Abstract
Background
Interferon (IFN) regulatory factors (IRFs), as transcriptional regulatory factors, play important roles in regulating the expression of type I IFN and IFN- stimulated genes (ISGs) in innate immune responses. In addition, they participate in cell growth and development and regulate oncogenesis.
Results
In the present study, the cDNA sequence of IRF10 in common carp (Cyprinus carpio L.) was characterized (abbreviation, CcIRF10). The predicted protein sequence of CcIRF10 shared 52.7–89.2% identity with other teleost IRF10s and contained a DNA-binding domain (DBD), a nuclear localization signal (NLS) and an IRF-associated domain (IAD). Phylogenetic analysis showed that CcIRF10 had the closest relationship with IRF10 of Ctenopharyngodon idella. CcIRF10 transcripts were detectable in all examined tissues, with the highest expression in the gonad and the lowest expression in the head kidney. CcIRF10 expression was upregulated in the spleen, head kidney, foregut and hindgut upon polyinosinic:polycytidylic acid (poly I:C) and Aeromonas hydrophila stimulation and induced by poly I:C, lipopolysaccharide (LPS) and peptidoglycan (PGN) in peripheral blood leucocytes (PBLs) and head kidney leukocytes (HKLs) of C. carpio. In addition, overexpression of CcIRF10 was able to decrease the expression of the IFN and IFN-stimulated genes PKR and ISG15.
Conclusions
These results indicate that CcIRF10 participates in antiviral and antibacterial immunity and negatively regulates the IFN response, which provides new insights into the IFN system of C. carpio.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12917-020-02674-z.
Keywords: Common carp (Cyprinus carpio L.), Interferon regulatory factor 10 (IRF10), poly I:C, Aeromonas hydrophila, IFN response
Background
Interferon (IFN) regulatory factors (IRFs), as transcriptional regulatory factors, play multiple important roles in host immune responses and other physiological processes; for example, they regulate the expression of type I IFN and IFN-stimulated genes (ISGs) [1], the activation and differentiation of distinct immune cell populations, cell growth, differentiation and apoptosis [2, 3]. The N-terminal regions of IRFs share a highly conserved DNA-binding domain (DBD), which contains five or six tryptophan repeats [4]. The C-terminal regions of IRFs generally possess an IRF-associated domain (IAD), which mediates the interactions between IRFs and other proteins to form transcriptional complexes [5].
To date, 11 kinds of IRFs have been identified in vertebrates [6], which can be divided into four subfamilies, IRF1 (IRF1, 2, and 11), IRF3 (IRF3 and 7), IRF4 (IRF4, 8, 9, and 10) and IRF5 (IRF5 and 6), according to their differences in the C-terminal region [7]. IRF10 is present only in non-mammals and was first identified in the chicken (Gallus gallus). Phylogenetic analysis has shown that GgIRF10 clusters into the IRF4 subfamily and plays a crucial role in the later stages of the immune response to invading pathogens in G. gallus [8]. Functional characterization of fish IRF10 has also begun recently, and this gene has been identified in teleosts including zebrafish (Danio rerio) [9], orange spotted grouper (Epinephelus coioides) [10, 11], Japanese flounder (Paralichthys olivaceus) [12], Atlantic cod (Gadus morhua) [13, 14], grass carp (Ctenopharyngodon idella) [15], rainbow trout (Oncorhynchus mykiss) [15, 16], Asian swamp eel (Monopterus albus) [15, 17], mandarin fish (Siniperca chuatsi) [18], Senegalese sole (Solea senegalensis) [19] and blunt snout bream (Megalobrama amblycephala) [20].
The teleost IRF10 genes are constitutively expressed in a variety of tissues and can be upregulated by the viral mimic polyinosinic:polycytidylic acid (poly I:C) [10–15, 17, 18, 21], lipopolysaccharide (LPS) [12, 17], viruses [12, 18, 19] or bacteria [10, 12, 13, 17, 20], suggesting that IRF10 plays roles in immune responses to both viral and bacterial infections. Furthermore, in zebrafish and orange spotted grouper, IRF10 inhibits IFNφ1 and IFNφ3 promoter activation and negatively regulates fish antiviral gene expression to prevent an excessive immune response, which is a unique regulatory mechanism of IFN responses in teleosts [10, 11].
As a pivotal aquaculture fish species widely cultured in both Asia and Europe, common carp (Cyprinus carpio L.) is also a good model for exploring the immune system of teleosts [22–24]. Several IRFs have been identified in common carp, including IRF1, IRF3, IRF5, IRF7 and IRF9 [25–28]. In this study, we identified the full-length cDNA sequence of IRF10 in C. carpio (named CcIRF10) and investigated its function. We examined the tissue distribution of CcIRF10 in healthy common carp and then evaluated the expression level of CcIRF10 upon viral or bacterial stimulation both in vivo and in vitro to determine its function in the immune response against pathogens. Furthermore, the regulatory role of CcIRF10 in the IFN signalling pathway was also determined in this study. The results will contribute to understanding of the innate immune system of fish.
Methods
Fish feeding and sampling
C. carpio specimens were purchased from a local fish farm (Jinan, Shandong, China), selected for size (approximately 200 g per fish), maintained in recirculating tap water at 20 °C and fed daily for more than 1 week before challenge and sampling. After treatment, the fish were euthanized by immersion in a solution of tricaine methane sulfonate (MS222, Sigma-Aldrich) at a concentration of 100 mg/l of water.
Cloning and analysis of CcIRF10 cDNA
The full-length cDNA sequence of CcIRF10 was obtained using RT-PCR and the rapid amplification of cDNA ends (RACE) method. First, the primers IRF10-F/IRF10-R were used to amplify a partial sequence of CcIRF10. The primers were designed based on the known fish IRF10 cDNA sequences. Then, 3′ and 5′ Full RACE Core Sets (TaKaRa) were used to obtain the full-length cDNA sequence. The PCR products were ligated into the pMD18-T vector (TaKaRa) and transformed into competent E. coli DH-5α for sequencing (Invitrogen). The domains of the protein sequence were analysed using the Simple Modular Architecture Research Tool (SMART, http://smart.embl-heidelberg.de). Multiple alignment and phylogenetic analysis were performed using MEGA 5.0. The primers used in this study are listed in Table 1.
Table 1.
