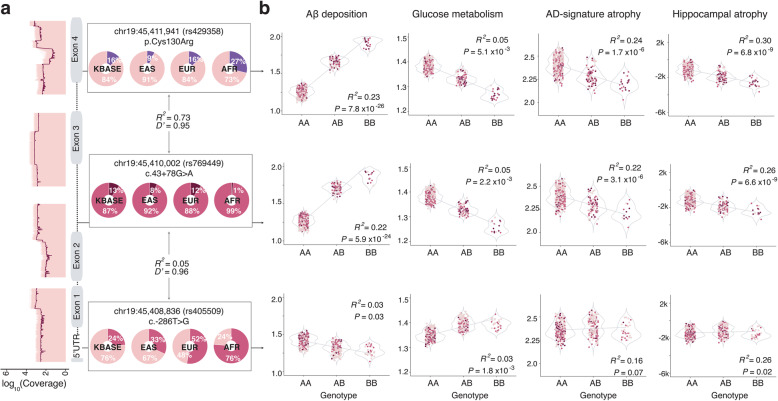
The APOE4 allele possesses the strongest association with AD susceptibility [8], but comprehensive analysis of its impact on structural and functional changes in a large number of aging brains remains understudied. In addition, our study design allowed association assessment of other APOE variants. Therefore, we sought to identify association between APOE common variants and the imaging features. Rare variants were not detected. We tested three common variants with an AF > 0.1. Two variants, rs429358 and rs769449, are located in the same LD block (R2 = 0.73, D′ = 0.95; Fig. 3a) and are strongly correlated with all five imaging traits (Table 2; Fig. 3b). An additional variant (rs405509) located in the promoter is in weak LD with rs429358 (R2 = 0.05, D′ = 1.00) and rs769449 (R2 = 0.06, D′ = 0.96; Fig. 3a). rs405509 possessed minimal association with the brain features (Table 2), with the association specifically residing in the 3′-portion of the gene.
Fig. 3.
Association of APOE variants with AD imaging biomarkers. a Log10-scaled coverage map of APOE in the KBASE cohort, along with the gene structure shown with gray boxes indicating exons. Black lines indicate the average coverage depths. On the right, AF of the three APOE variants in KBASE and major ethnic groups are displayed. b Regression plots for the three variant genotypes and imaging traits after adjusted with age and sex
Table 2.
Effects of common APOE SNVs on imaging biomarkers and conditional analysis of the SNVs controlling for each imaging biomarkers
| APOE SNP | Imaging biomarkers | Association (Quant.) | Cognitive function association | |||
|---|---|---|---|---|---|---|
| β | P | Padj | Unconditioned | Controlling for imaging biomarker (Quant.) | ||
| rs429358 (chr19:45411941) | Aβ deposition | 0.37 | 2.85 × 10−27 | 2.89 × 10−24 | 2.03 × 10−9 | 0.047 |
| AD-Cm | − 0.06 | 5.61 × 10−6 | 2.85 × 10−3 | 2.18 × 10−5 | ||
| PCC-Cm | − 0.10 | 3.07 × 10−10 | 3.12 × 10−7 | 3.16 × 10−3 | ||
| AD-Ct | − 0.11 | 8.70 × 10−13 | 8.80 × 10−10 | 2.82 × 10−3 | ||
| Hv | − 0.78 | 2.23 × 10−18 | 2.25 × 10−15 | 0.150 | ||
| rs769449 (chr19:45410002) | Aβ deposition | 0.39 | 1.51 × 10−25 | 7.65 × 10−23 | 7.29 × 10−8 | 0.260 |
| AD-Cm | − 0.06 | 2.78 × 10−6 | 2.82 × 10−3 | 3.08 × 10−3 | ||
| PCC-Cm | − 0.10 | 8.58 × 10−9 | 4.36 × 10−6 | 0.052 | ||
| AD-Ct | − 0.11 | 2.16 × 10−9 | 1.09 × 10−6 | 0.022 | ||
| Hv | − 0.73 | 3.72 × 10−13 | 1.88 × 10−10 | 0.210 | ||
| rs405509 (chr19:45408836) | Aβ deposition | − 0.09 | 5.78 × 10−3 | 0.84 | 1.94 × 10−3 | 0.020 |
| AD-Cm | 0.03 | 3.15 × 10−3 | 0.40 | 0.029 | ||
| PCC-Cm | 0.03 | 0.017 | 0.83 | 0.032 | ||
| AD-Ct | 0.02 | 0.16 | 0.99 | 0.011 | ||
| Hv | 0.14 | 0.089 | 0.96 | 0.017 | ||
Conditional analysis was performed to test if the influence of these APOE variants on neuroimaging features is causal to ADD risk (Table 2). Adjusting for each brain imaging feature effectively reduced the association between the two SNVs (rs429358 and rs769449) and AD susceptibility by approximately 105- to 108-fold in P values. This result suggests that the APOE variants modulate neuroimaging features that contributes to AD susceptibility. Conversely, the significance of another variant (rs405509) did not change after conditioning, indicating that brain imaging features are independent of its association with AD susceptibility.
