Abstract
Spike and nucleocapsid proteins of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2-SP, SARS-CoV-2-NP) are the main immunogenic targets for antibodies. We herein demonstrate that the glycosylation of SARS-CoV-2-NP masks some of its antibody epitopes. In many cases, this can lead to false-negative serological tests. Deglycosylation of SARS-CoV-2-NP significantly increased the number of positive tests. The glycosylation pattern analysis of this protein revealed that the putative N-linked glycosylation sites, at the amino acid positions 48 and 270, co-located with two of the main immunodominant B cell epitopes.
Keywords: SARS-CoV-2, Glycosylation, Nucleocapsid protein (NP)
1. Introduction
The most used SARS-CoV-2 proteins as antigens in the serological assays are nucleocapsid protein (SARS-CoV-2-NP), full trans-membrane spike protein (SARS-CoV-2-SP), and spike protein subunits RBD due to their ability to trigger a dominant and long-lasting immune response [1]. The NP is the most expressed of these immunodominant proteins. Antibodies (Abs) directed against the SARS-CoV-2-NP are longer-lived and occur in greater abundance in COVID-19 patients than antibodies targeting other viral proteins [2,3].
Lack of anti-SARS-CoV-2 Abs has been reported from clinically confirmed cases, raising issues about sensitivity and reproducibility of serological tests. One reason for it may be the quality and composition of the antigen used in serological assays. Unfortunately, the nature of the antigens utilized for in vitro serological assays is generally not accurately documented.
Several serological assays have been recently developed to detect serum immunoglobulins (Ig) directed against SARS-CoV-2, mainly based on enzyme-linked immunosorbent assays (ELISA). Serological assays are critical to assess the proportion of the population that has been exposed to the virus, the heterogeneity of the Ab response, and to determine its duration. The correlation between the presence of Abs against the virus in the serum and the protection against secondary infection is also critical.
In addition to the high proportion of false-negative RT-qPCR tests due to difficult swab sampling [4], a surprising lack of anti-SARS-CoV-2 Abs has been reported, even from some clinically confirmed cases. These observations raised issues about sensitivity and reproducibility of serological tests, as well as about the quality of Ab response induced by the infection. Most persons infected with SARS-CoV-2 display an Ab response between day 10 and day 21 after infection [5,6]. Although the duration of the Ab response to SARS-CoV-2 is still unknown, Ab levels to other coronaviruses wane over time in the range of 12–52 weeks from the onset of symptoms [7]. It should be noted, the fact that Ab levels are falling does not mean that our immune system is not capable of inducing a protective immune response. Reinfections with seasonal coronaviruses (HCoV-OC43 and HCoV-HKU1) occur in nature, usually within three years [8]. Although the primary infection with SARS-CoV-2 was shown to protect Rhesus macaques from the subsequent challenge [9], demonstration of long-term protection in humans from the same virus will require future studies.
The SARS-CoV-2-NP is highly basic and forms a helical ribonucleoprotein complex with viral RNA [10]. It has a high degree of sequence homology between different beta coronavirus proteins [11], regulates the replication and packaging of the viral genome that is essential for the viability of the virus. The SARS-CoV-2-NP is highly immunogenic and well expressed in antigen-presenting cells [15], initiating a good T cell response. Using the conservation profile of the protein, Forcelloni et al. identified several conserved potentially immunogenic regions of SARS-CoV-2-SP and SARS-CoV-2-NP. The SARS-CoV-2 derived B cell and T cell epitopes of those two proteins were further studied, and the location of the main immunodominant epitopes was finally determined [12,13].
In this work, we have developed a dual ELISA test against SARS-CoV-2-NP and showed the critical importance of epitope unmasking by deglycosylation when using a protein produced in a mammalian system. Indeed, the N glycosylation sites located at positions 48 and 270 coincide with important B cell epitopes. Our data also illustrates the potential importance of controls for contaminants in antigen preparation from the bacterial extract.