Primers used in the present study
| Name of primer | Sequence(5′-3′) | GenBank accession No. |
|---|---|---|
| CcIRF10-F | TAGCGCAGATAGACAGCG | MT646905 |
| CcIRF10-R | CACACCTTTCTCCAGGTG | |
| CcIRF10 ORF-F | CCGGAATTCATGGAAGACAGGTCGAGGCA | |
| CcIRF10 ORF-R | TCCCCGCGGTTCACTGGTTTTCCTGTGTGG | |
| CcIRF10 RT-F | GCTGTTGGATGGAGTGTGAATGG | |
| CcIRF10 RT-R | CCAGGTTCCCGTGATAGAACAAAC | |
| CcS11-F | CCGTGGGTGACATCGTTACA | |
| CcS11-R | TCAGGACATTGAACCTCACTGTCT | |
| EPC-IFN-F | TCAATCTCATGGATGCCTCAGAGC | FN178457 |
| EPC-IFN-R | TGGTATTGGGCCACGCATTCTT | |
| EPC-TNFα-F | ACAGGTGATGGTGTCGAGGAGGA | JN412133 |
| EPC-TNFα-R | TCTGAGACTTGTTGAGCGTGAAG | |
| EPC-ISG15-F | GTGAGCGGTGAAGCCACAGTTG | KM099174 |
| EPC-ISG15-R | GCGAACCGTTATCGGCAGACAG | |
| EPC-PKR-F | AGGCTTGATCCACAGAGACCTGAA | KM099176 |
| EPC-PKR-R | CGTTCCAGAAGTTGCACGTCATTG | |
| EPC-EF1α-F | AAGAGCGTTGAGAAGAAAG | AY643400 |
| EPC-EF1α-R | GAGTGCCCAGGTTTAGAG |
Experimental challenge of C. carpio with poly I:C and Aeromonas hydrophila
Poly I:C and A. hydrophila challenge experiments were performed in C. carpio as previously described [28–30]. In the poly I:C challenge experiments, 50 fish were intraperitoneally injected with 500 μl of poly I:C solution (2.6 mg/ml in PBS, Sigma). In the A. hydrophila challenge experiment, the bacteria were inactivated in 0.5% formalin at 37 °C for 36 h and then resuspended in PBS. Then, 50 fish were intraperitoneally injected with 500 μl of A. hydrophila (at a dose of 2.0 × 108 cells). At 0, 3, 6, 12, 24, 48 and 72 h post injection (hpi), the spleen, head kidney, foregut and hindgut were collected from three fish at every time points (n = 3). Total RNA was extracted using TRIzol reagent (TIANGEN), and cDNA was synthesized using a FastQuant RT Kit (TIANGEN).
In vitro stimulation of PBLs and HKLs
Peripheral blood leucocytes (PBLs) and head kidney leukocytes (HKLs) were isolated from C. carpio according to a previous report [28, 31]. In brief, C. carpio peripheral blood and head kidney cell suspensions were loaded onto freshly prepared 34%/51% Percoll (Sigma) density gradients and separated via centrifugation at 650×g for 30 min. The cells were resuspended in cold Leibovitz’s L-15 medium with 10% foetal bovine serum (FBS), 100 UI/ml penicillin and 100 mg/ml streptomycin. PBLs or HKLs (1 × 106) were maintained in a 24-well cell culture plate with 500 μl in each well and treated with 5 μl poly I:C (500 μg/ml), LPS (1 mg/ml) or peptidoglycan (PGN, 1 mg/ml). At 0, 3, 6, 12 and 24 h post stimulation, triplicate cell samples were harvested, and total RNA was extracted (n = 3).
Construction and transfection of CcIRF10 overexpression vectors
The ORF of CcIRF10 was amplified using Phusion High-Fidelity DNA polymerase (PrimeSTAR) with specific primers. Purified fragments were digested with the SacII and EcoRI restriction enzymes, ligated into the pcDNA3.1-EGFP vector, and transformed into E. coli Top10 cells. The overexpression vector pcDNA3.1-EGFP-CcIRF10 (abbreviation, pcIRF10) was verified by sequencing. The plasmids were extracted using an endotoxin-free plasmid isolation kit (TIANGEN) following the manufacturer’s instructions.
Epithelioma papulosum cyprini (EPC) cells were seeded in 24-well plates with 500 μl in each well at a concentration of 4 × 105 cells/ml and maintained at 25 °C in M199 medium (HyClone) supplemented with 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco) for 1 day so that they reached approximately 80% confluency. Each well of cells was transfected with 1 μg of plasmids using 2 μl of X-tremeGENE HP DNA Transfection Reagent (Roche). EPC cells transfected with empty plasmids served as controls, and the gene expression of IFN, PKR, ISG15 and TNFα was detected after overexpression of CcIRF10. The primers used in this study are listed in Table 1.
Real-time PCR analysis
Real-time PCR was performed as previously described in a Rotor-Gene Q PCR instrument (Qiagen) with TransStart Tip Green qPCR SuperMix (TransGen) [28]. All samples were analysed in triplicate, and the expression values of all genes were calculated relative to those of the 40S ribosomal protein S11 or the β-actin gene using the 2(−∆∆CT) method. The primers used are listed in Table 1.
Statistical analysis
Comparisons between the experimental group and the control group were performed using one-way analysis of variance (ANOVA) in GraphPad Prism 5, and a value of P < 0.05 was considered to indicate significance.
Results
cDNA cloning and molecular characterization of CcIRF10
The full-length cDNA of CcIRF10 was found to consist of 1274 bp. The CcIRF10 cDNA (GenBank accession no. MT646905) contains a 78 bp 5′-untranslated region (UTR), a 26 bp 3′-UTR containing mRNA instability motifs (1244AATAA1249), and an ORF of 1170 bp that translates into a 390-amino acid putative peptide with a predicted molecular mass of 44.4 kDa. The theoretical isoelectric point is 8.314. The predicted protein sequence contains a DBD (M1-R119) that possesses five tryptophans (Trp13, Trp28, Trp40, Trp60 and Trp79), an IAD (P186-L367) and a nuclear localization signal (NLS) in the DBD (Fig. S1).
