Abstract
The global pandemic of Coronavirus Disease 2019 (COVID-19) has brought the world to a grinding halt. A major cause of concern is the respiratory distress associated mortality attributed to the cytokine storm. Despite myriad rapidly approved clinical trials with repurposed drugs, and time needed to develop a vaccine, accelerated search for repurposed therapeutics is still ongoing. In this review, we present Nitazoxanide a US-FDA approved antiprotozoal drug, as one such promising candidate. Nitazoxanide which is reported to exert broad-spectrum antiviral activity against various viral infections, revealed good in vitro activity against SARS-CoV-2 in cell culture assays, suggesting potential for repurposing in COVID-19. Furthermore, nitazoxanide displays the potential to boost host innate immune responses and thereby tackle the life-threatening cytokine storm. Possibilities of improving lung, as well as multiple organ damage and providing value addition to COVID-19 patients with comorbidities, are other important facets of the drug. The review juxtaposes the role of nitazoxanide in fighting COVID-19 pathogenesis at multiple levels highlighting the great promise the drug exhibits. The in silico data and in vitro efficacy in cell lines confirms the promise of nitazoxanide. Several approved clinical trials world over further substantiate leveraging nitazoxanide for COVID-19 therapy.
Keywords: Antiviral, Clinical trials, COVID-19, Immunomodulation, Nitazoxanide, SARS-CoV-2
Highlights
-
•
Nitazoxanide holds great promise as it demonstrated a low IC50 against SARS-CoV-2, in vitro.
-
•
Nitazoxanide exhibits potential to block SARS-CoV-2 entry and prevent its multiplication.
-
•
Nitazoxanide proposes curbing the cytokine storm and also potentiating host innate immunity.
-
•
Nitazoxanide reveals possibility of lung protection and prevention of multiple organ failures.
-
•
Nitazoxanide may also provide therapeutic benefits in COVID-19 patients with comorbidities.
1. Introduction
The COVID-19 pandemic has created huge panic and alarm in mankind across the globe. The absence of an assured cure, increase in number of afflicted and the fear of fatality (over half a million deaths worldwide) has created a chaotic ambiance (Sahebnasagh et al., 2020; WHO, 2020). The concept of repurposing drugs has become the norm for COVID-19 therapy, and clinical trials with repurposed drugs are being approved across the globe at unheard of speeds to find an immediate solution (Guy et al., 2020; Lam et al., 2020). Direct-acting antivirals, antimalarials, antiparasitics, corticosteroids and biologics singly or in combination are the major candidates being evaluated. The proposed convalescent plasma therapy would find application only in severe emergencies, but is certainly not an alternative for drugs, while vaccines need time to develop (Jin et al., 2020; Lam et al., 2020; Li et al., 2020). The antiviral drugs remdesivir (GS 5734) and favipiravir (Avigan®) have received restricted emergency use approval against COVID-19 (Ethiraj, 2020), however search for promising therapeutics is still ongoing.
The singular cause for fear is the mortality rate of up to 5%, which is ascribed majorly to the cytokine storm, a hyperimmune host reaction to the virus, which leads to acute respiratory distress syndrome (ARDS) with other major organ failures (Cao, 2020; Khan et al., 2020). The cytokine storm attack, known to occur generally in the second week is so sudden and intense, that patients could unexpectedly turn from mild/moderate to severe, needing oxygen support and hospitalisation. If not tackled immediately severe lung damage that follows, often results in death. The approach of combining antiviral efficacy with interventions to curb the cytokine storm, with a single drug or drug combinations appears to be rational, promising and encouraging (Jamilloux et al., 2020; Nile et al., 2020). In this review we focus on one such promising drug candidate Nitazoxanide (Alinia®) for repurposing in COVID-19.
1.1. Repurposing “Nitazoxanide”
Nitazoxanide, a small-molecule (nitrothiazolyl-salicylamide) antiprotozoal drug marketed as tablets (500 mg) and suspension (100 mg/5 ml) (Rossignol et al., 2006), is currently approved for treating diarrhoea caused by the protozoa Cryptosporidium or Giardia in immunocompetent adults and children (Anderson and Curran, 2007; Fox and Saravolatz, 2005; Halsey, 2009). It has demonstrated broad spectrum antiviral efficacy in vitro, against a range of viruses including the respiratory syncytial virus, parainfluenza virus, coronavirus (CoV), rotavirus, norovirus, hepatitis B virus (HBV), hepatitis C virus (HCV), dengue virus, yellow fever virus, Japanese encephalitis virus, and human immunodeficiency virus (HIV). Furthermore, clinical trials suggested the potential role of nitazoxanide in gastroenteritis, hepatitis and influenza (Rossignol, 2014). A significant finding is the ability of nitazoxanide to promote balance between pro-inflammatory and anti-inflammatory responses in humans, which could play a crucial role in COVID-19 by curbing the hyperinflammatory cytokine storm (Jasenosky et al., 2019; Martins-Filho et al., 2020; Rossignol, 2016; Shakya et al., 2018a). Repurposing nitazoxanide against COVID-19 has been suggested in many reports (Alonso and Farina, 2020; Arshad et al., 2020; Bobrowski et al., 2020; Chibber et al., 2020; Mahmoud et al., 2020; Martins-Filho et al., 2020; Padmanabhan, 2020; Padmanabhan and Padmanabhan, 2020; Pepperrell et al., 2020; Rajoli et al., 2020). The approval of a number of clinical trials with nitazoxanide further substantiates this hypothesis (https://clinicaltrials.gov/) (Nitazoxanide clinical trials, 2020). This review presents mechanistic insights and unravels the promise of nitazoxanide as a repurposed drug candidate for comprehensively addressing COVID-19 pathogenesis.
2. Nitazoxanide
2.1. Structural features
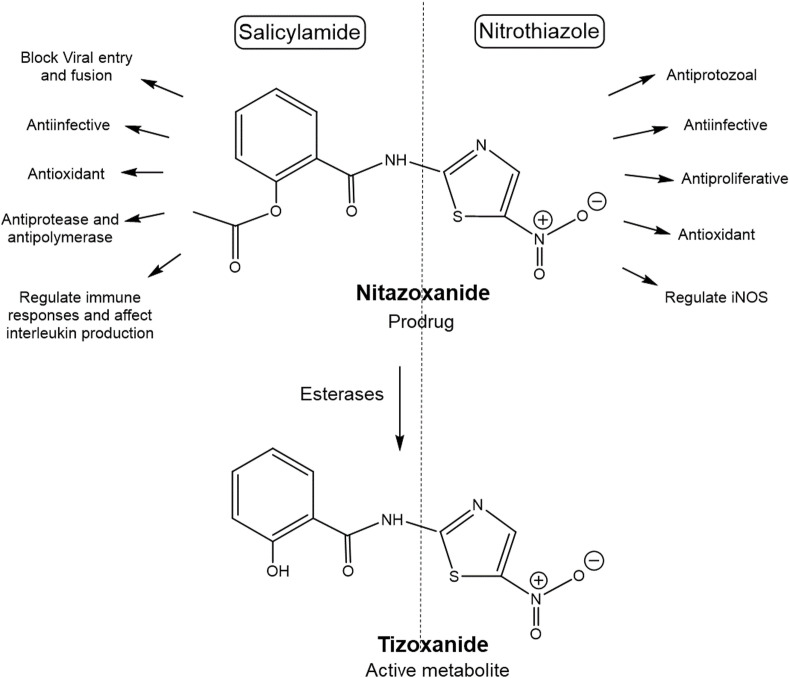
Nitazoxanide [2-acetyloxy-N-(5-nitro-2thiazolyl)benzamide] a prodrug, consists of a nitrothiazole moiety and salicylamide moiety. In the body nitazoxanide is rapidly deacetylated to tizoxanide [2-hydroxy-N-(5-nitro-2thiazolyl)benzamide] (TIZ), the active form (Hemphill et al., 2006; Rossignol, 2009). Multiple activities of nitazoxanide are attributed to the structural moieties (Fig. 1 ), for instance, acetylbenzamide or salicylamide moiety acts as an analgesic, antioxidant, anti-infective (against bacteria, fungi, parasite and viruses), affects interleukin production, regulates immune responses, antipolymerase activity against hepatitis virus, blocks influenza viral entry and host cell membrane fusion (Krátký and Vinšová, 2011). The nitrothiazole moiety exhibits antiprotozoal, antiproliferative, antiinfective effect (Colín-Lozano et al., 2017), control oxidative stress (El-Kowrany et al., 2019) and disrupts pyruvate: ferredoxin oxidoreductase dependent redox reactions essential for anaerobic protozoa and bacteria for their energy metabolism and survival (Hemphill et al., 2006; Kumar et al., 2014; Rossignol, 2009). Thus it can be postulated that nitazoxanide can inhibit various stages of the virus multiplication cycle and also potentiate host innate immunity (Martins-Filho et al., 2020; Padmanabhan, 2020). Furthermore, the potential of nitazoxanide to enhance natural host antiviral mechanisms, suggests lower possibility of developing antiviral resistance in comparison with drugs directly acting only on viral proteins (Krátký and Vinšová, 2011; Tilmanis et al., 2020).
Fig. 1.
Nitazoxanide structural features. Nitazoxanide a nitrothiazolyl benzamide is a prodrug metabolized to deacetylated active tizoxanide (TIZ). Both have the two major structural components a) salicylamide moiety and b) nitrothiazole moiety which are responsible for the multiple therapeutic actions. iNOS, instrinsic nitric oxide synthase.
2.2. Mechanism of action against SARS-CoV-2 pathogenesis
We now explain the pathways of SARS-CoV-2 entry and pathogenesis, systematically juxtaposing at each stage the role of nitazoxanide elucidating thereby the ability of the drug to exhibit direct antiviral action and host beneficial anti-inflammatory response to curb the cytokine storm. Importantly nitazoxanide is also reported to be useful in patients with co-morbidities (Elaidy et al., 2018; Fan et al., 2019; Ghusson and Vasquez, 2018; Nitazoxanide in fibrosis, 2020).
2.2.1. Entry and fusion
SARS-CoV-2 can enter the host cell using two pathways, the endosomal pathway or the fusion based cell surface non-endosomal pathway. As early availability of cell surface proteases is important to facilitate fusion-based entry (Kupferschmidt and Cohen, 2020; Tang et al., 2020), the endosomal pathway appears to be more favoured.
2.2.1.1. Non-endosomal pathway:
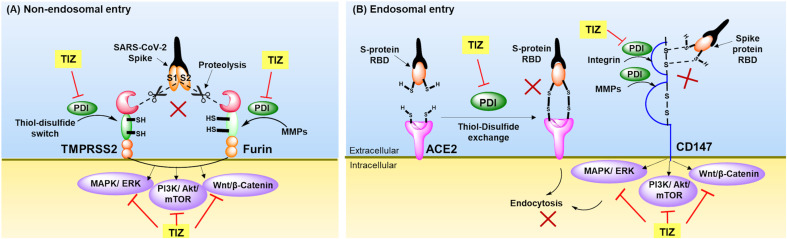
Fusion between virus and host cell membrane through the non-endosomal pathway results in the viral genetic material being released directly into the host cytosol. This route of entry involves cleavage of viral spike glycoproteins at (S1/S2) interface by host cell surface anchored proteases, transmembrane-protease-serine-2 (TMPRSS2) and furin, exposing fusion S2 peptide available for fusion with host cell membrane (Bestle et al., 2020; Jin et al., 2020; Omolo et al., 2020; Ou et al., 2020; Xiu et al., 2020). Such proteolytic cleavage is dependent on the thiol-disulfide exchange mechanism catalyzed by protein disulfide isomerase (PDI) (Diwaker et al., 2013; Hati and Bhattacharyya, 2020). PDI also regulates disulfide switch activity in the ectodomains of TMPRSS2 due to presence of cysteine-rich residues (Antalis et al., 2011; Curry and Rosewell, 2008; Rosewell et al., 2011; Turano et al., 2002). Nitazoxanide block this oxidoreductase mechanism (Fig. 2 A) by interacting with cysteine residues of surface-bound PDI through S-nitrosylation (Bekendam et al., 2018; Flaumenhaft et al., 2015; Müller et al., 2008a; Piacentini et al., 2018). Further, inhibition of furin (Bonacci et al., 2011; Denault et al., 2002; McCarthy et al., 2008) by nitazoxanide can facilitate blocking of other protease signalling pathways, including Wnt/β-catenin (Miner et al., 2019b; Qu et al., 2018), MAPK/ERK (Shou et al., 2019), and PI3K/Akt/mTOR (Senkowski et al., 2015; Shou et al., 2020).
Fig. 2.
Schematic representation of possible role of tizoxanide (TIZ)-inhibition of SARS-CoV-2 fusion and entry.
(A) Non-endosomal entry involves the proteolytic cleavage of SARS-CoV-2 spike proteins (S1/S2) by host cell proteases TMPRSS2 and furin. TIZ inhibits PDI and affects thiol-disulfide oxidoreductase switch in the ectodomains of TMPRSS2, and inhibition of PDI also inhibits MMPs activation required for furin action, (B) Endosomal entry involves the recognition and binding of RBD of SARS-CoV-2 spike proteins to the host cell receptors ACE2 and CD147 enabling receptor mediated endocytic entry. TIZ inhibits thiol-disulfide exchange mechanism essential for receptor interaction with RBD by inhibiting PDI. This PDI inhibition also hampers MMPs and intergrins activation required for CD-147 activity. TIZ also inhibits intracellular signalling MAPK/ERK, PI3K/Akt/mTOR and Wnt/β-catenin affecting SARS-CoV-2 fusion and entry. ACE2, Angiotensin converting enzyme 2; CD147, Cluster of differentiation 147; MAPK/ERK, Mitogen-activated protein kinase/extracellular signal-regulated kinase; MMPs, Matrix metalloproteinases; PDI, Protein disulfide isomerase; PI3K/Akt/mTOR, phosphatidylinositol-3-kinase/protein kinase B/mammalian target of rapamycin; RBD, Receptor binding domain; TIZ, Tizoxanide; TMPRSS2, Transmembrane protease serine 2.
2.2.1.2. Endosomal pathway:
The endosomal route of entry occurs through the angiotensin converting enzyme 2 (ACE2) receptor (Sharifkashani et al., 2020). The thiol-disulfide redox system governed by PDI assists disulfide bond (-S-S-) exchange between receptor binding domain (S-RBD of S1 fragment) of SARS-CoV-2 spike protein and ACE2 facilitating entry via the endosomal pathway. Reduction of disulfide bonds to thiol (–SH–SH-) impaired such binding (Hati and Bhattacharyya, 2020). Hence inhibition of the disulfide exchange can inhibit the interaction of S-protein RBD to ACE2, thereby hampering the receptor-mediated endocytosis route of SARS-CoV-2 entry (Fig. 2B) (Di Santo and Ehrisman, 2013; Diwaker et al., 2013; Müller et al., 2008a; Piacentini et al., 2018).
A newer entry pathway identified is the Cluster of differentiation 147 (CD147) receptor-mediated entry which proceeds through the activation of its catalytic domain by integrins and matrix metalloproteinases (MMPs), essential for S-protein binding (Gabison et al., 2005; Han et al., 2008; Liu and Zhu, 2020; Ulrich and Pillat, 2020). Nitazoxanide through its effect on PDI could inhibit activation of MMPs by blocking the cysteine switch mechanism (Bonacci et al., 2011; McCarthy et al., 2008; Van Wart and Birkedal-Hansen, 1990) to prevent such entry. Nitazoxanide can also inhibit other CD-147 mediated signalling pathways, like Wnt/β-catenin signalling (Miner et al., 2019a; Müller et al., 2008a; Qu et al., 2018), PI3K/Akt/mTOR signalling (Senkowski et al., 2015; Shou et al., 2020) and MAPK/ERK signalling (Shou et al., 2019).
