Abstract
Hydrogel nanoparticles (also known as nanogels) have been utilized for a wide range of applications including analytics, sensors, drug delivery, immune engineering, and biotechnology. While these types of nanoparticles can be characterized using standard colloidal characterization techniques, degradation profiles typically must be inferred from those of bulk gels with the same formulation, typically by applying swelling ratios and rheological measurements that tend to severely underestimate nanoparticle degradation rates. Herein, we present an analysis of the degradation via ester hydrolysis of poly(ethylene glycol) diacrylate (PEGDA)-based hydrogel nanoparticles in water, varied pH conditions, and redox environments. We perform this characterization using thermogravimetric analysis and mass spectrometry to analyze rates of degradation and products released, respectively, and compare results to those for equivalent bulk gel formulations. Our findings show that PEGDA-based nanoparticles display significant mass loss over time accompanied by negligible changes in hydrodynamic diameter, indicating a bulk mode of degradation. Nanoparticle mass loss occurs at a much higher rate than for bulk gels under comparable incubation conditions, confirming that bulk gel degradation serves as a poor surrogate for nanoparticle degradation. We further demonstrate that the incorporation of other diacrylate-based co-monomers drastically accelerates nanoparticle degradation rates. Through formulation considerations of co-monomer content and weight percent of PEGDA, we demonstrate the ability to tune the degradation rates of PEGDA-based nanoparticles on a range of hours to weeks. These findings highlight critical design considerations for enhancing the tunability and utility of PEGDA hydrogel nanoparticles and introduce a rigorous framework for the characterization of nanogel degradation.
Graphical abstract
Introduction
Hydrogel nanoparticles continue to attract attention for a variety of applications, including superabsorbent materials, chemical separations, sensors, and pharmaceutical drug delivery.1–6 These nanoparticles (also known as nanogels) are on the order of tens to hundreds of nanometers in size and can be readily optimized both chemically and physically for their intended application.7 Such nanoparticles are typically synthesized using hydrophilic and even degradable crosslinkers, leading to a high degree of swelling in aqueous environments both initially and over time.8,9 A wide variety of synthetic approaches and polymer architectures have been developed to generate nanogels capable of encapsulating cells and small molecular cargoes, directing self-assembly, and exhibiting stimuli-responsive behavior, all of which are critical for tailoring the nanoparticles for their intended application.6,10–12
For biomedical applications in particular, hydrogel nanoparticles offer considerable advantages over alternative, “hard” nanoparticle materials. Studies of biocompatible nanogels (e.g. poly(ethylene glycol) diacrylate [PEGDA]) have demonstrated the importance of particle modulus in tuning particle circulation time within the bloodstream, where softer nanoparticles have greatly extended circulation half-times as compared to more rigid nanoparticles of the same chemical species.13,14 PEGDA nanoparticles have also been shown to be immunologically inert,15,16 which contrasts with properties of commonly used biodegradable polyesters such as poly(lactic-co-glycolic acid) [PLGA] that are hydrophobic and yield acidic degradation products17 that can have immunomodulatory effects. PEGDA nanogels are readily formed through chain-growth polymerization and can accommodate acrylate co-monomers that provide additional functionality including pendant chemical handles for post-synthetic modification of the nanoparticle and degradation control using enzyme-sensitive, acid-labile, photolabile, and redox-sensitive crosslinkers.10,18,19 Inclusion of redox-sensitive crosslinkers in particular can aid in increasing nanoparticle degradability because of the abundance of glutathione (GSH), a biological reducing agent, in intracellular compartments. This can dramatically increase the otherwise lengthy degradation rate of PEGDA gels, which degrade by bulk erosion via the hydrolysis of ester linkages within the gel network.
Characterization of nanogel properties used for these applications is particularly important for establishing key structure-property relationships that ultimately dictate their function and performance; yet, effective characterization of these properties remain a challenge for many nanogel systems. This is especially true for the determination of the degradation profile of hydrogel nanoparticles for their use in drug delivery and immune engineering, where control of degradation rate and small molecule release kinetics is paramount in tuning the particle lifetime and intended biological response. Using standard colloidal techniques such as dynamic light scattering (DLS) and gel permeation chromatography (GPC), it is relatively straightforward to observe nanoparticle degradation from widely used materials that exhibit surface erosion, such as polycaprolactone (PCL);20 however, it is much more difficult to observe nanogel degradation since their mechanism of degradation is typically hydrolytic bulk degradation.21 Characterizing this type of degradation on the nanoscale remains analytically challenging, leaving nanogel degradation profiles to be inferred from the degradation profiles of bulk gel with the same formulation using swelling ratios and rheological measurements.9,22 Accordingly, the degradation characterization of many commonly employed hydrogel nanoparticles, including the widely used PEGDA platform, remains based on the assumption that bulk material characterization is representative of the nanoparticles, which has been shown not to be the case for other systems.23,24 There remains a need to directly characterize the rate and nature of the degradation of PEGDA nanoparticle systems and establish their changing physiochemical properties during this process.
Herein, we utilize a range of analytical techniques to directly probe the degradation of PEGDA-based hydrogel nanoparticles and determine degradation timescales for commonly utilized PEGDA formulations. We employ a series of colloidal nanoparticle characterization techniques and evaluate common approaches to infer hydrogel nanoparticle degradation profiles, including thermogravimetric analysis (TGA) and dynamic light scattering (DLS). Notably, we evaluate degradation in relevant pH and redox-reactive environments and compare nanoparticle findings to bulk gel characterization. Lastly, we utilize matrix-assisted laser desorption ionization (MALDI) to determine the nature of the degradation products from both nanoparticles and bulk gels to elucidate the nature of the resultant species present following breakdown of these materials. Through our detailed analysis, we aim to provide a comprehensive understanding of PEGDA hydrogel nanoparticle degradation that will be useful for similar formulations using these polymers and also establish a characterization framework that can be implemented for a wide range of nanogels.
Experimental
Nanoparticle Synthesis
PEGDA-based nanoparticles were generated by mixing a pre-particle polar phase, composed of PEGDA 700 (Mn = 700), charge-stabilizing functional monomer 2-carboxyethyl acrylate (CEA), and photoinitiator diphenyl(2,4,6-trimethylbenzoyl)phosphine oxide (PI) in methanol with 1 mL of a continuous, nonpolar phase of silicone oil AP1000. The amounts of pre-particle phase varied based on the desired properties of the particles as noted in Table 1 with the total volume of the polar phase being 100 μL. Additional formulations included 1,6-hexanediol dimethacrylate (HDDMA) or bis(2-methacryloyl)oxyethyl disulfide (DSDMA) in addition to the aforementioned components as shown in Table 1. For each formulation, we controlled the weight percentage of the solids (combination of PEGDA, CEA, and PI, as well co-monomer) in polar phase of the emulsion to evaluate either 50 wt% or 30 wt% solids.14,25 We also evaluated 50 wt% formulations that had either 10% or 30% of the moles of PEGDA replaced by the alternative crosslinkers of HDDMA and DSDMA. The polar phase was vortex mixed with the nonpolar phase for 1 minute and then a reverse mini-emulsion generated by sonicating the mixture for the times denoted in Table S1. The emulsion was then irradiated with UV light (from an APM LED UV Cube with a wavelength of 365 nm at a distance of ~28 cm from the light source, ~5–10 mW/cm2) for the times denoted in Table S1. Following polymerization, 800 μL of hexanes were added to the emulsion. The particles were centrifuged at 18,200 RCF for 10 minutes and sequentially washed two more times with ethanol (2x). The final suspension was resuspended in the solution of choice, either water, a 0.5 M sodium phosphate monobasic solution (NaH2PO4) at pH 5, a 0.5 M sodium phosphate dibasic solution (Na2HPO4) at pH 10, or a 0.5 M solution composed of 50% sodium phosphate monobasic and 50% sodium phosphate dibasic (NaH2PO4/Na2HPO4) at pH 7.4.
Table 1. PEGDA-Based Hydrogel Formulations.
