Abstract
The recently emerged COVID-19 has been declared a pandemic by the World Health Organization as to date; no therapeutic drug/vaccine is available for the treatment. Due to the lack of time and the urgency to contain the pandemic, computational screening appears to be the best tool to find a therapeutic solution. Accumulated evidence suggests that many phyto-compounds possess anti-viral activity. Therefore, we identified possible phyto-compounds that could be developed and used for COVID-19 treatment. In particular, molecular docking was used to prioritize the possible active phyto-compounds against two key targets namely RNA dependent RNA polymerase (RdRp) and main protease (Mpro) of SARS-CoV-2. In this study, an antiviral drug- Remdesivir (RdRp inhibitor) and Darunavir (Mpro inhibitor) are used as reference drugs. This study revealed that phyto-molecules- Mulberroside-A/C/E/F, Emblicanin A, Nimbolide, and Punigluconin showed high binding affinity against RdRp while Andrographolides, Mulberrosides, Anolignans, Chebulic acid, Mimusopic acid, and Punigluconin showed better binding affinity against Mpro as compared with the reference drug. Furthermore, ADME profiles validated the drug-likeness properties of prioritized phyto-compounds. Besides, to assess the stability, MD simulations studies were performed along with reference inhibitors for Mpro (Darunavir) and RdRp (Remdesivir). Binding free energy calculations (MM-PBSA) revealed the estimated value (ΔG) of Mpro_Darunavir; Mpro_Mulberroside E; RdRp_Remdesivir and RdRp_Emblicanin A were −111.62 ± 6.788, −141.443 ± 9.313, 30.782 ± 5.85 and −89.424 ± 3.130 kJmol−1, respectively. Taken together, the study revealed the potential of these phyto-compounds as inhibitors of RdRp and Mpro inhibitor that could be further validated against SARS-CoV-2 for clinical benefits.
Communicated by Ramaswamy H. Sarma
Keywords: COVID-19, phyto-compounds, molecular docking, SARS-CoV-2, molecular dynamics, MM-PBSA
1. Introduction
Novel Coronavirus disease (COVID-19) - an ongoing pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) responsible for causing severe acute respiratory symptoms such as pneumonia, fever, dyspnea, and asthenia (Hui et al., 2020; Lu et al., 2020; Zhu et al., 2020). According to the information on real-time world statics- worldometer, as of September 10, 2020, the virus has infected more than 28,181,355 people and caused 910,686 deaths (https://www.worldometers.info/coronavirus). A recent study suggests that SARS-CoV-2 is a positive-sense, single-stranded RNA virus belonging to the betacoronavirus genus of the coronaviridae family (Gorbalenya et al., 2020).
The genome of SARS-CoV-2 is much more similar to SARS and MERS (Middle East Respiratory Syndrome) that encodes structural proteins namely S (spike glycoprotein), E (envelope), M (membrane), and N (nucleocapsid) and non-structural proteins- main protease (Mpro), papain-like protease, RNA dependent RNA polymerase (RdRp). The structural proteins are chiefly responsible for the interactions between virus and host cells during viral entry events whereas the non-structural proteins are involved in the transcription and replication process during the virus life cycle. (Elmezayen et al., 2020; Kalita et al., 2020; Khan et al., 2020; Padhi et al., 2020; Zumla et al., 2016).
Among two proteases, main protease (Mpro)/3CLpro is a key enzyme for virus replication and has a dominant role in the post-translational process responsible for its maturation. Inhibition of Mpro activity can effectively block the virus replication process. Also, Mpro inhibitors are likely to be non-toxic to humans due to the lack of analogous cleavage specificity sites of human proteases. Mpro also plays an important role in host immune regulation (Liu et al., 2017; Liu & Wang, 2020; Zhang et al., 2020). Furthermore, a highly conserved three-dimensional structure of Mpro among all the known coronaviruses (CoVs), makes it a promising therapeutic target for the development of broad-spectrum anti-COVID drugs (Morse et al., 2020).
Besides, RNA-dependent RNA polymerase (RdRp) is another highly conserved anti-COVID-19 drug target. RdRp, also known as nsp12, acts as a catalyst for the CoV RNA synthesis and is a crucial member of corona viral replication/transcription machinery complex and importantly possesses no host cell homolog (Gao et al., 2020). This paves the way for the development of antiviral drugs with less toxicity to human cells. As viral RdRp lacks proofreading activity therefore, drugs such as chain terminators or mutagenic nucleoside analog inhibitors targeting RdRp have been investigated (Campagnola et al., 2011). Favipiravir and remdesivir are two such nucleoside analogs that function by blocking viral RNA synthesis and are currently being approved for emergency use for the COVID-19 treatment (Li & De Clercq, 2020).
Since CoVs are subjected to extensive mutations during their life cycle, but the probability of getting mutations in the highly conserved key proteins i.e. Mpro and RdRp is rare, as these mutations are usually lethal to the virus itself (Zhang et al., 2010). Therefore, in the current study, we hypothesized that targeting Mpro and RdRp offers a much more promising therapeutic strategy as it performs a dual function, one that prevents virus replication and proliferation and the other that reduces the risk of mutation mediating drug resistance.
Targeting the DNA/RNA synthesis or inhibiting the viral entry or their propagation has been the main mechanism of anti-viral agents derived from phyto-compounds. We know nature is a vast reservoir of diverse therapeutic agents and a large number of modern drugs are based upon either natural molecules or their derivatives (Cragg & Newman, 2001; Mathur & Hoskins, 2017). Scientific studies suggested that various phyto-compounds belong to flavonoids, phenolic, terpenoids, etc. groups have been found to possess therapeutic implementation against various diversified viruses (Ben-Shabat et al., 2020; Naithani et al., 2008). Therefore, in this study, we selected major bioactive phyto-compounds of traditionally used plants reported against different viral diseases.
There is an urgent need to prevent the outbreak by interrupting the viral infections. The computational screening method of drug discovery is a rapid and economic screening tool for screening of potential hits against selected targets (Sliwoski et al., 2014). The interaction between ligands and target proteins were analyzed using molecular docking method followed by the prediction of absorption, distribution, metabolism, and excretion (ADME) drug properties using the computational method (Berry et al., 2015). Molecular dynamics simulations of top docked protein-ligand complexes were performed to evaluate trajectories and interaction between receptor and drug molecules. The MM-PBSA approach subsequently estimated binding free energy calculations.
2. Materials and methods
2.1. Selection and preparation of a ligand library
The ligand library of 22 major bioactive molecules was prepared from 10 medicinal plants (Supplementary Table 1). Based on the literature search, we selected these 10 medicinal plants which possessed anti-viral activities against different groups of viruses and also available in Gujarat. Based on the recent articles of COVID-19, Remdesivir (Al-Tawfiq et al., 2020; Gao et al., 2020), Thymoquinone (Elfiky, 2020), Hydroxychloroquine (M. Wang et al., 2020), Favipiravir (Furuta et al., 2013), Darunavir and Nelfinavir (Chandel et al., 2020) were selected for a ligand library of positive control.
Table 1.
The values of binding affinity (kcal/mol) of molecular docking between various targets of SARS-CoV-2 and all selected phyto-compounds with comparison to reference compounds.
| Sr. No. | Ligand | Source Plant | PubChem ID | Chemical Group | Binding affinity (kcal/mol) |
|
|---|---|---|---|---|---|---|
| 6M71 (RNA polymerase) |
6LU7 (Main Protease/3CLpro) |
|||||
| 1 | Andrographolide | Andrographis paniculata | 5318517 | Diterpenoid | −6.3 | −6.5 |
| 2 | Hydro andrographolide | 72191643 | Diterpenoid | −6.3 | −6.1 | |
| 3 | Iso andrographolide | 101563021 | Diterpenoid | −6.3 | −6.1 | |
| 4 | Neo andrographolide | 9848024 | Diterpenoid | −7.0 | −6.5 | |
| 5 | Oxo andrographolide | 101593061 | Diterpenoid | −7.1 | −6.9 | |
| 6 | Mulberroside A | Morus alba | 6443484 | Phenols | −9.1 | −6.4 |
| 7 | Mulberroside C | 190453 | Phenols | −8.1 | −6.3 | |
| 8 | Mulberroside E | 10030502 | Phenols | −9.0 | −7.0 | |
| 9 | Mulberroside F | 60208818 | Phenols | −8.8 | −6.7 | |
| 10 | Mimusopic acid | Mimusops elengi | 6712545 | Triterpene | −6.5 | −6.2 |
| 11 | Glycyrrhizin | Glycyrrhiza glabara | 46783814 | Triterpenoid | −7.1 | −5.3 |
| 12 | Curcumin | Curcuma longa | 969516 | Phenols | −7.0 | −5.2 |
| 13 | Nimbidinin | Azardirachta indica | 101306757 | Triterpene | −7.1 | −5.7 |
| 14 | Nimbolide | 86287562 | Triterpene | −7.8 | −5.3 | |
| 15 | Emblicanin A | Phyllanthus emblica | 119058016 | Tannins | −9.2 | −5.7 |
| 16 | Emblicanin B | 119058017 | Tannins | −7.8 | −3.9 | |
| 17 | Punigluconin | 44631480 | Tannins | −8.7 | −6.5 | |
| 18 | Chebulic acid | Terminalia chebula | 12302892 | Phenols | −6.3 | −6.5 |
| 19 | Arjunolic acid | Terminalia arjuna | 73641 | Triterpenoid | −7.3 | −5.4 |
| 20 | Anolignan A | Terminalia bellerica | 72391 | Lignan | −7.0 | −6.5 |
| 21 | Anolignan B | 72388 | Lignan | −6.0 | −6.5 | |
| 22 | Anolignan C | 454714 | Lignan | −6.0 | −6.5 | |
| 23 | Remdesivir (RdRp) | – | 121304016 | −7.5 | −5.1 | |
| 24 | Thymoquinone | – | 10281 | −5.6 | −5.1 | |
| 25 | Hydroxychloroquine | – | 3652 | −5.2 | −5.7 | |
| 26 | Favipiravir | – | 492405 | −5.2 | ||
| 27 | Darunavir(Main Protease/3CLpro) | – | 213039 | – | −5.9 | |
| 28 | Nelfinavir | – | 64143 | – | −5.8 | |
Bold values signifies higher binding affinity as compared to reference drug.
