Abstract
INTRODUCTION:
We prospectively studied the frequency, spectrum, and predictors of gastrointestinal (GI) symptoms among patients with coronavirus disease-19 (COVID-19) and the relationship between GI symptoms and the severity and outcome.
METHODS:
Consecutive patients with COVID-19, diagnosed in a university hospital referral laboratory in northern India, were evaluated for clinical manifestations including GI symptoms, their predictors, and the relationship between the presence of these symptoms, disease severity, and outcome on univariate and multivariate analyses.
RESULTS:
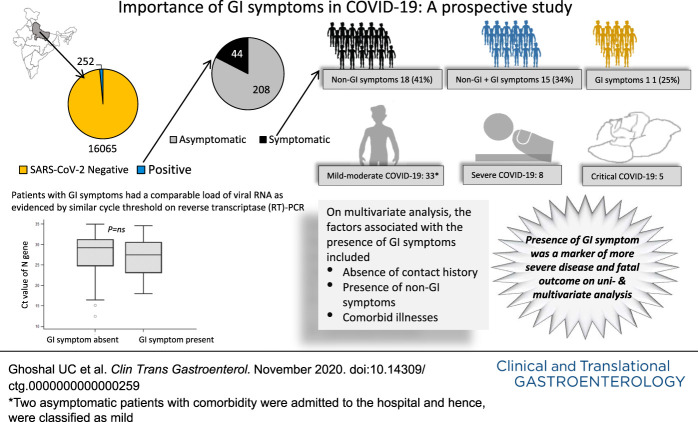
Of 16,317 subjects tested for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in their oropharyngeal and nasopharyngeal swabs during April–May 2020, 252 (1.5%) were positive. Of them, 208 (82.5%) were asymptomatic; of the 44 symptomatic patients, 18 (40.9%) had non-GI symptoms, 15 (34.1%) had a combination of GI and non-GI symptoms, and 11 (25.0%) had GI symptoms only. Thirty-three had mild-to-moderate disease, 8 severe, and 5 critical. Five patients (1.98%) died. On multivariate analysis, the factors associated with the presence of GI symptoms included the absence of contact history and presence of non-GI symptoms and comorbid illnesses. Patients with GI synptoms more often had severe, critical illness and fatal outcome than those without GI symptoms.
DISCUSSION:
Eighty-two percent of patients with COVID-19 were asymptomatic, and 10.3% had GI symptoms; severe and fatal disease occurred only in 5% and 2%, respectively. The presence of GI symptoms was associated with a severe illness and fatal outcome on multivariate analysis. Independent predictors of GI symptoms included the absence of contact history, presence of non-GI symptoms, and comorbid illnesses.
INTRODUCTION
Infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a RNA virus belonging to the β Coronavirus family, began in Wuhan, Hubei province, China, in December 2019 that caused the current pandemic of the disease (1). On May 17, 2020, the day when this analysis was undertaken, 4,717,079 subjects in 214 countries have been diagnosed to have COVID-19, of whom 312,388 (6.6%) died. The first patient with coronavirus disease (COVID-19) was confirmed in India on January 30,, 2020 (2). On the day of this analysis, 90,648 were diagnosed having COVID-19 in India, of whom 2,871 (3.2%) died. The infected subjects may also remain asymptomatic or may develop a minor flu-like illness or potentially fatal pneumonia and respiratory failure (3).
The SARS-CoV-2 virus enters into the host cells through transmembrane serine protease and angiotensin receptor 2 (ACE2) receptors, which have a high affinity to the spike (S) protein of the virus (4–6). The ACE2 receptors are not only expressed in the pulmonary epithelial cells but also in the esophageal, small intestinal, and colonic epithelial cells (7). Moreover, the virus is also excreted in the feces in about 50% of patients with COVID-19 (8), and some studies showed that the fecal excretion of the virus might continue in both adults and children even after the SARS-CoV-2 RNA turned negative in upper respiratory tract specimens (9,10). One study from China showed gastrointestinal (GI) lesions in the form of esophageal ulcers and the presence of the virus in the biopsies of the GI tract (11). Hence, SARS-CoV-2 infection affects not only the respiratory but also the GI tract. Although systemic and respiratory manifestations such as fever, cough, myalgia, fatigue, pneumonia, respiratory distress, coagulopathy, and even pulmonary hemorrhage are well-recognized manifestations of COVID-19, GI manifestations such as anorexia, nausea, vomiting, diarrhea, abdominal pain, and discomfort are being reported increasingly among these patients (3,6,12–14). A Chinese study reported GI symptoms to occur even in the absence of respiratory symptoms in 28% of patients (15). A few studies, mostly retrospective, did suggest that the presence of GI symptoms may be associated with the severity of the COVID-19, although the others refuted this observation (13,15,16). Factors related to the presence of GI symptoms have hardly been studied. Accordingly, we undertook a prospective study with the following aims: (i) frequency and spectrum of GI symptoms among patients with SARS-CoV-2 infection, (ii) factors associated with GI symptoms, and (iii) relationship between the presence of GI symptoms and severity and outcome of COVID-19.
