Abstract
Background:
Small-cell lung cancer (SCLC) accounts for almost 15% of lung cancer cases in the United States. Nomogram prognostic models could greatly facilitate risk stratification and treatment planning, as well as more refined enrollment criteria for clinical trials. We developed and validated a new nomogram prognostic model for SCLC patients using a large SCLC patient cohort from the National Cancer Database (NCDB).
Methods:
Clinical data of 24,680 SCLC patients diagnosed from 2004 to 2011 were used to develop the nomogram prognostic model. The model was then validated using an independent cohort of 9,700 SCLC patients diagnosed from 2012 to 2013. The prognostic performance was evaluated using p value, concordance index and integrated Area Under the (time-dependent Receiver Operating Characteristic) Curve.
Results:
The following variables were contained in the final prognostic model: age, gender, race, ethnicity, Charlson/Deyo Score, TNM Stage (assigned according to the AJCC 8th edition), treatment type (combination of surgery, radiation therapy and chemotherapy), and laterality. The model was validated in an independent testing group with a concordance index of 0.722 ± 0.004 and an integrated AUC of 0.79. The nomogram model has a significantly higher prognostic accuracy than previously developed models, including the AJCC 8th edition TNM-staging system. We implemented the proposed nomogram and four previously published nomograms in an online webserver.
Conclusions:
We developed a nomogram prognostic model for SCLC patients, and validated the model using an independent patient cohort. The nomogram performs better than earlier models, including models using AJCC staging.
Keywords: SCLC, Patient prognosis, Nomogram prognostic model
Introduction
Lung cancer is the leading cause of death from cancer in the United States and worldwide. Small-cell lung cancer (SCLC) accounts for 13.6% of all lung cancer cases 1, 2. Compared to non-small-cell lung cancer (NSCLC), in which the 5-year survival rate is 18.0%, SCLC has only a 6.2% 5-year survival rate, and is characterized by a more rapid tumor growth rate and death from recurrent disease 3, 4. Over the last several decades, there have been only modest improvements in patient survival 5 and no molecularly targeted therapy has proven beneficial for SCLC patients 6. Nomogram prognostic models that predict patient outcomes may facilitate better treatment stratification and outcome evaluation, as well as more refined patient enrollment criteria for clinical trials in SCLC. Furthermore, a recent study in breast cancer 7 showed that user-friendly online prognostic tools could greatly enhance patient care. However, currently there are no such online tools available for prognosis of SCLC.
To date there are three studies of nomograms in SCLC, published by Xie et al 4, Pan et al 8, and Xiao et al9. The nomograms developed from those studies provide useful tools for clinicians and researchers to stratify the risk of SCLC patients. However, two of the studies simply classified patients as limited or extensive stage without using the more accurate TNM staging proposed by the International Association for the Study of Lung Cancer (IASLC) 10. Furthermore, there is a lack of independent validation for these models, probably due to the limited sample size (n = 9384, 2758, and 6479 separately). Other non-nomogram prognostic models include the Manchester score and Spain score. However, both of these were developed on small sample sets (n = 407 for Manchester score and n = 341 for Spain score) and divide patients into only three risk groups 11, 12.
The goal of this study was to identify prognostic factors for SCLC patients, and then develop and validate a new nomogram prognostic model in a large SCLC patient cohort. The National Cancer Database (NCDB) includes over 200,000 patients diagnosed with SCLC from 2004 to 2013 in the United States, of which 34,380 SCLC patients without any missing values were used to develop and validate our nomogram prognostic model. The SCLC cases in the NCDB dataset were separated into a training cohort and a validation cohort based on the year of diagnosis. The model was developed from the training cohort of 24,680 SCLC patients diagnosed from 2004 to 2011, and then validated in the validation cohort of 9,700 SCLC patients diagnosed from 2012 to 2013. The prognostic performance was evaluated using p value, concordance index and integrated Area Under the Curve. In order to facilitate public usage, we implemented our nomogram and the previous ones by Xie et al. in an online webserver. Compared to the previously published models, our model has the following advantages: 1) it was validated in an independent set; 2) it was developed and validated with a much larger sample size; 3) it was developed across multiple facilities and facility types, which greatly diminishes sample selection bias; 4) it utilizes accurate SCLC staging criteria: the AJCC 8th edition TNM staging system proposed by IASLC 13, 14; and 5) it provides an online webserver so that clinicians can use the nomogram model easily.
Methods
Source of data
202,194 SCLC cases were identified from NCDB and 34,380 of them met our inclusion criterion that they do not contain any missing data for selected variables. The source of missing values is listed in Supplementary Table 1. The cases are independent and recorded by annual reports from all the CoC-accredited programs from 2004 to 2013. 24,680 cases that were diagnosed from 2004 to 2011 were assigned to the training group and used to develop the nomogram prognostic model. The 9,700 cases diagnosed from 2012 to 2013 were assigned to the testing group and used to validate the model.
Nomogram development
The nomogram was developed using the training cohort of 24,680 patients diagnosed from 2004 to 2011. Overall survival was defined as the length of time from diagnosis to death or last contact, and used as the primary outcome. Two extra variables were first constructed based on NCDB variables: treatment was defined as the stratification result of surgery, chemotherapy and radiation therapy; and TNM stage was defined according to the coding guidelines of the Collaborative Staging Manual and Coding Instructions for the new 8th edition lung cancer staging system defined by the American Joint Committee on Cancer (AJCC) and the Union for International Cancer Control (UICC) 15–18, and followed Yang et al’s method 19. Stages IA1, IA2, and IA3 were combined together in our study as stage IA, since no significant prognostic differences were detected among the three sub-stages 14. The assumptions were made here that the timing and sequence of the treatments were interchangeable, and none of these are salvage treatment due to recurrence/progression. The input variables were age, gender, race, Hispanic origin, Charlson/Deyo Score, sequence number, primary site, laterality, grade (tumor’s resemblance to normal tissue), 8th edition TNM stage and treatment type.
