Abstract
Metal ion homeostasis is essential for all forms of life. However, the breadth of intracellular impacts that arise upon dysregulation of metal ion homeostasis remain to be elucidated. Here, we used cadmium, a non-physiological metal ion, to investigate how the bacterial pathogen, Streptococcus pneumoniae, resists metal ion stress and dyshomeostasis. By combining transcriptomics, metabolomics and metalloproteomics, we reveal that cadmium stress dysregulates numerous essential cellular pathways including central carbon metabolism, lipid membrane biogenesis and homeostasis, and capsule production at the transcriptional and/or functional level. Despite the breadth of cellular pathways susceptible to metal intoxication, we show that S. pneumoniae is able to maintain viability by utilizing cellular pathways that are predominately metal-independent, such as the pentose phosphate pathway to maintain energy production. Collectively, this work provides insight into the cellular processes impacted by cadmium and how resistance to metal ion toxicity is achieved in S. pneumoniae.
Subject terms: Bacteriology, Metals, Metabolic pathways, Pathogens
Neville et al. investigate how Streptococcus pneumoniae mitigates metal ion stress. Despite cadmium induced dysregulation of central carbon metabolism and lipid membrane homeostasis, they find that S. pneumoniae can remain viable by selectively utilizing predominately metal-independent cellular pathways. This study provides insights into how bacteria overcome metal ion toxicity.
Introduction
Cadmium (Cd) is a naturally abundant metal in the Earth’s crust. However, industrialization has dramatically increased its flux into the biosphere, driven by processes including urban waste disposal, phosphate-based fertilizer usage, non-ferrous iron ore processing and battery disposal1,2. The ecological impact of anthropogenic Cd release is highlighted by increased accumulation of the metal in topsoils, and contamination of marine and terrestrial ecosystems3,4. The flux and accumulation of Cd in these environments has increased its rate of entry into the food chain with food consumption serving as a major route of human Cd intake (10–26 µg/day3) with substantial bioaccumulation (0.6–1.3 µg/day5). Cadmium is acutely toxic in biological systems, with an estimated biological half-life of 17–30 years6. The physiological consequences of Cd exposure are well-documented7, but despite this, the molecular basis of toxicity remains to be fully understood8. Current models propose that Cd, which occurs as the divalent cation Cd2+ in biological systems, exerts toxicity via the generation of reactive oxygen species that mediate DNA damage and lipid peroxidation9–11. However, this may be an indirect effect arising from the thiophilicity of the metal ion and its ability to coordinate with nitrogen and oxygen-contributing side chains present in the metal-binding sites of metalloproteins12. Cadmium competition for magnesium, calcium, manganese (Mn2+) or zinc (Zn2+) binding sites could perturb or abrogate metalloprotein function due to the acquisition of a non-cognate metal cofactor13. Recent bioinorganic chemical studies have provided support for this inference, with specific examples of Cd2+-mismetallation including Zn2+-finger DNA-binding proteins, leading to perturbation of DNA repair mechanisms14,15, and metalloregulatory proteins, leading to dysregulation of metal ion homeostasis16–18. Succinctly, the ability of Cd2+ to readily accumulate within cellular systems and inappropriately interact with metalloproteins contributes to its toxicity. Thus, investigation of Cd2+ toxicity provides a unique framework to elucidate the cellular impacts associated with a break down in the homeostasis of biologically important metal ions.
Here, we have used Cd2+ to investigate how metal intoxication impacts essential cellular processes in Streptococcus pneumoniae (the pneumococcus). S. pneumoniae is a Gram-positive human pathogen that is exposed to multiple inorganic chemical stresses during infection, such as Mn2+-limitation and Cu2+- and Zn2+-intoxication19–22. Although Cd2+ is a non-physiological metal ion, recent studies have indicated Cd2+ stress may also occur during colonization and infection of the lungs of tobacco smokers. Cigarettes contain high levels of Cd2+ (0.5–1 μg per cigarette23) due to the Cd2+ hyper-accumulative properties of Nicotiana tabacum leaves. Aerosolization of Cd2+ facilitates more rapid absorption via the lungs than through food consumption24 with recent studies showing persistent bioaccumulation of Cd2+ in lung tissues and bronchoalveolar lavage fluids25 in tobacco smokers. Moreover, S. pneumoniae exposed to tobacco smoke manifests a transcriptional response consistent with Cd2+ exposure26. Although questions remain regarding the effect of inorganic chemical insults during pneumococcal infection, the identity and importance of the metal homeostasis mechanisms are well-defined20,22,27–30. Prior work from our group has shown that S. pneumoniae acquires Cd2+ via the Mn2+-specific ABC permease, PsaBCA, and rapidly accumulates in the cytoplasm due to the absence of an efflux system18. In addition, S. pneumoniae has a comparatively limited metabolic capacity compared to many other bacterial pathogens, lacking an electron transport chain, a complete tricarboxylic acid cycle and an Entner-Dourdoroff pathway. These factors, in combination with the single cellular compartment of the organism and the lack of any native Cd2+ utilization by S. pneumoniae, provide a simplified system in which to dissect the complex molecular impacts of dysregulated metal ion homeostasis.
We used an integrated multi-omics approach that combined transcriptomics, metabolomics, and metalloproteomics to address this question. This revealed the global effects of Cd2+-mediated dysregulation of metal homeostasis on gene expression, metabolism, and metalloprotein metallation status. Essential cellular processes that were perturbed included inorganic ion (Mn2+ and Zn2+) homeostasis, corroborating our previous findings18, but also carbon metabolism and fatty acid (FA) biosynthesis. Notably, Cd2+-intoxication exerted a profound inhibitory effect on glycolysis, mediated by reduced gene expression and impaired protein function. Glucose catabolism proceeded via the pentose phosphate pathway (PPP), circumventing the impaired glycolytic enzymes, and resulting in a shift to mixed acid fermentation. Intoxication with Cd2+ also impacted FA biosynthesis and manifested as an alteration in membrane phospholipid composition and reduced membrane fluidity. The shift in glucose flux and upregulation of alternative, metal-independent enzymes, such as GapN, during exposure to Cd2+-intoxication suggest the presence of alternate metabolic pathways that facilitate survival during exposure to conditions that disrupt cellular metal homeostasis. Collectively, this study provides new insights into the molecular basis of Cd2+ toxicity, the breadth of impacts that arise from disruption of cellular metal ion homeostasis, and reveals the mitigation strategies that bacteria, such as S. pneumoniae, can employ to survive these chemical stress insults.
Results
Cadmium stress impacts S. pneumoniae growth and exerts global transcriptome changes
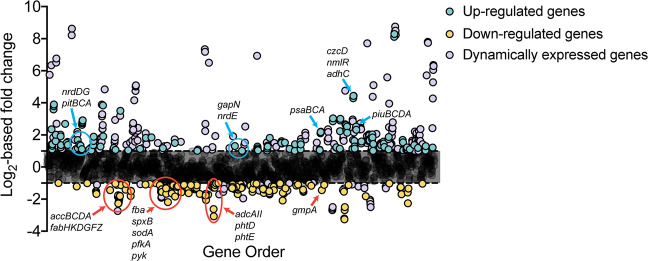
Exposure to 30 μM Cd2+ perturbs pneumococcal growth resulting in an extended lag time (0.11 vs. 0.19 h) and a reduction in the maximal growth rate (1.10 vs. 0.66 h−1) (Supplementary Fig. 1a, Supplementary Table 1). This effect has previously been ascribed to Cd2+-induced perturbation of cellular accumulation of manganese (Mn2+) and zinc (Zn2+) ions18, but the extent of the cellular impact of Cd2+ was not fully defined. Accordingly, we investigated the global transcriptional response of S. pneumoniae to 30 μM Cd2+ stress. Here, we observed that exposure to 30 μM Cd2+ resulted in transcriptional changes of >2-fold for 544 (26%) genes and >4-fold to 152 (7%) genes (Fig. 1) relative to untreated cells. Of the genes that showed differences of more than two-fold, 357 (17%) were upregulated, and 187 (9%) were downregulated. Validation of the RNA sequencing data were performed by qRT-PCR analysis using a subset of differentially expressed genes (Supplementary Fig. 2, Supplementary Table 2).
Fig. 1. S. pneumoniae transcriptome in response to 30 µM Cd2+ stress.
Gene expression profile of mid-log phase S. pneumoniae D39 grown in the presence of Cd2+ stress (30 μM Cd2+) compared to untreated. Genes are plotted along the x-axis, ordered according to locus tag (1–2069). Differential expression of genes in response to Cd2+ stress is displayed in Log2 values along the y-axis. Genes upregulated >2-fold have been colored blue. Genes downregulated by >2-fold have been colored yellow. Dynamically expressed genes as defined by Aprianto et al.46 have been colored purple. Annotated genes have been discussed further.
The largest proportion of significantly (>2-fold) upregulated genes belonged to the Clusters of Orthologous Groups (COG) defined categories of carbohydrate metabolism (30.1%) and inorganic ion homeostasis (24.4%), while genes associated with lipid metabolism represented the largest downregulated group (29.7%) (Supplementary Fig. 3). Genes upregulated to the greatest extent were associated with pneumococcal competence, within both the early (ComE) and late (ComX) competence regulons, and bacteriocin production (BlpR regulon) (Table 1)31. Genes comprising the pneumococcal competence and bacteriocin production regulons have highly dynamic transcriptional profiles and are readily induced by a variety of environmental factors including pH, oxygen tension, chemical and nutrient stress28,32–36. Our data show that Cd2+ stress also activates these regulons, which can likely be attributed to the depletion of cellular Mn2+ and Zn2+, consistent with our prior report18, although a direct response to Cd2+ cannot be excluded.
Table 1.
Summary of genes highly upregulated in S. pneumoniae upon exposure to 30 μM Cd2+.
| Locus Tag | Gene/Locus | Predicted function | Log2-fold changea | Regulon |
|---|---|---|---|---|
| SPD_1860 | comGD | Competence protein | +8.75 | ComX |
| SPD_0133 | cibA | Class IIb bacteriocin | +8.62 | ComX |
| SPD_1862 | comGB | Competence protein | +8.50 | ComX |
| SPD_1859 | comGE | Putative membrane protein | +8.46 | ComX |
| SPD_1861 | comGC | Competence protein | +8.38 | ComX |
| SPD_1857 | comGG | Uncharacterized protein | +8.37 | ComX |
| SPD_1858 | comGF | Competence protein | +8.36 | ComX |
| SPD_1863 | comGA | Competence protein | +8.23 | ComX |
| SPD_0132 | cibB | Putative bacteriocin | +8.21 | ComX |
| SPD_2034 | comFC | Competence protein | +7.80 | ComX |
| SPD_2035 | comFA | Competence protein | +7.74 | ComX |
| SPD_1711 | ssbB | Single-strand DNA-binding protein | +7.71 | ComX |
| SPD_0843 | comEA | Competence protein | +7.34 | ComX |
| SPD_0844 | comEC | Competence protein | +7.18 | ComX |
| SPD_1122 | dprA | DNA processing protein | +6.93 | ComX |
| SPD_0050 | comB | Competence factor transport protein | +6.76 | ComE |
| SPD_0049 | comA | Competence factor transporting protein | +6.51 | ComE |
| SPD_0865 | coiA | Competence protein | +6.50 | ComX |
| SPD_2064 | comD | Sensor histidine kinase | +6.36 | RpoD, ComE |
| SPD_0023 | comW | Uncharacterized protein | +6.34 | ComE |
| SPD_2065 | comC1 | Competence-stimulating peptide type 1 | +6.29 | RpoD, ComE |
| SPD_2063 | comE | Response regulator | +6.05 | RpoD, ComE |
| SPD_0014 | comX1 | Transcriptional regulator | +5.75 | ComE |
| SPD_2028 | cbpD | Choline binding protein D | +5.71 | ComX |
| SPD_1818 | comX2 | Transcriptional regulator | +5.59 | ComE |
| SPD_1744 | comM | Competence self-immunity protein | +5.10 | ComE |
| SPD_0475 | pncP | Bacteriocin self-immunity protein | +5.09 | ComE, BlpR |
| SPD_0474 | blpZ | Uncharacterized protein | +5.02 | ComE, BlpR |
| SPD_0308 | clpL | ATP-dependent Clp protease | +4.84 | CtsR, RpoD |
| SPD_1593 | cclA | Type IV prepilin peptidase | +4.76 | ComX |
| SPD_0473 | blpY | Bacteriocin self-immunity protein | +4.74 | ComE, BlpR |
| SPD_1638 | czcD | Cation diffusion facilitator protein | +4.44 | SczA |
| SPD_1637 | nmlR | MerR family transcriptional regulator | +4.43 | NmlR, SczA |
| SPD_1740 | cinA | Competence-damage inducible protein | +4.38 | ComX |
| SPD_1636 | adhC | Zn2+-containing alcohol dehydrogenase | +4.29 | NmlR, SczA |
| SPD_0466 | blpT | Uncharacterized bacteriocin protein | +4.16 | ComE, BlpR |
aLog2-fold change in gene expression comparing S. pneumoniae D39 in cation-defined media (CDM) relative to CDM supplemented with 30 μM Cd2+.
