Abstract
Background
Elevated plasma or serum troponin, indicating perioperative myocardial injury (PMI), is common after noncardiac surgery. However, underlying mechanisms remain unclear. Acute coronary syndrome (ACS) is associated with the early appearance of circulating microRNAs, which regulate post-translational gene expression. We hypothesised that if PMI and ACS share pathophysiological mechanisms, common microRNA signatures should be evident.
Methods
We performed a nested case control study of samples obtained before and after noncardiac surgery from patients enrolled in two prospective observational studies of PMI (postoperative troponin I/T>99th centile). In cohort one, serum microRNAs were compared between patients with or without PMI, matched for age, gender, and comorbidity. Real-time polymerase chain reaction quantified (qRT–PCR) relative microRNA expression (cycle quantification [Cq] threshold <37) before and after surgery for microRNA signatures associated with ACS, blinded to PMI. In cohort two, we analysed (EdgeR) microRNA from plasma extracellular vesicles using next-generation sequencing (Illumina HiSeq 500). microRNA-messenger RNA-function pathway analysis was performed (DIANA miRPath v3.0/TopGO).
Results
MicroRNAs were detectable in all 59 patients (median age 67 yr [61–75]; 42% male), who had similar clinical characteristics independent of developing PMI. In cohort one, serum microRNA expression increased after surgery (mean fold-change) hsa-miR-1-3p: 3.99 (95% confidence interval [CI: 1.95–8.19]; hsa-miR-133-3p: 5.67 [95% CI: 2.94–10.91]; P<0.001). These changes were not associated with PMI. Bioinformatic analysis of differentially expressed microRNAs from cohorts one (n=48) and two (n=11) identified pathways associated with adrenergic stress and calcium dysregulation, rather than ischaemia.
Conclusions
Circulating microRNAs associated with cardiac ischaemia were universally elevated in patients after surgery, independent of development of myocardial injury.
Keywords: microRNA, noncardiac surgery, perioperative period, perioperative myocardial injury, postoperative complications
Editor's key points.
-
•
Elevated troponin I, indicating perioperative myocardial injury, is common after noncardiac surgery by unclear mechanisms.
-
•
The authors hypothesised that perioperative myocardial injury and acute coronary syndrome share pathophysiological mechanisms and common microRNA signatures.
-
•
Serum microRNAs were compared between patients with or without perioperative myocardial injury in a nested control study of samples obtained before and after noncardiac surgery.
-
•
Circulating microRNAs associated with cardiac ischaemia were elevated in patients after surgery, independent of myocardial injury.
-
•
These microRNA changes after noncardiac surgery may indicate a generalised cardiac stress response with myocardial injury promoted by deficient cardioprotective mechanisms.
Perioperative myocardial injury (PMI), as defined by elevated plasma troponin after noncardiac surgery, occurs in up to 25% of patients1 and precedes subsequent noncardiac morbidity.2 Preoperative computerised tomography coronary angiogram shows that PMI does not appear to be associated with the severity of pre-existing coronary artery disease.3 These data suggest that there are likely to be several pathophysiologic mechanisms that contribute to asymptomatic elevations in troponin after surgery.4
Acute coronary syndrome (ACS) is associated with the release of microRNAs from cardiac tissue, frequently before increases in plasma troponin are detectable.5 MicroRNAs are small noncoding RNA molecules (approximately 23 nucleotides) that modulate gene expression through the degradation of messenger RNA and/or inhibit messenger RNA translation.6 MicroRNAs regulate cardiomyocyte differentiation and survival, calcium regulation, apoptosis, conduction, inflammation, and necrosis.7 MicroRNAs also regulate intercellular communication via extracellular vesicles released from the surface of the cell during activation and/or exosomes, which are smaller luminal vesicles (30–100 nm in diameter) originating from intracellular endosomes.8 MicroRNA-1, -21, -146, -133, -208, and -499 are associated with acute myocardial infarction and ACS in humans,5,9 although meta-analyses find wide heterogeneity between studies.5 Nevertheless, microRNAs -499, -1-3, and -21 have been proposed as biomarkers that may identify patients with ACS even more rapidly than high-sensitivity troponin.10 Thus, profiling circulating microRNA in patients undergoing elective noncardiac surgery may provide genomic signatures that add mechanistic insight into PMI.11,12
We hypothesised that if PMI and ACS share pathophysiological mechanisms, then microRNA profiles should be similar in both. Our specific objective was to assess whether the sub-group of patients with postoperative troponin concentrations indicative of myocardial injury, but who did not sustain myocardial infarction, share the same microRNA profile as seen in acute myocardial ischaemia. We therefore quantified expression of microRNAs in samples of serum and plasma extracellular vesicles obtained before and after noncardiac surgery in patients who developed PMI, compared with matched controls.
Methods
Study populations
This was a nested case control study of samples obtained from UK patients enrolled in two separate studies of myocardial injury in noncardiac surgery, specifically excluding patients with clinically confirmed myocardial infarction and/or pulmonary embolus, those receiving renal replacement therapy, and participants with elevated troponin-I (TnI) >99th centile before surgery. Both studies were conducted in accordance with the principles of the Declaration of Helsinki and the Research Governance Framework. The Measurement of Exercise Tolerance before Surgery (METS) study was approved by the UK Medical Research Ethical Committee (MREC: 13/LO/0135). Six hundred and twenty patients were recruited into the UK arm of the METS study cohort between March 1, 2013 and March 25, 2016.13 The second study, approved by the UK Medical Research Ethical committee MREC:16/LO/06/35, recruited 189 patients at University College London Hospital from October 5, 2016 to January 14, 2019.14 The protocols, including original inclusion/exclusion criteria, have been published but are also provided in the Supplementary data.
