Abstract
Background
ERBB2 exon 16 skipping is an alternatively spliced isoform of ERBB2, which was reported to lead to oncogenic activation of ERBB2 and could potentially cause tyrosine kinase inhibitor (TKI) resistance in non-small cell lung cancer (NSCLC) in case studies. In this study, we aimed to evaluate the frequency of ERBB2 exon 16 skipping in a large patient cohort and its function in cancer development.
Methods
A total of 38 680 Chinese patients with cancer whose tumour specimens and/or circulating cell-free DNA underwent targeted nextgeneration sequencing of cancer-related genes were retrospectively reviewed. Clinicopathological features and treatment history of patients harbouring ERBB2 exon 16 skipping were evaluated. RNA-sequencing was performed to validate the presence of exon 16 skipping in ERBB2 at the transcriptional level.
Results
ERBB2 exon 16 skipping is rare and was identified in a total of 18 patients (0.046% of total patients), including 12 lung cancers, which were caused by large fragment deletion spanning the whole or partial region of exon 16 (13/18, 72.2%) and/or splice site variants (6/18, 33.3%). The majority of these variants have not been previously reported and three of them were confirmed by RNA-sequencing. Among the 12 patients with lung cancer, 9 had coexisting activating EGFR mutations (exon 19 deletions or L858R) and received prior-treatment with epidermal growth factor receptor TKIs. Further analysis of matched pre-treatment and post-treatment samples in three EGFR-mutated NSCLC patients confirmed that ERBB2 exon 16 skipping was newly acquired on resistance to TKI therapies. In 6 out of 18 patients, including colorectal, gastric and ovarian cancers, there were no mutations in known cancer driver genes detected, indicating that ERBB2 exon 16 skipping might be the oncogenic driver in these patients.
Conclusions
Our data suggest that ERBB2 exon 16 skipping is another mechanism of TKI resistance in EGFR-mutated patients with lung cancer, in addition to its role of being an oncogenic driver in other solid malignancies.
Keywords: ERBB2, exon skipping, lung cancer, TKI resistance, oncogenesis
Key question.
What is already known about this subject?
The splice variant of ERBB2 resulting from exon 16 skipping was first identified as an oncogenic driver in breast cancer.
ERBB2 exon 16 skipping was rarely reported in other cancer types.
What does this study add?
We identified 18 patients harbouring ERBB2 exon 16 skipping from a large pan-cancer population and confirmed the presence of ERBB2Δ16 transcripts in three patients by RNA sequencing.
ERBB2 exon 16 skipping may underlie the acquired resistance to epidermal growth factor receptor (EGFR)-targeted therapies in patients with lung cancer who received EGFR tyrosine kinase inhibitors (TKIs).
No other known oncogenic drivers were detected in the remaining six patients with non-lung cancer, highlighting the role of ERBB2 exon 16 skipping as an oncogenic driver in these solid malignancies.
How might this impact on clinical practice?
This study emphasised the concurrence of ERBB2 exon 16 skipping beyond breast cancer.
The dual role of ERBB2 exon 16 skipping in TKI resistance and oncogenesis suggested its great potential to be a promising therapeutic target for drugs.
Introduction
The erb-b2 receptor tyrosine kinase 2 (ERBB2/HER2) is a member of human epidermal growth factor receptor (EGFR) family. In breast cancer, ERBB2 has drawn extensive attention and become a crucial biomarker as 20%–30% of breast cancer are onset through the amplification of ERBB2 gene and/or protein overexpression.1 Several targeted agents such as trastuzumab, ado-trastuzumab emtansine and pertuzumab have been approved as ERBB2-targeted therapies which are monoclonal antibodies. However, trastuzumab resistance has been widely observed in ERBB2-positive patients with breast cancer within a longer period of treatment.2 Several point mutations (eg, ERBB2 V659E and G660D) and in-frame insertion in exon 20 were identified as driver mutations as well and able to escape from trastuzumab targeting as they promoted the protein dimerisation causing the constitutive activation of ERBB2.3
More recently, an ERBB2 splicing variant skipping the exon 16 was reported and studied in breast cancer. ERBB2 exon 16 encodes a transmembrane domain, deletion of which changes the conformation of the extracellular domain and further promotes the protein dimerisation.4 In breast cancer, ERBB2 exon 16 skipping was identified as a major oncogenic driver that increases tumour proliferation and mediates trastuzumab resistance in vitro.5 ERBB2 exon 16 skipping was also reported in gastric, colorectal and ovarian cancers.6 However, there was barely any further investigation on its role in these cancer types due to the low incidence and it was only hypothesised to be a candidate oncogenic mutation in non-breast cancers. Nevertheless, ERBB2 exon 16 skipping was first reported in non-small cell lung cancer (NSCLC) in 2019 and proven to be a novel mechanism of osimertinib resistance cooperating with EGFR T790M/L858R mutants in vitro.7 It is worth noting that the patient was detected with ERBB2 exon 16 skipping in his first plasma sample collected after gefitinib and erlotinib treatment. Therefore, it is very likely that ERBB2 exon 16 skipping was acquired during the primary EGFR tyrosine kinase inhibitors (TKIs) treatment.