The next most highly upregulated group of genes were czcD, nmlR, and adhC, which comprise the overlapping SczA/NmlR regulons and belong to the inorganic ion homeostasis COG (Table 1). SczA is a cation-dependent, TetR family transcriptional regulator, which is primarily activated by Zn2+ in S. pneumoniae, but can also be regulated by other metals including cobalt, nickel and Cd2+ 18,37. Transcriptional activation by SczA leads to the expression of czcD, a cation diffusion facilitator pathway that can export Zn2+ and Co2+ ions from S. pneumoniae. However, CzcD does not export Cd2+ and, as a consequence, induction of czcD by Cd2+-SczA results in excessive Zn2+ efflux. Thus, our transcriptional observations are concordant with Zn2+ accumulation studies of Cd2+-intoxicated S. pneumoniae18. NmlR is a MerR family transcriptional regulator that controls expression of the Zn2+-dependent class III alcohol dehydrogenase, AdhC. Although the precise functional roles of NmlR and AdhC remain poorly defined, NmlR has been suggested to be an aldehyde-responsive regulator, while AdhC has been proposed to be involved in a variety of roles including carbohydrate metabolism, detoxification of reactive aldehydes and dicarbonyl compounds produced from triose sugars during carbon metabolism, and/or regenerating glutathione from glutathione-aldehyde adducts38,39. In addition, other genes involved in inorganic ion homeostasis also showed altered expression (Supplementary Table 3). Consistent with our earlier studies18, the genes involved in Mn2+ uptake were upregulated. Intriguingly, although Cd2+-stress did not impact cellular iron abundance, we observed upregulation of two iron import gene clusters, the pitCBDA and piuBCDA permeases, while the genes associated with a putative hemin importer, SPD_1588-1591, and piaB, a component of the Fe-hydroxamate import pathway, were downregulated. These changes in iron acquisition pathway expression can most likely be attributed to the metalloregulatory protein RitR responding to the reduced Mn2+ abundance during exposure to Cd2+-stress40,41.
In summary, the transcriptomics showed that cellular Cd2+ accumulation results in broad transcriptional changes in S. pneumoniae and consistent with our previous findings18, impacts pathways that employ metalloproteins and/or are associated with regulation of cellular metal homeostasis. Thus, we sought to elucidate how Cd2+ stress influenced metal-cofactor abundance in the pneumococcus.
Cadmium stress impacts the S. pneumoniae metalloproteome
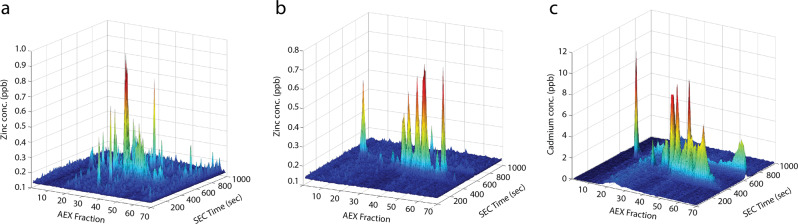
Zinc and Mn2+ are the most abundant first-row transition metal ions in S. pneumoniae18,42. It therefore follows that Cd2+-mediated dysregulation of Zn2+ and Mn2+ accumulation would have substantial effects on metalloproteins and metal-cofactor abundance. Here, we investigated the impact of Cd2+ accumulation by combining liquid chromatography-inductively coupled plasma-mass spectrometry (LC-ICP-MS) with protein mass spectrometry (MS) to generate proteomic maps of the cellular distribution of metal ions, i.e., the metalloproteome. The metalloproteome was visualized by two-dimensional separation of the proteome with metal ions detected by ICP-MS and represented as metal concentration in parts per billion (ppb). The two-dimensional separation of the proteome was performed by anion exchange (AEX) and size exclusion chromatography (SEC), which provided chromatographic distribution by resolving proteins of differing charge and mass, respectively. At equivalent total protein, variation in the concentration of metal (in ppb) and distribution of metal peaks can then be compared between Cd2+-treated and untreated samples to determine the impact of Cd2+ stress and its cellular accumulation on metalloprotein cofactor abundance.
Using this framework, the distribution of Zn2+ in the pneumococcal proteome was assessed (Fig. 2a) and contrasted with Zn2+ distribution in Cd2+-treated S. pneumoniae (Fig. 2b). Metalloproteomic maps of the cytoplasmic proteome show that protein-associated Zn2+ was directly affected with the number of protein-Zn2+ interactions (denoted by peaks) decreasing ~30% in Cd2+-treated cells compared to untreated (428 peaks, untreated vs. 306 peaks, Cd2+-treated; Fig. 2a, b). The decreased quantity and spatial distribution of Zn2+ peaks (protein-associated Zn2+) in the Cd2+-treated map suggests that there are fewer protein-Zn2+ interactions across a comparatively limited subset of cytoplasmic proteins during Cd2+ stress. This may be due to the overall reduction in cellular Zn2+ that occurs during Cd2+ stress18, Cd2+ competition for Zn2+ binding sites12, or a combination of these processes. To address this, the distribution of Cd2+-associated proteins in the treated pneumococcal proteome was assessed (Fig. 2c). Concordant with Cd2+-treatment, the pneumococcal proteome revealed 395 individual peaks, indicative of protein-Cd2+ interactions, and comparatively higher Cd2+ abundance across the cytoplasmic proteome (Supplementary Fig. 4). Comparison of the Cd2+ peaks with the Zn2+ proteome showed overlapping spatial distribution of the peaks, suggesting that Cd2+ ions were interacting with some of the fractions previously associated with Zn2+ ions (Supplementary Fig. 4). The distribution of Mn2+ ions in the metalloproteome was also investigated. However, due to the extent of Mn2+-depletion in Cd2+-treated cells18, robust maps could not be generated for meaningful comparisons. We then sought to ascertain the identities of the proteins putatively mismetallated by Cd2+ ions.
Fig. 2. Metalloproteomic maps of untreated and Cd2+-treated S. pneumoniae.
Detection and distribution of Zn2+- and Cd2+-associated proteins throughout the pneumococcal metalloproteome. a Zn2+-associated proteins in untreated S. pneumoniae, b Zn2+-associated proteins in Cd2+-treated S. pneumoniae, c Cd2+-associated proteins in Cd2+-treated S. pneumoniae. Anion exchange (AEX) separation of proteins is shown on the x-axis and size exclusion chromatography (SEC) separation of proteins (time in seconds) is shown on the y axis. The concentration of metal detected (in parts per billion [ppb]) is shown on the z-axis. All metalloproteomic maps are representative of 2 mg total cytoplasmic protein.
Streptococcus pneumoniae does not contain Cd2+-dependent proteins. Thus, it logically follows that protein association with Cd2+ is indicative of mismetallation. Mass spectrometry performed on the fractionated cytoplasmic proteome revealed the identities of many cellular proteins (Supplementary Data 1), which were data-mined for both characterized (Uniprot, BRENDA) and uncharacterized metalloproteins (NCBI Conserved Domains). The definitively identified metalloproteins (score ≥ 24) contained within specific Cd2+-enriched peaks are detailed in Table 2. The major cellular processes with apparent Cd2+-mismetallation impacts were carbohydrate metabolism, inorganic ion homeostasis and nucleotide biosynthesis. For inorganic ion homeostasis, the proteins identified within Cd2+-enriched peaks were the Mn2+-recruiting protein PsaA and the Zn2+-dependent transcriptional regulator AdcR. Previously, we have shown that PsaA can bind Cd2+ with high affinity and contributes to its cellular import18. Thus, our metalloproteomics data support our prior conclusions and highlight the utility of this technique to identify metalloproteins which may be susceptible to Cd2+ mismetallation. These data also provide additional insight into the molecular basis of Cd2+-mediated dysregulation of Zn2+ homeostasis. AdcR is a MarR family regulator that binds to DNA in the Zn2+-bound state, negatively regulating gene expression. Under conditions of Zn2+-depletion AdcR dissociates from DNA thereby permitting upregulation of the adc regulon, which encodes genes involved in Zn2+ uptake27,43,44. In our previous work, we reported that AdcR function was dysregulated during Cd2+ stress, potentially arising from Zn2+ being displaced from the thiol buffering pool onto AdcR or by Cd2+ mismetallation of AdcR18. Here, we report that AdcR was identified within a Cd2+ peak (Table 2) providing support for the formation of a mismetallated Cd2+-bound AdcR complex. We propose that the resultant depletion of cellular Zn2+ can be attributed to Cd2+-bound AdcR mimicking the Zn2+-bound state leading to repression of the adc regulon consistent with the downregulation of the Zn2+-recruiting genes adcAII, phtB, phtD, and phtE, (Supplementary Table 3). Taken together, the transcriptomic and metalloproteomic data provide insights into the mechanistic basis for Cd2+-mediated dysregulation of Zn2+ homeostasis.
Table 2.
Metalloproteins identified in Cd2+-enriched peaks.