Study 1: METS serum microRNA study
Inclusion/exclusion criteria
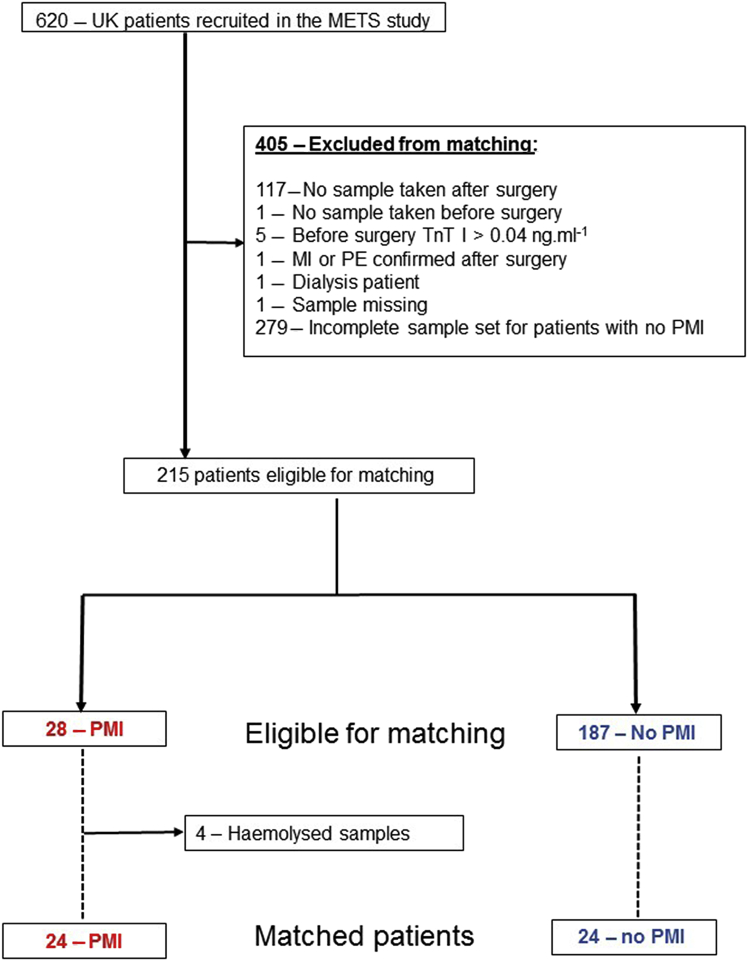
Inclusion criteria: PMI was defined as TnI >99th centile (>0.04 ng ml−1; Troponin-I Centaur CP Assay, Siemens, Tarrytown, NY, USA). Patients with TnI>99th centile after surgery were matched with patients in whom TnI remained ≤99th centile after surgery. Samples were matched by age, sex, Revised Cardiac Risk Index (RCRI) score and day after surgery. All patients with confirmed PMI had an electrocardiogram reviewed by an independent adjudication committee. We excluded patients with clinically confirmed myocardial infarction and/or pulmonary embolus, those receiving renal replacement therapy and participants with elevated TnI >99th centile before surgery (Fig 1).
Fig 1.
METS serum microRNA study flow diagram, illustrating patient/sample selection.
METS study sample preparation
Samples were obtained once before surgery and then again on each day after surgery until postoperative day 3. Whole blood (10 ml) was collected in a plain tube, allowed to clot at room temperature before centrifugation (3500 × g for 10 min). Serum was then aliquoted into two 2-ml RNA-free tubes. The two aliquoted samples were then frozen at -70°C and subsequently used separately for troponin quantification and microRNA analysis.
METS study microRNA isolation
Detailed methods and references are provided in the Supplementary data. In brief, 200 μl serum samples were screened for red discolouration (haemolysis) before isolation using the miReasy Serum/Plasma Advanced Kit (Qiagen, København, Denmark) in accordance with the manufacturer's instructions. For quality control, 1 μl of spike-in mixture containing UniSp2, UniSp4, and UniSp5 synthetic microRNAs (RNA Spike-in kit, Qiagen Copenhagen, Denmark) was added to each sample (Supplementary data), and 20 μl RNA was isolated using RNeasy UCP MiniElute columns and stored at -20°C for subsequent batched analysis.
Circulating microRNAs
We selected microRNA-1, -21, -146, -133, -208, and -499 as established reported microRNA signatures for ACS (Supplementary Table). Meta-analysis of microRNA discovery data has highlighted these microRNAs as potential biomarkers of myocardial infarction.15 The isoforms selected were chosen from the clinical studies that had isolated microRNA from plasma or serum. The nomenclature was crosschecked using the miRBase database16 (Supplementary material).
Quantitative real time polymerase chain reaction (RT–PCR)
Detailed methods are provided in the Supplementary material. Two microlitres of RNA were reversed transcribed in a 10 ul final reaction volume using the miRcury LNA RT kit (Qiagen). In accordance with manufacturer's recommendation, serum microRNA amounts are standardised by starting sample volume rather than RNA concentration. The reactions were performed in a Primus 96 plus thermal cycler (MWG Biotech, Ebersberg, Germany) for 60 min at 42°C, followed by incubation (inactivation of reaction) at 95°C for 5 min and then allowed to cool at 4°C. cDNA was then stored at -20°C. For qPCR cDNA was diluted (1:40) and added to a master mix before plating on miRNA PCR custom-made plates containing predesigned miRCURY LNA miRNA PCR Assays in triplicate (Supplementary data). Amplification was performed using the Applied Biosystem StepOneplus RT–PCR system (Applied Biosystems, Foster City, CA, USA). Cycle quantification (Cq) values were determined using the second derivative method (Applied Biosystem StepOneplus software).
Quality control
Cq values were calibrated using an interplate calibrating control assay, UniSp3 (Qiagen), pre-aliquoted into each plate. The calibrated Cq data generated from the spike controls were used to compare each sample and identify outlier samples to be excluded from analysis. If the Cq values of the technical control produce similar values for each sample, isolation efficiency across all samples can be considered consistent. For this study, UniSp5 was used as the isolation technical control. If the Cq value was ≥37, then the sample and patient was flagged as an outlier and excluded from analysis. The synthetic RNA UniSp6 was added to the RT reaction before cDNA synthesis. The outlier Cq values for UniSp6 were used to evaluate RT efficiency. Both hsa-miR-23a and hsa-miR-451 were used to detect haemolysis, which interferes with miR detection.17 Samples that breached published limits of detection for haemolysis were excluded. Amplification efficiency was calculated (LinRegPCR software, Version 2017.1; Heart Failure Research Centre, Amsterdam, Netherlands)18 and individual melt curves for each PCR reaction were inspected (Supplementary data).
Cq normalisation and microRNA expression
The Qiagen miRcury LNA miRNA Serum/Plasma Focus Panel was used to determine two reference gene(s) for this study as recommended by consensus guidelines, since there are no universal reference microRNAs defined for microRNA expression studies.19 The focus panel has 179 microRNA assays commonly found in the serum and plasma. The NormFinder algorithm20 was used to determine the most suitable reference microRNA (Supplementary material). MicroRNA miR-152-3p and hsa-miR-361-5p had the lowest combined stability value (0.077) and were selected as reference microRNA. Expression concentrations were compared using the relative Cq method, where microRNA Cq values were normalised to the geometric mean of the reference microRNA Cq values to give ΔCq values. The ΔCq values were converted to a fold change (FC) by applying the formula of 2−ΔCq.21
Study 2: microRNA concentrations in extracellular vesicles
Extracellular vesicles
To examine microRNA content in extracellular vesicles, a separate nested cohort of patients with troponin release and matched controls were assessed. MicroRNA content in extracellular vesicles requires the preparation of larger volumes of plasma which required specific different preparation to the samples obtained in METS. Postoperative plasma samples were prepared from 11 patients who underwent elective noncardiac surgery.14 PMI was defined as plasma high-sensitivity troponin T assay ≥99th centile (≥15 ng L−1) within 2 days of surgery (Cobas Assay; Roche Diagnostics, Mannheim, Germany). At 48 h after surgery whole blood was collected into ethylenediaminetetraacetic acid (EDTA) anticoagulant bottles, centrifuged at 3500 × g for 10 min (Cepr-AL6 Flexicool Rota; Capricorn Laboratory Equipment, Fordingbridge, UK). The plasma layer was sterile filtered (0.8 μm membrane filter; Starlabs, Hamburg, Germany) and transferred to Eppendorf tubes. The samples were immediately stored at -80°C until batch processing. At the same time blood samples were collected, an ECG was reviewed independently to ensure no patient with myocardial infarction was included.
