Abstract
Brassica oleracea var. acephala (kale) is a cruciferous vegetable widely cultivated for its leaves and flower buds in Atlantic Europe and the Mediterranean area, being a food of great interest as a "superfood" today. Little has been studied about the diversity of endophytic fungi in the Brassica genus, and there are no studies regarding kale. In this study, we made a survey of the diversity of endophytic fungi present in the roots of six different Galician kale local populations. In addition, we investigated whether the presence of endophytes in the roots was beneficial to the plants in terms of growth, cold tolerance, or resistance to bacteria and insects. The fungal isolates obtained belonged to 33 different taxa. Among those, a Fusarium sp. and Pleosporales sp. A between Setophoma and Edenia (called as Setophoma/Edenia) were present in many plants of all five local populations, being possible components of a core kale microbiome. For the first time, several interactions between endophytic fungus and Brassica plants are described and is proved how different interactions are beneficial for the plant. Fusarium sp. and Pleosporales sp. B close to Pyrenophora (called as Pyrenophora) promoted plant growth and increased cold tolerance. On the other hand, isolates of Trichoderma sp., Pleosporales sp. C close to Phialocephala (called as Phialocephala), Fusarium sp., Curvularia sp., Setophoma/Edenia and Acrocalymma sp. were able to activate plant systemic resistance against the bacterial pathogen Xanthomonas campestris. We also observed that Fusarium sp., Curvularia sp. and Setophoma/Edenia confered resistance against Mamestra brassicae larvae.
Subject terms: Microbiology, Applied microbiology, Microbial communities, Fungi, Plant sciences, Plant stress responses, Plant symbiosis
Introduction
Brassica crops represent one of the 10 most economically important vegetables in world agricultural and food markets. The most important crop species in this genus are Brassica oleracea (i.e. cauliflower, broccoli, Brussels sprouts, kale, cabbage, …), Brassica napus (i.e. rapeseed and leaf rape), and Brassica rapa (i.e. turnip, Chinese cabbage, pak choi…), being mainly cultivated in temperate regions of the Northern Hemisphere1. Kale, B. oleracea var. acephala, is a leafy vegetable which has gained a great popularity as a “superfood” in recent years, due to its anticancerogenic and antioxidant potential associated with the presence of various compounds from the polyphenol, glucosinolate, terpenoid or carotenoid group, and contents of Ca, folate, riboflavin, vitamin C, K and A2. Furthermore, kale is an important vegetable crop in Iberian Peninsula traditional farming systems, grown for their leaves and flower buds3.
The role of microbes in determining the health of soils and plants is increasingly acknowledged. Plants harbour a diversity of microorganisms that may engage in a continuum of interactions ranging from beneficial to adverse interactions. Some of these interactions may be transient and occur during a specific life stage of the plants regardless of its beneficial, detrimental or neutral impact4. Endophytes are microbes that can be isolated from asymptomatic plant tissue, including neutral, commensal and/or beneficial microorganisms as well as dormant saprobes and latent pathogens5. Some fungal endophytes are well known to contribute to plant fitness, improving the host adaptation to biotic and abiotic stress conditions6. For instance, some endophytes are able to reduce the damage of plant pathogens thanks to antagonism via hyperparasitism, competition or antibiosis, or by means of the activation of plant defences7,8; others act as entomopathogens (e.g. Beauveria bassiana or Metarhizium anisopliae), protecting host plants against herbivore pests by direct exposure or through the production of insecticidal compounds9. Moreover, endophytic fungi are able to increase the plant’s tolerance against abiotic stress factors such as drought, salinity or high temperature through the activation of host stress-responses, allowing the plants to avoid or mitigate the impact of the stress10.
As far as the Brassica genus is concerned, there are some studies on the diversity of endophytic fungi and their possible role in improving plant productivity11, although none has been done in kale. The inoculation of B. napus with Piriformospora indica promoted plant growth, seed yield and quality12. Several works focused on the impact of endophytic fungi in the resistance against biotic stresses. Aspergillus flavipes, Chaetomium globosum, Clonostachys rosea and Leptosphaeria biglobosa isolated from B. napus suppressed leaf blight of oilseed rape caused by Sclerotina sclerotiorum13. Aspergillus capensis had antifungal activity against the plant pathogenic fungi Botrytis cinerea, Monilinia fructicola, Sclerotinia trifoliorum and S. sclerotiorum14. Moreover, the inoculation of B. napus with a Metarhizium anisopliae endophyte provided the plants with greater resistance against Plutella xylostella larvae15.
Compared to other plant species, it is considered that the diversity of endophytic fungi present in brassicas is lower than that of other taxonomic groups, probably because of the presence of secondary metabolites derived from tryptophan and other aminoacids, known as glucosinolates16. Despite this, the use of endophytic fungi could have an impact on cultivation and the improvement of Brassica-crops within a more environmentally friendly agriculture.
On the other hand, the diversity of endophytic fungi associated with different species within the same genus can be abysmal, and even within the varieties of the same species, as already observed in Uncaria gambier gambir udang and gambir nasi varieties17, or in Rosa multiflora and var. carnea18.
At present, there are no diversity studies of endophytic fungi present in the roots of B. oleracea and very few in other species of the genus. Therefore, the main objective of this study was to estimate the diversity of root endophytes associated with different accessions of kale. Secondly, we evaluated the biological effect of some dominant endophytes in the promotion of plant growth, tolerance against cold, and resistance against pests and diseases.