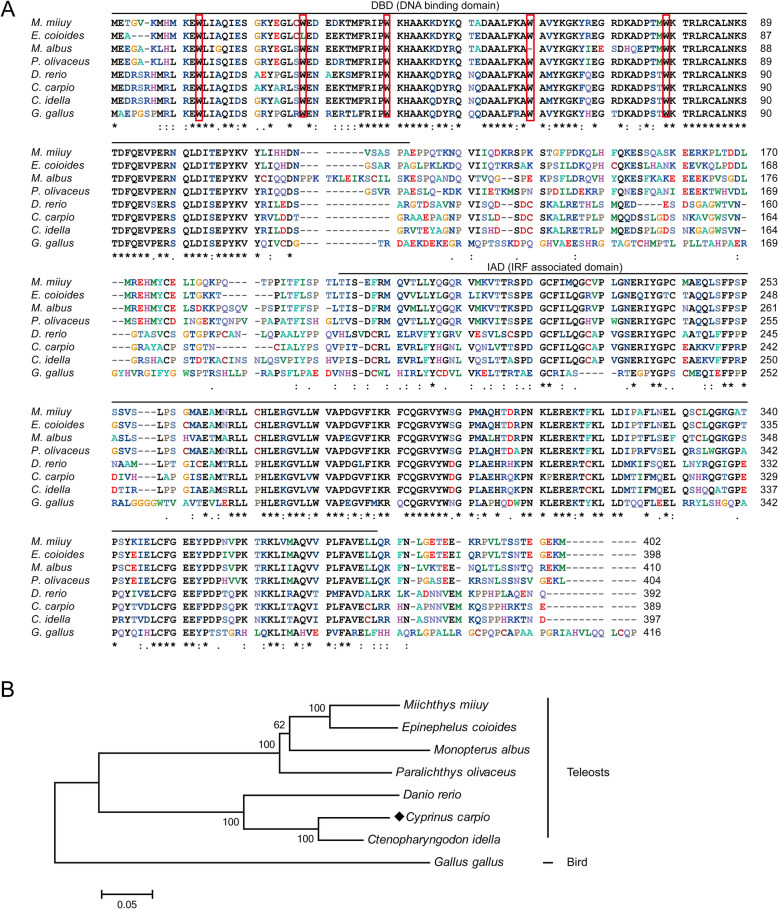
The predicted protein of CcIRF10 shares 89.2% identity with that of C. idella IRF10, 52.7–75.6% identity with those of other teleost IRF10s and 50.1% identity with a bird (G. gallus) protein (Table 2). Multiple alignments of CcIRF10 with the amino acid sequences from other vertebrates revealed areas of amino acids conserved in all vertebrates. Significant homology was found in the putative DBD (Fig. 1a). To explore the phylogenetic relationships of IRF10 in vertebrates, a phylogenetic tree including IRF10s from all known species was constructed using the neighbour-joining method. The tree was divided into two branches, teleost and bird. CcIRF10 had the closest relationship with C. idella IRF10 (Fig. 1b).
Table 2.
Protein length and GenBank accession numbers of IRF10
Fig. 1.
Multiple alignments (a) and phylogenetic analysis (b) of IRF10 protein sequences in different species. Identical (*) and similar (: or .) residues are indicated. The DBD and IAD are indicated by black lines. Five tryptophan (W) residues are boxed in red. The phylogenetic tree was produced by the neighbour-joining method in MEGA 5.0. C. carpio IRF10 is marked with a solid diamond (♦). The GenBank accession numbers of the genes are listed in Table 2
Tissue distribution of CcIRF10
The expression patterns of CcIRF10 in 11 tissues of healthy common carp, including the liver, spleen, head kidney, foregut, hindgut, gills, gonad, skin, muscle, buccal epithelium and brain, were detected by real-time PCR. The results showed that CcIRF10 mRNA was detected in all examined tissues, with the highest expression in the gonad, the lowest expression in the head kidney, and moderate expression in the other nine tissues (Fig. 2).
Fig. 2.
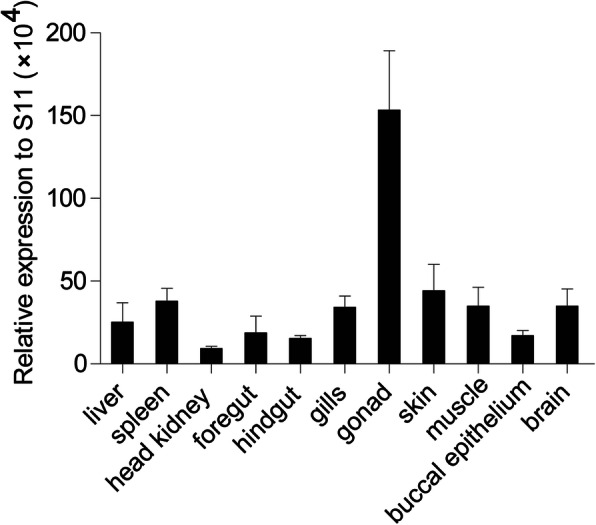
Tissue-specific expression of CcIRF10 under normal physiological conditions. CcIRF10 mRNA expression in the liver, spleen, head kidney, gills, skin, foregut, hindgut, buccal epithelium, gonad, muscle and brain was determined by real-time PCR. The gene expression levels were normalized using 40S ribosomal protein S11 mRNA. (n = 3)
Gene expression of CcIRF10 in response to poly I:C and A. hydrophila stimulation in vivo
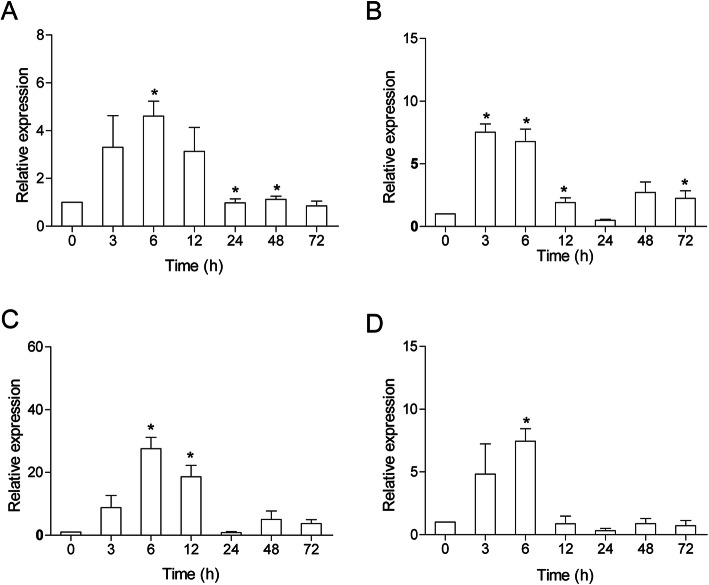
To determine the function of CcIRF10 in immune defence in common carp, CcIRF10 expression was examined in some immune-related tissues after viral or bacterial challenge. Upon poly I:C stimulation, the peak expression of CcIRF10 appeared at 6 hpi in the spleen (4.5-fold, P < 0.05), foregut (27.5-fold, P < 0.05) and hindgut (7.5-fold, P < 0.05). In the head kidney, CcIRF10 expression peaked at 3 hpi (7.5-fold, P < 0.05) (Fig. 3).
Fig. 3.