MicroRNA isolation from extracellular vesicles
Plasma samples were shipped to Exiqon (Vedbaek, Denmark) to isolate microRNA in extracellular vesicles and exosomes for next generation sequencing, masked to troponin values and clinical details. Quality control checks ensured high isolation efficiency and lack of haemolysis17 (Supplementary material). Library preparation was done using the NEBNext® Small RNA Library preparation kit (New England Biolabs, Ipswich, MA, USA). The library pool was sequenced on a NextSeq 500 sequencing instrument (Illumina, San Diego, CA, USA). Raw data were de-multiplexed and FASTQ files for each sample were generated using the bcl2fastq software (Illumina). The microRNA sequence reads (FASTQ) files were then mapped to the Homo sapiens genome (GRCh37) and the microRNA sequence database, MiRBase V20, to generate microRNA sequence counts using Bowtie 2 (Version 2.2.4.) software.22 MicroRNA counts were normalised for library size using the trimmed mean of M (TMM) method and differential expression of TMM analysis performed using the EdgeR statistical software package.23
Gene pathway analysis
We used pathway analysis software to identify related proteins within a pathway to infer biological mechanisms likely to be involved with myocardial injury. This approach enables unbiased identification of potential biological pathways associated with the microRNA changes measured. Pathway analysis helps to understand or interpret genomic data from canonical prior knowledge thereby identifying distinct cellular processes, diseases, or signalling pathways that are statistically associated with the selection of differentially expressed genes between compared conditions. For cohort 1, we used DNA Intelligent Analysis (DIANA)-miRPath v.3.0 pathway analysis software to assess in an unbiased manner which transcriptomic processes may be altered by microRNAs (microT-CDS v5.0).24
For cohort 2, differentially expressed microRNA were used to perform gene ontology enrichment analysis using topGO.25
Statistical analysis
Clinical characteristics of the patients were stratified according to the presence or absence of troponin elevation >99th centile values. Categorical data are summarised as absolute values (%). Continuous data are presented as mean (standard deviation [sd]) or median (inter-quartile range [IQR]) unless stated otherwise. ΔCq values were checked for normality using the Shapiro–Wilk test. For microRNA assays that were expressed in >95% across all samples, ΔCq values were analysed using a multi-level (mixed error-component) model, examining the presence/absence of PMI (>99th centile) and sampling time (before or after surgery). Fold-change in circulating serum microRNA expression were correlated with log TnI values after surgery (linear regression).26
Individual comparisons between groups were calculated using post hoc Bonferroni tests. For microRNA expressed in <95% samples, McNemar χ2 analysis was performed. P<0.05 was considered statistically significant. All statistical analyses were undertaken using NCSS 12 (Kaysville, UT, USA). Data have been deposited in NCBI's Gene Expression Omnibus27 and are accessible through GEO Series accession number GSE144777.
Sample size calculation
Receiver operating characteristic (ROC) curve analysis has shown that miR-499, miR-1-3 and miR-21 can be used to identify patients with ACS, area under the curve (AUC)=0.89.10 If these microRNAs were present and/or elevated in patients at the time of detecting PMI, >11 patients would be required to detect a difference in relative expression serum microRNA after surgery (α=0.01; 1-β=0.9). The sample size calculation was performed using the pROC package (R software, v3.4.3).28
Results
Cohort 1: METS serum microRNA study
Patient characteristics
Twenty-four patients identified from METS who developed myocardial injury (median TnI: 0.08 ng ml−1 [IQR: 0.05–0.13 ng ml−1]) were clinically similar to 24 matched patients in whom troponin remained within normal limits (Table 1).
Table 1.
Patient characteristics for METS study cohort.
| Whole cohort n=48 | No PMI n =24 | PMI n=24 | P-value | |
|---|---|---|---|---|
| Troponin concentration ng·ml−1 | ≤0.04 | 0.08 (0.05–0.13) | <0.001 | |
| Clinical characteristics | ||||
| Age (yr) | 67 (61–75) | 67 (60–74) | 66 (61–76) | 0.154 |
| Male sex (n; %) | 22 (45.8%) | 11 (45.8%) | 11 (45.8%) | 1.000 |
| Body mass index (kg m−2) | 27 (5) | 27 (5) | 27 (5) | 0.900 |
| Before surgery eGFR (ml min−1 1.73 m2) | 87 (76–96) | 84 (72–94) | 90 (79–103) | 0.192 |
| Haemoglobin (g dl−1) | 130 (22) | 128 (16) | 131 (26) | 0.520 |
| Co-morbidities (n; %) | ||||
| RCRI≥2 | 4 (8.3) | 2 (8.3) | 2 (8.3) | 1.000 |
| Coronary artery disease | 5 (10.4) | 3 (12.5) | 2 (8.3) | 0.637 |
| Congestive heart failure | 0 (0.0) | 0 (0.0) | 0 (0.0) | - |
| Cerebrovascular disease | 1 (2.1) | 1 (4.2) | 0 (0.0) | 0.312 |
| Diabetes mellitus | 10 (20.8) | 5 (20.8) | 5 (20.8) | 1.000 |
| Peripheral vascular disease | 1 (2.1) | 0 (0.0) | 1 (4.2) | 0.312 |
| Hypertension | 24 (50.0) | 11 (45.8) | 13 (54.2) | 0.564 |
| Obstructive lung disease# | 5 (10.4) | 3 (12.5) | 2 (8.3) | 0.636 |
| Significant malignancy∗ | 34 (70.8) | 18 (75.0) | 16 (66.7) | 0.524 |
| Preoperative medications (n; %) | ||||
| Beta-blocker | 6 (12.5) | 4 (16.7) | 2 (8.3) | 0.383 |
| Calcium channel blocker | 5 (10.4) | 3 (12.5) | 2 (8.3) | 0.637 |
| ARB/ACE-I | 19 (39.5) | 8 (33.3) | 11 (45.8) | 0.376 |
| Aspirin | 8 (16.7) | 4 (16.7) | 4 (16.7) | 1.000 |
| Surgical procedure type (n; %) | ||||
| Intraperitoneal or retroperitoneal | 30 (62.5) | 17 (70.8) | 13 (54.2) | 0.223 |
| Urological or gynaecological | 15 (31.3) | 6 (25.0) | 9 (37.5) | 0.350 |
| Orthopaedic | 3 (6.2) | 1 (4.2) | 2 (8.3) | 0.551 |
| Anaesthesia type (n; %) | ||||
| General anaesthesia alone | 24 (50.0) | 13 (54.2) | 11 (45.8) | 0.564 |
| General + regional anaesthesia | 24 (50.0) | 11 (45.8) | 13 (54.2) | 0.564 |
Data presented as mean (sd) for parametric data and as median (25th–75th inter-quartile range, IQR) for non-parametric data. Frequencies are presented with percentages (%). Age is rounded to the nearest year. Perioperative myocardial injury (PMI) was defined as troponin I (TnI) >99th centile (>0.04 ng·ml−1) after surgery. eGFR: estimated glomerular filtration rate; RCRI: Revised Cardiac Risk Index score; ACE-I: angiotensin converting enzyme inhibitor; ARB: angiotensin receptor blocker. Other units as indicated.
