Abstract
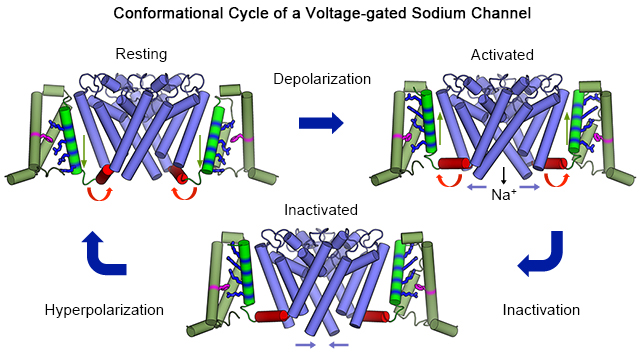
Electrical signaling was a dramatic development in evolution, allowing complex single-cell organisms like Paramecium to coordinate movement and early metazoans like worms and jellyfish to send regulatory signals rapidly over increasing distances. But how are electrical signals generated in biology? In fact, voltage-gated sodium channels conduct sodium currents that initiate electrical signals in all kingdoms of life from bacteria to man. They are responsible for generation of the action potential in vertebrate nerve and muscle, neuroendocrine cells, and other cell types1,2. Because of the high level of conservation of their core structure, it is likely that their fundamental mechanisms of action are conserved as well. Here we describe the complete cycle of conformational changes that a bacterial sodium channel undergoes as it transitions from resting to activated/open to inactivated/closed states, based on high-resolution structural studies of a single sodium channel. We further relate this conformational cycle of the ancestral sodium channel to the function of its vertebrate orthologs. The strong conservation of amino acid sequence and three-dimensional structure suggests that this model, at a fundamental level, is relevant for both prokaryotic and eukaryotic sodium channels and for voltage-gated calcium and potassium channels as well.
Graphical Abstract
Voltage-gated sodium channels in vertebrates consist of a large pore-forming α subunit of 220-250 kDa in association with one or two smaller β subunits of 30-40 kDa3. The α subunits are composed of ~2000 amino acid residues organized in four repeated domains (I-IV) having six transmembrane segments each (S1-S6)4. The S1-S4 segments in each domain form the voltage sensor. The S4 segment contains 4-8 repeats of a three-residue motif with a positively charged residue (usually Arg) flanked by two hydrophobic residues. These Arg residues serve as the gating charges that sense changes in the membrane potential. The S5-S6 segment and the P loop between them form the pore. The ion selectivity filter is located near the extracellular end of the pore, formed by the P loops. The pore is gated at its intracellular end by an activation gate formed by the intracellular ends of the four S6 segments (reviewed in 5).
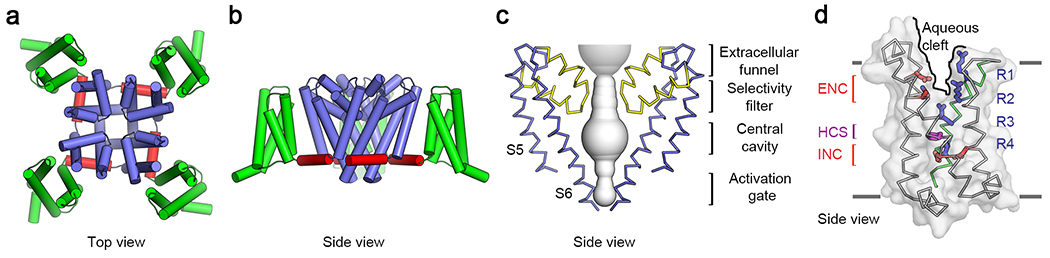
Bacterial sodium channels have a very similar architecture to vertebrate sodium channels, but they are homotetramers of identical pore-forming subunits having six transmembrane segments each, divided into a voltage sensor (S1-S4) and a pore-forming module (S5-S6)6,7. These ancestral sodium channels are ideal models for structural studies aimed at understanding the fundamental mechanisms of sodium channel function because they are four-fold symmetric and lack the large, unstructured intracellular and extracellular loops that are characteristic of vertebrate sodium channels7. The basic structure of the bacterial sodium channel NaVAb is illustrated in Figure 1 7. When viewed from the extracellular side, the ion-conducting pore is located in the center, surrounded by the four pore-forming modules (Figure 1a, blue). The four voltage sensors are located on the periphery of the pore domain (Figure 1a, green). When viewed from the membrane-facing side (Figure 1b), the S5 and S6 transmembrane segments that form the pore are located in the center (Figure 1b, blue), the four voltage sensors containing the S1-S4 segments are on the sides (Figure 1b, green), and they are connected to the pore domain by the S4-S5 linker (Figure 1B, red). As sodium ions move inward through the water-filled pore (Figure 1c, gray), they pass through the open extracellular funnel, the narrow ion selectivity filter, the large central cavity, and the activation gate, which is closed in this structure. A closer look at the voltage sensor reveals several important structural elements (Figure 1d). The hydrophobic constriction site (HCS) seals the voltage sensor, preventing transmembrane movement of water and ions. The gating charges in the S4 segment (R1-R4) are arrayed across the membrane, with some on the extracellular side of the HCS and the rest on the intracellular side. The gating charge(s) on the extracellular side of the HCS interact with the extracellular negative cluster (ENC) of Glu sidechains (Figure 1d, red), while the gating charge(s) on the intracellular side of the HCS interacts with the intracellular negative cluster (INC; Figure 1d, red). In response to membrane potential changes, the gating charges are expected to translocate across the membrane, thereby moving the S4 segment inward or outward relative to the remainder of the voltage sensor domain. Detailed studies using X-ray crystallography and cryo-electron microscopy (cryoEM), coupled with functional analyses, have now elucidated the structures of resting/closed, activated, open, and inactivated/closed states of the bacterial sodium channel NaVAb7–10. Based on these high-resolution (2.7 Å to 4.0 Å) structures of the same channel, we have developed a complete model of the structural changes that drive sodium channel function, as presented below.