Expression analysis of CcIRF10 in response to poly I:C challenge in vivo. Total RNA was extracted from spleen (a), head kidney (b), foregut (c) and hindgut (d) tissues at 0 (as a control), 3, 6, 12, 24, 48 and 72 h post injection for real-time PCR. The expression was normalized using that of the 40S ribosomal protein S11. (n = 3, mean ± SD, *P < 0.05)
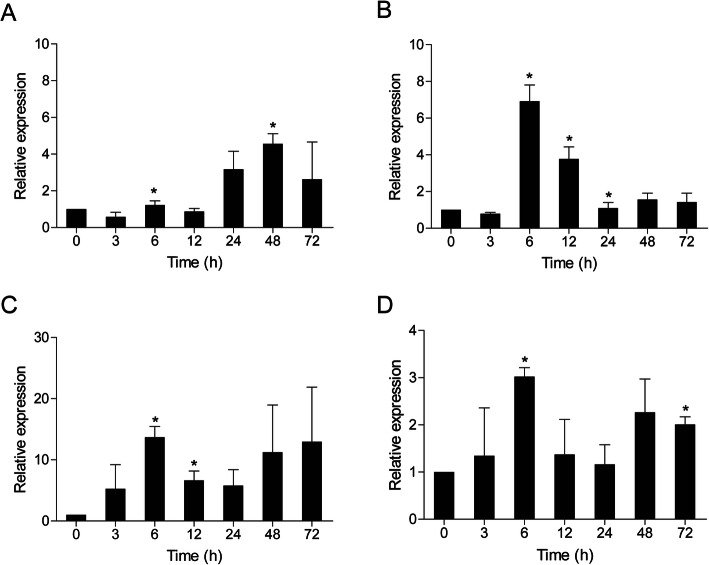
In A. hydrophila-infected fish, CcIRF10 was upregulated in the head kidney (6.8-fold, P < 0.05), foregut (13.7-fold, P < 0.05) and hindgut (3.0-fold, P < 0.05) at 6 hpi. In the spleen, CcIRF10 reached its peak at 48 hpi, with a 4.6-fold induction (P < 0.05) (Fig. 4).
Fig. 4.
Expression analysis of CcIRF10 in response to A. hydrophila challenge in vivo. Total RNA was extracted from spleen (a), head kidney (b), foregut (c) and hindgut (d) tissues at 0 (as a control), 3, 6, 12, 24, 48 and 72 h post injection for real-time PCR. The expression was normalized using that of the 40S ribosomal protein S11. (n = 3, mean ± SD, *P < 0.05)
Expression of CcIRF10 upon poly I:C, LPS and PGN stimulation in vitro
To examine the CcIRF10 transcription level in response to poly I:C, LPS or PGN challenge in vitro, we isolated leukocytes from the peripheral blood and head kidneys of C. carpio. As shown in Fig. 5a, real-time PCR data showed that the expression of CcIRF10 in the PBLs was upregulated by poly I:C (2.0-fold, P < 0.05) and PGN (2.2-fold, P < 0.05) at 12 h, but not by LPS. In HKLs, CcIRF10 was induced by all PAMPs. Upon PGN stimulation, the expression of CcIRF10 reached its peak at 12 h (3.8-fold, P < 0.05), and after stimulation by poly I:C (1.7-fold, P < 0.05) and LPS (4.1-fold, P < 0.05), the highest expression was found at 24 h (Fig. 5b).
Fig. 5.
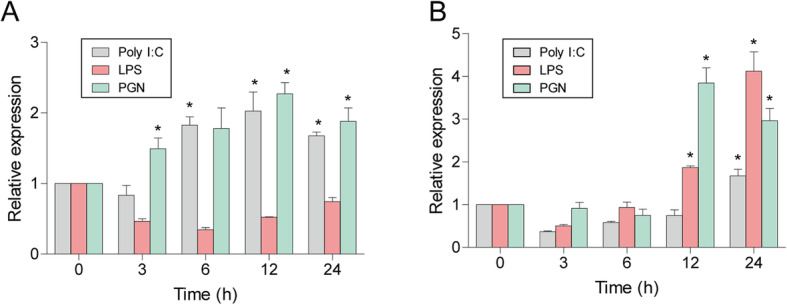
Expression levels of CcIRF10 in PBLs (a) and HKLs (b) upon poly I:C and LPS stimulation. Cells were collected at 0 (as a control), 3, 6, 12 and 24 h post-infection for RNA extraction and real-time PCR analysis. The expression was normalized using that of the 40S ribosomal protein S11. (n = 3, mean ± SD, *P < 0.05)
mRNA expression of cytokines in EPC cells overexpressing CcIRF10
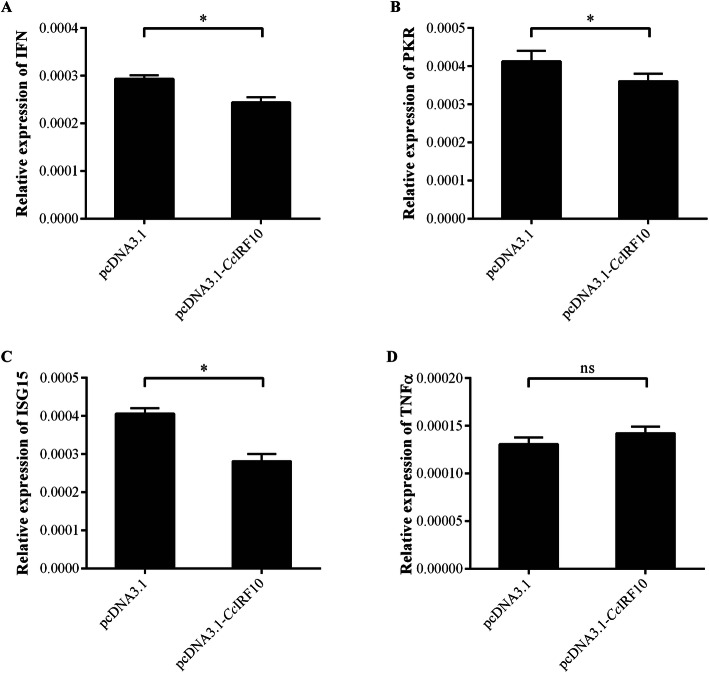
To investigate the regulatory role of CcIRF10 in the IFN signalling pathway, the gene expression of IFN, two IFN-stimulated genes (PKR and ISG15) and TNFα was detected after overexpression of CcIRF10 in EPC cells (Fig. S2 and S3). The results showed that the gene expression of IFN, PKR and ISG15 was reduced in the cells transfected with pcDNA3.1-EGFP-CcIRF10, which were 83, 0.87 and 0.69% of the expression in control cells, respectively (P < 0.05, Fig. 6a-c). However, the expression of TNFα, a non-ISG, was not changed in the cells (Fig. 6d).
Fig. 6.