2.2.2. Endosome vesicle formation and maturation
Receptor mediated entry follows the clathrin mediated endocytic pathway which typically results in early endosome formation and maturation to the late endosome before the contents are released in the cytosol. Such cytosolic release is a crucial step in furthering viral pathogenesis (Bayati et al., 2020; Glebov, 2020). Nitazoxanide can delay cytosolic release through its effect on multiple targets (Rossignol, 2014). Receptor mediated endocytosis dependent on oligomeric clathrin and regulated by two host cell adaptor protein complexes serine−threonine protein kinases, namely AP-2-associated protein kinase 1 (AAK1) and cyclin G-associated kinase (GAK) (Bekerman et al., 2017; Chibber et al., 2020; Stebbing et al., 2020). Nitazoxanide can inhibit Wnt/β-catenin signalling (El-Kersh et al., 2019; Qu et al., 2018; Shakya et al., 2018b), which in turn downregulates AAK1 thereby hampering clathrin mediated endocytosis (Agajanian et al., 2019). Another proposed mechanism is the ability of nitazoxanide to interact with cysteine residues through S-nitrosylation (Andrade and Reed, 2015; Bekendam et al., 2018; Goldman, 2010), which can hinder the irreversible inhibition of the kinases AAK1 and GAK by interaction with their cysteine residues Cys193 and Cys190 respectively (Sorrell et al., 2016). These two kinases along with phosphatidylinositol 3-phosphate5-kinase (PIKFYVE) initiates early endosome formation and causes synthesis of phosphatidylinositol-3,5-bisphosphate [PI(3,5)P2] (Phosphoinositides) (Kang et al., 2020; Ou et al., 2020), which activates two pore segment channel 2 (TPC2), a calcium channel expressed in lysosomal membranes (Filippini et al., 2020; Grimm and Tang, 2020). TPC2 maintains Ca2+ homeostasis and the Ca2+ release from this channel is controlled by nicotinic acid adenine dinucleotide phosphate (NAADP), phosphoinositide and mTOR (Jin et al., 2020; Petersen et al., 2020). The endolysosomal lumen serves as an important type of intracellular Ca2+ store, containing high Ca2+ concentration of ~600 μM (Alharbi and Parrington, 2019). In the presence of high endosomal Ca2+, the fusion proteins of coronavirus induces better membrane ordering, essential for fusion (Brailoiu and Brailoiu, 2016; Grimm and Tang, 2020). Nitazoxanide causes depletion of such ATP sensitive intracellular Ca2+ stores, thus delaying endosome maturation (Ashiru et al., 2014). Additionally, TPC2 is also regulated by Mg2+, the mitogen activated protein kinases (MAPKs), c-jun N-terminal kinase (JNK) and P38 (Alharbi and Parrington, 2019; Jha et al., 2014), which are inhibited by nitazoxanide (Senkowski et al., 2015; Shou et al., 2019; Tchouaffi-Nana et al., 2010), disturbing endosome maturation. The PIKFYVE and TPC2 functions are directly linked to Wnt/β-catenin (Miner et al., 2019a; Qu et al., 2018) and PI3K/Akt/mTOR signalling (Senkowski et al., 2015; Shou et al., 2020) which are via Vacuolar-type H+ ATPase (V-ATPase) activates Cathepsin L, which promotes viral-host cell membrane fusion, causing viral genome release into the host cytosol (Collins and Forgac, 2020; Kupferschmidt and Cohen, 2020; Liu et al., 2020; Madadlou, 2020; Yang and Shen, 2020). Nitazoxanide can interact with cysteine residues (Andrade and Reed, 2015; Aqeel et al., 2015; Goldman, 2010) of Cathepsin L, preventing its activation, which leads to inhibition of viral and host cell membrane fusion and consequently the viral genome release (Madadlou, 2020; Nga et al., 2014; Otto and Schirmeister, 1997; Verma et al., 2016). Furthermore, TIZ which possesses weak acidic pKa~5.8 and lipophilic logP~3.15 (Jurgeit et al., 2012; Miner et al., 2019a) can also induce endosome neutralization analogous to niclosamide (Homolak and Kodvanj, 2020; Jurgeit et al., 2012; Xu et al., 2020).
2.2.3. Translation and proteolysis
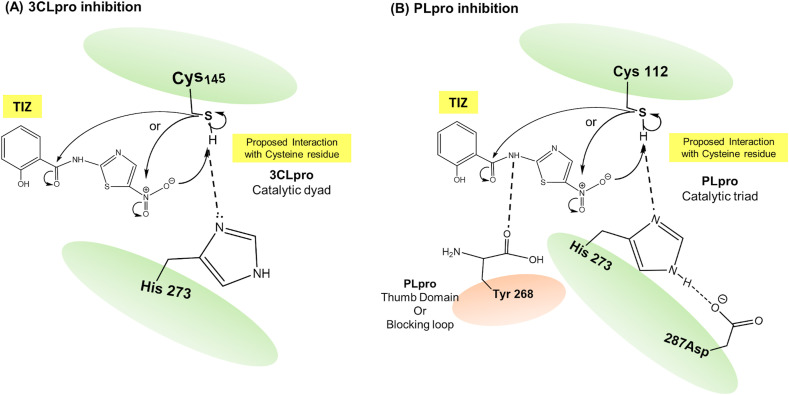
Once in the cytosol, the viral genome undergoes translation of open reading frames ORF1a/b through host ribosomes into the large replicase polyprotein 1a (pp1a) and pp1ab, while other ORFs encode structural proteins. These polypeptides (PP1a/PP1ab) are proteolytically cleaved by 3-chymotrypsin like protease (3CLpro or Mpro) and papain-like protease (PLpro) to form several nonstructural proteins (nsp1-16) (Astuti, 2020; Omolo et al., 2020). The PLpro (nsp3) causes proteolytic processing of nsp1, nsp2, and nsp3 (Klemm et al., 2020), while 3CLpro (nsp5) processes nsp12, nsp13 and other subunits of the replicase–transcriptase complex (RTC) (Fong, 2020). Both proteases are key targets for virus multiplication and governing the host directed immune cell response; therefore, they are attractive targets for the antiviral interventions (Shereen et al., 2020). 3CLpro and PLpro are cysteine rich proteases, involved in peptide bond hydrolysis (Gil et al., 2020; Pillaiyar et al., 2016). The schematic representation of binding and interaction of the active metabolite TIZ with these proteases is depicted in Fig. 3 .
Fig. 3.
Schematic representation of nitazoxanide antiprotease action against 3CLpro and PLpro of SARS-CoV-2. Tizoxanide (TIZ) (active metabolite of nitazoxanide) interacts with cysteine residue and thus inhibits the enzyme activity, (A) 3CLpro inhibition and (B) PLpro inhibition. Nucleophilic cysteine thiol interacts with electrophilic moieties of tizoxanide (TIZ). 3CLpro, 3-chymotrypsin like protease; PLpro, Papain like protease.
Inhibition of 3CLpro: The active site of 3CLpro of SARS-CoV-2 contains Cys145 and His41 residues forming a catalytic dyad (He et al., 2020; Tahir ul Qamar et al., 2020), where cysteine thiol (-SH) functions as a common nucleophile (electron donating group) responsible for proteolytic cleavage (Yang et al., 2003). Structure activity relationship studies of pyrazolones and pyrimidines revealed that the presence of aromatic nitro (-NO2) enhances cysteine protease inhibition activity, where the oxygen of the NO2 forms a H-bond with side chains of Gly143 and Cys145, while the phenyl ring forms hydrophobic interactions (Pillaiyar et al., 2016). It is therefore proposed that the NO2 group or the carbonyl carbon of amide bond of TIZ can interact with the catalytic cysteine nucleophile, while phenyl ring could exhibit hydrophobic interactions with the 3CLpro active site (Fig. 3A) (Piacentini et al., 2018). Recent molecular screening experiments based on linear free energy relationships reported activity of nitazoxanide against 3CLpro with a predicted IC50 value of 4.36 μM which was comparable with known 3CLpro inhibitors ledipasvir, saquinavir and superior to lopinavir and ritonavir (Fong, 2020).
Inhibition of PLpro: The PLpro proteolytic site possess a catalytic triad of Cys112–His273–Asp287 (Báez-Santos et al., 2015; Klemm et al., 2020) where Cys112 acts as thiol nucleophile forming a H-bond with His273 which is paired with Asp287 causing deprotonation of Cys112 (Foster et al., 2003; Stoermer, 2020). Interaction of electrophilic NO2 group of nitazoxanide with nucleophilic Cys112 residue at active site through S-nitrosylation could reversibly inhibit PLpro function (Foster et al., 2003; Hess et al., 2005; Xian et al., 2000). The SARS-CoV-2 PLpro has the thumb domain or a blocking loop, containing a critical tyrosine residue Tyr268 (Báez-Santos et al., 2015; Kandeel et al., 2020; Klemm et al., 2020) which forms H-bond with the amide nitrogen of PLpro inhibitors, anchoring the inhibitor in the binding site (Báez-Santos et al., 2015; Xian et al., 2000; Yu et al., 2020). Nitazoxanide exhibits potential as a PLpro inhibitor as the amide nitrogen can form H-bond with Tyr268 (Fig. 3B) (Piacentini et al., 2018). Antiprotease activity of nitazoxanide against SARS-CoV-2 is further substantiated by recent in silico molecular modelling study which reported a binding energy score of −110 kJ/mol indicating strong binding and furthermore nitazoxanide shared >70% binding similarity with remdesivir (Abdul Kadhim et al., 2020).
2.2.4. Viral genome synthesis
Synthesis of new genomic viral RNA relies on the multi-subunit replication transcription complex (RTC) which contains multiple enzymes viz. viral RNA-dependent RNA polymerase (RdRp) (nsp12), helicase (nsp13), exonucleases (nsp14) and endonuclease (nsp15) (Omolo et al., 2020; Ulferts et al., 2010). Such new viral genome synthesis occurs in the protective environment of double membrane vesicles (DMVs) or autophagosomes (Blanchard and Roingeard, 2015; Cottam et al., 2011; Omolo et al., 2020; Wong and Sanyal, 2020). Nitazoxanide not only exhibits the potential to inhibit all these major enzymes, but can also induce autophagy to disrupt protective environment of autophagosomes (Lam et al., 2012; Senkowski et al., 2015; Shou et al., 2020). Among the enzymes RdRp plays major role in replication of genomic viral RNA, and RdRp of SARS-CoV-2 shares a 96% sequence identity with SARS-CoV (Aftab et al., 2020; Gao et al., 2020). Recent molecular docking studies identified nitazoxanide amongst 10 most effective antipolymerase compounds with a docking score −9.76 indicating good binding affinity. Although the score was lower when compared with remdesivir (−14.06), galidesivir (−12.53), ribavirin (−11.88) and sofosbuvir (−11.13) it was comparable to nafamostat (−9.76), interferon (−9.62) and favipiravir (−9.36) (Aftab et al., 2020) proposing great promise. Helicase (nsp13) promotes the separation of double stranded RNA to single stranded RNA mediated through nucleotide triphosphate (NTP) hydrolysis-dependent manner (NTPase) (Shu et al., 2020). Helicase activity can be inhibited by hampering ATP binding or NTPase hydrolysis action, inhibiting nucleic acids interaction and helicase translocation, etc (Briguglio et al., 2011; Habtemariam et al., 2020). Reports suggests that, nitazoxanide inhibits oxidative phosphorylation leading to decreased ATP levels and causes viral protein misfolding, thereby inhibiting helicase ATP dependent action (Hemphill et al., 2006; Piacentini et al., 2018). The nsp14, exoribonuclease (ExoN) is a proofreading 3′–5′ exoribonuclease which help overcome errors caused in RNA replication, and helps modulate dsRNA levels and innate immune sensing (da Silva et al., 2020). ExoN activity depends on divalent metal ions Mg2+ and low concentration of Zn2+, while Ca2+ inactivates ExoN (Chen et al., 2007; Minskaia et al., 2006). Loss of ExoN proofreading has been found to enhance the antiviral effect of remdesivir (Agostini et al., 2018; Shannon et al., 2020). As nitazoxanide affects Mg2+ (Tchouaffi-Nana et al., 2010) and enhances intracellular Ca2+ release (Ashiru et al., 2014), its action on ExoN is proposed. A recent molecular docking study revealed the partial binding affinity of nitazoxanide to NendoU with glide score of −5.5, which is comparable with known antiviral drug remdesivir score −5.6 (Joshi and Poduri, 2020) proposing action on the endoribonuclease (nsp15). Nsp15 encodes a specific protein called NendoU, which acts by cleaving single and double-stranded RNA catalyzed by Mn2+ dependent manner (Senanayake, 2020), and helps the virus evade the host innate immune reaction (Kim et al., 2020). However, mutations in the nsps are an important cause of drug resistance, a major therapeutic challenge. Notably, nitazoxanide which is reportedly found to be less prone to such resistance, could thus serve as a safe, and effective antiviral intervention (Tilmanis et al., 2020). In vitro drug screening assay against coronavirus identified that nitazoxanide can inhibit viral N- protein expression, enhanced green fluorescent protein (EGFP) reporter gene expression and viral production (Cao et al., 2015). Artificial intelligence study exhibited better interaction score of nitazoxanide with the coatomer protein complex 2 subunit (COPB2) gene, involved in replication of SARS-CoV-2, confirming potential of nitazoxanide for repurposing against COVID-19 (Avchaciov et al., 2020).
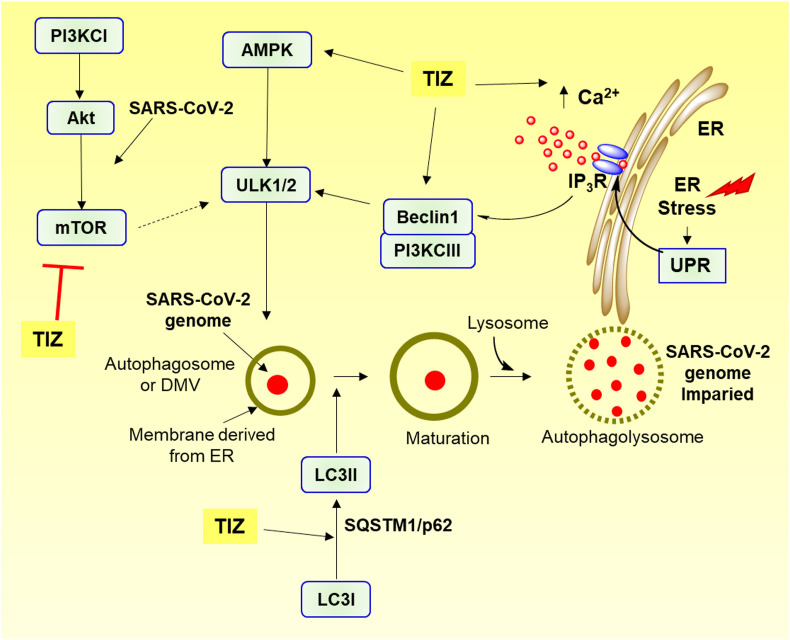
Protective environment of DMVs or autophagosomes concealing viral RNA is formed by the attachment of RTC with membrane derived from ER (Blanchard and Roingeard, 2015; Knoops et al., 2008), enabling it to escape the dsRNA-triggered host antiviral response (Astuti, 2020; Kumar et al., 2020; Margaritopoulos et al., 2017). SARS-CoV-2 escapes autophagy by inhibiting the activation of AMP-protein activated kinase (AMPK), and by promoting the mammalian activation target of rapamycin complex 1 (mTORC1) (Shojaei et al., 2020; Wong and Sanyal, 2020). Nitazoxanide promotes autophagy leading to degradation of DMVs contents by activating AMPK and inhibiting mTORC1, thereby adversely affecting virus genome synthesis (Fig. 4 ) (Cottam et al., 2011; Senkowski et al., 2015; Shou et al, 2019, 2020). The ability of nitazoxanide also increases intracellular Ca2+ levels, induces ER stress and thereby induces protective unfolded protein response (UPR) activation, leading to autophagy (Ashiru et al., 2014; Di Santo and Ehrisman, 2013, 2014).