The table below includes the weight percentages used for each of the respective components for both PEGDA-based nanoparticle ( ,
,  ,
,  ,
,  ,
,  ) and bulk gel (
) and bulk gel ( ,
,  ,
,  ,
,  ,
,  ,
,  ) formulations used, including formulations with and without co-monomers.
) formulations used, including formulations with and without co-monomers.
| Formulation (wt%) |
PEGDA | CEA | PI | Co-monomer | Methanol |
|---|---|---|---|---|---|
30 wt% ( , ,  ) ) |
26.7 | 3.0 | 0.3 | 0 | 70 |
50 wt% ( , ,  ) ) |
44.5 | 5.0 | 0.5 | 0 | 50 |
| 50 wt% with 10 mol% HDDMA (  , ,  ) ) |
42.4 | 5.4 | 0.5 | 1.7 | 50 |
| 50 wt% with 30 mol% HDDMA (  , ,  ) ) |
37.5 | 6.1 | 0.6 | 5.8 | 50 |
| 50 wt% with 10 mol% DSDMA (  , ,  ) ) |
42.2 | 5.3 | 0.5 | 2.0 | 50 |
| 50 wt% with 30 mol% DSDMA (  , ,  ) ) |
36.9 | 6.0 | 0.6 | 6.5 | 50 |
Nanoparticle Degradation Studies
PEGDA nanoparticle suspensions were concentrated or diluted to 3 mg/mL in 1 mL of the medium of choice (either deionized water, 0.5M NaH2PO4 pH 5, 0.5 M Na2HPO4 pH 10, or 50/50 by mole mixture of 0.5 M NaH2PO4/Na2HPO4 pH 7.4). These media will be referred to as the water, pH 5, pH 7.4, and pH 10 conditions. Nanoparticles then were incubated in a shaker kept at 37°C and 1000 rpm. All particle formulations were degraded in water, the pH 5 condition, pH 7.4 condition, and pH 10 condition; particles with crosslinkers were also degraded in three different phosphate buffered saline (PBS) solutions that either had 0 mM glutathione (GSH), 1 mM GSH, or 10 mM GSH. At designated timepoints, 100 μL aliquots were removed for analysis after vortex mixing, leaving the remaining sample to continue degrading and ensuring uniform concentration. The 100 μL aliquot was then analyzed via thermogravimetric analysis (TGA), dynamic light scattering (DLS), and matrix-assisted laser desorption ionization (MALDI); select samples were also analyzed via cryo-scanning electron microscopy (see sections below).
Bulk Gel Synthesis
PEGDA bulk gels were synthesized with both the 30 wt% and 50 wt% formulations, the latter of which included samples both with no co-monomer and with the four co-monomer formulations (six formulations total). A volume of 30 μL of the polar pre-particle components of the same weight percentages as noted in Table 1 were added to a cylindrical mold (4.78 mm in diameter). The mold was then exposed to UV light (from an APM LED UV Cube with a wavelength of 365 nm at a distance of ~18 cm from the light source, ~8–10 mW/cm2) for varying times as denoted in Table S2. The resulting gels were ejected from the mold using a plunger and sequentially washed in ethanol and water.
Bulk Gel Degradation Studies
All six gel formulations were incubated at 37°C in 1 mL of pH 5, pH 7.4, and pH 10 salt solutions for various amounts of time until swelling ratio analysis at the desired timepoint. An additional set of gels were also kept in 1 mL of water to allow for analysis of supernatant products using MALDI (see section below), for which 50 μL of supernatant was extracted per time point. At the designated time points for swelling ratio analysis, swollen gels were removed from the solutions and then patted gently with a Kimwipe to remove any excess liquid. The gels were then weighed using a Mettler Toledo XSE105 DualRange to record the swollen mass. Thermogravimetric analysis (TGA) was them performed using a TA TGA 550 to remove the water from the gels and measure their dry masses, which was then confirmed using the Mettler Toledo XSE105 DualRange balance.
Thermogravimetric Analysis (TGA)
Initial concentrations of non-degraded nanoparticles were determined via TGA using a TA Instruments TGA 550. Samples of 100 μL taken at each time point were spun down at 18,200 RCF for two hours and the supernatant carefully removed. The remaining pellet was resuspended in water via sonication. From the pellet resuspension, 70 μL was removed, diluted three times to 210 μL, and two technical replicates of the dilution at 50 μL added to sample pans on the TGA. The mass of the PEGDA nanoparticle was determined by degrading the nanoparticle via a temperature ramp to 430°C following water evaporation at 120°C. The mass fraction remaining was determined by normalizing to the total amount of mass in each sample determined from the initial Day 0 time point. Details of the temperature ramp as well as information on mass correction because of salt decomposition can be found in the supporting information in on pages S4–S5.
Dynamic Light Scattering (DLS)
DLS was performed using a Malvern Zetasizer Nano S. Resuspended PEGDA nanoparticle pellet samples were prepared for DLS measurement by diluting to ~0.1 mg/mL in water. Hydrodynamic diameters (Dh) and polydispersity indices (PDIs) were measured from two independently synthesized and degraded samples under the same conditions.
Matrix-Assisted Laser Desorption Ionization Time-of-Flight (MALDI-TOF) Mass Spectrometry
For samples extracted from water, 1 μL aliquots for nanoparticles or 50 μL aliquots for gel supernatants were removed for MALDI analysis of products. The nanoparticle aliquots were diluted to 30 μL prior to analysis. 2, 5, and 7 μL aliquots of the samples were spotted onto a 96-spot MSP grounded steel plate respectively after being mixed with 10 μL of alpha-cyano-4-hydroxycinnamic acid (alpha-CHCA). MALDI mass spectrometric analysis was then performed using a Bruker MicroFlex MALDI-TOF mass spectrometer.
Cryo-Scanning Electron Microscopy (Cryo-SEM)
Select PEGDA nanoparticle samples 10 μL in volume were added to a sample holder for cryo-SEM and flash frozen with liquid nitrogen. Day 0 samples were prepared at 10 mg/mL, while day 42 samples were prepared at 0.5 mg/mL. The samples were then sputter-coated for 60 seconds with platinum and imaged using an Apreo VolumeScope Scanning Electron Microscope at 2 kV from 10,000x to 40,000x magnifications under high vacuum.
Rate Constant Determination
Rate constants for the degradation of the PEGDA nanoparticles were obtained by assuming pseudo-first order kinetics in the hydrolysis of ester bonds, as has been reported previously.26 To calculate the first order rate constant, we used the following expression:
where [NP]/[NP0] represents the mass fraction of particles remaining at time t relative to time 0, k is the rate constant, and t is time. Standard deviation was obtained by performing a linear regression between the natural log of the fraction of mass remaining at time t using the GraphPad software.
Statistical Analysis
Statistical analysis of DLS, TGA, and swelling ratio results was performed using a two-way analysis of variance (ANOVA) with the Graphpad Prism 8 software. This software was also used to perform multiple comparisons for the aforementioned results at each of the respective timepoints and determined significance of each of the relevant variables. Analysis of groups was performed as indicated in figures, where asterisks indicating P values of * ≤ 0.05, ** ≤ 0.01, *** ≤ 0.0001 and n.s. not significant. Nanoparticle and gel degradation studies were repeated in triplicate unless noted, with the number of replicates indicated within each figure. Measurements are reported as mean +/− standard deviation throughout.