2. Material and methods
2.1. Ethics statement
Blood samples used in the current study were obtained from healthy donors in accordance with the principles of the Helsinki Declaration of 1975 and subsequent amendments by the World Medical Assembly. Permission No. 314 was issued to Sirje Rüütel Boudinot on May 14, 2020 by the Ethics Review Committee on Human Research of the National Institute for Health Development.
2.2. Blood samples
We tested 423 sera derived from blood donors and volunteers. Our set of samples does not represent well the whole population of Estonia. This sampling leads to a set of people aged from 19 to 95 years and about 90% of them live or work in Tallinn. Blood samples were collected from May to July 2020, possible infection time (when it was known) and clinical signs were noted, since there was a tight correlation between positivity of a serological test and time passed since Covid19 till blood sampling. Specifically, most serum samples collected after 2–3 months had lost the capacity to be detected positive in our ELISA test.
2.3. ELISA
The total Ab response against SARS-CoV-2 in plasma samples was tested using in-house ELISA. Briefly, the ELISA for total Ab detection was developed based on direct immunoassay (Ab-ELISA), using recombinant SARS-CoV-2-NP produced either in mammalian cells (Icosagen AS) or E. coli (MyBioSource). Microtiter plates were coated with SARS-CoV-2-NP (2 μg/mL) in a bicarbonate buffer (15 mM Na2CO3, 35 mM NaHCO3, pH 9.6) overnight at 4 °C. Plates were blocked using 2% (w/v) fat-free milk in PBS. Plates were then washed with PBS-T and decreasing concentrations of human sera were added starting from the dilution of 1:2000 and ending with the dilution 1:16000. Secondary antibody anti-human all IgH + L polyclonal antibody-HRP conjugate was incubated for 1 h at 37 °C. Plates were subsequently washed six times with PBS-T; TMB chromogenic substrate solution and stop solution were then added and the absorbance at 450 was determined.
2.4. Deglycosylation of recombinant SARS-CoV-2-NP
Deglycosylation under denaturing conditions was carried out by adding 1 μl of 0.2% SDS and 100 mM DTT mixture to 9 μl SARS-CoV-2-NP (1 mg/mL) in PBS (pH 7.4) buffer. The protein was denatured for 10 min at 99 °C, and the solution was subsequently allowed to cool to room temperature. Following, 1 μl of 15% Triton X-100 and 1 μl of PNGase F (500 U/ml, Sigma-Aldrich) were added. Deglycosylation was carried out for 2 h at 37 °C, after which the solution was incubated at 99 °C for 5 min. For deglycosylation under native conditions, 2.5 μl of PNGase F was added to 10 μl SARS-CoV-2 NP and the mixture was incubated for 20 h at 37 °C.
3. Results and discussion
3.1. Serological analysis of targeted sets of donors using dual anti SARS-CoV-2-NP ELISA
We tested 423 sera using ELISA based on 2 different recombinants and His-tagged SARS-CoV-2-NPs: one produced in Chinese hamster ovarian (CHO) cells, and the other one produced in E. coli. Strikingly, the results of these ELISA tests were generally inconsistent. Namely, among 423 tested sera, 125 (29,6%) were positive if the protein produced in E. coli was used, while only 43 (10.2%) were positive if the protein produced in CHO was used. In fact, two sera were positive with the protein from CHO, but were negative with the protein produced in E. coli.
These differences could be caused by: (1) a possible impact of the glycosylation of the recombinant protein produced in CHO cells, (2) the absence of S-S bridge in bacterially produced SARS-CoV-2, possibly altering the accessibility of epitopes and (3) a reactivity of human sera to contaminant proteins from E. coli.
3.2. Epitope masking by glycosylation on SARS-CoV-2-NP produced in CHO cells
Next, we analysed the glycosylation pattern of SARS-CoV-2-NP using NetNGlyc 1.0 Server prediction of results (Technical University of Denmark). Our study showed that SARS-CoV-2-NP had three potential and two actual N-linked glycosylation positions, located at 48 NNTA, 197 NSTP and 270 NVTQ. Sites 48 and 270 were positively proved with N-Glyc.