Fig. 4.
Schematic representation of tizoxanide induced autophagy hindering SARS-CoV-2 genome synthesis. Tizoxanide (TIZ) activates Beclin1, PI3KCIII and AMPK which leads to activation of ULK1 inducing autophagosome formation. TIZ also increases cytosolic Ca2+ concentration through IP3R by depleting ER Ca2+ store inducing ER stress and activating protective UPR signalling. TIZ causes conversion of LC3I to LC3II, degradation of p62/SQSTM1 and inhibits PI3KCI/Akt/mTOR signalling, all together promoting autophagy. AMPK, AMP-protein activated kinase; ER, Endoplasmic reticulum; IP3R, Inositol trisphosphate receptor; LC3, Light chain 3; p62/SQSTM1, cargo receptor p62/sequestome1; PI3KCI, Phosphatidylinositol-3-kinase class I; PI3KCIII, Phosphatidylinositol-3-kinase class III; PI3KCI/Akt/mTOR, phosphatidylinositol-3-kinase/protein kinase B/mammalian target of rapamycin; ULK1/2, Unc-51-like kinase-1/2; UPR, Unfolded protein response.
Out of many kinases, Beclin-1 acts as a master regulator of autophagy, which promotes the phagophore sequestration and enables the fusion of autophagosomes with lysosomes (Gassen et al., 2019; Lundin et al., 2014). While the conversion of Light chain LC3-I to LC3-II induces autophagosome maturation, the cargo receptor p62/sequestome1 (p62/SQSTM1) induces autophagic flux (Rubinsztein et al., 2012; Vakifahmetoglu-Norberg et al., 2015). TIZ enhanced Beclin1 expression, promoted the transformation of LC3-I to LC3-II, and the concentration-time based degradation of p62/SQSTM1 in RAW264.7, Vero, 293T, and HepG2 cells, inducing autophagosome maturation (Lam et al., 2012; Shou et al., 2020). Nitazoxanide therefore exhibits the ability to inhibit various targets of viral genome synthesis.
2.2.5. Packaging and virion release
The newly synthesized mRNAs are rapidly translated to the ER for producing and processing structurally associated proteins and budded in association with golgi bodies in the form of compartments called ER-golgi compartment. This compartment then encloses viral genome and promotes virion assembly. Finally, virions assembly are enclosed within smooth vesicles causing exocytosis of virion particles ready to infect neighbouring cells and other tissues (Fung and Liu, 2014; Zumla et al., 2016). Ca2+ induces major post-translational changes in the recently synthesized viral proteins within ER lumen (Stutzmann and Mattson, 2011). Nitazoxanide induces reduction of ER Ca2+ affecting post translational-modifications in proteins, and thereby hampering their release from ER (Ashiru et al., 2014; Perelygina et al., 2017; Rossignol et al., 2009).
2.2.6. Immunopathogenesis
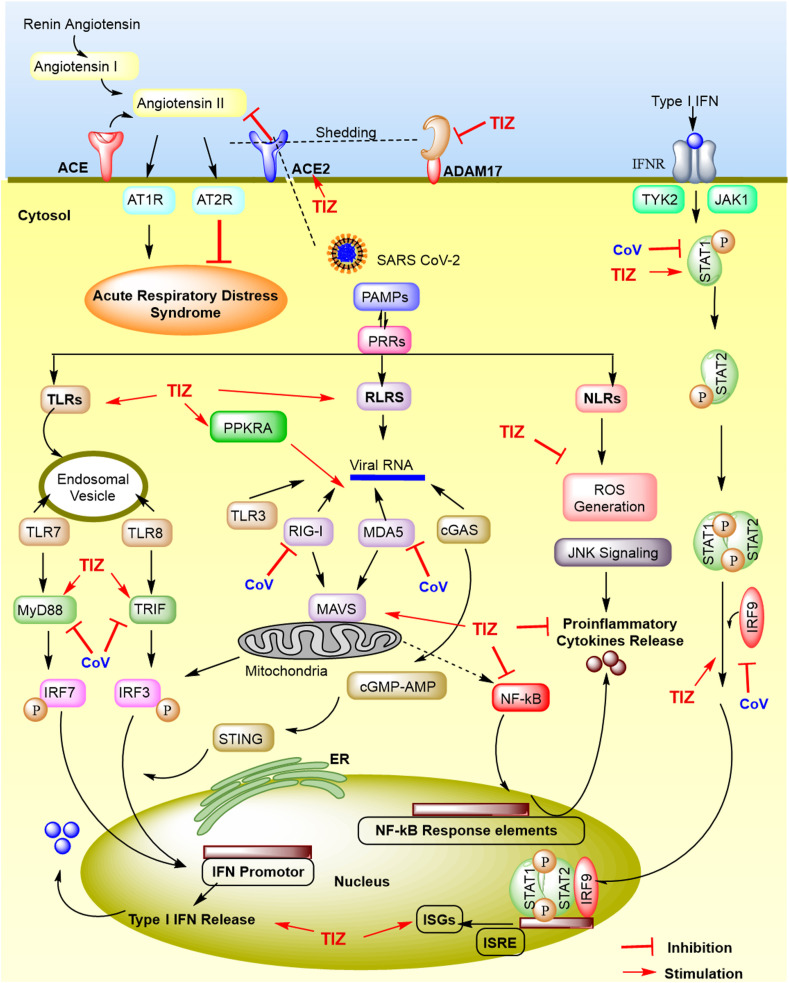
COVID-19 causes a complicated clash between host antiviral innate immunity and continuous upsurge of inflammatory markers, the latter being responsible for a fatal condition called “Cytokine storm” (Coperchini et al., 2020; Samudrala et al., 2020). High systemic levels of interleukins, interferons, interferon inducible protein, tumour necrosis factor, growth factors, colony-stimulating factors, macrophage inflammatory protein, monocyte chemoattractant protein, chemokines are a hallmark of the SARS-CoV-2 cytokine storm (Costela-Ruiz et al., 2020; Henderson et al., 2020; Pearce et al., 2020; Samudrala et al., 2020). Nitazoxanide causes activation of innate immune responses and induction of host-directed antiviral mechanisms, thereby exhibiting great promise against the SARS-CoV-2 cytokine storm (Fig. 5 ) (Mahmoud et al., 2020; Padmanabhan, 2020; Padmanabhan and Padmanabhan, 2020).
Fig. 5.
Evasion of host innate immune responses by SARS-CoV-2 and stimulation of host antiviral immune responses by tizoxanide (TIZ). SARS-CoV-2 entry via ACE2 is hampered by TIZ which regulates renin-angiotensin system by blocking angiotensin II binding to its receptors (AT1R and AT2R), to provide protective responses. ADAM-17 mediated ACE2 shedding is prevented by TIZ due to PDI inhibition, thus curbing lung inflammation and consequently ARDS. Upon entry, SARS-CoV-2 pathogen-associated molecular patterns (PAMPs) evades recognition by host cells pattern-recognition-receptors (PRRs), like RLRs, TLRs and NLRs, which is opposed by TIZ. These PRRs cause downstream activation of NF-κB, IRF3/7 signalling, which leads to the secretion of proinflammatory cytokines and type I-IFN respectively. Secreted IFN binds to IFN receptor activating JAK/STAT signalling, which forms a complex with IRF9 and produces interferon stimulated genes (ISGs) promoted under IFN-stimulated response element (ISRE), boosting antiviral innate immune responses, enabling virus clearance. TIZ activates Type-I IFN secretion and curb proinflammatory responses. ACE2, Angiotensin converting enzyme 2; ADAM-17, A disintegrin and metallopeptidase domain 17; ARDS, Acute respiratory distress syndrome; AT1R, Angiotensin II receptor type 1; AT2R, Angiotensin II receptor type 2; cGAS, cyclic GMP-AMP synthase; IFN, Interferon; IFNR, IFNα/β receptor; IRF3/7, Interferon regulatory factor 3/7; JAK, Janus kinase; MAVS, Mitochondrial antiviral-signalling protein; MDA5, Melanoma differentiation-associated gene 5; NF-κB, Nuclear factor-κB; NLRs, Nucleotide-binding oligomerization domain-(NOD)-like receptors; PDI, Protein disulfide isomerase; PPKRA, Phopsphorylated protein kinase receptor; RLRs, Retinoic acid-inducible gene I protein (RIG-I) like receptors; ROS, Reactive oxygen species; STAT, Signal transducer and activator of transcription; STING, stimulator of interferon genes protein; TLRs, Toll-like receptors; TRIF, TIR-domain-containing adaptor protein including IFN-β; TYK2, Tyrosine kinase 2.
The viral RNA expresses pathogen-associated molecular patterns (PAMPs) sensed by pattern recognition receptors (PRRs) of host cells, including Toll-like receptors (TLR-3, -7, -8, -9), Retinoic acid Inducible Gene-I-(RIG-I) like receptors (RLRs), and the nucleotide-binding oligomerization domain-(NOD) like receptors (NLRs) (Fung and Liu, 2019; Lim et al., 2016). The TLR-7/8 senses single-stranded viral RNA in the endosomes, while the cytoplasmic viral RNA is noticed by TLR-3, RIG-I, melanoma differentiation-associated gene 5 (MDA5) and cyclic GMP-AMP synthase (cGAS) (Dixit and Kagan, 2013; Vanpouille-Box et al., 2019). Upon viral RNA recognitions these transducer molecules trigger adaptive proteins such as TIR-domain-containing adaptor protein including Interferon-β (TRIF), mitochondrial antiviral-signalling protein (MAVS) and stimulator of interferon genes protein (STING) to activate downstream signalling effectors, i.e. MyD88, the transcription factor nuclear factor-κB (NF-κB) and interferon regulatory factor 3/7 (IRF3/7) (Frieman and Baric, 2008; Guo et al., 2020; Lei and Hilgenfeld, 2017). SARS-CoV-2 prevents IRF3/7 signalling, suppresses Type I Interferon (IFN–I) release and inhibits JAK/STAT signalling (Gordon et al., 2020; Jamilloux et al., 2020; Zumla et al., 2016). While NF-ĸB activation leads to the secretion of pro-inflammatory markers (TNF-α, IL-1β, IL-6, IFN-γ, IL-17), IRF3/7 activation induces secretion of IFN-I, which suppresses viral replication and spread, helps cell recovery and boosts adaptive immunity (Costela-Ruiz et al., 2020). The IFN secretion triggers the JAK1/TYK2-STAT1/2 pathway, inducing production of numerous IFN stimulated genes (ISGs) in the nucleus (Jamilloux et al., 2020). Nitazoxanide treatment thus results in broad amplification of the host immune responses, including an increase in RLR activation, enhanced MAVS and IRF3 activities, and induction of the antiviral phosphatase GADD34 (Jasenosky et al., 2019). Also Nitazoxanide mediates phosphorylation of protein kinase RNA (PKR) and eukaryotic initiation factor 2α (eIF2α) leading to the enhanced IFN-I secretion (Ashiru et al., 2014; Haas et al., 2020). Moreover, the viral genome proteins ORF3a, ORF8b and E activate NLR signalling and JNK signalling, which then triggers caspase-1, NF-kB signalling, stimulate pro-inflammatory cytokines and induces cell apoptosis (Freeman and Swartz, 2020; Shah, 2020). Nitazoxanide inhibited the production of pro-inflammatory cytokines TNF-α, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10 in peripheral blood mononuclear cells (Rossignol, 2016), exhibited downregulation of IL-6/JAK2/STAT3 pathway and significantly modulated the p53/caspases-dependent signalling pathways affecting apoptosis (Tantawy et al., 2020). TIZ treatment reduced inflammation in macrophage cell lines by suppressing NF-κB, MAPK and PI3K/Akt/mTOR pathway (Shou et al., 2019), whereas in case of LPS induced inflammatory microglia, TIZ treatment decreased the release of pro-inflammatory cytokines including TNF-α, IL-1β, IL-6, chemokines like CCL-2 and CCL-3, intrinsic nitric oxide synthase (iNOS) and cyclooxygenase2 (COX2) expression (Li et al., 2020; Trabattoni et al., 2016). Thus in the case of COVID-19 patients, nitazoxanide could serve as a potential candidate, curbing the inflammation and activating host innate immune responses crucial for addressing SARS-CoV-2 pathogenesis (Martins-Filho et al., 2020).
2.2.7. Lung damage
In COVID-19 patients, lung damage is attributed to shedding of SARS-CoV-2 entry receptor ACE2 in the extracellular milieu mediated by A disintegrin and metallopeptidase domain 17 (ADAM17) (Costela-Ruiz et al., 2020; Krossa et al., 2018; Palau et al., 2020; Vitiello and Ferrara, 2020). ADAM17 activation, impaired IFN-I response and persistent lymphopenia (lymphocyte count < 1.5 × 109/L) increases accumulation of lymphocytes in the lungs and other lymphoid organs with circulating higher levels of inflammatory cytokines (Venet et al., 2009; Yuki et al., 2020). The resulting inflammation, vascular leakage and enormous lung epithelial and endothelial cells damage, causes the acute respiratory distress syndrome (ARDS) (Costela-Ruiz et al., 2020; Hoffmann et al., 2020; Jamilloux et al., 2020; Venet et al., 2009; Vitiello and Ferrara, 2020; Zhang et al., 2020). This is further aggravated by downregulation of innate immunity and T-cell exhaustion, which makes COVID-19 patients immunocompetent (Diao et al., 2020; Samudrala et al., 2020; Tay et al., 2020). In one clinical case report, nitazoxanide treatment provided better relief to immunocompetent host infected with lung cryptosporidiosis, who presented with fever, cough, breathlessness, followed by diarrhoea, and had lung consolidation conditions similar to COVID-19 (Kumar et al., 2016).
In inflammatory lung conditions, there is downregulation of UPR and upregulation of PDI. In allergic airway disease model, ablation of PDI in lung epithelial cells, reduced apoptotic, inflammatory and fibrotic reactions (Chamberlain et al., 2019; Chamberlain and Anathy, 2020; Hoffman et al., 2016; Mohan et al., 2019; Stolf et al., 2011). However, the prolonged UPR response, lead to cell death, secretion of proinflammatory cytokines and epithelial cell damage as observed in common lung diseases such as pulmonary fibrosis, asthma and chronic obstructive pulmonary disease (COPD) (Chamberlain and Anathy, 2020; Hoffman et al., 2016). Coronavirus can induce the UPR in cell culture models (Bechill et al., 2008; Siu et al., 2014). Nitazoxanide activates and or modulates UPR signalling and thus promotes antioxidant responses, curbs inflammatory cytokines, inhibits PDI and thereby prevents cell damage (Di Santo and Ehrisman, 2013, 2014). The UPR consists of three pathways regulating the ER stress, which decreases protein load by promoting folding or otherwise induces extended UPR to cause apoptosis. Each UPR pathway involves a particular signalling molecule, including inositol-requiring protein-1 (IRE1), protein kinase RNA (PKR)-like ER kinase (PERK) and activating transcription factor-6 (ATF6) (Ron and Walter, 2007; Sureda et al., 2020). The IRE1 causes mRNA splicing and activation of tumour necrosis factor associated factor 2 (TRAF2) triggering downstream inflammatory and apoptotic signalling (Rajapaksa et al., 2015). CoV induces prolonged IRE1 and activates TRAF2 (Fung et al., 2014; Fung and Liu, 2014; Lim et al., 2016). In contrast, nitazoxanide induces glutathione S-transferase P1 (GSTP1) which inhibits TRAF2 (Müller et al., 2008b). CoV induces ATF-6 to release ER chaperone proteins like PDI (Fung et al., 2014), whereas nitazoxanide being a PDI inhibitor, plays a crucial role in maintaining protective UPR response (Di Santo and Ehrisman, 2013, 2014). PERK which causes phosphorylation of the eIF2α and induces Activating Transcription Factor 4 (ATF4), protecting the cell against oxidative damage (Sureda et al., 2020), is inhibited by CoV, while nitazoxanide upregulates the ATF4 mechanism (Ashiru et al., 2014). Nitazoxanide mediated UPR signalling via PERK activation can exhibit antiprotease and antioxidant activity by stimulating nuclear factor erythroid 2-related factor 2 (Nrf2) through NADPH quinone oxidoreductase1, a critical response against respiratory viral infection, including coronaviruses (Antalis et al., 2011; Curry Jr and Rosewell, 2008; Rosewell et al., 2011; Victor et al., 2020). Furthermore, the calcium binding proteins, translationally controlled tumour protein (TCTP) and binding immunoglobulin protein (BiP) are activated within cytosol and ER respectively, during nitazoxanide therapy to balance Ca2+ concentration, thus inhibiting apoptosis (Ashiru et al., 2014; Di Santo and Ehrisman, 2014). Another important aspect is the potent bronchodilation activity of nitazoxanide comparable to β-agonist isoproterenol, attributed to its role as antagonist to the Ca2+-Activated-Cl‾ channel transmembrane member 16A (TMEM16A). This proposes a new mechanism of bronchodilation useful in treatment of severe lung inflammatory diseases, asthma and COPD. Nitazoxanide therefore exhibits great promise in alleviating lung damage (Miner et al., 2019a).