| Locus tag | Protein | Accession number | Predicted function | Metal cofactor | Score | Number of peptides found | Sequence coverage (%) | Containing Fraction(s) |
|---|---|---|---|---|---|---|---|---|
| SPD_0577 | ZmpB | ZMPB_STRR6 | Zinc metalloprotease (EC 3.4.24.-) | Zn2+ | 25 | 61 | 27 | 11 |
| SPD_0789 | PfkA | PFKA_STRPS | Phosphofructokinase (EC 2.7.1.11) | Mg2+ | 39 | 13 | 29 | 32,33 |
| SPD_2000 | AdcR | ADCR_STRP2 | MarR family transcriptional repressor | Zn2+ | 40 | 6 | 14 | 34 |
| SPD_0526 | Fba | ALF_STRPN | Fructose-1,6-bisphosphate aldolase (EC 4.1.2.13) | Zn2+ | 430 | 57 | 63 | 32,33,34 |
| SPD_0636 | SpxB | POXB_STRPN | Pyruvate oxidase (EC 1.2.3.3) | Mg2+ | 111 | 24 | 29 | 32,34,37 |
| SPD_1390 | GlmM | GLMM_STRP4 | Phosphoglucosamine mutase (EC 5.4.2.10) | Mg2+ | 137 | 23 | 32 | 47,49,51,53 |
| SPD_0309 | LuxS | LUXS_STRPI | S-ribosylhomocysteine lyase | Fe3+ | 102 | 15 | 51 | 47,49,51 |
| SPD_1463 | PsaA | MTSA_STRPN | Mn2+ ABC transporter substrate-binding protein | Mn2+ | 27 | 8 | 25 | 47,49,51 |
| SPD_0874 | GlmU | GLMU_STRP4 | UDP-N-acetylglucosamine pyrophosphorylase | Mg2+ | 100 | 87 | 46 | 49,51,53,55 |
| SPD_1012 | Eno | ENO_STRPI | Enolase (EC 4.2.1.11) | Mg2+ | 152 | 28 | 32 | 55,57,59 |
| SPD_0389 | AccD | ACCD_STRPN | Acetyl-CoA carboxylase subunit D (EC 6.4.1.2) | Zn2+ | 24 | 18 | 32 | 59 |
| SPD_0012 | Hpt | A0A0H2ZP27 | Hypoxanthine-guanine phosphoribosyltransferase (EC:2.4.2.8) | Mg2+ | 38 | 6 | 34 | 57 |
| SPD_0024 | PurA | PURA_STRP2 | Adenylosuccinate synthetase (EC:6.3.4.4) | Mg2+ | 30 | 20 | 43 | 32,33,34 |
| SPD_1839 | Tkt/UlaH | A0A0H2ZLU9 | Transketolase (EC:2.2.1.1) | Mg2+ | 33 | 21 | 32 | 37,38,41 |
| SPD_0724 | DeoB | DEOB_STRP2 | Phosphopentomutase (EC:5.4.2.7) | Mn2+ | 59 | 9 | 19 | 38,41,47 |
| SPD_1484 | Ddl | DDL_STRP2 | D-alanine--D-alanine ligase (EC:6.3.2.4) | Mg2+/Mn2+ | 55 | 18 | 48 | 38 |
| SPD_0667 | SodA | A0A0H2ZLU4 | Superoxide dismutase (EC:1.15.1.1) | Fe2+/Mn2+ | 128 | 14 | 62 | 41 |
| SPD_1363 | PpaC | PPAC_STRP2 | Probable manganese-dependent inorganic pyrophosphatase (EC:3.6.1.1) | Mn2+ | 330 | 26 | 47 | 49,51 |
| SPD_0894 | PepT | PEPT_STRP2 | Peptidase T (EC:3.4.11.4) | Zn2+ | 40 | 9 | 11 | 49,51,53 |
| SPD_1434 | YgbL | GCH1L_STRPN | GTP cyclohydrolase 1 type 2 homolog | Mg2+ | 28 | 6 | 19 | 57 |
| SPD_0013 | FtsH | A0A0H2ZMA2 | ATP-dependent zinc metalloprotease FtsH (EC:3.4.24.-) | Zn2+ | 25 | 12 | 15 | 26 |
Glucose catabolism is disrupted by cellular Cd2+ accumulation
Building on the transcriptional and metalloproteomic profiling, we investigated the impact of Cd2+ intoxication on carbohydrate metabolism and FA biosynthesis by metabolomics. S. pneumoniae is a fermentative organism that generates energy via substrate-level phosphorylation since it lacks an electron transport chain. Similar to many other lactic acid bacteria S. pneumoniae uses glycolysis (Embden-Meyerhof pathway) to catabolize glucose45. Transcriptomics revealed that Cd2+ stress was associated with a significant downregulation (>2-fold) in the expression of numerous genes involved in glycolysis and the primary glucose PTS importer, manLMN (Table 3). Of particular note was the downregulation of the sugar phosphotransferase genes ptsH and ptsI, and the metabolic genes fructose-1,6-bisphosphate aldolase (fba), enolase (eno) and pyruvate oxidase (spxB). These genes are amongst the most highly expressed in the pneumococcus and are refractory to transcriptional regulation in most environmental conditions46. PtsHI has an essential role in mediating phosphoryl transfer from phosphoenolpyruvate (PEP) to the phosphotransferase system (PTS) sugar importers. Thus, downregulation of ptsHI would be predicted to broadly impact PTS-mediated carbohydrate import and may be responsible for the reduced expression of the glucose PTS importer manLMN. Downregulation of ptsH may also indirectly influence the coordination of carbon metabolism due to its interaction with carbon catabolite repression47. Downregulation of the highly expressed glycolytic enzymes, fba, eno and spxB would also be expected to alter glycolytic flux and thereby impact energy generation. Notably, the primary carbon catabolite repressor (ccpA) was not differentially expressed in response to Cd2+, and the differential expression of the aforementioned metabolic genes appears inconsistent with carbon catabolite control via CcpA48. This suggests that the altered metabolic response of S. pneumoniae to Cd2+ is not CcpA-coordinated.
Table 3.
Differential expression of carbon catabolism and fatty acid biosynthesis genes associated with 30 μM Cd2+ stress.
| Locus tag | Gene/Locus | Predicted function | Log2-fold changea |
|---|---|---|---|
| Increased relative expression | |||
| Glucose catabolism/glycolysis | |||
| SPD_1004 | gapN | Glyceraldehyde-3-phosphate dehydrogenase, NADP-dependent (EC 1.2.1.9) | +1.31 |
| Pentose phosphate pathway | |||
| SPD_0289 | eda | 2-deydro-3-deoxyphosphogluconate aldolase (EC 4.1.2.14/4.1.3.16) | +1.82 |
| SPD_0290 | SPD_0290 | Carbohydrate kinase, PfkB family protein | +1.60 |
| SPD_0291 | SPD_0291 | Ribose 5-phosphate isomerase, putative | +1.80 |
| Leloir pathway | |||
| SPD_1633 | galT-2 | Galactose-1-phosphate uridylyltransferase (EC 2.7.7.12) | +2.96 |
| SPD_1634 | galK | Galactokinase (EC 2.7.1.6) | +2.85 |
| SPD_1635 | galR | Galactose operon repressor | +1.68 |
| Pyruvate metabolism | |||
| SPD_0235 | pfl | Putative pyruvate formate lyase | +1.86 |
| SPD_1636 | adhC | Zn2+-containing alcohol dehydrogenase (EC:1.1.1.1) | +4.29 |
| Decreased relative expression | |||
| Glucose catabolism/glycolysis | |||
| SPD_0262 | manN | PTS-system, IID component | −1.50 |
| SPD_0263 | manM | PTS-system, IIC component | −1.59 |
| SPD_0264 | manL | PTS-system, IIAB components | −1.68 |
| SPD_0526 | fba | Fructose-1,6-bisphosphate aldolase, class II (EC 4.1.2.13) | −1.09 |
| SPD_0789 | pfkA | Phosphofructokinase (EC 2.7.1.11) | −1.39 |
| SPD_0790 | pyk | Pyruvate kinase (EC 2.7.1.40) | −1.31 |
| SPD_1012 | eno | Enolase (EC 4.2.1.11) | −1.22 |
| SPD_1039 | ptsI | Phosphocarrier protein HPr | −1.30 |
| SPD_1040 | ptsH | Phosphoenolpyruvate-protein phosphotransferase (EC 2.7.3.9) | −1.31 |
| SPD_1468 | gpmA | 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase (EC 5.4.2.11) | −1.44 |
| Pyruvate metabolism | |||
| SPD_0420 | pflB | Formate acetyltransferase (EC 2.3.1.54) | −1.41 |
| SPD_0636 | spxB | Pyruvate oxidase (EC 1.2.3.3) | −1.33 |
| Fatty acid biosynthesis | |||
| SPD_0378 | fabM | Enoyl-CoA hydratase/isomerase family protein (EC:5.3.3.14) | −2.74 |
| SPD_0379 | fabT | Transcriptional repressor | −1.86 |
| SPD_0380 | fabH | 3-oxoacyl-[acyl-carrier-protein] synthase 3 (EC:2.3.1.180) | −1.76 |
| SPD_0381 | acaP | Acyl carrier protein | −2.27 |
| SPD_0382 | fabK | Trans-2-enoyl-ACP reductase II (EC:1.3.1.9) | −2.27 |
| SPD_0383 | fabD | Malonyl CoA-acyl carrier protein transacylase (EC:2.3.1.39) | −2.18 |
| SPD_0384 | fabG | 3-oxoacyl-[acyl-carrier-protein] reductase (EC:1.1.1.100) | −2.19 |
| SPD_0385 | fabF | 3-oxoacyl-[acyl-carrier-protein] synthase 2 (EC:2.3.1.179) | −2.04 |
| SPD_0386 | accB | Acetyl-CoA carboxylase, biotin carboxyl carrier protein | −1.99 |
| SPD_0387 | fabZ | 3-hydroxyacyl-[acyl-carrier-protein] dehydratase (EC:4.2.1.59) | −1.92 |
| SPD_0388 | accC | Acetyl-CoA carboxylase, biotin carboxylase (EC:6.4.1.2 6.3.4.14) | −1.86 |
| SPD_0389 | accD | Acetyl-CoA carboxylase carboxyl transferase subunit β (EC:6.4.1.2 2.1.3.15) | −1.79 |
| SPD_0390 | accA | Acetyl-CoA carboxylase carboxyl transferase subunit α (EC:6.4.1.2 2.1.3.15) | −1.81 |
| SPD_0684 | bioY | Biotin transporter, putative | −1.84 |
aLog2-fold change in gene expression comparing S. pneumoniae D39 in CDM supplemented with 30 μM Cd2+ relative to CDM alone.
Glucose catabolism also relies upon the enzymatic activity of numerous metalloproteins. Here, we identified the Mg2+-dependent metalloenzymes phosphofructokinase A (PfkA) and enolase (Eno), and the Zn2+-dependent fructose-1,6-bisphosphate aldolase (Fba) in Cd2+-enriched peaks of the metalloproteomics indicating possible mismetallation. To address whether the cellular accumulation of Cd2+ affected glucose catabolism, we performed metabolomic analyses on S. pneumoniae exposed to 30 μM Cd2+. Metabolomics detected 416 metabolites and revealed that 162 (39%) were significantly less abundant and 63 (15%) significantly more abundant in Cd2+-treated cells, by comparison with untreated cells (P < 0.05) (Fig. 3). With respect to glucose catabolism, we observed an increase in the glycolytic precursors glucose, glucose-6-phosphate (Glu6P) and fructose 6-phosphate (F6P), indicating that flux into glycolysis was reduced or impaired prior to the formation of fructose 1,6 bisphosphate (FBP) in Cd2+-treated cells (Fig. 4 and Table 4). This finding is consistent with Cd2+-mismetallation of PfkA and/or Fba resulting in perturbation or abrogation of their cellular function. Despite the impaired flux into glycolysis, the pneumococcus could circumvent the blockage as the abundance of products from the latter stages of glycolysis, i.e., dihydroxyacetone phosphate (DHAP), 3-phosphoglycerate (3PGA) and PEP, were all observed to increase in abundance.
Fig. 3. Cadmium induced changes to the S. pneumoniae metabolome.
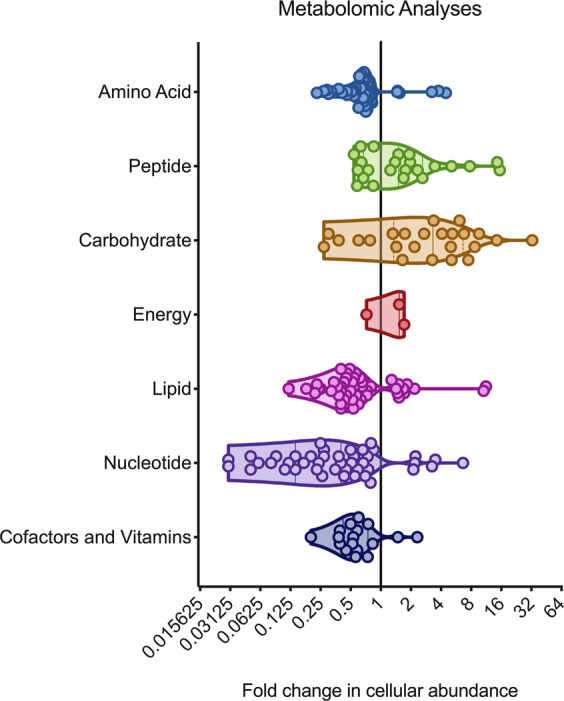
Changes in metabolite abundance in S. pneumoniae grown in the presence of 30 μM Cd2+ stress compared to untreated. Metabolites have been grouped by broad metabolic pathway with fold-change in cellular abundance denoted along the x-axis. Each spot represents an individual metabolite, with the shaded areas illustrating violin plots of the frequency distribution of the data. Data presented are the mean fold change in abundance of individual metabolites with statistically significant changes in abundance across six independent biological replicates (n = 6). Metabolite listings are provided in Supplementary Data 2.
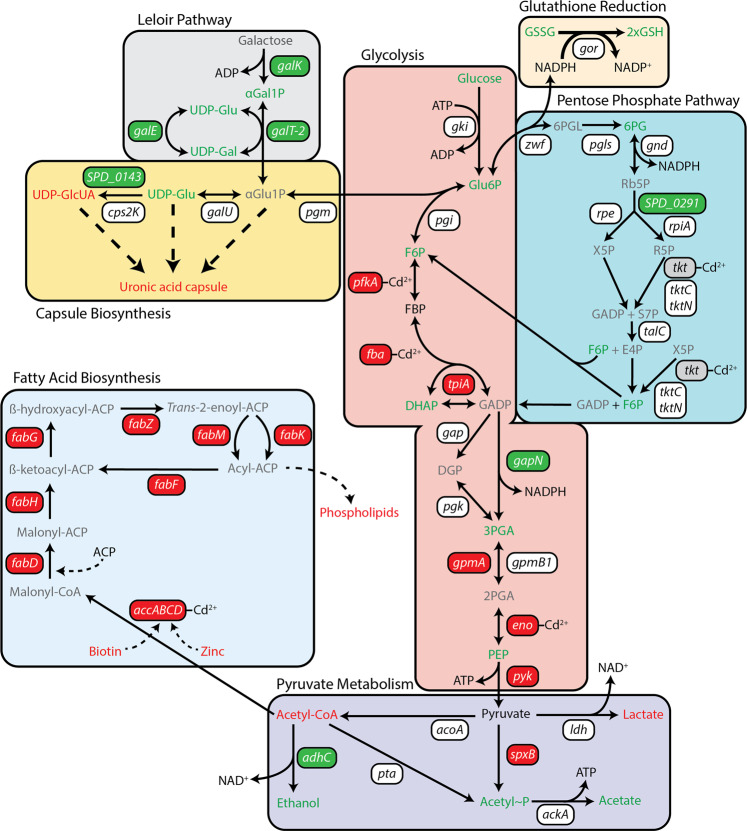
Fig. 4. Overview of Cd2+-induced impacts to central carbon metabolism in S. pneumoniae.
