Highlights
-
•
We evaluated a bead-based Luminex assay for the detection of SARS-CoV-2 antibodies.
-
•
A combination of the nucleocapsid protein and the receptor-binding domain results in a highly sensitive (96 %) and specific (99 %) assay that can detect positive cases at least five months after infection.
-
•
Because we find that IgG and neutralizing antibody levels can differ significantly between severe/mild and recent/old cases, we argue that serological tests should be validated on samples from all groups before use in serosurveillance studies.
Keywords: Sars-CoV-2, Bead-based assay, Serosurveillance, Luminex antibody test, Virus neutralization test
Abstract
Large-scale serosurveillance of severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) will only be possible if serological tests are sufficiently reliable, rapid and affordable. Many assays are either labour-intensive and require specialised facilities (e.g. virus neutralization assays), or are expensive with suboptimal specificity (e.g. commercial ELISAs and RDTs). Bead-based assays offer a cost-effective alternative and allow for multiplexing to test for antibodies against multiple antigens and against other pathogens. Here, we compare the performance of spike (S) and nucleocapsid (NP) antigens for the detection of SARS-CoV-2 specific IgG, IgM and IgA antibodies in a panel of sera that includes recent (up to six weeks after symptom onset, severe n = 44; and mild cases n = 52) and old infections (five months after symptom onset, mild n = 104), using a Luminex-bead based assay and comparison to a virus neutralization test. While we show that neutralizing antibody levels are significantly lower in mild than in severe cases, we demonstrate that a combination of the recombinant nucleocapsid protein (NP) and receptor-binding domain (RBD) results in highly specific (99 %) IgG antibody detection five months after infection in 96 % of cases. Although most severe Covid-19 cases developed a clear IgM and IgA response, titers fell below the detection threshold in more than 20 % of mild cases in our bead-based assay. In conclusion, our data supports the use of RBD and NP for the development of SARS-CoV-2 serological IgG bead-based assays.
1. Introduction
Since December 2019, severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) has spread at an unprecedented speed and scale, resulting in 1,460,000 deaths, 62,800,000 diagnosed (as of 30/11/2020) (Johns Hopkins University, 2020) and likely many more undiagnosed cases with mild or no symptoms (Centers for disease control and prevention, 2020). While it is now clear that most people develop a protective antibody response after recovery, it remains unknown how long the antibodies remain detectable (Zhang et al., 2020a; Lassaunière et al., 2020; Okba et al., 2020). Such serological data can be used to determine the overall attack rate and level of herd immunity in a given population, and lay the foundation for control and prevention policies.
Serological tests exist in a variety of different formats but not all are appropriate for large-scale serosurveillance (Zhang et al., 2020a; Krammer and Simon, 2020). The gold standard for serological testing remains the virus neutralization test (VNT) (Lassaunière et al., 2020). This assay format is very specific and directly assesses the neutralizing capacity of antibodies in serum (Xun et al., 2020). However, VNTs are also labour-intensive and generally require highly trained staff to work in BSL3 laboratory conditions. In contrast, enzyme-linked immunosorbent assays (ELISAs) require less trained operators and allow high-throughput screening, but are usually less specific due to cross-reactivity with other coronaviruses or other pathogens. Furthermore, ELISAs are relatively expensive since sufficient recombinant antigens needs to be produced (Amanat et al., 2020). Nevertheless, many commercial ELISAs became rapidly CE-labelled and are currently used for serosurveillance studies (Lassaunière et al., 2020). Since ELISAs as well as VNTs have important limitations for large-scale serosurveillance, microsphere bead-based assays using the Xmap Luminex technology have been increasingly developped (Dobaño et al., 2020; Rosado et al., 2020). This high-throughput platform allows the simultaneous detection of antibodies against different antigens from SARS-CoV-2, which can significantly increase the specificity of serological testing, in contrast to ELISA that usually includes only one antigen. Additional advantages of the bead-based assays are the need for lower serum amounts (<1 μl) and the lower cost (as less recombinant antigen is required) (Kerkhof et al., 2015).