2.2.8. Nitazoxanide use in comorbidities
In COVID-19 patients, the elevated levels of C-reactive protein, erythrocyte sedimentation rate, serum amyloid A and ferritin, respiratory distress, vascular leakage and coagulation, neuronal damage result in fatalities often associated with multiple organ failures (Chibber et al., 2020; Kermali et al., 2020; Nile et al., 2020; Samudrala et al., 2020; Zeng et al., 2020). Interestingly, inhibition of PDI by nitazoxanide can play an important role in preventing multiple organ damage reported in COVID-19 patients. In case of cardiovascular diseases, PDI supports plaque formation and causes coagulation (Li et al., 2016), whereas in case of diabetes it hampers insulin release and prevents lowering of blood glucose (Mohan et al., 2019; Shergalis and Neamati, 2016). Similarly, in cancer patients, cell migration, invasion and metastasis is mediated through PDI activity (Lee, 2017; Xu et al., 2014). Moreover, in case of brain disorders, nitazoxanide inhibited neurodegeneration (Perri et al., 2016; Uehara et al., 2006; Verkhratsky and Petersen, 2002). Further, in the case of liver diseases activation of prolonged UPR signalling and PDI plays a crucial role in disease progression (Maiers and Malhi, 2019; Mohan et al., 2019). Renal fibrosis also exhibits high secretory level of PDI (ERP57) causing kidney damage (Dihazi et al., 2013). Yet another mechanism by which nitazoxanide exhibits multiple organ protection is via phosphodiesterases (PDEs) inhibition (WO2018173069A1) (Deshpande et al., 2018). PDEs causes phosphodiesteric bond cleavage in cyclic nucleotides, cAMP and cGMP, leading towards fibrosis, memory loss, pulmonary hypertension, vasoconstriction, renal dysfunction and various other multiple organ damage (Mondaini, 2020; Santing et al., 2001; Sharma et al., 2013; Thomas et al., 2001). Thus inhibiting PDI, PDEs and simultaneously activating protective UPR signalling could be critical in effective therapy of patients with comorbidities like cardiovascular disease, cancer, diabetes, kidney, liver and brain disorders (Di Santo and Ehrisman, 2013; Müller et al., 2008a).
3. Nitazoxanide promising in vitro efficacy against SARS-CoV-2
Nitazoxanide has demonstrated superior efficacy against coronavirus replication in cell culture assays at low micromolar concentrations (Rossignol, 2014). Recent reports on comparative in vitro efficacy of nitazoxanide hampering SARS-CoV-2 in Vero E6 cells at 48 h post-infection revealed low 50% effective concentration (EC50) 2.12 μM. The in vitro antiviral efficacy found promising and was comparable with antiviral drug remdesivir (EC50 0.77 μM), antimalarial chloroquine (EC50 1.13 μM) and superior to favipiravir (EC50 61.88 μM) (Wang et al., 2020).
4. Nitazoxanide dosing and safety
Nitazoxanide is a hydrophobic drug with low aqueous solubility (~7.5 μg/ml) and high permeability, which dictates its dose-dependent therapeutic efficacy. Nitazoxanide exhibits oral bioavailability of about 30%, which can rise up to 50% with food (Rossignol, 2014; Stachulski et al., 2018). Its low bioavailability and poorly discovered lung concentrations, are major challenges that need to be addressed (Anderson and Curran, 2007; Padmanabhan, 2020; Padmanabhan and Padmanabhan, 2020; Rajoli et al., 2020). In adults, nitazoxanide dose of 500 mg twice-daily achieved peak concentration of 10 μg/ml, at approximately 2–4 h for TIZ, while 1 g twice daily dose achieved the peak concentration of 24 μg/ml (Fox and Saravolatz, 2005; Jr, 2004). Moreover, nitazoxanide controlled-release tablets 300 mg twice-daily dose exhibited peak plasma concentrations of 4.6 μg/ml which is well above its in vitro antiviral concentration (Haffizulla et al., 2014; Rossignol, 2014). The estimated IC50 and IC90 of nitazoxanide against SARS-CoV-2 are 0.68 μg/ml and 1.84 μg/ml respectively, which can be readily achieved using frequent dosing (Padmanabhan and Padmanabhan, 2020). The physiologically-based pharmacokinetic (PBPK) modelling reported the optimal doses of nitazoxanide as 1200 mg four times, 1600 mg three times, 2900 mg twice daily in the fasted state and 700 mg four times, 900 mg three times and 1400 mg twice daily when given with food, to provide its plasma and lung concentrations well above its reported in vitro 90% effective concentration (EC90) against SARS-CoV-2 (4.64 μM or 1.43 μg/ml) (Rajoli et al., 2020). Furthermore, one study utilized the ratio of Cmax/EC90 based on approved human dose of nitazoxanide, which exhibited the ratio value above 1 indicating plasma Cmax concentrations exceeded those necessary to inhibit 90% of SARS-CoV-2 replication (Arshad et al., 2020). Interestingly, nitazoxanide is well tolerated up to 4 g daily dose and its LD50 is higher than 10,000 mg/kg, indicating high safety. Nitazoxanide high dose of 1 g two times daily administered in AIDS patients suffering from diarrhoea, devoid of any severe side effects (Fox and Saravolatz, 2005; Jr, 2004). Thus nitazoxanide is effective and safe therapeutic intervention and its estimated generic pricing is $1.41 for a 14-day COVID-19 treatment at 500 mg twice daily dose (Mahmoud et al., 2020; Martins-Filho et al., 2020; Pepperrell et al., 2020), which makes therapy inexpensive.
5. Nitazoxanide and COVID-19 severity
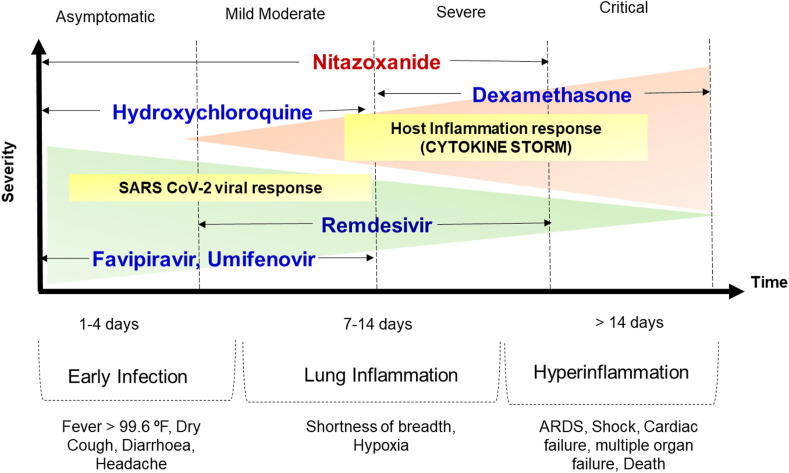
The clinical pathology of COVID-19 is characterized as mild, moderate, severe and critical (Siddiqi and Mehra, 2020). The life-threatening challenges emerge in severe and critical phases attributed to the cytokine storm (Chibber et al., 2020; Samudrala et al., 2020; Soy et al., 2020; Wang et al., 2020). While, favipiravir, remdesivir, umifenovir and hydroxychloroquine may act at early and mild to moderate phases, nitazoxanide holds promise to act even in the severe phase due to the potential to curb the cytokine storm (Krishna et al., 2020; Nyarko et al., 2020; Omolo et al., 2020; Sanders et al., 2020). Dexamethasone also found effective against cytokine storm (Horby et al., 2020; Robinson, 2020). Furthermore, the possibility of nitazoxanide in therapy of patients with comorbidities is certainly exciting. Fig. 6 depicts the various stages of COVID-19 and compares nitazoxanide with some of the promising emergency use approved drug candidates for COVID-19.
Fig. 6.
Different clinical phases of COVID-19 infection based on severity and recommended drug use. ARDS, Acute respiratory distress syndrome.
6. Clinical trials of nitazoxanide against COVID-19
Number of clinical trials registered using nitazoxanide as the only drug or in combination with other antivirals further substantiates the hypothesis on the high promise of nitazoxanide (Mahmoud et al., 2020; Martins-Filho et al., 2020). A total of 19 new clinical trials for COVID-19, have been registered in the USA, Egypt, Mexico, Argentina, Nigeria and Brazil in the past five months, between April to August 2020 (https://clinicaltrials.gov/) (Nitazoxanide clinical trials, 2020). Of these registered trials ten trials are nitazoxanide alone while the other nine are combinations of nitazoxanide with antivirals (Table 1 ). Notably, in the majority of registered clinical trials, the nitazoxanide dose is increased to 500 mg or 600 mg twice a day and more. Moreover, nitazoxanide has also been proposed in combination with other potential drugs such as hydroxychloroquine, ivermectin and ribavirin. These trials indeed emphasise the promise of the drug for COVID-19. A recent study identified 73 combinations of potential 32 drugs against SARS-CoV-2 using in silico modelling and further validated them through in vitro studies. The study reported high synergy of nitazoxanide with three antivirals remdesivir, amodiaquine and umifenovir. The study also reports that the combination of remdesivir and hydroxychloroquine demonstrated strong antagonism (Bobrowski et al., 2020). The combination use of nitazoxanide with macrolide antibiotic azithromycin (Kelleni, 2020) and antimalarial drug hydroxychloroquine has been proposed (Padmanabhan, 2020).
Table 1.
Clinical studies registered on Nitazoxanide/Nitazoxanide combinations against COVID-19 (Updated August 28, 2020).
| Sr. no. | Trial Number | Study Proposed | Drug Used & Dose | No of patients, Status & Date of Posting |
|---|---|---|---|---|
| A | Nitazoxanidealone | |||
| 1 | NCT04486313 | Efficacy and safety in mild to moderate patients | Two nitazoxanide 300 mg tablets administered orally twice daily with food for 5 days with vitamin B complex versus Placebo and vitamin B complex | 800 Recruiting July 24, 2020 |
| Romark Laboratories L.C. USA | ||||
| Phase III | ||||
| 2 | NCT04463264 | Efficacy and safety in mild patients | Nitazoxanide orally 500 mg every 6 h for 14 days) orally with food versus Placebo |
135 Recruiting July 9, 2020 |
| Austral University, Argentina | ||||
| Phase II/III | ||||
| 3 | NCT04441398 | Efficacy and safety to treat mild ambulatory patients | Nitazoxanide 600 mg three times a day (total dose 1800 mg/day) for 7 days versus Placebo | 300 Not yet recruiting June 22, 2020 |
| Azidus Brazil | ||||
| Phase II/III | ||||
| 4 | NCT04435314 | Efficacy and safety of post exposure prophylaxis | Nitazoxanide 600 mg three times a day (total dose 1800 mg/day) for 7 days versus Placebo | 200 Not yet recruiting June 17, 2020 |
| Azidus Brazil | ||||
| Phase II | ||||
| 5 | NCT04423861 | Efficacy in non-critical patients | Nitazoxanide 600 mg three times a day (total dose 1800 mg/day) for 7 days | 50 Not yet recruiting June 9, 2020 |
| Hospital Casa de Saúde - Vera Cruz - Campinas - SP – Brazil | ||||
| Phase II | ||||
| 6 | NCT04406246 | Prophylactic treatment | Nitazoxanide 500 mg every 6 h for two days and then every 12 h for four days. | 150 Recruiting May 28, 2020 |
|
Materno-Perinatal Hospital of the State of Mexico, Mexico | ||||
| Phase IV | ||||
| 7 | NCT04359680 | Efficacy and safety | Nitazoxanide 600 mg orally twice daily for six weeks versus Placebo | 800 Recruiting April 24, 2020 |
| Romark Laboratories L.C. USA | ||||
| Phase III | ||||
| 8 | NCT04348409 | Safety and efficacy in moderate condition | Nitazoxanide 600 mg twice daily for 6 weeks versus Placebo | 50 Recruiting April 16, 2020 |
| Hospital Vera Cruz Campinas, São Paulo, Brazil | ||||
| Proof of Concept | ||||
| 9 | NCT04343248 | Efficacy and safety | Nitazoxanide 600 mg twice daily for 6 weeks | 300 Recruiting April 13, 2020 |
| Romark Laboratories L. C. USA | ||||
| Phase III | ||||
| 10 | NCT04343248 | Efficacy and safety | Nitazoxanide 600 mg orally twice daily for 6 weeks versus Placebo | 600 Not yet recruiting April 13, 2020 |
| Romark Laboratories L.C. USA | ||||
| Phase III | ||||
| B | Nitazoxanide combination | |||
| 11 | NCT04498936 | Efficacy and safety | Nitazoxanide (500 mg, orally) four times per day for 14 days, plus the standard care treatment versus regimen combination of Sofosbuvir/Ledipasvir (400 mg and 90 mg, orally) once daily for 14 days, plus the standard care treatment | 240 Recruiting August 5, 2020 |
| Assiut University Hospital, Assiut, Egypt | ||||
| Phase IV | ||||
| 12 | NCT04459286 | Efficacy and safety | Nitazoxanide 1000 mg tablets twice daily and 300/100 mg atazanavir/ritonavir tablets once daily with meal for 28 days versus Standard of care treatment | 98 Not yet recruiting July 7, 2020 |
| Infectious Disease Hospital, Olodo | ||||
| Ibadan, Oyo State, Nigeria | ||||
| Phase II | ||||
| 13 | NCT04392427 | Combination efficacy | Nitazoxanide, ivermectin and ribavirin 200 mg or 400 mg for 7 days + zinc supplement versus Placebo | 100 Not yet recruiting May 18, 2020 |
| Mansoura University, Mansoura, Egypt | ||||
| Phase III | ||||
| 14 | NCT04382846 | Efficacy novel regimens |
Nitazoxanide, Ivermectin, Chloroquine, Azithromycin dose not mentioned |
80 Not yet recruiting May 11, 2020 |
| Tanta University, Tanta, Egypt | ||||
| Phase III | ||||
| 15 | NCT04360356 | Combination therapy | Ivermectin 200 mcg/kg once orally on empty stomach plus Nitazoxanide 500 mg twice daily orally with meal for 6 days versus standard care oxygen via ventilators |
100 Not yet recruiting April 24, 2020 |
| Tanta University, Tanta, Egypt | ||||
| Phase II/III | ||||
| 16 | NCT04361318 | Combination therapy | 200 mg of Hydroxychloroquine orally three times daily for 10 days plus 500 mg of Nitazoxanide orally twice daily for 6 days versus Standard care |
100 Not yet recruiting April 24, 2020 |
| Tanta University, Tanta, Egypt | ||||
| Phase II/III | ||||
| 17 | NCT04351347 | Efficacy | Drug: Chloroquine Drug: Nitazoxanide + Chloroquine Drug: Ivermectin + Chloroquine |
60 Recruiting April 17, 2020 |
| Tanta University, Tanta, Egypt | ||||
| Phase II/III | ||||
| 18 | NCT04345419 | Efficacy and safety | Chloroquine versus Favipiravir versus Nitazoxanide versus Ivermectin versus Niclosamide dose not mentioned |
100 Not yet recruiting April 14, 2020 |
| Tanta university hospital | ||||
| Tanta, Egypt | ||||
| Phase II/III | ||||
| 19 | NCT04341493 | Safety and efficacy | Hydroxychloroquine 200 mg twice daily for 10 days versus Nitazoxanide 500 mg twice daily + Hydroxychloroquine 200 mg twice daily for 10 days |
86 Recruiting April 10, 2020 |
| Materno-Perinatal Hospital of the State of Mexico, Mexico | ||||
| Phase IV | ||||
Trials accessed from https://clinicaltrials.gov/.