Previous diagnosis of asthma, reactive airways disease, chronic obstructive lung disease, chronic bronchitis, or emphysema.
Indication for surgery was for treatment of cancer.
Circulating microRNA expression
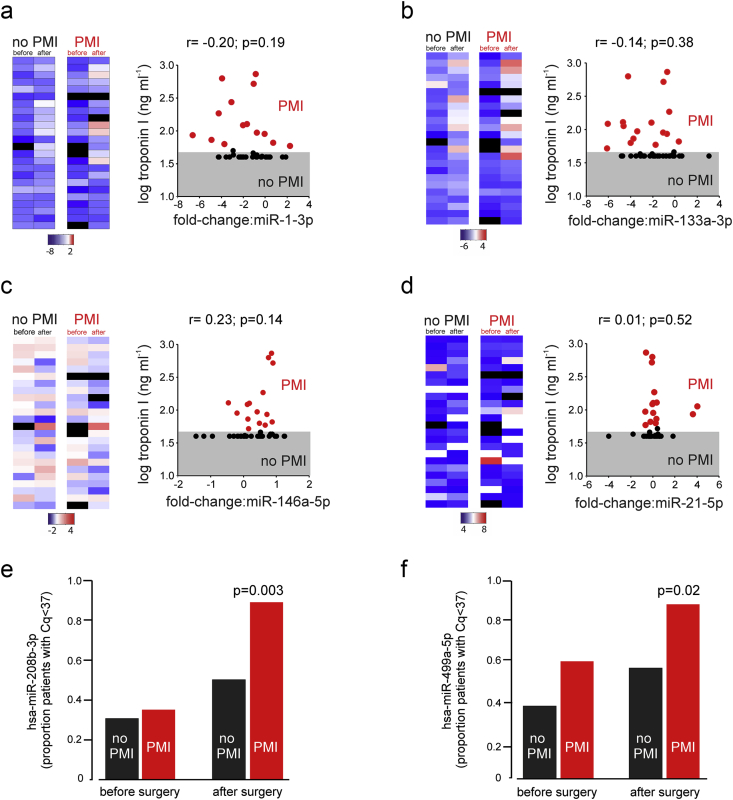
After surgery, circulating microRNA expression concentrations in serum increased for hsa-miR-1-3p (mean fold-change: 3.99 (95% confidence interval [CI]: 1.95–8.19); F1,47.5=24.16; P<0.001) and hsa-miR-133-3p (fold-change: 5.67 (95% CI: 2.94–10.91); F1,42=31.13; P<0.001) but these changes did not correlate with postoperative troponin elevation (Fig 2a and b; Supplementary Fig 1). Expression concentrations of hsa-miR146a-5p decreased after surgery only in patients with elevated troponin after surgery (F1,34.5=6.43, P=0.03; Fig 2c). There was no change in expression for hsa-miR-21-5p after surgery (Fig 2d). In patients with troponin elevation after surgery, hsa-miR-208b-3p was detected in 20/22 (91%) patients, compared with 12/24 (50%) patients with troponin concentrations below the 99th centile after surgery (odds ratio [OR]: 10.00 (95% CI: 1.90–52.55); P=0.003; Fig 2e). hsa-miR-499a-5p was also detected in a higher proportion (19/22; 86.3%) of patients with elevated troponin concentrations after surgery, compared with 13/24 (54.2%) patients with troponin concentrations below the 99th centile after surgery (OR: 5.36 [95% CI 1.32–24.17]; P=0.02; Fig 2f).
Fig 2.
Detectable circulating serum microRNAs before and after noncardiac surgery, in relation to the development of perioperative myocardial injury (PMI) (defined as troponin I [TnI] >99th centile [>0.04 ng ml−1]). (a–d) Heatmaps showing relative serum expression concentrations for each matched sample using the relative cycle quantification (Cq) method. Each row represents an individual patient. MicroRNA Cq values were normalised to the geometric mean of the reference microRNA Cq values to give ΔCq values. The ΔCq values were converted to a fold change (FC) by applying the formula of 2-ΔCq. Bars below each microRNA heatmap indicate range of fold-change expression (log2 scaled). Black bars denote sample that failed strict quality control threshold. Each heatmap is accompanied by a plot showing the degree of correlation between fold-change in microRNA expression after surgery and log serum troponin I concentration (grey shaded area denotes <99th centile). R value derived from linear regression; P-values derived from t-test to reject/accept that the slope is zero. (a) hsa-miR-1-3p. (b) hsa-miR-133a-3p. (c) hsa-miR-146a-5p. (d) hsa-miR-21-5p. (e) Proportion of patient serum samples in whom hsa-miR-208b-3p were detected. P-value refers to PMI vs no PMI after surgery (odds ratio [OR]:10.0 (95% CI 1.9–52.6); P=0.003; χ2test). (f) Proportion of patient serum samples wherein whom hsa-miR-499a-5p were detected. P-value refers to PMI vs no PMI after surgery (OR:5.4 [95% CI 1.3–24.2]; P=0.02; χ2test).