Cartoon representation of metabolic pathways associated with central carbon metabolism, capsule biosynthesis and fatty acid biosynthesis. Pathways have been labeled and shaded accordingly. Genes are presented in boxes, with transcriptionally upregulated (>2-fold) genes shown in green boxes, transcriptionally downregulated (>2-fold) genes in red boxes, and genes with changes of <2-fold in white boxes. Metabolomic data are presented as the differential coloring of metabolites (text only) found in the corresponding metabolic pathways. Metabolites in green were accumulated to a significantly higher abundance in Cd2+-stressed cells, compared to untreated (P < 0.05). Metabolites in red were significantly depleted in abundance in Cd2+-stressed cells, compared to untreated (P < 0.05). Metabolites in black were not significantly different from untreated cells. Metabolites in gray were not detected in the metabolomic analysis. Metalloproteomic identification of proteins associated within a Cd2+ peak, have been annotated with ‘Cd2+’ next to the corresponding gene. Gene, protein, and metabolite names can be found on Tables 2, 3 and 4.
Table 4.
Differential accumulation of cellular metabolites associated with central carbon metabolism.
| Metabolite | Full name | Fold change in abundancea |
|---|---|---|
| Glucose catabolism/glycolysis | ||
| Glu | Glucose | 6.64 |
| Glu6P | Glucose-6-phosphate | 8.69 |
| F6P | Fructose-6-phosphate | 6.79 |
| FBP | Fructose 1,6-bisphosphate | 0.89 |
| DHAP | Dihydroxyacetone phosphate | 1.32 |
| 3PGA | 3-phosphoglycerate | 1.43 |
| PEP | Phosphoenolpyruvate | 5.02 |
| Pentose phosphate pathway | ||
| 6PG | 6-phosphogluconate | 9.59 |
| F6P | Fructose-6-phosphate | 6.79 |
| Pyruvate metabolism | ||
| Pyruvate | Pyruvate | 1.68b |
| Acetyl-P | Acetyl-phosphate | 1.71 |
| Acetyl-CoA | Acetyl-CoA | 0.23 |
| Ethanol | Ethanol | 2.5 |
| Lactate | Lactate | 0.27 |
| Acetate | Acetate | 1.1 |
| Glutathione recycling | ||
| GSH | Glutathione (reduced) | 4.47 |
| GSSG | Glutathione (oxidized) | 1.46 |
| Leloir pathway/capsule biosynthesis | ||
| αGal1P | α-galactose-1-phosphate | 5.09 |
| UDP-Glu | UDP-glucose | 3.27 |
| UDP-Gal | UDP-galactose | 2.71 |
| UDP-GlcUA | UDP-glucuronate | 0.78 |
aFold change in total abundance comparing S. pneumoniae D39 in CDM supplemented with 30 μM Cd2+ relative to CDM alone.
bDid not meet statistical significance (P > 0.05).
Disruption of glycolysis can be circumvented using the Pentose Phosphate Pathway
Manganese limitation in S. pneumoniae has previously been shown to influence glucose catabolism, diverting flux into the PPP49,50. This metabolic shift has been proposed to aid in mitigating the impact of decreased Mn2+-superoxide dismutase (SodA) expression and activity49 by regenerating NADPH consumed in maintaining the reduced glutathione pool of the cell. Here, our collective data are consistent with glucose flux into the PPP. We observed that, in addition to Cd2+-induced cellular depletion of Mn2+, sodA was significantly downregulated (−1.64-fold, P < 0.05) and the expressed protein was potentially mismetallated by Cd2+ (Table 2). In contrast to glycolysis, no genes in the PPP were downregulated by Cd2+ stress, and only transketolase (Tkt) was identified in the Cd2+-enriched peaks by metalloproteomics. Disruption of Tkt activity is predicted to be compensated for by the metal-independent alternative transketolase, TktCN (Fig. 4). Metabolomic analyses indicated glucose flux into the PPP with a significant increase in 6-phosphogluconate (6PG), a key PPP intermediate generated by glucose-6-phosphate 1-dehydrogenase (Zwf) during NADP+ reduction (P < 0.05, Table 4). Consistent with activation of this pathway, the metabolomics also showed that the glutathione pool was expanded in Cd2+-treated cells and predominantly present in the form of reduced glutathione. Collectively, these data are consistent with glucose flux into the PPP and provide a plausible pathway allowing the pneumococcus to circumvent the Cd2+-mediated blockages in the initial stages of glycolysis.
Glucose catabolism via the PPP can release F6P and GADP back to glycolysis, although given the putative impairment of PfkA and/or Fba, only GADP would be available for further consumption in Cd2+-treated cells (Fig. 4). However, the next key metabolic enzyme, glyceraldehyde 3-phosphate dehydrogenase (gap), has been shown to be readily inhibited by Cu2+ and Zn2+ ions51,52. Although Gap was not identified by metalloproteomics in the Cd2+-enriched fractions, it contains the same metal-binding residues as the Zn2+-susceptible Gap from S. pyogenes (92% identity)51, suggesting that it may be susceptible to Cd2+-mismetallation. However, S. pneumoniae also encodes an alternative, non-phosphorylating Gap variant, GapN, that can convert GADP to 3PG via the generation of NADPH, albeit at the expense of ATP production53,54. This alternate enzyme provides a pathway to circumvent inhibition of Gap and regenerate NADPH for maintenance of the expanded cellular glutathione pool. Consistent with this model, we observed that gapN was significantly upregulated (P < 0.05. Table 3) in Cd2+-treated S. pneumoniae compared to untreated. In addition, targeted metabolite analyses of Cd2+-treated cells revealed that cellular NADPH levels were approximately two-fold higher compared to untreated (P = 0.056), while ATP levels were significantly depleted (60%; P < 0.0001) in these cells (Supplementary Fig. 5). Thus, this adaptive metabolic response enables S. pneumoniae to utilize glucose during metal ion intoxication, but at the expense of ATP generation.
Collectively, these data provide insights into the way in which Cd2+ accumulation disrupts glucose metabolism leading to a reduction in the bacterial growth rate. Our findings reveal a multifactorial process involving depletion of cellular Mn2+, altered transcription of glycolytic pathway genes and possible Cd2+-mismetallation of glycolytic enzymes. However, this work also highlights how the pneumococcus can overcome these impediments to achieve growth in the presence of severe metal ion dyshomeostasis.
Galactose does not protect S. pneumoniae from Cd2+-induced perturbation of energy production
Despite the primacy of glucose in S. pneumoniae carbon metabolism, the pneumococcus retains the capacity to utilize at least 32 other known sugars for energy production55. We observed that genes associated with galactose metabolism in the Leloir pathway were upregulated >2-fold (Table 3) in Cd2+-treated cells, by comparison with untreated. This was of particular interest as galactose utilization in S. pyogenes conferred resistance against Zn2+-intoxication51. In S. pyogenes, Zn2+-inhibited glycolysis at F6P and Gap, but this could be overcome by tagatose-6-phosphate pathway-mediated galactose catabolism. Accordingly, we investigated the capacity for galactose to protect S. pneumoniae from Cd2+-intoxication. This was addressed using a carbon-source free cation-defined media (CDM) supplemented with either 0.5% glucose (CDM-Glu) or 0.5% galactose (CDM-Gal). We observed that S. pneumoniae grown in CDM-Gal had reduced growth by comparison with CDM-Glu (Supplementary Fig. 1, Supplementary Table 1). Upon treatment with Cd2+, we observed that the maximal growth rates were highly similar for both carbon sources (0.54 for CDM-Glu vs. 0.46 for CDM-Gal h−1). This indicated that galactose was not protective against Cd2+. Unexpectedly, it suggested the Cd2+-intoxicated pneumococci metabolize both carbon sources at similar rates.
Carbon metabolism in S. pneumoniae can occur either via homolactic or mixed acid fermentation pathways. Catabolism of glucose and glycosaminoglycan disaccharides proceeds via homolactic fermentation, producing predominately lactate from pyruvate, via the action of lactate dehydrogenase (Ldh)56,57. In contrast, galactose is metabolized by mixed acid fermentation resulting in the production of acetate, formate, and ethanol58. The fermentation profile associated with galactose has been attributed to the slower metabolic flux of this sugar permitting carbon to be diverted to acetate production via the action of pyruvate formate-lyase which generates acetyl-CoA59. Metabolomic analyses of the terminal metabolic products of glucose in Cd2+-treated S. pneumoniae showed a significant reduction in lactate production, by comparison with untreated cells (Fig. 4 and Table 4). This occurred concomitantly with significant increases in the mixed acid fermentation end-products acetate (~15%, P = 0.0002) and ethanol (~250%, P = 0.0003) (Supplementary Fig. 5). Taken together, these data show that Cd2+ accumulation shifts the metabolic profile of glucose catabolism in S. pneumoniae from homolactic fermentation to mixed acid fermentation. This is most likely due to disruption of glycolysis and the impaired flux through FBP, which allosterically regulates Ldh activity. However, this catabolic pathway may play a crucial role for Cd2+-treated cells as it enables the generation of one ATP molecule per pyruvate molecule, through the concerted action of phosphotransacetylase (Pta) and acetate kinase (AckA) (Fig. 4).
Although galactose catabolism by the Leloir pathway was not protective against Cd2+-induced perturbation of energy production, it may have other effects on S. pneumoniae. Capsule biosynthesis requires precursors, such as UDP-glucose and UDP-glucuronate, which may be affected by the upregulation of the Leloir pathway genes. Metabolomics revealed that the Leloir pathway intermediates, UDP-galactose, and galactose-1-phosphate showed significantly increased accumulation (P < 0.05) during growth in Cd2+-supplemented CDM (Table 4). This suggested that in Cd2+-treated cells, the upregulated Leloir pathway genes might be siphoning essential precursors away from capsule biosynthesis. We assayed pneumococcal capsule production via uronic acid detection and observed significantly (P < 0.05) less cell-associated capsule in Cd2+-treated cells, by comparison to untreated (Fig. 5). Taken together, these data suggest that activation of the Leloir pathway in response to cellular Cd2+ accumulation is not protective and results in reduced capsule production.
Fig. 5. Capsule production during Cd2+-stress.
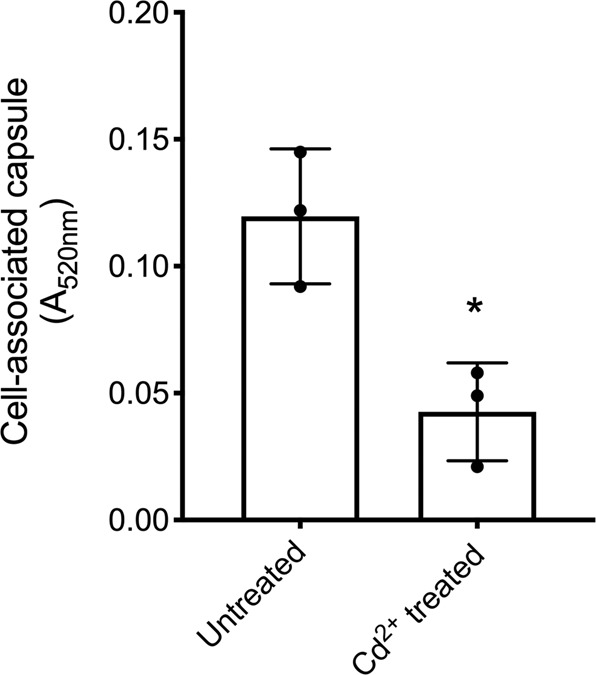
Determination of capsule production via detection of uronic acid from S. pneumoniae D39 untreated and treated with 30 µM Cd2+. Data presented are the mean ± SD of three independent biological replicates (n = 3). The statistical significance of the differences in the mean data were determined by two-tailed unpaired t-tests (*P = 0.0153).