Results & Discussion
Degradation of PEGDA-Based Nanoparticles
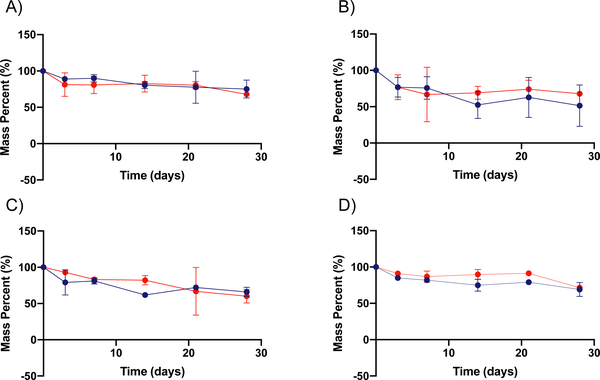
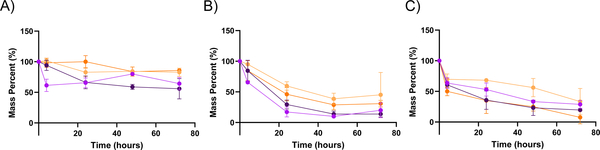
Ester bonds generated during the free radical polymerization of PEGDA are susceptible to hydrolysis, unlike non-functionalized PEG material. PEGDA hydrogel degradation is expected to proceed as a function of the molecular weight of the polymer crosslinker used, the density of crosslinks, the number of linkages susceptible to hydrolysis, and the reactivity of those linkages.27 Common methods for measuring the degradation of bulk hydrogels are either via modulus or equilibrium swelling ratio measurements. In the case of nanogels, however, there is great difficulty in making these measurements. In the case of modulus determination, a wetted AFM is necessary, which has very low throughput and requires particle-by-particle measurements, making aggregate determinations difficult. Equilibrium swelling ratio measurements cannot effectively be made for nanoparticles at all, as the dry and wet masses cannot be readily determined or distinguished from one another. Accordingly, we elected to utilize thermogravimetric analysis (TGA) to measure the degradation of nanoparticles and compared this degradation to traditional bulk gel approaches. Using TGA, we determine the mass of PEGDA nanoparticles remaining at given timepoints following centrifugation to separate the nanoparticles from degraded polymeric material, which will remain in the supernatant. From this approach, we determined the mass fraction of non-degraded nanoparticles and thus their respective rates of degradation in the four aforementioned environments. To investigate the degradation rates of PEGDA-based nanoparticles, we incubated the 30 wt% and 50 wt% nanoparticle formulations in four different solutions: water, pH 5 condition (0.5 M NaH2PO4), pH 7.4 condition (50/50 by mole mixture of NaH2PO4 and Na2HPO4, 0.5 M overall), and pH 10 condition (0.5 M Na2HPO4). The pH 5 and pH 7.4 environments were selected to be parallel to those found in the phagosome of a cell and in standard physiological environments, while the pH 10 and water conditions were selected for a basic environment comparison and a non-salt-containing comparison, respectively.28 The TGA results of 50 wt% and 30 wt% PEGDA nanoparticles degrading over a 4-week time course are shown in Figure 1.
Figure 1. Degradation of PEGDA-based nanoparticles as determined via TGA.
A through D show the degradation in terms of mass percent for both 30 wt% ( ) and 50 wt% (
) and 50 wt% ( ) PEGDA nanoparticles respectively in different solvents. This was determined by TGA of the remaining nanoparticles over time (n = 3). Degradation reported for 30% and 50% PEGDA nanoparticles in (A) water, (B) pH 5 condition, (C) pH 7.4 condition, and (D) pH 10 condition. Error bars present on all points; where not visible, the standard deviation is smaller than the data symbol.
) PEGDA nanoparticles respectively in different solvents. This was determined by TGA of the remaining nanoparticles over time (n = 3). Degradation reported for 30% and 50% PEGDA nanoparticles in (A) water, (B) pH 5 condition, (C) pH 7.4 condition, and (D) pH 10 condition. Error bars present on all points; where not visible, the standard deviation is smaller than the data symbol.
As can be seen from Figure 1, both PEGDA nanoparticle formulations degrade slowly over the month timeframe, with only the pH 5 condition achieving mass loss beyond half of the original sample. The rates of degradation of the 30 wt% and 50 wt% PEGDA nanoparticles (Table 2) are comparable to each other under all conditions, with generally increasing rates of degradation as pH is reduced. This may indicate that the generation of carboxylic acid groups from ester hydrolysis are auto-catalytic in the further degradation of nanoparticles under lower pH conditions. This effect is more predominant in the case of the 30 wt% nanoparticle formulation, which may be a result of a greater swelling ratio in the 30 wt% particle formulation, which is seen for the bulk gel formulation (Figure 2). The rate constants for the degradation of the PEGDA nanoparticles in each of the four conditions, shown below in Table 2, were determined by assuming pseudo-first order kinetics in the hydrolysis of ester bonds, as has been reported previously.26 These constants reflect the trends noted above, particularly that the degradation occurs relatively slowly and the rate in the pH 5 condition is notably greater than that in the other three conditions.
Table 2. Pseudo-first order rate constants for 30 wt% and 50 wt% PEGDA-based nanoparticles.
The table shows rate constants in units of inverse days for the two PEGDA-based nanoparticle formulations in each of the four respective incubation conditions. The rate constants were determined assuming pseudo-first order kinetics.
| NP Formulation | 30 wt% ( ) (days−1) ) (days−1) |
50 wt% ( ) (days−1) ) (days−1) |
|---|---|---|
| Degradation Condition | ||
| Water | 0.0095 ± 0.0032 | 0.0170 ± 0.0065 |
| pH 5 Condition | 0.0320 ± 0.0057 | 0.0165 ± 0.0027 |
| pH 7.4 Condition | 0.0120 ± 0.0041 | 0.0197 ± 0.0061 |
| pH 10 Condition | 0.0096 ± 0.0022 | 0.0088 ± 0.0023 |
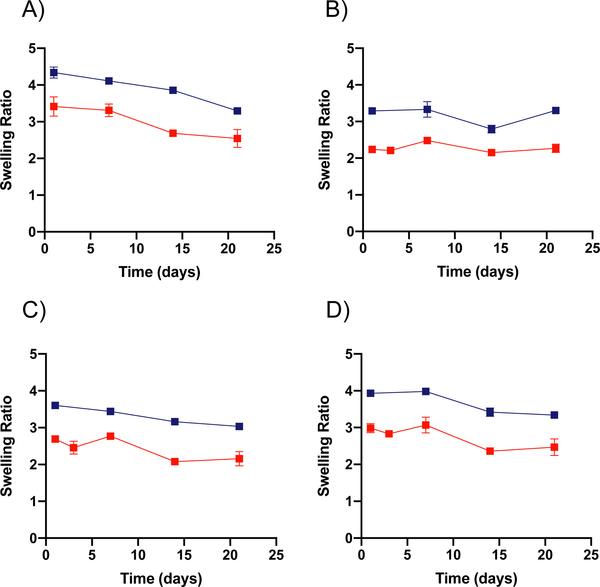
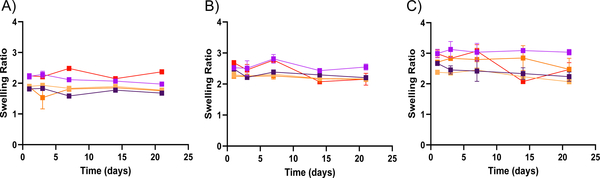
Figure 2. Swelling ratio measurements for bulk gel PEGDA formulations.
A through D show the change in swelling ratio over time for both 30 wt% ( ) and 50 wt% (
) and 50 wt% ( ) PEGDA bulk gels respectively in different solvents (n = 2). Swelling ratio for 30 wt% and 50 wt% PEGDA bulk gels in (A) water, (B) pH 5 condition, (C) pH 7.4 condition, and (D) pH 10 condition. Error bars present on all points; where not visible, the standard deviation is smaller than the data symbol.
) PEGDA bulk gels respectively in different solvents (n = 2). Swelling ratio for 30 wt% and 50 wt% PEGDA bulk gels in (A) water, (B) pH 5 condition, (C) pH 7.4 condition, and (D) pH 10 condition. Error bars present on all points; where not visible, the standard deviation is smaller than the data symbol.