Evaluating a set of compatible immunogenic antigens is essential for the development of these multiplex bead-based assays. For many SARS-CoV-2 antibody tests, the main targets are the large spike glycoprotein (S) and the nucleocapsid protein (NP) (Taskin Tok et al., 2017). The S protein is a trimeric class I fusion protein that consists of two subunits, namely S1 and S2 (Wang et al., 2020; Walls et al., 2020). The S1 protein mediates binding to host cells via interactions with the human receptor angiotensin converting enzyme 2 (ACE2) and is very immunogenic with its receptor-binding domain (RBD) as the main target for neutralizing antibodies (Premkumar et al., 2020). The S2 subunit regulates fusion of the viral and host cellular membrane. The S protein, therefore, is an important target for the development of medical treatments and vaccines due to its role in cell binding and entry (Padron-Regalado, 2020). The NP plays a vital role in the transcription and replication of the virus, and is suggested to be more sensitive than the S protein for detecting early infections (Taskin Tok et al., 2017; Burbelo et al., 2015). Here, we evaluate the performance of these four antigens (NP, RBD, S1, S1S2) for the detection of SARS-CoV-2 IgG, IgM and IgA antibodies in a bead-based assay using sera from severe and mild cases in the early convalescent phase (<6weeks) and later during the infection (>5 months). We also examine correlations between levels of SARS-CoV-2 neutralizing antibodies and those of antigen-binding antibodies measured by our Luminex assay.
2. Methods
2.1. Human serum samples
We used a panel of 296 serum samples: 44 samples from 22 hospitalized COVID-19 patients (severe cases) at the University Hospital of Antwerp (UZA), 156 samples from COVID-19 positive healthcare workers confirmed to have antibodies by VNT (mild/asymptomatic cases), and 96 leftover samples from the travel clinic of the Institute of Tropical Medicine (ITM) in Antwerp that we expect to be antibody negative because they were sampled prior to the pandemic in November 2019 (negative cases). Samples from severe cases were collected between March,3th and April, 6th from RT-qPCR confirmed COVID-19 cases (median days post symptom onset = 17, Q1 = 10 and Q3 = 22). Of these, 22 severe cases were sampled two times over a period of one or two weeks. Samples from 52 healthcare workers were collected <6 weeks post symptom onset between April,22th and April,26th in 17 different hospitals in Belgium (median days post symptom onset = 22, Q1 = 18 and Q3 = 35). Samples from 104 healthcare workers were collected five months post symptom onset (Aug 2020). Infectivity of health care workers from the second group was also confirmed via a nasopharyngeal swap and RT-qPCR (Corman et al., 2020). All samples from healthcare workers belonged to different individuals.
2.2. SARS-CoV-2 neutralization test
Serial dilutions of heat-inactivated serum (1/50-1/1600 in assay medium consisting of EMEM supplemented with 2 mM l-glutamine, 100 U/ml - 100 μg/ml of Penicillin-Streptomycin and 2 % foetal bovine serum) were incubated with 3xTCID100 of a primary isolate of SARS-CoV-2 during 1 h (37 °C / 7 % CO2). Sample-virus mixtures and virus/cell controls were added to Vero cells (18.000cells/well) in a 96well plate and incubated for 5 days (37 °C / 7 % CO2). The cytopathic effect caused by viral growth was scored microscopically. The Reed-Muench method was used to calculate the neutralising antibody titre that reduced the number of infected wells by 50 % (NT50) or 90 % (NT90) (Reed and Muench, 1938), these values were used as proxy for the neutralizing antibody concentration in each sample.
2.3. Luminex bead-based immunoassay
We ordered commercially available recombinant NP, RBD, S1 and the complete Spike unit (S1S2) antigens derived from SARS-CoV-2 at Sino Biological (BIOCONNECT, Huissen, The Netherlands). The lyophilized proteins were resuspended in a buffer according to the manufacturer’s instructions and stored until use. Each antigen was coupled to a maximum of 1.25 × 10^6 (Krammer and Simon, 2020) paramagnetic MAGPLEX COOH-microsphere beads from Luminex Corporation (Austin, TX), as described previously (Kerkhof et al., 2015; Ambrosino et al., 2010). Different concentrations of binding-antigens were titrated and we choose a concentration that yield 70–80 % of the maximum signal, as this should provide the largest linear dynamic range for the assay (Angeloni et al., 2013). Consequently, we coupled 1 μg of RBD, S1 and S1S2 and 2 μg of NP to 1.25 × 10^6 (Krammer and Simon, 2020) beads for IgG detection. Because we obtained the best results with RBD and NP (and observed high correlations between RBD and S1/S1S2), we continued only with these antigens for the further analyses. We increased the antigen concentrations for IgA detection to 2 μg for RBD and 4 μg for NP.