The 3D structures of all 22 natural test ligands and all positive control ligands were retrieved from the PubChem database (https://pubchem.ncbi.nlm.nih.gov). All ligands were minimized by default parameter i.e. Universal Force Field (Rappe et al., 1992) as implemented in Open Babel software package (O’Boyle et al., 2011), followed by conversion of ligands to PDBQT format using graphical user interface version of PyRx virtual screening tool-python prescription 0.8.
2.2. Receptor preparation
The X-ray crystallographic structures of targeted proteins-3C-like protease (PDB ID-6LU7) and RNA dependent RNA polymerase (PDB ID- 6M71) were retrieved from RCSB-Protein data bank (https://www.rcsb.org.) Proteins were refined and prepared by the charge assignment, solvation parameters, and fragmental volumes using Autodock tool 4.2.6 (Morris et al., 2009).
2.3. Molecular docking between receptor and ligands
Molecular docking analysis was carried out using Auto dock Vina Wizard of PyRx software (Dallakyan & Olson, 2015; Pagadala et al., 2017). The Auto-Grid engine in PyRx was used to generate the configuration files. The grid box dimensions (Å) are X = 74.8063, Y = 84.5421, Z = 106.0308 for RdRp and X = 51.7507, Y = 66.9737, Z = 88.0470 for Mpro for vina parameter exhaustiveness was set to by default value i.e. 8.After the virtual screening was completed, PyRx automatically advances to analyze result are giving us by default, 9 best binding models for each docking run. The highest binding energy (most negative) was considered as the ligand with maximum binding affinity. The 2D ligand-protein interaction diagrams were generated by Maestro version 11.6.013 (Schrödinger Suite, LLC, NY) to find out the involved amino acids with their interactive position in the docked molecule. To figure out the drug surface hotspot from the docked structures of RdRp and 3CL-pro with their prioritized inhibitors using PyMOL software (Seeliger & de Groot, 2010).
2.4. Drug likeness properties-ADME prediction
Prediction of the drug ability of potential hits was tested using Lipinski’s rule of five and ADME properties. Different molecular parameters such as molecular weight, numbers of hydrogen bond acceptors, number of hydrogen bond donors, and LogP values were analyzed as per Lipinski’s rule of five. Further, absorption, distribution, metabolism, and excretion (ADME) properties were evaluated using Swiss ADME program (Daina et al., 2017).
2.5. Molecular Dynamics (MD) simulation of top docked complexes
The top docked compounds with Mpro and RdRp were further subjected to MD simulation. The top docked compound Mulberroside E and reference compound-Darunavir were selected for Mpro protein and, Emblicanin A and reference drug-Remdesivir were selected for RdRp for MD simulations studies. A set of all four MD simulations systems was prepared and subjected for MD simulation with 50 ns time duration. The MD simulations were performed by the reported method of Patel et al. (Patel et al., 2017; 2018) using GROMACS ver.2016.4 (Van Der Spoel et al., 2005) with Amber99SB force-field (Lindorff-Larsen et al., 2010). ACPYPE server was used for generating inhibitor parameters and topology (Sousa da Silva & Vranken, 2012). All the MD simulations systems were solvated using Three-site (TIP3P) water model with a dodecahedron box configuration setting with a distance of 1 nm from edges of protein in all the directions. The MD systems were then neutralized with an equal number of counter ions (Na+/Cl-) and further energy minimization with the steepest descent algorithm was used to remove any steric clashes and bad contacts and generate maximum force below 1000 kJmol−1nm−1 (50000 steps max). Post energy minimization, equilibration with position restraint was carried out under NVT (constant number [N], constant volume [V] and constant temperature [T]) and NPT (constant number [N], constant pressure [P] and constant temperature [T]) ensemble for 1 ns each. Berendsen thermostat algorithm (Berendsen et al., 1984) was used for maintaining the system at constant volume (100 ps) and at a constant temperature (300 K) in NVT equilibration. Further, NPT equilibration was performed at a constant pressure (1 bar) for 100 ps maintained by Parrinello-Rahmanbarostat (Parrinello & Rahman, 1980). The Particle Mesh Ewald approximation was applied with 1 nm cut-off for calculating long-range electrostatic interactions, computing coulomb & the van der Waals interactions (Darden et al., 1993). The bond length was constraint using the LINCS algorithm (Hess et al., 1997). Finally, a 50 ns simulation run was carried out with the default parameters and saving the coordinates at every 2 fs time frame. The MDS trajectories were visualized using VMD (Humphrey et al., 1996) and Chimera (Pettersen et al., 2004). For calculation of root mean square deviation (RMSD), root mean square fluctuation (RMSF), and hydrogen bonds (H-bonds), etc. in-built ‘gmx’ commands were used in GROMACS and the plotting tool GRACE was used for the generation and visualization of the plots (http://plasma-gate.weizmann.ac.il/Grace) as reported in our previous study (Patel et al., 2018).
2.6. Clustering of conformations and PCA/essential dynamics
The 50 ns entire MDS trajectories were subjected to RMSD based clustering using ‘gmx cluster’ that explores the conformational landscape among the ensemble of protein structures. The GROMOS algorithm (Daura et al., 1999) was used to determine the dominant conformation with Cα RMSD cut-off value 0.15 nm. To understand the collective and overall motion for all the MDS systems, Principal Component Analysis (PCA) or Essential Dynamics (ED) analysis was carried out as reported in our previous study using ‘gmxcovar’ and ‘gmxanaeig’ tools (Manhas et al., 2019; Patel et al., 2020). PCA reduces the complexity of the data and results in the concerted motion in the MDS which are correlated and significant for biological functions. The set of eigen vectors and eigen values were computed by diagonalizing the covariance matrix after removing the translational and rotational motions. The amplitude of the eigenvector is represented by eigen values along with the multidimensional space, while the Cα displacement along each eigenvector shows the concerted motions of the protein along each direction.
2.7. Computation of binding free energy using MM/PBSA
The Poisson–Boltzmann or generalized Born and surface area continuum solvation (MM/PBSA and MM/PBSA) are routinely used and widely-accepted methods for computing the protein-inhibitor affinity (Genheden & Ryde, 2015; Sun et al., 2018; Wang et al., 2016). For calculating the binding free energy of Mpro and RdRp docked complex with Mulberroside E and Emblicanin A, along with selected reference inhibitor, Darunavir, and Remdesivir respectively. The binding free energy calculations and energy contribution by individual residues were used to quantitatively estimate the inhibitor affinity for Mproand RdRp. The ‘g_mmpbsa’ tool (Kumari et al., 2014) with default parameters was used for molecular mechanics potential energy (electrostatic + Van der Waals interactions) and solvation free energy (polar + non-polar solvation energies) calculations. The last stable 30 ns (150 frames) trajectories assess by the RMSD plot from each docked complex were used to estimate binding free energy. The frames were selected at a regular interval of 200 ps covering a wide range of trajectory to cover different conformational space of the docked complexes for better structure-function correlation (Padhi et al., 2020).
3. Results and discussion
In the present study, we prepared a ligand library of 22 phyto-compounds of 10 medicinal plants and subjected for molecular docking to predict hits for two key targets namely RNA dependent RNA polymerase (RdRp) and main protease (Mpro) of SARS-CoV-2 that are responsible for current pandemic COVID-19. The results of molecular docking and ADME analysis, molecular dynamics simulation and binding free energy calculations of the present study have been discussed below:
3.1. Molecular docking of phyto-compounds with RdRp and mpro
Molecular docking is one of the vital method in the in silico drug design and discovery process. It provides the details of protein-ligand interaction in the form of binding affinity score and also generates the binding pose (Khan et al., 2020). The positive control is very important in every type of in silico or wet-lab experiments. Based on the literature survey of the recently published articles, we selected a few anti-viral compounds for the preparation of a library of positive control (references is already mentioned in methodology) against RdRp and Mpro. The results of the docking study revealed that Remdesivir showed the best binding affinity −7.5 kcal/mol against RdRp and HIV protease inhibitor drug- Darunavir showed the best binding affinity −5.9 kcal/mol amongst positive control ligand library. Interestingly, more than 30% ligands and 65% ligands from test ligand library exhibited strong binding affinity to RdRp and Mpro respectively compared to these positive molecules. Our results also revealed that most of the bioactive compounds that showed a significant score against different targets of SARS-CoV-2 belong to phenol, terpenes, and tannins groups. Target specific efficacies of potential phyto-compounds are discussed as below.