The degradation shown in Figure 1 stands in contrast to the rates found for bulk gel formulations of the 50 wt% and 30 wt% PEGDA hydrogels. These bulk gels do not show any signs of degradation up to three weeks, as can be seen by the lack of change in swelling ratio of the bulk hydrogels shown in Figure 2. Here, the swelling ratio seen for bulk gels of the 50 wt% formulation averages ~2.5, while the 30 wt% formulation averages ~3.5. Though swollen to degrees similar to those found previously, these bulk gels do not have any statistical increase in swelling ratio over time. They would be expected to reach swelling ratios of 20 or greater when showing signs of significant degradation.25,29 One might hypothesize that the lower swelling ratio observed for the 50 wt% gel formulation would lead to slower degradation of the equivalent nanoparticle formulation; however, we observe increased degradation rate for nanoparticles in the pH 5 condition. This supports the hypothesis that the greater number of ester bonds per nanoparticle in the 50 wt% formulation causes a more rapid degradation even though its swelling ratio is lower. These results also indicate that use of bulk gels as surrogates for PEGDA nanoparticle degradation gives an incomplete picture of the nanoparticle degradation and that independent degradation rate determination is necessary for nanoparticle formulations. We hypothesize that this may have resulted from differences in the swelling ratio of the nanoparticles relative to bulk hydrogels, possibly arising from the high degree of curvature. Additional variation in degradation rate may arise from the high surface to volume ratio present in the nanogel, which is not the case for the bulk gels. Furthermore, this indicates that for applications in which gradual degradation is desirable over a one to two-month time scale in slightly acidic conditions, as it would for drug delivery applications, the PEGDA nanoparticulate formulations have clear advantages.
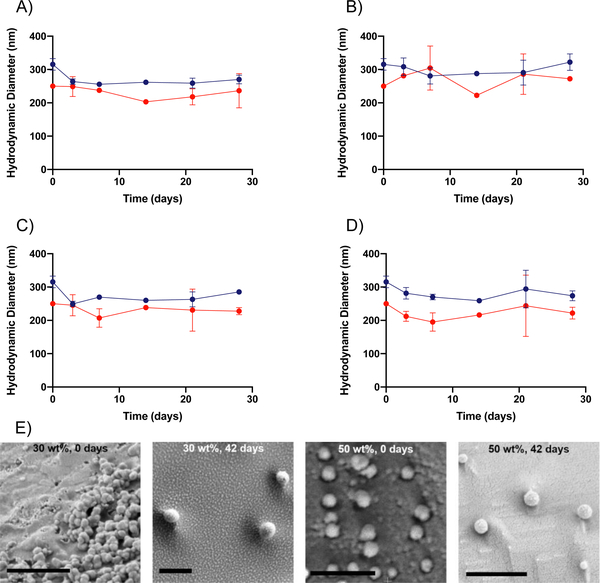
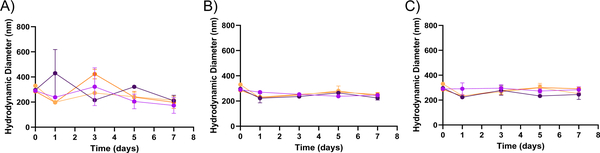
In addition to the rates of degradation, we also sought to gain insight into the mechanism of degradation of the PEGDA hydrogel nanoparticles utilized in this study. For nanoparticles, surface erosion is dominant when breakdown of the material is limited by the progression of the hydration front into the matrix because of limited ability of the solvent to penetrate into the matrix. Bulk erosion is dominant when the polymer matrix is hydrated rapidly, and water is readily imbibed by the matrix.21 In the case of bulk PEGDA hydrogels, bulk erosion has been shown to be the primary mechanism of breakdown, as would be expected because of PEG’s hydrophilic nature.30 This bulk degradation mechanism is likely present for the PEGDA hydrogel nanoparticles as well, as is supported by DLS and cryo-SEM imaging results. As can be seen in Figure 3A–3D, the DLS results indicate that the average hydrodynamic diameters (Dh) of the hydrogel nanoparticles do not change significantly over time for any of the four conditions. These results indicate that Dh of the nanoparticles is not changing over time and suggest that bulk degradation is taking place when considered in tandem with the TGA results that indicate that mass loss is occurring. With constant size and mass loss both being observed, the nature of mass loss must be through bulk degradation as opposed to surface erosion, a case for which Dh would decrease over time. These results are further supported by the cryo-SEM results, shown in Figure 3E, which indicate that for both the 50 wt% and 30 wt% formulations, the hydrogel nanoparticles do not change in diameter significantly over the course of 42 days, despite TGA results indicating that 30% or more of the nanoparticles has degraded. Select samples were also observed to have a ruptured appearance, which may be indicative of the degradation of the nanoparticles, as well. This can be seen in Figures S18 – S20.
Figure 3. DLS sizing and cryo-SEM imaging of PEGDA nanoparticles.
A through D show the change in hydrodynamic diameter (Dh) over time for both 30 wt% ( ) and 50 wt% (
) and 50 wt% ( ) PEGDA nanoparticles respectively in different solvents (n = 2). Dh DLS measurements of 30 wt% and 50 wt% PEGDA nanoparticles in (A) water, (B) pH 5, (C) pH 7.4, and (D) pH 10 conditions. (E) Cryo-SEM images of 30 wt% and 50 wt% PEGDA nanoparticles in water at 0-day and 42-day time points. For all images, the scale bar indicates a distance of 1 μm. Error bars present on all points; where not visible, the standard deviation is smaller than the data symbol.
) PEGDA nanoparticles respectively in different solvents (n = 2). Dh DLS measurements of 30 wt% and 50 wt% PEGDA nanoparticles in (A) water, (B) pH 5, (C) pH 7.4, and (D) pH 10 conditions. (E) Cryo-SEM images of 30 wt% and 50 wt% PEGDA nanoparticles in water at 0-day and 42-day time points. For all images, the scale bar indicates a distance of 1 μm. Error bars present on all points; where not visible, the standard deviation is smaller than the data symbol.
These DLS results highlight a key aspect of nanogels, in that they maintain a constant hydrodynamic diameter throughout the course of degradation. This contrasts with commonly employed polymers for nanoparticle drug delivery, such as PCL, which degrades via surface erosion and thus changes in diameter over time.20 Changes in the size of the particle may impact its function for intended application. For example, in the case of drug delivery, control over the particle size is paramount for control over on cellular internalization, further highlighting the potential benefit of utilizing these hydrogel nanoparticles for this application.31,32
Collectively, the TGA, DLS, and cryo-SEM direct analysis of PEGDA nanogels demonstrates unique aspects of PEGDA degradation from the nanoparticle form. The accelerated degradation observed in the nanogels as compared to that for the bulk gels may have arisen because of the difference in the surface area to volume ratio and/or possible edge effects from the different shape, curvature, and size scales. Additionally, PEGDA is known to form a heterogeneous hydrogel network33; likely heterogeneities generated during the free radical polymerization of the PEGDA nanogel have much more significant effects on the polymer network when restricted to nanometer dimensions. The exact nature of the network micro- and nano- structure warrants further investigation to elucidate differences between bulk gel and nanogel networks and explain potential variation in degradation profiles. Knowledge of the precise network microstructure would contribute to predictions of the loading and diffusion of molecules through the gel, which was found to be greatly impacted by the size reduction to nanometer lengths. We observe rapid release of rhodamine B, a model cargo, from the hydrogel nanoparticles (Figure S21), which was not observed in the bulk gels (Figure S22). This rapid release can be attributed to the significantly lower theoretical diffusion times for networks with average mesh sizes of ~ 1 nm (see SI page 17).29 As a result, encapsulation of a freely diffusing model drug (rhodamine B) was unable to serve as a surrogate for nanogel degradation, as has been previously shown for bulk gel and microparticle systems.19
PEGDA-Based Hydrogel Degradation with Co-Monomer Formulations
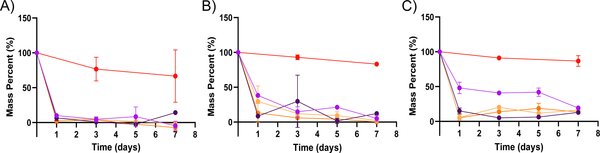
A common approach to altering the degradation kinetics of PEGDA-based hydrogel nanoparticles is to incorporate stimuli-responsive crosslinkers into their formulations. Disulfide-containing linkers are especially amenable to intracellular degradation, owing to the drop in pH and increase in GSH concentration within the endosomal compartments of the cell in which the nanoparticle will be internalized. To investigate the effect of these labile crosslinkers on the degradation rates of the PEGDA-based nanoparticles, we elected to incorporate HDDMA and DSDMA co-monomers in 50 wt% formulations by replacing 10 and 30 mol% of the PEGDA with the desired co-monomer. DSDMA was selected as a co-monomer because of the presence of the disulfide linkage within its backbone, while HDDMA was chosen as a presumed non-degradable control, lacking the disulfide linkage. Nanoparticles formulated with PEGDA in addition to HDDMA or DSDMA co-monomers were incubated in the pH 5, pH 7.4, and pH 10 conditions and their degradation was monitored via TGA as shown in Figure 4.