Figure 1. Structure of NaVAb7.
a, NaVAb structure in top view. Pore module, blue; voltage sensor, green; S4-S5 linker, red. b, NaVAb structure in side view, colored as in panel a. c, Architecture of the NaVAb pore. Glu177 side-chains (purple sticks); pore volume, grey; P loops, yellow. d, Voltage sensor. Side view illustrating the extracellular negative cluster (red, ENC), the intracellular negative cluster (red, INC), hydrophobic constriction site (purple, HCS). The Arg gating charges (R1-R4) are in blue.
Structures of The Resting State and Closed Pore
Determination of the structure of the resting state of a sodium channel was very challenging because the resting state exists in vivo at the resting membrane potential of ~−80 to −90 mV in vertebrate nerve and muscle and at ~−160 mV in bacteria. We introduced mutations that shifted the voltage dependence of activation by >+200 mV, such that the mutant sodium channel was stabilized in the resting state at 0 mV for structural work10. Because the resting state is inherently unstable, we conducted an unbiased screen of paired substituted Cys residues that could be specifically crosslinked by disulfide bonds in the Cys-free background of NaVAb, as assessed in bacteria at the very negative resting membrane potential of −160 mV to favor the resting state10. The final construct is trapped in the resting state under basal conditions in cultured eukaryotic cells, but it is released by reduction of disulfide bonds with dithiothreitol, demonstrating directly that the functional resting state has indeed been captured10.
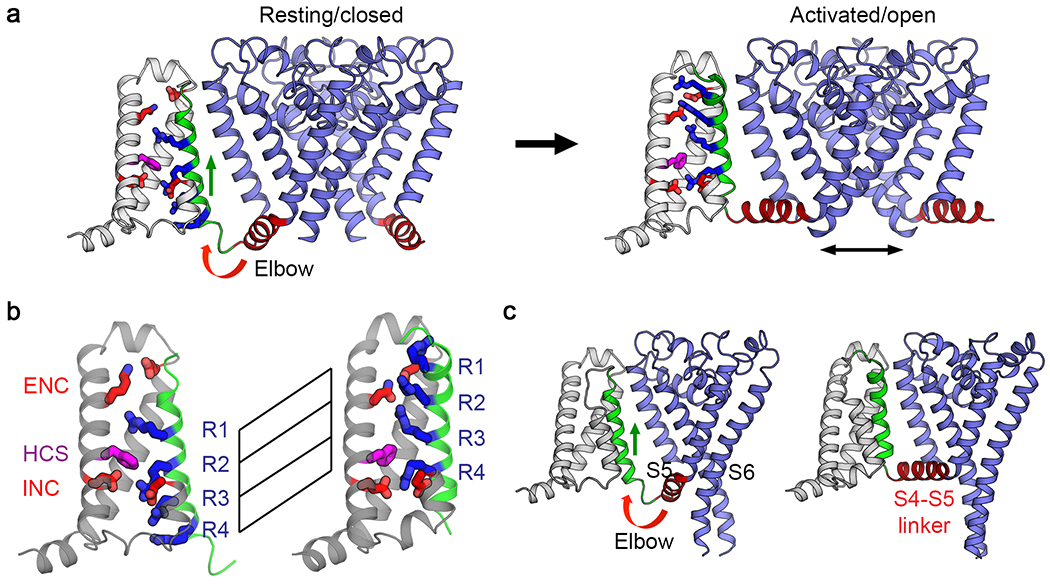
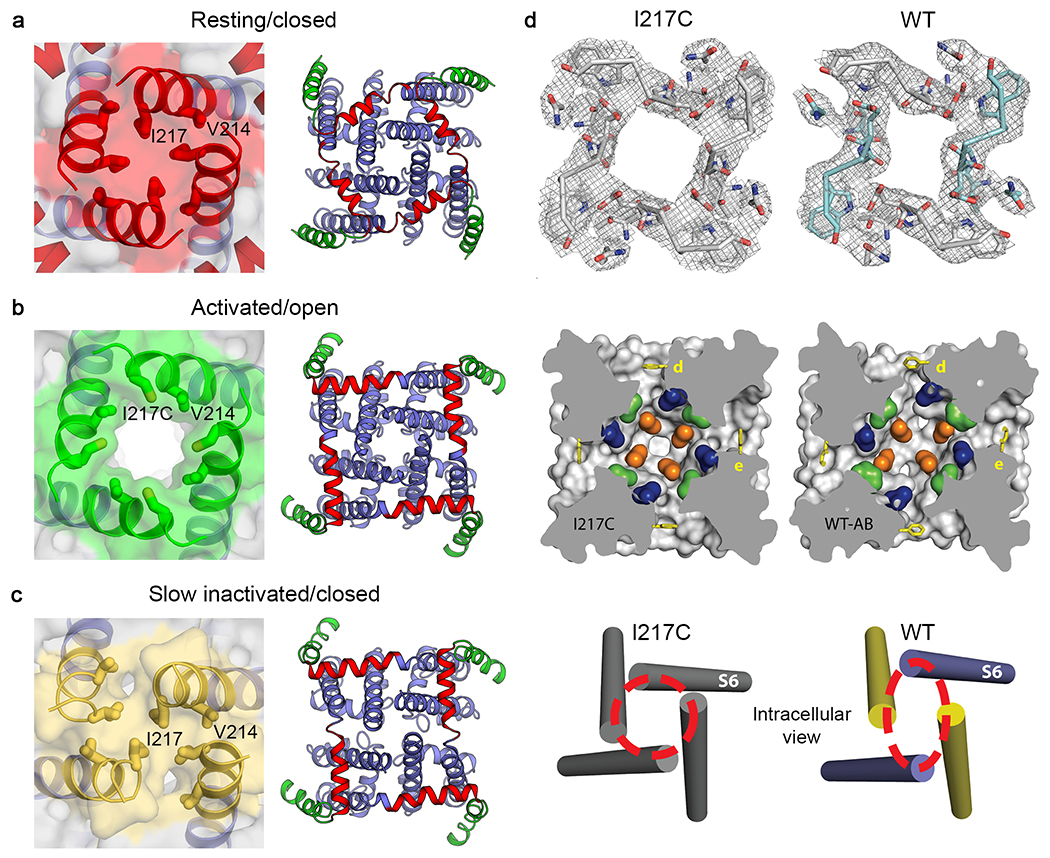
The resting state structure is illustrated in Figure 2a (left), as determined by cryoEM. The hydrophobic constriction site (Figure 2b; HCS, purple) forms a band of hydrophobic residues that seals the voltage sensor and prevents movement of water and ions through it. Remarkably, the entire membrane electric field falls across this narrow (~5 Å) band of protein. The S4 segment with the four Arg gating charges is arrayed across the membrane and penetrates the HCS. Only one Arg gating charge (Figure 2b; R1, blue) is located on the extracellular side of the HCS. The remaining three Arg gating charges (Figure 2b; R2-R4, blue) are on the intracellular side of the HCS making ion pair interactions with negatively charged Asp and Glu sidechains in the INC (Figure 2b; red). The inward position of the S4 segment has pushed the S4-S5 linker into the cytoplasm in a sharply bent ‘elbow’ conformation (Figure 2a, c, left). The central pore is tightly closed at the activation gate by the sidechains of four Ile217 residues in the resting state (Figure 3a). There is no space for movement of water or ions (Figure 3a, left). This closed conformation of the activation gate is enforced by a tight ‘collar’ formed by the four S4-S5 linkers (Figure 3a, right, blue helices). This position of the tight collar of S4-S5 segments is generated by formation of the elbow at the intracellular end of the S4 segment (Figure 2a).
Figure 2. Comparison of NaVAb structures in the resting state and the activated state7,10.