Univariate Cox regression and Wald test were then used to screen for variables that were significantly correlated with overall survival in the training group. Predictors with a p-value less than 0.05 were fed to a multivariate Cox regression model. Backward stepwise selection based on Bayesian Information Criterion (BIC) was used to further eliminate redundant variables. The resulting multivariate Cox regression model was used to calculate risk score and build the final nomogram prognostic model.
Model validation
To validate our model, four criteria were used to evaluate prediction performance in the testing set. First, the cases were grouped according to their predicted risk score, and Kaplan-Meier survival curves and Wald test were used to compare survival differences among the groups. Second, a concordance index (c-index) was calculated to estimate the similarity between the ranking of true survival time and of predicted risk score. The theoretical value of the c-index is between 0 and 1; a c-index larger than 0.5 indicates prediction performance better than random guessing. When evaluating the performances of different models, c-indexes from different models were compared using z-test. Third, the area under the curve (AUC) of time-dependent receiver operating characteristics (ROC) 20, 21 was calculated at each month from the 1st to the 30th month. Integrated AUC was calculated by averaging the 30 AUC values. Fourth, calibration curves were plotted to evaluate the consistency between predicted survival probability and actual survival proportion at 1 and 2 years, separately22. A perfect prediction would result in a 45-degree calibration curve (i.e. the identity line).
The other two models, the AJCC 8th edition TNM staging system and the traditional limited/extensive staging system, were also tested for prognostic performance in the testing group. C-index and integrated AUC were used to compare this nomogram with the two staging systems. Here, extensive stage was defined based on the presence of distant metastases (M1 stage) 23, 24. All other cases (M0 stage) were grouped as limited stage. To compare performance of the proposed nomogram with TNM staging system and limited/extensive staging system, a nonparametric approach proposed by Kang et al was used to compare the correlated C-indexes with right-censored survival outcome25.
All computations were conducted in the R environment, version 3.3.2 26. R packages “survival” (version 2.40-1), “timeROC” (version 0.3), “rms” (version 5.1-2), and “compare” (version 1.3.1) were used. Results with p-value ≤ 0.05 were considered statistically significant.
Implementation of this and previously published models
To facilitate researchers’ and clinicians’ usage of our model, we created a user-friendly webserver for our nomogram and the models from Pan et al8, Xiao et al9, and the two models from Xie et al4. The nomogram from this study calculates the risk score, plots the survival curve and provides survival probabilities for 120 months at 6-month increments. The Pan et al model provides 1-year and 2-year survival probabilities. The Xiao et al model provides 3-year and 5-year survival probabilities. The Xie et al models for both extensive and limited stage cases provide 6-month and 12-month survival probabilities and predicted median survival time. Data points were read from Figure 1 of the Pan et al publication8, Figure 1B of the Xiao et al publication9, and Figures 1 and 2 of the Xie et al publication 4, and the corresponding survival probability for a given score was calculated by linear interpolation.
Figure 1.
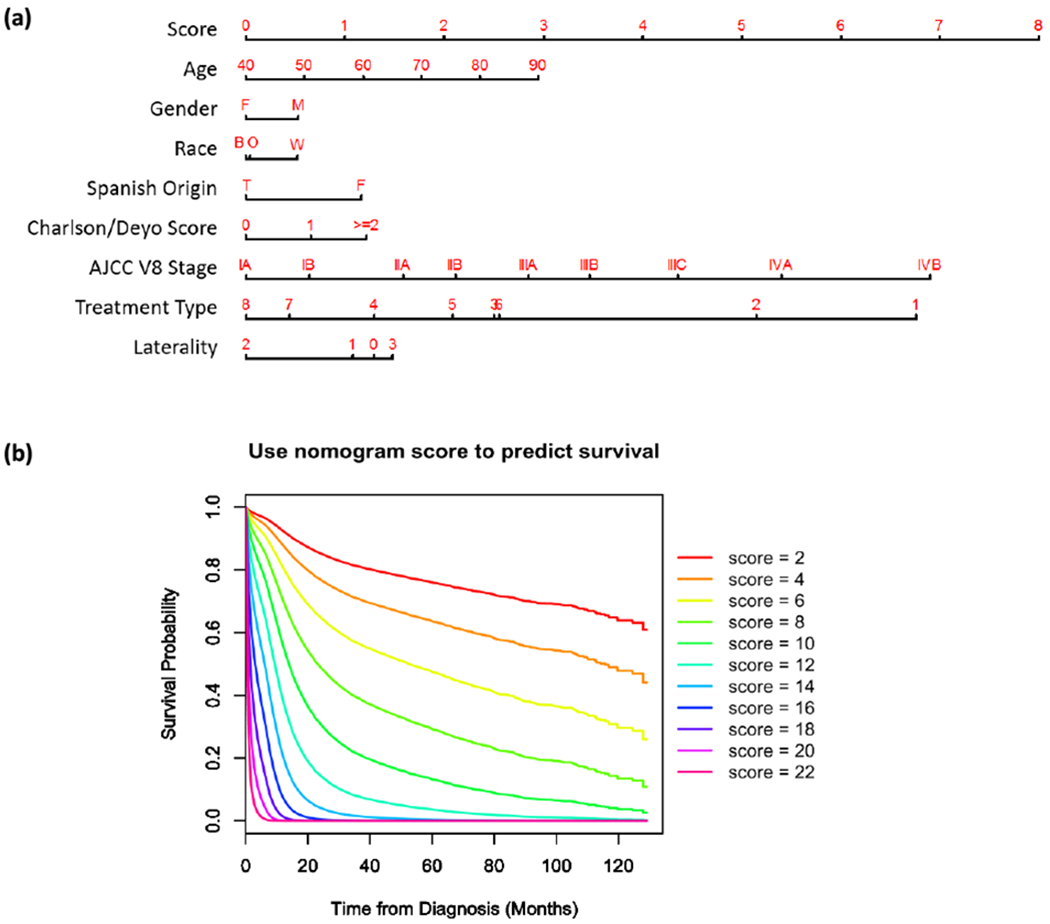
Nomogram to calculate risk score and predict survival probability. (a) Race includes black (B), white (W) and other (O). Treatment types include: no surgery, no chemo, no radiation (1); no surgery, no chemo, radiation done (2); no surgery, chemo done, no radiation (3); no surgery, chemo done, radiation done (4); surgery done, no chemo, no radiation (5); surgery done, no chemo, radiation done (6); surgery done, chemo done, no radiation (7); and surgery done, chemo done, radiation done (8). Laterality of tumor origin includes: not a paired site (0), only one side (either left or right) is involved (1), bilateral involvement (2), paired site with unknown origin side or midline tumor (3). (b) Predicted patient survival probability curve corresponding to risk scores ranging from 2 to 22.