Figure 4. Degradation of PEGDA-based nanoparticles with co-monomers as determined via TGA.
A through C show the degradation in terms of mass percent for 50 wt% PEGDA nanoparticles with co-monomers: No co-monomer ( ), 10 mol% HDDMA (
), 10 mol% HDDMA ( ), 30 mol% HDDMA (
), 30 mol% HDDMA ( ), 10 mol% DSDMA (
), 10 mol% DSDMA ( ), and 30% DSDMA (
), and 30% DSDMA ( ) in different solvents. This was determined by TGA of the remaining nanoparticles over time (n = 2 or 3). Degradation reported in (A) pH 5 condition, (B) pH 7.4 condition, and (C) pH 10 condition. Error bars present on all points; where not visible, the standard deviation is smaller than the data symbol.
) in different solvents. This was determined by TGA of the remaining nanoparticles over time (n = 2 or 3). Degradation reported in (A) pH 5 condition, (B) pH 7.4 condition, and (C) pH 10 condition. Error bars present on all points; where not visible, the standard deviation is smaller than the data symbol.
As seen in Figure 4, the incorporation of both HDDMA and DSDMA co-monomers drastically increases the rate of degradation of PEGDA hydrogel nanoparticles in all three conditions relative to the 50 wt% formulation without co-monomer. Unlike the 50 wt% nanoparticles without co-monomer, those with the co-monomers show rapid mass loss occurring between one and three days. This stands in contrast to the base 50 wt% formulation, which shows significant degradation after multiple weeks. It has been demonstrated previously that the presence of heterogeneities in a hydrogel network can cause a reduction in the amount of crosslinks.9 Therefore, we hypothesize that the heterogeneities generated with the addition of both of these co-monomers contribute to the observed overall increase in degradation rates. Generally, the rate of mass loss after the addition of co-monomers seems to trend in a composition dependent manner; nanoparticles with 30 mol% of a co-monomer tend to degrade more rapidly than their 10 mol% counterparts, with minimal variation between the HDDMA and DSDMA formulations. However, all four co-monomer-containing formulations degrade more rapidly than the corresponding 50 wt% formulation with no co-monomer, as confirmed by thier calculated rate constants (Table 3). Of the three tested conditions, nanoparticle degradation occurred most rapidly in the pH 5 condition. Similar to the enhanced degradation rates seen in Figure 1, we hypothesize that the degradation products in the pH 5 condition may also autocatalyze ester hydrolysis in nanoparticles containing different co-monomers.
Table 3. Pseudo-first order rate constants for co-monomer formulations.
The table shows rate constants in units of inverse days for the four PEGDA-based nanoparticle formulations with co-monomers as well as for the 50 wt% formulation with no co-monomer in each of the three respective incubation conditions. The rate constants were determined assuming pseudo-first order kinetics.
| Formulation (NP) |
50 wt% ( ) (days−1) ) (days−1) |
50 wt% with 10% HDDMA ( ) (days−1) ) (days−1) |
50 wt% with 30% HDDMA ( ) (days−1) ) (days−1) |
50 wt% with 10% DSDMA ( ) (days−1) ) (days−1) |
50 wt% with 30% DSDMA ( ) (days−1) ) (days−1) |
|---|---|---|---|---|---|
| Degradation Condition | |||||
| pH 5 Condition | 0.017 ± 0.003 | 0.699 ± 0.254 | 1.490 ± 0.345 | 0.669 ± 0.224 | 1.039 ± 0.560 |
| pH 7.4 Condition | 0.020 ± 0.006 | 0.357 ± 0.088 | 0.139 ± 0.171 | 0.547 ± 0.136 | 0.605 ± 0.174 |
| pH 10 Condition | 0.009 ± 0.002 | 0.167 ± 0.037 | 0.169 ± 0.123 | 0.031 ± 0.136 | 0.023 ± 0.130 |
Furthermore, we used DLS to monitor any potential changes in the mode of degradation following the incorporation of HDDMA or DSDMA co-monomers. As seen from Figure 5, there are minimal changes to the average Dh, especially when compared to corresponding changes in mass percentage shown in Figure 4. As in Figure 3, the constant Dh, indicates that bulk degradation is a likely mechanism for degradation of all of the tested nanoparticle compositions in the pH 5, pH 7.4, and pH 10 conditions.
Figure 5. DLS sizing of 50 wt% nanoparticles containing HDDMA and DSDMA co-monomers.
A through C show the change in hydrodynamic diameter (Dh) over time for 50 wt% PEGDA nanoparticles with co-monomers: 10 mol% HDDMA ( ), 30 mol% HDDMA (
), 30 mol% HDDMA ( ), 10 mol% DSDMA (
), 10 mol% DSDMA ( ), and 30% DSDMA (
), and 30% DSDMA ( ) in different solvents. (n = 2). Dh DLS measurements in (A) pH 5, (B) pH 7.4, and (C) pH 10 conditions. Error bars present on all points; where not visible, the standard deviation is smaller than the data symbol.
) in different solvents. (n = 2). Dh DLS measurements in (A) pH 5, (B) pH 7.4, and (C) pH 10 conditions. Error bars present on all points; where not visible, the standard deviation is smaller than the data symbol.
Analogous to our previous studies, we sought to investigate the effect of heterogeneities on hydrogel swelling and degradation to compare to nanoparticle results. We measured the equilibrium swelling ratio of bulk gels synthesized with the same HDDMA and DSDMA co-monomer formulations as the nanoparticles and compared results to those obtained for 50 wt% PEGDA hydrogels lacking these co-monomers (Figure 6). The results show no significant changes in swelling ratios of bulk gels containing HDDMA or DSDMA co-monomers as compared to the bulk gels containing no co-monomers in the pH 5, pH 7.4, and pH 10 conditions. The swelling ratio is constant up to three weeks, which indicates that there is no evidence of degradation in bulk gels. These results contrast significantly with the trends observed for the degradation of nanoparticles of the same formulations, which show drastic mass loss as early as 24 hours. This result again indicates that bulk gel formulations serve as poor surrogates for exploration of co-monomer-containing PEGDA nanoparticle degradation, as was the case for the nanoparticle formulations without co-monomers.
Figure 6. Equilibrium swelling ratio of 50 wt% bulk gels containing HDDMA and DSDMA co-monomers.
A through C show the change in swelling ratio over time for 50 wt% PEGDA nanoparticles with or without co-monomers: No co-monomer ( ), 10 mol% HDDMA (
), 10 mol% HDDMA ( ), 30 mol% HDDMA (
), 30 mol% HDDMA ( ), 10 mol% DSDMA (
), 10 mol% DSDMA ( ), and 30% DSDMA (
), and 30% DSDMA ( ) in different solvents. (n = 2). Swelling ratio reported in (A) pH 5 condition, (B) pH 7.4 condition, and (C) pH 10 conditions. Error bars present on all points; where not visible, the standard deviation is smaller than the data symbol.