Gene-level analysis for rare variants
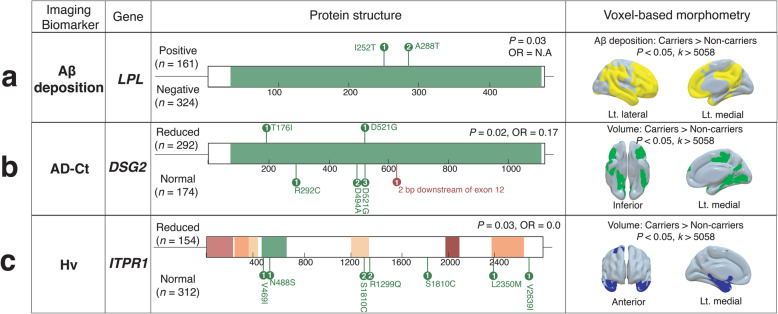
To investigate the associations of rare variants with AD imaging biomarkers and cognitive impairment, we collected functional rare variants (AF < 0.05, LoF or evolutionary conserved missense variants). A total of eight genes appeared to be significant at a gene-level analysis (P < 0.05; Fig. 4; Supplementary Table S6 and Supplementary Fig. S5). Variants in LPL were detected only in cases with high Aβ deposition (n = 3, P = 0.03; Fig. 4a). No rare variants were significantly associated with AD-Cm hypometabolism, but FERMT2 (OR 2.37 [1.07–5.61]; P = 0.02) and NFAT5 (OR 4.24 [1.15–23.32]; P = 0.02) showed significant enrichment for the presence of PCC hypometabolism (Table. S5). DSG2 showed significant enrichment in controls with normal AD-Ct and Hv (OR 0.17 [0.02–0.89], P = 0.02, and OR 0.0 [0.00–1.02], P = 0.03; Fig. 4b; Supplementary Fig. S5a). Interestingly, the observed rare variants were mainly located in the sites that bind calcium ions or other proteoglycans, such as N-acetylglucosamine (Supplementary Fig. S6c). Also, functional variants in ITPR1 were observed only in controls with normal Hv (n = 9; Fig 4c). These results demonstrate novel associations between rare variants and brain features that can be documented by imaging techniques.
Fig. 4.
Genes with rare variants that are significantly associated with in vivo AD pathologies. Observed rare functional variants in the case or control groups defined by each clinical parameter are shown for each gene. a LPL with Aβ deposition in the global brain regions. b DSG2 with AD-signature cortical thickness. c ITPR1 with hippocampal volume. The right panel displays the exploratory voxel-based analyses of brain imaging to demonstrate the regional pattern differences in AD imaging biomarker phenotype between carriers and non-carriers
Discussion
We investigated the associations between genetic variants and various brain features that are accessible via imaging techniques. We identified a group of variants that are linked to Aβ deposition, AD-Cm, PCC-cm, AD-Ct, and Hv (Fig. 1b). Our results bolster the role of APOE alleles in contributing to AD pathogenesis susceptibility.