METHODS
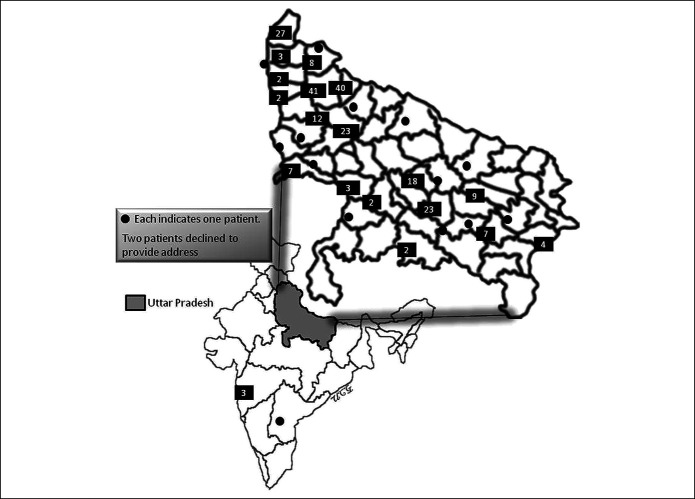
Consecutive patients with SARS-CoV-2 infection diagnosed in a referral laboratory of a university hospital in northern India during a 1-month period (between April and May 2020) were prospectively studied after their consent and clearance from the ethics committee of the institute (2020-117-EMP-EXP-17). Subjects visited the institute directly or the oropharyngeal and nasopharyngeal swab samples were obtained from the various districts of the state of Uttar Pradesh (Figure 1) through the Integrated Disease Surveillance Program (IDSP); IDSP is a scheme under the Ministry of Health and Family Welfare, government of India, to quickly respond for surveillance of infectious disease outbreak with a central and multiple state surveillance units, one from each state. The demographic and clinical data were obtained by face-to-face interviews for the patients directly visiting the institute and by an electronic online questionnaire using Survey Monkey (SurveyMonkey Enterprise, San Mateo, CA) assisted by telephonic interviews for remote patients. The questionnaire was designed to obtain data on demographic parameters, dietary and contact histories, clinical parameters including the presence of comorbidity, medication history, addiction, previous and current GI symptoms, and current systemic and respiratory symptoms. Patients were assessed for GI symptoms such as anorexia, nausea, vomiting, diarrhea, abdominal pain, and discomfort. Patients were quarantined either at home or hospitals depending on the severity of illness and followed-up either physically or telephonically. The severity of the COVID-19 was assessed as described previously—(i) critical (required ventilator), (ii) severe (needed oxygen), (iii) moderate (although pneumonia was present, the patient did not require oxygen), and (iv) mild (only upper respiratory symptoms) (17). Those without symptoms were classified as asymptomatic.
Figure 1.
A map of India showing the state of Uttar Pradesh from where the study subjects (except 4 participants) were included through the Integrated Disease Surveillance Program. The number within each black box within the enlarged map of Uttar Pradesh represents the number of patients from that particular region. The filled circle indicates one patient in each area. Two patients declined to provide their addresses.
Testing for SARS-CoV-2 RNA
The naso- and oro-pharyngeal swab samples were obtained as per standard protocol wearing personal protective equipment (18). The indications of testing for SARS-CoV-2 RNA included the following: (i) all subjects with suggestive symptoms, (ii) all contacts of confirmed COVID-19 patients, (iii) high-risk occupational workers (e.g., healthcare and security staff members), (iv) people traveling back to the country from an international destination, where the infection was endemic, and (v) on the discretion of the physicians.
RNA extraction from the clinical specimens
RNA was extracted from the oro- and naso-pharyngeal swab samples with a QIAamp viral RNA mini kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. The specimens were tested under standard biosafety guidelines (18).
SARS-CoV-2 detection
The real-time reverse transcriptase polymerase chain reaction (PCR) for SARS-CoV-2 was performed by the TaqPathTM COVID-19 Combo Kit (Applied Biosystems, CA) as per instructions of the manufacturer. Briefly, the PCR condition included uracil-DNA glycosylases enzyme (mixed in the master mix to prevent laboratory carryover contamination) incubation at 25°C for 2 minutes, followed by reverse transcription at 53°C for 10 minutes and activation at 95°C for 2 minutes. PCR amplification was performed for 40 cycles at 95°C for 03 seconds and 60°C for 30 seconds using an Applied Biosystems 7500 instrument (Applied Biosystems, Singapore).
Determination of the cycle threshold
Cycle threshold (Ct) gives a measure of viral load in the sample, a lower meaning higher load, and vice versa. The results of Ct are known to correlate with that of digital droplet PCR for quantification (19). During reverse transcriptase polymerase chain reaction, open reading frame 1ab, nucleocapsid protein (N), and spike (S) genes were amplified and tested. The Ct-value was defined as the number of cycles required for the fluorescence signal to cross the threshold (i.e., exceed the background level).
Statistical analysis
The data were checked for normal distribution using the Shapiro-Wilk test. Parametric and nonparametric continuous data were presented as mean and SD and median and interquartile range, respectively. Categorical data were presented as numbers and percentages. Categorical and continuous data were analyzed using χ2, unpaired t, and Mann–Whitney U tests depending on the distribution. Stepwise logistic regression and likelihood ratio tests were used for multivariate analyses. P values less than 0.05 were considered significant. R, Epicalc, and R-studio software (R development core team, Vienna, Austria) and Statistical Package for Social Science version 15 (SPSS, Inc.), Epi Info (Centers for Disease Control and Prevention, Atlanta, Georgia, USA), and MedCalc version 14 (Warandeberg 3, 1000 Brussels, Belgium) were used for the statistical analysis.