) in different solvents. (n = 2). Swelling ratio reported in (A) pH 5 condition, (B) pH 7.4 condition, and (C) pH 10 conditions. Error bars present on all points; where not visible, the standard deviation is smaller than the data symbol.
The incorporation of redox-sensitive monomers within the PEGDA network is especially relevant in biomedical applications because of the abundance of reducing agents in critical physiological environments. For example, GSH, a tripeptide that is abundant in cancerous tissues and intracellular compartments, is able to cleave disulfide bonds within a polymer network through a thiol-disulfide exchange mechanism.34 Accordingly, the incorporation of GSH-sensitive crosslinkers in the PEGDA network would be presumed to greatly increase nanoparticle degradability in GSH-containing environments, which is ultimately advantageous in achieving efficient drug delivery, especially in cancer-related applications. To investigate the sensitivity of PEGDA nanoparticles with disulfide bond-bearing DSDMA co-monomers to a reducing environment, we incubated nanoparticles in PBS both the absence and presence of GSH, respectively. Two GSH concentrations were used, 1 and 10 mM, which are representative of GSH concentrations found in different intracellular compartments.35 The results of mass loss (as determined via TGA) from the degradation of PEGDA-based nanoparticles formulated with HDDMA or DSDMA co-monomers in PBS with added GSH are shown in Figure 7.
Figure 7. Degradation of PEGDA-based nanoparticles with co-monomers in GSH as determined via TGA.
A through C show the degradation in terms of mass percent for 50 wt% PEGDA nanoparticles with co-monomers: 10 mol% HDDMA ( ), 30 mol% HDDMA (
), 30 mol% HDDMA ( ), 10 mol% DSDMA (
), 10 mol% DSDMA ( ), and 30% DSDMA (
), and 30% DSDMA ( ) in PBS with different amounts of GSH. This was determined by TGA of the remaining nanoparticles over time (n = 2). Degradation reported in (A) PBS, (B) 1 mM GSH, and (C) 10 mM GSH. Error bars present on all points; where not visible, the standard deviation is smaller than the data symbol.
) in PBS with different amounts of GSH. This was determined by TGA of the remaining nanoparticles over time (n = 2). Degradation reported in (A) PBS, (B) 1 mM GSH, and (C) 10 mM GSH. Error bars present on all points; where not visible, the standard deviation is smaller than the data symbol.
As seen in Figure 7, there is rapid degradation of nanoparticles containing DSDMA in the presence of different amounts of GSH, with 30% DSDMA nanoparticles degrading faster than 10% DSDMA nanoparticles, as expected. The difference in the rate of degradation of 10% DSDMA and 30% DSDMA nanoparticles seems to increase at higher levels of GSH. Interestingly, HDDMA-containing nanoparticles incubated with GSH show faster degradation than disulfide bond-bearing DSDMA nanoparticles. This is especially true for low GSH conditions, while 30% DSDMA nanoparticles appear to exceed 10% HDDMA and 30% HDDMA nanoparticles only at high GSH levels. Unexpected degradation patterns of HDDMA and DSDMA nanoparticles may be attributed to two competing degradation mechanisms in DSDMA nanoparticles: thiol-disulfide exchange and ester hydrolysis. HDDMA nanoparticles, however, only degrade through ester hydrolysis. We hypothesize that GSH plays a role in ester hydrolysis through protonating the carbonyl in the ester making it more electrophilic and thus more susceptible to nucleophilic attack. In addition, ester hydrolysis is hypothesized to be a more dominant mechanism given that the number of ester bonds is much greater than the number of disulfide bonds in DSDMA nanoparticles. Though surprising, the results shown in Figure 7 show the susceptibility of both DSDMA and HDDMA nanoparticles to degradation in the presence of GSH and provide insight on nanoparticle and hydrogel design considerations for intracellular and tumoral drug delivery.
Degradation Product Characterization
In addition to use in drug delivery, in which the rate of degradation is often critical to cargo release, PEGDA hydrogels also have potential in immune engineering and regenerative medicine applications. For these applications, the products released from the degradation of the PEGDA-based gels and nanoparticles themselves can have effects on the cells that uptake these products. Accordingly, we decided to explore the nature of the products released from the degradation of both the PEGDA nanoparticles and bulk gels over time via MALDI-TOF. Representative results of the spectra obtained are shown below in Figure 8, which show the common products obtained from the degradation of each nanoparticle or bulk gel formulation degraded in water. Additional spectra may be found in supplementary Figures S23–S50, which show the change in the products over time, which are minimal within each formulation.
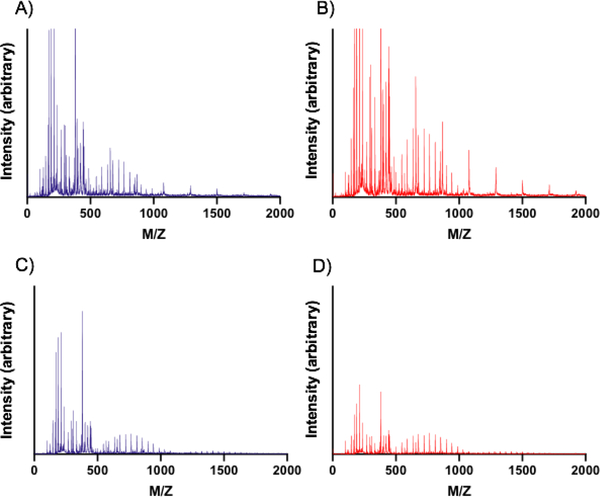
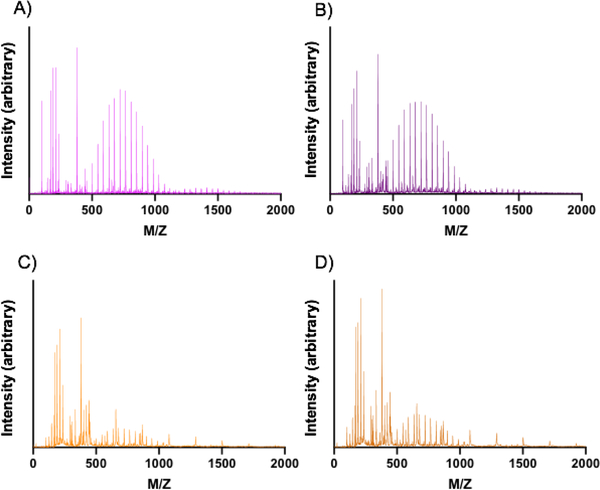
Figure 8. MALDI-TOF of products of PEGDA hydrogel nanoparticle and bulk gel degradation.
Product distribution as determined by MALDI-TOF from samples incubated in water at 37°C for 7 days. (A) 30 wt% PEGDA nanoparticles, (B) 50 wt% PEGDA nanoparticles, (C) 30 wt% PEGDA bulk gels, and (D) 50 wt% PEGDA bulk gels.
As Figure 8A and 8B show, the primary products of the degradation of the 30 wt% and 50 wt% PEGDA-based nanoparticles occur in the peaks around 700 MW, likely indicating recovery of PEDGA. The spacing of 44 Daltons (Da) of mass difference between the peaks supports this hypothesis since this is the mass of a repeat unit of PEG and that the monomer used was a distribution, not monodisperse. The lower molecular weight products around 350 and 250 Da that are also present likely correspond to PEGDA as well, but with greater charge states (Z values) of +2 and +3, respectively, while the mass remains the same. Other potential products in the low MW region around 120 Da could be CEA that has cleaved via ester hydrolysis or, less likely, the MEHQ inhibitor included in some of the precursors. The feature of these mass spectrometry results which contrast most with their bulk gel counterparts (Figures 8C and 8D) is the presence of peaks spaced ~210 Da apart at 1100 Da, 1310 Da, and 1520 Da. These peaks are not found in the bulk gel formulations; we propose that the peaks present in the 30 wt% and 50 wt% hydrogel nanoparticles at 1100, 1310, and 1520 Da are PEGDA chains that have adjacent chains cleaved via ester hydrolysis and thus will contain extra mass from the adjacent ester groups that end as diols following nucleophilic attack during hydrolysis. In comparison, we observed peaks centered around 1400 Da for the bulk gels. These peaks likely correspond to dimers of PEGDA that had been cleaved from the bulk hydrogel network. This conclusion is further supported by peak spacing of 44 Da, as was the case with the PEGDA monomer.