Fatty acid biosynthesis is impaired during Cd2+ stress
Essential FAs required for the S. pneumoniae membrane are synthesized by the type II FA synthase (FASII) system60, which is encoded by the fab gene cluster (fabMTHKDGFZ). De novo FA biosynthesis also relies upon the acetyl-CoA carboxylase genes (accBCDA) and the acyl carrier protein (acpP), which is responsible for the transport of FAs. Transcriptional control of the FA biosynthetic gene cluster is primarily regulated by FA sensing of the negative regulator FabT, with FA biosynthesis genes upregulated in a ΔfabT strain61. We observed that S. pneumoniae lipid metabolism and FA biosynthesis were strongly affected by cellular Cd2+ accumulation with the FA biosynthesis genes downregulated between 3.3 and 6.7-fold, when compared to untreated cells (Table 3). Notably, expression of FabT was also reduced by 3.5-fold, suggesting that transcription of the fab cluster may be repressed via an alternative mechanism.
The first committed step in FA biosynthesis is the conversion of acetyl-CoA to malonyl-CoA. Metabolomic and transcriptomic analyses revealed a significant four-fold decrease in the abundance of acetyl CoA (P = 0.01) in Cd2+-treated cells, by comparison with untreated cells. This occurred concomitantly with a 19-fold increase in adhC transcription and increased ethanol production (Fig. 4 and Tables 3, 4). Conversion of acetyl CoA to malonyl CoA occurs via acetyl CoA carboxylase (AccBCDA), a heteromeric enzyme requiring biotin and Zn2+ cofactors62. We observed that Cd2+ exposure resulted in a two-fold decrease (P = 0.0025) in biotin attributable to the 3.6-fold downregulation of the biotin importer (bioY) (Tables 3, 4). Further, AccD, the Zn2+-dependent subunit of AccBCDA, was detected by metalloproteomics within a Cd2+-enriched peak (Table 2) suggesting mismetallation. Taken together, these data show that Cd2+-accumulation disrupts the FASII system at a transcriptional and functional level. This manifests through perturbation of the initial step in the FA biosynthesis pathway and a generalized downregulation of the FASII genes. Accordingly, we sought to assess the impact of this disruption on membrane composition.
Membrane composition and fluidity is altered by Cd2+ stress
In addition to de novo FA biosynthesis by the FASII system, the pneumococcus can incorporate exogenous FAs into its membrane via FA phosphorylation using the FakAB1B2B3 and PlsXY systems61,63,64. This facilitates phospholipid acquisition without use of the energy intensive FASII system. We observed that expression of the plsXY and fakAB1B2 systems were not affected by Cd2+ exposure. In contrast, fakB3 (SPD_0646), which is responsible for incorporation of polyunsaturated FAs, was downregulated ~4-fold relative to untreated cells. This suggests acquisition of exogenous unsaturated FAs may be impaired during exposure to Cd2+ 63. Thus, we next examined acyl chain composition of the S. pneumoniae membrane via LC-MS. In Cd2+-treated cells, the abundance of saturated acyl chains (14:0, 16:0 and 18:0) was reduced 1.4–2.5-fold and monounsaturated acyl chains (16:1 and 18:1) were extensively depleted (2.7–3.4-fold) (Fig. 6a). The physiological impact of the increased abundance of saturated acyl chains, relative to unsaturated acyl chains, on the pneumococcal membrane was then assessed. Using the fluorescent probe diphenylhexatriene, we observed that Cd2+-treated cells had significantly increased membrane rigidity relative to untreated cells (P = 0.0005) (Fig. 6b). This indicated that the greater ratio of saturated to unsaturated acyl chains resulted in tighter packing of saturated phospholipids in the cell membrane during metal ion dyshomeostasis.
Fig. 6. Cadmium induced changes to the S. pneumoniae membrane.
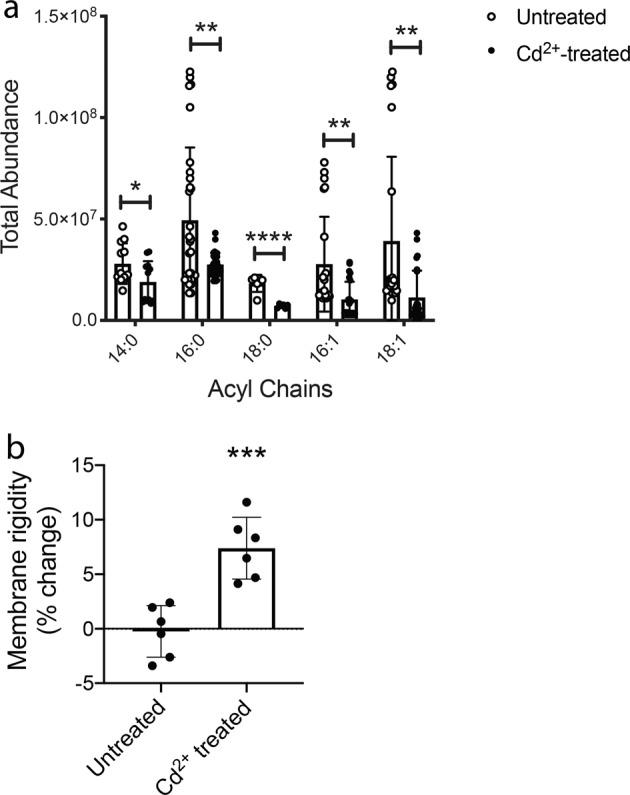
Biophysical changes to the S. pneumoniae membrane in response to 30 µM Cd2+ stress. a Total abundance of saturated (14:0, 16:0, 18:0) and unsaturated (16:1, 18:1) acyl chains as determined by LC/MS metabolomic analyses. Acyl chain abundance in untreated S. pneumoniae is shown in white circles and acyl chain abundance from 30 µM Cd2+-treated S. pneumoniae is shown in black circles. Data presented are the mean ± SD of six independent biological replicates (n = 6). The statistical significance of the differences in the mean data were determined by two-tailed unpaired t-tests (*P = 0.0383, **P = 0.0016, 0.0013, 0.0029, and ****P < 0.0001). b Membrane rigidity as determined by diphenylhexatriene fluorescence normalized to untreated signal and expressed as % change. Data presented are the mean ± SD of six independent biological replicates (n = 6). The statistical significance of the differences in the mean data were determined by two-tailed unpaired t-tests (***P = 0.0005).
Discussion
Transition metal ion homeostasis is crucial for the viability of all organisms. In the human pathogen S. pneumoniae, we have probed the molecular and cellular impacts of metal ion dysregulation by using the non-physiological metal ion Cd2+. This exploits the extensive chemical similarity of Cd2+ with the biologically essential metal ion Zn2+ 8,12. However, the pneumococcus lacks Cd2+-specific regulatory and efflux pathways most likely due to limited environmental exposure and an absence of selective pressures to evolve resistance. As a consequence, Cd2+ is readily accumulated by S. pneumoniae resulting in dysregulation of Mn2+ and Zn2+ homeostasis mechanisms18. Thus, Cd2+ is an ideal probe to study the broader cellular impacts that arise from disruptions in metal ion homeostasis. Although the core pathways and processes impacted by Cd2+ stress in S. pneumoniae are conserved in many bacteria, we predict that as organism complexity increases as a function of genome size, this will provide both additional mechanisms of resistance to Cd2+ and targets of susceptibility.
The FASII biosynthetic pathway and the exogenous FA acquisition systems are essential for membrane biogenesis in S. pneumoniae65. Our investigations showed that exposure to Cd2+ was associated with a downregulation of the genes in the FASII pathway, albeit via an unknown mechanism. Studies in other bacteria, notably E. coli66 and B. subtilis67, have previously shown that acc gene transcription, and therefore the first committed step in the FASII pathway, is directly related to the growth rate of the organism. Although this has not been established to occur in S. pneumoniae, the Cd2+-induced reduction in the growth rate may impact the expression of the FASII pathway. Alternatively, downregulation of the FASII pathway may be due to an additional regulatory mechanism, such as YycFG, which has been found to modulate the expression of the FASII system in S. pneumoniae68,69, and was not transcriptionally impacted by Cd2+ exposure. Although further studies will be required to elucidate the precise mechanisms for how these biosynthetic pathways are impacted, our findings highlight how Cd2+-induced changes in central carbon metabolism directly influence other essential cellular biosynthetic pathways.
The effect of Cd2+ treatment on phospholipid production resulted in significant changes to membrane composition. Notably, an increase in membrane rigidity due to the greater ratio of saturated:unsaturated membrane acyl chains. Unsaturated FAs contain double carbon bonds that are highly susceptible to reactive oxygen species such as H2O2 and OH⦁70. Prior studies have shown that exposure of S. pneumoniae to H2O2 resulted in a reduction in the abundance of double bonds within the membrane phospholipids71. This has been attributed to FabF, which has a thiol-reactive cysteine that serves as a sensor of cellular oxidative stress72. Accordingly, in Cd2+-treated S. pneumoniae the depletion of cellular Mn2+ and concomitant loss of Mn-SOD may serve as a trigger for shifting the ratio of saturated:unsaturated FAs in the membrane in order to minimize susceptibility to lipid peroxidation73. Thus, the physiological consequence of this change is decreased membrane fluidity and increased resistance to intracellular oxidative stress.
Central carbon metabolism was prominently impacted by Cd2+ accumulation with multiple disruptions in glycolysis, activation of the PPP, and a switch to mixed acid fermentation of pyruvate. Glucose is primarily catabolized by glycolysis in S. pneumoniae. However, glucose flux can be diverted into the PPP by depletion of NADPH, as shown in Corynbacterium glutamicum74. Cellular depletion of Mn2+, which occurs in S. pneumoniae during Cd2+ stress18, also activates this metabolic switch to enable regeneration of NADPH required to maintain the cellular pool of reduced GSH49. This is crucial for Mn-SOD independent resistance against oxidative stress and, during exposure to Cd2+, for expansion of the reduced GSH pool to buffer intracellular Cd2+ ions. Thus, we infer that activation of the PPP is an adaptive metabolic change that enhances resistance to metal stress, regulated via NADPH abundance. Further, we speculate that the PPP is finely tuned to be a metal-intoxication resistant pathway, comprised of metal ion independent catabolic enzymes such as Zwf, Gnd, RpiA, TalC and TktCN75–77. In this way, the PPP enables glucose catabolism in the presence of metal ion intoxication, albeit with less efficiency than the metalloenzyme-dominated pathway of glycolysis, with metabolites entering the latter half of glycolysis and continuing their catabolic progression via metal-independent alternative enzymes such as GapN53. Hence, this work highlights the evolutionary cost of employing metal-dependent proteins. Although metal ions can be used to enhance protein activity and thereby enable more efficient enzymatic functionality, as exemplified in glycolysis, this dependency renders metalloproteins susceptible to inactivation upon interaction with non-cognate metals, such as Cd2+. Nevertheless, we cannot exclude the possibility that activation of the PPP arises from impaired flux through glycolysis due to downregulation of glycolytic genes, possibly as a result of the Cd2+-induced growth delay, and/or impaired function of glycolytic metalloenzymes. In the absence of Cd2+ the glycolytic metabolite FBP regulates the direction of carbon flux via activation of the enzymes PfkA and Pyk and inhibition of Gnd, which prevents flux into the PPP74,78,79. FBP also allosterically activates Ldh and thereby regulates the fermentation pathway of pyruvate80. Treatment of S. pneumoniae with Cd2+ reduced the abundance of FBP relative to other glycolytic intermediates, attributable to decreased expression of pfkA and potential Cd2+-mismetallation of the enzyme. As a consequence, this impairment in glycolysis may also trigger activation of the PPP. Irrespective of the mechanistic basis, glucose appears to be catabolized by the PPP during Cd2+-induced metal ion dyshomeostasis enabling regeneration of NADPH, but at the expense of ATP production. The overall rate of glucose flux through the PPP appears to be lower than that observed with glycolysis as suggested by the increased abundance of metabolic intermediates from the latter half of glycolysis. Despite the primacy of CcpA in regulating optimal carbon catabolism in S. pneumoniae48,81, the observed transcriptional changes in key metabolic genes appear inconsistent with CcpA-dependent regulation of glucose catabolism48. However, it is important to note that studies of CcpA in S. pneumoniae have used a variety of distinct nutritional parameters48,81. The resultant differences in growth rates, gene expression and analytical approaches complicate direct comparisons between studies. Nevertheless, our findings suggest that the observed shunt toward the PPP is not coordinated by a general carbon catabolite repression mechanism, but is instead an adaptive response to the cellular impacts of Cd2+ exposure. Collectively, these observations highlight the capacity of S. pneumoniae to undergo an adaptative metabolic change in response to environmental stress.