Pathway analysis
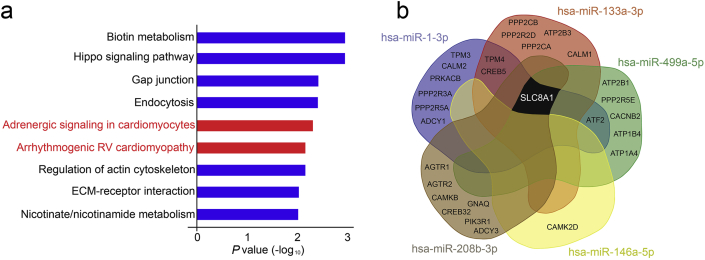
Nine signalling pathways were identified by DIANA in silico pathway analysis (false-discovery rate <0.01) for microRNA that changed after surgery (Fig 3a). This agnostic analysis approach highlighted involvement of adrenergic signalling in cardiomyocytes (hsa 04261; P=0.005) and arrhythmogenic right ventricular cardiomyopathy (hsa 05412; P=0.006). All microRNAs that were elevated in serum after surgery (n=4) regulated expression of SLC8A1, a gene that controls Ca2+overload through the NCX1 antiporter membrane protein by removing Ca2+ from cells (Fig 3b). Subunits of protein phosphatases (that regulate cardiac excitability through dephosphorylation of cardiac ion channels), angiotensin receptors, and the sarcoplasmic/endoplasmic reticulum calcium ATPase (SERCA) pump were also identified as targets of microRNAs that changed after surgery (Fig 3b).
Fig 3.
Bioinformatic analysis identifying pathways and genes regulated by serum microRNAs that change after noncardiac surgery. (a) Signalling pathways (false-discovery rate threshold P-value<0.01), as derived from DIANA mir-Path v3.0, which bioinformatically links microRNA with their regulatory roles in biological and signalling pathways. Cardiac-specific pathways are highlighted in red. (b) Shared genes regulated by serum microRNA identified as changing after noncardiac surgery. The four microRNAs identified as changing after surgery regulate mRNA expression of SLC8A1 (NCX1, sodium–calcium exchanger) and subunits of the protein phosphatase enzyme.
Cohort 2: microRNA concentrations in extracellular vesicles
Patient characteristics
Characteristics of the 11 patients from whom samples were obtained 48 h after surgery are presented in Supplementary Table 1. The median (IQR) troponin-T values were 19 (17–23) ng L−1 in patients with troponin elevation (n=6), compared with patients in whom plasma troponin remained lower than the 99th centile value (n=5; median: 6 [5–9] ng L−1; P=0.006). The two groups were clearly matched for age, sex, co-morbidities, and type of surgery.
Next generation sequencing
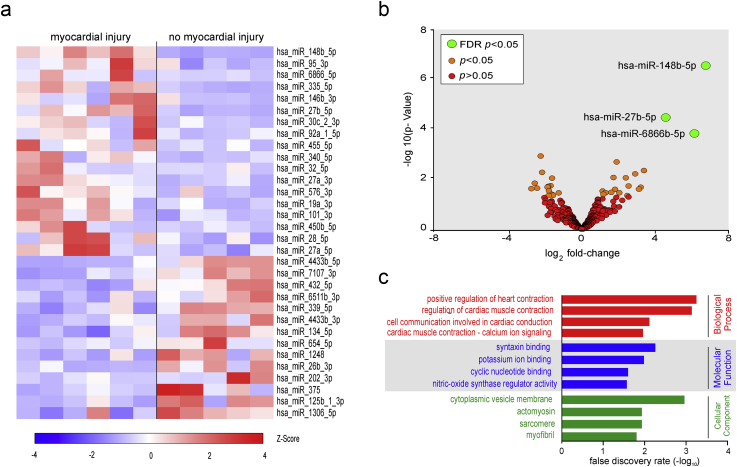
A mean of 9.8 million reads were obtained for each sample. Mean mapping rate to the genome and microRNA annotation was 62.9%. Mapping to miRbase, with normalisation for differences in the sequences depth within each sample, revealed 345 unique microRNA within all samples where tags per million (TPM) ≥1 and 164 unique microRNAs where TPM ≥10. Thirty-two microRNAs were differentially expressed in patients with troponin elevation after surgery (Fig 4a). After correcting for false discovery rate <0.05, three microRNAs remained differentially expressed; hsa-miR-148b-5p, hsa-miR-27b-5p, and hsa-miR-6866b-5p (Fig 4b). The top two biological processes identified through gene ontology were associated with adrenergic stressor pathways involved in cardiac muscle physiology (Fig 4c; Supplementary material).
Fig 4.
Next generation sequencing of microRNAs detected in extracellular vesicles. (a) Heat map showing all differentially expressed microRNA (P<0.05), comparing patients with PMI (hsTnT >14 ng L−1) vs no troponin elevation. Values are trimmed mean of M values. (Z score transformation; red indicates higher expression concentrations). (b) Volcano plot of microRNAs identified in all samples. Green dots denote differentially expressed microRNA with adjusted P-value (FDR; false discovery rate) <0.05; hsa-miR-148b- 5p, hsa-miR-27b-5p, hsa-miR-6866b-5p. Orange dots denote differentially expressed microRNA with P-value <0.05 and red dots denote microRNA that were not differentially expressed. (c) GO terms that were significantly enriched among the differentially expressed microRNA, comparing patients with PMI (hsTnT >14 ng L−1) vs no troponin elevation.
Discussion
We found that microRNA expression in patients with postoperative myocardial injury was independent of troponin concentrations, suggesting that in this population the microRNA expressed after surgery reflect a generalised stress/injury response rather than a type II myocardial infarction. Further, next generation sequencing of microRNA content in extracellular vesicles (Study 2) supports this contention, as this analysis suggest that this sub-type of myocardial injury is consistent with adrenergic activation.
Although these exploratory data are limited by the small sample size, we provide further mechanistic insight into PMI by quantifying circulating microRNAs from samples obtained serially in closely matched patients undergoing noncardiac surgery. This exploratory study shows that microRNAs are readily detected in serum and extracellular vesicles obtained from patients undergoing noncardiac surgery. Using technical spike-in quality controls and standardised reproducible sampling protocols, we report that circulating concentrations of cardiac-specific microRNAs that increase after ACS10 29 were also elevated after noncardiac surgery. MicroRNA expression concentrations were independent of troponin concentrations, suggesting strongly that several microRNA signatures described in the ACS literature reflect a generalised stress/injury response. In support of our circulating microRNA data, agnostic identification of microRNA expression in extracellular vesicles (which was also masked to serial troponin values) revealed pathways from differentially expressed microRNAs that predominantly regulate cardiac muscle contraction. Our findings are consistent with the activation of G protein-coupled receptor signalling mediators, which have an established role through β-arrestins in regulating processing of microRNAs.30 G-protein-coupled receptor-mediated stress signalling appears likely to be a dominant factor that contributes to troponin elevation after surgery, as microRNA findings described for myocardial ischaemia/infarction were not recapitulated by our agnostic bioinformatic analyses.