The association analysis of common variants led to the discovery of seven significant associations with AD brain imaging biomarkers. Particularly, we observed an association between CASS4 common variants and Hv (Table 1). Previous association studies suggested that CASS4, encoding a cytoskeletal protein that provides a physical connection with the extracellular matrix, contributes to the risk of ADD [8, 27]. Our observation implies that the previously reported association between CASS4 and ADD may be through neurodegeneration of the hippocampus via an Aβ-independent pathway. Voxel-wise analysis of MRI demonstrated a region-specific GM atrophy in the bilateral hippocampi and adjacent regions in CASS4 variant carriers (Fig. 2d). The presence of hippocampus-specific H3K27me3 signals (Supplementary Fig. S3a) suggests that these variants are related to a region-specific epigenetic modification in the hippocampus, although future studies will be necessary to clarify the function of these CASS4 variants.
PIWIL1 common variants are related to both Aβ deposition and atrophy in the AD-signature region and the hippocampus (Table 1 and Supplementary Table S7). Voxel-wise analysis of PiB-PET consistently revealed that PIWIL1 variant carriers displayed greater levels of cerebral Aβ deposition as compared to non-carriers, and regional GM atrophy mainly in the bilateral temporal and PC-PCC areas and medial and inferior frontal regions, which were reported to be vulnerable to the AD process [19]. PIWIL1 is a member of the PIWI subfamily that plays important roles in stem cell self-renewal, RNA silencing, translational regulation, and neuron development [29], which potentially implicates multiple aspects of brain degeneration. Moreover, the PIWIL1 variants also co-localize with a brain-specific H3K27me3 signal (Supplementary Figure S3b), which may provide insight into the underlying function of PIWIL1 in neurodegeneration of AD-vulnerable regions. Additionally, significant associations between common variants in PSEN1 and PSEN2 and neurodegeneration biomarkers of AD (i.e., PSEN1-rs7523 with AD-Ct and PSEN2-rs75733498 with AD-Cm) were observed. Pathogenic rare variants in these genes cause early-onset familial AD. These genes encode major components of the γ-secretase of APP synthesis and proteolysis, leading to Aβ production [30]. It is not clear how these variants are associated with AD-type neurodegeneration, but this observation demonstrates that dysregulation of the genes confers susceptibility to AD via alterations of regional cerebral glucose metabolism or regional cortical thickness, which demonstrates their versatile roles in AD pathogenesis beyond altering Aβ production.
Unlike common or rare variants with association, we found associations between APOE variants and all brain imaging features of AD. This finding is consistent with previous reports [31–35], conferring a partial explanation for the genetic link between APOE and AD susceptibility. An association test of a variant in the other LD block revealed a marginal association, suggesting a specific functional association between the cell surface receptor binding region and the imaging features. Conditional analysis demonstrated a causal relationship between the imaging features and the association of APOE variants with AD susceptibility (Table 2).
The gene-level rare variant association test revealed several genes previously related to AD brain features with cognitive impairment. Notably, carriers of DSG2 rare variants harbor increased AD-Ct, Hv, and cognitive function, suggesting that these variants play protective roles in neurodegeneration in AD-related regions (Fig 4b and Supplementary Fig. S5a). DSG2 encodes a calcium-binding glycoprotein components of the desmosome that binds to plaque proteins and intermediate filaments [36] and was reported to confer AD risk based on GWAS [8]. Although a molecular link explaining this association remains unclear, we observed that the variants were located in calcium ion or proteoglycan binding sites (Supplementary Fig. S5c). These sites are evolutionally conserved, and the binding of these molecules is essential for protein function [37].
The KBASE protocol ensures unified subject assessment and standardization of all samples and data collection, processing, and quality control. It enables multi-trait analysis of genetic loci, explicitly targeting each in vivo AD pathology rather than relying on clinical diagnoses with limited accuracy.