Figure 2.
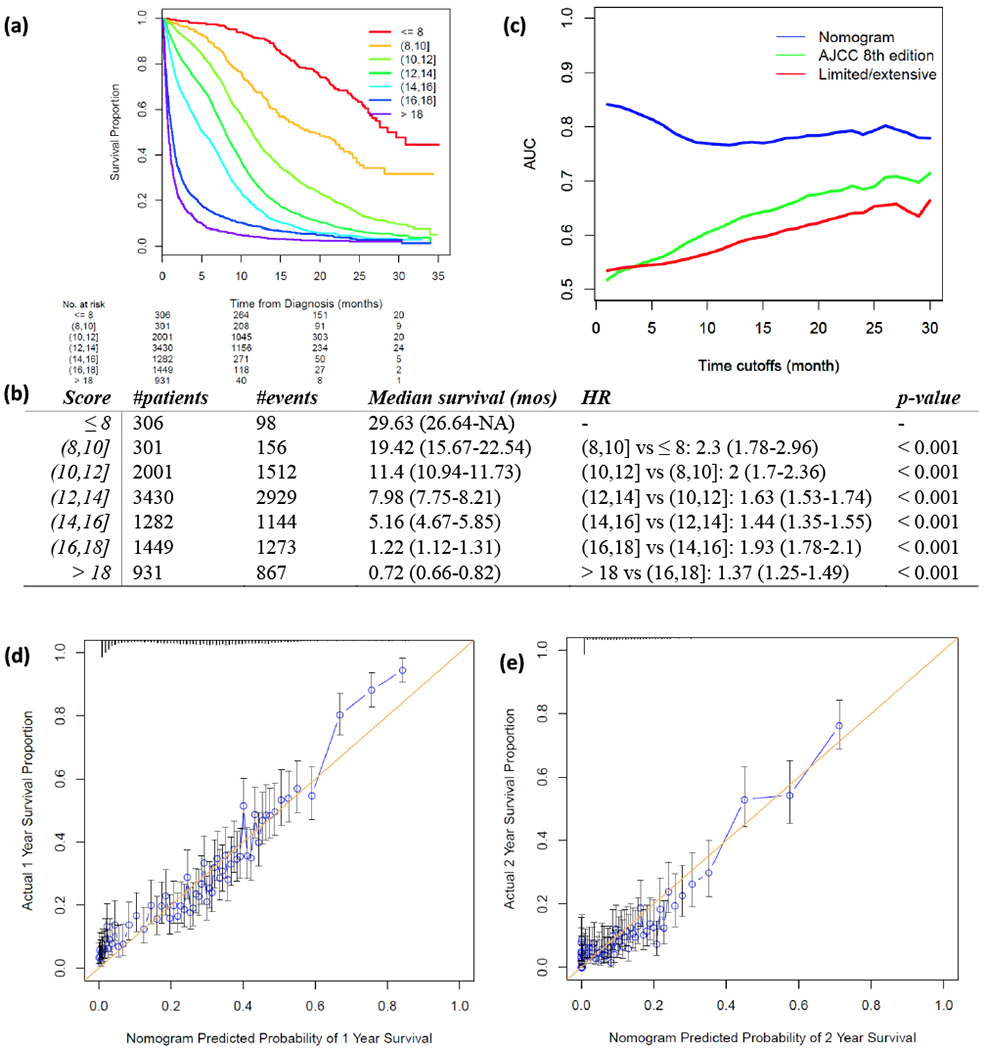
Validation of proposed nomogram prognostic model in the testing set. (a) Risk scores of testing set cases were calculated according to the model in Figure 1 and grouped into 8 subgroups. K-M plot was depicted for each group. (b) Summary of groups in (a). Hazard Ratio (HR) was calculated using Coxph regression model between each two adjacent lines. P-value was calculated using Wald test. (c) Area under the curve (AUC) was calculated for three prognostic models for every month from the 1st to the 30th month. Blue: nomogram developed in this study; green: AJCC 8th TNM staging system; red: limited/extensive staging system. (d, e) Calibration curves compare predicted and actual survival proportions at 1 year (d) and 2 years (e), separately. Each point in the plot refers to a group of patients, with the nomogram predicted probability of survival shown on x-axis and actual survival proportion shown on y-axis. Distributions of predicted survival probabilities are plotted at the top. Error bars represent 95% confidence intervals.
Results
Characteristics of the training and validation cohorts
In total, 202,194 SCLC cases were identified in NCDB, among which, 34,380 cases that did not contain any missing variables were included in this study. Based on year of diagnosis, included cases were divided into two distinct groups: cases that were diagnosed from 2004 to 2011 (n = 24,680) were used as the training cohort, while cases that were diagnosed from 2012 to 2013 (n = 9,700) were used as the validation cohort. The follow-up time ranged from 0 to 10.76 years (median 0.64 year) for the training cohort, and from 0 to 2.92 years (median 0.53 year) for the testing cohort. Characteristics of the two sets are shown in Table 1. In comparing the training and testing sets, the demographic variables were similar, while the clinical variables, including Charlson/Deyo score, 8th AJCC stage, and laterality, were significantly different.
