Abstract
The SARS-CoV-2 genome encodes for approximately 30 proteins. Within the international project covid19-nmr, we distribute the spectroscopic analysis of the viral proteins and RNA. Here, we report NMR chemical shift assignments for the protein nsp7. The 83 amino acid nsp7 protein is an essential cofactor in the RNA-dependent RNA polymerase. The polymerase activity and processivity of nsp12 are greatly enhanced by binding 1 copy of nsp7 and 2 copies of nsp8 to form a 160 kD complex. A separate hexadecameric complex of nsp7 and nsp8 (8 copies of each) forms a large ring-like structure. Thus, nsp7 is an important component of several large protein complexes that are required for replication of the large and complex coronavirus genome. We here report the near-complete NMR backbone and sidechain resonance assignment (1H,13C,15N) of isolated nsp7 from SARS-CoV-2 in solution. Further, we derive the secondary structure and compare it to the previously reported assignments and structure of the SARS-CoV nsp7.
Keywords: SARS-CoV-2, Non-structural protein, Solution NMR-spectroscopy, COVID19-NMR
Biological context
SARS-CoV-2, the causative agent of COVID-19, emerged late in 2019 to cause a global pandemic. This novel coronavirus is highly related to SARS-CoV that emerged in 2002. While several coronaviruses routinely infect humans causing mild respiratory symptoms (van der Hoek 2007), SARS-CoV-2 is capable of causing severe respiratory symptoms or even death (Bchetnia et al. 2020). Coronaviruses are enveloped viruses with large positive-sense RNA genomes. At 30 kb or even greater, some of the genomes of the Nidovirales order, which includes coronaviruses, are among the largest RNA genomes known, requiring a highly processive RNA synthesis process for their replication.
Upon entering host cells, the coronavirus RNA genome acts as an mRNA to be translated by host ribosomes to produce the viral polyproteins pp1a and pp1ab. These polyproteins are cleaved to produce a suite of 16 non-structural proteins (nsp) that are responsible for replication and transcription of the viral RNA genome (Snijder et al. 2016). The viral nsp assemble membrane enclosed compartments containing the virus RNA-dependent RNA-polymerase. The minimal polymerase complex required for activity in vitro is composed of nsp7, nsp8 and nsp12. While the polymerase active site is contained wholly within nsp12, nsp7 and nsp8 act as essential co-factors for enzyme activity enabling processive RNA synthesis (Subissi et al. 2014). Nsp7 has also been proposed to act with nsp8 as a part of an RNA primase to generate RNA primers for viral RNA synthesis (Imbert et al. 2006; te Velthuis et al. 2012). However, the activity of the primase or its proposed additional role to extend RNA primer has not been universally reproduced (Subissi et al. 2014).
Previous structures of nsp7 show the protein to be composed of four helical regions where the positioning of the N- and C-terminal helical regions are altered upon binding to nsp8 (Johnson et al. 2010; Zhai et al. 2005). Structural determination of the polymerase complexes of SARS-CoV (Kirchdoerfer and Ward 2019) and SARS-CoV-2 (Gao et al. 2020), show that two subunits of nsp8 bind to nsp12 and that one of these nsp8 subunits interacts with nsp7 giving a 1:2:1 nsp7:nsp8:nsp12 stoichiometry. A separate crystal structure of SARS-CoV nsp7 bound to nsp8, while resembling the nsp7-nsp8 interactions in the nsp7-nps8-nsp12 cryoEM structures, showed the assembly of a large 8:8 protein complex (Zhai et al. 2005). Lacking solution evidence for this large complex and alternate assemblies for nsp7 and nsp8 observed in crystal structures of feline coronavirus (Xiao et al. 2012) and SARS-CoV-2 (Konkolova et al. 2020) leaves ambiguity as to the biological role of these large nsp7-nsp8 complexes in the virus life cycle. As a well-conserved component of the virus replication machinery, a greater understanding of nsp7 structure and dynamics will accelerate our understanding of this essential protein complex improving models of protein–protein interactions and laying an important foundation for the development of antiviral therapeutics.