In this study, we identified 18 patients with ERBB2 exon 16 skipping from a cohort of 38 680 patients undergoing next generation sequencing (NGS) analysis. Our data revealed that ERBB2 exon 16 skipping may be an acquired mutation contributing to the general TKIs resistance in lung cancer, in addition to being an oncogenic driver in colorectal, gastric and ovarian cancers as previously accepted in breast cancer.
Methods
Patient sample collection
A series of 38 680 consecutive cancer cases were analysed using comprehensive genomic profiling targeting 400+ cancer-relevant genes conducted by a Clinical Laboratory Improvement Amendments-certified, College of American Pathologists-accredited laboratory (Nanjing Geneseeq Technology, Jiangsu, China) as previously described.8 Written informed consent was collected from each patient on sample collection according to the protocols approved by the ethical committee of each hospital. We identified patients with ERBB2 alterations in the LIMS database by using natural language search programme. Relevant demographic and clinical data were extracted from the database for these cases, including age, gender, date of diagnosis, histology type, pathological stage and evaluation of treatment response as per reports by clinical investigators. For tumour tissue samples, the pathological diagnosis and tumour content of each case was confirmed by pathologists. 8–10 mL of peripheral blood was collected in EDTA-coated tubes and centrifuged at 1800×g for 10 min within 2 hours of collection to separate the plasma for circulating tumour DNA (ctDNA) extraction and white blood cells for genomic DNA extraction as germline control.
DNA extraction and targeted enrichment ctDNA from plasma was purified using the Circulating Nucleic Acid Kit (Qiagen) following the manufacturer’s protocol. Genomic DNA from the white blood cells was extracted using the DNeasy Blood & Tissue Kit (Qiagen), while FFPE (Formalin-Fixed Paraffin-Embedded) genomic DNA was purified using the QIAamp DNA FFPE Tissue Kit (Qiagen). All DNA was quantified using the dsDNA HS Assay Kit on a Qubit Fluorometer (Life Technologies). Sequencing libraries were prepared using the KAPA Hyper Prep Kit (KAPA Biosystems), as described previously.8 Indexed DNA libraries were pooled together for probe-based hybridisation capture of the targeted gene regions covering over 400 cancer-related genes for all solid tumours.
Sequencing data processing
Sequencing was performed using the Illumina HiSeq4000 platform, followed by data analysis as previously described.9 In brief, sequencing data were analysed by Trimmomatic10 to remove low-quality (quality <15) or N bases, and were then mapped to the human reference genome, hg19, using the Burrows-Wheeler Aligner (https://github.com/lh3/bwa/tree/master/bwakit). PCR duplicates were removed by Picard (available at: https://broadinstitute.github.io/picard/). The Genome Analysis Toolkit (GATK) (https://software.broadinstitute.org/gatk/) was used to perform local realignments around indels and base quality reassurance. Gene fusions were identified by Fusion And Chromosomal Translocation Enumeration and Recovery Algorithm (FACTERA).11 SNPs and indels were analysed by VarScan212 and HaplotypeCaller/UnifiedGenotyper in GATK, with the mutant allele frequency cut-off at 0.5% for tissue samples, 0.1% for cell-free DNA samples and a minimum of three unique mutant reads. Common SNPs were excluded if they were present in >1% population frequency in the 1000 Genomes Project or the Exome Aggregation Consortium (ExAC) 65000 exomes database. The resulting mutation list was further filtered by an in-house list of recurrent artefacts based on a normal pool of whole blood samples.
RNA-seq
Total RNA from FFPE samples was extracted using the RNeasy FFPE kit (QIAGEN). Total RNA amount was quantified by the Bioanalyzer 2100 (Agilent Technologies). Ribosomal RNA and residual genomic DNA were depleted by the KAPA Standard RNA-Seq Kit with RiboErase (HMR) and DNase digestion, followed by purification using the Agencourt RNA Clean XP Beads according to the manufacturers’ protocol. KAPA Stranded RNA-Seq Library Preparation Kit was used to construct Illumina-compatible sequencing libraries including RNA fragmentation and priming, double-stranded cDNA synthesis, adaptor ligation and PCR amplification.