RESULTS
During the 1-month study period, a total of 16,317 subjects were tested for SARS-CoV-2 RNA in their oro- and naso-pharyngeal swab samples. Although 3,355 subjects (20.6%) visited the institute directly, 12,962 subjects' (79.4%) samples were obtained from remote centers through IDSP (Figure 1). Two hundred fifty-two of 16,317 subjects (1.5%) showed positive test results for SARS-CoV-2 RNA (15/252, 5.9% among the institute subjects and 237, 94.1% in IDSP samples). The rate of positivity in Institute samples was lower than that obtained from the IDSP subjects (15/3,355 [0.4%] vs 237/12,962 [1.8%]; P < 0.00001).
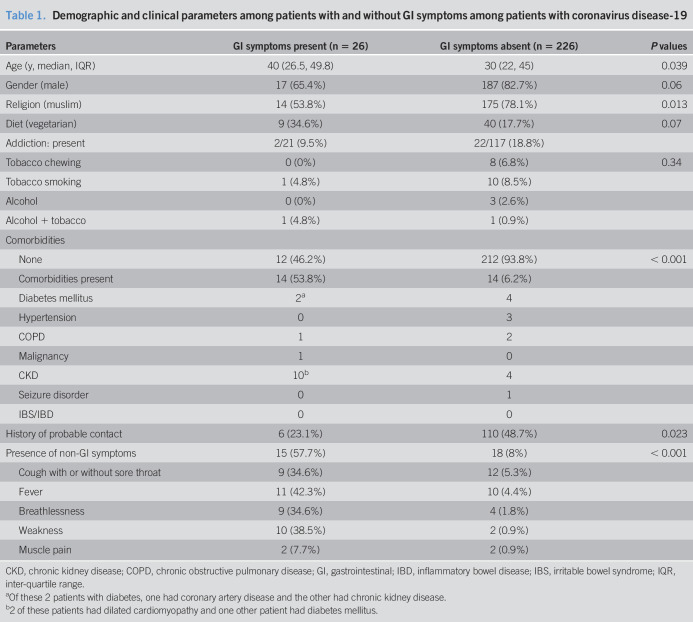
Of 252 subjects with COVID-19 (mean age 34.3 ± 15.6-years, 204 [81%] male), 28 (11%) had comorbid illnesses (Table 1). None of the patients had a previous history of irritable bowel syndrome (by Rome criteria), inflammatory bowel disease, malabsorption syndrome, or were on treatment for such disorders (Table 1). Of 138 subjects providing information on addiction, 24 (17.4%) had addictions to either alcohol, tobacco chewing, or smoking (Table 1). One hundred eighty-nine patients were of Muslim and 63 of Hindu religions, and the dietary practice of none of the former group and 49 of the latter group was vegetarianism. One hundred sixteen subjects (46%) had previous contact with a patient with COVID-19.
Table 1.
Demographic and clinical parameters among patients with and without GI symptoms among patients with coronavirus disease-19
| Parameters | GI symptoms present (n = 26) | GI symptoms absent (n = 226) | P values |
| Age (y, median, IQR) | 40 (26.5, 49.8) | 30 (22, 45) | 0.039 |
| Gender (male) | 17 (65.4%) | 187 (82.7%) | 0.06 |
| Religion (muslim) | 14 (53.8%) | 175 (78.1%) | 0.013 |
| Diet (vegetarian) | 9 (34.6%) | 40 (17.7%) | 0.07 |
| Addiction: present | 2/21 (9.5%) | 22/117 (18.8%) | |
| Tobacco chewing | 0 (0%) | 8 (6.8%) | 0.34 |
| Tobacco smoking | 1 (4.8%) | 10 (8.5%) | |
| Alcohol | 0 (0%) | 3 (2.6%) | |
| Alcohol + tobacco | 1 (4.8%) | 1 (0.9%) | |
| Comorbidities | |||
| None | 12 (46.2%) | 212 (93.8%) | < 0.001 |
| Comorbidities present | 14 (53.8%) | 14 (6.2%) | |
| Diabetes mellitus | 2a | 4 | |
| Hypertension | 0 | 3 | |
| COPD | 1 | 2 | |
| Malignancy | 1 | 0 | |
| CKD | 10b | 4 | |
| Seizure disorder | 0 | 1 | |
| IBS/IBD | 0 | 0 | |
| History of probable contact | 6 (23.1%) | 110 (48.7%) | 0.023 |
| Presence of non-GI symptoms | 15 (57.7%) | 18 (8%) | < 0.001 |
| Cough with or without sore throat | 9 (34.6%) | 12 (5.3%) | |
| Fever | 11 (42.3%) | 10 (4.4%) | |
| Breathlessness | 9 (34.6%) | 4 (1.8%) | |
| Weakness | 10 (38.5%) | 2 (0.9%) | |
| Muscle pain | 2 (7.7%) | 2 (0.9%) |
CKD, chronic kidney disease; COPD, chronic obstructive pulmonary disease; GI, gastrointestinal; IBD, inflammatory bowel disease; IBS, irritable bowel syndrome; IQR, inter-quartile range.