Strengths and limitations
All individuals enrolled in the KBASE cohort underwent careful clinical and genetic characterization. The KBASE protocol adopts unified subject assessment, standardization of all imaging, biofluid, DNA and RNA data collection, and processing followed by meticulous data quality control. We analyzed variants of wide range of allele frequencies on AD-associated genes. Therefore, 132 AD-associated genes for LOAD were covered for genetic variant associations with the AD brain imaging features. Our next step will be to scrutinize the molecular biological mechanisms of variants and their functions on AD pathology.
Several limitations of our study warrant discussions. First, we tried to replicate our results using the ADNI database, which includes unified clinical information, brain imaging data, and genomic data of participants. We adjusted raw brain imaging data of ADNI database using our methodology and conducted association analyses. However, replication was not feasible due to the allele frequency differences originated from the ethnic discrepancies and differences in phenotyping methods, except the APOE SNVs. We also collected a large-scale elderly Korean cohort (n = 4683) and conducted a sequencing analysis using our customized targeted LOAD gene panel. Associations between FDG uptake and two novel SNPs were replicated—rs75733498 on PSEN2 (P = 1.6 × 10−4) and rs2722372 on NME8 (P = 8.6 × 10−3)—although this cohort lacks brain imaging assessment and cognitive impairment trait was accessible.
Conclusions
Here, we explored the KBASE data and conducted integrative analysis that revealed novel associations between AD-related gene variants and various brain features. Thus, scrutinizing the biological mechanisms of genetic variants and their functions in AD pathology by interrogating multiple aspects of brain morphology and in vivo AD pathologies will lead to a better understanding of the mechanism of AD pathogenesis.
Supplementary Information
Additional file 1. Supplementary methods, tables, and figures.
Acknowledgements
We thank the participants and members of the KBASE consortium. Professor Han Buhm provided us with critical comments.
The coinvestigators of the KBASE Research Group are listed elsewhere (http://kbase.kr).
Authors’ contributions
Conceptualization: MC, IM-J, and DYL. Formal analysis: JS and MSB. Methodology: JS and MC. Genome data acquisition: JS, J-CP, S-HH, JB, IM-J, and MC. Clinical data acquisition: MSB, DY, JHL, SYJ, SAS, YKK, KMK, C-HS, GJ, and DYL. Supervision: MC and DYL. Writing—original draft: JS and MC. Writing, review, and editing: JS, MSB, DYL, and MC. The authors read and approved the final manuscript.
Funding
The study was supported by grants from the National Research Foundation of Korea (2014M3C7A1046049 and 2018M3C9A5064708 for Choi M and 2014M3C7A1046042 for Lee DY) and grants from the Ministry of Health and Welfare of Korea (HI18C0630 for Mook-Jung IH and Lee DY, and HI19C0149 for Lee DY).
Availability of data and materials
All data needed to evaluate the conclusions in the paper are present in the paper. Additional data related to this paper may be requested from the authors.
Ethics approval and consent to participate
The Institutional Review Boards of Seoul National University Hospital and SMG-SNU Boramae Medical Center approved the study (1401-027-547 and 26-2015-60). This study was conducted in accordance with the recommendations of the current version of the Declaration of Helsinki. All participants and/or their legal representatives provided written informed consent.This action is appropriate.
Consent for publication
All authors consented for the publication of the manuscript.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jieun Seo and Min Soo Byun contributed equally to this work.
Contributor Information
Dong Young Lee, Email: selfpsy@snu.ac.kr.
Murim Choi, Email: murimchoi@snu.ac.kr.