7. Limitations and challenges
Although nitazoxanide demonstrated efficacy in clinical studies against Influenza, HBV, HCV, rotavirus, rhinovirus/enterovirus infections, it was never approved/marketed for these indications, due to poor significance over the placebo group. Although the clinical study against uncomplicated influenza demonstrated efficacy, the results were not convincing given only 1–2 log reduction in viral titer and minor difference in time for discharge between placebo (116.7 h) and test arms (nitazoxanide 300 mg twice daily 95.5 h and nitazoxanide 300 mg once daily 109.1 h) (Haffizulla et al., 2014). We contend that the dose was probably low. The differences would have probably been evident with further increase in dose to 500 mg twice a day or through the design of a bioenhanced formulation. The Mexico trial involving nitazoxanide against severe acute respiratory illness (SARI), found discouraging, with highlighted reasons for the same. It said, negative results might be associated with low plasma drug levels, which attributed to low dose and poor bioavailability. They also highlighted other limitations of their study, which included the heterogeneous patient population, a variety of factors affecting decisions related to the primary endpoint (hospital discharge), and limited sample size. Also, the maximum dose selected was 300 mg twice daily for five days (Gamiño-Arroyo et al., 2019).
The phase 3 global study data on uncomplicated influenza involved a higher dose of nitazoxanide 600 mg twice daily (1.2 gm/day) alone or in combination with oseltamavir 75 mg twice daily (NCT01610245). The results are however not available yet. This study substantiates our premise on the need for a higher dose. Nitazoxanide requires further translational development and de-risking studies to conclude its effective use in clinical situation to provide benefit to COVID-19 victims.
8. Conclusion
Nitazoxanide an FDA approved antiprotozoal, safe, inexpensive and well-tolerated old drug, proposes great potential for repurposing against COVID-19. In this review we highlight proposed mechanism of nitazoxanide against numerous targets involved in SARS-CoV-2 pathogenesis affecting viral entry and multiplication. The potential of nitazoxanide to inhibit the inflammatory cytokine storm while simultaneously stimulating innate immune responses presents great promise to prevent ARDS. The ability to protect the lung, avert associated multiple organ damage, and the beneficial effects in SARS-CoV-2 patients with comorbidities proposes nitazoxanide as a promising candidate for repurposing in COVID-19.
9. Future perspectives
Nitazoxanide presents exciting possibilities for repurposing in COVID-19. The drug is approved as an antiprotozoal and for localized action in the gastro-intestinal tract (GIT). Hence considering strategies to enhance bioavailability could be an important approach to enhance efficacy at possibly lower doses (Omolo et al., 2020). Furthermore, directing the drug in high concentration to the target site following oral administration could be achieved by lymphatic targeting of oral particulate carriers (Bachhav et al, 2017, 2018). With the myriad targets the drug exhibits potential to attack in the pathogenesis of COVID-19, let us hope it is the panacea the world seeks to overcome the fatality and the fear of COVID-19.
Funding
Chhatrapati Shahu National Research Fellowship (CSNRF fellowship) from BARTI, Pune, India to Amit S. Lokhande (CSNRF/2015-16/2567).
Conflicts of interest/competing interests
The authors declare no competing financial interest.
Ethical approval
Not required.
CRediT authorship contribution statement
Amit S. Lokhande: Conceptualization, Data curation, Data analysis and/or interpretation, Writing- original draft, Visualization, Writing - review & editing. Padma V. Devarajan: Conceptualization, Supervision, Writing - review & editing. All authors have reviewed the manuscript and approved the submission.
Acknowledgements
Dr Babasaheb Ambedkar Research and Training Institute (BARTI), Pune, India.
References
- Abdul Kadhim A.H., Hadi N.R., Abdulhussein M. Preprocessing of the candidate antiviral drugs against COVID-19 in models of SARS cov2 targets. La Prensa Medica Argentina. 2020;106:2. doi: 10.1002/ddr.21701. [DOI] [Google Scholar]
- Aftab S.O., Ghouri M.Z., Masood M.U., Haider Z., Khan Z., Ahmad A., Munawar N. Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. J. Transl. Med. 2020;18:275. doi: 10.1186/s12967-020-02439-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agajanian M.J., Walker M.P., Axtman A.D., Ruela-de-Sousa R.R., Serafin D.S., Rabinowitz A.D., Graham D.M., Ryan M.B., Tamir T., Nakamichi Y., Gammons M.V., Bennett J.M., Couñago R.M., Drewry D.H., Elkins J.M., Gileadi C., Gileadi O., Godoi P.H., Kapadia N., Müller S., Santiago A.S., Sorrell F.J., Wells C.I., Fedorov O., Willson T.M., Zuercher W.J., Major M.B. WNT activates the AAK1 kinase to promote clathrin-mediated endocytosis of LRP6 and establish a negative feedback loop. Cell Rep. 2019;26:79–93. doi: 10.1016/j.celrep.2018.12.023. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., Ray A.S., Cihlar T., Siegel D., Mackman R.L., Clarke M.O., Baric R.S., Denison M.R. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9 doi: 10.1128/mBio.00221-18. e00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alharbi A.F., Parrington J. Endolysosomal Ca2+ signaling in cancer: the role of TPC2, from tumorigenesis to metastasis. Front. Cell Dev. Biol. 2019;7:302. doi: 10.3389/fcell.2019.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso D.F., Farina H.G. Repurposing of host-based therapeutic agents for the treatment of coronavirus disease 2019 (COVID-19): a link between antiviral and anticancer mechanisms? Int. J. Antimicrob. Agents. 2020;56:106125. doi: 10.1016/j.ijantimicag.2020.106125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson V.R., Curran M.P. Nitazoxanide: a review of its use in the treatment of gastrointestinal infections. Drugs. 2007;67:1947–1967. doi: 10.2165/00003495-200767130-00015. [DOI] [PubMed] [Google Scholar]
- Andrade R.M., Reed S.L. New drug target in protozoan parasites: the role of thioredoxin reductase. Front. Microbiol. 2015;6:975. doi: 10.3389/fmicb.2015.00975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antalis T.M., Bugge T.H., Wu Q. Membrane-anchored serine proteases in health and disease. Prog. Mol. Biol. Transl. Sci. 2011;99:1–50. doi: 10.1016/B978-0-12-385504-6.00001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aqeel Y., Siddiqui R., Farooq M., Khan N.A. Anaerobic respiration: in vitro efficacy of Nitazoxanide against mitochondriate Acanthamoeba castellanii of the T4 genotype. Exp. Parasitol. 2015;157:170–176. doi: 10.1016/j.exppara.2015.08.007. [DOI] [PubMed] [Google Scholar]
- Arshad U., Pertinez H., Box H., Tatham L., Rajoli R.K., Curley P., Neary M., Sharp J., Liptrott N.J., Valentijn A. Prioritisation of potential anti-SARS-CoV-2 drug repurposing opportunities based on ability to achieve adequate target site concentrations derived from their established human pharmacokinetics. medRxiv. 2020 doi: 10.1101/2020.04.16.20068379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashiru O., Howe J.D., Butters T.D. Nitazoxanide, an antiviral thiazolide, depletes ATP-sensitive intracellular Ca2+ stores. Virology. 2014;462:135–148. doi: 10.1016/j.virol.2014.05.015. [DOI] [PubMed] [Google Scholar]
- Astuti I. Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab. Syndr. 2020;14:407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avchaciov K., Burmistrova O., Fedichev P.O. AI for the repurposing of approved or investigational drugs against COVID-19. 2020. https://www.researchgate.net/deref/ [DOI]
- Bachhav S.S., Dighe V.D., Devarajan P.V. Exploring Peyer's patch uptake as a strategy for targeted lung delivery of polymeric rifampicin nanoparticles. Mol. Pharm. 2018;15:4434–4445. doi: 10.1021/acs.molpharmaceut.8b00382. [DOI] [PubMed] [Google Scholar]
- Bachhav S.S., Dighe V.D., Kotak D., Devarajan P.V. Rifampicin Lipid-Polymer hybrid nanoparticles (LIPOMER) for enhanced Peyer's patch uptake. Int. J. Pharm. 2017;532:612–622. doi: 10.1016/j.ijpharm.2017.09.040. [DOI] [PubMed] [Google Scholar]
- Báez-Santos Y.M., John S.E.S., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayati A., Kumar R., Francis V., McPherson P.S. SARS-CoV-2 uses clathrin-mediated endocytosis to gain access into cells. bioRxiv. 2020 doi: 10.1101/2020.07.13.201509. 2020.07.13.201509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechill J., Chen Z., Brewer J.W., Baker S.C. Coronavirus infection modulates the unfolded protein response and mediates sustained translational repression. J. Virol. 2008;82:4492–4501. doi: 10.1128/jvi.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekendam R.H., Iyu D., Passam F., Stopa J.D., De Ceunynck K., Muse O., Bendapudi P.K., Garnier C.L., Gopal S., Crescence L. Protein disulfide isomerase regulation by nitric oxide maintains vascular quiescence and controls thrombus formation. J. Thromb. Haemost. 2018;16:2322–2335. doi: 10.1111/jth.14291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekerman E., Neveu G., Shulla A., Brannan J., Pu S.-Y., Wang S., Xiao F., Barouch-Bentov R., Bakken R.R., Mateo R., Govero J., Nagamine C.M., Diamond M.S., De Jonghe S., Herdewijn P., Dye J.M., Randall G., Einav S. Anticancer kinase inhibitors impair intracellular viral trafficking and exert broad-spectrum antiviral effects. J. Clin. Invest. 2017;127:1338–1352. doi: 10.1172/JCI89857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestle D., Heindl M.R., Limburg H., Pilgram O., Moulton H., Stein D.A., Hardes K., Eickmann M., Dolnik O., Rohde C. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci. Alliance. 2020;3 doi: 10.26508/lsa.202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard E., Roingeard P. Virus-induced double-membrane vesicles. Cell Microbiol. 2015;17:45–50. doi: 10.1111/cmi.12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrowski T., Chen L., Eastman R.T., Itkin Z., Shinn P., Chen C., Guo H., Zheng W., Michael S., Simeonov A. Discovery of synergistic and antagonistic drug combinations against SARS-CoV-2 in vitro. BioRxiv. 2020 doi: 10.1101/2020.06.29.178889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonacci G., Schopfer F.J., Batthyany C.I., Rudolph T.K., Rudolph V., Khoo N.K., Kelley E.E., Freeman B.A. Electrophilic fatty acids regulate matrix metalloproteinase activity and expression. J. Biol. Chem. 2011;286:16074–16081. doi: 10.1074/jbc.m111.225029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brailoiu G.C., Brailoiu E. Modulation of calcium entry by the endo-lysosomal system. Adv. Exp. Med. Biol. 2016;898:423–447. doi: 10.1007/978-3-319-26974-0_18. [DOI] [PubMed] [Google Scholar]
- Briguglio I., Piras S., Corona P., Carta A. Inhibition of RNA helicases of ssRNA+ virus belonging to Flaviviridae, Coronaviridae and Picornaviridae families. Int. J. Med. Chem. 2011;2011 doi: 10.1155/2011/213135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Forrest J.C., Zhang X. A screen of the NIH Clinical Collection small molecule library identifies potential anti-coronavirus drugs. Antiviral Res. 2015;114:1–10. doi: 10.1016/j.antiviral.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020;20:269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain N., Anathy V. Pathological consequences of the unfolded protein response and downstream protein disulphide isomerases in pulmonary viral infection and disease. J. Biochem. 2020;167:173–184. doi: 10.1093/jb/mvz101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain N., Korwin-Mihavics B.R., Nakada E.M., Bruno S.R., Heppner D.E., Chapman D.G., Hoffman S.M., van der Vliet A., Suratt B.T., Dienz O. Lung epithelial protein disulfide isomerase A3 (PDIA3) plays an important role in influenza infection, inflammation, and airway mechanics. Redox Biol. 2019;22:101129. doi: 10.1016/j.redox.2019.101129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Jiang M., Hu T., Liu Q., Chen X.S., Guo D. Biochemical characterization of exoribonuclease encoded by SARS coronavirus. J. Biochem. Mol. Biol. 2007;40:649–655. doi: 10.5483/bmbrep.2007.40.5.649. [DOI] [PubMed] [Google Scholar]
- Chibber P., Haq S.A., Ahmed I., Andrabi N.I., Singh G. Advances in the possible treatment of COVID-19: a review. Eur. J. Pharmacol. 2020;883:173372. doi: 10.1016/j.ejphar.2020.173372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colín-Lozano B., León-Rivera I., Chan-Bacab M.J., Ortega-Morales B.O., Moo-Puc R., López-Guerrero V., Hernández-Núñez E., Argüello-Garcia R., Scior T., Barbosa-Cabrera E., Navarrete-Vázquez G. Synthesis, in vitro and in vivo giardicidal activity of nitrothiazole-NSAID chimeras displaying broad antiprotozoal spectrum. Bioorg. Med. Chem. Lett. 2017;27:3490–3494. doi: 10.1016/j.bmcl.2017.05.071. [DOI] [PubMed] [Google Scholar]
- Collins M.P., Forgac M. Regulation and function of V-ATPases in physiology and disease. Biochim. Biophys. Acta Biomembr. 2020;183341 doi: 10.1016/j.bbamem.2020.183341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The cytokine storm in COVID-19: an overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costela-Ruiz V.J., Illescas-Montes R., Puerta-Puerta J.M., Ruiz C., Melguizo-Rodríguez L. Cytokine Growth Factor Rev. 2020. SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottam E.M., Maier H.J., Manifava M., Vaux L.C., Chandra-Schoenfelder P., Gerner W., Britton P., Ktistakis N.T., Wileman T. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011;7:1335–1347. doi: 10.4161/auto.7.11.16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curry Jr T.E., Rosewell K. A potential role of MMP2 and MMP9 in regulation of hepsin and protein disulfide isomerase from granulosa cells. Biol. Reprod. 2008;78:127–128. doi: 10.1093/biolreprod/78.s1.127d. [DOI] [Google Scholar]
- Curry T.E., Rosewell K. A potential role of MMP2 and MMP9 in regulation of hepsin and protein disulfide isomerase from granulosa cells. Biol. Reprod. 2008;78:127–128. doi: 10.1093/biolreprod/78.s1.127d. [DOI] [Google Scholar]
- da Silva S.J.R., da Silva C.T.A., Mendes R.P.G., Pena L. Role of nonstructural proteins in the pathogenesis of SARS-CoV-2. J. Med. Virol. 2020 doi: 10.1002/jmv.25858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denault J.-B., Bissonnette L., Longpré J.-M., Charest G., Lavigne P., Leduc R. Ectodomain shedding of furin: kinetics and role of the cysteine-rich region. FEBS (Fed. Eur. Biochem. Soc.) Lett. 2002;527:309–314. doi: 10.1016/S0014-5793(02)03249-0. [DOI] [PubMed] [Google Scholar]
- Deshpande S.K., Kulkarni S.A., Aslekar A.S. 2018. Therapeutic Agent for Phosphodiesterase Inhibition and its Related Disorders. WO2018173069A1. [Google Scholar]
- Di Santo N., Ehrisman J. A functional perspective of nitazoxanide as a potential anticancer drug. Mutat. Res. 2014;768:16–21. doi: 10.1016/j.mrfmmm.2014.05.005. [DOI] [PubMed] [Google Scholar]
- Di Santo N., Ehrisman J. Research perspective: potential role of nitazoxanide in ovarian cancer treatment. Old drug, new purpose? Cancers. 2013;5:1163–1176. doi: 10.3390/cancers5031163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao B., Wang C., Tan Y., Chen X., Liu Ying, Ning L., Chen L., Li M., Liu Yueping, Wang G. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dihazi H., Dihazi G.H., Bibi A., Eltoweissy M., Mueller C.A., Asif A.R., Rubel D., Vasko R., Mueller G.A. Secretion of ERP57 is important for extracellular matrix accumulation and progression of renal fibrosis, and is an early sign of disease onset. J. Cell Sci. 2013;126:3649–3663. doi: 10.1242/jcs.125088. [DOI] [PubMed] [Google Scholar]
- Diwaker D., Mishra K.P., Ganju L. Potential roles of protein disulphide isomerase in viral infections. Acta Virol. 2013;57:293–304. [PubMed] [Google Scholar]
- Dixit E., Kagan J.C. Intracellular pathogen detection by RIG-I-like receptors. Adv. Immunol. 2013;117:99–125. doi: 10.1016/b978-0-12-410524-9.00004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elaidy S.M., Hussain M.A., El-Kherbetawy M.K. Time-dependent therapeutic roles of nitazoxanide on high-fat diet/streptozotocin-induced diabetes in rats: effects on hepatic peroxisome proliferator-activated receptor-gamma receptors. Can. J. Physiol. Pharmacol. 2018;96:485–497. doi: 10.1139/cjpp-2017-0533. [DOI] [PubMed] [Google Scholar]
- El-Kersh W.M., Bahaa E.-D.W., El-Deen S.A.S., Ammar A.I., Matar A.M. Prolonged Cryptosporidium parvum infection can Be a risk factor for intestinal malignancy even in immunocompetent host. Egyptian Journal of Medical Microbiology. 2019;28:63–70. [Google Scholar]
- El-Kowrany S.I., El Ghaffar A.E.-S.A., Shoheib Z.S., Mady R.F., Gamea G.A.M. Evaluation of nitazoxanide as a novel drug for the treatment of acute and chronic toxoplasmosis. Acta Trop. 2019;195:145–154. doi: 10.1016/j.actatropica.2019.04.013. [DOI] [PubMed] [Google Scholar]
- Ethiraj G. How will newly-approved antiviral drugs help in the fight against Covid-19? An internist explains Remdesivir and favipiravir can now be administered for restricted emergency use. Scroll.in. 2020 https://scroll.in/article/966000/how-will-newly-approved-antiviral-drugs-help-in-the-fight-against-covid-19-an-internist-explains [Google Scholar]
- Fan L., Qiu X., Zhu Z., Lv J., Lu J., Mao F., Zhu J., Wang J., Guan X., Chen J., Ren J., Ye J., Zhao Y., Li J., Shen X. Nitazoxanide, an anti-parasitic drug, efficiently ameliorates learning and memory impairments in AD model mice. Acta Pharmacol. Sin. 2019;40:1279–1291. doi: 10.1038/s41401-019-0220-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippini A., D'Amore A., Palombi F., Carpaneto A. Could the inhibition of endo-lysosomal two-pore channels (TPCs) by the natural flavonoid naringenin represent an option to fight SARS-CoV-2 infection? Front. Microbiol. 2020;11:970. doi: 10.3389/fmicb.2020.00970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaumenhaft R., Furie B., Zwicker J.I. Therapeutic implications of protein disulfide isomerase inhibition in thrombotic disease. Arterioscler. Thromb. Vasc. Biol. 2015;35:16–23. doi: 10.1161/ATVBAHA.114.303410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong C.W. Screening potential anti-virals for the main protease of the Coronaviridae family including SARS-CoV-2, SARS-CoV, MERS. 2020. https://hal.archives-ouvertes.fr/hal-02897882 Research Report No. Eigenenergy South Australia.