3.1.1. RNA dependent RNA polymerase (RdRp)
Coronaviruses (CoVs) are reported to utilize a multiple-subunit transcriptional structure. A set of non-structural proteins (nsp) come together to assist in viral replication and transcription. A key element, the RNA-dependent RNA polymerase (RdRp, also known as nsp12), drive the synthesis of viral RNA and hence plays a critical role in the replication and transcription cycle of SARS-CoV-2 (Subissi et al., 2014). RdRp is considered as a potential target for the inhibition of SARS-CoV-2 and subsequent inhibition of COVID-19. Therefore we aimed to dock all the ligands against RdRp. The docking results of the positive controls revealed that Remdesivir possessed the highest binding affinity energy of −7.5 kcal/mol and Remdesivir also mentioned as RdRp inhibitor in the Gao et al. report (Gao et al., 2020). Total 7 phyto-compounds namely Mulberroside A, Mulberroside C, Mulberroside E, Mulberroside F, Nimbolide, Emblicanin A, and Punigluconin possessed more binding affinity than Remdesivir. All four Mulberrosides that are bioactive molecules of Morus alba plant showed good binding interaction and binding affinity score (Table 1). Accumulated evidence also suggested that these compounds or the plant extract of Morus alba containing these compounds also possessed potential anti-viral activity and immunomodulatory activity (Chan et al., 2016). The results of Vora et al. also showed that Mulberrosides were potentially effective against multi targets of HIV (Vora et al., 2019). Emblicanin A and Punigluconin of Phyllanthus emblica also showed good binding affinity −9.2 kcal/mol and −8.7 kcal/mol respectively. These molecules and the extracts of Phyllanthus emblica that containing these molecules are also reported for antiviral activities against different groups of viruses and as an immunomodulator (Belapurkar et al., 2014; Nisar et al., 2018; Salehi et al., 2018). Therefore, these molecules could be tested and developed as RdRp inhibitors.
Furthermore, Hydroxychloroquine (HCQ) – an anti-malarial drug (Lim et al., 2009) that is used in COVID-19 treatment. In our ligand library, a phyto-molecule-Nimbolide from Azardirachta indica plant is reported for anti-malarial activity (Udeinya et al., 2004) and also exhibited good binding affinity −7.8 kcal/mol against RdRp. Hence, this molecule could be further used against RdRp target of the COVID-19.
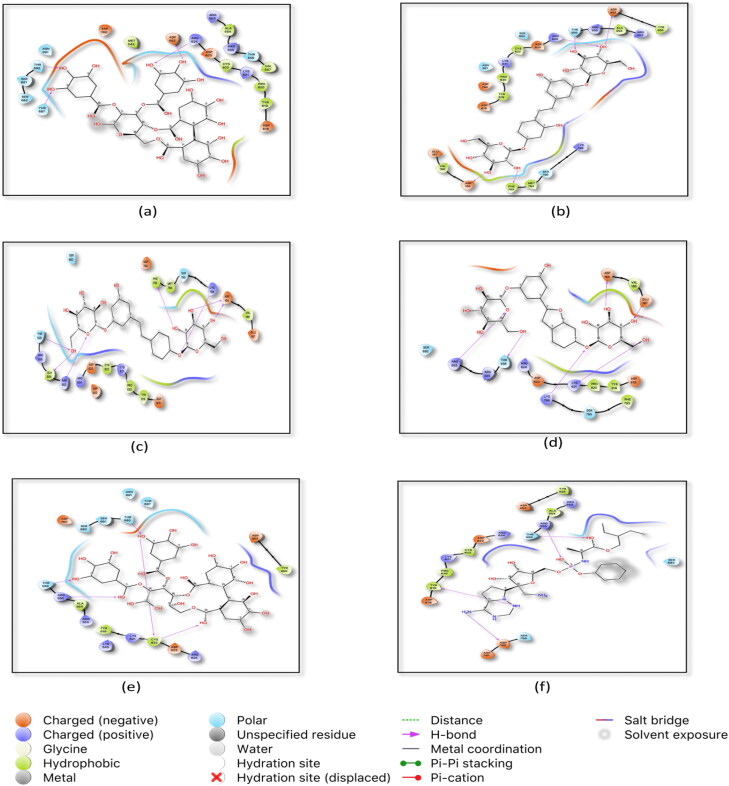
The active site residues Arg 553, Arg 555, Arg 624 and Asp 760 were common in both the positive control and the top-ranked ligands (Table 2). Hydrogen bond is considered as a crucial type of interaction in drug discovery and development process as of their strong influence on drug likeliness properties ( Sinha et al., 2019; Vora, Patel et al., 2020 ). The active residues which interact by hydrogen bonding are shown in Figures 1 and 3D pocket images are given in Supplementary Figure 1.
Table 2.
Active amino acid residues participating in interaction.
| Ligands | Amino acid residues interacting with |
|
|---|---|---|
| RNA dependent RNA polymerase | Main protease/ Mpro | |
| Andrographolide | – | GLN107,THR292 |
| Hydro andrographolide | – | GLN107,GLN110 |
| Iso andrographolide | – | ARG105 |
| Neo andrographolide | – | GLU290 |
| Oxo andrographolide | – | THR111,THR292 |
| Anolignan A | – | GLN107,THR111,THR292 |
| Anolignan B | – | THR111 |
| Anolignan C | – | THR111 |
| Chebulic acid | – | GLN110,THR111,ASN151,SER158 |
| Emblicanin A | ASP452,ARG624,THR687,THR680 | – |
| Emblicanin B | ARG555,THR556,SER682,SER759,ASP760 | – |
| Mimusopic acid | – | LYS5,GLU290 |
| Mulberroside A | ASP164,ASP452,TYR455,THR556,ARG624,PHE793 | ASN151,SER158,ASP176 |
| Mulberroside C | TYR619,LYS621,CYS622,ASP623,ASP760 | PHE103,GLN110 |
| Mulberroside E | ASP164,ARG553,ALA554,THR556,LYS798,PHE793 | THR111,HIS246 |
| Mulberroside F | ASP164,GLU167,ARG553,THR556,LYS621,LYS798 | PHE103,ARG105 |
| Nimbolide | ARG553,ARG555,ASN691 | – |
| Punigluconin | ARG555,THR556,CYS622,THR680 | LYS5,GLY138 |
| Darunavir(Mpro/3CLpro) | – | LYS5,LYS137,ASP289 |
| Remdesivir( RNA polymerase) | ARG553,ARG555,ARG619,ASP760 | – |
Figure 1.
Top docked natural ligands and standard inhibitor with RNA dependent RNA Polymerase (RdRp). (a) 2D ligand interaction of Emblicanin A in the active site of RdRp (b) interaction of Mulberroside A in RdRp active site (c) interaction of Mulberroside E in RdRp active site (d) interaction of Mulberroside F in RdRp active site (e) interaction of Punigluconin in RdRp active site (f) interaction of Remdesivir in RdRp active site.
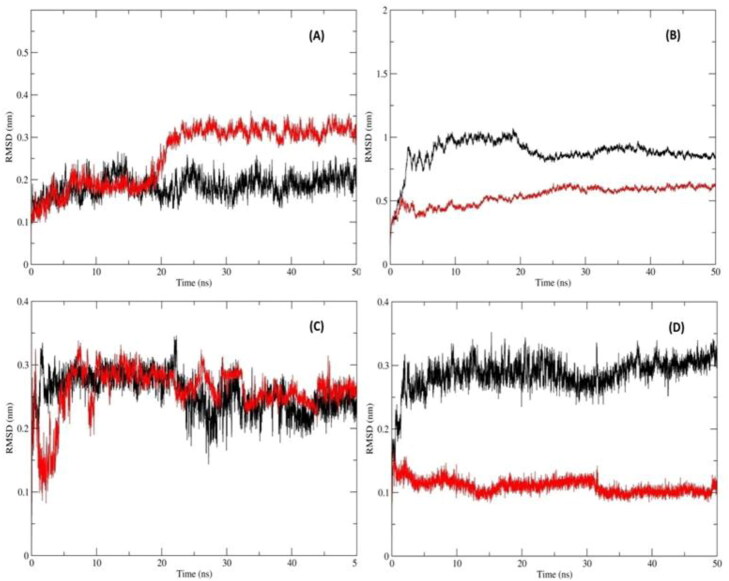
Figure 3.
RMSD of Mpro and RdRp with inhibitors computing the deviation (nm) vs. function of time (50 ns):(A) RMSD of the protein Cα backbone atoms Mpro_Darunavir (Black); Mpro_MulberrosideE (Red) (B) RMSD of the protein Cα backbone atoms RdRp_Remdesivir (Black) and RdRp_EmblicaninA (Red) (C) RMSD of the inhibitor atoms of Mpro_Darunavir (Black) and Mpro_MulberrosideE (Red)(D) RMSD of the inhibitor atoms of RdRp_Remdesivir (Black)and RdRp_EmblicaninA (Red).
3.1.2. Main protease (mpro)/3CLpro
Reports suggest that coronavirus protease enzyme Mpro is also known as 3-chymotrypsin-like protease (3CLpro) or the main protease that is necessary for proteolytic maturation of the SARS-CoV-2. This Mpro plays a critical part in the immune regulation and cleaving the polyproteins pp1a and pp1ab. Active enzymes such as RNA polymerase, endo-ribonuclease, and exo-ribonuclease are produced by cleavage of polyproteins by Mpro (Zhou et al., 2019). This makes Mpro an attractive target for anti-COVID-19 drugs, as the inhibition of the main protease of SARS-CoV-2 would inhibit the viral maturation process as wells as enhance the patient’s immune response against the disease. Prompted by this we docked all the ligands against main protease Mpro)/3CLpro. Darunavir showed maximum binding affinity −5.9 kcal/mol against main protease from all the selected reference drugs. Since the results of Chandel et al. (2020) suggested that Nelfinavir, a HIV protease inhibitor exhibited the highest binding affinity against main protease of COVID-19 (Chandel et al., 2020). Therefore, we also docked a HIV protease inhibitor-Darunavir along with Nelfinavir and the docking results revealed that Darunavir possessed greater binding affinity than Nelfinavir against main protease of SARS-CoV-2. Hence, Darunavir was further selected as a positive reference. Lys5, Lys137, and Asp 289 are active aminoacid residues participating in the hydrogen bond interaction (Table 2). Around 65% ligands of our ligand library exhibited more binding affinity in compare to positive reference drug. Different diterpenoids of Andrographis paniculata showed higher binding affinity than Darunavir (Table 1).The in silico results of Enmozhi et al. (2020) reported that Andrographolide showed binding affinity −3 kcal/mol against main protease of SARS-CoV-2 (Enmozhi et al., 2020). In our study, Neo andrographolide, Oxo andrographolide, Iso andrographolide, and Hydro andrographolide along with Andrographolide showed better binding affinity (Table 1). Various studies also prove that these molecules containing plant Andrographis paniculata possessed antiviral and immunostimulant activities (Churiyah et al., 2015). Therefore, these molecules could become a prominent drug as main protease inhibitor of COVID-19 virus. The active site residues of these molecules were Gln107, Thr292, Glu290, Gln110, and Arg105 which showed hydrogen bond interaction (Figure 2). Mulberrosides of Morus alba plants also exhibited a more binding affinity ranged from −6.2 to −7.0 kcal/mol. Interestingly, Mulberrosides also showed better binding affinity against RdRp, hence these molecules could be developed as a multi-target hit for SARS-CoV-2. Punigluconin, Chebulic acid, and Anolignans that are present in Phyllanthus emblica, Terminalia chebula and Terminalia bellerica respectively also offer higher binding affinity against main protease. These three plant components are present in Triphala which have various therapeutic activities including immunostimulant (Belapurkar et al., 2014; Peterson et al., 2017). Hence, these molecules are individually or in combination could be developed as a protease inhibitor of the COVID-19 virus.