Relative expression of IFN (a), PKR (b), ISG15 (c) and TNFα (d) in CcIRF10-transfected EPC cells. The expression was normalized to that of EF1α. (n = 3, mean ± SD, *P < 0.05)
Discussion
IRF10, which has been reported to play important roles in immune responses to both viral and bacterial infections in teleosts, can inhibit the activation of IFN promoters and negatively regulate fish antiviral gene expression to prevent an excessive immune response [10, 11, 32]. In the present study, a non-mammalian IRF10 gene was cloned from common carp, and the antiviral and antibacterial immune functions of CcIRF10 were investigated. The predicted CcIRF10 protein contains two conserved functional domains, the N-terminal DBD and the C-terminal IAD, suggesting that the functions of IRF10 may be conserved throughout vertebrates. The DBD contains a highly conserved five-tryptophan repeat that can bind to IFN-stimulating response elements (ISREs) and IRF-regulatory elements (IRF-Es) in target promoters [4, 33]. Notably, the five well-conserved tryptophan residues in D. rerio, C. idella, G. gallus, and C. carpio are all at positions 10, 25, 37, 57 and 76 in the N-terminus. The IAD, which is responsible for homo−/heterodimer interactions of the IRFs and association with other transcription factors, is less conserved than the DBD [5]. Phylogenetic analysis of predicted IRF10 protein sequences of C. carpio and other vertebrate species supported the division of IRF10 into two branches: teleost and bird. These results match the established evolutionary relationships among teleost and other vertebrate species and support the authenticity of the nomenclature for IRF10.
IRF10 was first identified in G. gallus and is highly expressed in white blood cells and splenic lymphocytes, whereas low expression levels are found in other tissues [8]. In the present study, constitutive expression of the CcIRF10 gene was detectable in all 11 tissues of C. carpio when analysed by real-time PCR, although there were differences in the levels of expression. This ubiquitous tissue expression pattern supports the findings of previous studies on IRFs in teleosts, including M. albus [17], G. morhua [13], D. rerio [34], O. mykiss [35], turbot (Scophthalmus maximus) [36], P. olivaceus [12, 37], rock bream (Oplegnathus fasciatus) [38], blunt snout bream (Megalobrama amblycephala) [39] and half-smooth tongue sole (Cynoglossus semilaevis) [40]. CcIRF10 was found to be most highly expressed in gonads (Fig. 2), which is different from the expression patterns in P. olivaceus, C. idella and M. albus. In P. olivaceus, the expression of IRF10 has been found to be very high in the gills, intestine, trunk kidney, heart, stomach, head kidney and PBLs; in C. idella, IRF10 expression has been found to be high in all tested tissues, with the highest expression in the thymus and gills; and in M. albus, the highest expression level has been observed in intestine, whereas the lowest level has been found in the liver [12, 15, 17]. However, D. rerio IRF10 is highly expressed in the testis, which is also a reproductive organ [9]. These results suggest that IRF10 may not only play an important role in the immune system but also likely participate in the regulation of the reproductive system in teleosts.
Previous studies on C. carpio have shown that the expression of IRF1, IRF3, IRF5 and IRF7 is upregulated upon stimulation with poly I:C or viruses [25–27]. In the present study, following poly I:C injection, the induction of CcIRF10 in the foregut (27.5-fold) was much stronger than that in the other tissues (4.5- to 7.5-fold), revealing the important role of CcIRF10 in the mucosal immune system response to poly I:C (Fig. 3). Moreover, similar results were observed in PBLs and HKLs with poly I:C stimulation (Fig. 5). The expression of IRF10 in P. olivaceus is also upregulated by poly I:C stimulation in PBLs [12]. In G. morhua, two isoforms of IRF10 have been identified, and the expression of the long isoform reaches its peak at 6 hpi at 16 °C and 24 hpi at 10 °C, whereas the highest expression of the short isoform is observed at 6 hpi at both 16 °C and 10 °C after poly I:C stimulation [13, 41]. Moreover, IRF10 in P. olivaceus, C. idella, M. albus and zebrafish embryo fibroblast-like ZF4 cells can be upregulated by viruses (viral haemorrhagic septicaemia virus [VHSV] or grass carp haemorrhagic virus [GCHV]) or poly I:C [9, 12]. The observed induction of IRF10 expression by various viruses and poly I:C suggests that the fish IRF10 may play a crucial role in protecting the host from viral infection.
A. hydrophila, a well-known fish-pathogenic bacterium, is primarily found in temperate and freshwater environments and causes infections in various organisms. Fish are becoming increasingly susceptible to A. hydrophila because of the increasingly intensive rearing methods used in aquaculture [42]. Moreover, A. hydrophila breakouts have caused great economic losses around the world [43]. To gain insights into the immune mechanism of CcIRF10 in the antibacterial response, its expression pattern in response to A. hydrophila was investigated using real-time PCR. When fish were challenged with A. hydrophila, the levels of CcIRF10 were upregulated in all four tissues, with the highest induction in the foregut (Fig. 4). IRF10 of P. olivaceus can also be induced by Edwardsiella tarda and Streptococcus iniae [37]. Upon Aeromonas salmonicida infection, the greatest expression of the long isoform of G. morhua IRF10 occurs at 24 hpi, whereas the highest expression of the short isoform is observed at 6 hpi at 10 °C and 16 °C, suggesting the distinct roles of the two isoforms in the immune system of G. morhua [13, 41]. It should be noted that E. coioides IRF10 is responsive to both poly I:C stimulation and Vibrio parahaemolyticus infection but increases to a greater extent after poly I:C stimulation [10]. In accordance with these results, our study showed that the fold change in CcIRF10 induced by poly I:C stimulation (4.5- to 27.5-fold) was greater than that induced by A. hydrophila infection (3.0- to 13.7-fold). LPS is the major component of the outer membranes of gram-negative bacteria, but it has been reported that lower vertebrates (such as fish) may be resistant to the toxic effects of LPS [44]. Therefore, it was unsurprising that CcIRF10 was not upregulated by LPS in the PBLs (Fig. 5a). PGN, a major component of the bacterial cell wall, consists of sugars and amino acids. Similar to the findings of a previous study regarding head kidney macrophages of O. mykiss, CcIRF10 was upregulated by PGN stimulation in vitro [15]. Hence, our in vivo and in vitro findings, together with the previous analogous results, suggest that CcIRF10 is more susceptible to poly I:C than to A. hydrophila infection and plays a substantial role in the foregut, which is a mucosal immune organ. Moreover, fish IRF10 may play essential roles in both antiviral and antibacterial defence, as reported for G. gallus IRF10.
In mammals, IFNs are natural glycoproteins produced by cells of the innate and adaptive immune systems in most vertebrates in response to challenge by viruses, bacteria, fungi, parasites, and tumour cells [45]. In addition, IFNs can also be produced by non-immune cells such as fibroblasts and epithelial cells [45]. Similarly, in fish, IFNs play a crucial role in innate immunity [46, 47]. To investigate the regulatory role of CcIRF10 in the IFN response, we detected the mRNA expression of type I IFN, ISGs (PKR and ISG15) and TNFα in CcIRF10-transfected EPC cells. The transcription of IFN, PKR, ISG15 was downregulated in EPC cells transfected with the pcDNA3.1-CcIRF10 plasmid (Fig. 6). This result is in line with the findings of a previous study that overexpression of D. rerio IRF10 downregulated the expression of IFN-stimulated genes induced by poly I:C and promoted the replication of spring viremia of carp virus (SVCV) in EPC cells [9]. Moreover, this study found that the ISRE site in the promoter was responsible for DrIRF10-mediated inhibition of gene expression of IFNs and ISGs [9, 48]. The mechanism involved in the inhibition of IFN signalling pathway in common carp may be similar, which needs our further study. However, G. gallus IRF10 can upregulate the expression of MHC class I and GBP [8], suggesting that the function of IRF10 might be different between fish and birds.