a, Structural transition between resting/closed and activated/open states. Structures in the resting/closed state (left) and activated/open state (right) are captured from Movie S1 and illustrated as backbone cartoons with only one voltage sensor shown for clarity. S4 is green, and important sidechains in the voltage sensor are shown as sticks colored as in Figure 1. b, Gating charge movement. Four Arg gating charges R1-R4 are shown in blue, the extracellular negative cluster (ENC) of E32 and N49 and intracellular negative cluster (INC) of E59 and E80 in red. Phe in the hydrophobic constriction site (HCS) is shown in purple. S4 (green) moves outward by 11.5 Å, passing two gating charges through the HCS. Part of S3 is omitted for clarity. c, Side view of the structures focusing on S4 (green) and the S4-S5 linker (red). The S4 segment moves outward across the membrane from the resting to the activated states while the S1 to S3 segments rotate slightly but remain essentially unchanged with respect to their original transmembrane position. The S4-S5 linker is illustrated as an elbow that connects the S4 movement to gating of the pore.
Figure 3. The activation gate and the pore in resting, activated, and inactivated states7–10.
a, Solvent accessible surface of NaVAb in the resting/closed state (left), adapted from10. Interactions between the S4-S5 linker and S6 near the activation gate in the resting state (right). b, Solvent accessible surface of NaVAb in the activated/open state (left), adapted from10. Interactions between the S4-S5 linker and S6 near the activation gate in the activated state (right). The pore is tightly closed at the activation gate in the resting/closed state (red, panel a) but wide open in the activated/open state (green, panel b). The movement of the S4-S5 linker from the resting/closed state to the activated/open state unbends the elbow and causes an exchange of interactions between the S4-S5 linker residues (red) and the S6 residues (blue), which surround the activation gate formed by the S6 segments. c, Solvent accessible surface of NaVAb in the slow-inactivated/closed state (left). Interactions between the S4-S5 linker and S6 near the activation gate in the slow-inactivated state (right). d, Structural transition between pre-open and inactivated states viewed in cross-section at three levels of the pore: top, ion selectivity filter; center S4 is green, central cavity; bottom, activation gate, adapted from8.
Transition to The Activated State of the Voltage Sensor
The initial crystal structure of NaVAb revealed the pre-open state, with the four voltage sensors activated at 0 mV but the pore not yet open10. This is an expected state of a homotetrameric sodium channel, just the instant before the activation gate swings open. We speculate that this inherently unstable state is trapped in these structures by a single mutation (I217C) at the activation gate that holds the pore closed in the crystal structure10. In the activated voltage sensor, three Arg gating charges (Figure 2a, b; R1, R2, and R3, red) are located on the extracellular side of the HCS interacting with the ENC, whereas only R4 remains on the intracellular side interacting with the INC. The voltage-driven translocation of these gating charges through the membrane electric field across the narrow HCS is the essence of voltage sensing, in which a change in electric field is converted into a protein conformational change. This translocation of the S4 segment and its gating charges pulls the S4-S5 linker upward to lie along the inner surface of the membrane and removes the sharp inward bend in the elbow (Figure 2c and Movie 1).