Sequencing was performed on Illumina HiSeq NGS platforms (Illumina). To generate sequence reads in the FASTQ format, base calling was performed on bcl2fastq V.2.16.0.10 (Illumina) and quality control was performed with Trimmomatic (V.0.33).10 RNA-seq reads were mapped to the human genome (hg19, Genome Reference Consortium GRCh37) using STAR (V.2.5.3a)13 to identify individual exon, intron and intergenic features. The average coverage of mapped reads across base positions of the feature coordinates were calculated. Exon junctions were detected and visualised on the Integrative Genomics Viewer.14
Results
Overall incidence of ERBB2 exon 16 skipping patients in human cancer
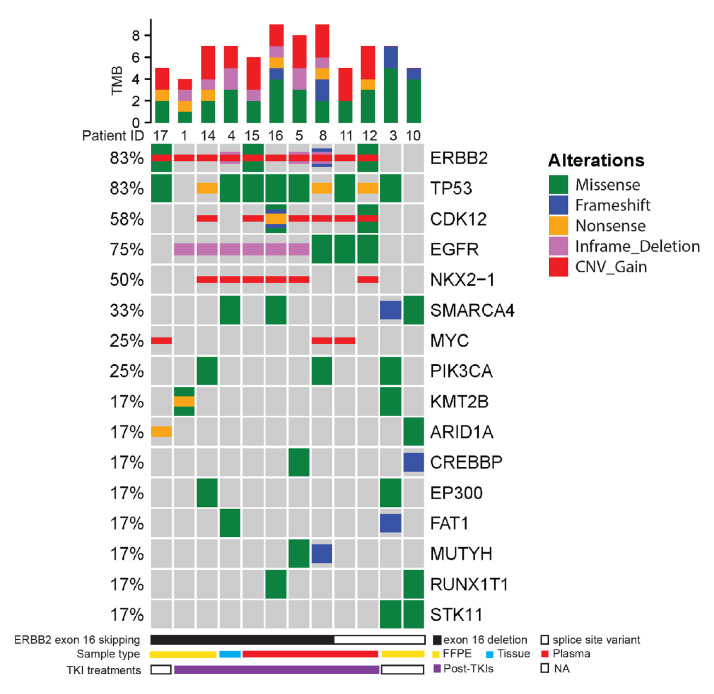
From January 2014 to December 2019, a total of 38 680 individual cancers were successfully evaluated by comprehensive genomic profiling using hybrid capture based NGS by a Clinical Laboratory Improvement Amendments-certified, College of American Pathologists-accredited laboratory (Nanjing Geneseeq Technology, Jiangsu, China). From the NGS analysis database, we identified 18 patients with ERBB2 exon 16 skipping (0.047%, 18/38680). The 18 patients were 11 women and 7 men. The age of diagnosis ranged from 28 to 80 years old (median=53) (table 1). The majority of the 18 patients were diagnosed as lung cancer (12/18, 66.67%) and particularly 10 of them were identified as NSCLC. Genomic analysis revealed that 9 of the 12 patients with lung cancer harboured activating EGFR mutations as oncogenic drivers (six with EGFR exon 19 deletion and three with EGFR L858R) (figure 1). These EGFR+ patients with lung cancer received one or multiple TKIs during their clinical treatments and the detailed treatment history was provided in online supplemental table S1. There were three patients without detectable oncogenic EGFR mutations, two of which were never exposed to TKIs and the treatment history of the third was unavailable. In addition to lung cancer, we also identified two gastric cancer, two rectal cancer, one ovarian cancer and one gallbladder cancer patients with ERBB2 exon 16 skipping. The two patients with rectal cancer underwent similar treatment procedures including surgery, chemotherapy and multiple types of monoclonal antibody (see online supplemental table S2 for details). The gallbladder case received surgical removal and adjuvant chemotherapy. However, the clinical history of three patients with ovarian or gastric cancer was unavailable.
Table 1.
Patient characteristics and mutational information
| Clinical information | Total (N=18) | Lung (N=12) | Non-lung (N=6) |
| Median age (range) | 53 (28–80) | 52 (28–70) | 62 (49–80) |
| Sex | |||
| Male | 7 (38.9%) | 4 (33.3%) | 3 (50%) |
| Female | 11 (61.1%) | 8 (66.7%) | 3 (50%) |
| Stage | |||
| IV | 9 (50%) | 7 (58.3%) | 2 (33.3%) |
| Unknown | 9 (50%) | 5 (41.7%) | 4 (66.7%) |
| Treatment | |||
| TKI-involved | 10 (55.6%) | 9 (75%) | 1 (16.7%) |
| TKI-exclusive | 4 (22.2%) | 2 (16.7%) | 2 (33.3%) |
| Unknown | 4 (22.2%) | 1 (8.3%) | 3 (50%) |
| Mutation type | |||
| Ex16 deletion | 13 (72.2%)* | 8 (66.7%)* | 5 (83.3%) |
| Splice site deletion | 2 (11.1%) | 1 (8.3%) | 1 (16.7%) |
| Splice site point mutation | 4 (22.2%)* | 4 (33.3%)* | 0 |
*One patient harboured both Ex16 deletion and splice site point mutation.
Ex16, exon 16; TKI, tyrosine kinase inhibitor.
Figure 1.