Of these 2 patients with diabetes, one had coronary artery disease and the other had chronic kidney disease.
2 of these patients had dilated cardiomyopathy and one other patient had diabetes mellitus.
Symptoms and illness severity
Although 44 of 252 (17.5%) had some symptoms, all of the remaining 208 patients (82.5%) were asymptomatic. Of the 44 symptomatic patients, 18 (40.9%) had non-GI symptoms only, 15 (34.1%) had a combination of GI and non-GI symptoms, and 11 (25.0%) had GI symptoms only. Thirty-three had mild-to-moderate disease, 8 severe, and 5 critical. Five patients (1.98%) (4 [80%] male, 4 [80%] with comorbidities) died. Two asymptomatic patients with comorbidity were admitted to the hospital and hence, were classified as mild. Comorbid illnesses included one malignancy and 3 chronic renal failure with hypertension, one of whom had dilated cardiomyopathy as well. One patient each in the age group from 25 to 30 years, 35–40 years, 40–45 years, 60–65 years, and 65–70 years had a fatal illness.
Non-GI symptoms
Non-GI symptoms included cough with or without sore throat (n = 21), fever (n = 21), weakness (n = 12), and muscle pain (n = 4).
GI symptoms
Of 26/252 patients (10.3%) with GI symptoms, these were the only clinical manifestations in 11 subjects. GI symptoms included anorexia (n = 14), nausea (n = 14), vomiting (n = 16), abdominal pain (n = 6), and diarrhea (n = 3).
Univariate analysis of the relationship between GI symptoms and demographic, clinical, and laboratory parameters
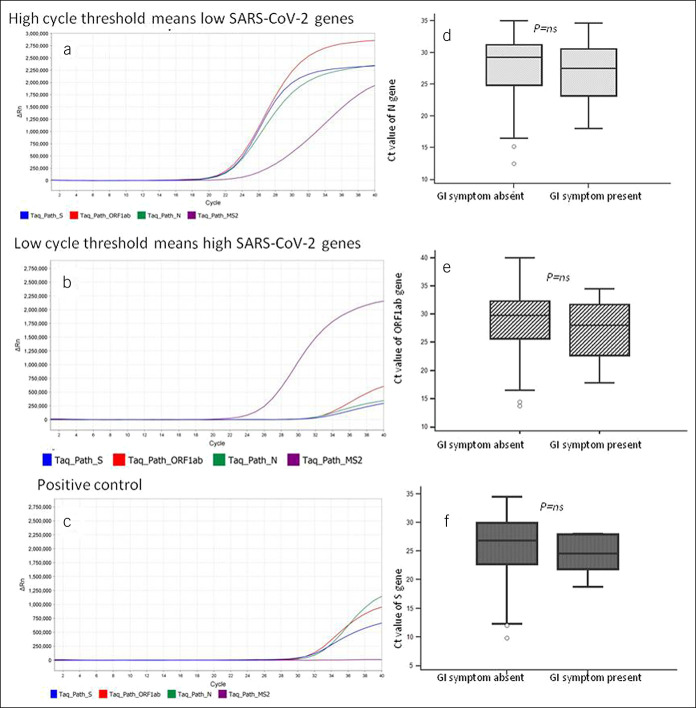
Patients with GI symptoms were older, more often vegetarian, had comorbid illnesses, more likely to have non-GI symptoms, less often had a history of contact, and Muslim religion (Table 1). Of 116 patients reporting a previous history contact, 17 (14.6%) were symptomatic (GI symptoms in 4 [3.4%], non-GI symptoms in 11 [9.5%], and both in 2 [1.7%]) and 99 (85.3%) were asymptomatic. Of 136 patients without previous history of contact, 27 (19.8%) were symptomatic (GI symptoms in 7 [5.1%], non-GI symptoms in 7 [5.1%], and both 13 [9.5%]) and 109 (80.1%) were asymptomatic. Patients with a previous history of contact were less likely to have GI symptoms either alone or in combination with non-GI symptoms than those without a history of contact (6/116 [5.2%] vs 20/136 [14.7%], respectively; P = 0.02). Of the 13 patients who were younger than 15 years, none had any GI or non-GI symptoms. By contrast, of the 239 patients older than 15 years, 26 (10.8%) had GI symptoms (P = 0.4). Patients with GI symptoms as compared to those without GI symptoms had comparable median Ct values of all the studied SARS-CoV-2 genes, suggesting similar oro- and naso-pharyngeal RNA levels in both the groups (Figure 2).
Figure 2.
Amplification plots of real-time polymerase chain reactions of 2 representative patients with severe acute respiratory distress syndrome coronavirus-2 (SARS-CoV-2) infection. (a) High Ct, (b). Low Ct, (c) Positive control. Comparison of Ct values of nucleocapsid protein (N) gene (d), open reading frame 1ab gene (e), and spike protein (S) gene (f) among patients with and without gastrointestinal (GI) symptoms. The P values were not significant (NS) for any of the comparisons. MS2 represents control indicating the adequacy of RNA extraction. Ct, Cycle threshold; GI, gastrointestinal.