Opening the Activation Gate at the Intracellular End of the Pore
To capture NaVAb with its pore open, we truncated the large C-terminal domain at the membrane, removing 40 intracellular amino acid residues that are unique to bacterial sodium channels and therefore are unlikely to have conserved functions across all sodium channels9. NaVAbΔ40 functions normally, except that it activates at 12 mV more negative membrane potential than wild-type, consistent with stabilization of the open state9. Structural analysis by X-ray crystallography revealed a striking new conformation of the activation gate, in which the collar of S4-S5 segments is loosened and the activation gate is open to an internal van der Waals diameter of ~8.5 Å (Figure 3b) 9, which is ~2 Å wider than the orifice of the activation gate of another bacterial sodium channel NaVMs in its proposed open state11. Although the activation gate opens wide, the movements of the S6 segments are subtle, primarily rotating to move the side chains of the four Ile217 residues away from the lumen of the pore without much movement of the helical backbone. Importantly, molecular dynamics analysis revealed that hydrated Na+ ions move unimpeded through this opening, consistent with designation of this structure as an open activation gate9.
The Sliding Helix Mechanism of Voltage Sensing
The structures of the resting/closed, activated, and open states of NaVAb elucidate the first half of its conformational cycle. Although there has been much active debate about the molecular mechanisms of voltage-dependent gating, these structures unequivocally support the ‘sliding helix’ model of voltage sensing12,13, which posits that the S4 gating charges move across the membrane electric field through the voltage sensor structure, facilitated by specific interactions with hydrophilic and negatively charged sidechains of amino acid residues in the surrounding S1-S3 segments. The electrostatic force of the resting membrane potential pulls the S4 gating charges inward in the resting state (Figure 2b, R1-R4), poised like a cocked gun. Release of this electrostatic force allows the S4 segments containing the gating charges to shoot rapidly outward in the activated state (Figure 2b, R1-R4). Comparison of the structures of the resting and activated states clearly indicates that the S4 segment moves outward by ~11.5 Å and rotates significantly upon activation (Figure 2a, b). This movement is accompanied by a partial conformational transition from 310 helix to alpha helix near the extracellular end of S4, which occurs as the gating charges move outward through the HCS in a linear manner. The S4 gating charges are stabilized and their movement is catalyzed by exchange of ion pair partners from the INC to the ENC and by interactions with a key highly conserved Phe residue in the HCS (Figure 2a, b), which has been termed the ‘charge transfer center’ 14. In this way, the huge energetic cost of placing the Arg gating charges in a transmembrane position is dissipated, which stabilizes them in their transmembrane position and allows them to move in response to voltage changes and begin the process of electromechanical coupling (Movie 1).
Electromechanical Coupling to Pore Opening
The structures of the resting and open states of NaVAb define the mechanism of electromechanical coupling that opens the pore5. In the activated state (Figure 2a, b), the outward and rotational movements of the S4 segment pull the S4-S5 linker upward toward the inner surface of the membrane, removing the sharp bend in the elbow, and induce a bending and rotation of the S6 segments. The removal of the sharp bend in elbow of the S4-S5 linkers relieves the tight grip of the collar on the activation gate and allows its opening movements (Figure 3a, b). The subtle rotation of the S6 segment turns the sidechains of the four Ile217 residues away from the lumen of the pore and increases its inner van der Waals diameter from essentially 0 Å to ~8.5 Å (Figure 3a, b; Movie 2). This size is sufficient for rapid Na+ conductance as assessed by molecular dynamics9,15.
In contrast to the intracellular activation gate, the ion selectivity filter near the outer membrane boundary of the pore does not move significantly during the activation process10. Molecular dynamics analysis shows that the selectivity filter is open to Na+ conductance in resting and activated states, even when the intracellular activation gate is closed9,15. Thus, there is a clear division of labor between these two distinct components of the pore. Ions are selected for rapid conductance at the selectivity filter near the extracellular end of the pore, which is always open. Ion conductance is controlled by the activation gate at the intracellular end of the pore, which opens and closes in response to membrane potential and voltage-driven conformational changes in the voltage sensor.
Transition to the Slow Inactivated State
Bacterial and vertebrate sodium channels undergo a slow-inactivation process, which takes place on the time scale of 10 msec to sec in different channel types6,16. In NaVAb, we have characterized three distinct slow-inactivated states in electrophysiological recordings: two reversible inactivated states that are engaged in the time frame of 10 to 100 msec and one use-dependent inactivated state that requires prolonged repetitive stimulation and is not reversible on the time scale of our electrophysiological experiments17. The conformational changes that drive this progression of inactivated states involve a key, highly conserved Thr residue in the S6 segments just on the intracellular side of the ion selectivity filter, as well as the intracellular C-terminal domain, which forms a coiled-coil tail that is required for slow use-dependent inactivation17.
Because the final inactivated state is essentially irreversible on accessible experimental time scales, it is likely that wild-type NaVAb would crystallize in this state. After much difficulty, we were eventually able to determine the crystal structure of wild-type NaVAb, without the I217C mutation that we used to improve resolution in our original work8. This structure revealed a striking conformational change in the pore (Figure 3c, d). At each level of the pore, including the selectivity filter, central cavity, and activation gate, two of the S6 segments have moved toward the central axis of the pore and two have moved away, resulting in a dimer-of-dimers conformation having two-fold symmetry rather than four-fold symmetry (Figure 3c, d). In this process, the activation gate adopts a closed configuration in which the Cα’s of Ile217 are organized in a parallelogram or rhomboid shape rather than a square (Figure 3c, d; Movies 3a and 3b). Thus, slow inactivation is mediated by a partial asymmetric collapse of the pore and de-wetting of the opposing faces of two S6 helices, plausibly explaining why it is slow and difficult to reverse.