We followed the protocol as described in Ayouba et al. 2020 (Ayouba et al., 2020) for the assay. In brief, a microsphere working mixture was prepared in a hypertonic phosphate buffered saline-BSA (PBS-BN) solution with a concentration of 2000 beads/antigen/well. To choose an appropriate serum concentration, titers were measured in serial dilutions (1/150-1/6400). We observed that a 1/300 serum dilution gave the best signal to noise ratio for IgG and IgM and 1/150 for IgA (Supplementary Figs. 1 and 2). For the IgM detection in serum, we added Gullsorb™ IgG inactivation reagent in a 1/10 dilution. Beads and diluted sera were added to each well in a final volume of 150 μl. Plates were incubated at room temperature for two hours in the dark for IgG and IgA and for four hours for IgM, and then washed with 200 μl/well of PBS-BN buffer. Reactions were read after incubation (30 min) with a biotin-labelled anti-human IgG, IgA or IgM secondary antibody and streptavidin-R-phycoerythrin conjugate (10 min) using a Luminex® 100/200 analyzer. Results were expressed as median fluorescent intensities (MFI).
2.4. Statistical evaluation and diagnostic performance
We assessed the sensitivity and specificity of the Luminex bead-based assay by looking at Receiver Operating Characteristics (ROC) curves for the different groups of infected COVID-19 patients: Recent-Severe (RS), Recent-Mild (RM) and Old-Mild (OM). First, sensitivity and specificity were traded-off to calculate cut-off values for SARS-CoV2 antigens in single-plex. To test which combinations of antigens could best improve the accuracy of the test in multiplex, we checked ROC curves calculated by supervised machine learning Random Forest (RF) algorithm models, as implemented in the R-package ‘randomForest’ (Breiman and Cutler, 2018). Variable (antigen) importance was assessed using the ‘varImplot’ function of the same package. Antigens with the lowest ‘mean decrease in accuracy’ and ‘mean decrease in Gini’ were removed from the bead set. Selection of antigens was performed based on the Area Under a Curve (AUC) values. Because it was clear that including S1 and S1S2 antigens to the bead set did not improve the classification performance of the IgG test (Table 1 ), we excluded these antigens for the statistical analyses.
Table 1.
Performance of the IgG, IgA and IgM assays. AUC and sensitivities (sens) are presented for different categories of infection for a low (>90 %) and high (>99 %) specificity (spec) target.
| Recent infections (<6 weeks) |
Old infections (>5 months) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Severe |
Mild |
Mild |
||||||||
| Isotype | Antigen | AUC | Sens (at 90 % Spec) | Sens (at 99 % Spec) | AUC | Sens (at 90 % Spec) | Sens (at 99 % Spec) | AUC | Sens (at 90 % Spec) | Sens (at 99 % Spec) |
| IgG | RBD | 0.94 | 99 | 91 | 0.93 | 98 | 98 | 0.94 | 99 | 96 |
| S1 | 0.90 | 92 | 84 | 0.92 | 94 | 93 | 0.93 | 95 | 47 | |
| S1S2 | 0.92 | 92 | 90 | 0.91 | 92 | 79 | 0.94 | 99 | 95 | |
| NP | 0.92 | 95 | 85 | 0.93 | 98 | 70 | 0.68 | 95 | 85 | |
| RBD + NP + S1+S1/2 | 0.98 | 99 | 95 | 0.99 | 99 | 99 | 0.99 | 99 | 97 | |
| RBD + NP | 0.98 | 99 | 94 | 0.99 | 99 | 99 | 0.99 | 99 | 96 | |
| IgA | RBD | 0.94 | 97 | 66 | 0.88 | 84 | 47 | 0.66 | 42 | 5 |
| NP | 0.94 | 97 | 92 | 0.77 | 62 | 42 | 0.53 | 15 | 3 | |
| RBD + NP | 0.98 | 97 | 92 | 0.92 | 84 | 60 | 0.69 | 42 | 17 | |
| IgM | RBD | 0.94 | 100 | 72 | 0.79 | 68 | 30 | 0.69 | 65 | 10 |
| NP | 0.94 | 97 | 90 | 0.71 | 53 | 35 | 0.68 | 46 | 19 | |
| RBD + NP | 0.98 | 100 | 90 | 0.79 | 68 | 35 | 0.82 | 65 | 20 | |
We then investigated if mean MFI (NP and RBD) for all isotypes and neutralizing antibody levels differ between the different groups of COVID-19 cases using a Tukey’s test. We corrected for a potential sampling time effect and dependency bias by excluding all samples that were taken from severe COVID-19 cases in the first week after hospitalization. All antibody levels were log-transformed to meet normality assumptions. All P-values were corrected for multiple testing bias using the Bonferroni method. To assess the relationship between the different antigen levels and the neutralizing antibody response, we correlated the NT50 measurements to MFI values using a nonparametric Spearman’s rank (rs) test. All analyses were performed in the statistical software R.3.6.1.