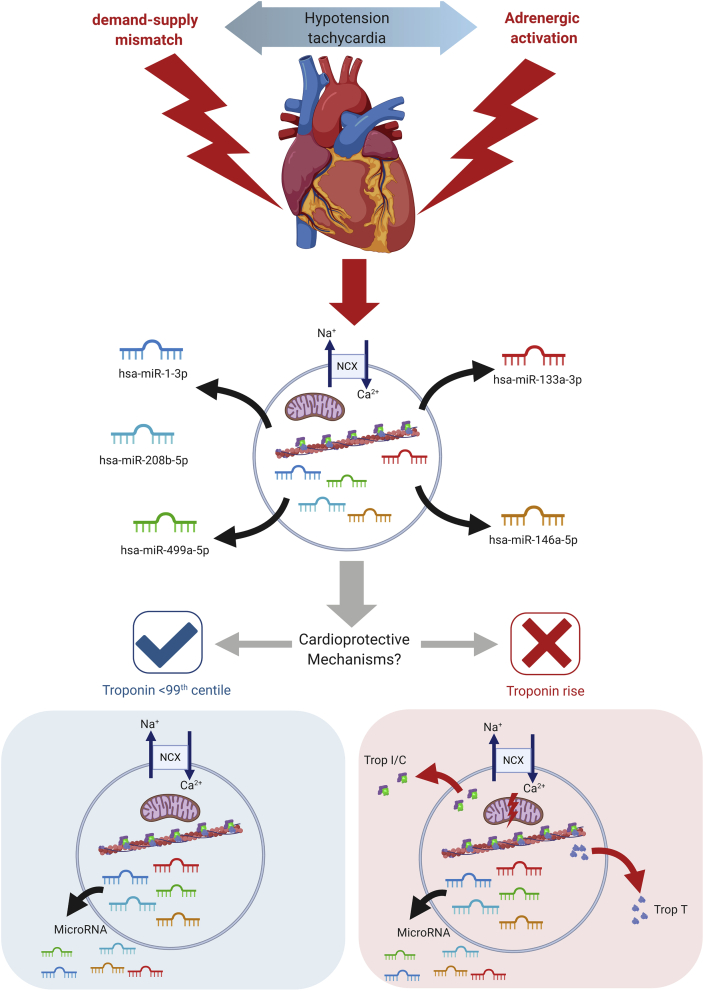
Together, microRNA data from both plasma extracellular vesicles and serum suggest that a cardiac-stressor phenotype is common amongst patients after noncardiac surgery regardless of the development of troponin release and myocardial injury (summarised in Fig 5).
Fig 5.
Refined hypothesis of potential mechanisms contributing to perioperative myocardial injury. This figure summarises our microRNA findings and their potential roles in cardioprotection. Trop I, troponin I; Trop C, troponin C; Trop T, troponin T; NCX, sodium–calcium exchanger. Adrenergic stress is a common feature of the perioperative period, leading to the release of microRNA from cardiac cells, which may be cardiomyocytes, fibroblasts, or other cell types. In individuals at highest risk of elevated troponin after surgery (e.g. older patients, patients with cardiac failure, diabetes mellitus, renal disease), pre-existing (preoperative), or acquired, loss of cardioprotective signalling mechanisms promotes the likelihood of calcium overload from adrenergic stress, resulting in mitochondrial dysfunction, bioenergetic compromise and myocardial injury manifest clinically by the measurement (at low concentrations) of troponin T in plasma/serum.
Several microRNA have been reported as specific biomarkers for ACS, including miR-499, miR-1, miR-133a/b, and miR-208a/b. In a prospective single-centre study, concentrations of cardiac miRNAs (miR-1, -208a, and -499), miR-21, and miR-146a increased in 106 patients diagnosed with ACS, even in patients with initially negative high-sensitive troponin or symptom onset <3 h.10 The combination of miR-1, miR-499, and miR-21 increased the diagnostic value in all suspected ACS patients, over and above that for hs-troponin T.10 However, in contrast to microRNA studies in ACS, we found that troponin elevation after surgery was not related to ACS-specific microRNAs, including key players such as miR-1 and miR-133a. In the case of hsa-miR-146a-5p, expression concentrations were decreased in patients who developed PMI, in direct contrast to elevated circulating concentrations reported after myocardial infarction.10,31 MicroRNA-208, a cardiac-specific microRNA that regulates cardiac myosin heavy chain expression,32 increases in patients after acute myocardial infarction,33 but we detected miR-208a in only ∼10% of patients. Similarly, the low level of detection rates for miR-208b in our study are in contrast to expression concentrations correlating with troponin concentrations34 including after acute myocardial infarction.31 Although hsa-miR-499a-5p was detected in a higher proportion of patients who developed PMI, the detection rates for both miR-208b-3p and miR499a-5p we found are consistent with low detection rates reported in the literature10,29 and low constitutive concentrations in plasma or serum.35
Our two independent studies analysing microRNA derived from separate sources (serum, extracellular vesicles) suggested common mechanistic targets. Agnostic bioinformatic analyses highlighted microRNA changes in both serum and extracellular vesicles that converge on established gene transcription targets that regulate cardiomyocyte cell Ca2+ homeostasis,36 predominantly involving the sodium-calcium exchanger (NCX1) encoded by the SLC8A1 gene.37 Calcium enters cardiac myocytes through voltage-gated Ca2+ channels during excitation, and is extruded from myocytes primarily by the NCX1.28 Ischaemia/reperfusion reverses the sodium/calcium exchange mechanism, leading to an increase in intracellular Ca2+, mitochondrial dysfunction and cardiomyocyte death. However, sustained stress also inhibits NCX1 activity, an effect that requires corticosteroid release in concert with adrenergic activation.38
Reversible protein phosphorylation is essential for excitation–contraction coupling, Ca2+ handling, cell metabolism, myofilament regulation, and cell–cell communication.39 About 90% of dephosphorylation is catalysed by just three protein phosphatases, PP1, PP2A, and PP2B (calcineurin), all of which are regulated by microRNA we identified as changing after surgery. We identified multiple potential PP2A regulatory subunits potentially related to microRNA changes after surgery, which likely reflects the diversity of expression between cardiac cell types, subcellular domains, and disease phenotypes.39 Cardiac myosin binding protein-C and the inhibitory subunit of TnI are also dephosphorylated by both PP1 and PP2A. Targeting of PP2A to TnI involves the PP2A regulatory subunit B56α, which is lost after β-adrenergic stimulation. Aberrant regulation of PP2A signalling is a dominant feature in experimental sympathetic-mediated tachycardia and generation of reactive oxygen species.