OncoPrint showing the distribution of genomic alterations in patients with lung cancer. OncoPrint provides an overview of genomic alterations (legend) in particular genes (rows) affecting individual samples (columns). The missense, frameshift, non-sense, inframe deletion and copy number gain are shown as green, blue, orange, purple and red, respectively. The mutational frequency of each gene is labelled on the left. The relative TMB of each patient is shown on top of the patient ID. The bottom annotation indicates the mutation type (solid bar: exon 16 deletion; open bar: splice site variant) causing the ERBB2 exon 16 skipping, NGS sample type (yellow: FFPE; blue: tissue; red: plasma) and TKI treatments (white: no TKI treatments; purple: post-TKIs). NGS, next-generation sequencing; TKI, tyrosine kinase inhibitor; TMB, tumor mutational burden; FFPE, formalin-fixed paraffin-embedded.
esmoopen-2020-000985supp001.pdf (50.1KB, pdf)
RNA-seq validation of ERBB2 exon 16 skipping caused by large fragment deletions and splice site variations
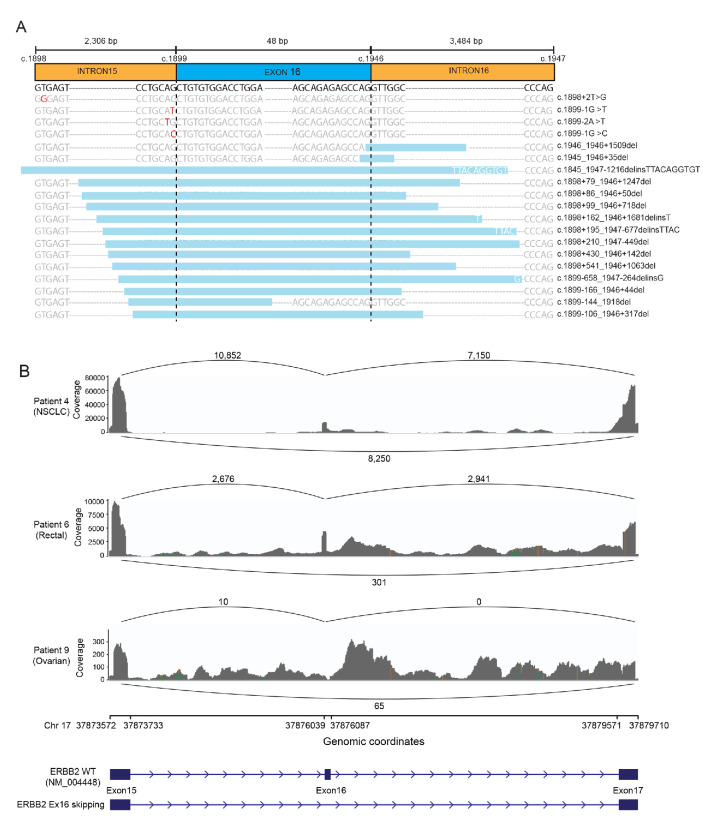
Since we identified the ERBB2 exon 16 skipping based on NGS analysis which reflected the genomic changes, it is still debatable whether the DNA alterations could cause the skip of exon 16 at the RNA or protein level. Therefore, we first subgrouped the ERBB2 exon 16 skipping events according to the mutation types. Among the 18 identified patients, 12 (66.67%) had the whole or partial region of exon 16 deleted (Ex16del full or partial), 5 (27.78%) had splice site variants (three with point mutation and two with splice site deletion; table 1 and figure 2A). Only one patient (patient 5) was detected with both exon 16 deletion and splice site variant. To validate the ERBB2 exon 16 skipping at the RNA level, we did the RNA-seq in three patients with available samples (figure 2B). Patient 4 was diagnosed as NSCLC with the whole exon 16 deletion at the DNA level detected in a post-afatinib tumour tissue sample. RNA-seq confirmed this exon-16-skipping event at an allele frequency of approximately 50%. Similarly, another whole exon 16 deletion in patient 6 with rectal cancer was validated by RNA-seq. In addition, RNA-seq also supported the occurrence of exon 16 skipping when the splice site was deleted as shown in patient 9 who had ovarian cancer. Therefore, both direct deletion of the coding sequence of exon 16 and variants at splice sites could result in the ERBB2 exon 16 skipping.
Figure 2.
Schematic diagram of ERBB2 exon 16 skipping and RNA-seq validation. (A) The positions of all ERBB2 exon 16 skipping variations identified in 18 patients are displayed in relation to the ERBB2 gene. Point mutations are highlighted in red. Deleted regions are shown by the light blue boxes with inserted nucleotides labelled in white if applicable. (B) SASHIMI plots for reads alignment at ERBB2 locus of patient 4 with NSCLC; patient 6 with rectal cancer; and patient 9 with ovarian cancer. The number of reads corresponding to specific exon–exon junctions (shown as loops) is labelled for each junction. The coverage of RNA-seq for each sample is indicated on the y-axis. Genomic coordinates of chromosome 17 show the location of ERBB2 exon 15–17. Schematic illustrations of wild-type ERBB2 and exon-16-skipping ERBB2 are shown corresponding to the genomic coordinates. NSCLC, non-small cell lung cancer.