3. Results
3.1. Performance assessment of the Luminex bead-based assay
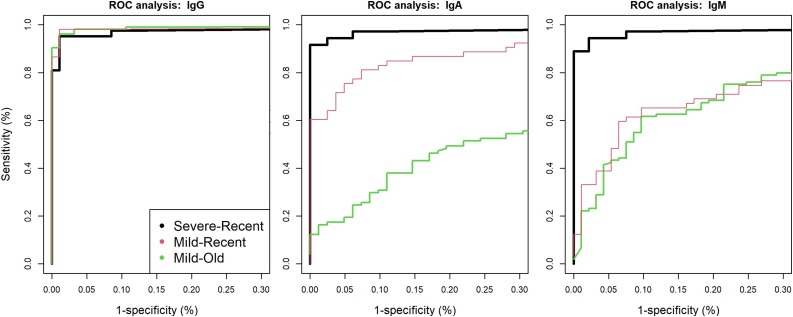
Sensitivity and specificity were first calculated for individual antigens using a ROC analysis (Fig. 1 and Table 1). For the IgG assay, variations in AUC values suggest differences in classification performance for each antigen in single-plex. IgG-RBD had the highest classification performance and IgG-S1 the lowest, while IgG-S1S2 and IgG-NP had a similar classification performance. Subsequently, we used a random forest algorithm to test if combinations of different antigens could improve the classification performance of the assay. While multiplexing all antigens gave the best specificity (99 %) and sensitivity (95–100 %), the removal of IgG-S1 and IgG-S1S2 resulted in an almost similar AUC. The variable importance scores (‘mean decrease in accuracy’ and ‘mean decrease in Gini’) also suggested that IgG-S1 and IgG-S1S2 can be removed from the multiplex without reduction in classification performance (Supplementary Fig. 3). We detected four negative control samples for which the IgG-NP MFI values clearly fell above the 99 % specificity cut-off and one sample for which the IgG-RBD MFI value exceeded the cut-off value. However, the positive IgG-RBD sample was different from the four positive IgG-NP samples, showing that combining IgG-RBD and IgG-NP can significantly increase the specificity of the IgG test (to 100 %). Because the negative serum samples were anonymized, it was impossible to link these non-specific reactivity to any co-infection.
Fig. 1.
Receiver Operating Characteristic (ROC) curves for combinations of RBD and NP antigens in multiplex as calculated by a random forest algorithm for IgG, IgA and IgM isotypes. Different colours represent different groups of recent/old and severe/mild infections.
The sensitivity of the IgA assay was high for recent severe COVID-19 cases at both high and low specificity cut-offs (92 and 97 %, respectively), but lower in recent mild (60 and 84 %, respectively) and old mild cases (<17 % and 42 %, respectively). Similarly, the sensitivity of the IgM assay was high for recent severe COVID-19 cases (90 and 100 %, respectively), but much lower in recent (35 and 68 %, respectively) and old mild cases (20 and 65 %, respectively). These results suggest that IgA and IgM are not useful for follow up beyond the acute phase of the infection, but could be applied to detect re-infection of a patient.