- Foster M.W., McMahon T.J., Stamler J.S. S-nitrosylation in health and disease. Trends Mol. Med. 2003;9:160–168. doi: 10.1016/S1471-4914(03)00028-5. [DOI] [PubMed] [Google Scholar]
- Fox L.M., Saravolatz L.D. Nitazoxanide: a new thiazolide antiparasitic agent. Clin. Infect. Dis. 2005;40:1173–1180. doi: 10.1086/428839. [DOI] [PubMed] [Google Scholar]
- Freeman T.L., Swartz T.H. Targeting the NLRP3 inflammasome in severe COVID-19. Front. Immunol. 2020;11:1518. doi: 10.3389/fimmu.2020.01518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M., Baric R. Mechanisms of severe acute respiratory syndrome pathogenesis and innate immunomodulation. Microbiol. Mol. Biol. Rev. 2008;72:672–685. doi: 10.1128/mmbr.00015-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Huang M., Liu D.X. Coronavirus-induced ER stress response and its involvement in regulation of coronavirus–host interactions. Virus Res. 2014;194:110–123. doi: 10.1016/j.virusres.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Human coronavirus: host-pathogen interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014;5:296. doi: 10.3389/fmicb.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabison E.E., Hoang-Xuan T., Mauviel A., Menashi S. EMMPRIN/CD147, an MMP modulator in cancer, development and tissue repair. Biochimie. 2005;87:361–368. doi: 10.1016/j.biochi.2004.09.023. [DOI] [PubMed] [Google Scholar]
- Gamiño-Arroyo A.E., Guerrero M.L., McCarthy S., Ramírez-Venegas A., Llamosas-Gallardo B., Galindo-Fraga A., Moreno-Espinosa S., Roldán-Aragón Y., Araujo-Meléndez J., Hunsberger S., Ibarra-González V., Martínez-López J., García-Andrade L.A., Kapushoc H., Holley H.P., Smolskis M.C., Ruiz-Palacios G.M., Beigel J.H., Mexico Emerging Infectious Diseases Clinical Research Network (LaRed) Efficacy and safety of nitazoxanide in addition to standard of care for the treatment of severe acute respiratory illness. Clin. Infect. Dis. 2019;69:1903–1911. doi: 10.1093/cid/ciz100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassen N.C., Niemeyer D., Muth D., Corman V.M., Martinelli S., Gassen A., Hafner K., Papies J., Mösbauer K., Zellner A. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat. Commun. 2019;10:1–16. doi: 10.1038/s41467-019-13659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghusson N., Vasquez G. Successfully treated norovirus- and sapovirus-associated diarrhea in three renal transplant patients. Case Rep. Infect. Dis. 2018;2018 doi: 10.1155/2018/6846873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil C., Ginex T., Maestro I., Nozal V., Barrado-Gil L., Cuesta-Geijo M.A., Urquiza J., Ramírez D., Alonso C., Campillo N.E. COVID-19: drug targets and potential treatments. J. Med. Chem. 2020 doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- Glebov O.O. Understanding SARS-CoV-2 endocytosis for COVID-19 drug repurposing. FEBS J. 2020 doi: 10.1111/febs.15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman R.C. Maximizing bactericidal activity with combinations of bioreduced drugs. Future Med. Chem. 2010;2:1253–1271. doi: 10.4155/fmc.10.215. [DOI] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., O’meara M.J., Guo J.Z., Swaney D.L., Tummino T.A., Huttenhain R. A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing. BioRxiv. 2020 doi: 10.1101/2020.03.22.002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C., Tang R. Could an endo-lysosomal ion channel be the Achilles heel of SARS-CoV2? Cell Calcium. 2020;88:102212. doi: 10.1016/j.ceca.2020.102212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J., Tan K.-S., Wang D.-Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Military Medical Research. 2020;7:1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy R.K., DiPaola R.S., Romanelli F., Dutch R.E. Rapid repurposing of drugs for COVID-19. Science. 2020;368:829–830. doi: 10.1126/science.abb9332. [DOI] [PubMed] [Google Scholar]
- Haas M.J., Feng V., Gonzales K., Onstead-Haas L., Mooradian A.D. High-throughput analysis identifying drugs that reduce oxidative and ER stress in human coronary artery endothelial cells. Eur. J. Pharmacol. 2020;879:173119. doi: 10.1016/j.ejphar.2020.173119. [DOI] [PubMed] [Google Scholar]
- Habtemariam S., fazel Nabavi S., Banach M., Berindan-Neagoe I. Should we try SARS-CoV-2 helicase inhibitors for COVID-19 therapy? Arch. Med. Res. 2020;S0188–4409(20):30863–30868. doi: 10.1016/j.arcmed.2020.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffizulla J., Hartman A., Hoppers M., Resnick H., Samudrala S., Ginocchio C., Bardin M., Rossignol J.-F., Group, U.N.I.C.S Effect of nitazoxanide in adults and adolescents with acute uncomplicated influenza: a double-blind, randomised, placebo-controlled, phase 2b/3 trial. Lancet Infect. Dis. 2014;14:609–618. doi: 10.1016/s1473-3099(14)70717-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halsey J.L. Current approaches to the treatment of gastrointestinal infections: focus on nitazoxanide. Clin. Med. Therapeut. 2009;1 doi: 10.4137/CMT.S2297. CMT–S2297. [DOI] [Google Scholar]
- Han Z., He H., Bi X., Dai Q., Zhu G., Ye Y., Liang Y., Qin W., Zhang Z., Zeng G. CD147, MMP-1, MMP-2 and MMP-9 protein expression as significant prognostic factors in human prostate cancer. Oncology. 2008;75:230–236. doi: 10.1159/000163852. [DOI] [PubMed] [Google Scholar]
- Hati S., Bhattacharyya S. Impact of thiol–disulfide balance on the binding of covid-19 spike protein with angiotensin-converting enzyme 2 receptor. ACS Omega. 2020;5:16292–16298. doi: 10.1021/acsomega.0c02125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Hu L., Huang X., Wang C., Zhang Z., Wang Y., Zhang D., Ye W. Potential of coronavirus 3C-like protease inhibitors for the development of new anti-SARS-CoV-2 drugs: insights from structures of protease and inhibitors. Int. J. Antimicrob. Agents. 2020;56:106055. doi: 10.1016/j.ijantimicag.2020.106055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemphill A., Mueller J., Esposito M. Nitazoxanide, a broad-spectrum thiazolide anti-infective agent for the treatment of gastrointestinal infections. Expert Opin. Pharmacother. 2006;7:953–964. doi: 10.1517/14656566.7.7.953. [DOI] [PubMed] [Google Scholar]
- Henderson L.A., Canna S.W., Schulert G.S., Volpi S., Lee P.Y., Kernan K.F., Caricchio R., Mahmud S., Hazen M.M., Halyabar O. On the alert for cytokine storm: immunopathology in COVID-19. Arthritis Rheumatol. 2020;72:1059–1063. doi: 10.1002/art.41285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess D.T., Matsumoto A., Kim S.-O., Marshall H.E., Stamler J.S. Protein S -nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- Hoffman S.M., Chapman D.G., Lahue K.G., Cahoon J.M., Rattu G.K., Daphtary N., Aliyeva M., Fortner K.A., Erzurum S.C., Comhair S.A. Protein disulfide isomerase–endoplasmic reticulum resident protein 57 regulates allergen-induced airways inflammation, fibrosis, and hyperresponsiveness. J. Allergy Clin. Immunol. 2016;137:822–832. doi: 10.1016/j.jaci.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homolak J., Kodvanj I. Widely available lysosome targeting agents should be considered as potential therapy for COVID-19. Int. J. Antimicrob. Agents. 2020;56:106044. doi: 10.1016/j.ijantimicag.2020.106044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horby P., Lim W.S., Emberson J., Mafham M., Bell J., Linsell L., Staplin N., Brightling C., Ustianowski A., Elmahi E. Effect of dexamethasone in hospitalized patients with COVID-19: preliminary report. Drug Ther. Bull. 2020;58:133. doi: 10.1136/dtb.2020.000045. [DOI] [PubMed] [Google Scholar]
- Jamilloux Y., Henry T., Belot A., Viel S., Fauter M., El Jammal T., Walzer T., François B., Sève P. Should we stimulate or suppress immune responses in COVID-19? Cytokine and anti-cytokine interventions. Autoimmun. Rev. 2020;102567 doi: 10.1016/j.autrev.2020.102567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasenosky L.D., Cadena C., Mire C.E., Borisevich V., Haridas V., Ranjbar S., Nambu A., Bavari S., Soloveva V., Sadukhan S. The FDA-approved oral drug nitazoxanide amplifies host antiviral responses and inhibits Ebola virus. iScience. 2019;19:1279–1290. doi: 10.1016/j.isci.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha A., Ahuja M., Patel S., Brailoiu E., Muallem S. Convergent regulation of the lysosomal two-pore channel-2 by Mg2+, NAADP, PI(3,5)P2 and multiple protein kinases. EMBO J. 2014;33:501–511. doi: 10.1002/embj.201387035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Zhang Y., Alharbi A., Hanbashi A., Alhoshani A., Parrington J. Targeting two-pore channels: current progress and future challenges. Trends Pharmacol. Sci. 2020;41:582–594. doi: 10.1016/j.tips.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Liu J.-Y., Feng R., Ji L., Jin Z.-L., Li H.-B. Drug treatment of coronavirus disease 2019 (COVID-19) in China. Eur. J. Pharmacol. 2020;173326 doi: 10.1016/j.ejphar.2020.173326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi G., Poduri R. Virtual screening enabled selection of antiviral agents against Covid-19 disease targeting coronavirus endoribonuclease NendoU: plausible mechanistic interventions in the treatment of new virus strain. ChemRxiv preprint. 2020 doi: 10.26434/chemrxiv.12198966.v2. [DOI] [Google Scholar]
- Jr A.C.W. Nitazoxanide: a new broad spectrum antiparasitic agent. Expert Rev. Anti Infect. Ther. 2004;2:43–49. doi: 10.1586/14787210.2.1.43. [DOI] [PubMed] [Google Scholar]
- Jurgeit A., McDowell R., Moese S., Meldrum E., Schwendener R., Greber U.F. Niclosamide is a proton carrier and targets acidic endosomes with broad antiviral effects. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandeel M., Abdelrahman A.H.M., Oh-Hashi K., Ibrahim A., Venugopala K.N., Morsy M.A., Ibrahim M.A.A. Repurposing of FDA-approved antivirals, antibiotics, anthelmintics, antioxidants, and cell protectives against SARS-CoV-2 papain-like protease. J. Biomol. Struct. Dyn. 2020;1–8 doi: 10.1080/07391102.2020.1784291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.-L., Chou Y.-Y., Rothlauf P.W., Liu Z., Piccinotti S., Soh T.K., Cureton D., Case J.B., Chen R.E., Diamond M.S., Whelan S.P.J., Kirchhausen T. Inhibition of PIKfyve kinase prevents infection by EBOV and SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.04.21.053058. 2020.04.21.053058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleni M. Nitazoxanide/azithromycin combination for COVID-19: a suggested new protocol for COVID-19 early management. 2020. Preprint. [DOI] [PMC free article] [PubMed]
- Kermali M., Khalsa R.K., Pillai K., Ismail Z., Harky A. The role of biomarkers in diagnosis of COVID-19–A systematic review. Life Sci. 2020;254:117788. doi: 10.1016/j.lfs.2020.117788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S., Tombuloglu H., Hassanein S.E., Rehman S., Bozkurt A., Cevik E., Abdel-Ghany S., Nabi G., Ali A., Sabit H. Coronavirus diseases 2019: current biological situation and potential therapeutic perspective. Eur. J. Pharmacol. 2020;886:173447. doi: 10.1016/j.ejphar.2020.173447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Jedrzejczak R., Maltseva N.I., Wilamowski M., Endres M., Godzik A., Michalska K., Joachimiak A. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Sci. 2020;29:1596–1605. doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm T., Ebert G., Calleja D.J., Allison C.C., Richardson L.W., Bernardini J.P., Lu B.G., Kuchel N.W., Grohmann C., Shibata Y. Mechanism and inhibition of SARS-CoV-2 PLpro. bioRxiv. 2020 doi: 10.1101/2020.06.18.160614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., Van Den Worm S.H., Zevenhoven-Dobbe J.C., Van Der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krátký M., Vinšová J. Antiviral activity of substituted salicylanilides--a review. Mini Rev. Med. Chem. 2011;11:956–967. doi: 10.2174/138955711797068382. [DOI] [PubMed] [Google Scholar]
- Krishna G., Pillai V.S., Veettil M.V. Approaches and advances in the development of potential therapeutic targets and antiviral agents for the management of SARS-CoV-2 infection. Eur. J. Pharmacol. 2020;885:173450. doi: 10.1016/j.ejphar.2020.173450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krossa S., Scheidig A.J., Grötzinger J., Lorenzen I. Redundancy of protein disulfide isomerases in the catalysis of the inactivating disulfide switch in A Disintegrin and Metalloprotease 17. Sci. Rep. 2018;8:1103. doi: 10.1038/s41598-018-19429-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H., Singh V.B., Meena B.L., Agrawal J., Beniwal S., Swami T. Pulmonary cryptosporidiosis in an immunocompetent host treated successfully with nitazoxanide. Lung India: Official Organ of Indian Chest Society. 2016;33:69–71. doi: 10.4103/0970-2113.173085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Adhikari S., Hurdle J.G. Action of nitroheterocyclic drugs against Clostridium difficile. Int. J. Antimicrob. Agents. 2014;44:314–319. doi: 10.1016/j.ijantimicag.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Nyodu R., Maurya V.K., Saxena S.K. Coronavirus Disease 2019 (COVID-19) Springer; 2020. Morphology, genome organization, replication, and pathogenesis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pp. 23–31. [Google Scholar]
- Kupferschmidt K., Cohen J. Race to find COVID-19 treatments accelerates. Science. 2020;367:1412–1413. doi: 10.1126/science.367.6485.1412. [DOI] [PubMed] [Google Scholar]