Univariate analysis of the relationship between GI symptoms and disease severity and outcome
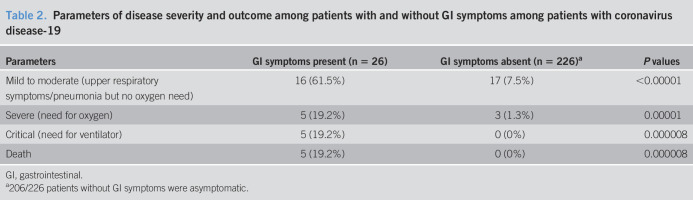
Patients with GI symptoms than those without GI symptoms more often had a severe and critical illness (Table 2). All the 5 patients who succumbed to the disease had GI symptoms.
Table 2.
Parameters of disease severity and outcome among patients with and without GI symptoms among patients with coronavirus disease-19
| Parameters | GI symptoms present (n = 26) | GI symptoms absent (n = 226)a | P values |
| Mild to moderate (upper respiratory symptoms/pneumonia but no oxygen need) | 16 (61.5%) | 17 (7.5%) | <0.00001 |
| Severe (need for oxygen) | 5 (19.2%) | 3 (1.3%) | 0.00001 |
| Critical (need for ventilator) | 5 (19.2%) | 0 (0%) | 0.000008 |
| Death | 5 (19.2%) | 0 (0%) | 0.000008 |
GI, gastrointestinal.
206/226 patients without GI symptoms were asymptomatic.
Results of multivariate analysis
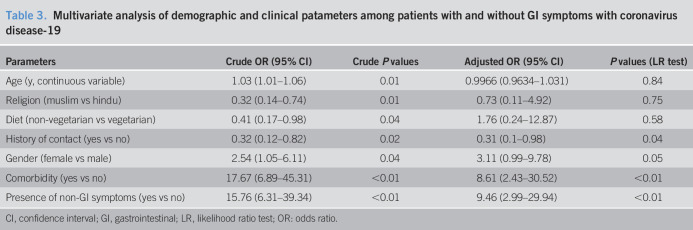
On multivariate analysis, the independent factors associated with the presence of GI symptoms included the absence of contact history, presence of non-GI symptoms, and comorbid illnesses (Table 3).
Table 3.
Multivariate analysis of demographic and clinical patameters among patients with and without GI symptoms with coronavirus disease-19
| Parameters | Crude OR (95% CI) | Crude P values | Adjusted OR (95% CI) | P values (LR test) |
| Age (y, continuous variable) | 1.03 (1.01–1.06) | 0.01 | 0.9966 (0.9634–1.031) | 0.84 |
| Religion (muslim vs hindu) | 0.32 (0.14–0.74) | 0.01 | 0.73 (0.11–4.92) | 0.75 |
| Diet (non-vegetarian vs vegetarian) | 0.41 (0.17–0.98) | 0.04 | 1.76 (0.24–12.87) | 0.58 |
| History of contact (yes vs no) | 0.32 (0.12–0.82) | 0.02 | 0.31 (0.1–0.98) | 0.04 |
| Gender (female vs male) | 2.54 (1.05–6.11) | 0.04 | 3.11 (0.99–9.78) | 0.05 |
| Comorbidity (yes vs no) | 17.67 (6.89–45.31) | <0.01 | 8.61 (2.43–30.52) | <0.01 |
| Presence of non-GI symptoms (yes vs no) | 15.76 (6.31–39.34) | <0.01 | 9.46 (2.99–29.94) | <0.01 |
CI, confidence interval; GI, gastrointestinal; LR, likelihood ratio test; OR: odds ratio.
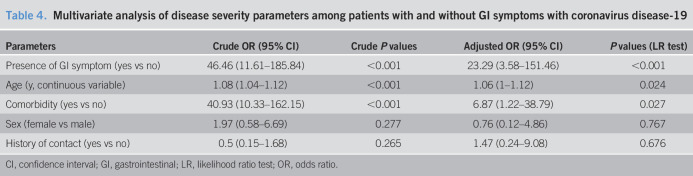
As shown in Table 4, the presence of GI symptoms was associated with more severe illness on multivariate analysis. The other independent factors related to the presence of severe illness included older age and the presence of comorbid diseases. However, the female sex and the history of contact had no association with the severity of the illness on multivariate analysis.
Table 4.
Multivariate analysis of disease severity parameters among patients with and without GI symptoms with coronavirus disease-19
| Parameters | Crude OR (95% CI) | Crude P values | Adjusted OR (95% CI) | P values (LR test) |
| Presence of GI symptom (yes vs no) | 46.46 (11.61–185.84) | <0.001 | 23.29 (3.58–151.46) | <0.001 |
| Age (y, continuous variable) | 1.08 (1.04–1.12) | <0.001 | 1.06 (1–1.12) | 0.024 |
| Comorbidity (yes vs no) | 40.93 (10.33–162.15) | <0.001 | 6.87 (1.22–38.79) | 0.027 |
| Sex (female vs male) | 1.97 (0.58–6.69) | 0.277 | 0.76 (0.12–4.86) | 0.767 |
| History of contact (yes vs no) | 0.5 (0.15–1.68) | 0.265 | 1.47 (0.24–9.08) | 0.676 |
CI, confidence interval; GI, gastrointestinal; LR, likelihood ratio test; OR, odds ratio.