ERBB2 exon 16 skipping as a resistance mechanism acquired during TKI therapy in lung cancer
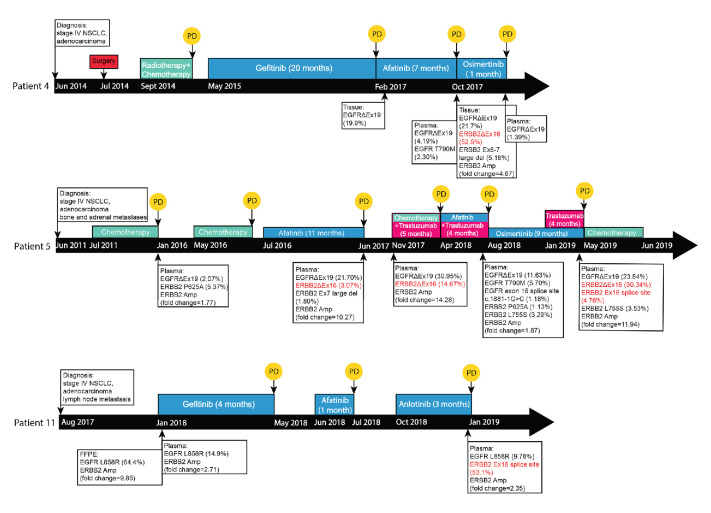
To investigate the occurrence mechanism of ERBB2 exon 16 skipping in lung cancer, we analysed the mutational profile of each patient at different time points of the clinical history. Among the 12 patients with lung cancer, 9 had coexisting activating EGFR mutations (exon 19 deletions or L858R) who received prior treatment with one or multiple TKIs (online supplemental table S1). The remaining three cases were absent of canonical oncogenic driver mutations and did not receive any targeted therapies (online supplemental table S1). Particularly, three NSCLC patients underwent multiple NGS testing with both pre-EGFR and post-EGFR TKI treatment samples for analysis, while other patients had post-TKI samples only. As illustrated in figure 3, for patient 4, ERBB2 exon 16 skipping, which was caused by the full deletion of ERBB2 exon 16 as validated by RNA-seq (figure 2B), was acquired post-afatinib and most likely conferred osimertinib insensitivity (figure 3 and online supplemental table S1). For patient 5, a 9-month radiographic response was observed when on osimertinib and an ERBB2 exon 16 full deletion was detected in the post-osimertinib sample (figure 3 and online supplemental table S1). In patient 11, an ERBB2 exon 16 splice site variant was acquired after a serial TKI treatment (figure 3 and online supplemental table S1). All these evidence suggested that ERBB2 exon 16 skipping mediated acquired resistance to EGFR TKI therapy in patients with lung cancer.
Figure 3.
Clinical history and sample collection time points of three NSCLC patients Clinical treatments and primary diagnoses of each patient are located on top of the black time arrow which are coloured by treatment type (blue: TKI treatments; green: chemotherapy; pink: trastuzumb; and red: surgery). Disease progressions are indicated by the yellow circle with ‘PD’. The time points of plasma, FFPE or tissue samples collected from each patient are shown along the time arrow. For each detected sample, EGFR-related and ERBB2-related mutations are listed in the box with mutational allele frequencies. ERBB2 exon 16 skipping, both large fragment deletion (ERBB2ΔEx16) and splice site mutations are highlighted in red. EGFR, epidermal growth factor receptor; NSCLC, non-small cell lung cancer; TKI, tyrosine kinase inhibitor; FFPE, formalin-fixed paraffin-embedded; PD, progressed disease.
Besides the three patients in whom ERBB2 exon 16 skipping was validated by RNA-seq (figure 2), other ERBB2 splice site mutations (including c.1899-2A>T, c.1899–1G>T and c.1899–1G>C, online supplemental table S1) have also been reported in lung and breast cancers on the Catalogue Of Somatic Mutations In Cancer database indicating they were recurrent in cancers. Furthermore, we observed a high frequency (10/12, 83.33%) of the co-occurrence of ERBB2 amplification (figure 1). TP53 (10/12, 83.33%) represented the most frequently mutated gene in addition to ERBB2 in these patients (figure 1). Recurrent copy number variations were detected in CDK12 (6/12, 50%), NKX2.1 (6/12, 50%) and MYC (3/12, 25%) (figure 1). Taken together, we believed that ERBB2 exon 16 skipping is an acquired mutation potentially contributing to TKI resistance.
Exon 16 skipping ERBB2 as an oncogenic driver in diverse cancer types other than lung cancer
In breast cancer, ERBB2 exon 16 skipping is generally considered as an important oncogenic event driving the resistance to trastuzumab which has been widely studied both in vitro and in vivo.5 7 15–17 To our best knowledge, very limited studies reported the ERBB2 exon 16 skipped isoform in gastric, colorectal and ovarian cancers and considered it as a candidate oncogenic mutation as well.6 18–20 In our cohort, six patients were detected with ERBB2 exon 16 skipping in non-lung cancers resulting from either whole exon 16 coding sequence deletion or deletions at the splice donor site. We have confirmed the skipping of ERBB2 exon 16 at the RNA level in patient 6 and 9 (figure 2B). Three patients were treatment-naïve, one patient received surgery plus chemotherapy, and the other two rectal patients were treated with anti-HER2 therapies including lapatinib and HER2 monoclonal antibody trastuzumab (online supplemental table S2). More specifically, patient 7 showed partial response to the combination therapy of trastuzumab plus lapatinib at last follow-up, while patient 6 progressed on trastuzumab after two cycles of the therapy.