Recovery from Slow Inactivation
To complete its conformational cycle, NaVAb must return to the resting state, ready for activation and pore opening. This transition requires two major conformational changes. First, repolarization must re-establish the resting membrane potential, and the electrostatic force of the electric field must pull the S4 gating charges inward to their positions in the resting state. Second, the four-fold symmetry of the pore must be re-established. These are both energetically costly transitions because of the electrostatic energy required to move the gating charges inward and the chemical energy needed to separate and re-hydrate the surfaces of the S6 segments and thereby restore a water-filled passage in the pore. We consider it essential to restore four-fold symmetry and re-wet the pore-lining surfaces of the S6 transmembrane segments before rapid voltage-dependent activation and pore opening can occur. It is likely that there are multiple intermediate steps in this recovery process (Movies 3a and b), which remain undefined at the structural level at this stage of our analysis.
Relevance for Vertebrate Sodium Channels
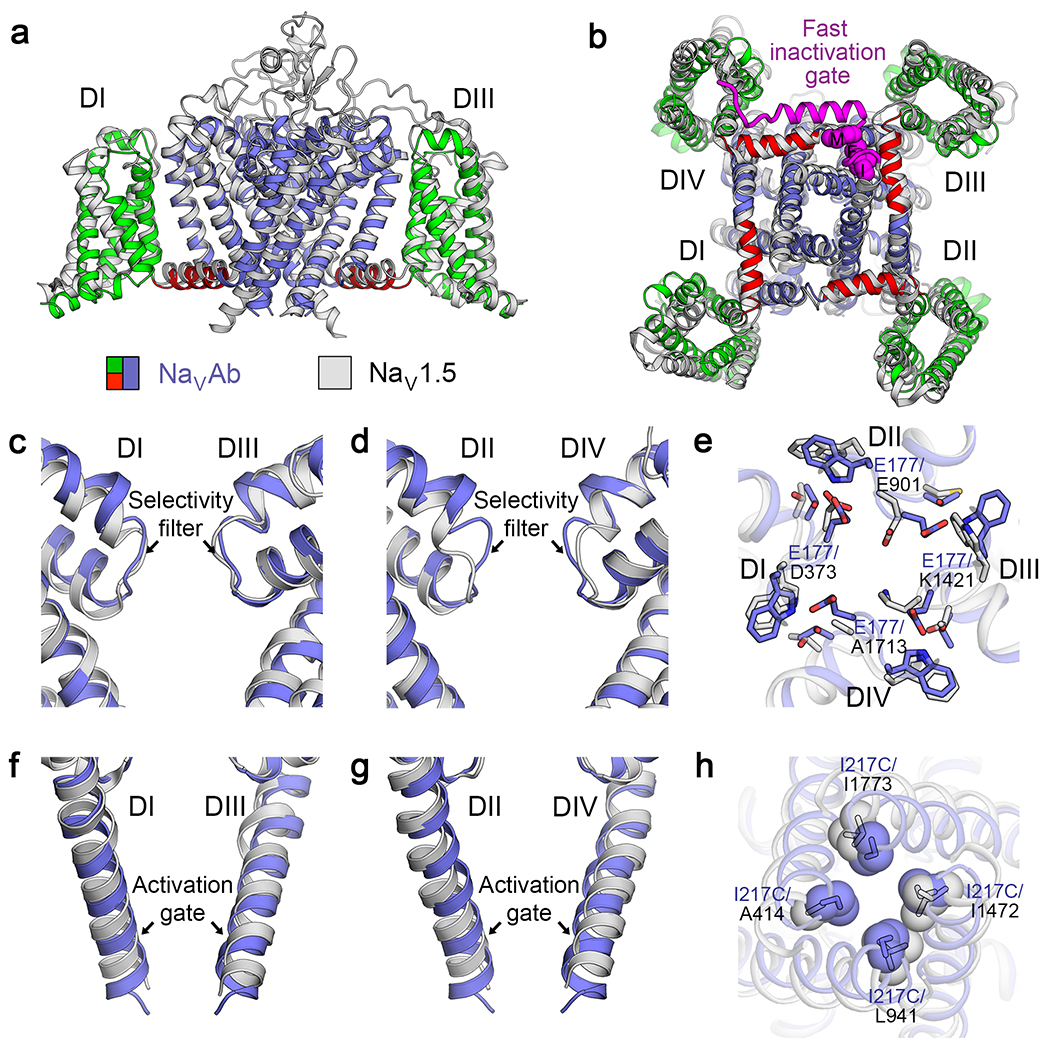
The amino acid sequences and the high-resolution structures of the core transmembrane regions of the vertebrate sodium channels are very similar to NaVAb. For example, comparison of the backbone structure of NaVAb to domain I of NaV1.518 has an RMSD of 2.7 Å, comparable to the resolution limit of these structures (Figure 4a, b). The backbone structure is also virtually identical at the highly conserved ion selectivity filter (Figure 4c, d). On the other hand, close inspection of the side chain positions reveals important changes in the shape of the selectivity filter, from square to rhombic, and in the location and pose of the key Lys side chain in the “DEKA” motif (vs. “EEEE” in NaVAb) that is crucial for sodium selectivity in eukaryotes (Figure 4e). Similarly, the activation gate has a nearly identical fold in NaVAb and NaV1.5 (Figure 4f, g), but the shape of the orifice is changed by the different side chains in the eukaryotic structure (Figure 4h). Thus, the molecular adaptations that enhance sodium selectivity and pore opening in vertebrate sodium channels reflect substitutions of a small number of crucial amino acid residues in the context of a conserved core transmembrane structure.