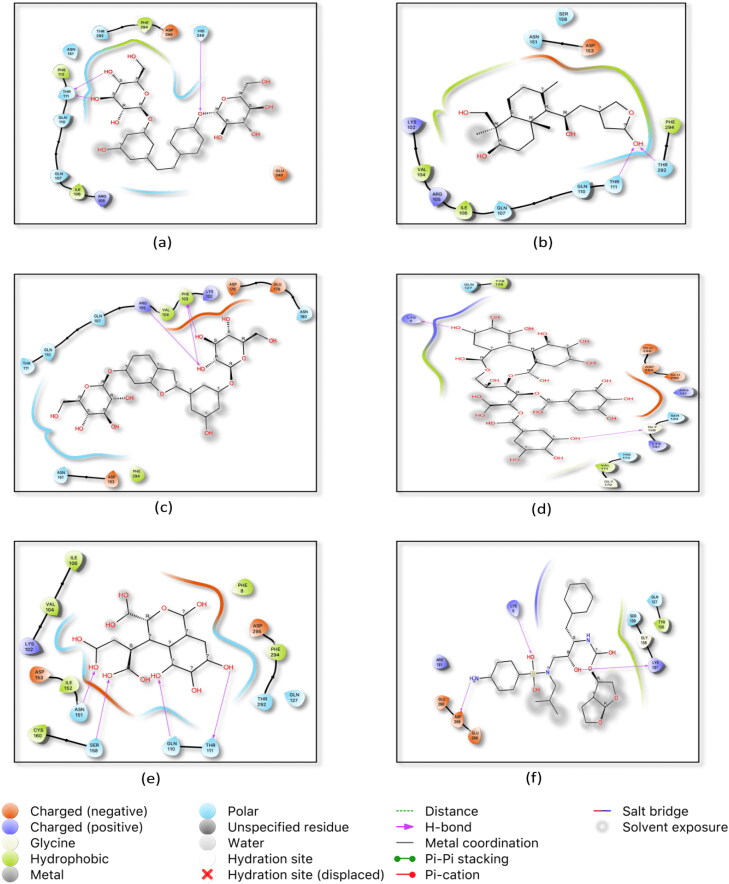
Figure 2.
Top docked natural ligands and standard inhibitor with Main Protease (Mpro). (a) 2 D ligand interaction of Mulberroside E in the active site of Mpro (b) interaction of Oxo-andrographolide in Mpro active site (c) interaction of Mulberroside F in Mpro active site (d) interaction of Punigluconin in Mpro active site (e) interaction of Chebulic acid in Mproactive site (f) interaction of Darunavir in Mpro active site.
The active residues which possessed hydrogen bond interaction are given in Table 2 and the 3D images are given in Supplementary Figure 2.
3.2. Drug likeness properties-ADME prediction
Drug likeness is a crucial criterion in selection of hits at the early stage of drug discovery process. The pharmacokinetics properties of potential RdRp inhibitors and protease inhibitors were tested by SWISS ADME online server and the results are shown in Table 3. The results of aqueous solubility depicted that all potent hits are fit in soluble to moderately soluble range. All potential compounds have no Blood brain barrier (BBB) permeability and CYP2D6 inhibitors except Anolignans and Iso andrographolide. P-Glycoprotein (P-gp) plays a major role to protect the central nervous system from xeno-biotics. Most of the hits of our study pass in P-gp substrate criteria. Of 17, total 11 hits have high gastrointestinal (GI) absorption capacity and remaining have low GI absorption activity that could be modified and developed as a drug like candidate.
Table 3.
Predicted pharmacokinetics properties for druglikeliness of top ranked ligands.
| Compound Name | Aqueous Solubility | Solubility class | BBB permeant | CYP2D6 inhibitor | P Glycoprotein substrate | GI absorption |
|---|---|---|---|---|---|---|
| Andrographolide | −3.18 | Soluble | No | No | Yes | High |
| Hydro andrographolide | −3 | Soluble | No | No | Yes | High |
| Iso andrographolide | −3.35 | Soluble | No | Yes | Yes | High |
| Neo andrographolide | −4.01 | Moderately soluble | No | No | Yes | High |
| Oxo andrographolide | −2.79 | Soluble | No | No | Yes | High |
| Mulberroside A | −2.53 | Soluble | No | No | No | High |
| Mulberroside C | −3.79 | Soluble | No | No | No | Low |
| Mulberroside E | −2.66 | Soluble | No | No | No | Low |
| Mulberroside F | −2.73 | Soluble | No | No | Yes | Low |
| Mimusopic acid | −5.91 | Moderately soluble | No | No | No | High |
| Nimbolide | −3.94 | Soluble | No | No | Yes | High |
| Emblicanin A | −5.63 | Moderately soluble | No | No | Yes | Low |
| Punigluconin | −5.17 | Moderately soluble | No | No | Yes | Low |
| Chebulic acid | −1.39 | Soluble | No | No | No | Low |
| Anolignan A | −5.17 | Moderately soluble | Yes | Yes | No | High |
| Anolignan B | −5.07 | Moderately soluble | Yes | Yes | No | High |
| Anolignan C | −4.28 | Moderately soluble | Yes | Yes | Yes | High |
3.3. Molecular dynamics simulations of mpro and RdRp with inhibitors
To investigate the stability, dynamics and conformational changes of docked Mpro and RdRp inhibitor complexes, MD simulations were carried out which reveals the interaction and stability of inhibitor complexes with protein. Mulberroside E was the top docked compound against Mpro and Emblicanin A against RdRp. Therefore, these two docked complex were subjected for MD simulation studies. The comparisons of MD results of these potential compounds were made with the MD results of the reference drugs used in the study. The RMSD and RMSF values of protein and ligands, involved hydrogen bond in interactions, radius of gyration, solvent-accessible surface area (SASA), and PCA component analysis was carried out to check the stability of protein-ligand complexes. Finally, the binding free energy of all complexes was computed for the last stable 30 ns trajectory.
3.3.1. Assessment of stability using RMSD and RMSF values of protein and ligands
The Root mean square deviation (RMSD) and root mean square fluctuation (RMSF) was calculated for all frames in the trajectory with respect to reference frame for 50 ns simulation period. The protein RMSD values give insights into its structural conformation throughout the simulation and ligand RMSD values gives the idea about the higher stability with respect to the protein (Vora et al., 2020). The lower RMSD values indicated the higher stability of the simulation system. The observations and discussion for all four complexes are as below:
3.3.1.1. Reference compound-Darunavir with Mpro
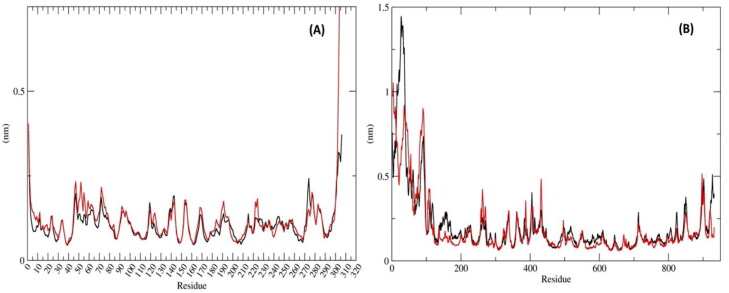
The mean RMSD of protein Mpro was 0.184 ± 0.02 nm (Figure 3(A)) and Darunavir was 0.259 ± 0.03 nm (Figure 3(C)). Ideally, the RMSD values should be less than 0.3 nm. Hence, these RMSD values were within the acceptable range. The mean RMSF of the protein was 0.106 ± 0.048 nm (Figure 4(A)) which denoted that less residual fluctuation occurred during the entire simulation.
Figure 4.
Residue-wise RMSF deviations (nm) of Mpro and RdRp with inhibitors: (A) RMSF of the protein Cα backbone atoms of Mpro_Darunavir (black); Mpro_MulberrosideE (red) (B) RMSF of the protein Cα backbone atoms ofRdRp_Remdesivir (black) and RdRp_EmblicaninA (red).
3.3.1.2. Mulberroside E with Mpro
In this complex, the mean RMSD of protein was 0.258 ± 0.06 nm (Figure 3(A) and the RMSD value of Mulberroside E was 0.255 ± 0.03 nm (Figure 3(C)). Both the mean RMSD values were within the standard range. The RMSD value of Mpro protein with respect to Mulberroside E was slightly higher than reference complex. The mean RMSF value of the protein-Mpro was 0.121 ± 0.06 nm (Figure 4(A)) which was almost similar to the reference complex.