3.2. Differences in antibody levels between severe and mild cases
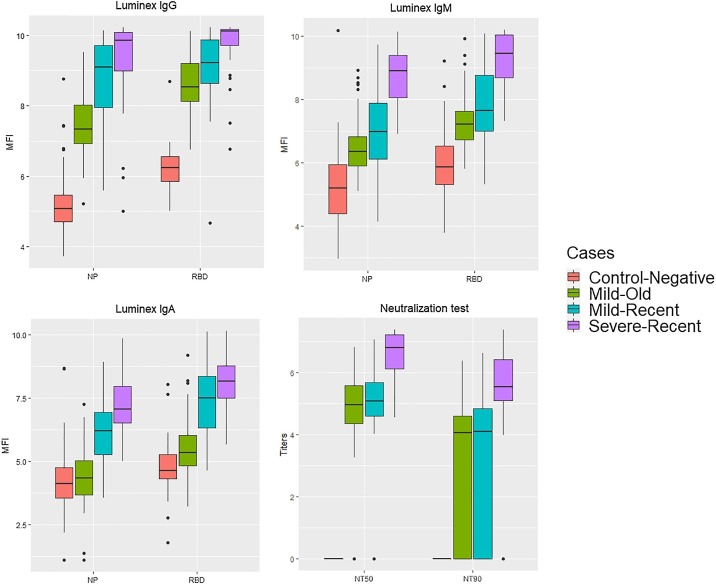
We investigated whether MFI (NP and RBD) and neutralizing antibody levels differ between severe and mild cases, and whether they change over time (Fig. 2 and Table 2 ). Neutralizing antibody levels (NT50 and NT90) were significantly higher for severe than for mild cases, but did not differ between recent and old mild cases. Interestingly, neutralizing antibodies (at NT50) were absent in only two samples five months after infection. IgG antibody levels were significantly lower in the old compared to the recent infections for IgG-NP, while IgG-RBD levels only differed between the severe-recent and mild-old infection groups. Similar as for NT50, all levels for IgG-RBD and most (>96 %) IgG-NP levels remained higher than the cut-off values (at 99 % sensitivity) of the negative controls five months after infection. IgA and IgM antibody levels decreased significantly over time for both IgA- and IgM-RBD and NP, but IgA-NP did not differ significantly between recent severe and mild. In general, antibody levels for both RBD and NP were the highest for recent-severe cases and the lowest for mild-old cases for all isotypes.
Fig. 2.
Boxplots representing differences in log(neutralizing antibodies) and log(MFI levels) for NP and RBD between recent-severe (n = 22), recent-mild (52), old-mild cases (n = 104) and the negative control (n = 96) group for all isotypes.
Table 2.
Bonferroni corrected P-values expressing the effects of disease severity (severe, mild) and time since symptom onset (recent, old) on antibody levels as measured by the Luminex or VNT.
| Antigen | Group | IgG | IgA | IgM | Titer | VNT |
|---|---|---|---|---|---|---|
| RBD | MO vs MR | 0.264 | <0.001 | 0.288 | NT50 | 1 |
| SR vs MR | 0.156 | 0.723 | <0.001 | <0.001 | ||
| SR vs MO | <0.001 | <0.001 | <0.001 | <0.001 | ||
| NP | MO vs MR | 0.006 | <0.001 | 1 | NT90 | 1 |
| SR vs MR | 1 | <0.001 | <0.001 | 0.007 | ||
| SR vs MO | <0.001 | <0.001 | <0.001 | 0.005 |
MO = mild old; MR = mild recent; SR = severe recent.
3.3. Correlations between neutralizing antibody titers and Luminex RBD and NP levels
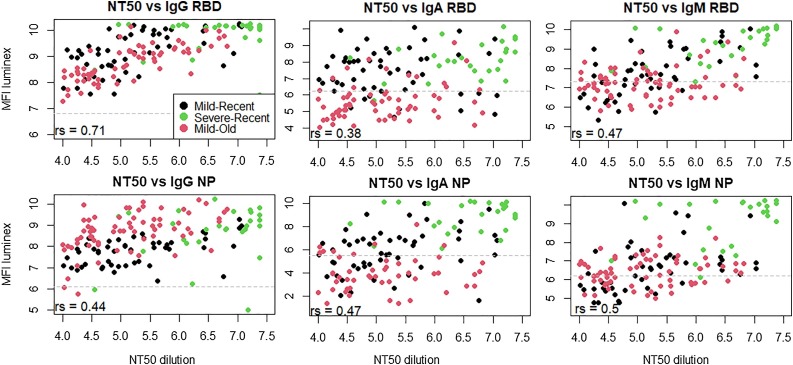
To assess if MFI levels can be used as proxy for the neutralizing capacity of serum, we correlated these levels to the NT50 (Fig. 3 ). We found strong correlations between NT50 and IgG-RBD (rs = 0.71, p < 0.0001). Weaker correlations were observed for IgA and IgM-RBD (rs = 0.38-0.47, p < 0.0001) and for IgG, IgA and IgM-NP (rs = 0.40-0.50, p < 0.0001).