DISCUSSION
The present prospective study on a large sample of patients with COVID-19 from India showed that (i) as high as 82% patients with COVID-19 were asymptomatic, (ii) 10.3% patients had GI symptoms, (iii) 25% of the symptomatic patients had GI symptoms as the only manifestation, (iv) the presence of GI symptoms was associated with the absence of contact history, presence of non-GI symptoms, and comorbid illnesses on multivariate analysis, (v) patients with GI symptoms had a comparable load of viral RNA as evidenced by similar Ct values on reverse transcriptase polymerase chain reaction, (vi) severe and critical illness occurred in 5% patients, and less than 2% died, and (vii) the presence of GI symptoms was associated with more severe disease and fatal outcome.
Infection with SARS-CoV-2 is increasingly recognized to cause GI symptoms in addition to the respiratory symptoms (13,20). In a meta-analysis from Hong Kong, including 60 studies on 4,243 patients, the pooled prevalence of GI symptoms was 17.6% (95% confidence interval [CI], 12.3%–24.5%); this frequency is somewhat higher than that observed in our study (13). It is important to note that many of the earlier studies have been conducted in hospitalized patients receiving multiple drugs, including antibiotics. The prevalence of GI symptoms reported in our study may be a true reflection of the frequency because most of these patients were not hospitalized and were not on drugs that might be associated with GI symptoms. In another recent systematic review and meta-analysis of 62 studies, including 8,301 patients, GI symptoms were reported to be present in 10% of patients (20). In yet another recent meta-analysis from the United States of 47 studies, including 10,890 patients, pooled prevalence of GI symptoms such as diarrhea, nausea, vomiting was 7.8% (21). Our finding showing that 25% of symptomatic patients with COVID-19 had only GI symptoms has been reported earlier (15). In 5 studies from China, the frequency of GI symptoms alone in patients with COVID-19 varied between 3.4% and 28%, although an American study contradicted this observation (13,19).
A few studies, some with perhaps a small sample size or retrospective design, suggested that GI symptoms may be associated with more severe illness and worse outcome (15,16); however, this suggestion has not been borne out in a meta-analysis (pooled prevalence in severe disease: 17.1% [95% CI 6.9%–36.7%]) vs in less severe disease: 11.8% [95% CI, 4.1%–29.1%]) and an original study from the United States. (13,22). Therefore, this issue needs studies on a larger sample of patients recruited prospectively. Moreover, most of the earlier studies were on hospitalized patients in whom other factors such as drugs (antibiotic, chloroquine etc.) could have contributed to GI symptoms. In this prospective study on a relatively large number of patients, most of whom were not hospitalized, we found that the severity of the COVID-19 and mortality were associated with the presence of GI symptoms on both univariate and multivariate analyses.
We also found several factors to be associated with the presence of GI symptoms; these included the absence of contact history, presence of non-GI symptoms, and comorbid illnesses on multivariate analysis. To the best of our knowledge, no previous study addressed the association between these factors and the presence of GI symptoms. The possible explanation for such an association between absence of contact history and presence of GI symptoms might be related to the fact that in the absence of a contact history, subjects are likely to be tested for SARS-CoV-2 RNA only when they report symptoms; by contrast, those with a contact history are tested even in the absence of symptoms. Patients older than 15 years of age were more often symptomatic (including GI symptoms) than the younger patients. This is quite expected. The young children get less severe COVID-19 than the older population because the ACE2 gene expression, which is required for viral entry in nasal epithelial cells, is lower in young children and it increases with age (23).
Interestingly, the current study showed that illness was less severe (5% vs 14%–31% in the United States and 38.6% in Italy (24)) and fatal (1.98% vs 19% in the United States (25) and 26% in Italy (26)) in India than that described in other countries. What could be the possible explanation for these contradictory observations? The severity of illness in patients with SARS-CoV-2 infection is related mainly to the host's immune response to the virus resulting in a cytokine storm (3,27). In tropical and subtropical countries, particularly in areas with a less hygienic condition, autoimmune and allergic diseases are less likely, possibly due to a robust T regulatory response (hygiene hypothesis) (28,29). Could this play a role in causing a less severe cytokine storm in Indian patients? Could good gut microbiota play a role in reducing the illness severity? A single study on this issue from Hong Kong on a small number of patients showed that baseline abundance of Coprobacillus, Clostridium ramosum, and Clostridium hathewayi had a positive correlation and Faecalibacterium prausnitzii (an anti-inflammatory bacterium) had a negative correlation with the COVID-19 severity (30). Similar viral RNA load among patients with and without GI symptoms may suggest that the host rather than agent factors might be more important in disease severity and outcome of SARS-CoV-2 infection. Could a different (less virulent) genotype of the SARS-CoV-2 in India be responsible for the lesser severity of the disease? Earlier studies did show different subtypes of SARS-CoV-2 in different geographical areas. Of the 3 different subtypes of the virus (A [the ancestral type such as the bat coronavirus], B, and C), “A” and “C” are prevalent in the United States and Europe and “B” subtype is common in East Asia (31). Could dietary differences in India than the rest of World be important? These possible explanations need to be studied in the future. Another reason for lesser fatality in India might be related to a lower proportion of the older population in developing countries and the presence of associated comorbid illnesses, which is known to influence disease severity and outcome (32). Although some experts suggested that the high frequency of Bacillus Calmette-Guerin vaccination among the Indian people in their childhood might be one of the reasons for less severe illness (33), this might be only a surrogate marker of a high frequency of infective diseases leading to a better T regulatory response.