Figure 4. Mammalian NaV and bacterial NaVAb are structurally conserved.
a-b, Superposition between rat NaV1.5 (light gray) and NaVAb (blue) from side view (a) and bottom view (b). The fast inactivation gate and its key Isoleucine-Phenylalanine-Methionine (IFM) motif are highlighted in purple. c-d, Superposition of the selectivity filter of NaV1.5 domains I and III (c), and domains II and IV (d) with the selectivity filter of NaVAb from side view. e, Key specialized and conserved side chains at the selectivity filter viewed from extracellular side. NaV1.5 uses specific “DEKA” side chains while NaVAb uses four glutamates to partially dehydrate and conduct sodium ions. f-g, Superposition of the activation gate of NaV1.5 domains I and III (f), and domains II and IV (g) with the activation gate of NaVAb from side view. h, Key specialized side chains in NaV1.5 and NaVAb that modulate the shape and diameter of the activation gate.
The close structural similarity of the basic functional elements of prokaryotic and eukaryotic sodium channels indicates that their backbone conformational movements during voltage sensing, activation, pore opening, and slow inactivation are likely to be similar in bacteria and vertebrates. The vertebrate sodium channels are more complex because of differences in their amino acid sequences in their four domains and the addition of large intracellular and extracellular linkers that serve regulatory functions. However, it is likely that these complexities are simply variations on the structural and functional themes established originally in the ancestral bacterial sodium channels. Structure-function studies of the voltage sensors of vertebrate NaV channels provide direct support for this view. Although their conformation is very similar to NaVAb (Figure 4a, b), voltage sensors in vertebrate NaV channels activate sequentially in both voltage and time, with domains I and II activating before domain III, and domain IV activating last19. These differences in voltage sensor function change the stochastic sequence of activation of bacterial NaV channels into a specific ordered sequence in vertebrates, which may provide more precise channel opening and faster rising action potentials. Substitutions of single amino acid residues adjacent to gating charges in the S4 segment control this functional difference in domains I-III of NaV1.420. These results support our general model that specializations of voltage sensor function in vertebrate NaV channels are fine-tuned by a few specific side-chain alterations in the context of a nearly identical core structure and a conserved series of voltage-dependent conformational changes.
One key functional element of vertebrate sodium channels that is missing in the bacterial sodium channels is fast inactivation, first described by Hodgkin and Huxley in the squid giant axon1. Fast inactivation is mediated by the fast-inactivation gate and its key IFM motif located in the intracellular linker between domains III and IV of vertebrate sodium channels5 (Figure 4b, purple). This structural motif is absent in bacterial sodium channels, which do not have a corresponding fast-inactivation process on the time scale of 1 ms16,17. This evolutionary add-on is crucial for vertebrate physiology, in order to terminate the sodium current within 1-2 msec to support the rapid action potential firing required for nerve and muscle function. Fast inactivation is triggered by the voltage sensors in domains III and IV of vertebrate NaV channels19, and the outward movements of their S4 segments are coupled to closure of the IFM motif of the fast inactivation gate into its receptor at the edge of the activation gate and the intracellular mouth of the pore (Figure 4b, purple)18,21,22. The exact timing of fast inactivation is fine-tuned by small numbers of amino acid substitutions in these transmembrane segments23. These results provide further support for our model that the core conformational changes in the voltage sensors of vertebrate NaV channels are conserved from their bacterial ancestors, whereas evolutionary specializations like insertion of the fast inactivation gate plus substitution of selected amino acid side chains in the voltage sensors lead to new and physiologically important adaptations of function.
Drug Binding and Access to Sodium Channels
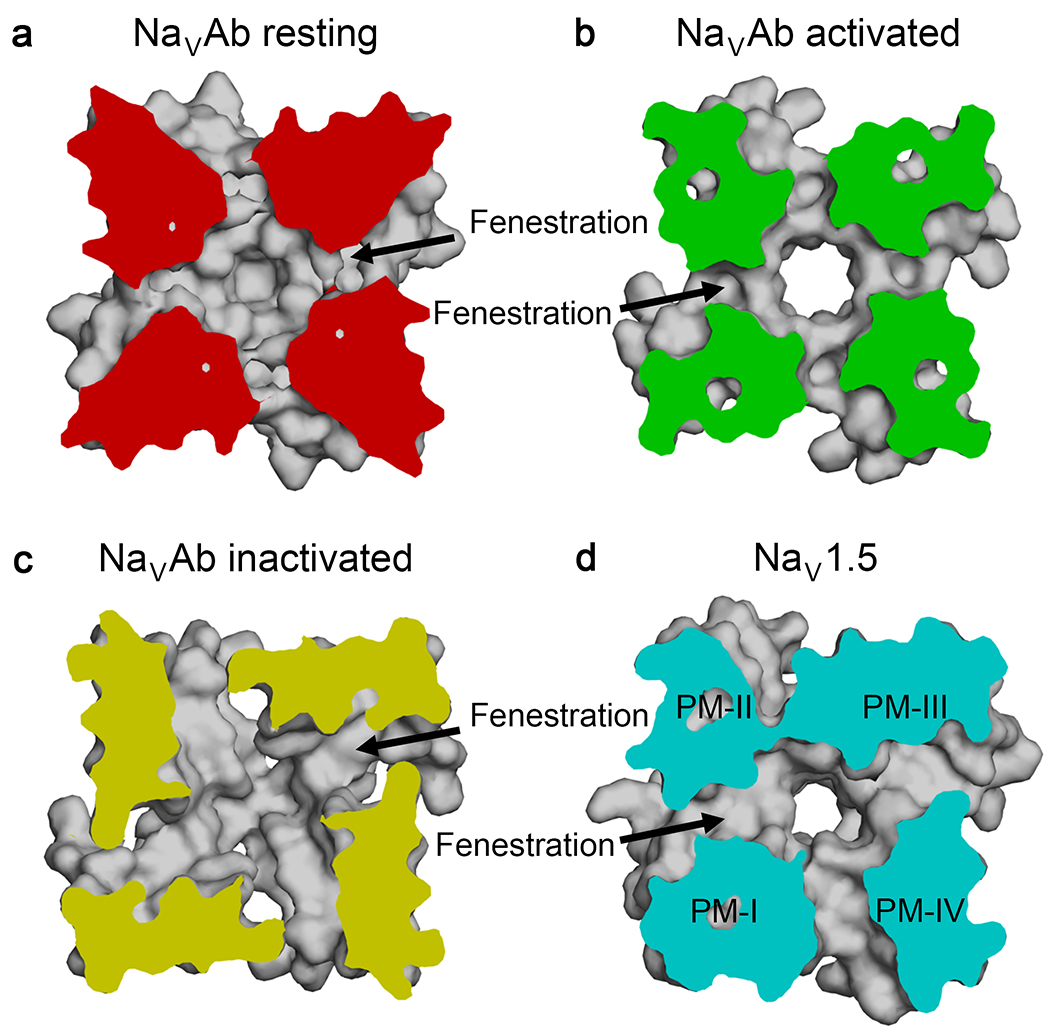