Table 1.
Characteristics of training set and testing set. P-values were calculated by Chi-square test.
| Training set (%) | Testing set (%) | p-value | |
|---|---|---|---|
| No. of cases | 24,680 | 9,700 | |
| Year of diagnosis | 2004-2011 | 2012-2013 | |
| Age | 0.09 | ||
| < 65y | 9,559 (38.7) | 3,855 (39.7) | |
| ≥ 65y | 15,121 (61.3) | 5,845 (60.3) | |
| Gender | 0.9 | ||
| Male | 12,240 (49.6) | 4,803 (49.5) | |
| Female | 12,440 (50.4) | 4,897 (50.5) | |
| Race | 0.73 | ||
| White | 22,276 (90.3) | 8,779 (90.5) | |
| Black | 1,912 (7.7) | 727 (7.5) | |
| Other | 492 (2) | 194 (2) | |
| Hispanic origin | 0.91 | ||
| Non-Hispanic | 24,084 (97.6) | 9,463 (97.6) | |
| Hispanic | 596 (2.4) | 237 (2.4) | |
| Charlson/Deyo score | <0.001 | ||
| 0 | 13,288 (53.8) | 5,031 (51.9) | |
| 1 | 7,629 (30.9) | 3,061 (31.6) | |
| ≥ 2 | 3,763 (15.2) | 1,608 (16.6) | |
| Sequence number | 0.82 | ||
| 0 | 24,084 (97.6) | 9,463 (97.6) | |
| 1 | 527 (2.1) | 213 (2.2) | |
| ≥ 2 | 69 (0.3) | 24 (0.2) | |
| AJCC V8 TNM stage | <0.001 | ||
| IA | 1,207 (4.9) | 160 (1.6) | |
| IB | 463 (1.9) | 74 (0.8) | |
| IIA | 140 (0.6) | 18 (0.2) | |
| IIB | 853 (3.5) | 97 (1) | |
| IIIA | 1,548 (6.3) | 156 (1.6) | |
| IIIB | 902 (3.7) | 89 (0.9) | |
| IIIC | 208 (0.8) | 27 (0.3) | |
| IVA | 14,699 (59.6) | 6,655 (68.6) | |
| IVB | 4,660 (18.9) | 2,424 (25) | |
| Treatment | <0.001 | ||
| No surgery, no chemo, no radiation | 5,025 (20.4) | 2,213 (22.8) | |
| No surgery, no chemo, radiation done | 1,230 (5) | 520 (5.4) | |
| No surgery, chemo done, no radiation | 7,668 (31.1) | 3,473 (35.8) | |
| No surgery, chemo done, radiation done | 7,901 (32) | 3,050 (31.4) | |
| Surgery done, no chemo, no radiation | 856 (3.5) | 116 (1.2) | |
| Surgery done, no chemo, radiation done | 64 (0.3) | 8 (0.1) | |
| Surgery done, chemo done, no radiation | 1,000 (4.1) | 165 (1.7) | |
| Surgery done, chemo done, radiation done | 936 (3.8) | 155 (1.6) | |
| Primary site | <0.001 | ||
| C340 | 2,298 (9.3) | 911 (9.4) | |
| C341 | 11,019 (44.6) | 4,152 (42.8) | |
| C342 | 968 (3.9) | 368 (3.8) | |
| C343 | 4,959 (20.1) | 1,923 (19.8) | |
| C348 | 485 (2) | 200 (2.1) | |
| C349 | 4,951 (20.1) | 2,146 (22.1) | |
| Laterality | <0.001 | ||
| Not a paired site | 2,298 (9.3) | 911 (9.4) | |
| Only one side involved | 20,447 (82.8) | 8,016 (82.6) | |
| Bilateral involvement | 624 (2.5) | 154 (1.6) | |
| Paired site but lateral origin unknown; midline tumor | 1,311 (5.3) | 619 (6.4) | |
| Grade | <0.001 | ||
| Well differentiated | 88 (0.4) | 8 (0.1) | |
| Moderately differentiated | 179 (0.7) | 39 (0.4) | |
| Poorly differentiated | 2,795 (11.3) | 899 (9.3) | |
| Undifferentiated | 5,037 (20.4) | 1,457 (15) | |
| Cell type not determined, not stated or not applicable | 16,581 (67.2) | 7,297 (75.2) | |
Building nomogram prognostic model in training cohort
In univariate analysis, age, gender, race, Hispanic origin, Charlson/Deyo score, TNM stage by AJCC 8th edition, treatment type, primary site, laterality, and grade were significantly associated with overall survival in the training group (Table 2). After stepwise selection to further remove potential redundancy, age, sex, race, ethnicity, Charlson/Deyo score, TNM stage by AJCC 8th edition, treatment type, and laterality were used in the final nomogram model (coefficients summarized in Table 3). The final risk score was calculated by adding up the score of each item using the nomogram depicted in Figure 1a. The TNM stage defined by the AJCC 8th edition showed the largest range of risk scores, followed by the treatment type and age. The predicted survival probability using the Cox regression model of risk scores was plotted in Figure 1b.
Table 2.