References
- 1.Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63(2):168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- 2.Chouraki V, Seshadri S. Chapter five - Genetics of Alzheimer’s disease. In: Friedmann T, Dunlap JC, Goodwin SF, editors. Advances in Genetics. 87: Academic Press; 2014. p. 245–94. [DOI] [PubMed]
- 3.Bekris LM, Yu CE, Bird TD, Tsuang DW. Genetics of Alzheimer disease. J Geriatr Psychiatry Neurol. 2010;23(4):213–227. doi: 10.1177/0891988710383571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu CC, Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9(2):106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278(16):1349–1356. [PubMed] [Google Scholar]
- 6.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43(5):429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45(12):1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303(18):1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bis JC, Jian X, Kunkle BW, Chen Y, Hamilton-Nelson KL, Bush WS, et al. Whole exome sequencing study identifies novel rare and common Alzheimer's-Associated variants involved in immune response and transcriptional regulation. Mol Psychiatry. 2020;25(8):1859–75. [DOI] [PMC free article] [PubMed]
- 11.Khera AV, Chaffin M, Wade KH, Zahid S, Brancale J, Xia R, et al. Polygenic prediction of weight and obesity trajectories from birth to adulthood. Cell. 2019;177(3):587–596. doi: 10.1016/j.cell.2019.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wainschtein P, Jain DP, Yengo L, Zheng Z, Cupples LA, Shadyab AH, et al. Recovery of trait heritability from whole genome sequence data. bioRxiv; 2019. 10.1101/588020. [DOI]
- 13.Byun MS, Yi D, Lee JH, Choe YM, Sohn BK, Lee JY, et al. Korean brain aging study for the early diagnosis and prediction of Alzheimer’s disease: methodology and baseline sample characteristics. Psychiatry Investig. 2017;14(6):851–863. doi: 10.4306/pi.2017.14.6.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jack CR, Jr, Albert MS, Knopman DS, McKhann GM, Sperling RA, Carrillo MC, et al. Introduction to the recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):257–262. doi: 10.1016/j.jalz.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jack CR, Jr, Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, et al. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol. 2010;9(1):119–128. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jack CR, Jr, Wiste HJ, Weigand SD, Rocca WA, Knopman DS, Mielke MM, et al. Age-specific population frequencies of cerebral beta-amyloidosis and neurodegeneration among people with normal cognitive function aged 50-89 years: a cross-sectional study. Lancet Neurol. 2014;13(10):997–1005. doi: 10.1016/S1474-4422(14)70194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reiman EM, Chen K, Liu X, Bandy D, Yu M, Lee W, et al. Fibrillar amyloid-beta burden in cognitively normal people at 3 levels of genetic risk for Alzheimer’s disease. Proc Natl Acad Sci U S A. 2009;106(16):6820–6825. doi: 10.1073/pnas.0900345106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wenham PR, Price WH, Blandell G. Apolipoprotein E genotyping by one-stage PCR. Lancet. 1991;337(8750):1158–1159. doi: 10.1016/0140-6736(91)92823-k. [DOI] [PubMed] [Google Scholar]
- 19.Wirth M, Villeneuve S, Haase CM, Madison CM, Oh H, Landau SM, et al. Associations between Alzheimer disease biomarkers, neurodegeneration, and cognition in cognitively normal older people. JAMA Neurol. 2013;70(12):1512–1519. doi: 10.1001/jamaneurol.2013.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arfanakis K, Wilson RS, Barth CM, Capuano AW, Vasireddi A, Zhang S, et al. Cognitive activity, cognitive function, and brain diffusion characteristics in old age. Brain Imaging Behav. 2016;10(2):455–463. doi: 10.1007/s11682-015-9405-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyall DM, Cox SR, Lyall LM, Celis-Morales C, Cullen B, Mackay DF, et al. Association between APOE e4 and white matter hyperintensity volume, but not total brain volume or white matter integrity. Brain Imaging Behav. 2020;14(5):1468-76. [DOI] [PMC free article] [PubMed]