Sodium channels are the molecular targets of drugs used for local anesthesia, cardiac arrhythmia, and epilepsy2. Recent structural studies on bacterial and vertebrate sodium channels confirm the classical model that these clinically important drugs bind in the lumen of the pore and physically block it18,24. Moreover, fenestrations in the sidewalls of the pore provide a hydrophobic pathway from the lipid phase of the membrane into the drug receptor site in both bacterial and vertebrate NaV channels, as illustrated in cross sections of the central cavity at the level of the drug receptor site viewed from the selectivity filter (Figure 5)7,8,18,21,22,24,25. The fenestrations are in similar conformations in the resting and open states of NaVAb (Figure 5a, b)7,9,10,24. In contrast, they are significantly changed in the two-fold symmetric structure of the slow-inactivated state of NaVAb, where two fenestrations are larger and two are smaller than in the resting and activated states (Figure 5c)8. In the primary conformation visualized in vertebrate NaV channels to date, the fenestrations between domains I/IV and III/IV are largest, domain I/II is intermediate, and domain II/III is smallest (Figure 5d)18,26. It is remarkable that the mechanisms of drug action and the structures of the drug receptor site and fenestrations are so well conserved all the way from bacterial to vertebrate NaV channels.
Figure 5. Drug access to the pore via the hydrophobic fenestrations.
Top view of a cross section below the selectivity filter of the pore module shows fenestrations and hydrophobic access to the drug receptor site in the central cavity of the pore. a, Fenestrations of NaVAb in resting state, b, activated state, and c, inactivated state. d, Fenestrations of rat apo-NaV1.518. PM, pore module.
Relevance for Voltage Gated Calcium and Potassium Channels
The amino acid sequence of NaVAb is as similar to mammalian calcium channels as it is to mammalian sodium channels6, and the backbone of its transmembrane core overlays that of CaV1.1 27 with an RMSD of 2.7 Å . The ion selectivity of NaVAb can be converted to that of CaV1.2 cardiac calcium channels with only three amino acid substitutions28. It is likely that bacterial sodium channels are the ancestors of both mammalian NaV and CaV channels29. Therefore, the sequence of conformational changes we describe here is most likely recapitulated in voltage sensing, activation, coupling to pore opening, and voltage-dependent inactivation of calcium channels.
Voltage-gated potassium channels are a distinct clade of ion channels from voltage-gated sodium and calcium channels based on comparison of their pore sequences29. The conformation of their ion selectivity filter is completely different 7,30. However, the voltage sensor of KV1.2 can be superimposed with the voltage sensor of NaVAb with an RMSD of 2.3 Å7,30, the structures of its S4-S5 linker and activation gate are very similar, and fenestrations are observed that lead from the lipid bilayer to the inner pore31. Therefore, we consider it highly likely that the sequence of conformational changes we describe here is also recapitulated in voltage sensing, activation, coupling to pore opening, and voltage-dependent C-type inactivation of potassium channels.
Looking Ahead
A major goal looking into the future is to define a comparable series of functional states of mammalian sodium channels that are the targets for disease mutations and therapeutic drugs in order to visualize the detailed structural adaptations that underlie the cell-specific functional specializations of these highly evolved NaV channels. So far, the structures of mammalian sodium channels from nerve22,32, skeletal muscle21, and heart18 have been captured by cryo-electron microscopy in similar states. The voltage sensors are partially activated, and the fast inactivation gate is partially closed, consistent with intermediate inactivated states. Understanding disease mutations requires determination of their structures in multiple functional states. Structure-based drug design would be greatly advanced by channel structures with drugs bound in different functional states. The series of functional states of NaVAb that we describe here at high resolution provides templates and methods for capturing mammalian sodium channels in multiple functional states and determining their structures with disease mutations inserted and/or drugs bound. We look forward to significant advances in understanding ion channelopathies and treating diseases of hyperexcitability like chronic pain, epilepsy, and cardiac arrhythmia more effectively with sodium channel-blocking drugs based on this structural approach.