Fig. 3.
Correlations between log(NT50) and log(MFI) levels to RBD and NP for IgG, IgA and IgM, as calculated by the nonparametric Spearman correlation test (rs). Seropositivity cut-off levels are indicated by the dashed grey lines.
4. Discussion
While a plethora of commercial serological tests became available months after the discovery of SARS-CoV-2, researchers have already experienced problems in terms of sensitivity and specificity with many early market commercial tests (Krammer and Simon, 2020). Problems may arise because tests were initially evaluated on sera from severe COVID-19 patients, who are suggested to develop a much higher immune response than mild or asymptomatic cases (Zhang et al., 2020b). Here, we show that neutralizing and binding IgG, IgA and IgM antibody levels are indeed higher for severe than mild cases in the early convalescent phase (<6 weeks) (Fig. 1). Furthermore, given that antibody levels directed to other human coronaviruses are suggested to decrease rapidly after infection (usually after 0.5−1 year) (Edridge et al., 2020a,b) and seroreversion (antibody positive to negative) has already been shown/demonstrated for asymptomatic SARS-CoV-2 infections (Long et al., 2020; Seow et al., 2020; Perreault et al., 2020), it was expected that antibody levels of mild/asymptomatic cases might drop rapidly below the cut-off value of many diagnostic tests within months after infection. Our data, however, show that both neutralizing and IgG antibodies remain detectable in the majority (>96 %) of PCR-confirmed cases at least five months after symptom onset of mild/asymptomatic infections. While it cannot be excluded that seroreversion will happen after this time window and the first re-infections have been noted (Tillett et al., 2020; Selhorst et al., 2020), we highlight that the absence of detectable antibody levels does not necessarily mean that the majority of recovered people will become completely susceptible for COVID-19 (immunity also depends on the T-cell mediated immune responses that might last much longer) (Gutierrez et al., 2020). Nevertheless, a potential decrease in IgG antibody levels might complicate the epidemiological assessment and should be taken into account by mathematical models that aim to understand/predict the transmission dynamics in the population (Rosado et al., 2020; Borremans et al., 2020). Because of the much faster decline in IgM and IgA antibodies (half of them were not detectable five months after infection in our study), these isotypes seem to be useless for serosurveillance purposes. However, they could be relevant to better detect recent infections or to assess if re-infection occurred.
The ROC analyses showed that NP and RBD antigens can be used to develop a highly specific (99 %) and sensitive (96 %) Luminex SARS-CoV-2 antibody detection assay. Although these antigens performed well in single-plex (Table 1), a combination of the two antigens clearly increased the assay’s prediction performance in comparison to the best performing single-plex for the IgG assay (RBD, ΔAUC = 0.04). The increase in specificity is explained by the multiplexing of two antigens in relation to the random forest algorithm. For example, a few non-specific bindings were noted in the negative control samples (4x NP and 1x RBD) (Fig. 2). While these samples would clearly be categorized as positives in a single-plex assay, they were actually categorized as negative by the random forest as the aspecific binding did not happen in the same sample. This aspect makes a multiplex assay appealing for serosurveillance in sub-Saharan Africa, where more infectious diseases occur that can cause false positive results in serological assays (Esbroeck et al., 2016). We highlight that the exclusion of IgG-S1S2 and IgG-S1 is not due to their bad prediction performance, but rather because they behave almost identical as RBD (i.e. they are redundant in the multiplex). The observation that RBD slightly outperformed the other spike antigens in a single-plex can be explained by the fact that this region is less conserved between different coronaviruses in contrast to the whole spike protein, which might increase RBD’s specificity (Edridge et al., 2020b; Premkumar et al., 2011; Du et al., 2009; Ng et al., 2020). Consequently, a combination of NP with RBD/S1 is also put forward in two independent Luminex bead-based assays that were recently published (Dobaño et al., 2020; Ayouba et al., 2020).