The strength of our study is a prospective design, large sample size, multivariate analysis, and more wide coverage because of the inclusion of the patients from the community through the IDSP program (Figure 1) mitigating the possible recruitment bias. We also excluded the presence of previous chronic GI diseases, such as irritable bowel syndrome and inflammatory bowel disease, and the effect of concurrent drug treatment, which might contribute to the development of GI symptoms (34). However, our study is not free from limitations. Though the previously published data from other countries on morbidity and mortality with which our data have been compared are mostly hospital based, most of our patients are from the community. Because this is an observational study, we did not evaluate the mechanistic aspects of pathogenesis of GI symptoms in patients with COVID-19. Both direct viral cytopathic damage on the mucosa and an indirect effect of the systemic cytokine storm may contribute to the pathogenesis of GI symptoms in patients with COVID-19. We do not report on fecal viral RNA or systemic cytokine response in this study. The Ct value is not the gold standard method for quantification of SARS-CoV-2 RNA, although the Ct value does correlate with the value of viral quantification by digital droplet PCR (19).
We conclude that as high as 82% of patients with COVID-19 were asymptomatic and severe and fatal diseases occurred only in 5% and 2% patients, respectively, in our population. 10.3% of patients had GI symptoms, 25% of the symptomatic patients had GI symptoms as the only manifestation, and GI symptoms were associated with more severe illness and fatal outcome. We found some factors such as the absence of contact history, presence of non-GI symptoms, and comorbid diseases to be the predictors of presence of GI symptoms on multivariate analysis. We believe that determinants of disease severity and outcome would have important clinical implications, mainly because an effective treatment and vaccine are yet to be available for therapy and prevention of COVID-19.
CONFLICTS OF INTEREST
Guarantor of the article: Uday C. Ghoshal, MD, DNB, DM, FACG, FAMS, RFF.
Specific author contributions: Uday C. Ghoshal, MD, DNB, DM, FACG, FAMS, RFF and Ujjala Ghoshal, MD, MNAMS, contributed equally to this work and are therefore, the joint first authors. U.C.G. designed the study, collected clinical data electronically using an online questionnaire, analyzed the data, wrote the first draft of the study; U.G., A.G., D.S., S.S., J.S., and A.P. did microbiological works and helped clinical data collection, A.M. collected clinical data remotely as well on-site, R.K.S. and A.N. cared for the admitted patients, S.R. helped online data collection and analysis, S.V. collected clinical specimens and helped microbiological works, and R.K.D. supervised the whole study.
Financial support: Department of Biotechnology, Government of India, for the funding of the study (project No. BT/PR40311/COD/139/9/2020).
Potential competing interests: None to report.
Study Highlights.
WHAT IS KNOWN
✓ GI symptoms are not uncommon in patients with coronavirus disease-19 (COVID-19).
✓ Although a few studies, mostly retrospective and on hospitalized patients, showed that the presence of GI symptoms was associated with severe COVID-19 and fatal outcome, the others refuted it.
WHAT IS NEW HERE
✓ As high as 82% of patients with COVID-19 were asymptomatic and 10.3% of patients (most from the community) had GI symptoms; 25% of the symptomatic patients had GI symptoms as the only manifestation.
✓ The presence of GI symptoms was associated with the absence of contact history, presence of non-GI symptoms, and comorbid illnesses on multivariate analysis.
✓ Patients with GI symptoms had a comparable load of viral RNA as evidenced by similar Ct values on reverse transcriptase (RT)-PCR.
✓ Severe and critical illness occurred in 5% of patients, and less than 2% died.
✓ The presence of GI symptoms was associated with more severe disease and fatal outcome.
TRANSLATIONAL IMPACT
✓ Determinants of disease severity and outcome, including the GI symptoms, would have important clinical implications mainly because effective treatment and vaccine are yet to be available for therapy and prevention of COVID-19, respectively.
✓ Less severity of the disease in this population might be related to (i) less virulent viral genotype, (ii) good T regulatory response because of infective illness being common in this population (hygiene hypothesis), and (iii) better gut microbiota.
Supplementary Material
ACKNOWLEDGMENT
The authors thank the Department of Biotechnology, Government of India, for the funding for the study through (project No. BT/PR40311/COD/139/9/2020) to Uday C Ghoshal as the Principal Investigator.
The authors thank Virendra K Misra and Hemant Verma from the Dept. of Microbiology for their technical support. The authors also thank the patients and the staff members of the COVID-19 Laboratory, Dept. of Microbiology, SGPGI, Lucknow.