3.3.1.4. Reference compound-Remdesivir with RdRp
The mean RMSD value of RdRp protein was 0.879 ± 0.11 nm (Figure 3(B)) and Remdesivir was 0.286 ± 0.02 nm (Figure 3(D)) that was within the acceptable range. The RMSF of RdRp with respect to Remdesivir was 0.208 ± 0.090 nm (Figure 4(B)).
3.3.1.5. Emblicanin a with RdRp
The protein RMSD was 0.538 ± 0.07 nm (Figure 3(B)) and the value of RMSD of Emblicanin A was 0.109 ± 0.01 nm (Figure 3(D)). The RMSD value of RdRp with respect to Emblicanin A was found to be substantially lower than that of the reference complex. This confirmed that, this docked complex was stable during the simulations as compared with the reference inhibitors. The RMSF value of RdRp with respect to Emblicanin A was 0.191 ± 0.47 nm (Figure 4(B)). The RMSF was of RdRP was similar in pattern as compared to the reference complex.
The deviation values for Mprocomplex with Mulberroside E and Darunavir were in a similar range but values for RdRp with Emblicanin A were less than half of RdRp with Remdesivir indicating a stable conformational behaviour during the dynamics. From the above observation with RMSD deviations, it can be concluded that Emblicanin A behaves well within the active site of the RdRp protein.
3.3.2. Radius of gyration
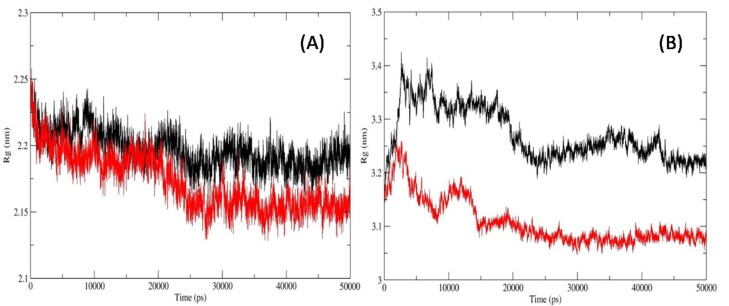
We also computed Radius of Gyration (RoG) which measure the shape of the protein at each time-point by comparing it to the experimentally obtainable hydro-dynamic radius. The average ROG values of protein Cα backbones for Mpro_Darunavir; Mpro_Mulberroside E; RdRp_Remdesivir and RdRp_Emblicanin A were 2.19 ± 0.014, 2.17 ± 0.021, 3.27 ± 0.048, and 3.1 ± 0.043 nm, respectively (Figure 5(A) and (B)). The RoG values of Mpro were similar in binding with reference inhibitor as well as Mulberroside E. Similarly, the RoG values of RdRp were same in binding with the reference inhibitor and Emblicanin A.
Figure 5.
Radius of Gyration of the protein Cα backbone atoms of (A) Mpro_Darunavir(black); Mpro_MulberrosideE (red) (B) RdRp_Remdesivir (black) and RdRp_EmblicaninA (red).
3.3.3. Assessment of surface accessible solvent area (SASA)
To assess the surface area of protein that is accessible to solvent in which it is simulated, we calculated the surface accessible solvent area (SASA) variable for the entire trajectory for all four systems. Total SASA calculated for Mpro_Darunavir; Mpro_Mulberroside E; RdRp_Remdesivir and RdRp_Emblicanin A were 150.37 ± 2.16, 149.24 ± 2.54, 443.78 ± 11.47 and 441.26 ± 10.06 (Supplementary Figure 3). The SASA values were significantly similar in case of both the protein docked with respective reference inhibitor and the molecules under investigation in this study.
3.3.4. Hydrogen bond interactions of amino acid residues of mpro and RdRp with inhibitors
For the protein-inhibitor complex, Hydrogen bond formation is the key indicator of specificity and molecular interactions between the protein and inhibitor complexes. The mean values for H-bonds formed between Mpro and RdRp with respective inhibitors were calculated for entire trajectories. The average H-bonds formed for Mpro_Darunavir; Mpro_Mulberroside E; RdRp_Remdesivir and RdRp_Emblicanin A were 2.55 ± 1.27, 3.29 ± 0.751, 2.34 ± 1.01, and 8.21 ± 1.13, respectively (Figure 6(A) and (B)). RdRp_Emblicanin A complex showed a dramatic increase in H-bond formation compared to known reference inhibitor Remdesivir for RdRp. The intra H-bonds in Mpro and RdRp were also computed for entire trajectories for all inhibitor complexes. The average intra H-bonds formed for Mpro_Darunavir; Mpro_Mulberoside E; RdRp_Remdesivir and RdRp_Emblicanin A were 215.9 ± 6.152, 215.7 ± 7.129, 682.4 ± 15.5, and 699.83 ± 15.46 respectively. The values for inhibitor complexes of Mpro and RdRp are almost identical, indicating a stable molecular dynamic simulations system in the absence and presence of inhibitors.
Figure 6.
H-bonds of Mpro and RdRp with inhibitors: (A)Inter H-bond formation between Mpro and Darunavir (black) and Mulberroside E (red); (B) Inter H-bond formation between RdRp and Remdesivir (black) and Emblicanin A (red).
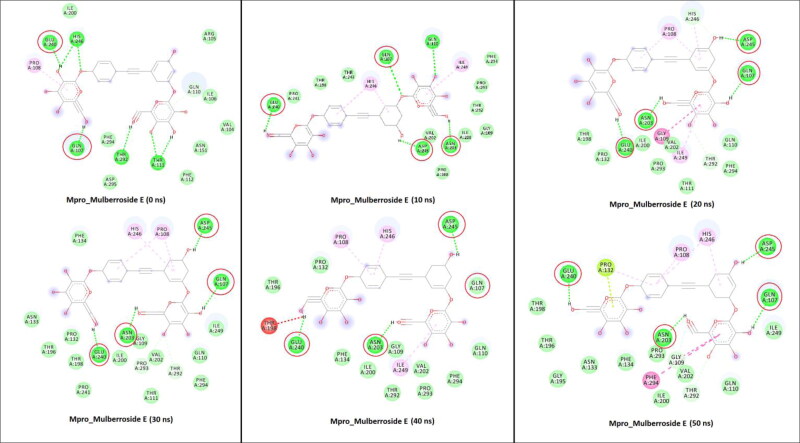
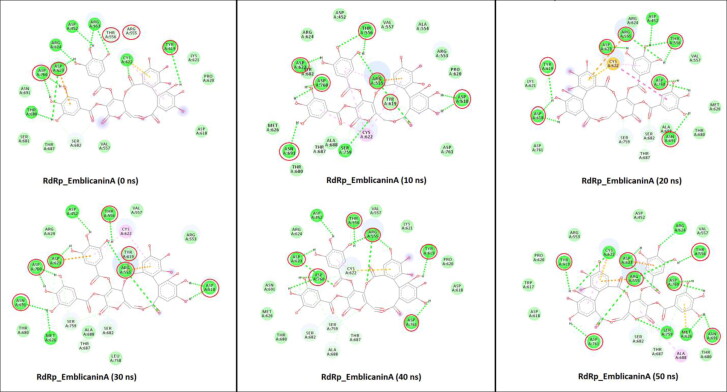
The GLN107, ASN203, GLU240, and ASP245 amino acid residues of Mpro were formed a hydrogen bonds with Mulberroside E during simulation (Figure 7). These residues were involved in the tighter binding of the Mulberroside E. While in the case of RdRp_Emblicanin A, the major residues involved in H-bond interactions were ARG555, THR556, ASP618, TYR619, ASP623, ASN691, ASP760, ASP761, and ASP245 (Figure 8). The reported study suggested that Remdesivir binds with RdRp and that the blocking of the template entry site and arginine, serine and aspartate residues plays a crucial role in this binding (Koulgi et al., 2020). Similarly our potential compound Emblicanin A also binds with arginine and aspartate residues which give the information about the possible mechanism. The molecular determinants of H-bond were identified by visualization of coordinates at every 10 ns time stamp as shown in Figures 7 and 8, as well as by calculating percentage H-bond existence throughout the entire simulation (Table S2).
Figure 7.
2D representation of Mpro interactions with Mulberroside E. M pro_Mulberroside E interactions analyzed at every 10 ns interval. Residues mainly contributing in H-bond formation with the Mulberroside E are circled in each inset diagram with red colour.
Figure 8.
2D representation of RdRp interactions with Emblicanin A. RdRp_ Emblicanin A interactions analyzed at every 10 ns interval. Residues mainly contributing in H-bond formation with the Emblicanin A are circled in each inset diagram with red colour.