Observing that the two glycosylation sites of SARS-CoV-2-NP located at positions 48 and 270 matched the main B cell epitopes previously reported for SARS-CoV-2 (Fig. 1 A), we hypothesised that epitope masking might be the main reason for the discrepancies observed in our double ELISA. We, therefore, repeated the assay using protein antigen deglycosylated by PNGase F for a number of sera selected to represent the different types of situations.
Fig. 1.
N-glycosylation analysis of recombinant SARS-CoV-2-NP. A: Predicted N-glycosylation sites in SARS-CoV-2-NP. Green – the asparagine residues predicted by NetNGlyc 1.0 to be N-glycosylated, red – T-cell epitope, blue – B-cell epitopes. B: Deglycosylation of recombinant Covid NP using treatment with PNGase F followed by SDS-PAGE analysis. (a) Deglycosylation of SARS-CoV-2-NP under denaturing conditions. (b) Deglycosylation of SARS-CoV-2-NP under native conditions. (c) SARS-CoV-2-NP expressed in mammalian cells (CHO based cell line; SARS-CoV-2-NP expressed by QMCF Technology and purified by Metal-affinity chromatography gel filtration). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Treatment of SARS-CoV-2-NP by PNGase F resulted in reducing its molecular weight by 2 kDa, indicating glycosylation of the protein (Fig. 1B). A similar decrease in molecular weight was observed for both SDS denatured and native SARS-CoV-2-NP, confirming the deglycosylation of SARS-CoV-2-NP.
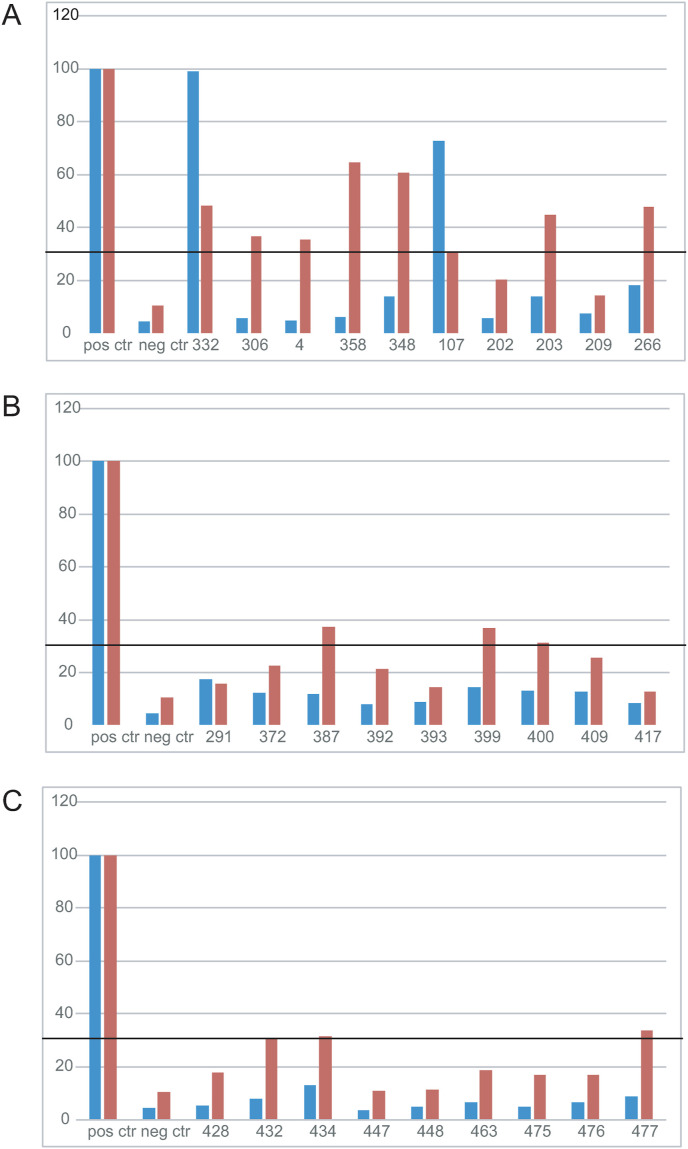
We next tested the impact of deglycosylation on the ELISA. Fig. 2 shows that epitope unmasking by deglycosylation generally led to positive ELISA test when using serum samples that were negative with the glycosylated SARS-CoV-2-NP from CHO, and positive with the SARS-CoV-2-NP produced in E. coli. Interestingly, the overall reactivity of used sera was much improved after deglycosylation. Among 28 selected sera previously showing inconsistent results, 10 sera showed positive results and 14 sera raised their positivity 2–3 times after deglycosylation of SARS-CoV-2-NP, although the value stayed below the conservative significance threshold (Fig. 2). However, we had two cases in which the trend was the opposite: sample nr 107 reduced its value after deglycosylation by 50% and sample nr 291 by about 5%, although this change did not affect the final result: sample nr 107 remained positive and sample nr 291 - negative (as it was before deglycosylation).