Supplementary Material
Acknowledgements
Research from the authors’ laboratories presented here was supported by Research Grants from the National Institutes of Health (R01 HL112808 to W.A.C. and N.Z.; R01 NS15751 to W.A.C., and R35 NS111573 to W.A.C.) and by the Howard Hughes Medical Institute (N.Z.). We thank J. Li (Pharmacology, University of Washington) for editorial assistance.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Hodgkin AL & Huxley AF A quantitative description of membrane current and its application to conduction and excitation in nerve. J.Physiol 117, 500–544 (1952). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hille B Ionic Channels of Excitable Membranes, 3rd Ed. (Sinauer Associates Inc., 2001). [Google Scholar]
- 3.Catterall WA The molecular basis of neuronal excitability. Science 223, 653–661 (1984). [DOI] [PubMed] [Google Scholar]
- 4.Numa S & Noda M Molecular structure of sodium channels. Ann N Y Acad Sci. 479, 338–355. (1986). [DOI] [PubMed] [Google Scholar]
- 5.Catterall WA, Wisedchaisri G & Zheng N The chemical basis for electrical signaling. Nature Chem. Biol. 13, 455–463, doi: 10.1038/nchembio.2353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ren D et al. A prokaryotic voltage-gated sodium channel. Science 294, 2372–2375, doi: 10.1126/science.1065635 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Payandeh J, Scheuer T, Zheng N & Catterall WA The crystal structure of a voltage-gated sodium channel. Nature 475, 353–358 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Payandeh J, Gamal El-Din TM, Scheuer T, Zheng N & Catterall WA Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature 486 135–139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lenaeus MJ et al. Structures of closed and open states of a voltage-gated sodium channel. Proc Natl Acad Sci U S A 114, E3051–E3060, doi: 10.1073/pnas.1700761114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wisedchaisri G et al. Resting-state structure and gating mechanism of a voltage-gated sodium channel. Cell 178, 993–1003, doi: 10.1016/j.cell.2019.06.031 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCusker EC et al. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat Commun 3, 1102, doi: 10.1038/ncomms2077 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Catterall WA Molecular properties of voltage-sensitive sodium channels. Annu.Rev.Biochem 55, 953–985 (1986). [DOI] [PubMed] [Google Scholar]
- 13.Yarov-Yarovoy V et al. Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proc. Natl. Acad. Sci. U.S.A 109 E93–E102, doi:1118434109 [pii] 10.1073/pnas.1118434109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tao X, Lee A, Limapichat W, Dougherty DA & MacKinnon R A gating charge transfer center in voltage sensors. Science 328, 67–73, doi: 10.1126/science.1185954 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakrabarti N et al. Catalysis of Na+ permeation in the bacterial sodium channel NaVAb. Proc Natl Acad Sci U S A 110, 11331–11336, doi: 10.1073/pnas.1309452110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pavlov E et al. The pore, not cytoplasmic domains, underlies inactivation in a prokaryotic sodium channel. Biophys J 89, 232–242, doi:S0006-3495(05)72673-4 [pii] 10.1529/biophysj.104.056994 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gamal El-Din TM, Lenaeus MJ, Ramanadane K, Zheng N & Catterall WA Molecular dissection of multiphase inactivation of the bacterial sodium channel NaVAb. J. Gen. Physiol 151, 174–185, doi: 10.1085/jgp.201711884 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang D et al. Structure of the cardiac sodium channel. Cell 180, 122–134, doi: 10.1016/j.cell.2019.11.041 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chanda B & Bezanilla F Tracking voltage-dependent conformational changes in skeletal muscle sodium channel during activation. J. Gen. Physiol 120, 629–645 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lacroix JJ, Campos FV, Frezza L & Bezanilla F Molecular bases for the asynchronous activation of sodium and potassium channels required for nerve impulse generation. Neuron 79, 651–657, doi: 10.1016/j.neuron.2013.05.036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan X et al. Structure of the human voltage-gated sodium channel NaV1.4 in complex with beta1. Science 362, pii: eaau2486 doi: 10.1126/science.aau2486 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Shen H, Liu D, Wu K, Lei J & Yan N Structures of human NaV1.7 channel in complex with auxiliary subunits and animal toxins. Science 363, 1303–1308, doi: 10.1126/science.aaw2493 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Capes DL, Goldschen-Ohm MP, Arcisio-Miranda M, Bezanilla F & Chanda B Domain IV voltage-sensor movement is both sufficient and rate limiting for fast inactivation in sodium channels. J. Gen. Physiol 142, 101–112, doi: 10.1085/jgp.201310998 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gamal El-Din TM, Lenaeus MJ, Zheng N & Catterall WA Fenestrations control resting-state block of a voltage-gated sodium channel. Proc. Natl. Acad. Sci. U. S. A 115, 13111–13116, doi: 10.1073/pnas.1814928115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boiteux C et al. Local anesthetic and antiepileptic drug access and binding to a bacterial voltage-gated sodium channel. Proc Natl Acad Sci U S A 111, 13057–13062, doi: 10.1073/pnas.1408710111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen PT, DeMarco KR, Vorobyov I, Clancy CE & Yarov-Yarovoy V Structural basis for antiarrhythmic drug interactions with the human cardiac sodium channel. Proc Natl Acad Sci U S A 116, 2945–2954, doi: 10.1073/pnas.1817446116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu J et al. Structure of the voltage-gated calcium channel CaV1.1 at 3.6 A resolution. Nature 537, 191–196, doi: 10.1038/nature19321 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Tang L et al. Structural basis for Ca2+ selectivity of a voltage-gated calcium channel. Nature 505, 56–61, doi: 10.1038/nature12775 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu FH & Catterall WA The VGL-chanome: a protein superfamily specialized for electrical signaling and ionic homeostasis. Sci STKE 2004, re15 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Long SB, Tao X, Campbell EB & MacKinnon R Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450, 376–382, doi:nature06265 [pii] 10.1038/nature06265 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Jorgensen C et al. Lateral Fenestrations in K(+)-Channels Explored Using Molecular Dynamics Simulations. Mol Pharm 13, 2263–2273, doi: 10.1021/acs.molpharmaceut.5b00942 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Pan X et al. Molecular basis for pore blockade of human Na+ channel NaV1.2 by the mu-conotoxin KIIIA. Science 363, 1309–1313, doi: 10.1126/science.aaw2999 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.