The Cd2+-mediated impact on carbon flux also provides a mechanistic explanation for how intracellular metal abundance can directly influence biosynthetic pathways, such as capsule production. In Lactococcus acidophilus it has been shown that Fe2+ and Cu2+ supplementation increases the production of the primary capsule component hyaluronic acid (HA) via interaction with PPP enzymes and pathway intermediates82. In contrast, Zn2+ intoxication of S. pyogenes decreased HA capsule production due to inhibition of phosphoglucomutase (Pgm)51. Here, we observed that Cd2+accumulation also resulted in decreased capsule levels, although this appeared to be independent of pgm expression and function. This indicated an alternative mechanism of capsule biosynthesis disruption in Cd2+-treated pneumococci. A possible explanation is that inappropriate activation of the Leloir pathway during Cd2+ exposure results in siphoning of αGlu1P, thereby preventing its flux into capsular polysaccharide production and leading to the reduced capsule.
Collectively, this study has revealed the extensive breadth of cellular and metabolic adaptations required for the pneumococcus to maintain viability during metal ion dyshomeostasis. The accumulation of Cd2+ in the cytoplasm exerts substantial deleterious impacts upon the cell by disrupting energy production, likely through a combination of cellular Mn2+ and Zn2+ depletion and possible competition between Cd2+ and native metal cofactors for crucial metalloproteins. Importantly, this work has shown how adaptive changes in central carbon metabolism may be used to mitigate the loss of metal ion homeostasis and provide continued energy production during metal ion intoxication.
Methods
Growth of S. pneumoniae D39
S. pneumoniae was routinely grown in cation-defined media (CDM), which corresponded to C + Y media without supplementation of transition metals83. The base transition metal concentration of the media was determined by ICP-MS on an Agilent 7500cx (Adelaide Microscopy, University of Adelaide) to be: 0.23 µM manganese, 5.6 µM iron, 0.15 µM cobalt, 0.16 µM nickel, 2.8 µM copper, and 12.1 µM zinc. CDM was routinely prepared with 0.2% [w/v] glucose for microbiological analyses83. Carbon-source comparison growth assays were conducted in 0.5% [w/v] glucose or 0.5% [w/v] galactose where specified. All growth experiments were conducted in CDM supplemented with 1 μM MnSO4. Thirty micromolar CdCl2 was added where specified. All chemicals used in this study were purchased from Sigma Aldrich unless otherwise specified. Cultures of S. pneumoniae D39 were routinely prepared from overnight growth on blood agar, resuspended, and inoculated into CDM to an absorbance at 600 nm (A600) of 0.05. The culture was incubated at 37 °C + 5% CO2 and grown to A600 = 0.3. Growth kinetic assays were conducted in 96-well plate format using a FLUOstar Omega spectrophotometer (BMG Labtech). S. pneumoniae were inoculated in a final volume of 200 μL CDM (± supplementation where indicated) to a starting A600 = 0.01 in a clear 96-well plate (Greiner). Plate was incubated at 37 °C + 5% CO2 for >16 h with readings taken every 30 min. Growth assay data were analyzed using GraphPad Prism.
RNA sequencing and qRT-PCR sample preparation
S. pneumoniae was grown as detailed above. At mid-log phase (A600 = 0.3), 500 μL of culture was mixed with 1 mL of RNA Protect (Qiagen) and cells were harvested via centrifugation before storage at −80 °C. Bacterial pellets were RNA extracted and purified using RNeasy Protect Bacterial Mini kit (Qiagen) after enzymatic lysis using lysozyme and mutanolysin, all according to the manufacturer’s instructions. DNase treatment was performed on-column during RNA extraction using RNase-free DNase (Qiagen).
qRT-PCR
Quantitative reverse transcription PCR was conducted using Superscript III (Invitrogen) on a QuantStudio 7 Real-time PCR system (Applied Biosystems). The transcription levels of genes analyzed were normalized to those obtained for 16s rRNA. Primer sequences are listed in Supplementary Table S2.
RNA sequencing
RNA was extracted and prepared as above from biological quadruplicates of S. pneumoniae D39. RNA was pooled and analyzed on a Bioanalyzer 2100 (Agilent) to confirm a RIN value > 8 according to the manufacturer’s instructions. RNA was then submitted to the Australian Genome Research Facility (AGRF) for sequencing. In brief, the Epicentre Bacterial Ribozero Kit (Illumina) was used to deplete ribosomal RNA content before the generation of barcoded libraries using Ultra Directional RNA kit (New England Biolabs). Prepared libraries were then sequenced using an Illumina HiSeq2500 with Version 3 SBS reagents and 2 × 100 bp single-end chemistry. Reads were aligned to the S. pneumoniae D39 genome (GenBank accession number NC_008533) using BOWTIE2 version 2.2.684. Counts for each gene were obtained using SAMtools version 1.285 and BEDtools version 2.24.086, and differential gene expression was determined using R (DESeq Library) version 3.2.287. Transcriptomic data have been deposited in the NCBI Gene Expression Omnibus databank under submission identifier GSE141681.
Bioinformatic COG analysis
Clusters of Orthologous Groups (COG) analysis was performed using the S. pneumoniae D39 reference sequence (NC_008533.gbk) from the National Centre for Biotechnology Information (NCBI). COG classifications of proteins (as designated by NCBI) are applied based on computational analysis of sequenced genomes to predict protein functionality and/or biochemical behavior based on homology to currently characterized proteins88. All available COG classifications for the S. pneumoniae D39 genome were aligned to RNA sequencing data and clustered according to the functional category. Calculations were then performed to determine the percentage of genes in each cluster that showed differential expression of greater than two-fold.
Metabolomic sample preparation
S. pneumoniae cultures for metabolomic analyses were grown as detailed above in biological sextuplicate. At A600 = 0.3, cultures were harvested by centrifugation at 7000 × g for 7 mins at 4 °C. Once pelleted, cells were flash-frozen using liquid nitrogen to preserve the metabolite profile and stored at −80 °C. Cultures were processed and analyzed via LC/MS by Metabolon, North Carolina, USA.
Targeted metabolite analyses
For metabolites that were not detected using metabolomics, targeted analyses using commercially available kits were undertaken for the detection of ethanol (Megazyme), acetate (Biovision), ATP (BacTiter-Glo; Promega) and NAD(P)H/NAD(P) (Abcam). All assays were carried out according to the manufacturer’s instructions. In brief, S. pneumoniae were grown in 7 mL cultures of CDM ± 30 µM Cd2+ as previously described in a minimum of biological triplicate. Cells were harvested via centrifugation at 7000 × g for 7 min at 4 °C and resuspended in assay-appropriate buffer. All statistical analyses were conducted using a two-tailed, unpaired t-test (GraphPad Prism).
Metalloproteomic sample preparation
S. pneumoniae cultures for metalloproteomic analyses were prepared as detailed above. At A600 = 0.3, 45 mL cultures were harvested by centrifugation at 4 °C for 7 min at 7000 × g. Cell pellets were then washed thrice with 20 mL PSB, 5 mM EDTA, followed by three washes with 20 mL PBS at 4 °C for 7 min at 7000 × g. The harvested cell pellets were then stored at −80 °C for LC-ICP-MS analysis.
LC-ICP-MS
Metalloproteomic culture cell pellets were resuspended in 200 μL 20 mM Tris-HCl pH 8.0 and sonicated using a Bioruptor system (Diagenode) for 25 cycles (30 s on, 30 s off). Sonicated cultures were then centrifuged at 4 °C for 15 min at 18,000 × g to remove insoluble material. The supernatant was harvested and 2 mg total protein was fractionated via AEXchromatography using a Bio IEX 3 μm column (Agilent) on an Infinity 1260 HPLC system (Agilent). AEX fractions were collected in 1-min intervals, corresponding to 400 μL fractions. AEX fractions were then independently separated via SEC using a Bio SEC-3 column (Agilent) on an Infinity 1260 HPLC system (Agilent), directly hyphenated to an ICP-MS 7500cx (Agilent) to determine metal content. Detection of metals (recorded in counts per second) were normalized to an internal standard (antinomy [Sb]) and metal concentration (in ppb) was interpolated from a calibration curve of known metal concentrations. Determination of protein-metal interactions (denoted by number of peaks with increased metal abundance) was conducted using baseline-corrected area under curve analysis (GraphPad Prism 8.0). Metalloproteomic maps were generated using MATLAB 2020a.
Mass spectrometry
MS samples were prepared essentially according to89. In brief, selected AEX fractions were chosen for MS analysis based on the presence of Cd2+ and Zn2+ peaks in the metalloproteomic maps. The protein concentrations of the chosen AEX fractions were determined by A280. Approximately 0.5 μg of protein was added to an equal volume of HPLC grade acetonitrile (ACN), and trypsin digested according to protocol89. Proteins were harvested through evaporation of the liquid phase in a vacuum concentrator centrifuge for ~2 h. Samples were individually resuspended in 250 μL of 0.1% v/v trifluoroacetic acid (TFA) in ddH2O, and C18 treated using the default ‘Peptide Clean-up’ method on the automated BRAVO liquid handling robot (Agilent). Eluted peptides were harvested through evaporation of the liquid phase and resuspended in 10 μL 2% v/v ACN, 0.1% v/v TFA in ddH20. Final protein concentration was determined by A280, with all samples having a final concentration of ~1.5 mg mL−1.
Samples were then run on the Q ExactiveTM Plus Hybrid Quadrupole-OrbitrapTM Mass Spectrometer (ThermoFisher Scientific) at the Bio21 Institute, Melbourne, Australia.
MS data analysis
MS data analysis was conducted using Matrix Scientific MASCOT Server. Data files (.mgf) were uploaded into MASCOT MS/MS ion search and analyzed using SwissProt database for matches to Streptococcus pneumoniae taxonomy. Parameters were as follows: Enzymatic cleavage by trypsin, allowing one missed cleavage site. Fixed Carbamidomethyl (C) modification and variable Oxidation (M) modification. Peptide tolerance of 1.2 Da, MS/MS tolerance of 0.6 Da with monoisotopic mass values. Peptide charges of +1, +2, +3 were allowed with an unrestricted protein mass. The significance threshold was set at P < 0.05. Ion scores of ≥24 were indicative of protein identity or extensive homology. Only proteins with >2 peptide hits with ion scores ≥ 24 were considered true identifications in accordance with90.
Uronic acid capsule assay
S. pneumoniae was grown as previously described to a final A600 = 0.2, harvested via centrifugation and resuspended in 500 µL 150 mM Tris-HCl pH 7.0, 1 mM MgSO4. Subsequent sample processing and quantification of capsular uronic acid was conducted as described in refs. 91,92. In brief, resuspended cultures were incubated at 37 °C for 30 min with 0.01% [v/v] deoxycholate to lyse the cells, followed by overnight incubation at 37 °C with 100 U mutanolysin, 50 µg DNase and 25 µg RNase. Samples were then subsequently incubated with 100 µg proteinase K for 4 h at 56 °C. One hundred microlitres of each sample were then added to 600 µL 12.5 mM sodium tetraborate in 98% [v/v] H2SO4. Samples were vortexed and heated at 95 °C for 5 min and cooled on ice before the addition of 3-phenylphenol solution (10 µL). Samples were then immediately added to a clear 96-well plate and absorbance was read at 520 nm using a PHERAstar spectrophotometer (BMG Labtech). The amount of cell-associated capsule was determined from 3 independent experiments and the statistical difference was assessed by a two-tailed, unpaired t-test (GraphPad Prism).
Membrane rigidity assay
Membrane rigidity was determined as described in93. In brief, bacterial cultures were grown as previously described to A600 = 0.3 and cells were washed in PBS. Bacteria were incubated with 1,6-diphenyl-1,3,5-hexatriene dissolved in tetrahydrofuran. After incubation at 37 °C for 30 min, cells were washed and fluorescence polarization (excitation 350/emission 450 nm) was determined on a PHERAstar spectrophotometer (BMG Labtech). The relative change in membrane rigidity was determined from six independent experiments and the statistical difference was assessed by a two-tailed, unpaired t-test (GraphPad Prism).