Fig. 2.
Reactivity of the same sera to glycosylated versus deglycosylated SARS-CoV-2-NPs as measured by ELISA. Normalized optical densities (OD) for ELISA analysis using 1 : 8000 serum dilutions from selected blood donors. Panels A, B and C: ELISA results showing SARS-CoV-2-NP specific Ab response of chosen blood donors (n = 28) before (blue bars) and after (red bars) deglycosylation of eukaryotic SARS-CoV-2-NP. Black line shows a pass of conservative significance threshold if the absorbance value was 3 times above the value of negative control. All values are presented as the percentage of the positive control sample value. All sera were tested 2 to 3 times. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
To investigate the origin of the other possible difference between the two tests we performed ELISA using three irrelevant proteins produced in E. coli. We prepared two controls of ELISA that were coated with irrelevant recombinant viral proteins, one from Porcine Circovirus Type 2, protein ORF3 and the other one from Potyvirus coat protein. Twenty three tested human sera out of 24 showed negative results to irrelevant viral proteins produced in E. coli (data not shown). However, approximately 60% of the human autoantigens are significantly homologous to the microbial proteins in their amino acid sequences. The homologous sequences between the humans and the microbes also suggest a probability that the autoantibodies in autoimmune diseases are derived from the host immune response to the commensal microbes [14]. This may partly explain the reason for the presence of anti-E. coli Abs in circulation in humans and the need to have respective controls when bacterially expressed proteins are used in serological tests.
4. Conclusions
In this report, we have shown the interest of dual ELISA in serological analysis against SARS-CoV-2 proteins and addressed the possible mechanism leading to false-negative tests. Dual ELISA using in parallel two of the SARS-CoV-2-NPs produced in bacteria and eukaryotic cells can be useful for increasing the accuracy of the serological analyses of anti-SARS-CoV-2 Abs. However, epitope unmasking by N glycosylation has to be performed by proteins produced in eukaryotic cells. Mutation of N glycosylation can also be considered, but it may change the epitope.
The results of this study also suggest that many Covid-19 infected people produce Abs whose epitopes are located at or near to the glycosylation sites of SARS-CoV-2-NP.
Funding
This work was supported by the Estonian Research Council grant COVSG34.
Author contributions statement
SRB designed experiments, AR, RR and M-LK performed wet-lab experiments. AR performed primary data analysis. All authors (AR, RR, M-LK, JR, VS, AL, SRB) contributed substantially to the conception and design of the study; revising the article critically for important intellectual content and made the final approval of the version to be submitted.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
We thank Dr. Ave Lellep, Director of Blood Centre, North Estonia Medical Centre, Dr Ülle Uustalu and Annely Rüütel for the donor blood samples and we would like to thank all our blood donors and volunteers.