Methods and experiments
Construct design
This study uses the SARS-CoV-2 NCBI reference genome entry NC_045512.2, identical to GenBank entry MN908947.3 (Wu et al. 2020). This sequence was inserted into a pET46 vector, containing an N-terminal His6-tag, Ek protease and a tobacco etch virus (TEV) protease cleavage sites. Due to the nature of the TEV protease cleavage site, the purified protein contained one artificial N-terminal residue (G0) preceding the native protein sequence.
Sample preparation
Uniformly 13C,15N-labelled Nsp7 protein was expressed in Eschirichia coli strain Rosetta2 pLysS in M9 minimal medium containing 1 g/L 15NH4Cl (Cambridge Isotope Laboratories), 4 g/L 13C6-D-glucose (Cambridge Isotope Laboratories) and 100 μg/mL ampicillin. Bacterial cultures were grown to an O.D. 600 nm of 0.8 at 37 °C and induced with 0.5 mM IPTG for 14–16 h at 16 °C. The cell pellet was resuspended in buffer A (10 mM HEPES, pH 7.4, 300 mM NaCl, 30 mM imidazole and 2 mM dithiothreitol). The cells were lysed using a microfluidizer operating at 20,000 psi. The lysate was clarified by centrifugation at 25,000 × g for 30 min and then filtration using a 0.45 µm vacuum filter. Clarified supernatant was bound to Ni–NTA agarose (Qiagen), washed with buffer A and then eluted with buffer A containing 300 mM total imidazole. Protein containing fractions were pooled and cleaved with 1% (w/w) TEV protease over night at room temperature while dialyzing against 10 mM MOPS, pH 7.0, 150 mM NaCl, 2 mM dithiothreitol. TEV protease and tag were removed via a second IMAC purification. Protein was further purified with a Superdex200 column (GE Life Sciences) using a buffer containing 10 mM MOPS, pH 7.0, 150 mM NaCl, 2 mM dithiothreitol. Fractions containing the purified proteins were concentrated using Amicon Ultra concentrators (Millipore Sigma). The final NMR sample contained 1.7 mM 13C,15N-nsp7, 10 mM MOPS, pH 7.0, 150 mM NaCl, 2 mM DTT, 0.025% NaN3, 7% D2O.
NMR experiments
All experiments for the backbone and side chain assignments of nsp7 were recorded at 298 K using 600 MHz Varian VNMRS and Bruker Avance III spectrometers, equipped with an H/C/N Cryoprobe. All spectra were acquired using standard pulse sequences optimized to achieve the best performance on cryogenic probes and with non-uniform sampling. The set of NMR experiments used for resonance assignments is summarized in Table 1. Proton resonances were calibrated with respect to the signal of 2,2-dimethylsilapentane-5-sulfonic acid (DSS). Nitrogen and carbon chemical shifts were referenced indirectly to the 1H standard using a conversion factor derived from the ratio of NMR frequencies (Wishart et al. 1995). Spectra were processed using NMRPipe (Delaglio et al. 1995) with SMILE (Ying et al. 2017) for NUS reconstruction and analyzed using NMRFAM-Sparky (Lee et al. 2015).