3.3.5. Mpro and RdRp conformation clustering and principle component analysis (PCA)
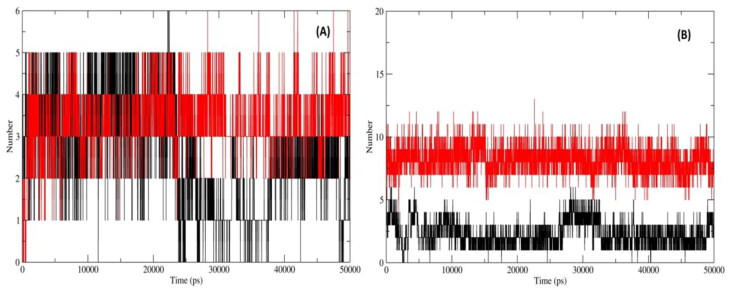
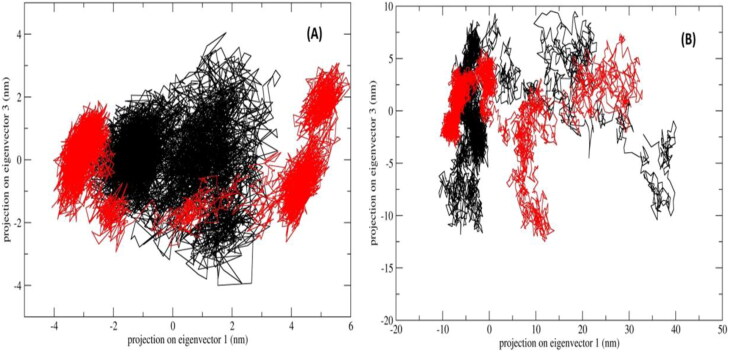
Principle component analysis (PCA) was performed to assess the conformational space and transition dynamics of Mpro with Mulberoside E and RdRp with Emblicanin A which was compared with known inhibitors of Mpro (Darunavir) and RdRp (Remdesivir). The PCA is a statistical calculation to decrease the complexity of MDS trajectory data by extracting only the collective motion of Cα backbone atoms while preserving most of the other variations that are significantly essential to assess the complexes stability. The 2D projection of the trajectories for two major principal components PC1 and PC3 for Mpro_Darunavir; Mpro_Mulberroside E; RdRp_Remdesivir and RdRp_Emblicanin A which represents different conformations in 2D space is shown in Figure 9. The PCA results revealed the following observations. First, the 2D projections of Mpro_Mulberroside E showed occupies lesser phase space and more well-defined clusters i.e. 5 compared to 3 clusters found in Mpro_Darunavir (Figure 9(A)). Second, the 2D projections of RdRp_Emblicanin A occupy much lesser phase space compared to RdRp_Remdesivir (Figure 9(B)). We have also plotted the projection of eigenvector 2 and eigenvector 1 (Supplementary Figure 4). The observations were similar as mentioned above of eigenvector 1 and 3. The positive and negative limits are depicted by the co-variance plots where; positive values are related to the motion of the atoms occurring along the same direction (correlated), whereas the negative values indicate motion of the atoms in the opposite direction (anti-correlated). Our PCA analysis from MD simulations (50 ns) revealed that for both the protein Mpro and RdRp, the inhibitors had more same direction (correlated) motion. The flexibility of all proteins-inhibitor complexes was analysed by calculating the trace value for diagonalized covariance matrix which is the sum of Eigen values. As revealed by the projection of trajectory along PC1 and PC2, Mpro_Darunavir, Mpro_Mulberroside E, RdRp_Emblicanin A and RdRp_Remdesivir showed a trace values of covariance matrix as 11.32, 22.68, 187.66, and 234.59 (nm2) respectively. The trace value of covariance matrix for Mpro_Mulberroside E was found to be doubled compared to the Mpro_Darunavir, while the trace value for RdRp_Remdesivir was found significantly higher than the RdRp_Emblicanin A. The data corroborates well with the lower flexibility in collective motion of reference protein-inhibitor complexes compared to the molecules proposed in this studies. GROMOS based clustering analysis was carried out to assess the dominant and number of cluster transitions occurring during the entire MDS. For Mpro_Mulberroside E, a total of 10 clusters were formed with average RMSD 0.21 nm and 264 transitions, whereas for reference inhibitor Mpro_Darunavir, a total of 9 clusters were formed with average RMSD 0.14 nm and 516 transitions. Similarly for RdRp_Emblicanin A, a total of 21 clusters were formed with average RMSD 0.33 and 490 transitions, whereas for reference inhibitor RdRp_Remdesivir, a total of 36 clusters were formed with average RMSD 0.33 and 544 transitions. The number of clusters formed (distinct conformations), as well as the transitions in RdRp_Emblicanin A, is lesser compare to RdRp_Remdesivir. This reflects the overall stability of RdRp_Emblicanin A during MD simulations. It was concluded that the presence of Emblicanin A complex with RdRp indicated more stable and less flexible dynamics.
Figure 9.
PCA, 2D projection scatter plot (A) Overlay of 2D scatter plot projection the motion of the proteins in phase space for the two principle components, PC1 and PC3 derived for of Mpro_Darunavir (black); Mpro_Mulberroside E (red); (B) Overlay of 2D scatter plot projection the motion of the proteins in phase space for the two principle components, PC1 and PC3 derived for of RdRp_Remdesivir (black) and RdRp_Emblicanin A (red).
3.3.6. Binding free energy estimation and energy decomposition of SARS-CoV-2 mpro and RdRp complexes with inhibitors
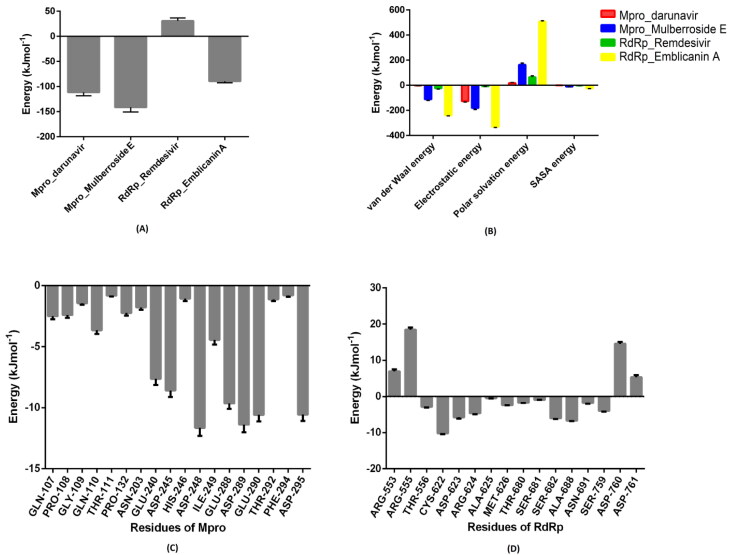
The binding free energy (ΔG) between Mpro_Darunavir; Mpro_Mulberroside E; RdRp_Remdesivir and RdRp_Emblicanin A complexes were calculated using the MM-PBSA method for the last 30 ns stable trajectories. It is an estimation of the non-bonded interaction energies and, a total of 150 frames at every 200ps from the last 30 ns trajectories were considered for computation. The estimated value of ΔG calculated Mpro_Darunavir; Mpro_Mulberoside E; RdRp_Remdesivir and RdRp_Emblicanin A were −111.62 ± 6.788, −141.443 ± 9.313, 30.782 ± 5.85, −89.424 ± 3.130 kJmol−1, respectively (Figure 10(A)). Furthermore, the individual component for binding energy, the electrostatic interactions, the Van der Waals, and non-polar solvation energy except the polar solvation energy had contributed negatively to the overall interaction as shown in Figure 10(B). For molecular insights into key residues involved in Mpro_Mulberroside E and RdRp_Emblicanin A interactions with respective inhibitor, the residue-wise energy decomposition plot was computed which shows the total binding energy contribution for each residue for all the MDS (Figure 10(C) and (D)). The binding energy for Mulberroside E was higher compare to reference inhibitor Darunavir for Mpro, while binding energy of Remdesivir was positive and Emblicanin A was negative for RdRp.
Figure 10.
MM-PBSA Calculation of binding free energy.(A) The total binding free energy for all the Mpro and RdRp inhibitor complexes calculated for last 30 ns stable trajectory for a total of 150 frames, each at 200 ps interval. (B) Representative contributions of each energy component for binding free energy for Mpro and RdRp interactions with inhibitors. (C) The contribution of important binding residues of Mpro with Mulberroside E to the total binding free energy. (D) The contribution of important binding residues of RdRp with Emblicanin A to the total binding free energy. The (-ve) values indicate stable complex formation for PfCDPK2_inhibitor complexes, while the (+ve) values indicate a destabilizing effect.
4. Conclusion
In the present study, we used in silico approach to identify potential phyto-compounds that inhibit the RNA dependent RNA polymerase and main protease of SARS-CoV-2 which play a crucial role in COVID-19. Of 22 phyto-compounds, total 7 displayed as RdRp inhibitors and 15 showed as main protease inhibitors. These hits belong to phenol, terpenes and tannins groups. Interestingly, Mulberroside A/C/E/F and Punigluconin exhibited the best binding affinity score against both the key targets of COVID-19 virus. ADME/T prediction of prioritized ligands revealed the high bioavailability and drug likeliness properties. MD simulations studies of Mpro and RdRp in complex with selected phyto-compounds (Mulberroside E and Emblicanin A) with the highest docking score revealed the formation of a stable complex during the entire simulations. Particularly, RMSD, RMSF and PCA analysis revealed that the Emblicanin A interaction with RdRp was much more stabilizing. Binding free energy calculations (MM-PBSA) Mpro_Mulberroside E and RdRp_Emblicanin A were found to be better than the reference inhibitor used in the study. Overall findings of these in silico studies concluded that Mulberroside E and Emblicanin A gave better interaction and more stable in comparison to currently approved Remdesivir drug for COVID-19. Therefore, these molecules could be developed as either a main drug or as a combination therapy for multi-targeted drug development against SARS-CoV-2. The extracts of the source plants of these potential multi-targeted molecules namely Morus alba and Phyllanthus emblica could be an alternative remedy for COVID-19. However, further experimental studies are required to prove these in silico findings.
Supplementary Material
Disclosure statement
There are no conflicts of interest.