References
- 1.Tilocca B., Soggiu A., Sanguinetti M., Musella V., Britti D., Bonizzi L., Urbani A., Roncada P. Comparative computational analysis of SARS-CoV-2 nucleocapsid protein epitopes in taxonomically related coronaviruses. Microb. Infect. 2020;22:188–194. doi: 10.1016/j.micinf.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang M.-S., Lu Y.-T., Ho S.-T., Wu C.-C., Wei T.-Y., Chen C.-J., Hsu Y.-T., Chu P.-C., Chen C.-H., Chu J.-M., Jan Y.-L., Hung C.-C., Fan C.-C., Yang Y.-C. Antibody detection of SARS-CoV spike and nucleocapsid protein. Biochem. Biophys. Res. Commun. 2004;314:931–936. doi: 10.1016/j.bbrc.2003.12.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., Chia W.N., Chen M.I.-C., Wang L.-F., Ooi E.E., Kalimuddin S., Tambyah P.A., Low J.G.-H., Tan Y.-J., Bertoletti A. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 4.Woloshin S., Patel N., Kesselheim A.S. False negative tests for SARS-CoV-2 infection — challenges and implications. N. Engl. J. Med. 2020;383:e38. doi: 10.1056/NEJMp2015897. [DOI] [PubMed] [Google Scholar]
- 5.Woelfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Mueller M.A., Niemeyer D., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Bruenink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Clinical presentation and virological assessment of hospitalized cases of coronavirus disease 2019 in a travel-associated transmission cluster. MedRxiv. 2020 doi: 10.1101/2020.03.05.20030502. 2020.03.05.20030502. [DOI] [Google Scholar]
- 6.Zhao J., Yuan Q., Wang H., Liu W., Liao X., Su Y., Wang X., Yuan J., Li T., Li J., Qian S., Hong C., Wang F., Liu Y., Wang Z., He Q., Li Z., He B., Zhang T., Ge S., Liu L., Zhang J., Xia N., Zhang Z. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. MedRxiv. 2020 doi: 10.1101/2020.03.02.20030189. 2020.03.02.20030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kellam P., Barclay W. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J. Gen. Virol. 2020;101:791–797. doi: 10.1099/jgv.0.001439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edridge A.W., Kaczorowska J.M., Hoste A.C., Bakker M., Klein M., Jebbink M.F., Matser A., Kinsella C., Rueda P., Prins M., Sastre P., Deijs M., van der Hoek L. Coronavirus protective immunity is short-lasting. MedRxiv. 2020 doi: 10.1101/2020.05.11.20086439. 2020.05.11.20086439. [DOI] [PubMed] [Google Scholar]
- 9.Bao L., Deng W., Gao H., Xiao C., Liu J., Xue J., Lv Q., Liu J., Yu P., Xu Y., Qi F., Qu Y., Li F., Xiang Z., Yu H., Gong S., Liu M., Wang G., Wang S., Song Z., Zhao W., Han Y., Zhao L., Liu X., Wei Q., Qin C. Reinfection could not occur in SARS-CoV-2 infected rhesus macaques. BioRxiv. 2020 doi: 10.1101/2020.03.13.990226. 2020.03.13.990226. [DOI] [Google Scholar]
- 10.Wang Q., Li C., Zhang Q., Wang T., Li J., Guan W., Yu J., Liang M., Li D. Interactions of SARS Coronavirus Nucleocapsid Protein with the host cell proteasome subunit p42. Virol. J. 2010;7:99. doi: 10.1186/1743-422X-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12:254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forcelloni S., Benedetti A., Dilucca M., Giansanti A. Identification of conserved epitopes in SARS-CoV-2 spike and nucleocapsid protein. BioRxiv. 2020 doi: 10.1101/2020.05.14.095133. 2020.05.14.095133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang P. Autoantibodies and anti-microbial antibodies: homology of the protein sequences of human autoantigens and the microbes with implication of microbial etiology in autoimmune diseases. BioRxiv. 2018 doi: 10.1101/403519. [DOI] [Google Scholar]
- 15.Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.-Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]