Table 1.
List of experiments collected to perform the sequence specific assignment of nsp7
| Experiments | Time Domain Data Size (complex points) | Spectral width (ppm) | ns | Delay time (s) | |||||
|---|---|---|---|---|---|---|---|---|---|
| t1 | max t2 | max t3 | NUS points | F1 | F2 | F3 | |||
| 1H-15 N-HSQC | 1024 | 192 | 20.0 (1H) | 29.6 (15 N) | 16 | 1 | |||
| HNCACB | 1024 | 60 | 52 | 1116 | 20.0 (1H) | 66.3 (13C) | 29.6 (15 N) | 32 | 1 |
| CBCA(CO)NH | 1024 | 76 | 52 | 1400 | 20.0 (1H) | 66.3 (13C) | 29.6 (15 N) | 32 | 1 |
| HNCO | 1024 | 52 | 52 | 961 | 20.0 (1H) | 9.9 (13C) | 29.6 (15 N) | 16 | 1 |
| HN(CA)CO | 1024 | 52 | 52 | 961 | 20.0 (1H) | 9.9 (13C) | 29.6 (15 N) | 48 | 1 |
| 1H-13C-HSQC aliphatic | 1024 | 268 | 20.0 (1H) | 66.3 (13C) | 48 | 1 | |||
| C(CO)NH | 1024 | 64 | 48 | 999 | 20.0 (1H) | 66.3 (13C) | 29.6 (15 N) | 48 | 1 |
| HBHA(CO)NH | 1024 | 64 | 48 | 1064 | 20.0 (1H) | 5.3 (1H) | 29.6 (15 N) | 32 | 1 |
| H(CCO)NH | 1024 | 64 | 48 | 1064 | 20.0 (1H) | 6.3 (1H) | 29.6 (15 N) | 64 | 1 |
| H(C)CH-TOCSY aliphatic | 1024 | 64 | 76 | 1536 | 20.0 (1H) | 8.3 (1H) | 72.9 (13C) | 32 | 1 |
| NOESY 1H-13C-HSQC aliphatic | 1024 | 64 | 64 | 1513 | 20.0 (1H) | 12.7 (1H) | 72.9 (13C) | 32 | 1 |
| 1H-13C-HSQC aromatic | 1024 | 34 | 16.7 (1H) | 33.1 (13C) | 128 | 1 | |||
| (HB)CB(CGCD)HE | 1024 | 48 | 16.7 (1H) | 26.5 (13C) | 256 | 1 | |||
| H(C)CH-TOCSY aromatic | 1024 | 34 | 48 | 512 | 16.7 (1H) | 3.0 (1H) | 33.1 (13C) | 64 | 1 |
| NOESY 1H-13C-HSQC aromatic | 1024 | 64 | 48 | 1131 | 20.0 (1H) | 12.7 (1H) | 33.1 (13C) | 32 | 1 |
Assignments and data deposition
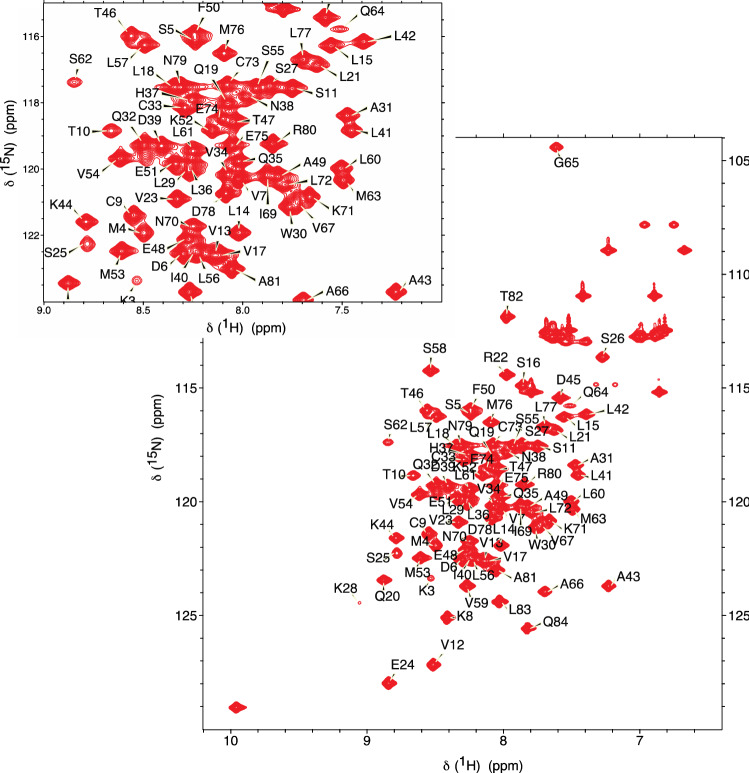
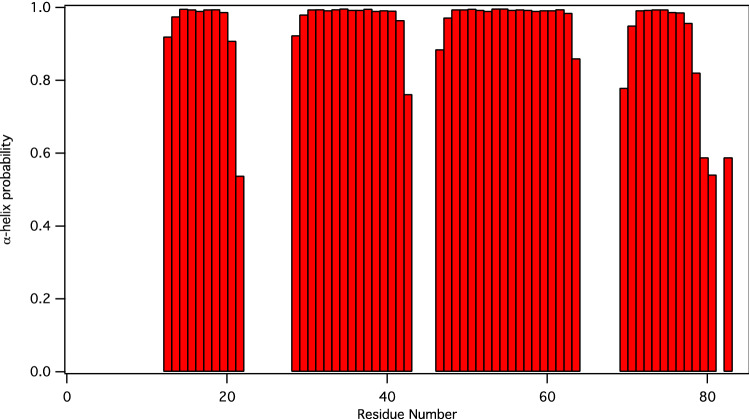
The 1H-15N HSQC spectrum of nsp7 shows well-dispersed amide signals (Fig. 1). Assignments were performed with i-PINE (Lee et al. 2019) using PINE-Sparky2 automated (Lee and Markley 2018) interface and manual confirmation in NMRFAM-Sparky (Lee et al. 2015). Backbone assignments are 96% complete with G0, S1, and D67 not visible in the 15N-HSQC spectrum. For the nsp7 sequence (S1-Q83), assignments are 99% complete for Cα, Cβ, and CO (only V66 unassigned), and 98% complete for HN and N (D67 and S1 unassigned). Secondary structure prediction was performed using chemical shift assignments of five atoms (HN, Cα, Cβ, CO, N) for a given residue in the sequence with TALOS-N (Shen and Bax 2013). The results for nsp7 are shown in Fig. 2.