Statistics and reproducibility
Sample sizes were derived based on the accepted conventions for life sciences research, i.e., for independent biological replicates, n = 3 is sufficient for performing statistical comparisons between experimental groups. Experiments contributing to a single dataset were performed across multiple days/weeks to ensure that analyses were being conducted across independent bacterial cultures and batches of reagents. For the -omics analyses, sample sizes were guided by acceptable industry standards. RNA sequencing used four pooled biological replicates (Australian Genome Research Facility), metabolomic analyses used six biological replicates (Metabolon), metalloproteomic maps and MS data were generated from single biological replicates to allow direct comparison between metal ion determination and protein identity. Data reproducibility was ensured through temporal separation of replicate experiments.
All statistical analyses were conducted using GraphPad Prism. Statistical tests were selected based on the nature of the data. For direct comparison of untreated and Cd2+-treated groups, two-tailed, unpaired t-tests were routinely applied.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
This work was supported by the National Health and Medical Research Council (NHMRC) Program Grant 1071659 to J.C.P., Project Grants 1080784 and 1122582 to C.A.M., and the Australian Research Council (ARC) Discovery Project Grant DP170102102 to J.C.P. and C.A.M. S.L.N. is an NHMRC Early Career Research Fellow (1142695) and C.A.M. is an ARC Future Fellow (FT170100006). J.W.R is supported by 1U01AI124302, 1RO1AI110618, and by ALSAC. B.R.R acknowledges receiving partial support from the NHMRC (1061550 & 1138673), and the Motor Neuron Disease Research Institute of Australia. We thank Prof. A.G. McEwan for critical reading and discussions, and the Melbourne Mass Spectrometry and Proteomics Facility of the Bio21 Molecular Science and Biotechnology Institute, The University of Melbourne, mass spectrometry analysis support.
Author contributions
S.L.N., B.A.E., and C.A.M. were responsible for conceptualization, experimental design, sample preparation and executed all experiments. A.L. and B.R.R. provided expertise and assistance for all metalloproteomics and mass spectrometry data analyses. J.W.R. conducted metabolomic analyses. S.L.N. and C.A.M. drafted the manuscript. All authors (S.L.N., B.A.E., A.L., J.C.P., B.R.R., J.W.R., and C.A.M.) contributed to data analysis and reviewed the final manuscript.
Data availability
RNA sequencing data have been deposited in NCBI Gene Expression Omnibus databank under submission identifier GSE141681. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE94 partner repository with the dataset identifier PXD021643 and 10.6019/PXD021643. Raw data files for all other Figures are available from the corresponding authors upon reasonable request.
Competing interests
The authors declare the following competing interests: B.R.R. receives research support from Agilent Technologies and e-MSion, Inc. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Stephanie L. Neville, Email: stephanie.neville@unimelb.edu.au
Christopher A. McDevitt, Email: christopher.mcdevitt@unimelb.edu.au
Supplementary information
Supplementary information is available for this paper at 10.1038/s42003-020-01417-y.
References
- 1.Sarkar A, Ravindran G, Krishnamurthy V. A brief review on the effect of cadmium toxicity: from cellular to organ level. Int. J. Biotechnol. Res. 2013;3:17–36. [Google Scholar]
- 2.Srivastava, A., Siddiqui, N., Koshe, R. K. & Singh, V. K. Advances in Health and Environment Safety, pp. 279–296 (Springer, Singapore, 2018).
- 3.Pan J, Plant JA, Voulvoulis N, Oates CJ, Ihlenfeld C. Cadmium levels in Europe: implications for human health. Environ. Geochem. Health. 2010;32:1–12. doi: 10.1007/s10653-009-9273-2. [DOI] [PubMed] [Google Scholar]
- 4.Hutton M, Symon C. The quantities of cadmium, lead, mercury and arsenic entering the UK environment from human activities. Sci. Total Environ. 1986;57:129–150. doi: 10.1016/0048-9697(86)90018-5. [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization. Cadmium: environmental aspects. (World Health Organization, 1992).
- 6.Amadi CN, Igweze ZN, Orisakwe OE. Heavy metals in miscarriages and stillbirths in developing nations. Middle East Fertil. Soc. J. 2017;22:91–100. doi: 10.1016/j.mefs.2017.03.003. [DOI] [Google Scholar]
- 7.Mezynska M, Brzoska MM. Environmental exposure to cadmium—A risk for health of the general population in industrialized countries and preventive strategies. Environ. Sci. Pollut. Res. 2018;25:3211–3232. doi: 10.1007/s11356-017-0827-z. [DOI] [PubMed] [Google Scholar]
- 8.Dietrich N, Tan C-H, Cubillas C, Earley BJ, Kornfeld K. Insights into zinc and cadmium biology in the nematode Caenorhabditis elegans. Arch. Biochem. Biophys. 2016;611:120–133. doi: 10.1016/j.abb.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 9.Bertin G, Averbeck D. Cadmium: cellular effects, modifications of biomolecules, modulation of DNA repair and genotoxic consequences (a review) Biochimie. 2006;88:1549–1559. doi: 10.1016/j.biochi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Ercal N, Gurer-Orhan H, Aykin-Burns N. Toxic metals and oxidative stress part I: mechanisms involved in metal-induced oxidative damage. Curr. Top. Med. Chem. 2001;1:529–539. doi: 10.2174/1568026013394831. [DOI] [PubMed] [Google Scholar]
- 11.Lin A-j, Zhang X-h, Chen M-m, Qing C. Oxidative stress and DNA damages induced by cadmium accumulation. J. Environ. Sci. 2007;19:596–602. doi: 10.1016/S1001-0742(07)60099-0. [DOI] [PubMed] [Google Scholar]
- 12.Brzóska M, Moniuszko-Jakoniuk J. Interactions between cadmium and zinc in the organism. Food Chem. Toxicol. 2001;39:967–980. doi: 10.1016/S0278-6915(01)00048-5. [DOI] [PubMed] [Google Scholar]
- 13.Blum U, Schwedt G. Inhibition behavior of acid phosphatase, phosphodiesterase I and adenosine deaminase as tools for trace metal analysis and speciation. Analytica Chim. Acta. 1998;360:101–108. doi: 10.1016/S0003-2670(97)00717-4. [DOI] [Google Scholar]
- 14.Moulis, J.-M. & Thévenod, F. New Perspectives in Cadmium Toxicity: An Introduction. (Springer, 2010). [DOI] [PubMed]
- 15.Asmuss M, Mullenders LH, Eker A, Hartwig A. Differential effects of toxic metal compounds on the activities of Fpg and XPA, two zinc finger proteins involved in DNA repair. Carcinogenesis. 2000;21:2097–2104. doi: 10.1093/carcin/21.11.2097. [DOI] [PubMed] [Google Scholar]
- 16.Helbig K, Grosse C, Nies DH. Cadmium toxicity in glutathione mutants of Escherichia coli. J. Bacteriol. 2008;190:5439–5454. doi: 10.1128/JB.00272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng X, Tang J, Liu X, Jiang P. Response of P. aeruginosa E(1) gene expression to cadmium stress. Curr. Microbiol. 2012;65:799–804. doi: 10.1007/s00284-012-0224-2. [DOI] [PubMed] [Google Scholar]
- 18.Begg SL, et al. Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae. Nat. Commun. 2015;6:6418. doi: 10.1038/ncomms7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eijkelkamp BA, et al. Dietary zinc and the control of Streptococcus pneumoniae infection. PLoS Pathog. 2019;15:e1007957. doi: 10.1371/journal.ppat.1007957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shafeeq S, et al. The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Mol. Microbiol. 2011;81:1255–1270. doi: 10.1111/j.1365-2958.2011.07758.x. [DOI] [PubMed] [Google Scholar]
- 21.Johnson MD, et al. Role of copper efflux in pneumococcal pathogenesis and resistance to macrophage-mediated immune clearance. Infect. Immun. 2015;83:1684–1694. doi: 10.1128/IAI.03015-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDevitt CA, et al. A molecular mechanism for bacterial susceptibility to zinc. PLoS Pathog. 2011;7:e1002357. doi: 10.1371/journal.ppat.1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satarug S, Moore MR. Adverse health effects of chronic exposure to low-level cadmium in foodstuffs and cigarette smoke. Environ. Health Perspect. 2004;112:1099–1103. doi: 10.1289/ehp.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.U.S Department of Health and Human Services. (ed Agency for Toxic Substances and Dsiease Registry) (Atlanta, Georgia, US, 2012).
- 25.Sundblad B-M, et al. Extracellular cadmium in the bronchoalveolar space of long-term tobacco smokers with and without COPD and its association with inflammation. Int. J. Chronic Obstr. Pulm. Dis. 2016;11:1005. doi: 10.2147/COPD.S105234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manna S, et al. The transcriptomic response of Streptococcus pneumoniae following exposure to cigarette smoke extract. Sci. Rep. 2018;8:1–12. doi: 10.1038/s41598-017-17765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shafeeq S, Kloosterman TG, Kuipers OP. Transcriptional response of Streptococcus pneumoniae to Zn2+ limitation and the repressor/activator function of AdcR. Metallomics. 2011;3:609–618. doi: 10.1039/c1mt00030f. [DOI] [PubMed] [Google Scholar]
- 28.Rosch JW, Gao G, Ridout G, Wang YD, Tuomanen EI. Role of the manganese efflux system mntE for signalling and pathogenesis in Streptococcus pneumoniae. Mol. Microbiol. 2009;72:12–25. doi: 10.1111/j.1365-2958.2009.06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McAllister LJ, et al. Molecular analysis of the psa permease complex of Streptococcus pneumoniae. Mol. Microbiol. 2004;53:889–901. doi: 10.1111/j.1365-2958.2004.04164.x. [DOI] [PubMed] [Google Scholar]
- 30.Kloosterman TG, Van Der Kooi‐Pol MM, Bijlsma JJ, Kuipers OP. The novel transcriptional regulator SczA mediates protection against Zn2+ stress by activation of the Zn2+‐resistance gene czcD in Streptococcus pneumoniae. Mol. Microbiol. 2007;65:1049–1063. doi: 10.1111/j.1365-2958.2007.05849.x. [DOI] [PubMed] [Google Scholar]
- 31.Winkler ME, Morrison DA. Competence beyond genes: filling in the details of the pneumococcal competence transcriptome by a systems approach. J. Bacteriol. 2019;201:e00238–00219. doi: 10.1128/JB.00238-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Echenique JR, Chapuy‐Regaud S, Trombe MC. Competence regulation by oxygen in Streptococcus pneumoniae: involvement of ciaRH and comCDE. Mol. Microbiol. 2000;36:688–696. doi: 10.1046/j.1365-2958.2000.01891.x. [DOI] [PubMed] [Google Scholar]
- 33.Claverys J-P, Prudhomme M, Martin B. Induction of competence regulons as a general response to stress in Gram-positive bacteria. Annu. Rev. Microbiol. 2006;60:451–475. doi: 10.1146/annurev.micro.60.080805.142139. [DOI] [PubMed] [Google Scholar]
- 34.Chen J-D, Morrison DA. Modulation of competence for genetic transformation in Streptococcus pneumoniae. Microbiology. 1987;133:1959–1967. doi: 10.1099/00221287-133-7-1959. [DOI] [PubMed] [Google Scholar]
- 35.Claverys J-P, Havarstein LS. Extracellular-peptide control of competence for genetic transformation in Streptococcus pneumoniae. Front. Biosci. 2002;7:d1798–d1814. doi: 10.2741/claverys. [DOI] [PubMed] [Google Scholar]
- 36.Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys J-P. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science. 2006;313:89–92. doi: 10.1126/science.1127912. [DOI] [PubMed] [Google Scholar]
- 37.Kloosterman TG, van der Kooi-Pol MM, Bijlsma JJ, Kuipers OP. The novel transcriptional regulator SczA mediates protection against Zn2+ stress by activation of the Zn2+-resistance gene czcD in Streptococcus pneumoniae. Mol. Microbiol. 2007;65:1049–1063. doi: 10.1111/j.1365-2958.2007.05849.x. [DOI] [PubMed] [Google Scholar]
- 38.Potter AJ, Trappetti C, Paton JC. Streptococcus pneumoniae uses glutathione to defend against oxidative stress and metal ion toxicity. J. Bacteriol. 2012;194:6248–6254. doi: 10.1128/JB.01393-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okado-Matsumoto A, Fridovich I. The role of α, β-dicarbonyl compounds in the toxicity of short chain sugars. J. Biol. Chem. 2000;275:34853–34857. doi: 10.1074/jbc.M005536200. [DOI] [PubMed] [Google Scholar]