- Lam K.K., Zheng X., Forestieri R., Balgi A.D., Nodwell M., Vollett S., Anderson H.J., Andersen R.J., Av-Gay Y., Roberge M. Nitazoxanide stimulates autophagy and inhibits mTORC1 signaling and intracellular proliferation of Mycobacterium tuberculosis. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S., Lombardi A., Ouanounou A. COVID-19: a review of the proposed pharmacological treatments. Eur. J. Pharmacol. 2020;886:173451. doi: 10.1016/j.ejphar.2020.173451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. Emerging roles of protein disulfide isomerase in cancer. BMB reports. 2017;50:401–410. doi: 10.5483/bmbrep.2017.50.8.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J., Hilgenfeld R. RNA-virus proteases counteracting host innate immunity. FEBS Lett. 2017;591:3190–3210. doi: 10.1002/1873-3468.12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Liu S.-M., Yu X.-H., Tang S.-L., Tang C.-K. Coronavirus disease 2019 (COVID-19): current status and future perspectives. Int. J. Antimicrob. Agents. 2020;55:105951. doi: 10.1016/j.ijantimicag.2020.105951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.W., He R.Z., Li Y., Ruan Z.F. Tizoxanide mitigates inflammatory response in LPS-induced neuroinflammation in microglia via restraining p38/MAPK pathway. Eur. Rev. Med. Pharmacol. Sci. 2020;24:6446–6454. doi: 10.26355/eurrev_202006_21543. [DOI] [PubMed] [Google Scholar]
- Li Y., Chen L., Wang C., Chen J., Zhang X., Hu Y., Niu X., Pei D., He Z., Bi Y. Extracellular matrix metalloproteinase inducer enhances host resistance against pseudomonas aeruginosa infection through MAPK signaling pathway. Am. J. Transl. Res. 2016;8:5619–5627. [PMC free article] [PubMed] [Google Scholar]
- Lim Y.X., Ng Y.L., Tam J.P., Liu D.X. Human coronaviruses: a review of virus–host interactions. Diseases. 2016;4 doi: 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Zhu D. Cyclophilin A and CD147: novel therapeutic targets for the treatment of COVID-19. Med. Drug Discov. 2020;100056 doi: 10.1016/j.medidd.2020.100056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Luo S., Libby P., Shi G.-P. Cathepsin L-selective inhibitors: a potentially promising treatment for COVID-19 patients. Pharmacol. Ther. 2020;213:107587. doi: 10.1016/j.pharmthera.2020.107587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin A., Dijkman R., Bergström T., Kann N., Adamiak B., Hannoun C., Kindler E., Jónsdóttir H.R., Muth D., Kint J. Targeting membrane-bound viral RNA synthesis reveals potent inhibition of diverse coronaviruses including the middle East respiratory syndrome virus. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madadlou A. Food proteins are a potential resource for mining cathepsin L inhibitory drugs to combat SARS-CoV-2. Eur. J. Pharmacol. 2020;885:173499. doi: 10.1016/j.ejphar.2020.173499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud D.B., Shitu Z., Mostafa A. Drug repurposing of nitazoxanide: can it be an effective therapy for COVID-19? J. Genet. Eng. Biotechnol. 2020;18:1–10. doi: 10.1186/s43141-020-00055-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiers J.L., Malhi H. Seminars in Liver Disease. NIH Public Access; 2019. Endoplasmic reticulum stress in metabolic liver diseases and hepatic fibrosis; pp. 235–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margaritopoulos G.A., Lasithiotaki I., Antoniou K.M. Toll-like receptors and autophagy in interstitial lung diseases. Eur. J. Pharmacol. 2017;808:28–34. doi: 10.1016/j.ejphar.2016.09.032. [DOI] [PubMed] [Google Scholar]
- Martins-Filho P.R., Barreto-Alves J.A., Fakhouri R. Potential role for nitazoxanide in treating SARS-CoV-2 infection. Am. J. Physiol. Lung Cell Mol. Physiol. 2020;319:L35–L36. doi: 10.1152/ajplung.00170.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy S.M., Bove P.F., Matthews D.E., Akaike T., van der Vliet A. Nitric oxide regulation of MMP-9 activation and its relationship to modifications of the cysteine switch. Biochemistry. 2008;47:5832–5840. doi: 10.1021/bi702496v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner K., Labitzke K., Liu B., Wang P., Henckels K., Gaida K., Elliott R., Chen J.J., Liu L., Leith A. Drug repurposing: the anthelmintics niclosamide and nitazoxanide are potent TMEM16A antagonists that fully bronchodilate airways. Front. Pharmacol. 2019;10:51. doi: 10.3389/fphar.2019.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner K., Labitzke K., Liu B., Wang P., Henckels K., Gaida K., Elliott R., Chen J.J., Liu L., Leith A., Trueblood E., Hensley K., Xia X.-Z., Homann O., Bennett B., Fiorino M., Whoriskey J., Yu G., Escobar S., Wong M., Born T.L., Budelsky A., Comeau M., Smith D., Phillips J., Johnston J.A., McGivern J.G., Weikl K., Powers D., Kunzelmann K., Mohn D., Hochheimer A., Sullivan J.K. Drug repurposing: the anthelmintics niclosamide and nitazoxanide are potent TMEM16A antagonists that fully bronchodilate airways. Front. Pharmacol. 2019;10 doi: 10.3389/fphar.2019.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. Discovery of an RNA virus 3′→5′ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. Unit. States Am. 2006;103:5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan S., R P.R.M., Brown L., Ayyappan P., G R.K. Endoplasmic reticulum stress: a master regulator of metabolic syndrome. Eur. J. Pharmacol. 2019;860:172553. doi: 10.1016/j.ejphar.2019.172553. [DOI] [PubMed] [Google Scholar]
- Mondaini N. Phosphodiesterase type 5 inhibitors and COVID-19: are they useful in disease management? The World Journal of Men’s Health. 2020;38:254–255. doi: 10.5534/wjmh.200089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J., Naguleswaran A., Müller N., Hemphill A. Neospora caninum: functional inhibition of protein disulfide isomerase by the broad-spectrum anti-parasitic drug nitazoxanide and other thiazolides. Exp. Parasitol. 2008;118:80–88. doi: 10.1016/j.exppara.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Müller J., Sidler D., Nachbur U., Wastling J., Brunner T., Hemphill A. Thiazolides inhibit growth and induce glutathione-S-transferase Pi (GSTP1)-dependent cell death in human colon cancer cells. Int. J. Cancer. 2008;123:1797–1806. doi: 10.1002/ijc.23755. [DOI] [PubMed] [Google Scholar]
- Nga B.T.T., Takeshita Y., Yamamoto M., Yamamoto Y. Studies of inhibitory mechanisms of propeptide-like cysteine protease inhibitors. Enzym. Res. 2014 doi: 10.1155/2014/848937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nile S.H., Nile A., Qiu J., Li L., Jia X., Kai G. COVID-19: pathogenesis, cytokine storm and therapeutic potential of interferons. Cytokine Growth Factor Rev. 2020;53:66–70. doi: 10.1016/j.cytogfr.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitazoxanide clinical trials. 2020. https://clinicaltrials.gov/ct2/results?cond=&term=Nitazoxanide&cntry=&state=&city=&dist=
- Nitazoxanide in fibrosis GENFIT. 2020. https://www.genfit.com/pipeline/nitazoxanide-in-fibrosis/
- Nyarko R., Boateng E., Kahwa I., Boateng P. A comparison analysis on remdesivir, favipiravir, hydroxychloroquine, chloroquine and azithromycin in the treatment of corona virus disease 2019 (COVID-19)- A review. World J. Pharm. Pharmaceut. Sci. 2020;9:121–133. doi: 10.20959/wjpps20205-16143. [DOI] [Google Scholar]
- Omolo C.A., Soni N., Fasiku V.O., Mackraj I., Govender T. Update on therapeutic approaches and emerging therapies for SARS-CoV-2 virus. Eur. J. Pharmacol. 2020;883:173348. doi: 10.1016/j.ejphar.2020.173348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto H.-H., Schirmeister T. Cysteine proteases and their inhibitors. Chem. Rev. 1997;97:133–172. doi: 10.1021/cr950025u. [DOI] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan S. Potential dual therapeutic approach against SARS-CoV-2/COVID-19 with Nitazoxanide and Hydroxychloroquine. 2020. https://www.researchgate.net/deref/ [DOI]
- Padmanabhan S., Padmanabhan K. Nitazoxanide–a potential ally in the treatment of COVID-19. 2020. https://www.researchgate.net/deref/ [DOI]
- Palau V., Riera M., Soler M.J. ADAM17 inhibition may exert a protective effect on COVID-19. Nephrol. Dial. Transplant. 2020;35:1071–1072. doi: 10.1093/ndt/gfaa093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce L., Davidson S.M., Yellon D.M. The cytokine storm of COVID-19: a spotlight on prevention and protection. Expert Opin. Ther. Targets. 2020:1–8. doi: 10.1080/14728222.2020.1783243. [DOI] [PubMed] [Google Scholar]
- Pepperrell T., Pilkington V., Owen A., Wang J., Hill A.M. Review of safety and minimum pricing of nitazoxanide for potential treatment of COVID-19. J. Virus Erad. 2020;6:52. doi: 10.1016/S2055-6640(20)30017-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelygina L., Hautala T., Seppänen M., Adebayo A., Sullivan K.E., Icenogle J. Inhibition of rubella virus replication by the broad-spectrum drug nitazoxanide in cell culture and in a patient with a primary immune deficiency. Antiviral res. 2017;147:58–66. doi: 10.1016/j.antiviral.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perri E.R., Thomas C.J., Parakh S., Spencer D.M., Atkin J.D. The unfolded protein response and the role of protein disulfide isomerase in neurodegeneration. Front. Cell Dev. Biol. 2016;3:80. doi: 10.3389/fcell.2015.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen O.H., Gerasimenko O.V., Gerasimenko J.V. Endocytic uptake of SARS-CoV-2: the critical roles of pH, Ca2+, and NAADP. Function. 2020;1 doi: 10.1093/function/zqaa003. zqaa003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piacentini S., Frazia S.L., Riccio A., Pedersen J.Z., Topai A., Nicolotti O., Rossignol J.-F., Santoro M.G. Nitazoxanide inhibits paramyxovirus replication by targeting the Fusion protein folding: role of glycoprotein-specific thiol oxidoreductase ERp57. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-28172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.-H. An overview of severe acute respiratory syndrome–coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J. Med. Chem. 2016;59:6595–6628. doi: 10.1021/acs.jmedchem.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y., Olsen J.R., Yuan X., Cheng P.F., Levesque M.P., Brokstad K.A., Hoffman P.S., Oyan A.M., Zhang W., Kalland K.-H., Ke X. Small molecule promotes β-catenin citrullination and inhibits Wnt signaling in cancer. Nat. Chem. Biol. 2018;14:94–101. doi: 10.1038/nchembio.2510. [DOI] [PubMed] [Google Scholar]
- Rajapaksa G., Nikolos F., Bado I., Clarke R., Gustafsson J.-\A.A., Thomas C. ERβ decreases breast cancer cell survival by regulating the IRE1/XBP-1 pathway. Oncogene. 2015;34:4130–4141. doi: 10.1038/onc.2014.343. [DOI] [PubMed] [Google Scholar]
- Rajoli R.K., Pertinez H., Arshad U., Box H., Tatham L., Curley P., Neary M., Sharp J., Liptrott N.J., Valentijn A. Dose prediction for repurposing nitazoxanide in SARS-CoV-2 treatment or chemoprophylaxis. medRxiv. 2020 doi: 10.1101/2020.05.01.20087130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. Everything you need to know about the COVID-19 therapy trials. Pharmaceut. J. 2020 https://www.pharmaceutical-journal.com/news-and-analysis/features/everything-you-need-to-know-about-the-covid-19-therapy-trials/20208126.article [Google Scholar]
- Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rosewell K., Al-Alem L., Li F., Kelty B., Curry T.E. Identification of hepsin and protein disulfide isomerase A3 as targets of gelatinolytic action in rat ovarian granulosa cells during the periovulatory period. Biol. Reprod. 2011;85:858–866. doi: 10.1095/biolreprod.111.092072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol J., Kabil S.M., El–Gohary Y., Younis A.M. Effect of nitazoxanide in diarrhea and enteritis caused by Cryptosporidium species. Clin. Gastroenterol. Hepatol. 2006;4:320–324. doi: 10.1016/j.cgh.2005.12.020. [DOI] [PubMed] [Google Scholar]
- Rossignol J.-F. Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus. J. Infect. Public Health. 2016;9:227–230. doi: 10.1016/j.jiph.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol J.-F. Nitazoxanide: a first-in-class broad-spectrum antiviral agent. Antiviral res. 2014;110:94–103. doi: 10.1016/j.antiviral.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol J.-F. Thiazolides: a new class of antiviral drugs. Expert Opin. Drug Metab. Toxicol. 2009;5:667–674. doi: 10.1517/17425250902988487. [DOI] [PubMed] [Google Scholar]
- Rossignol J.F., Frazia S.L., Chiappa L., Ciucci A., Santoro M.G. Thiazolides, a new class of anti-influenza molecules targeting viral hemagglutinin at the post-translational level. J. Biol. Chem. 2009;284:29798–29808. doi: 10.1074/jbc.M109.029470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein D.C., Codogno P., Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 2012;11:709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahebnasagh A., Saghafi F., Avan R., Khoshi A., Khataminia M., Safdari M., Habtemariam S., Ghaleno H.R., Nabavi S.M. The prophylaxis and treatment potential of supplements for COVID-19. Eur. J. Pharmacol. 2020;173530 doi: 10.1016/j.ejphar.2020.173530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samudrala P.K., Kumar P., Choudhary K., Thakur N., Wadekar G.S., Dayaramani R., Agrawal M., Alexander A. Virology, pathogenesis, diagnosis and in-line treatment of COVID-19. Eur. J. Pharmacol. 2020;883:173375. doi: 10.1016/j.ejphar.2020.173375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders J.M., Monogue M.L., Jodlowski T.Z., Cutrell J.B. Pharmacologic treatments for coronavirus disease 2019 (COVID-19): a review. J. Am. Med. Assoc. 2020;323:1824–1836. doi: 10.1001/jama.2020.6019. [DOI] [PubMed] [Google Scholar]
- Santing R.E., de Boer J., Rohof A., van der Zee N.M., Zaagsma J. Bronchodilatory and anti-inflammatory properties of inhaled selective phosphodiesterase inhibitors in a Guinea pig model of allergic asthma. Eur. J. Pharmacol. 2001;429:335–344. doi: 10.1016/S0014-2999(01)01333-4. [DOI] [PubMed] [Google Scholar]
- Senanayake S.L. Overcoming nonstructural protein 15-nidoviral uridylate-specific endoribonuclease (nsp15/NendoU) activity of SARS-CoV-2. Future Drug Discovery FDD. 2020;42 doi: 10.4155/fdd-2020-0012. [DOI] [Google Scholar]