Furthermore, TP53 mutations (5/6, 83.3%) and ERBB2 amplification (5/6, 83.3%) were the most frequently concurrent mutations with ERBB2 exon 16 skipping. The frequency of CDK12 amplification (3/6, 50%) was the same as ZNF217 amplification (3/6, 50%), which were both the highest except ERBB2 amplification (5/6, 83.3%). Overall, there was no significant difference in the frequency of concurrent mutations between patients with lung cancer and non-lung cancer who carried ERBB2 exon 16 deletion. The occurrence of ERBB2 exon 16 skipping in multiple cancer types with completely different therapeutics indicated its potential to be oncogenic. However, further investigation needs to be done to come to a solid conclusion.
Discussion
In this paper, we have reported the occurrence of ERBB2 exon 16 skipping in 18 cases of non-breast cancers which is the largest cohort study till now due to its rarity. Particularly, in lung cancer, only one group reported an NSCLC patient harbouring ERBB2 exon 16 skipping and investigated its role in osimertinib resistance in vitro.21 Our study included 12 patients with lung cancer having ERBB2 exon 16 skipping with one being validated by RNA-seq. According to the analysis of matched samples prior and post-TKI treatments, we concluded that ERBB2 exon 16 skipping was an acquired mutation during TKI treatment which may also contribute to the TKI resistance. EGFR T790M is the most common secondary mutation acquired during the first-generation TKIs therapy.22 Other EGFR point mutations have also been reported to mediate the TKI resistance such as C797S, G796S and V802F.23 24 Other than that, EGFR-independent mechanisms of TKI resistance also caught broad attention in recent years including MET, MYC and ERBB2 amplification.25 Here, we observed the coexisting of ERBB2 amplification and exon 16 skipping in nine patients with lung cancer who received TKI treatments which indicated the possibility that ERBB2 exon 16 skipping cooperates with its amplification to mediate TKI resistance. Other concurrent genomic alterations included TP53 mutations, CDK12 amplification and MYC amplification which all contribute to carcinogenesis and drug resistance.26–28
ERBB2 overexpression is a well-known oncogenic driver in breast cancer targeted by Food and Drug Administration (FDA)-approved drugs such as trastuzumab. Though ERBB2 exon 16 skipping has been reported and studied in breast cancer, its response to trastuzumab is controversial. In vitro, several groups found that cell lines that transfected with exon-16-skipping ERBB2 were resistant to trastuzumab5 29 due to the conformational changes. However, in transgenic mice model, expression of exon-16-skipping ERBB2 was sufficient to improve the response to trastuzumab.17 Since we also noticed the frequent coexisting of ERBB2 exon 16 skipping and ERBB2 amplification, the high levels of wild-type ERBB2 may be enough to mediate trastuzumab’s targeting. Recently, an HER2 antibody-drug conjugate, trastuzumab deruxtecan, has launched in the USA and Japan as a third-line treatment for breast cancer. Meanwhile, several phase II trials reported promising responses for HER2-positive or HER2-mutated patients in gastric,30 31 NSCLC32 and colorectal33 cancer. As the exon-16-skipped HER2 forms a stable and constitutively active homodimer on the surface of tumour cells,34 it is worth investigating whether trastuzumab deruxtecan could benefit the patients with this unique isoform of ERBB2 in the future.
Exon-skipping events in oncogenes are not rare. One of the most well-studied examples is MET exon 14 skipping which is a targetable genomic alteration and has been reported mainly in NSCLC.35 In March 2020, tepotinib has been approved in Japan for specifically targeting MET exon 14 skipping alterations in NSCLC which is the first approved drug worldwide. Later in May, capmatinib was approved by FDA as a selective MET (MET proto-oncogene, receptor tyrosine kinase) inhibitor for treating MET exon 14 skipping-mutated NSCLC. Unfortunately, so far there are not any approved drugs or ongoing clinical trials that specifically target this exon-16-skipping ERBB2 isoform. However, it is still worth being examined and investigated as a potential biomarker to predict drug efficacy.
ERBB2 exon 16 skipping has been barely reported in other cancer types with no mechanism investigations. In this paper, we reported six cases of other cancers and hypothesised ERBB2 exon 16 skipping to be an oncogenic driver as proven in breast cancer. In our cohort, ERBB2 exon 16 skipping resulted from either exon 16 deletion or splice site variants. We had available samples from three patients to do RNA-seq validation and all of them had been confirmed. Due to the limitation of clinical sample availability, we could not repeat the RNA-seq for patient 9 whose sequencing coverage was lower than the other two (figure 2B). However, the skipping event of ERBB2 exon 16 in patient 9 was still confirmed at the transcriptional level. Therefore, both direct deletion of exon 16 coding region and splice site deletion could eventually cause the ERBB2 exon 16 skipping.
Above all, ERBB2 exon 16 skipping is a known oncogenic mutation in breast cancer, driving the resistance to trastuzumab in vitro.5 16 In NSCLC, ERBB2 exon 16 skipping was investigated in vitro as a novel mechanism of osimertinib resistance.21 In other cancer types, a few cases reported the detection of ERBB2 exon 16 skipping but the detailed mechanism remained to be investigated. We have shown here that ERBB2 exon 16 skipping is an acquired mutation contributing to the general TKIs resistance in lung cancer and also working as an oncogenic driver in colorectal, gastric and ovarian cancers as previously accepted in breast cancer.