In the universal presence of a stressor phenotype after surgery in patients with, and without elevations in plasma troponin, a plausible explanation for our findings is that loss of cardioprotective mechanisms promote myocardial injury (Fig 5). An aberrant inflammatory response, indicated by the decline in the anti-inflammatory microRNA hsa-miR-146a-5p, is one such plausible mechanism. By inhibiting the expression of IRAK1, TRAF6, and NFĸβ, hsa-miR-146a-5p suppresses the production of proinflammatory cytokines40 which promote myocardial injury.41 As inflammation is strongly associated with developing PMI,4 reduced expression of hsa-miR-146a-5p indicates that the dysregulation of inflammation after surgery may contribute to myocardial injury. Furthermore, loss of cardioprotective vagal tone,42 which occurs rapidly in many patients during noncardiac surgery,14 is associated with troponin elevation after noncardiac surgery. Experimental preservation of vagal activity after myocardial ischaemia maintains normal Ca2+ handling by restoring the protein and mRNA concentrations of sarcoplasmic reticulum Ca2+ ATPase (SERCA2a), NCX1, and phospholamban.43
The main strength of our study was investigating two separate populations in which we could obtain samples using optimised protocols that enabled microRNA detection in serum and plasma extracellular vesicles to address our overarching hypothesis. Moreover, we compared serial changes in microRNA masked to troponin concentrations before and after surgery. Reference microRNA were used for normalisation of Cq values. For microRNAs that were detected in all samples, the Cq values were consistent with reported values.31 In fact, the detection threshold in clinical studies using the same microRNA isolated from serum has been much higher, that is Cq<50.29 The utilisation of the spike-in microRNA at isolation, reverse transcription, and PCR stage allowed the identification of outliers and minimised technical variation. A further strength of this work is that the amplification efficiencies are reported in keeping with Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines. The low efficiency values for hsa-miR-208b-3p and hsa-miR499a-5p are consistent with reported low detection rates for both.44 Our limited targeting of circulating serum microRNAs that are established genomic biomarkers for ACS and myocardial infarction clearly cannot exclude other novel species that may be involved in PMI. In the absence of continuous electrocardiographic and/or haemodynamic monitoring, we cannot entirely exclude that undetected periods of supply–demand mismatch underpin the pathway analysis identification of adrenergic stress driving troponin release. However, this explanation seems unlikely as the same genomic targets were apparent in both patients with and without elevated troponin concentrations after surgery. However, the purpose of this initial exploratory study was to investigate whether established ischaemic and/or thrombotic microRNA signatures are present after troponin elevation in noncardiac surgery.
In summary, circulating microRNAs that are elevated in ACS also increase after noncardiac surgery, but this pattern does not appear to be specific for patients with PMI (as defined by high-sensitivity troponin). MicroRNA changes after noncardiac surgery appear to occur as a feature of generalised cardiac stress, suggesting that myocardial injury is promoted by deficient cardioprotective mechanisms that prevent troponin release from otherwise injured cardiomyocytes.
Authors' contributions
Study design: GLA, SMM
Study conduct: SMM, TEFA, AGDA, AR, GM, RCMS, DB, BHC, DNW, RMP, GLA
Experiments: SMM, AGDA, GLA
Data analysis: SMM, GLA
Drafting/revising manuscript: GLA, SMM
Declarations of interest
GLA is an editor for Intensive Care Medicine Experimental and the British Journal of Anaesthesia. GLA has undertaken consultancy work for GlaxoSmithKline; TEFA is a member of the associate editorial board of the British Journal of Anaesthesia. There are no other relationships or activities that could appear to have influenced the submitted work.
Funding
GLA is supported by the British Journal of Anaesthesia/Royal College of Anaesthetists basic science Career Development Award, the British Oxygen Company Research Chair Grant in Anaesthesia from the Royal College of Anaesthetists and the British Heart Foundation Programme Grant (RG/14/4/30736). The New Investigator Award from the Canadian Institutes of Health Research to DNW; the Endowed Chair in Translational Anesthesiology Research at St Michael's Hospital and University of Toronto to DNW; Merit Awards from the Department of Anesthesia at the University of Toronto to DNW, CDM, and BHC; a Medical Research Council (UK) Clinical Research Training Fellowship to TEFA. The METS study was supported by grants from the Canadian Institutes of Health Research, Heart and Stroke Foundation of Canada, Ontario Ministry of Health and Long-Term Care, Ontario Ministry of Research and Innovation, the UK National Institute of Academic Anaesthesia, the UK Clinical Research Collaboration, the Australian and New Zealand College of Anaesthetists, and Monash University (Melbourne, Victoria, Australia).