Univariate analysis results summary. HR: Hazard Ratio, CI: Confidence Interval.
| Variable | HR (95% CI) | p-value |
|---|---|---|
| Age | 1.023 (1.023-1.024) | < 0.001 |
| Sex (Female vs. Male) | 0.84 (0.83-0.85) | < 0.001 |
| Race | ||
| White | 1 (reference) | - |
| Black | 0.97 (0.95-0.99) | 0.006 |
| Other | 0.94 (0.90-0.97) | 0.001 |
| Hispanic origin (Yes vs. No) | 0.95 (0.92-0.99) | 0.028 |
| Charlson/Deyo score | ||
| 0 | 1 (reference) | - |
| 1 | 1.22 (1.20-1.24) | < 0.001 |
| ≥ 2 | 1.59 (1.56-0.61) | < 0.001 |
| Sequence number | ||
| 0 | 1 (reference) | - |
| 1 | 1 (0.98-1.01) | 0.82 |
| ≥ 2 | 1.01 (0.92-1.11) | 0.83 |
| AJCC V8 TNM stage | ||
| IA | 1 (reference) | - |
| IB | 1.22 (1.07-1.39) | < 0.001 |
| IIA | 1.63 (1.34-1.98) | < 0.001 |
| IIB | 1.6 (1.45-1.78) | < 0.001 |
| IIIA | 2.12 (1.94-2.31) | < 0.001 |
| IIIB | 2.55 (2.32-2.81) | < 0.001 |
| IIIC | 3.26 (2.81-3.78) | < 0.001 |
| IVA | 5.25 (4.88-5.65) | < 0.001 |
| IVB | 7.04 (6.51-7.61) | < 0.001 |
| Treatment | ||
| No surgery, no chemo, no radiation | 1 (reference) | - |
| No surgery, no chemo, radiation done | 0.72 (0.7-0.74) | < 0.001 |
| No surgery, chemo done, no radiation | 0.46 (0.45-0.47) | < 0.001 |
| No surgery, chemo done, radiation done | 0.26 (0.25-0.26) | < 0.001 |
| Surgery done, no chemo, no radiation | 0.19 (0.18-0.2) | < 0.001 |
| Surgery done, no chemo, radiation done | 0.28 (0.24-0.33) | < 0.001 |
| Surgery done, chemo done, no radiation | 0.13 (0.13-0.14) | < 0.001 |
| Surgery done, chemo done, radiation done | 0.13 (0.12-0.14) | < 0.001 |
| Primary site | ||
| C340 | 1 (reference) | - |
| C341 | 0.89 (0.88-0.91) | < 0.001 |
| C342 | 0.9 (0.87-0.92) | < 0.001 |
| C343 | 0.96 (0.94-0.98) | < 0.001 |
| C348 | 1.07 (1.03-1.11) | < 0.001 |
| C349 | 1.13 (1.11-1.16) | < 0.001 |
| Laterality | ||
| Not a paired site | 1 (reference) | - |
| Only one side involved | 0.94 (0.93-0.96) | < 0.001 |
| Bilateral involvement | 1.47 (1.4-1.54) | < 0.001 |
| Paired site but lateral origin unknown; midline tumor | 1.18 (1.15-1.21) | < 0.001 |
| Grade | ||
| Well differentiated | 1 (reference) | - |
| Moderately differentiated | 0.99 (0.86-1.14) | 0.86 |
| Poorly differentiated | 1.29 (1.15-1.46) | < 0.001 |
| Undifferentiated | 1.39 (1.23-1.56) | < 0.001 |
| Cell type not determined, not stated or not applicable | 1.44 (1.28-1.62) | < 0.001 |
Table 3.
Hazard Ratio (HR) and 95% confidence interval of nomogram parameters.
| HR (95% CI) | p-value | |
|---|---|---|
| Age | 1.01 (1.01-1.02) | < 0.001 |
| Sex (Female vs. Male) | 0.88 (0.85-0.9) | < 0.001 |
| Race | ||
| White | 1 (reference) | - |
| Black | 0.88 (0.84-0.92) | < 0.001 |
| Other | 0.89 (0.8-0.98) | 0.02 |
| Hispanic origin (Yes vs. No) | 0.75 (0.68-0.82) | < 0.001 |
| Charlson/Deyo score | ||
| 0 | 1 (reference) | - |
| 1 | 1.18 (1.14-1.21) | < 0.001 |
| >= 2 | 1.36 (1.31-1.41) | < 0.001 |
| AJCC V8 TNM stage | ||
| IA | 1 (reference) | - |
| IB | 1.17 (1.02-1.35) | 0.02 |
| IIA | 1.49 (1.2-1.84) | < 0.001 |
| IIB | 1.7 (1.52-1.9) | < 0.001 |
| IIIA | 2.04 (1.83-2.26) | < 0.001 |
| IIIB | 2.38 (2.11-2.68) | < 0.001 |
| IIIC | 2.97 (2.5-3.54) | < 0.001 |
| IVA | 3.86 (3.48-4.27) | < 0.001 |
| IVB | 5.62 (5.06-6.24) | < 0.001 |
| Treatment | ||
| No surgery, no chemo, no radiation | 1 (reference) | - |
| No surgery, no chemo, radiation done | 0.67 (0.63-0.71) | < 0.001 |
| No surgery, chemo done, no radiation | 0.35 (0.33-0.36) | < 0.001 |
| No surgery, chemo done, radiation done | 0.25 (0.24-0.26) | < 0.001 |
| Surgery done, no chemo, no radiation | 0.31 (0.28-0.35) | < 0.001 |
| Surgery done, no chemo, radiation done | 0.35 (0.27-0.46) | < 0.001 |
| Surgery done, chemo done, no radiation | 0.21 (0.19-0.23) | < 0.001 |
| Surgery done, chemo done, radiation done | 0.18 (0.17-0.2) | < 0.001 |
| Laterality | ||
| Not a paired site | 1 (reference) | - |
| Only one side involved | 0.95 (0.91-0.99) | 0.02 |
| Bilateral involvement | 0.72 (0.66-0.79) | < 0.001 |
| Paired site but lateral origin unknown; midline tumor | 1.05 (0.98-1.13) | 0.19 |
Validation in testing cohort and sensitivity analysis in regards to missing data
The proposed nomogram was validated in the independent testing set (n=9,700). The survival difference between any two adjacent groups, which were grouped by predicted risk score, was significant (p-value < 0.05, Figure 2a & 2b). The median survival times of score groups ranged from 0.7 months (when risk score > 18) to 30.9 months (when risk score < 6). The c-index was 0.722 ± 0.004 and the integrated AUC was 0.79 from the 1st month to the 30th month (Figure 2c, Supplementary Table 2). A calibration curve at 1 year (Figure 2d) or 2 years (Figure 2e) also showed high consistency between predicted survival probability and actual survival proportion.