Fig. 1.
Assigned 1H,15 N-HSQC spectrum of the 13C,15 N-labelled SARS-CoV-2 nsp7 at 1.7 mM concentration in 10 mM MOPS, pH 7.0, 150 mM NaCl, 2 mM DTT, 0.025% NaN3 and 7% D2O measured at 298 K on a 600 MHz Agilent NMR Spectrometer with backbone NH chemical shift assignments shown. The inset shows the central region of the spectrum enlarged for clarity
Fig. 2.
Display of TALOS-N predicted secondary structure for nsp7. Helical probability shown in red, residues that are highly dynamic, predicted to be coil, or for which there is not a consistent prediction are not shown
With the exception of G0, the aliphatic and aromatic side chain C-H groups for all residues were assigned (> 99% completeness overall, 100% for nsp7 sequence). In addition, the Nδ2-Hδ2 groups of N residues, the Nε2-Hε2 groups of Q residues, the Nε-Hε groups of R residues and the Nε1-Hε1 group of W29 were also assigned. The Nζ-Hζ groups of K residues and the Nδ1-Hδ1 and Nε2-Hε2 groups of H36 were not assigned.
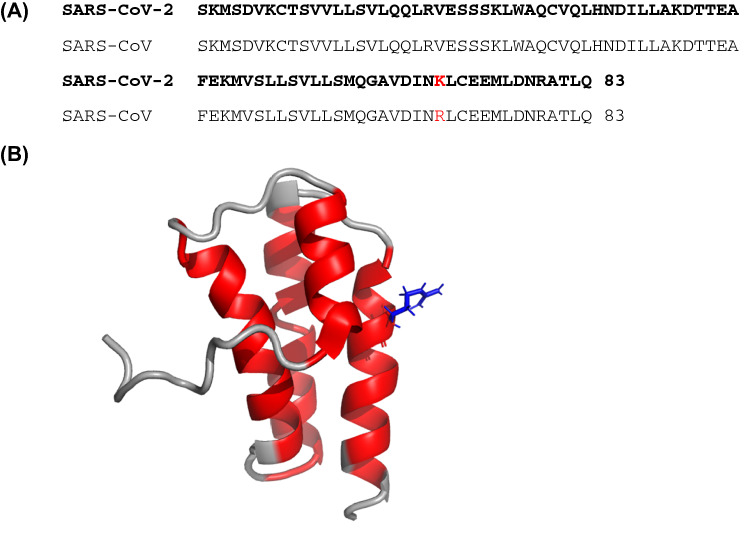
The structure of nsp7 from SARS coronavirus was previously determined by NMR by the Wüthrich lab, first at pH 7.5 and high ionic strength (Peti et al. 2005) (BMRB ID 6513, PDB ID 1YSY) and later at pH 6.5 (Johnson et al. 2010) (BMRB ID 16,981, PDB ID 2KYS). The sequence of nsp7 from SARS-2 and SARS are nearly identical, with only a single conservative amino acid difference at position 70 (Fig. 3). The backbone chemical shifts for nsp7 from SARS-2 and SARS are very similar, as might be expected given the very high sequence identity. The dihedral angles predicted by TALOS-N for nsp7 from SARS are in good agreement with the previous SARS coronavirus nsp7 NMR structure (Fig. 3).