References
- Al-Tawfiq, J. A., Al-Homoud, A. H., & Memish, Z. A. (2020). Remdesivir as a possible therapeutic option for the COVID-19. Travel Medicine and Infectious Disease, 34, 101615. 10.1016/j.tmaid.2020.101615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belapurkar, P., Goyal, P., & Tiwari-Barua, P. (2014). Immunomodulatory effects of triphala and its individual constituents: A review. Indian Journal of Pharmaceutical Sciences, 76(6), 467–475. [PMC free article] [PubMed] [Google Scholar]
- Ben-Shabat, S., Yarmolinsky, L., Porat, D., & Dahan, A. (2020). Antiviral effect of phytochemicals from medicinal plants: Applications and drug delivery strategies. Drug Delivery and Translational Research, 10(2), 354–367. 10.1007/s13346-019-00691-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen, H. J. C., Postma, J. P. M., Van Gunsteren, W. F., Dinola, A., & Haak, J. R. (1984). Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics, 81(8), 3684–3690. 10.1063/1.448118 [DOI] [Google Scholar]
- Berry, M., Fielding, B. C., & Gamieldien, J. (2015). Potential broad spectrum inhibitors of the coronavirus 3CLpro: A virtual screening and structure-based drug design study. Viruses, 7(12), 6642–6660. 10.3390/v7122963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagnola, G., Gong, P., & Peersen, O. B. (2011). High-throughput screening identification of poliovirus RNA-dependent RNA polymerase inhibitors. Antiviral Research, 91(3), 241–251. 10.1016/j.antiviral.2011.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, E., Lye, P.-Y., & Wong, S.-K. (2016). Phytochemistry, pharmacology, and clinical trials of Morus alba. Chinese Journal of Natural Medicines, 14(1), 17–30. 10.3724/SP.J.1009.2016.00017 [DOI] [PubMed] [Google Scholar]
- Chandel, V., Raj, S., Rathi, B., & Kumar, D. (2020). In silico identification of potent COVID-19 main protease inhibitors from FDA approved antiviral compounds and active phytochemicals through molecular docking: A drug repurposing approach. Chemical Biology Letters, 7(3), 166–175. 10.20944/preprints202003.0349.v1 [DOI] [Google Scholar]
- Churiyah, Pongtuluran, O. B., Rofaani, E. & Tarwadi, (2015). Antiviral and immunostimulant activities of Andrographis paniculata. HAYATI Journal of Biosciences, 22(2), 67–72. 10.4308/hjb.22.2.67 [DOI] [Google Scholar]
- Cragg, G. M., & Newman, D. J. (2001). Natural product drug discovery in the next millennium. Pharmaceutical Biology, 39(sup1), 8–17. 10.1076/phbi.39.s1.8.0009 [DOI] [PubMed] [Google Scholar]
- Daina, A., Michielin, O., & Zoete, V. (2017). SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports, 7, 42717 10.1038/srep42717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallakyan, S., & Olson, A. (2015). Small-molecule library screening by docking with PyRx. Methods in Molecular Biology (Clifton, N.J.), 1263, 243–250. 10.1007/978-1-4939-2269-7_19 [DOI] [PubMed] [Google Scholar]
- Darden, T., York, D., & Pedersen, L. (1993). Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. The Journal of Chemical Physics, 98(12), 10089–10092. 10.1063/1.464397 [DOI] [Google Scholar]
- Daura, X., Gademann, K., Jaun, B., Seebach, D., van Gunsteren, W. F., & Mark, A. E. (1999). Peptide folding: When simulation meets experiment. Angewandte Chemie International Edition, 38(1-2), 236–240. [Google Scholar]
- Elfiky, A. (2020). Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sciences, 248, 117477 10.1016/j.lfs.2020.117477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmezayen, A. D., Al-Obaidi, A., Şahin, A. T., & Yelekçi, K. (2020). Drug repurposing for coronavirus (COVID-19): in silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. Journal of Biomolecular Structure and Dynamics, 0(ja), 1–12. 10.1080/07391102.2020.1758791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enmozhi, S. K., Raja, K., & Sebastine, I. (2020). Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: An in silico approach. Journal of Biomolecular Structure and Dynamics, 0(0), 000. 10.1080/07391102.2020.1760136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta, Y., Gowen, B. B., Takahashi, K., Shiraki, K., Smee, D. F., & Barnard, D. L. (2013). Favipiravir (T-705), a novel viral RNA polymerase inhibitor. Antiviral Research, 100(2), 446–454. 10.1016/j.antiviral.2013.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Y., Yan, L., Huang, Y., Liu, F., Zhao, Y., Cao, L., Wang, T., Sun, Q., Ming, Z., Zhang, L., Ge, J., Zheng, L., Zhang, Y., Wang, H., Zhu, Y., Zhu, C., Hu, T., Hua, T., Zhang, B., … Rao, Z. (2020). Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science (New York, N.Y.), 368(6492), 779–779. 10.1126/science.abb7498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genheden, S., & Ryde, U. (2015). The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opinion on Drug Discovery, 10(5), 449–461. 10.1517/17460441.2015.1032936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya, A. E., Baker, S. C., Baric, R. S., de Groot, R. J., Drosten, C., Gulyaeva, A. A., & Ziebuhr, J. (2020). The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology, 5(4), 536–544. 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess, B., Bekker, H., Berendsen, H. J. C., & Fraaije, J. G. E. M. (1997). LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry, 18(12), 1463–1472. [Google Scholar]
- Hui, D. S., I Azhar, E., Madani, T. A., Ntoumi, F., Kock, R., Dar, O., Ippolito, G., Mchugh, T. D., Memish, Z. A., Drosten, C., Zumla, A., & Petersen, E. (2020, February). The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health – The latest 2019 novel coronavirus outbreak in Wuhan, China. International Journal of Infectious Diseases: IJID: Official Publication of the International Society for Infectious Diseases, 91, 264–266. 10.1016/j.ijid.2020.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., & Schulten, K. (1996). VMD: Visual molecular dynamics. Journal of Molecular Graphics, 14(1), 33–38. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Kalita, P., Padhi, A. K., Zhang, K. Y. J., & Tripathi, T. (2020). Design of a peptide-based subunit vaccine against novel coronavirus SARS-CoV-2. Microbial Pathogenesis, 145, 104236 10.1016/j.micpath.2020.104236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, R. J., Jha, R. K., Amera, G. M., Jain, M., Singh, E., Pathak, A., Singh, R. P., Muthukumaaran, J., & Singh, A. K. (2020). Targeting SARS-CoV-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. Journal of Biomolecular Structure and Dynamics, 0(0), 1–14. 10.1080/07391102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulgi, S., Jani, V., Uppuladinne, M. V. N., Sonavane, U., & Joshi, R. (2020). Remdesivir-bound and ligand-free simulations reveal the probable mechanism of inhibiting the RNA dependent RNA polymerase of severe acute respiratory syndrome coronavirus 2. RSC Advances, 10(45), 26792–26803. 10.1039/D0RA04743K [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari, R., Kumar, R., Lynn, A., & Lynn, A. (2014). g_mmpbsa-a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model, 54(7), 1951–1962. 10.1021/ci500020m [DOI] [PubMed] [Google Scholar]
- Li, G., & De Clercq, E. (2020). Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nature Reviews. Drug Discovery, 19(3), 149–150. 10.1038/d41573-020-00016-0 [DOI] [PubMed] [Google Scholar]
- Lim, H.-S., Im, J.-S., Cho, J.-Y., Bae, K.-S., Klein, T. A., Yeom, J.-S., Kim, T.-S., Choi, J.-S., Jang, I.-J., & Park, J.-W. (2009). Pharmacokinetics of hydroxychloroquine and its clinical implications in chemoprophylaxis against malaria caused by Plasmodium vivax. Antimicrobial Agents and Chemotherapy, 53(4), 1468–1475. 10.1128/AAC.00339-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindorff-Larsen, K., Piana, S., Palmo, K., Maragakis, P., Klepeis, J. L., Dror, R. O., & Shaw, D. E. (2010). Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins: Structure, Function, and Bioinformatics, 78(8), 1950–1958. 10.1002/prot.22711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Zhu, J., Xue, J., Qin, Z., Shen, F., Liu, J., Chen, X., Li, X., Wu, Z., Xiao, W., Zheng, C., & Wang, Y. (2017). In silico-based screen synergistic drug combinations from herb medicines: A case using Cistanche tubulosa. Scientific Reports, 7(1), 1–12. 10.1038/s41598-017-16571-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., & Wang, X. J. (2020). Potential inhibitors against 2019-nCoV coronavirus M protease from clinically approved medicines. Journal of Genetics and Genomics = Yi Chuan Xue Bao, 47(2), 119–121. 10.1016/j.jgg.2020.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H., Stratton, C. W., & Tang, Y.-W. (2020). Outbreak of pneumonia of unknown etiology in Wuhan, China: The mystery and the miracle. Journal of Medical Virology, 92(4), 401–402. 10.1002/jmv.25678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manhas, A., Patel, D., Lone, M. Y., & Jha, P. C. (2019). Identification of natural compound inhibitors against PfDXR: A hybrid structure-based molecular modeling approach and molecular dynamics simulation studies . Journal of Cellular Biochemistry, 120(9), 14531–14543. 10.1002/jcb.28714 [DOI] [PubMed] [Google Scholar]