Because VNTs are labour-intensive, require biosafety level-3 (BSL-3) conditions and specific training skills, simple serological tests that could predict individual levels of protecting-immunity are highly needed to assess the effect of vaccine campaigns (Premkumar et al., 2020). For this reason, we correlated neutralizing titers (NT50) to antibody levels obtained by this Luminex assay. The highest correlations were observed to the IgG-RBD, suggesting that IgG-RBD levels can be used as proxy to predict levels of neutralizing antibodies. Although IgA and IgM were suggested to contribute to a much larger extend to virus neutralization compared to IgG in the early convalescent phase (Sterlin et al., 2020), we found moderate-to-weak correlations to IgA and IgM in general. This pattern is likely explained by the absence of IgA and IgM later during the infection, while neutralizing antibodies remain clearly present.
In conclusion, we have evaluated a highly sensitive and specific serological assay for the detection of SARS-CoV- 2 IgG antibodies using a robust and high-throughput technology. The assay predicts relatively accurately neutralizing antibody levels in the early convalescent phase and later in the infection. We also found that neutralizing and binding IgG antibodies remain detectable at least five months after the infection, but that titers differ between severe and mild/asymptomatic COVID-19 cases. The latter highlights that serological tests are best evaluated on serum panels that include mild and asymptomatic cases before use in large-scale serosurveillance settings.
Ethics statement
Approval to sample from COVID-19 cases was obtained from the Ethical committee of the University of Ghent (BC-07587), local committees of each participating hospital and all participants provided consent to participate. All serum samples from severe and mild/asymptomatic cases underwent a viral inactivation protocol by heating at 56 °C for 30 min. The use of prepandemic leftover samples was approved by ITM’s internal review board. We declare that the planning conduct and reporting of the study was in line with the Declaration of Helsinki, as revised in 2013.
Funding
The work was funded by a European & Developing Countries Clinical Trials Partnership (EDCTP) project (Africover: RIA2020EF-3031), the Research Foundation Flanders (FWO) (G0G4220N and G054820N), the Health Care Worker seroprevalence study (Sciensano/ITM), NCT04373889 and intramural funds from the Institute of Tropical Medicine Antwerp. Joachim Mariën is a member of the Outbreak Research Team which is funded by the Department of Economy, Science and Innovation of the Flemish government in Belgium (EE145 4150).
Author statement
Conceived the study: JMa, MAW and KA. Wrote the paper: JMa, KA. Performed the lab experiments: JMa, JMi, LH, AC, KK, and NF. Performed the statistical analyses: JMa. Supervised data collection and laboratory work: ID, HJ, ED, LM, MVE and KA. All authors read and approved the final manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.jviromet.2020.114025.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- Amanat F. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1101/2020.03.17.20037713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosino E. A multiplex assay for the simultaneous detection of antibodies against 15 Plasmodium falciparum and Anopheles gambiae saliva antigens. Malar. J. 2010;9:1–12. doi: 10.1186/1475-2875-9-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeloni S. 2013. Luminex xMAP Cookbook; pp. 1–116.www.luminexcorp.com [Google Scholar]
- Ayouba A. Multiplex detection and dynamics of IgG antibodies to SARS-CoV2 and the highly pathogenic human Coronaviruses SARS-CoV and MERS-CoV. J. Clin. Virol. 2020;104521 doi: 10.1016/j.jcv.2020.104521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borremans B. Quantifying antibody kinetics and RNA shedding during early-phase SARS-CoV-2 infection. eLife. 2020;9 doi: 10.1101/2020.05.15.20103275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L., Cutler A. 2018. Package ‘randomForest’. Cran Repos. [DOI] [Google Scholar]
- Burbelo P.D. Detection of Nucleocapsid Antibody to SARS-CoV-2 is More Sensitive than Antibody to Spike Protein in COVID-19 Patients. arXiv. 2015;53:1689–1699. [Google Scholar]
- Centers for disease control and prevention . 2020. Commercial Laboratory Seroprevalence Survey Data.https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/commercial-lab-surveys.html Available at: [Google Scholar]