With regard to prognostic ability, the proposed nomogram performed better than the two commonly used SCLC staging systems, the AJCC TNM system and limited/extensive staging system (Figure 2c, Supplementary Table 2, Supplementary Figure 1 a&b). The AUC of the nomogram was the highest throughout the 1st to the 30th month, followed by the 8th edition TNM staging system. The integrated AUC of the proposed nomogram was 0.789, while those of the 8th edition TNM staging system and the limited/extensive staging system were 0.634 and 0.598, respectively. The c-index of this nomogram (0.722 ± 0.004) was also significantly higher than the c-indexes of the 8th edition TNM staging system (0.550 ± 0.003, p-value < 0.001) and the limited/extensive staging system (0.539 ± 0.002, p-value < 0.001), confirming the strong prognostic power of this proposed nomogram.
To evaluate the robustness of our model to missing data, a sensitivity analysis was performed on the excluded cases diagnosed from the year 2012 to 2013 (n = 11,020). The missed variables were imputed using corresponding modes in the training cohort (Table 1): missed stages (n = 10,416) were imputed as “stage IVA”; missed treatment types (n = 508) were imputed as “No Surgery, Chemo Done, Radiation Done”; missed Hispanic origins (n = 819) were imputed as “False”. Under the circumstance of having at least one variable imputed, the survival difference between any two adjacent predicted risk groups was still significant (Supplementary Figure 2 a&b). The c-index was 0.691 ± 0.004, and the integrated AUC was 0.734 (Supplementary Figure 2c). A calibration curve at 1 year (Supplementary Figure 2d) or 2 years (Supplementary Figure 2e) still showed high consistency between predicted survival probability and actual survival proportion, proving the robustness of this nomogram to missing data.
Development of webserver for easy access of our own and previously published models
An online version of our nomogram (Figure 3a) can be accessed at http://lce.biohpc.swmed.edu/lungcancer/sclc_nomogram, to assist researchers and clinicians. Online implementation of the other nomograms from Pan et al8, Xiao et al9, and Xie et al4 are also available (Figure 3b–d). Predicted survival probability across time can be easily determined by inputting clinical features and reading output figures and tables generated by the webserver.
Figure 3.
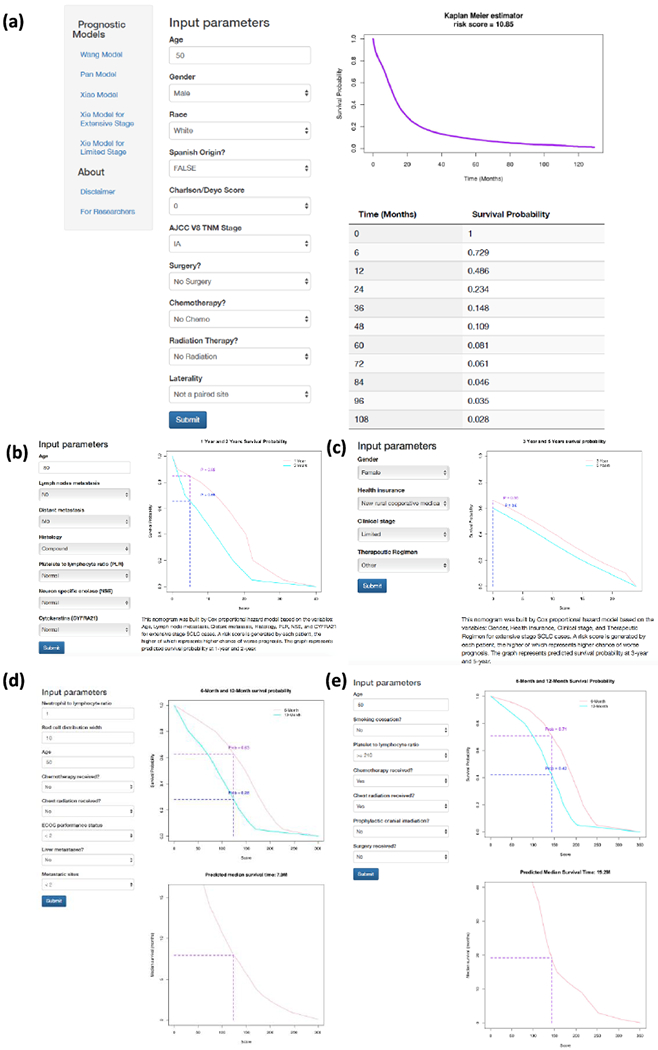
Online webserver interface for our nomogram as well as previous prognostic models. (a) The newly developed nomogram in this study (Wang model). (b-e) Published nomograms by Pan et al (b), Xiao et al (c), and Xie et al (d: Extensive Stage; e: Limited Stage).
Discussion
In this study, a nomogram prognostic model was developed and validated using a large cohort of SCLC cases across the United States. This nomogram, based on routinely available demographic, staging and treatment information, predicts the survival probability for individual SCLC patients. The publicly accessible online implementation will assist clinicians in making treatment decisions.
Compared with other prognostic indexes, such as the Manchester Score 11 and the Spain prognostic index 12, our model calculates individualized survival probability rather than assigning cases into a few risk groups, thus better capturing heterogeneity across patients. Compared with the previously published nomogram by Xie et al., this model used a much larger training dataset and involved multiple treatment facilities, which allowed for smaller sampling bias. The internal c-index of this model was 0.744 ± 0.002, higher than in previously published models (0.73 for both nomograms in 4). Independent validation of our model showed significantly different outcomes among different score groups (Figure 2a&b). A high concordance index (0.722 ± 0.004) and integrated AUC score (0.789, Figure 2c, Supplementary Table 2) in the testing set also indicated the strong predictive ability of our nomogram model. In addition, combining demographic, clinical and treatment information together produced a nomogram with better performance than using staging information alone (Figure 2b, Supplementary Table 2). Thus, this comprehensive and individualized risk score calculation method could be used as stratification criteria in randomized studies and clinical trials.