Fig. 3.
Comparison of nsp7 from SARS-CoV-2 and SARS-CoV. a Sequence comparison shows a single conservative amino acid substitution. b The SARS-CoV-2 helical regions (red) shown in Fig. 2 are plotted on the SARS-CoV nsp7 structure determined by NMR at pH 6.5 (PDB 2KYS). This structure is higher quality than the pH 7.5 structure (PDB 1YSY) because more complete assignments and a larger number of restraints were obtained at pH 6.5. The location of the single amino acid difference between SARS-CoV and SARS-CoV-2 nsp7 is highlighted in blue sticks
The chemical shift values for the 1H, 13C and 15N resonances of nsp7 have been deposited at the BioMagResBank (https://www.bmrb.wisc.edu) under accession number 50337. Raw data has been deposited in BMRbig (https://bmrbig.org/) under deposition ID bmrbig4.
Acknowledgements
Funding for this project was provided by the National Science Foundation EAGER MCB-2031269 and the National Institutes for Health, National Institute for Allergy and Infectious Disease AI123498. This study made use of the National Magnetic Resonance Facility at Madison, which is supported by NIH grant P41GM103399 (NIGMS), old number: P41RR002301. Equipment was purchased with funds from the University of Wisconsin-Madison, the NIH P41GM103399, S10RR02781, S10RR08438, S10RR023438, S10RR025062, S10RR029220), the NSF (DMB-8415048, OIA-9977486, BIR-9214394), and the USDA.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Bchetnia M, Girard C, Duchaine C, Laprise C. The outbreak of the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): a review of the current global status. J Infect Public Health. 2020 doi: 10.1016/j.jiph.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Gao Y, et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert I, et al. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 2006;25:4933–4942. doi: 10.1038/sj.emboj.7601368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Jaudzems K, Wuthrich K. NMR structure of the SARS-CoV nonstructural protein 7 in solution at pH 6.5. J Mol Biol. 2010;402:619–628. doi: 10.1016/j.jmb.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer RN, Ward AB. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkolova E, Klima M, Nencka R, Boura E. Structural analysis of the putative SARS-CoV-2 primase complex. J Struct Biol. 2020;211:107548. doi: 10.1016/j.jsb.2020.107548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Bahrami A, Dashti HT, Eghbalnia HR, Tonelli M, Westler WM, Markley JL. I-PINE web server: an integrative probabilistic NMR assignment system for proteins. J Biomol NMR. 2019;73:213–222. doi: 10.1007/s10858-019-00255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Markley JL. PINE-SPARKY.2 for automated NMR-based protein structure research. Bioinformatics. 2018;34:1586–1588. doi: 10.1093/bioinformatics/btx785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Tonelli M, Markley JL. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peti W, et al. Structural genomics of the severe acute respiratory syndrome coronavirus: nuclear magnetic resonance structure of the protein nsP7. J Virol. 2005;79:12905–12913. doi: 10.1128/JVI.79.20.12905-12913.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J Biomol NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder EJ, Decroly E, Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subissi L, et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci U S A. 2014;111:E3900–3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Velthuis AJ, van den Worm SH, Snijder EJ. The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension. Nucleic Acids Res. 2012;40:1737–1747. doi: 10.1093/nar/gkr893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoek L. Human coronaviruses: what do they cause? Antivir Ther. 2007;12:651–658. [PubMed] [Google Scholar]
- Wishart DS, et al. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- Wu F, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, et al. Nonstructural proteins 7 and 8 of feline coronavirus form a 2:1 heterotrimer that exhibits primer-independent RNA polymerase activity. J Virol. 2012;86:4444–4454. doi: 10.1128/JVI.06635-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying J, Delaglio F, Torchia DA, Bax A. Sparse multidimensional iterative lineshape-enhanced (SMILE) reconstruction of both non-uniformly sampled and conventional NMR data. J Biomol NMR. 2017;68:101–118. doi: 10.1007/s10858-016-0072-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Sun F, Li X, Pang H, Xu X, Bartlam M, Rao Z. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat Struct Mol Biol. 2005;12:980–986. doi: 10.1038/nsmb999. [DOI] [PMC free article] [PubMed] [Google Scholar]