- 40.Ulijasz AT, Falk SP, Weisblum B. Phosphorylation of the RitR DNA‐binding domain by a Ser–Thr phosphokinase: implications for global gene regulation in the streptococci. Mol. Microbiol. 2009;71:382–390. doi: 10.1111/j.1365-2958.2008.06532.x. [DOI] [PubMed] [Google Scholar]
- 41.Ong C-LY, et al. Interplay between manganese and iron in pneumococcal pathogenesis: role of the orphan response regulator RitR. Infect. Immun. 2013;81:421–429. doi: 10.1128/IAI.00805-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Couñago RM, et al. Imperfect coordination chemistry facilitates metal ion release in the Psa permease. Nat. Chem. Biol. 2014;10:35–41. doi: 10.1038/nchembio.1382. [DOI] [PubMed] [Google Scholar]
- 43.Reyes-Caballero H, et al. The metalloregulatory zinc site in Streptococcus pneumoniae AdcR, a zinc-activated MarR family repressor. J. Mol. Biol. 2010;403:197–216. doi: 10.1016/j.jmb.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ogunniyi AD, et al. Pneumococcal histidine triad proteins are regulated by the Zn2+-dependent repressor AdcR and inhibit complement deposition through the recruitment of complement factor H. FASEB J. 2009;23:731–738. doi: 10.1096/fj.08-119537. [DOI] [PubMed] [Google Scholar]
- 45.Tettelin H, et al. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 46.Aprianto R, Slager J, Holsappel S, Veening J-W. High-resolution analysis of the pneumococcal transcriptome under a wide range of infection-relevant conditions. Nucleic Acids Res. 2018;46:9990–10006. doi: 10.1093/nar/gky750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fleming E, Lazinski DW, Camilli A. Carbon catabolite repression by seryl phosphorylated HPr is essential to Streptococcus pneumoniae in carbohydrate‐rich environments. Mol. Microbiol. 2015;97:360–380. doi: 10.1111/mmi.13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carvalho SM, Kloosterman TG, Kuipers OP, Neves AR. CcpA ensures optimal metabolic fitness of Streptococcus pneumoniae. PLoS ONE. 2011;6:e26707. doi: 10.1371/journal.pone.0026707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ogunniyi AD, et al. Central role of manganese in regulation of stress responses, physiology, and metabolism in Streptococcus pneumoniae. J. Bacteriol. 2010;192:4489–4497. doi: 10.1128/JB.00064-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eggleston LV, Krebs HA. Regulation of the pentose phosphate cycle. Biochemical J. 1974;138:425–435. doi: 10.1042/bj1380425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ong C-lY, Walker MJ, McEwan AG. Zinc disrupts central carbon metabolism and capsule biosynthesis in Streptococcus pyogenes. Sci. Rep. 2015;5:10799. doi: 10.1038/srep10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krotkiewska B, Banaś T. Interaction of Zn2+ and Cu2+ ions with glyceraldehyde-3-phosphate dehydrogenase from bovine heart and rabbit muscle. Int. J. Biochem. 1992;24:1501–1505. doi: 10.1016/0020-711X(92)90078-F. [DOI] [PubMed] [Google Scholar]
- 53.Iddar A, Valverde F, Assobhei O, Serrano A, Soukri A. Widespread occurrence of non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase among Gram-positive bacteria. Int. Microbiol. 2005;8:251–258. [PubMed] [Google Scholar]
- 54.Spaans SK, Weusthuis RA, Van Der Oost J, Kengen SW. NADPH-generating systems in bacteria and archaea. Front. Microbiol. 2015;6:742. doi: 10.3389/fmicb.2015.00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bidossi A, et al. A functional genomics approach to establish the complement of carbohydrate transporters in Streptococcus pneumoniae. PloS ONE. 2012;7:e33320. doi: 10.1371/journal.pone.0033320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paixão L, et al. Transcriptional and metabolic effects of glucose on Streptococcus pneumoniae sugar metabolism. Front. Microbiol. 2015;6:1041. doi: 10.3389/fmicb.2015.01041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carvalho SM, et al. Pyruvate oxidase influences the sugar utilization pattern and capsule production in Streptococcus pneumoniae. PLoS ONE. 2013;8:e68277. doi: 10.1371/journal.pone.0068277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paixão L, et al. Host glycan sugar-specific pathways in Streptococcus pneumoniae: galactose as a key sugar in colonisation and infection. PLoS ONE. 2015;10:e0121042. doi: 10.1371/journal.pone.0121042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yesilkaya H, et al. Pyruvate formate lyase is required for pneumococcal fermentative metabolism and virulence. Infect. Immun. 2009;77:5418–5427. doi: 10.1128/IAI.00178-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y-M, Rock CO. Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 2008;6:222. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- 61.Lu YJ, Rock CO. Transcriptional regulation of fatty acid biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 2006;59:551–566. doi: 10.1111/j.1365-2958.2005.04951.x. [DOI] [PubMed] [Google Scholar]
- 62.Nakkaew A, Chotigeat W, Eksomtramage T, Phongdara A. Cloning and expression of a plastid-encoded subunit, beta-carboxyltransferase gene (accD) and a nuclear-encoded subunit, biotin carboxylase of acetyl-CoA carboxylase from oil palm (Elaeis guineensis Jacq.) Plant Sci. 2008;175:497–504. doi: 10.1016/j.plantsci.2008.05.023. [DOI] [Google Scholar]
- 63.Gullett, J. M., Cuypers, M. G., Frank, M. W., White, S. W. & Rock, C. O. A fatty acid binding protein of Streptococcus pneumoniae facilitates the acquisition of host polyunsaturated fatty acids. J. Biol. Chem. jbc. RA119. 010659 (2019). [DOI] [PMC free article] [PubMed]
- 64.Parsons JB, et al. A thioesterase bypasses the requirement for exogenous fatty acids in the plsX deletion of Streptococcus pneumoniae. Mol. Microbiol. 2015;96:28–41. doi: 10.1111/mmi.12916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fujita Y, Matsuoka H, Hirooka K. Regulation of fatty acid metabolism in bacteria. Mol. Microbiol. 2007;66:829–839. doi: 10.1111/j.1365-2958.2007.05947.x. [DOI] [PubMed] [Google Scholar]
- 66.Li S-J, Cronan J. Growth rate regulation of Escherichia coli acetyl coenzyme A carboxylase, which catalyzes the first committed step of lipid biosynthesis. J. Bacteriol. 1993;175:332–340. doi: 10.1128/JB.175.2.332-340.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marini PE, Perez CA, de Mendoza D. Growth-rate regulation of the Bacillus subtilis accBC operon encoding subunits of acetyl-CoA carboxylase, the first enzyme of fatty acid synthesis. Arch. Microbiol. 2001;175:234–237. doi: 10.1007/s002030100256. [DOI] [PubMed] [Google Scholar]
- 68.Mohedano ML, Amblar M, De La Fuente A, Wells JM, López P. The response regulator YycF inhibits expression of the fatty acid biosynthesis repressor FabT in Streptococcus pneumoniae. Front. Microbiol. 2016;7:1326. doi: 10.3389/fmicb.2016.01326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mohedano ML, et al. Evidence that the essential response regulator YycF in Streptococcus pneumoniae modulates expression of fatty acid biosynthesis genes and alters membrane composition. J. Bacteriol. 2005;187:2357–2367. doi: 10.1128/JB.187.7.2357-2367.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bielski B, Arudi RL, Sutherland MW. A study of the reactivity of HO2/O2-with unsaturated fatty acids. J. Biol. Chem. 1983;258:4759–4761. [PubMed] [Google Scholar]
- 71.Pesakhov S, et al. Effect of hydrogen peroxide production and the Fenton reaction on membrane composition of Streptococcus pneumoniae. Biochimica et. Biophysica Acta. 2007;1768:590–597. doi: 10.1016/j.bbamem.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 72.Benisty R, Cohen AY, Feldman A, Cohen Z, Porat N. Endogenous H2O2 produced by Streptococcus pneumoniae controls FabF activity. Biochimica et. Biophysica Acta. 2010;1801:1098–1104. doi: 10.1016/j.bbalip.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 73.Shah K, Kumar RG, Verma S, Dubey R. Effect of cadmium on lipid peroxidation, superoxide anion generation and activities of antioxidant enzymes in growing rice seedlings. Plant Sci. 2001;161:1135–1144. doi: 10.1016/S0168-9452(01)00517-9. [DOI] [Google Scholar]
- 74.Moritz B, Striegel K, de Graaf AA, Sahm H. Kinetic properties of the glucose‐6‐phosphate and 6 phosphogluconate dehydrogenases from Corynebacterium glutamicum and their application for predicting pentose phosphate pathway flux in vivo. Eur. J. Biochem. 2000;267:3442–3452. doi: 10.1046/j.1432-1327.2000.01354.x. [DOI] [PubMed] [Google Scholar]
- 75.Hoskins J, et al. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 2001;183:5709–5717. doi: 10.1128/JB.183.19.5709-5717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lanie JA, et al. Genome sequence of Avery’s virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 2007;189:38–51. doi: 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Slager J, Aprianto R, Veening J-W. Deep genome annotation of the opportunistic human pathogen Streptococcus pneumoniae D39. Nucleic Acids Res. 2018;46:9971–9989. doi: 10.1093/nar/gky725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kirtley ME, McKay M. Fructose-1,6-bisphosphate, a regulator of metabolism. Mol. Cell. Biochem. 1977;18:141–149. doi: 10.1007/BF00280279. [DOI] [PubMed] [Google Scholar]
- 79.Bridges RB, Palumbo MP, Wittenberger C. Purification and properties of an NADP-specific 6-phosphogluconate dehydrogenase from Streptococcus faecalis. J. Biol. Chem. 1975;250:6093–6100. [PubMed] [Google Scholar]
- 80.Wittenberger CL, Angelo N. Purification and properties of a fructose-1, 6-diphosphate-activated lactate dehydrogenase from Streptococcus faecalis. J. Bacteriol. 1970;101:717–724. doi: 10.1128/JB.101.3.717-724.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Iyer R, Baliga NS, Camilli A. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 2005;187:8340–8349. doi: 10.1128/JB.187.24.8340-8349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Choi S-B, Lew L-C, Hor K-C, Liong M-T. Fe2+ and Cu2+ increase the production of hyaluronic acid by Lactobacilli via affecting different stages of the pentose phosphate pathway. Appl. Biochem. Biotechnol. 2014;173:129–142. doi: 10.1007/s12010-014-0822-5. [DOI] [PubMed] [Google Scholar]
- 83.Lacks S, Hotchkiss RD. A study of the genetic material determining an enzyme activity in pneumococcus. Biochimica et. Biophysica Acta. 1960;39:508–518. doi: 10.1016/0006-3002(60)90205-5. [DOI] [PubMed] [Google Scholar]
- 84.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:1. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bluemlein K, Ralser M. Monitoring protein expression in whole-cell extracts by targeted label-and standard-free LC-MS/MS. Nat. Protoc. 2011;6:859–869. doi: 10.1038/nprot.2011.333. [DOI] [PubMed] [Google Scholar]
- 90.Carr S, et al. The need for guidelines in publication of peptide and protein identification data: Working Group on Publication Guidelines for Peptide and Protein Identification Data. Mol. Cell. Proteom. 2004;3:531–533. doi: 10.1074/mcp.T400006-MCP200. [DOI] [PubMed] [Google Scholar]
- 91.Morona JK, Morona R, Paton JC. Attachment of capsular polysaccharide to the cell wall of Streptococcus pneumoniae type 2 is required for invasive disease. Proc. Natl Acad. Sci. USA. 2006;103:8505–8510. doi: 10.1073/pnas.0602148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Blumenkrantz N, Asboe-Hansen G. New method for quantitative determination of uronic acids. Anal. Biochem. 1973;54:484–489. doi: 10.1016/0003-2697(73)90377-1. [DOI] [PubMed] [Google Scholar]
- 93.Eijkelkamp BA, et al. Arachidonic acid stress impacts pneumococcal fatty acid homeostasis. Front. Microbiol. 2018;9:813. doi: 10.3389/fmicb.2018.00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Perez-Riverol Y, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
RNA sequencing data have been deposited in NCBI Gene Expression Omnibus databank under submission identifier GSE141681. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE94 partner repository with the dataset identifier PXD021643 and 10.6019/PXD021643. Raw data files for all other Figures are available from the corresponding authors upon reasonable request.