- Corman V.M. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:1–8. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobaño C. Highly sensitive and specific multiplex antibody assays to quantify immunoglobulins M, A and G against SARS-CoV-2 antigens. bioRxiv. 2020 doi: 10.1101/2020.06.11.147363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L. The spike protein of SARS-CoV – a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edridge A.W.D. Human coronavirus reinfection dynamics: lessons for SARS-CoV-2. Medrxiv. 2020:1–10. doi: 10.1101/2020.05.11.20086439. [DOI] [Google Scholar]
- Edridge A.W.D. Coronavirus protective immunity is short-lasting. medRxiv. 2020 doi: 10.1101/2020.05.11.20086439. 05.11.20086439. [DOI] [PubMed] [Google Scholar]
- Esbroeck M., Meersman K., Michiels J., Ariën K., Bossche D., Van den Specificity of Zika virus ELISA: interference with malaria. Euro Surveill. 2016;29:252–262. doi: 10.2807/1560-7917.ES.2016.21.21.30237. [DOI] [PubMed] [Google Scholar]
- Gutierrez L., Beckford J., Alachkar H. Deciphering the TCR repertoire to solve the COVID-19 mystery. Trends Pharmacol. Sci. 2020:1–13. doi: 10.1016/j.tips.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns Hopkins University . 2020. COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE)https://coronavirus.jhu.edu/map.html [DOI] [Google Scholar]
- Kerkhof K. Implementation and application of a multiplex assay to detect malaria-specific antibodies: a promising tool for assessing malaria transmission in Southeast Asian pre-elimination areas. Malar. J. 2015;14:1–14. doi: 10.1186/s12936-015-0868-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F., Simon V. Serology assays to manage COVID-19. Science. 2020;1227:1–5. doi: 10.1126/science.abc1227. [DOI] [PubMed] [Google Scholar]
- Lassaunière R. Evaluation of nine commercial SARS-CoV-2 immunoassays. medRxiv. 2020:1–15. [Google Scholar]
- Long Q.X. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Ng K. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science (80-.) 2020;21:1–9. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okba N.M.A. SARS-CoV-2 specific antibody responses in COVID-19 patients. medRxiv. 2020 doi: 10.1101/2020.03.18.20038059. PP R118105. [DOI] [Google Scholar]
- Padron-Regalado E. Vaccines for SARS-CoV-2: lessons from other coronavirus strains. Infect. Dis. Ther. 2020;9:255–274. doi: 10.1007/s40121-020-00300-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault J. Waning of SARS-CoV-2 RBD antibodies in longitudinal convalescent plasma samples within four months after symptom onset. Blood. 2020;136(22) doi: 10.1182/blood.2020008367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premkumar L. The RBD of the spike protein of sars-group coronaviruses is a highly specific target of SARS-CoV-2 antibodies but not other pathogenic human and animal coronavirus antibodies. medxriv. 2011:10–12. [Google Scholar]
- Premkumar L. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5:1–9. doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L., Muench H.A. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 1938;27:493–497. [Google Scholar]
- Rosado J., Cockram C., Merkling S.H., Demeret C., Meola A. Serological signatures of SARS-CoV-2 infection: Implications for antibody-based diagnostics. medRxiv. 2020:1–30. [Google Scholar]
- Selhorst P. Symptomatic SARS-CoV-2 re-infection of a health care worker in a Belgian nosocomial outbreak despite primary neutralizing antibody response. Affiliations : Medrxiv. 2020;32 [Google Scholar]
- Seow Jeffrey, Graham C., Merrick B., Acors S., Doores K. Longitudinal evaluation and decline of antibody responses in SARS-CoV-2 infection 2. medRxiv. 2020:1–24. [Google Scholar]
- Sterlin A.D., Mathian A., Gorochov G. IgA dominates the early neutralizing antibody response to SARS-CoV-2. medRxiv. 2020:25–28. [Google Scholar]
- Taskin Tok T., Tatar G., Tugba T.T. Structures and functions of coronavirus proteins: molecular modeling of viral nucleoprotein-International Journal of Virology & Infectious Diseases International Journal of Virology & Infectious Diseases. Int. J. Virol. Infect. Dis. 2017;2:1–7. [Google Scholar]
- Tillett R.L. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect. Dis. 2020;3099:1–7. doi: 10.1016/S1473-3099(20)30764-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C. Structure, function, and Antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:1–6. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xun J. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered 2 patient cohort and their implications. Lancet. 2020 (preprint) [Google Scholar]
- Zhang W. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. Early viral clearance and antibody kinetics of COVID-19 among asymptomatic carriers. medRxiv. 2020 doi: 10.3389/fmed.2021.595773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.