Handling editor: Hugh C Hemmings Jr
Footnotes
This article is accompanied by an editorial: Aetiology of perioperative myocardial injury: a scientific conundrum with profound clinical implications by Howell et al., Br J Anaesth 2020:125:642–646, doi: 10.1016/j.bja.2020.08.007
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bja.2020.05.066.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Botto F., Alonso-Coello P., Chan M.T. Myocardial injury after noncardiac surgery: a large, international, prospective cohort study establishing diagnostic criteria, characteristics, predictors, and 30-day outcomes. Anesthesiology. 2014;120:564–578. doi: 10.1097/ALN.0000000000000113. [DOI] [PubMed] [Google Scholar]
- 2.Ackland G.L., Abbott T.E.F., Jones T.F. Early elevation in plasma high-sensitivity troponin T and morbidity after elective noncardiac surgery: prospective multicentre observational cohort study. Br J Anaesth. 2020;124:535–543. doi: 10.1016/j.bja.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sheth T., Chan M., Butler C. Prognostic capabilities of coronary computed tomographic angiography before non-cardiac surgery: prospective cohort study. BMJ. 2015;350:h1907. doi: 10.1136/bmj.h1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ackland G.L., Abbott T.E.F., Cain D. Preoperative systemic inflammation and perioperative myocardial injury: prospective observational multicentre cohort study of patients undergoing non-cardiac surgery. Br J Anaesth. 2019;122:180–187. doi: 10.1016/j.bja.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Navickas R., Gal D., Laucevicius A., Taparauskaite A., Zdanyte M., Holvoet P. Identifying circulating microRNAs as biomarkers of cardiovascular disease: a systematic review. Cardiovasc Res. 2016;111:322–337. doi: 10.1093/cvr/cvw174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Treiber T., Treiber N., Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20:5–20. doi: 10.1038/s41580-018-0059-1. [DOI] [PubMed] [Google Scholar]
- 7.Chistiakov D.A., Orekhov A.N., Bobryshev Y.V. Cardiac-specific miRNA in cardiogenesis, heart function, and cardiac pathology (with focus on myocardial infarction) J Mol Cell Cardiol. 2016;94:107–121. doi: 10.1016/j.yjmcc.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 8.Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J.J., Lotvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 9.Kaur A., Mackin S.T., Schlosser K. Systematic review of microRNA biomarkers in acute coronary syndrome and stable coronary artery disease. Cardiovasc Res. 2019 doi: 10.1093/cvr/cvz302. [DOI] [PubMed] [Google Scholar]
- 10.Oerlemans M.I., Mosterd A., Dekker M.S. Early assessment of acute coronary syndromes in the emergency department: the potential diagnostic value of circulating microRNAs. EMBO Mol Med. 2012;4:1176–1185. doi: 10.1002/emmm.201201749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Rooij E. The art of MicroRNA research. Circ Res. 2011;108:219–234. doi: 10.1161/CIRCRESAHA.110.227496. [DOI] [PubMed] [Google Scholar]
- 12.Creemers E.E., Tijsen A.J., Pinto Y.M. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110:483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 13.Wijeysundera D.N., Pearse R.M., Shulman M.A. Assessment of functional capacity before major non-cardiac surgery: an international, prospective cohort study. Lancet. 2018;391:2631–2640. doi: 10.1016/S0140-6736(18)31131-0. [DOI] [PubMed] [Google Scholar]
- 14.May S.M., Reyes A., Martir G. Acquired loss of cardiac vagal activity is associated with myocardial injury in patients undergoing noncardiac surgery: prospective observational mechanistic cohort study. Br J Anaesth. 2019;123:758–767. doi: 10.1016/j.bja.2019.08.003. [DOI] [PubMed] [Google Scholar]
- 15.Cheng C., Wang Q., You W., Chen M., Xia J. MiRNAs as biomarkers of myocardial infarction: a meta-analysis. PLoS One. 2014;9 doi: 10.1371/journal.pone.0088566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozomara A., Birgaoanu M., Griffiths-Jones S. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blondal T., Jensby Nielsen S., Baker A. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods. 2013;59:S1–S6. doi: 10.1016/j.ymeth.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 18.Ruijter J.M., Ramakers C., Hoogaars W.M. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peltier H.J., Latham G.J. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–852. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersen C.L., Jensen J.L., Orntoft T.F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Canc Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 21.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Langmead B., Wilks C., Antonescu V., Charles R. Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics. 2018;35:421–432. doi: 10.1093/bioinformatics/bty648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vlachos I.S., Zagganas K., Paraskevopoulou M.D. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43:W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexa A., Rahnenfuhrer J., Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 26.De Rosa S., Fichtlscherer S., Lehmann R., Assmus B., Dimmeler S., Zeiher A.M. Transcoronary concentration gradients of circulating microRNAs. Circulation. 2011;124:1936–1944. doi: 10.1161/CIRCULATIONAHA.111.037572. [DOI] [PubMed] [Google Scholar]
- 27.Barrett T., Wilhite S.E., Ledoux P. NCBI GEO: archive for functional genomics data sets – update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robin X., Turck N., Hainard A. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinform. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Widera C., Gupta S.K., Lorenzen J.M. Diagnostic and prognostic impact of six circulating microRNAs in acute coronary syndrome. J Mol Cell Cardiol. 2011;51:872–875. doi: 10.1016/j.yjmcc.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 30.Xiao K., McClatchy D.B., Shukla A.K. Functional specialization of beta-arrestin interactions revealed by proteomic analysis. Proc Natl Acad Sci USA. 2007;104:12011–12016. doi: 10.1073/pnas.0704849104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang G.K., Zhu J.Q., Zhang J.T. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31:659–666. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 32.van Rooij E., Sutherland L.B., Qi X., Richardson J.A., Hill J., Olson E.N. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 33.Xiao J., Shen B., Li J. Serum microRNA-499 and microRNA-208a as biomarkers of acute myocardial infarction. Int J Clin Exp Med. 2014;7:136–141. [PMC free article] [PubMed] [Google Scholar]
- 34.Corsten M.F., Dennert R., Jochems S. Circulating microRNA-208b and microRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet. 2010;3:499–506. doi: 10.1161/CIRCGENETICS.110.957415. [DOI] [PubMed] [Google Scholar]
- 35.Adachi T., Nakanishi M., Otsuka Y. Plasma microRNA 499 as a biomarker of acute myocardial infarction. Clin Chem. 2010;56:1183–1185. doi: 10.1373/clinchem.2010.144121. [DOI] [PubMed] [Google Scholar]
- 36.Harada M., Luo X., Murohara T., Yang B., Dobrev D., Nattel S. MicroRNA regulation and cardiac calcium signaling: role in cardiac disease and therapeutic potential. Circ Res. 2014;114:689–705. doi: 10.1161/CIRCRESAHA.114.301798. [DOI] [PubMed] [Google Scholar]
- 37.Shattock M.J., Ottolia M., Bers D.M. Na+/Ca2+ exchange and Na+/K+-ATPase in the heart. J Physiol. 2015;593:1361–1382. doi: 10.1113/jphysiol.2014.282319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hudecova S., Kubovcakova L., Kvetnansky R. Modulation of expression of Na+/Ca2+ exchanger in heart of rat and mouse under stress. Acta Physiol (Oxf) 2007;190:127–136. doi: 10.1111/j.1748-1716.2007.01673.x. [DOI] [PubMed] [Google Scholar]
- 39.Lubbers E.R., Mohler P.J. Roles and regulation of protein phosphatase 2A (PP2A) in the heart. J Mol Cell Cardiol. 2016;101:127–133. doi: 10.1016/j.yjmcc.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taganov K.D., Boldin M.P., Chang K.J., Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frangogiannis N.G. The inflammatory response in myocardial injury, repair, and remodelling. Nat Rev Cardiol. 2014;11:255–265. doi: 10.1038/nrcardio.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abbott T.E.F., Pearse R.M., Cuthbertson B.H., Wijeysundera D.N., Ackland G.L., investigators Ms. Cardiac vagal dysfunction and myocardial injury after non-cardiac surgery: a planned secondary analysis of the measurement of Exercise Tolerance before Surgery Study. Br J Anaesth. 2019;122:188–197. doi: 10.1016/j.bja.2018.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y., Chen A., Song L. Low-level vagus nerve stimulation reverses cardiac dysfunction and subcellular calcium handling in rats with post-myocardial infarction heart failure. Int Heart J. 2016;57:350–355. doi: 10.1536/ihj.15-516. [DOI] [PubMed] [Google Scholar]
- 44.Bustin S.A., Benes V., Garson J.A. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.