- Senkowski W., Zhang X., Olofsson M.H., Isacson R., Höglund U., Gustafsson M., Nygren P., Linder S., Larsson R., Fryknäs M. Three-dimensional cell culture-based screening identifies the anthelmintic drug nitazoxanide as a candidate for treatment of colorectal cancer. Mol. Cancer Ther. 2015;14:1504–1516. doi: 10.1158/1535-7163.mct-14-0792. [DOI] [PubMed] [Google Scholar]
- Shah A. Novel coronavirus-induced NLRP3 inflammasome activation: a potential drug target in the treatment of COVID-19. Front. Immunol. 2020;11:1021. doi: 10.3389/fimmu.2020.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakya A., Bhat H.R., Ghosh S.K. Update on nitazoxanide: a multifunctional chemotherapeutic agent. Curr. Drug Discov. Technol. 2018;15:201–213. doi: 10.2174/1570163814666170727130003. [DOI] [PubMed] [Google Scholar]
- Shakya A., Bhat H.R., Ghosh S.K. Update on nitazoxanide: a multifunctional chemotherapeutic agent. Curr. Drug Discov. Technol. 2018;15:201–213. doi: 10.2174/1570163814666170727130003. [DOI] [PubMed] [Google Scholar]
- Shannon A., Le N.T.-T., Selisko B., Eydoux C., Alvarez K., Guillemot J.-C., Decroly E., Peersen O., Ferron F., Canard B. Remdesivir and SARS-CoV-2: structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antiviral Res. 2020;178:104793. doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifkashani S., Bafrani M.A., Khaboushan A.S., Pirzadeh M., Kheirandish A., Yavarpour_Bali H., Hessami A., Saghazadeh A., Rezaei N. Angiotensin-converting enzyme 2 (ACE2) receptor and SARS-CoV-2: potential therapeutic targeting. Eur. J. Pharmacol. 2020;884:173455. doi: 10.1016/j.ejphar.2020.173455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Kumar K., Deshmukh R., Sharma P.L. Phosphodiesterases: regulators of cyclic nucleotide signals and novel molecular target for movement disorders. Eur. J. Pharmacol. 2013;714:486–497. doi: 10.1016/j.ejphar.2013.06.038. [DOI] [PubMed] [Google Scholar]
- Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shergalis A., Neamati N. Protein disulfide isomerase. In: Choi S., editor. Encyclopedia of Signaling Molecules. Springer New York; New York, NY: 2016. pp. 1–12. [DOI] [Google Scholar]
- Shojaei S., Suresh M., Klionsky D.J., Labouta H.I., Ghavami S. Autophagy and SARS-CoV-2 infection: apossible smart targeting of the autophagy pathway. Virulence. 2020;11:805–810. doi: 10.1080/21505594.2020.1780088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou J., Kong X., Wang X., Tang Y., Wang C., Wang M., Zhang L., Liu Y., Fei C., Xue F. Tizoxanide inhibits inflammation in LPS-Activated RAW264. 7 macrophages via the suppression of NF-κB and MAPK activation. Inflammation. 2019;42:1336–1349. doi: 10.1007/s10753-019-00994-3. [DOI] [PubMed] [Google Scholar]
- Shou J., Wang M., Cheng X., Wang X., Zhang L., Liu Y., Fei C., Wang C., Gu F., Xue F. Tizoxanide induces autophagy by inhibiting PI3K/Akt/mTOR pathway in RAW264. 7 macrophage cells. Arch Pharm. Res. (Seoul) 2020;43:257–270. doi: 10.1007/s12272-019-01202-4. [DOI] [PubMed] [Google Scholar]
- Shu T., Huang M., Di Wu Y.R., Zhang X., Han Y., Mu J., Wang R., Qiu Y., Zhang D.-Y., Zhou X. SARS-Coronavirus-2 Nsp13 possesses NTPase and RNA helicase activities that can Be inhibited by bismuth salts. Virol. Sin. 2020;35:321–329. doi: 10.1007/s12250-020-00242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi H.K., Mehra M.R. COVID-19 illness in native and immunosuppressed states: a clinical–therapeutic staging proposal. J. Heart Lung Transplant. 2020;39:405–407. doi: 10.1016/j.healun.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K.-L., Chan C.-P., Kok K.-H., Woo P.C., Jin D.-Y. Comparative analysis of the activation of unfolded protein response by spike proteins of severe acute respiratory syndrome coronavirus and human coronavirus HKU1. Cell Biosci. 2014;4:3. doi: 10.1186/2045-3701-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrell F.J., Szklarz M., Abdul Azeez K.R., Elkins J.M., Knapp S. Family-wide structural analysis of human numb-associated protein kinases. Structure. 2016;24:401–411. doi: 10.1016/j.str.2015.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy M., Keser G., Atagündüz P., Tabak F., Atagündüz I., Kayhan S. Cytokine storm in COVID-19: pathogenesis and overview of anti-inflammatory agents used in treatment. Clin. Rheumatol. 2020;39:405–407. doi: 10.1016/j.healun.2020.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachulski A.V., Santoro M.G., Piacentini S., Belardo G., Frazia S.L., Pidathala C., Row E.C., Berry N.G., Iqbal M., Allman S.A. Second-generation nitazoxanide derivatives: thiazolides are effective inhibitors of the influenza A virus. Future Med. Chem. 2018;10:851–862. doi: 10.4155/fmc-2017-0217. [DOI] [PubMed] [Google Scholar]
- Stebbing J., Phelan A., Griffin I., Tucker C., Oechsle O., Smith D., Richardson P. COVID-19: combining antiviral and anti-inflammatory treatments. Lancet Infect. Dis. 2020;20:400–402. doi: 10.1016/S1473-3099(20)30132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoermer M. Homology models of the papain-like protease PLpro from coronavirus 2019-nCoV. 2020. [DOI]
- Stolf B.S., Smyrnias I., Lopes L.R., Vendramin A., Goto H., Laurindo F.R., Shah A.M., Santos C.X. Protein disulfide isomerase and host-pathogen interaction. The Scientific World Journal. 2011;11:1749–1761. doi: 10.1100/2011/289182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutzmann G.E., Mattson M.P. Endoplasmic reticulum Ca2+ handling in excitable cells in health and disease. Pharmacol. Rev. 2011;63:700–727. doi: 10.1124/pr.110.003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureda A., Alizadeh J., Nabavi S.F., Neagoe I.B., Cismaru C.A., Jeandet P., Los M.J., Clementi E., Nabavi S.M., Ghavami S. Endoplasmic reticulum as a potential therapeutic target for covid-19 infection management? Eur. J. Pharmacol. 2020;882:173288. doi: 10.1016/j.ejphar.2020.173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahir ul Qamar M., Alqahtani S.M., Alamri M.A., Chen L.-L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020;10 doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang T., Bidon M., Jaimes J.A., Whittaker G.R., Daniel S. Coronavirus membrane fusion mechanism offers as a potential target for antiviral development. Antiviral Res. 2020 doi: 10.1016/j.antiviral.2020.104792. 104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantawy M.A., El-Sherbeeny N.A., Helmi N., Alazragi R., Salem N., Elaidy S.M. Synthetic antiprotozoal thiazolide drug induced apoptosis in colorectal cancer cells: implications of IL-6/JAK2/STAT3 and p53/caspases-dependent signaling pathways based on molecular docking and in vitro study. Mol. Cell. Biochem. 2020;469:143–157. doi: 10.1007/s11010-020-03736-4. [DOI] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Rénia L., MacAry P.A., Ng L.F. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchouaffi-Nana F., Ballard T.E., Cary C.H., Macdonald T.L., Sifri C.D., Hoffman P.S. Nitazoxanide inhibits biofilm formation by Staphylococcus epidermidis by blocking accumulation on surfaces. Antimicrob. Agents Chemother. 2010;54:2767–2774. doi: 10.1128/AAC.00901-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas N.J., Carcillo J.A., Herzer W.A., Mi Z., Jackson E.K. Chronic type IV phosphodiesterase inhibition protects glomerular filtration rate and renal and mesenteric blood flow in a zymosan-induced model of multiple organ dysfunction syndrome treated with norepinephrine. J. Pharmacol. Exp. Ther. 2001;296:168–174. [PubMed] [Google Scholar]
- Tilmanis D., Koszalka P., Barr I.G., Rossignol J.-F., Mifsud E., Hurt A.C. Host-targeted Nitazoxanide has a high barrier to resistance but does not reduce the emergence or proliferation of oseltamivir-resistant influenza viruses in vitro or in vivo when used in combination with oseltamivir. Antiviral Res. 2020;104851 doi: 10.1016/j.antiviral.2020.104851. [DOI] [PubMed] [Google Scholar]
- Trabattoni D., Gnudi F., Ibba S.V., Saulle I., Agostini S., Masetti M., Biasin M., Rossignol J.-F., Clerici M. Thiazolides elicit anti-viral innate immunity and reduce HIV replication. Sci. Rep. 2016;6:27148. doi: 10.1038/srep27148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turano C., Coppari S., Altieri F., Ferraro A. Proteins of the PDI family: unpredicted non-ER locations and functions. J. Cell. Physiol. 2002;193:154–163. doi: 10.1002/jcp.10172. [DOI] [PubMed] [Google Scholar]
- Uehara T., Nakamura T., Yao D., Shi Z.-Q., Gu Z., Ma Y., Masliah E., Nomura Y., Lipton S.A. S-nitrosylated protein-disulphide isomerase links protein misfolding to neurodegeneration. Nature. 2006;441:513–517. doi: 10.1038/nature04782. [DOI] [PubMed] [Google Scholar]
- Ulferts R., Imbert I., Canard B., Ziebuhr J. Molecular Biology of the SARS-Coronavirus. Springer; 2010. Expression and functions of SARS coronavirus replicative proteins; pp. 75–98. [Google Scholar]
- Ulrich H., Pillat M.M. CD147 as a target for COVID-19 treatment: suggested effects of azithromycin and stem cell engagement. Stem Cell Rev. Rep. 2020:1–7. doi: 10.1007/s12015-020-09976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakifahmetoglu-Norberg H., Xia H., Yuan J. Pharmacologic agents targeting autophagy. J. Clin. Invest. 2015;125:5–13. doi: 10.1172/jci73937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wart H.E., Birkedal-Hansen H. The cysteine switch: a principle of regulation of metalloproteinase activity with potential applicability to the entire matrix metalloproteinase gene family. Proc. Natl. Acad. Sci. U.S.A. 1990;87:5578–5582. doi: 10.1073/pnas.87.14.5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanpouille-Box C., Hoffmann J.A., Galluzzi L. Pharmacological modulation of nucleic acid sensors — therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 2019;18:845–867. doi: 10.1038/s41573-019-0043-2. [DOI] [PubMed] [Google Scholar]
- Venet F., Chung C.-S., Huang X., Lomas-Neira J., Chen Y., Ayala A. Lymphocytes in the development of lung inflammation: a role for regulatory CD4+ T cells in indirect pulmonary lung injury. J. Immunol. 2009;183:3472–3480. doi: 10.4049/jimmunol.0804119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhratsky A., Petersen O.H. The endoplasmic reticulum as an integrating signalling organelle: from neuronal signalling to neuronal death. Eur. J. Pharmacol. 2002;447:141–154. doi: 10.1016/S0014-2999(02)01838-1. [DOI] [PubMed] [Google Scholar]
- Verma S., Dixit R., Pandey K.C. Cysteine proteases: modes of activation and future prospects as pharmacological targets. Front. Pharmacol. 2016;7 doi: 10.3389/fphar.2016.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor P., Sarada D., Ramkumar K.M. Pharmacological activation of Nrf2 promotes wound healing. Eur. J. Pharmacol. 2020;886:173395. doi: 10.1016/j.ejphar.2020.173395. [DOI] [PubMed] [Google Scholar]
- Vitiello A., Ferrara F. Correlation between renin-angiotensin system and Severe Acute Respiratory Syndrome Coronavirus 2 infection: what do we know? Eur. J. Pharmacol. 2020;883:173373. doi: 10.1016/j.ejphar.2020.173373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Jiang M., Chen X., Montaner L.J. Cytokine storm and leukocyte changes in mild versus severe SARS-CoV-2 infection: review of 3939 COVID-19 patients in China and emerging pathogenesis and therapy concepts. J. Leukoc. Biol. 2020;108:17–41. doi: 10.1002/JLB.3COVR0520-272R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO WHO coronavirus disease (COVID-19) dashboard. 2020. https://covid19.who.int/?gclid=EAIaIQobChMIleGg_tKp6gIVCB4rCh26ggk4EAAYASAAEgKEhvD_BwE
- Wong H.H., Sanyal S. Manipulation of autophagy by (+) RNA viruses. Seminars in Cell & Developmental Biology, Autophagy in host-microbe interactions. 2020;101:3–11. doi: 10.1016/j.semcdb.2019.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xian M., Chen X., Liu Z., Wang K., Wang P.G. Inhibition of papain by S-nitrosothiols. Formation of mixed disulfides. J. Biol. Chem. 2000;275:20467–20473. doi: 10.1074/jbc.M001054200. [DOI] [PubMed] [Google Scholar]
- Xiu S., Dick A., Ju H., Mirzaie S., Abdi F., Cocklin S., Zhan P., Liu X. Inhibitors of SARS-CoV-2 entry: current and future opportunities. J. Med. Chem. 2020 doi: 10.1021/acs.jmedchem.0c00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Shi P.-Y., Li H., Zhou J. Broad spectrum antiviral agent niclosamide and its therapeutic potential. ACS Infect. Dis. 2020;6:909–915. doi: 10.1021/acsinfecdis.0c00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Sankar S., Neamati N. Protein disulfide isomerase: a promising target for cancer therapy. Drug Discov. Today. 2014;19:222–240. doi: 10.1016/j.drudis.2013.10.017. [DOI] [PubMed] [Google Scholar]
- Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Shen H.-M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int. J. Biol. Sci. 2020;16:1724–1731. doi: 10.7150/ijbs.45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R., Chen L., Lan R., Shen R., Li P. Computational screening of antagonists against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. Int. J. Antimicrob. Agents. 2020;56:106012. doi: 10.1016/j.ijantimicag.2020.106012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuki K., Fujiogi M., Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin. Immunol. 2020;215:108427. doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F., Guo Y., Yin M., Chen X., Deng G. Association of inflammatory markers with the severity of COVID-19: a meta-analysis. Int. J. Infect. Dis. 2020;96:467–474. doi: 10.1016/j.ijid.2020.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.-Y. Coronaviruses—drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]