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at https://links.lww.com/CTG/A430
References
- 1.Zhai P, Ding Y, Wu X, et al. he epidemiology, diagnosis and treatment of COVID-19. Int J Antimicrob Agents 2020;55:105955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar SU, Kumar DT, Christopher BP, et al. The rise and impact of COVID-19 in India. Front Med (Lausanne) 2020;7:250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang D, Comish P, Kang R. The hallmarks of COVID-19 disease. Plos Pathog 2020;16:e1008536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wan Y, Shang J, Graham R, et al. Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS coronavirus. J Virol 2020;94:e00127–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ding S, Liang TJ. Is SARS-CoV-2 also an enteric pathogen with potential fecal-oral transmission: A COVID-19 virological and clinical review. Gastroenterology 2020;159:53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dhar J, Samanta J, Kochhar R. Corona virus disease-19 pandemic: The gastroenterologists' perspective. Indian J Gastroenterol 2020;39:220–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gui M, Song W, Zhou H, et al. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res 2017;27:119–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu Y, Guo C, Tang L, et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol 2020;5:434–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao F, Tang M, Zheng X, et al. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 2020;158:1831–33.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santos VS, Gurgel RQ, Cuevas LE, et al. Prolonged fecal shedding of SARS-CoV-2 in pediatric patients. A quantitative evidence synthesis. J Pediatr Gastroenterol Nutr 2020;71:150–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin L, Jiang X, Zhang Z, et al. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut 2020;69:997–1001. [DOI] [PubMed] [Google Scholar]
- 12.Buja LM, Wolf DA, Zhao B, et al. The emerging spectrum of cardiopulmonary pathology of the coronavirus disease 2019 (COVID-19): Report of 3 autopsies from Houston, Texas, and review of autopsy findings from other United States cities. Cardiovasc Pathol 2020;48:107233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung KS, Hung IF, Chan PP, et al. Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from the Hong Kong cohort and systematic review and meta-analysis. Gastroenterology 2020;159:81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mucha SR, Dugar S, McCrae K, et al. Coagulopathy in COVID-19. Cleve Clin J Med 2020;87:461–8. [DOI] [PubMed] [Google Scholar]
- 15.Jin X, Lian JS, Hu JH, et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut 2020;69:1002–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan L, Mu M, Ren HG, et al. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: A descriptive, cross-sectional, multicenter study. Am J Gastroenterol 2020;115:766–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallis LA. COVID-19 severity scoring tool for low resourced settings. Afr J Emerg Med 2020. [Epub ahead of print April 2, 2020.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghoshal U, Vasanth S, Tejan N. A guide to laboratory diagnosis of corona virus disease-19 for the gastroenterologis. Indian J Gastroenterol 2020;39:236–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu F, Yan L, Wang N, et al. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin Infect Dis 2020;71:793–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar A, Arora A, Sharma P, et al. Gastro-intestinal and hepatic manifestations of COVID-19 and their relationship to severe clinical course: A systematic review and meta-analysis. Indian J Gastroenterol 2020;39:268–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sultan S, Altayar O, Siddique SM, et al. AGA institute rapid review of the GI and liver manifestations of COVID-19, meta-analysis of international data, and recommendations for the consultative management of patients with COVID-19. Gastroenterology 2020;159:320–34.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nobel YR, Phipps M, Zucker J, et al. Gastrointestinal symptoms and COVID-19: Case-control study from the United States. Gastroenterology 2020;159:373–5.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bunyavanich S, Do A, Vicencio A. Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. JAMA 2020;323:2427–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colaneri M, Sacchi P, Zuccaro V, et al. Clinical characteristics of coronavirus disease (COVID-19) early findings from a teaching hospital in Pavia, North Italy, 21 to 28 february 2020. Euro Surveill 2020;25:2000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aggarwal S, Garcia-Telles N, Aggarwal G, et al. Clinical features, laboratory characteristics, and outcomes of patients hospitalized with coronavirus disease 2019 (COVID-19): Early report from the United States. Diagnosis (Berl) 2020;7:91–6. [DOI] [PubMed] [Google Scholar]
- 26.Inciardi RM, Adamo M, Lupi L, et al. Characteristics and outcomes of patients hospitalized for COVID-19 and cardiac disease in Northern Italy. Eur Heart J 2020;41:1821–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun X, Wang T, Cai D, et al. Cytokine storm intervention in the early stages of COVID-19 pneumonia. Cytokine Growth Factor Rev 2020;53:38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scudellari M. News feature: Cleaning up the hygiene hypothesis. Proc Natl Acad Sci U S A 2017;114:1433–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hussain Z, El-Omar E, Lee YY. Dual infective burden of Helicobacter pylori and intestinal parasites: Good or bad news for the host? Indian J Gastroenterol 2020;39:111–6. [DOI] [PubMed] [Google Scholar]
- 30.Zuo T, Zhang F, Lui GCY, et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology 2020;159:944–55.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forster P, Forster L, Renfrew C, et al. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci U S A 2020;117:9241–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shalimar, Elhence A, Vaishnav M, et al. Poor outcomes in patients with cirrhosis and Corona virus disease-19. Indian J Gastroenterol 2020;39:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Neill LAJ, Netea MG. BCG-induced trained immunity: Can it offer protection against COVID-19? Nat Rev Immunol 2020;20:335–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baryah ANS, Midha V, Mahajan R, et al. Impact of Corona virus disease-19 (COVID-19) pandemic on gastrointestinal disorders. Indian J Gastroenterol 2020;39:214–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.