- 22.Xu G, Zheng S, Zhu Z, Yu X, Jiang J, Jiang J, et al. Association of tau accumulation and atrophy in mild cognitive impairment: a longitudinal study. Ann Nucl Med. 2020;34(11):815–23. [DOI] [PubMed]
- 23.Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee S, Seo J, Park J, Nam JY, Choi A, Ignatius JS, et al. Korean Variant Archive (KOVA): a reference database of genetic variations in the Korean population. Sci Rep. 2017;7(1):4287. doi: 10.1038/s41598-017-04642-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Raskin L, Samuels DC, Shyr Y, Guo Y. Genome measures used for quality control are dependent on gene function and ancestry. Bioinformatics. 2015;31(3):318–323. doi: 10.1093/bioinformatics/btu668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D’Aoust LN, Cummings AC, Laux R, Fuzzell D, Caywood L, Reinhart-Mercer L, et al. Examination of candidate exonic variants for association to Alzheimer disease in the Amish. PLoS One. 2015;10(2):e0118043. doi: 10.1371/journal.pone.0118043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sloan CA, Chan ET, Davidson JM, Malladi VS, Strattan JS, Hitz BC, et al. ENCODE data at the ENCODE portal. Nucleic Acids Res. 2016;44(D1):D726–D732. doi: 10.1093/nar/gkv1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao PP, Yao MJ, Chang SY, Gou LT, Liu MF, Qiu ZL, et al. Novel function of PIWIL1 in neuronal polarization and migration via regulation of microtubule-associated proteins. Mol Brain. 2015;8:39. doi: 10.1186/s13041-015-0131-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delabio R, Rasmussen L, Mizumoto I, Viani GA, Chen E, Villares J, et al. PSEN1 and PSEN2 gene expression in Alzheimer’s disease brain: a new approach. J Alzheimers Dis. 2014;42(3):757–760. doi: 10.3233/JAD-140033. [DOI] [PubMed] [Google Scholar]
- 31.Cacciaglia R, Molinuevo JL, Falcon C, Brugulat-Serrat A, Sanchez-Benavides G, Gramunt N, et al. Effects of APOE-epsilon4 allele load on brain morphology in a cohort of middle-aged healthy individuals with enriched genetic risk for Alzheimer’s disease. Alzheimers Dement. 2018;14(7):902–912. doi: 10.1016/j.jalz.2018.01.016. [DOI] [PubMed] [Google Scholar]
- 32.Kantarci K, Lowe V, Przybelski SA, Weigand SD, Senjem ML, Ivnik RJ, et al. APOE modifies the association between Abeta load and cognition in cognitively normal older adults. Neurology. 2012;78(4):232–240. doi: 10.1212/WNL.0b013e31824365ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Tan L, Wang HF, Liu Y, Hao XK, Tan CC, et al. Multiple effect of APOE genotype on clinical and neuroimaging biomarkers across Alzheimer’s disease spectrum. Mol Neurobiol. 2016;53(7):4539–4547. doi: 10.1007/s12035-015-9388-7. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Yu JT, Wang HF, Han PR, Tan CC, Wang C, et al. APOE genotype and neuroimaging markers of Alzheimer’s disease: systematic review and meta-analysis. J Neurol Neurosurg Psychiatry. 2015;86(2):127–134. doi: 10.1136/jnnp-2014-307719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mishra S, Blazey TM, Holtzman DM, Cruchaga C, Su Y, Morris JC, et al. Longitudinal brain imaging in preclinical Alzheimer disease: impact of APOE epsilon4 genotype. Brain. 2018;141(6):1828–1839. doi: 10.1093/brain/awy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Apostolova LG, Risacher SL, Duran T, Stage EC, Goukasian N, West JD, et al. Associations of the top 20 Alzheimer disease risk variants with brain amyloidosis. JAMA Neurol. 2018;75(3):328–341. doi: 10.1001/jamaneurol.2017.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harrison OJ, Brasch J, Lasso G, Katsamba PS, Ahlsen G, Honig B, et al. Structural basis of adhesive binding by desmocollins and desmogleins. Proc Natl Acad Sci U S A. 2016;113(26):7160–7165. doi: 10.1073/pnas.1606272113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary methods, tables, and figures.
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper. Additional data related to this paper may be requested from the authors.