- Mathur, S., & Hoskins, C. (2017). Drug development: Lessons from nature. Biomedical Reports, 6(6), 612–614. 10.3892/br.2017.909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of Computational Chemistry, 30(16), 2785–2791. 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse, J. S., Lalonde, T., Xu, S., & Liu, W. R. (2020). Learning from the past: Possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019-nCoV. Chembiochemistry: A European Journal of Chemical Biology, 21(5), 730–738. 10.1002/cbic.202000047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naithani, R., Huma, L. C., Holland, L. E., Shukla, D., McCormick, D. L., Mehta, R. G., & Moriarty, R. M. (2008). Antiviral activity of phytochemicals: A comprehensive review. Mini Reviews in Medicinal Chemistry, 8(11), 1106–1133. 10.2174/138955708785909943 [DOI] [PubMed] [Google Scholar]
- Nisar, M. F., He, J., Ahmed, A., Yang, Y., Li, M., & Wan, C. (2018). Chemical components and biological activities of the genus phyllanthus: A review of the recent literature. Molecules (Basel, Switzerland), 23(10), 2567. 10.3390/molecules23102567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle, N. M., Banck, M., James, C. A., Morley, C., Vandermeersch, T., & Hutchison, G. R. (2011). Open Babel: An open chemical toolbox. Journal of Cheminformatics, 3, 33. 10.1186/1758-2946-3-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhi, A. K., Kalita, P., Zhang, K. Y. J., & Tripathi, T. (2020). High throughput designing and mutational mapping of RBD-ACE2 interface guide non-conventional therapeutic strategies for COVID-19. BioRxiv, 10.1101/2020.05.19.104042 [DOI] [Google Scholar]
- Padhi, A., Seal, A., & Tripathi, T. (2020). How does arbidol inhibit the novel coronavirus SARS-CoV-2? Atomistic Insights from Molecular Dynamics Simulations. 10.26434/chemrxiv.12464576.v1 [DOI]
- Pagadala, N. S., Syed, K., & Tuszynski, J. (2017). Software for molecular docking: A review. Biophysical Reviews, 9(2), 91–102. 10.1007/s12551-016-0247-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello, M., & Rahman, A. (1980). Crystal structure and pair potentials: A molecular-dynamics study. Physical Review Letters, 45(14), 1196–1199. 10.1103/PhysRevLett.45.1196 [DOI] [Google Scholar]
- Patel, B., Patel, D., Parmar, K., Chauhan, R., Singh, D. D., & Pappachan, A. (2018). L. donovani XPRT: Molecular characterization and evaluation of inhibitors. Biochimica et Biophysica Acta - Proteins and Proteomics, 1866(3). 10.1016/j.bbapap.2017.12.002 [DOI] [PubMed] [Google Scholar]
- Patel, D., Athar, M., & Jha, P. C. (2020). Exploring ruthenium-based organometallic inhibitors against plasmodium calcium dependent kinase a combined ensemble docking, QM paramterization and molecular dynamics study funding sources. BioRxiv, 2(PfCDPK2). 10.1101/2020.03.31.017541 [DOI] [Google Scholar]
- Patel , P., Parmar, K., Patel, D., Kumar, S., Trivedi, M., & Das, M. (2018). Inhibition of amyloid fibril formation of lysozyme by ascorbic acid and a probable mechanism of action. International Journal of Biological Macromolecules, 114, 666–678. 10.1016/j.ijbiomac.2018.03.152 [DOI] [PubMed] [Google Scholar]
- Patel, P., Parmar, K., Vyas, V. K. V. K., Patel, D., & Das, M. (2017). Combined in silico approaches for the identification of novel inhibitors of human islet amyloid polypeptide (hIAPP) fibrillation. Journal of Molecular Graphics & Modelling, 77, 295–310. 10.1016/j.jmgm.2017.09.004 [DOI] [PubMed] [Google Scholar]
- Peterson, C. T., Denniston, K., & Chopra, D. (2017). Therapeutic uses of triphala in ayurvedic medicine. Journal of Alternative and Complementary Medicine (New York, N.Y.), 23(8), 607–614. 10.1089/acm.2017.0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera-a visualization system for exploratory research and analysis . Journal of Computational Chemistry, 25(13), 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Rappe, A. K., Casewit, C. J., Colwell, K. S., Goddard, W. A., & Skiff, W. M. (1992). UFF, a full periodic table force field for molecular mechanics and molecular dynamics simulations. Journal of the American Chemical Society, 114(25), 10024–10035. 10.1021/ja00051a040 [DOI] [Google Scholar]
- Salehi, B., Kumar, N., Şener, B., Sharifi-Rad, M., Kılıç, M., Mahady, G., Vlaisavljevic, S., Iriti, M., Kobarfard, F., Setzer, W., Ayatollahi, S., Ata, A., & Sharifi-Rad, J. (2018). Medicinal plants used in the treatment of human immunodeficiency virus. International Journal of Molecular Sciences, 19(5), 1459. 10.3390/ijms19051459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeliger, D., & de Groot, B. L. (2010). Ligand docking and binding site analysis with PyMOL and Autodock/Vina. Journal of Computer-Aided Molecular Design, 24(5), 417–422. 10.1007/s10822-010-9352-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha, S., Patel, S., Athar, M., Vora, J., Chhabria, M. T., Jha, P. C., & Shrivastava, N. (2019). Structure-based identification of novel sirtuin inhibitors against triple negative breast cancer: An in silico and in vitro study. International Journal of Biological Macromolecules, 140, 454–468. 10.1016/j.ijbiomac.2019.08.061 [DOI] [PubMed] [Google Scholar]
- Sliwoski, G., Kothiwale, S., Meiler, J., & Lowe, E. W. J. (2014). Computational methods in drug discovery. Pharmacological Reviews, 66(1), 334–395. 10.1124/pr.112.007336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa da Silva, A. W., & Vranken, W. F. (2012). ACPYPE - AnteChamber PYthon Parser interfacE. BMC Research Notes, 5(1), 367 10.1186/1756-0500-5-367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subissi, L., Posthuma, C. C., Collet, A., Zevenhoven-Dobbe, J. C., Gorbalenya, A. E., Decroly, E., Snijder, E. J., Canard, B., & Imbert, I. (2014). One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proceedings of the National Academy of Sciences of the United States of America, 111(37), E3900–E3909. 10.1073/pnas.1323705111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, H., Duan, L., Chen, F., Liu, H., Wang, Z., Pan, P., Zhu, F., Zhang, J. Z. H., & Hou, T. (2018). Assessing the performance of MM/PBSA and MM/GBSA methods. 7. Entropy effects on the performance of end-point binding free energy calculation approaches. Physical Chemistry Chemical Physics: PCCP, 20(21), 14450–14460. 10.1039/c7cp07623a [DOI] [PubMed] [Google Scholar]
- Udeinya, I. J., Mbah, A. U., Chijioke, C. P., & Shu, E. N. (2004). An antimalarial extract from neem leaves is antiretroviral. Transactions of the Royal Society of Tropical Medicine and Hygiene, 98(7), 435–437. 10.1016/j.trstmh.2003.10.016 [DOI] [PubMed] [Google Scholar]
- Van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A. E., & Berendsen, H. J. C. (2005). GROMACS: Fast, flexible, and free. Journal of Computational Chemistry, 26(16), 1701–1718. 10.1002/jcc.20291 [DOI] [PubMed] [Google Scholar]
- Vora, J., Athar, M., Sinha, S., Jha, P. C., & Shrivastava, N. (2020). Binding insight of anti-HIV phytocompounds with prime targets of HIV: A molecular dynamics simulation analysis. Current HIV Research, 18(2), 132–141. 10.2174/1570162X18666200129112509 [DOI] [PubMed] [Google Scholar]
- Vora, J., Patel, S., Athar, M., Sinha, S., Chhabria, M. T., Jha, P. C., & Shrivastava, N. (2020). Pharmacophore modeling, molecular docking and molecular dynamics simulation for screening and identifying anti-dengue phytocompounds. Journal of Biomolecular Structure & Dynamics, 38(6), 1726–1740. 10.1080/07391102.2019.1615002 [DOI] [PubMed] [Google Scholar]
- Vora, J., Patel, S., Sinha, S., Sharma, S., Srivastava, A., Chhabria, M., & Shrivastava, N. (2019). Molecular docking, QSAR and ADMET based mining of natural compounds against prime targets of HIV. Journal of Biomolecular Structure & Dynamics, 37(1), 131–146. 10.1080/07391102.2017.1420489 [DOI] [PubMed] [Google Scholar]
- Wang, C., Nguyen, P. H., Pham, K., Huynh, D., Le, T.-B N., Wang, H., Ren, P., & Luo, R. (2016). Calculating protein-ligand binding affinities with MMPBSA: Method and error analysis. Journal of Computational Chemistry, 37(27), 2436–2446. 10.1002/jcc.24467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M., Cao, R., Zhang, L., Yang, X., Liu, J., Xu, M., Shi, Z., Hu, Z., Zhong, W., & Xiao, G. (2020). Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Research, 30(3), 269–271. 10.1038/s41422-020-0282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D., Chen, J., Deng, L., Mao, Q., Zheng, J., Wu, J., Zeng, C., & Li, Y. (2010). Evolutionary selection associated with the multi-function of overlapping genes in the hepatitis B virus. Infection, Genetics and Evolution: Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases, 10(1), 84–88. 10.1016/j.meegid.2009.10.006 [DOI] [PubMed] [Google Scholar]
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L., Becker, S., Rox, K., & Hilgenfeld, R. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (New York, N.Y.), 368(6489), 409–412. 10.1126/science.abb3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Fang, L., Yang, Z., Xu, S., Lv, M., Sun, Z., Chen, J., Wang, D., Gao, J., & Xiao, S. (2019). Identification of novel proteolytically inactive mutations in coronavirus 3C-like protease using a combined approach. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology, 33(12), 14575–14587. 10.1096/fj.201901624RR [DOI] [PubMed] [Google Scholar]
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., Zhao, X., Huang, B., Shi, W., Lu, R., Niu, P., Zhan, F., Ma, X., Wang, D., Xu, W., Wu, G., Gao, G. F., & Tan, W. (2020). A novel coronavirus from patients with pneumonia in China, 2019. The New England Journal of Medicine, 382(8), 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla, A., Chan, J. F. W., Azhar, E. I., Hui, D. S. C., & Yuen, K.-Y. (2016). Coronaviruses - drug discovery and therapeutic options. Nature Reviews. Drug Discovery, 15(5), 327–347. 10.1038/nrd.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.