Acknowledgments
We would like to thank the patients and family members who gave their consent on presenting the data in this study, as well as the investigators and research staff involved in this study.
Footnotes
LS and CX contributed equally.
Contributors: LS: conceptualisation, methodology, writing—original draft, review and editing; CX: conceptualisation, methodology; YM and QO: formal analysis, investigation and writing—original draft, review and editing; XW: writing—review and editing; SL: data curation and validation; YS: project administration and resources; RG and JK: supervision.
Funding: This work was supported by Beijing Medical and Health Public Welfare Fund Medical Science Research Fund [grant number: B20151DS].
Competing interests: YM, QO and XW are employees of Geneseeq Technology, Canada. YS is an employee of Nanjing Geneseeq Technology, Nanjing, Jiangsu, China.
Patient consent for publication: Not required.
Ethics approval: This study was approved by the ethical committee of the First Affiliated Hospital of Guangxi Medical University. Written informed consent was collected from each patient uponon sample collection according to the protocols approved.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available upon reasonable request. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1.Le X-F, Pruefer F, Bast RC. HER2-targeting antibodies modulate the cyclin-dependent kinase inhibitor p27Kip1 via multiple signaling pathways. Cell Cycle 2005;4:87–95. 10.4161/cc.4.1.1360 [DOI] [PubMed] [Google Scholar]
- 2.Scaltriti M, Rojo F, Ocaña A, et al. . Expression of p95HER2, a truncated form of the HER2 receptor, and response to anti-HER2 therapies in breast cancer. J Natl Cancer Inst 2007;99:628–38. 10.1093/jnci/djk134 [DOI] [PubMed] [Google Scholar]
- 3.Ou S-HI, Schrock AB, Bocharov EV, et al. . HER2 transmembrane domain (TMD) mutations (V659/G660) that stabilize homo- and heterodimerization are rare oncogenic drivers in lung adenocarcinoma that respond to afatinib. J Thorac Oncol 2017;12:446–57. 10.1016/j.jtho.2016.11.2224 [DOI] [PubMed] [Google Scholar]
- 4.Robichaux JP, Elamin YY, Tan Z, et al. . Mechanisms and clinical activity of an EGFR and HER2 exon 20–selective kinase inhibitor in non–small cell lung cancer. Nat Med 2018;24:638–46. 10.1038/s41591-018-0007-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitra D, Brumlik MJ, Okamgba SU, et al. . An oncogenic isoform of HER2 associated with locally disseminated breast cancer and trastuzumab resistance. Mol Cancer Ther 2009;8:2152–62. 10.1158/1535-7163.MCT-09-0295 [DOI] [PubMed] [Google Scholar]
- 6.Ross JS, Fakih M, Ali SM, et al. . Targeting HER2 in colorectal cancer: the landscape of amplification and short variant mutations in ERBB2 and ERBB3. Cancer 2018;124:1358–73. 10.1002/cncr.31125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alajati A, Sausgruber N, Aceto N, et al. . Mammary tumor formation and metastasis evoked by a HER2 splice variant. Cancer Res 2013;73:5320–7. 10.1158/0008-5472.CAN-12-3186 [DOI] [PubMed] [Google Scholar]
- 8.Shu Y, Wu X, Tong X, et al. . Circulating tumor DNA mutation profiling by targeted next generation sequencing provides guidance for personalized treatments in multiple cancer types. Sci Rep 2017;7:583. 10.1038/s41598-017-00520-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Z, Yang N, Ou Q, et al. . Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor Osimertinib in non-small cell lung cancer patients. Clin Cancer Res 2018;24:3097–107. 10.1158/1078-0432.CCR-17-2310 [DOI] [PubMed] [Google Scholar]
- 10.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 2014;30:2114–20. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newman AM, Bratman SV, Stehr H, et al. . FACTERA: a practical method for the discovery of genomic rearrangements at breakpoint resolution. Bioinformatics 2014;30:3390–3. 10.1093/bioinformatics/btu549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koboldt DC, Zhang Q, Larson DE, et al. . VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568–76. 10.1101/gr.129684.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobin A, Davis CA, Schlesinger F, et al. . STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 2013;14:178–92. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marchini C, Gabrielli F, Iezzi M, et al. . The human splice variant Δ16HER2 induces rapid tumor onset in a reporter transgenic mouse. PLoS One 2011;6:e18727. 10.1371/journal.pone.0018727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turpin J, Ling C, Crosby EJ, et al. . The ErbB2ΔEx16 splice variant is a major oncogenic driver in breast cancer that promotes a pro-metastatic tumor microenvironment. Oncogene 2016;35:6053–64. 10.1038/onc.2016.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castagnoli L, Iezzi M, Ghedini GC, et al. . Activated d16HER2 homodimers and SRC kinase mediate optimal efficacy for trastuzumab. Cancer Res 2014;74:6248–59. 10.1158/0008-5472.CAN-14-0983 [DOI] [PubMed] [Google Scholar]