Conclusions
In the present study, the full-length cDNA sequence of IRF10 from common carp was identified and characterized. In vivo and in vitro studies indicated that CcIRF10 participates in both antiviral and antibacterial immune responses. Furthermore, overexpression of CcIRF10 was able to decrease the expression of the IFN and IFN-stimulated genes PKR and ISG15, indicating that CcIRF10 might negatively regulate the IFN response of C. carpio. The study will provide a valuable experimental foundation for future studies on the immune system of common carp and a theoretical basis for the prevention of fish disease.
Supplementary Information
Supplementary Fig. S1. Nucleotide sequence and the deduced amino acid sequence of C. carpio IRF10. Lowercase letters indicate the 5′ and 3′ UTR, while uppercase letters indicate the ORF or amino acid. The start codon (ATG) and stop codon (TGA) are boxed in red. The DBD and IAD are shaded in blue and pink, respectively. The NLS is underlined, and the five tryptophan (W) residues are boxed in black.
Supplementary Fig. S2. Overexpression of CcIRF10 in EPC cells. EPC cells were transfected with pcDNA3.1-EGFP empty plasmid or pcDNA3.1-EGFP-CcIRF10, and the gene expression of CcIRF10 was detected using real-time PCR. The expression was normalized to that of EF1α. (n = 3, mean ± SD, **P < 0.01).
Supplementary Fig. S3. Transfection efficiency of EPC cells. Bright field (A) and fluorescent image (B) of EPC cells transfected with pcDNA3.1-EGFP empty plasmid. Bright field (C) and fluorescent image (D) of EPC cells transfected with pcDNA3.1-EGFP-CcIRF10 plasmid. (original magnification × 40).
Acknowledgments
Not applicable.
Abbreviations
- IFN
Interferon
- IRF
Interferon regulatory factor
- ISG
IFN-stimulated gene
- DBD
DNA-binding domain
- NLS
Nuclear localization signal
- IAD
IRF associated domain
- poly I:C
polyinosinic:polycytidylic acid
- LPS
Lipopolysaccharide
- EPC
Epithelioma papulosum cyprinid
- RACE
Rapid amplification of the cDNA ends
- SMART
Simple modular architecture research tool
- PBL
Peripheral blood leucocyte
- HKL
Head kidney leukocyte
- PGN
Peptidoglycan
- ORF
Open reading frame
- ANOVA
One-way analysis of variance
- UTR
Untranslated region
- hpi
Hour post injection
- ISRE
IFN stimulating response element
- IRF-E
IRF regulatory element
- VHSV
Viral hemorrhagic septicemia virus
- GCHV
Grass carp haemorrhagic virus
- SVCV
Spring viremia of carp virus
- RLR
Retinoic acid-inducible gene I (RIG-I)-like receptor
Authors’ contributions
HL and GWY conceived and designed the experiments. YYZ, SJS, HPZ and HW performed the experiments. RRL and XPC analyzed the data. YYZ and HL wrote the paper. All authors read and approved the final manuscript.
Funding
This work was supported by the National Key R&D Program of China (2018YFD0900302–8), the National Natural Science Foundation of China (31972828) and the Shandong Provincial Natural Science Foundation of China (ZR2018BCE054). The funders were not involved in the study design; collection, analysis and interpretation of the data; or in writing the manuscript.
Availability of data and materials
The dataset supporting the conclusions of this article is available in the GenBank (https://www.ncbi.nlm.nih.gov/nuccore/MT646905) and the accession number is MT646905.
Ethics approval and consent to participate
The protocol was approved by the Animal Experimental Ethics Committee of Shandong Normal University (Permit Number: AEECSDNU2017007).
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yaoyao Zhu and Shijuan Shan contributed equally to this work.
Contributor Information
Guiwen Yang, Email: yanggw@sdnu.edu.cn.
Hua Li, Email: lihua@sdnu.edu.cn.
References
- 1.Mamane Y, Heylbroeck C, Genin P, Algarte M, Servant MJ, LePage C, DeLuca C, Kwon H, Lin R, Hiscott J. Interferon regulatory factors: the next generation. Gene. 1999;237(1):1–14. doi: 10.1016/s0378-1119(99)00262-0. [DOI] [PubMed] [Google Scholar]
- 2.Harada H, Taniguchi T, Tanaka N. The role of interferon regulatory factors in the interferon system and cell growth control. Biochimie. 1998;80(8–9):641–650. doi: 10.1016/s0300-9084(99)80017-0. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen H, Hiscott J, Pitha PM. The growing family of interferon regulatory factors. Cytokine Growth Factor Rev. 1997;8(4):293–312. doi: 10.1016/s1359-6101(97)00019-1. [DOI] [PubMed] [Google Scholar]
- 4.Escalante CR, Yie J, Thanos D, Aggarwal AK. Structure of IRF-1 with bound DNA reveals determinants of interferon regulation. Nature. 1998;391(6662):103–106. doi: 10.1038/34224. [DOI] [PubMed] [Google Scholar]
- 5.Eroshkin A, Mushegian A. Conserved transactivation domain shared by interferon regulatory factors and Smad morphogens. J Mol Med. 1999;77(5):403–405. doi: 10.1007/s001090050369. [DOI] [PubMed] [Google Scholar]
- 6.Huang B, Qi ZT, Xu Z, Nie P. Global characterization of interferon regulatory factor (IRF) genes in vertebrates: glimpse of the diversification in evolution. BMC Immunol. 2010;11:22. doi: 10.1186/1471-2172-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shu C, Sun Y, Xu T. Molecular characterization of three IRF1 subfamily members reveals evolutionary significance of IRF11 in miiuy croaker. Dev Comp Immunol. 2015;53(2):385–391. doi: 10.1016/j.dci.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Nehyba J, Hrdlickova R, Burnside J, Bose HR., Jr A novel interferon regulatory factor (IRF), IRF-10, has a unique role in immune defense and is induced by the v-Rel oncoprotein. Mol Cell Biol. 2002;22(11):3942–3957. doi: 10.1128/MCB.22.11.3942-3957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li S, Lu LF, Feng H, Wu N, Chen DD, Zhang YB, Gui JF, Nie P, Zhang YA. IFN regulatory factor 10 is a negative regulator of the IFN responses in fish. J Immunol. 2014;193(3):1100–1109. doi: 10.4049/jimmunol.1400253. [DOI] [PubMed] [Google Scholar]