In this nomogram, age, gender, race, ethnicity, Charlson/Deyo score, AJCC 8th edition stage, treatment type and laterality were kept after univariate Cox regression screening and backward stepwise selection. Age, gender, and Charlson/Deyo score have previously been shown significantly relevant to survival of SCLC patients 4, 27. Noticeably, AJCC 8th edition stage contributed the most to the final risk score (Figure 1a), with clear distinctions between each two adjacent TNM stages (Table 3), and showed better prognostic performance than the limited/extensive staging system with higher c-index and AUC (Figure 2b, Supplementary Table 2). The significant contribution of TNM stage to this nomogram externally validates the performance of the 8th edition TNM lung cancer classification system, and highlights the importance of applying this more accurate staging system to SCLC rather than using the traditional limited/extended staging 10, 13, 28.
This proposed nomogram also illustrates the prognostic implications of using different treatment methods (Figure 1a, Table 3). As expected, cases treated with both surgery and chemo-radiation therapy have the lowest risk score and cases not treated with any method have the highest risk score. Furthermore, the nomogram (Figure 1b) is consistent with current research in that it predicts better survival for surgery with chemo-radiation (treatment type 7 and 8) than for surgery with chemotherapy alone (type 3 and 4) [21]. However, the risk scores of different treatment methods are not recommended for direct use as a guideline for treatment selection, since clinical treatment decisions should be made based on multiple factors such as TNM stage and patient comorbidities (Supplementary Table 3)3.
There were several limitations in the development of this nomogram. The first limitation was a lack of some routinely available clinical data, such as the neutrophil to lymphocyte ratio (NLR) and platelet to lymphocyte ratio (PLR). The absence of this information prevented direct comparison of performance between our model and another published nomogram 4. Constructing a prognostic model using both the factors identified in our model and other lab tests such as NLR would thus be beneficial in creating an even more accurate prognostic prediction. The second limitation was the inability to capture interaction terms among the predictors. For example, patients with early stage disease (stage I & II) were more likely to receive surgery than patients with late stage disease (stage III and IV). The interactions between stage and treatment strategies are worth further investigation. To satisfy the requirement for convenience and interpretability of the nomogram, interaction terms were not considered in this model. However, a more complex model considering all potential interaction terms would be expected to have better prognostic performance. The third limitation was that the sequence of treatment was not considered. Since neither recurrence nor progression is recorded in the dataset, we have to consider the treatment as baseline variables instead of time-varying covariates. By including the treatment as baseline covariates, we assume that the exact treatment combination was decided and given at the time of diagnosis. This assumption is necessary in order to incorporate the treatment information into the model, when the exact time of the treatment is missing. Finally, out of 200,000 SCLC patients from the NCDB, there are only 34,380 patients without missing values. This large percent of missing data might introduce some selection bias.
Conclusion
We developed a nomogram prognostic model for SCLC patients, and validated the model using an independent patient cohort. The proposed nomogram shows better prognostic performance than other existing models. This nomogram and previously published prognostic models were implemented on an online webserver. Researchers, clinicians and patients can easily predict the survival probability for each individual patient using this webserver.
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health [5R01CA152301, P50CA70907, 5P30CA142543, 1R01GM115473, K24CA201543 and 1R01CA172211]; and the Cancer Prevention and Research Institute of Texas [RP120732]
The NCDB is a joint project of the Commission on Cancer of the American College of Surgeons and the American Cancer Society. The data used in this study is derived from a de-identified NCDB file. The American College of Surgeons and the Commission on Cancer have not verified and are not responsible for the analytic or statistical methodology employed, or the conclusions drawn from this data by the investigator.
Footnotes
Disclosure: The authors declare no conflicts of interest.
References
- 1.Howlader N, Noone AM, Krapcho M, Neyman N, Aminou R, Waldron W and Cho H. SEER cancer statistics review, 1975–2008. . Bethesda, MD: National Cancer Institute, 19. 2011. [Google Scholar]
- 2.Siegel RL, Miller KD and Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. [DOI] [PubMed] [Google Scholar]
- 3.Kalemkerian GP, Akerley W, Bogner P, Borghaei H, Chow LQ, Downey RJ, Gandhi L, Ganti AK, Govindan R, Grecula JC, Hayman J, Heist RS, Horn L, Jahan T, Koczywas M, Loo BW jr., Merritt RE, Moran CA, Niell HB, O’Malley J, Patel JD, Ready N, Rudin CM, Williams CC jr., Gregory K, Hughes M and National Comprehensive Cancer N. Small cell lung cancer. J Natl Compr Canc Netw. 2013;11:78–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xie D, Marks R, Zhang M, Jiang G, Jatoi A, Garces YI, Mansfield A, Molina J and Yang P. Nomograms Predict Overall Survival for Patients with Small-Cell Lung Cancer Incorporating Pretreatment Peripheral Blood Markers. J Thorac Oncol. 2015;10:1213–20. [DOI] [PubMed] [Google Scholar]
- 5.Govindan R, Page N, Morgensztern D, Read W, Tierney R, Vlahiotis A, Spitznagel EL and Piccirillo J. Changing epidemiology of small-cell lung cancer in the United States over the last 30 years: analysis of the surveillance, epidemiologic, and end results database. J Clin Oncol. 2006;24:4539–44. [DOI] [PubMed] [Google Scholar]