A similar analysis was performed using MALDI-TOF data for the degradation products of bulk PEGDA hydrogels containing co-monomers (shown in Figure 9). These results show the presence of PEGDA, as was the case for all other hydrogel formulations. Interestingly, the product profiles for the bulk gels containing the HDDMA co-monomer (Figure 9A and 9B) much more closely resemble those for the 30 wt% and 50 wt% bulk gels (Figure 9C and 9D), while the bulk gels containing DSDMA have product profiles (Figure 9C and 9D) much more closely resembling those for the nanoparticles (Figure 8A and 8B). The HDDMA-containing gels show presence of dimers in addition to the PEGDA monomers. The DSDMA-containing gels show the presence of the products at 1100, 1310, and 1520 Da, similar to those found in the nanoparticle formulations. It is possible that the products from the degradation are the same as those found from the nanoparticle degradation, but it is also possible that these products contain segments of DSDMA conjugated to PEGDA monomers, which would align closely with this mass difference. This is likely not the case, however, given that the HDDMA-containing gels did not show a similar product pattern.
Figure 9. MALDI-TOF of products of PEGDA bulk gel degradation with co-monomers.
Product distribution as determined by MALDI-TOF from samples incubated in water at 37°C for 7 days. (A) 50 wt% PEGDA bulk gels with 10% HDDMA co-monomer, (B) 50 wt% PEGDA bulk gels with 30% HDDMA co-monomer (C) 50 wt% PEGDA bulk gels with 10% DSDMA co-monomer, and (D) 50 wt% PEGDA bulk gels with 30% DSDMA co-monomer.
Overall, the range of molecular weight degradation components from MALDI-TOF data correlates well with previous reports of PEGDA bulk gels.36 Furthermore, our observations indicate that the majority of degradation components are relatively small molecular weight oligomers, in comparison to the complete particle. This was confirmed via GPC (Figure S51), which shows no additional peaks aside from those from the monomer and potentially other low molecular weight products. This confirmation indicates that analysis via MALDI-TOF was appropriate despite the technique’s difficulty with ionizing higher molecular weight molecules. For drug delivery applications, these oligomers fall well below the 70 kDa (~5.5 nm) range considered to be the limit for renal clearance. Thus, degradation components from the PEGDA-based nanoparticles studied here would all be expected to be cleared by the kidney following degradation inside the body.37,38 Further isolation of degradation products, characterization, and cellular assays will be needed to determine the immunological effects of these oligomers.
Conclusions
In this work, we provide an extensive analysis of the degradation products, rates, and profiles of PEGDA-based hydrogel nanoparticles both with and without co-monomers and perform comparisons to their bulk gel counterparts. The degradation rates for the PEGDA-based nanoparticles are determined via TGA and their mechanism of breakdown inferred from DLS results in tandem with the degradation observed via TGA. We conclude that the PEGDA-based nanogels degrade through a bulk degradation mechanism since their hydrodynamic diameters change minimally while they lose significant mass. The significant mass loss observed occurs at rates much greater than that for their bulk gel counterparts, which show no significant signs of degradation through three weeks under the same conditions. We also have noted that the degradation rates of co-monomer-containing PEGDA-based nanoparticles are much greater than those without the co-monomers, providing another handle with which degradation can be tuned. By altering the co-monomer content and wt% of PEGDA in the formulation, PEGDA-based nanoparticles were shown to have tunable degradability in time frames ranging from days to months in physiologically-relevant solvents. Notably, our findings demonstrate that common methods of estimating nanogel degradation through bulk gel analysis can dramatically underestimate the degradation rate of nanoparticles and must be applied with caution. We hope that the framework for hydrogel degradation analysis and consideration for PEGDA-based formulations presented here can be applied to future applications of PEGDA-based hydrogels for drug delivery, immune engineering, and biomaterials applications.
Supplementary Material
Acknowledgements
We thank C. Roberts for DLS access, D. Powell for assistance with Cryo-SEM, and A. Kloxin for helpful discussions. Research reported in this publication was supported by National Institutes of Health under Award Number P20GM104316 and T32GM008550 from the National Institute of General Medical Sciences. The mass spectroscopy in this publication was made possible by the Delaware COBRE program, supported by a grant from the National Institute of General Medical Sciences – NIGMS (5 P30 GM110758–02) from the National Institutes of Health. Thermo Scientific™ Apreo VS SEM microscope: Microscopy equipment was acquired with a shared instrumentation grant (S10 OD025165) and access was supported by the NIH-NIGMS (P20 GM103446), the NSF (IIA-1301765) and the State of Delaware. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflicts of Interest
n/a
References
- (1).Debord JD; Lyon LA Thermoresponsive Photonic Crystals. J. Phys. Chem. B 2002. 10.1021/jp001238c. [DOI] [Google Scholar]
- (2).Lee WF; Huang YC Swelling and Antibacterial Properties for the Superabsorbent Hydrogels Containing Silver Nanoparticles. J. Appl. Polym. Sci 2007. 10.1002/app.26906. [DOI] [Google Scholar]
- (3).DeForest CA; Anseth KS Advances in Bioactive Hydrogels to Probe and Direct Cell Fate. Annu. Rev. Chem. Biomol. Eng 2012, 3 (1), 421–444. 10.1146/annurev-chembioeng-062011-080945. [DOI] [PubMed] [Google Scholar]
- (4).Nayak S; Lyon LA Soft Nanotechnology with Soft Nanoparticles. Angew. Chemie Int. Ed 2005, 44 (47), 7686–7708. 10.1002/anie.200501321. [DOI] [PubMed] [Google Scholar]
- (5).Mu C; Zhang Y; Cui W; Liang Y; Zhu Y. Removal of Bisphenol A over a Separation Free 3D Ag3PO4-Graphene Hydrogel via an Adsorption-Photocatalysis Synergy. Appl. Catal. B Environ 2017. 10.1016/j.apcatb.2017.04.018. [DOI] [Google Scholar]
- (6).Tang JD; Mura C; Lampe KJ Stimuli-Responsive, Pentapeptide, Nanofiber Hydrogel for Tissue Engineering. J. Am. Chem. Soc 2019, 141 (12), 4886–4899. 10.1021/jacs.8b13363. [DOI] [PubMed] [Google Scholar]
- (7).Hamidi M; Rostamizadeh K; Shahbazi MA Hydrogel Nanoparticles in Drug Delivery. In Intelligent Nanomaterials: Processes, Properties, and Applications; 2012. 10.1002/9781118311974.ch15. [DOI] [Google Scholar]
- (8).Geever LM; Cooney CC; Lyons JG; Kennedy JE; Nugent MJD; Devery S; Higginbotham CL Characterisation and Controlled Drug Release from Novel Drug-Loaded Hydrogels. Eur. J. Pharm. Biopharm 2008. 10.1016/j.ejpb.2007.12.021. [DOI] [PubMed] [Google Scholar]
- (9).Anseth KS; Bowman CN; Brannon-Peppas L. Mechanical Properties of Hydrogels and Their Experimental Determination. Biomaterials. 1996. 10.1016/0142-9612(96)87644-7. [DOI] [PubMed] [Google Scholar]