- 10.Huang B, Jia QQ, Liang Y, Huang WS, Nie P. Interferon regulatory factor 10 (IRF10): cloning in orange spotted grouper, Epinephelus coioides, and evolutionary analysis in vertebrates. Fish Shellfish Immunol. 2015;46(2):669–677. doi: 10.1016/j.fsi.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 11.Huang B, Li WX, Wang ZX, Liang Y, Huang WS, Nie P. Identification of a novel splice variant isoform of interferon regulatory factor 10, IRF10, in orange spotted grouper Epinephelus coioides. Fish Shellfish Immunol. 2020;97:637–647. doi: 10.1016/j.fsi.2019.12.056. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki Y, Yasuike M, Kondo H, Aoki T, Hirono I. Molecular cloning and expression analysis of interferon regulatory factor 10 (IRF10) in Japanese flounder, Paralichthys olivaceus. Fish Shellfish Immunol. 2011;30(1):67–76. doi: 10.1016/j.fsi.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 13.Inkpen SM, Hori TS, Gamperl AK, Nash GW, Rise ML. Characterization and expression analyses of five interferon regulatory factor transcripts (Irf4a, Irf4b, Irf7, Irf8, Irf10) in Atlantic cod (Gadus morhua) Fish Shellfish Immunol. 2015;44(1):365–381. doi: 10.1016/j.fsi.2015.02.032. [DOI] [PubMed] [Google Scholar]
- 14.Eslamloo K, Xue X, Booman M, Smith NC, Rise ML. Transcriptome profiling of the antiviral immune response in Atlantic cod macrophages. Dev Comp Immunol. 2016;63:187–205. doi: 10.1016/j.dci.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 15.Xu Q, Jiang Y, Wangkahart E, Zou J, Chang M, Yang D, Secombes CJ, Nie P, Wang T. Sequence and expression analysis of interferon regulatory factor 10 (IRF10) in three diverse teleost fish reveals its role in antiviral defense. PLoS One. 2016;11(1):e0147181. doi: 10.1371/journal.pone.0147181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quesada-Garcia A, Encinas P, Valdehita A, Baumann L, Segner H, Coll JM, Navas JM. Thyroid active agents T3 and PTU differentially affect immune gene transcripts in the head kidney of rainbow trout (Oncorynchus mykiss) Aquat Toxicol. 2016;174:159–168. doi: 10.1016/j.aquatox.2016.02.016. [DOI] [PubMed] [Google Scholar]
- 17.Xu QQ, Yang DQ, Tuo R, Wan J, Chang MX, Nie P. Gene cloning and induced expression pattern of IRF4 and IRF10 in the Asian swamp eel (Monopterus albus) Dongwuxue Yanjiu. 2014;35(5):380–388. doi: 10.13918/j.issn.2095-8137.2014.5.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laghari ZA, Li L, Chen SN, Huo HJ, Huang B, Zhou Y, Nie P. Composition and transcription of all interferon regulatory factors (IRFs), IRF111 in a perciform fish, the mandarin fish, Siniperca chuatsi. Dev Comp Immunol. 2018;81:127–140. doi: 10.1016/j.dci.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 19.Carballo C, Castro D, Borrego JJ, Manchado M. Gene expression profiles associated with lymphocystis disease virus (LCDV) in experimentally infected Senegalese sole (Solea senegalensis) Fish Shellfish Immunol. 2017;66:129–139. doi: 10.1016/j.fsi.2017.04.028. [DOI] [PubMed] [Google Scholar]
- 20.Zhan FB, Jakovlic I, Wang WM. Identification, characterization and expression in response to Aeromonas hydrophila challenge of five interferon regulatory factors in Megalobrama amblycephala. Fish Shellfish Immunol. 2019;86:204–212. doi: 10.1016/j.fsi.2018.10.037. [DOI] [PubMed] [Google Scholar]
- 21.Peng D, Wang Z, Huang A, Zhao Y, Qin FX. A novel function of F-box protein FBXO17 in negative regulation of type I IFN signaling by recruiting PP2A for IFN regulatory factor 3 deactivation. J Immunol. 2017;198(2):808–819. doi: 10.4049/jimmunol.1601009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Y, Zhang S, Feng S, Sun J, Xu P. Genome wide identification, phylogeny and expression of zinc transporter genes in common carp. PLoS One. 2014;9(12):e116043. doi: 10.1371/journal.pone.0116043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Zhang F, Guo H, Zhu Y, Yuan J, Yang G, An L. Molecular characterization of hepcidin gene in common carp (Cyprinus carpio L.) and its expression pattern responding to bacterial challenge. Fish Shellfish Immunol. 2013;35(3):1030–1038. doi: 10.1016/j.fsi.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 24.Li T, Li H, Peng S, Zhang F, An L, Yang G. Molecular characterization and expression pattern of X box-binding protein-1 (XBP1) in common carp (Cyprinus carpio L.): indications for a role of XBP1 in antibacterial and antiviral immunity. Fish Shellfish Immunol. 2017;67:667–674. doi: 10.1016/j.fsi.2017.06.055. [DOI] [PubMed] [Google Scholar]
- 25.Shan S, Qi C, Zhu Y, Li H, An L, Yang G. Expression profile of carp IFN correlate with the up-regulation of interferon regulatory factor-1 (IRF-1) in vivo and in vitro: the pivotal molecules in antiviral defense. Fish Shellfish Immunol. 2016;52:94–102. doi: 10.1016/j.fsi.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 26.Feng H, Liu H, Kong R, Wang L, Wang Y, Hu W, Guo Q. Expression profiles of carp IRF-3/−7 correlate with the up-regulation of RIG-I/MAVS/TRAF3/TBK1, four pivotal molecules in RIG-I signaling pathway. Fish Shellfish Immunol. 2011;30(4–5):1159–1169. doi: 10.1016/j.fsi.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 27.Zhu Y, Qi C, Shan S, Zhang F, Li H, An L, Yang G. Characterization of common carp (Cyprinus carpio L.) interferon regulatory factor 5 (IRF5) and its expression in response to viral and bacterial challenges. BMC Vet Res. 2016;12(1):127. doi: 10.1186/s12917-016-0750-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Y, Shan S, Feng H, Jiang L, An L, Yang G, Li H. Molecular characterization and functional analysis of interferon regulatory factor 9 (irf9) in common carp Cyprinus carpio: a pivotal molecule in the Ifn response against pathogens. J Fish Biol. 2019;95(2):510–519. doi: 10.1111/jfb.14000. [DOI] [PubMed] [Google Scholar]
- 29.Shan SJ, Liu DZ, Wang L, Zhu YY, Zhang FM, Li T, An LG, Yang GW. Identification and expression analysis of irak1 gene in common carp Cyprinus carpio L.: indications for a role of antibacterial and antiviral immunity. J Fish Biol. 2015;87(2):241–255. doi: 10.1111/jfb.12714. [DOI] [PubMed] [Google Scholar]