- 6.Jett JR, Schild SE, Kesler KA and Kalemkerian GP. Treatment of small cell lung cancer: Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest. 2013;143:e400S–19S. [DOI] [PubMed] [Google Scholar]
- 7.Shachar SS and Muss HB. Internet tools to enhance breast cancer care. Npj Breast Cancer. 2016;2:16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan H, Shi X, Xiao D, He J, Zhang Y, Liang W, Zhao Z, Guo Z, Zou X, Zhang J and He J. Nomogram prediction for the survival of the patients with small cell lung cancer. J Thorac Dis. 2017;9:507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao HF, Zhang BH, Liao XZ, Yan SP, Zhu SL, Zhou F and Zhou YK. Development and validation of two prognostic nomograms for predicting survival in patients with non-small cell and small cell lung cancer. Oncotarget. 2017;8:64303–64316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The diagnosis and treatment of lung cancer (update). National Collaborating Centre for Cancer; (UK: 2011. [PubMed] [Google Scholar]
- 11.Cerny T, Blair V, Anderson H, Bramwell V and Thatcher N. Pretreatment prognostic factors and scoring system in 407 small-cell lung cancer patients. Int J Cancer. 1987;39:146–9. [DOI] [PubMed] [Google Scholar]
- 12.Maestu I, Pastor M, Gomez-Codina J, Aparicio J, Oltra A, Herranz C, Montalar J, Munarriz B and Reynes G. Pretreatment prognostic factors for survival in small-cell lung cancer: a new prognostic index and validation of three known prognostic indices on 341 patients. Ann Oncol. 1997;8:547–53. [DOI] [PubMed] [Google Scholar]
- 13.Rami-Porta R, Bolejack V, Giroux DJ, Chansky K, Crowley J, Asamura H, Goldstraw P, International Association for the Study of Lung Cancer S, Prognostic Factors Committee ABM and Participating I. The IASLC lung cancer staging project: the new database to inform the eighth edition of the TNM classification of lung cancer. J Thorac Oncol. 2014;9:1618–24. [DOI] [PubMed] [Google Scholar]
- 14.Abdel-Rahman O Validation of the AJCC 8th lung cancer staging system among patients with small cell lung cancer. Clin Transl Oncol. 2017. [DOI] [PubMed] [Google Scholar]
- 15.Eberhardt WE, Mitchell A, Crowley J, Kondo H, Kim YT, Turrisi A 3rd, Goldstraw P, Rami-Porta R, International Association for Study of Lung Cancer S, Prognostic Factors Committee ABM and Participating I. The IASLC Lung Cancer Staging Project: Proposals for the Revision of the M Descriptors in the Forthcoming Eighth Edition of the TNM Classification of Lung Cancer. J Thorac Oncol. 2015;10:1515–22. [DOI] [PubMed] [Google Scholar]
- 16.Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, Rami-Porta R, Postmus PE, Rusch V, Sobin L, International Association for the Study of Lung Cancer International Staging C and Participating I. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007;2:706–14. [DOI] [PubMed] [Google Scholar]
- 17.Rami-Porta R, Bolejack V, Crowley J, Ball D, Kim J, Lyons G, Rice T, Suzuki K, Thomas CF Jr, Travis WD, Wu YL, Staging I, Prognostic Factors Committee AB and Participating I. The IASLC Lung Cancer Staging Project: Proposals for the Revisions of the T Descriptors in the Forthcoming Eighth Edition of the TNM Classification for Lung Cancer. J Thorac Oncol. 2015;10:990–1003. [DOI] [PubMed] [Google Scholar]
- 18.Postmus PE, Brambilla E, Chansky K, Crowley J, Goldstraw P, Patz EF Jr., Yokomise H, International Association for the Study of Lung Cancer International Staging C, Cancer R, Biostatistics, Observers to the C and Participating I. The IASLC Lung Cancer Staging Project: proposals for revision of the M descriptors in the forthcoming (seventh) edition of the TNM classification of lung cancer. J Thorac Oncol. 2007;2:686–93. [DOI] [PubMed] [Google Scholar]
- 19.Yang L, Wang S, Zhou Y, Lai S, Xiao G, Gazdar A and Xie Y. Evaluation of the 7th and 8th editions of the AJCC/UICC TNM staging systems for lung cancer in a large North American cohort. Oncotarget. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J and Ma S. Time-dependent ROC analysis under diverse censoring patterns. Statistics in medicine. 2011;30:1266–77. [DOI] [PubMed] [Google Scholar]
- 21.Heagerty PJ, Lumley T and Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56:337–44. [DOI] [PubMed] [Google Scholar]
- 22.Iasonos A, Schrag D, Raj GV and Panageas KS. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol. 2008;26:1364–70. [DOI] [PubMed] [Google Scholar]
- 23.Slotman BJ, van Tinteren H, Praag JO, Knegjens JL, El Sharouni SY, Hatton M, Keijser A, Faivre-Finn C and Senan S. Use of thoracic radiotherapy for extensive stage small-cell lung cancer: a phase 3 randomised controlled trial. Lancet. 2015;385:36–42. [DOI] [PubMed] [Google Scholar]
- 24.Ganti AK, Zhen W and Kessinger A. Limited-stage small-cell lung cancer: therapeutic options. Oncology (Williston Park). 2007;21:303–12; discussion 312, 315-8, 323. [PubMed] [Google Scholar]
- 25.Kang L, Chen W, Petrick NA and Gallas BD. Comparing two correlated C indices with right-censored survival outcome: a one-shot nonparametric approach. Stat Med. 2015;34:685–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R: A language and environment for statistical computing. [computer program]. R Foundation for Statistical Computing, Vienna, Austria; 2016. [Google Scholar]
- 27.Combs SE, Hancock JG, Boffa DJ, Decker RH, Detterbeck FC and Kim AW. Bolstering the case for lobectomy in stages I, II, and IIIA small-cell lung cancer using the National Cancer Data Base. J Thorac Oncol. 2015;10:316–23. [DOI] [PubMed] [Google Scholar]
- 28.Vallieres E, Shepherd FA, Crowley J, Van Houtte P, Postmus PE, Carney D, Chansky K, Shaikh Z, Goldstraw P, International Association for the Study of Lung Cancer International Staging C and Participating I. The IASLC Lung Cancer Staging Project: proposals regarding the relevance of TNM in the pathologic staging of small cell lung cancer in the forthcoming (seventh) edition of the TNM classification for lung cancer. J Thorac Oncol. 2009;4:1049–59.19652623 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