- (10).Mellott MB; Searcy K; Pishko MV Release of Protein from Highly Cross-Linked Hydrogels of Poly(Ethylene Glycol) Diacrylate Fabricated by UV Polymerization. Biomaterials 2001. 10.1016/S0142-9612(00)00258-1. [DOI] [PubMed] [Google Scholar]
- (11).Peppas NA; Kim B. Stimuli-Sensitive Protein Delivery Systems. J. Drug Deliv. Sci. Technol 2006, 16 (1), 11–18. 10.1016/S1773-2247(06)50002-4. [DOI] [Google Scholar]
- (12).Li J; Li X; Ni X; Wang X; Li H; Leong KW Self-Assembled Supramolecular Hydrogels Formed by Biodegradable PEO-PHB-PEO Triblock Copolymers and Alpha-Cyclodextrin for Controlled Drug Delivery. Biomaterials 2006. 10.1016/j.biomaterials.2006.03.025. [DOI] [PubMed] [Google Scholar]
- (13).Merkel TJ; Jones SW; Herlihy KP; Kersey FR; Shields AR; Napier M; Luft JC; Wu H; Zamboni WC; Wang AZ; et al. Using Mechanobiological Mimicry of Red Blood Cells to Extend Circulation Times of Hydrogel Microparticles. Proc. Natl. Acad. Sci 2011, 108 (2), 586–591. 10.1073/pnas.1010013108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Anselmo AC; Zhang M; Kumar S; Vogus DR; Menegatti S; Helgeson ME; Mitragotri S. Elasticity of Nanoparticles Influences Their Blood Circulation, Phagocytosis, Endocytosis, and Targeting. ACS Nano 2015, 9 (3), 3169–3177. 10.1021/acsnano.5b00147. [DOI] [PubMed] [Google Scholar]
- (15).Roberts RA; Shen T; Allen IC; Hasan W; DeSimone JM; Ting JPY Analysis of the Murine Immune Response to Pulmonary Delivery of Precisely Fabricated Nano- and Microscale Particles. PLoS One 2013, 8 (4). 10.1371/journal.pone.0062115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Fromen CA; Rahhal TB; Robbins GR; Kai MP; Shen TW; Luft JC; DeSimone JM Nanoparticle Surface Charge Impacts Distribution, Uptake and Lymph Node Trafficking by Pulmonary Antigen-Presenting Cells. Nanomedicine Nanotechnology, Biol. Med 2016, 12 (3), 677–687. 10.1016/j.nano.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Allen RP; Bolandparvaz A; Ma JA; Manickam VA; Lewis JS Latent, Immunosuppressive Nature of Poly(Lactic-Co-Glycolic Acid) Microparticles. ACS Biomater. Sci. Eng 2018, 4 (3), 900–918. 10.1021/acsbiomaterials.7b00831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Fromen CA; Robbins GR; Shen TW; Kai MP; Ting JPY; DeSimone JM Controlled Analysis of Nanoparticle Charge on Mucosal and Systemic Antibody Responses Following Pulmonary Immunization. Proc. Natl. Acad. Sci 2015, 112 (2), 488–493. 10.1073/pnas.1422923112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Parrott MC; Finniss M; Luft JC; Pandya A; Gullapalli A; Napier ME; DeSimone JM Incorporation and Controlled Release of Silyl Ether Prodrugs from PRINT Nanoparticles. J. Am. Chem. Soc 2012, 134 (18), 7978–7982. 10.1021/ja301710z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Lin CC; Metters AT Hydrogels in Controlled Release Formulations: Network Design and Mathematical Modeling. Advanced Drug Delivery Reviews. 2006. 10.1016/j.addr.2006.09.004. [DOI] [PubMed] [Google Scholar]
- (21).Lin C; Metters AT Hydrogels in Controlled Release Formulations : Network Design and Mathematical Modeling ☆. 2006, 58, 1379–1408. 10.1016/j.addr.2006.09.004. [DOI] [PubMed] [Google Scholar]
- (22).Tibbitt MW; Kloxin AM; Sawicki LA; Anseth KS Mechanical Properties and Degradation of Chain and Step-Polymerized Photodegradable Hydrogels. Macromolecules 2013, 46 (7), 2785–2792. 10.1021/ma302522x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Park TG Degradation of Poly(Lactic-Co-Glycolic Acid) Microspheres: Effect of Copolymer Composition. Biomaterials 1995, 16 (15), 1123–1130. [DOI] [PubMed] [Google Scholar]
- (24).Jeong B; Bae YH; Kim SW Drug Release from Biodegradable Injectable Thermosensitive Hydrogel of PEG–PLGA–PEG Triblock Copolymers. J. Control. Release 2000, 63 (1), 155–163. [DOI] [PubMed] [Google Scholar]
- (25).Fish MB; Fromen CA; Lopez-Cazares G; Golinski AW; Scott TF; Adili R; Holinstat M; Eniola-Adefeso O. Exploring Deformable Particles in Vascular-Targeted Drug Delivery: Softer Is Only Sometimes Better. Biomaterials 2017, 124, 169–179. 10.1016/j.biomaterials.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Metters A; Hubbell J. Network Formation and Degradation Behavior of Hydrogels Formed by Michael-Type Addition Reactions. 2005, 290–301. 10.1021/bm049607o. [DOI] [PubMed] [Google Scholar]
- (27).Hoffman AS Hydrogels for Biomedical Applications. 2002, 54, 3–12. [DOI] [PubMed] [Google Scholar]
- (28).Hu Y-B; Dammer EB; Ren R-J; Wang G. The Endosomal-Lysosomal System: From Acidification and Cargo Sorting to Neurodegeneration. Transl. Neurodegener 2015, 4, 18 10.1186/s40035-015-0041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Zustiak SP; Leach JB Hydrolytically Degradable Poly ( Ethylene Glycol ) Hydrogel Scaffolds with Tunable Degradation and Mechanical Properties. 2010, 1348–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Parlato M; Reichert S; Barney N; Murphy WL Poly ( Ethylene Glycol ) Hydrogels with Adaptable Mechanical and Degradation Properties for Use in Biomedical Applications A. 2014, 687–698. 10.1002/mabi.201300418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Gratton SEA; Ropp PA; Pohlhaus PD; Luft JC; Madden VJ; Napier ME; DeSimone JM The Effect of Particle Design on Cellular Internalization Pathways. Proc. Natl. Acad. Sci 2008, 105 (33), 11613 LP – 11618. 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Champion JA; Walker A; Mitragotri S. Role of Particle Size in Phagocytosis of Polymeric Microspheres. Pharm. Res 2008, 25 (8), 1815–1821. 10.1007/s11095-008-9562-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Malo de Molina P; Lad S; Helgeson ME Heterogeneity and its Influence on the Properties of Difunctional Poly(ethylene glycol) Hydrogels: Structure and Mechanics. Macromolecules. 2015, 48 (15) 5402–5411. doi.org/10.1021/acs.macromol.5b01115 [Google Scholar]
- (34).Quinn JF; Whittaker MR; Davis TP Glutathione Responsive Polymers and Their Application in Drug Delivery Systems. Polymer Chemistry. 2017. 10.1039/c6py01365a. [DOI] [Google Scholar]
- (35).Smith CV; Jones DP; Guenthner TM; Lash LH; Lauterburg BH Compartmentation of Glutathione: Implications for the Study of Toxicity and Disease. Toxicology and Applied Pharmacology. 1996. 10.1006/taap.1996.0191. [DOI] [PubMed] [Google Scholar]
- (36).Xin AX; Gaydos C; Mao JJ In Vitro Degradation Behavior of Photopolymerized PEG Hydrogels as Tissue Engineering Scaffold. In 2006 International Conference of the IEEE Engineering in Medicine and Biology Society; 2006; pp 2091–2093. 10.1109/IEMBS.2006.260811. [DOI] [PubMed] [Google Scholar]
- (37).Deshmukh M; Kutscher HL; Gao D; Sunil VR; Malaviya R; Vayas K; Stein S; Laskin JD; Laskin DL; Sinko PJ Biodistribution and Renal Clearance of Biocompatible Lung Targeted Poly(Ethylene Glycol) (PEG) Nanogel Aggregates. J. Control. Release 2012, 164 (1), 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Caliceti P; Veronese FM Pharmacokinetic and Biodistribution Properties of Poly(Ethylene Glycol)–Protein Conjugates. Adv. Drug Deliv. Rev 2003, 55 (10), 1261–1277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.