- 30.Shan S, Wang L, Zhang F, Zhu Y, An L, Yang G. Characterization and expression analysis of Toll-interacting protein in common carp, Cyprinus carpio L., responding to bacterial and viral challenge. SpringerPlus. 2016;5:639. doi: 10.1186/s40064-016-2293-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu YY, Xing WX, Shan SJ, Zhang SQ, Li YQ, Li T, An L, Yang GW. Characterization and immune response expression of the rig-I-like receptor mda5 in common carp Cyprinus carpio. J Fish Biol. 2016;88(6):2188–2202. doi: 10.1111/jfb.12981. [DOI] [PubMed] [Google Scholar]
- 32.Zhang YB, Gui JF. Molecular regulation of interferon antiviral response in fish. Dev Comp Immunol. 2012;38(2):193–202. doi: 10.1016/j.dci.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 33.Honda K, Taniguchi T. IRFs: master regulators of signalling by toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6(9):644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 34.Ai K, Luo K, Li Y, Hu W, Gao W, Fang L, Tian G, Ruan G, Xu Q. Expression pattern analysis of IRF4 and its related genes revealed the functional differentiation of IRF4 paralogues in teleost. Fish Shellfish Immunol. 2017;60:59–64. doi: 10.1016/j.fsi.2016.11.038. [DOI] [PubMed] [Google Scholar]
- 35.Holland JW, Karim A, Wang T, Alnabulsi A, Scott J, Collet B, Mughal MS, Secombes CJ, Bird S. Molecular cloning and characterization of interferon regulatory factors 4 and 8 (IRF-4 and IRF-8) in rainbow trout, Oncorhynchus mykiss. Fish Shellfish Immunol. 2010;29(1):157–166. doi: 10.1016/j.fsi.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Chen X, Hu G, Dong X, Liu Q, Zhang S. Molecular cloning and expression analysis of interferon regulatory factor 8 (IRF8) in turbot, Scophthalmus maximus. Vet Immunol Immunopathol. 2012;149(1–2):143–150. doi: 10.1016/j.vetimm.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 37.Hu GB, Zhao MY, Lin JY, Liu QM, Zhang SC. Molecular cloning and characterization of interferon regulatory factor 9 (IRF9) in Japanese flounder, Paralichthys olivaceus. Fish Shellfish Immunol. 2014;39(2):138–144. doi: 10.1016/j.fsi.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 38.Bathige SD, Whang I, Umasuthan N, Lim BS, Park MA, Kim E, Park HC, Lee J. Interferon regulatory factors 4 and 8 in rock bream, Oplegnathus fasciatus: structural and expressional evidence for their antimicrobial role in teleosts. Fish Shellfish Immunol. 2012;33(4):857–871. doi: 10.1016/j.fsi.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 39.Zhan FB, Liu H, Lai RF, Jakovlic I, Wang WM. Expression and functional characterization of interferon regulatory factors (irf2, irf7 and irf9) in the blunt snout bream (Megalobrama amblycephala) Dev Comp Immunol. 2017;67:239–248. doi: 10.1016/j.dci.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 40.Zhang J, Li YX, Hu YH. Molecular characterization and expression analysis of eleven interferon regulatory factors in half-smooth tongue sole, Cynoglossus semilaevis. Fish Shellfish Immunol. 2015;44(1):272–282. doi: 10.1016/j.fsi.2015.02.033. [DOI] [PubMed] [Google Scholar]
- 41.Hori TS, Gamperl AK, Booman M, Nash GW, Rise ML. A moderate increase in ambient temperature modulates the Atlantic cod (Gadus morhua) spleen transcriptome response to intraperitoneal viral mimic injection. BMC Genomics. 2012;13:431. doi: 10.1186/1471-2164-13-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomas JM. The main Aeromonas pathogenic factors. ISRN Microbiol. 2012;2012:256261. doi: 10.5402/2012/256261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li C, Wang R, Su B, Luo Y, Terhune J, Beck B, Peatman E. Evasion of mucosal defenses during Aeromonas hydrophila infection of channel catfish (Ictalurus punctatus) skin. Dev Comp Immunol. 2013;39(4):447–455. doi: 10.1016/j.dci.2012.11.009. [DOI] [PubMed] [Google Scholar]
- 44.Rodkhum C, Hirono I, Stork M, Di Lorenzo M, Crosa JH, Aoki T. Putative virulence-related genes in Vibrio anguillarum identified by random genome sequencing. J Fish Dis. 2006;29(3):157–166. doi: 10.1111/j.1365-2761.2006.00692.x. [DOI] [PubMed] [Google Scholar]
- 45.Chevaliez S, Pawlotsky JM. Interferon-based therapy of hepatitis C. Adv Drug Deliv Rev. 2007;59(12):1222–1241. doi: 10.1016/j.addr.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 46.Langevin C, Aleksejeva E, Passoni G, Palha N, Levraud JP, Boudinot P. The antiviral innate immune response in fish: evolution and conservation of the IFN system. J Mol Biol. 2013;425(24):4904–4920. doi: 10.1016/j.jmb.2013.09.033. [DOI] [PubMed] [Google Scholar]
- 47.Zou J, Secombes CJ. Teleost fish interferons and their role in immunity. Dev Comp Immunol. 2011;35(12):1376–1387. doi: 10.1016/j.dci.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 48.Sun F, Zhang YB, Liu TK, Shi J, Wang B, Gui JF. Fish MITA serves as a mediator for distinct fish IFN gene activation dependent on IRF3 or IRF7. J Immunol. 2011;187(5):2531–2539. doi: 10.4049/jimmunol.1100642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Fig. S1. Nucleotide sequence and the deduced amino acid sequence of C. carpio IRF10. Lowercase letters indicate the 5′ and 3′ UTR, while uppercase letters indicate the ORF or amino acid. The start codon (ATG) and stop codon (TGA) are boxed in red. The DBD and IAD are shaded in blue and pink, respectively. The NLS is underlined, and the five tryptophan (W) residues are boxed in black.
Supplementary Fig. S2. Overexpression of CcIRF10 in EPC cells. EPC cells were transfected with pcDNA3.1-EGFP empty plasmid or pcDNA3.1-EGFP-CcIRF10, and the gene expression of CcIRF10 was detected using real-time PCR. The expression was normalized to that of EF1α. (n = 3, mean ± SD, **P < 0.01).
Supplementary Fig. S3. Transfection efficiency of EPC cells. Bright field (A) and fluorescent image (B) of EPC cells transfected with pcDNA3.1-EGFP empty plasmid. Bright field (C) and fluorescent image (D) of EPC cells transfected with pcDNA3.1-EGFP-CcIRF10 plasmid. (original magnification × 40).
Data Availability Statement
The dataset supporting the conclusions of this article is available in the GenBank (https://www.ncbi.nlm.nih.gov/nuccore/MT646905) and the accession number is MT646905.