- 18.Yu D-H, Tang L, Dong H, et al. . Oncogenic HER2 fusions in gastric cancer. J Transl Med 2015;13:116. 10.1186/s12967-015-0476-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunter SM, Anglesio MS, Ryland GL, et al. . Molecular profiling of low grade serous ovarian tumours identifies novel candidate driver genes. Oncotarget 2015;6:37663–77. 10.18632/oncotarget.5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Volpi CC, Pietrantonio F, Gloghini A, et al. . The landscape of d16HER2 splice variant expression across HER2-positive cancers. Sci Rep 2019;9:3545. 10.1038/s41598-019-40310-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu C-C, Liao B-C, Liao W-Y, et al. . Exon 16-Skipping HER2 as a novel mechanism of Osimertinib resistance in EGFR L858R/T790M-Positive non-small cell lung cancer. J Thorac Oncol 2020;15:50–61. 10.1016/j.jtho.2019.09.006 [DOI] [PubMed] [Google Scholar]
- 22.Balak MN, Gong Y, Riely GJ, et al. . Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res 2006;12:6494–501. 10.1158/1078-0432.CCR-06-1570 [DOI] [PubMed] [Google Scholar]
- 23.Thress KS, Paweletz CP, Felip E, et al. . Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med 2015;21:560–2. 10.1038/nm.3854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ou S-HI, Cui J, Schrock AB, et al. . Emergence of novel and dominant acquired EGFR solvent-front mutations at Gly796 (G796S/R) together with C797S/R and L792F/H mutations in one EGFR (L858R/T790M) NSCLC patient who progressed on osimertinib. Lung Cancer 2017;108:228–31. 10.1016/j.lungcan.2017.04.003 [DOI] [PubMed] [Google Scholar]
- 25.Ahsan A. Mechanisms of resistance to EGFR tyrosine kinase inhibitors and therapeutic approaches: an update. Adv Exp Med Biol 2016;893:137–53. 10.1007/978-3-319-24223-1_7 [DOI] [PubMed] [Google Scholar]
- 26.Rivlin N, Brosh R, Oren M, et al. . Mutations in the p53 tumor suppressor gene: important milestones at the various steps of tumorigenesis. Genes Cancer 2011;2:466–74. 10.1177/1947601911408889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paculová H, Kohoutek J. The emerging roles of CDK12 in tumorigenesis. Cell Div 2017;12:7. 10.1186/s13008-017-0033-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rihawi K, Alfieri R, Fiorentino M, et al. . MYC amplification as a potential mechanism of primary resistance to crizotinib in ALK-rearranged non-small cell lung cancer: a brief report. Transl Oncol 2019;12:116–21. 10.1016/j.tranon.2018.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castiglioni F, Tagliabue E, Campiglio M, et al. . Role of exon-16-deleted HER2 in breast carcinomas. Endocr Relat Cancer 2006;13:221–32. 10.1677/erc.1.01047 [DOI] [PubMed] [Google Scholar]
- 30.Shitara K, Bang Y-J, Iwasa S, et al. . Trastuzumab Deruxtecan in previously treated HER2-positive gastric cancer. N Engl J Med 2020;382:2419–30. 10.1056/NEJMoa2004413 [DOI] [PubMed] [Google Scholar]
- 31.Shitara K, Bang Y-J, Iwasa S, et al. . Trastuzumab deruxtecan (T-DXd; DS-8201) in patients with HER2-positive advanced gastric or gastroesophageal junction (GEJ) adenocarcinoma: a randomized, phase II, multicenter, open-label study (DESTINY-Gastric01). J Clin Oncol 2020;38:4513 10.1200/JCO.2020.38.15_suppl.4513 [DOI] [Google Scholar]
- 32.Smit EF, Nakagawa K, Nagasaka M, et al. . Trastuzumab deruxtecan (T-DXd; DS-8201) in patients with HER2-mutated metastatic non-small cell lung cancer (NSCLC): interim results of DESTINY-Lung01. J Clin Oncol 2020;38:9504 10.1200/JCO.2020.38.15_suppl.9504 [DOI] [Google Scholar]
- 33.Siena S, Di Bartolomeo M, Raghav KPS, et al. . A phase II, multicenter, open-label study of trastuzumab deruxtecan (T-DXd; DS-8201) in patients (pts) with HER2-expressing metastatic colorectal cancer (mCRC): DESTINY-CRC01. J Clin Oncol 2020;38:4000 10.1200/JCO.2020.38.15_suppl.4000 [DOI] [Google Scholar]
- 34.Kwong KY, Hung MC. A novel splice variant of HER2 with increased transformation activity. Mol Carcinog 1998;23:62–8. [DOI] [PubMed] [Google Scholar]
- 35.Lee GD, Lee SE, Oh D-Y, et al. . Met exon 14 skipping mutations in lung adenocarcinoma: clinicopathologic implications and prognostic values. J Thorac Oncol 2017;12:1233–46. 10.1016/j.jtho.2017.04.031 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
esmoopen-2020-000985supp001.pdf (50.1KB, pdf)