Results
Fungal diversity
From the 900 root-fragments (180 per B. oleracea accession), 376 fungal isolates were obtained, at a rate of 54–98 fungal isolates per accession. Isolates were obtained from 41.77% of the root fragments plated. All sampled plants harbored fungi in their roots, and on average, 13 isolates were obtained from the roots of each plant.
The isolates were grouped into morphotypes, obtaining 27 morphotypes for MBG-BRS0106, 27 for MBG-BRS0292, 45 for MBG-BRS0426, 33 for MBG-BRS0446 and 21 for MBG-BRS0468. After sequencing one or more isolates of each morphotype, 179 ITS1-5.8S-ITS2 nucleotide sequences were obtained. After those sequences were clustered, considering that those being 97% or more identical belonged to the same taxon, 33 different fungal taxa were identified (Table 1). A phylogenetic tree made with the sequences of these 33 taxa helped to accommodate into taxonomic orders isolates whose sequences were less than 95% identical to those of a type strain (Supplementary Fig S1).
Table 1.
Core and abundant fungal species isolated from surface sterilized roots of B. oleracea (kale) from 5 different accessions.
| Type strain with closest sequence identity | Identity to closet match (%) | Proposed taxon | ITS sequence accession number | Order | Incidence in plants (%) | Number of B. oleracea accesions |
|---|---|---|---|---|---|---|
| Fusarium foetens | 99.3 | Fusarium sp. | MT628384 | Hypocreales | 63.3 | 5 |
| Edenia gomezpompae | 92.5 | Pleosporales sp. A | MT628351 | Pleosporales | 56.7 | 5 |
| Acrocalymma fici | 95.9 | Acrocalymma sp. | MT626728 | Pleosporales | 46.7 | 3 |
| Alternaria destruens | 100 | Alternaria sp. A | MT628452 | Pleosporales | 20.0 | 3 |
| Pyrenophora nisikadoi | 94.7 | Pleosporales sp. B | MT628399 | Pleosporales | 13.3 | 3 |
| Alternaria mimicula | 100 | Alternaria sp. B | MT628543 | Pleosporales | 13.3 | 3 |
| Barrenia panicia | 98.5 | Barrenia sp. | MT636549 | Helotiales | 13.3 | 3 |
| Ceratobasidium ramicola | 93.1 | Basidiomycota A | MT629733 | unknown | 10.0 | 1 |
| Phialocephala hiberna | 94 | Helotiales sp. | MT628664 | Helotiales | 6.7 | 2 |
| Polyschema sclerotigenum | 98.4 | Polyschema sp. | MT628702 | Pleosporales | 6.7 | 2 |
| Mucor moelleri | 99.1 | Mucor sp. A | MT639934 | Mucorales | 6.7 | 1 |
| Curvularia coatesiae | 99.6 | Curvularia sp. | MT640053 | Pleosporales | 6.7 | 1 |
| Cladosporium spp. | 99.8 | Cladosporium sp. | MT641243 | Capnodiales | 6.7 | 2 |
| Phomopsis tuberivora | 99.4 | Diaporthe sp | MT636064 | Diaporthales | 6.7 | 1 |
| Phoma schachtii | 99.2 | Phoma sp. | MT628903 | Pleosporales | 3.3 | 1 |
| Rhizopus oryzae | 99.8 | Rhizopus sp. | MT635401 | Mucorales | 3.3 | 1 |
| Mucor hiemalis | 96.2 | Mucor sp. B | MT636070 | Mucorales | 3.3 | 1 |
| Penicillium cremeogriseum | 99.8 | Penicillium sp. | MT636161 | Eurotiales | 3.3 | 1 |
| Chaetomium novozelandicum | 99.8 | Chaetomium sp. | MT641231 | Sordariales | 3.3 | 1 |
| Trichoderma hamatum | 99.8 | Trichoderma sp. | MT641233 | Hypocreales | 3.3 | 1 |
| Codinaea acaciae | 98.1 | Codinaea sp. | MT640043 | Chaetosphaeriales | 3.3 | 1 |
| Dendryphion europaeum | 96 | Dendryphion sp. | MT641239 | Pleosporales | 3.3 | 1 |
| Minutisphaera aspera | 99.4 | Minutisphaera sp | MT636088 | Minutisphaerales | 3.3 | 1 |
| Ceratobasidium papillatum | 90.7 | Basidiomycota B | MT640104 | unknown | 3.3 | 1 |
| Ceratobasidium ramicola | 91.1 | Basidiomycota C | MT636101 | unknown | 3.3 | 1 |
| Phragmocephala garethjonesii | 92.8 | Pleosporales sp. C | MT641235 | Pleosporales | 3.3 | 1 |
| Trametes versicolor | 99.4 | Trametes sp. | MT635595 | Polyporales | 3.3 | 1 |
| Aaosphaeria arxii | 99 | Aaosphaeria sp. | MT645080 | Pleosporales | 3.3 | 1 |
| Mycofalcella calcarata | 96.9 | Mycofalcella sp. | MT636550 | Helotiales | 3.3 | 1 |
| Aspergillus aureolus | 99.6 | Aspergillus sp. B | MT639933 | Eurotiales | 3.3 | 1 |
| Aspergillus spp. | 98.4 | Aspergillus sp. A | MT641240 | Eurotiales | 3.3 | 1 |
| Tetraploa sasicola | 95.6 | Tetraploa sp. | MT641232 | Pleosporales | 3.3 | 1 |
| Plectosphaerella niemeijerarum | 99.4 | Plectosphaerella sp. | MT641266 | Glomerellales | 3.3 | 1 |
A core microbiome is defined as the group of microbes commonly found within a host's microbiome, using persistence of the association as the criterion to select microbes potentially providing critical function within the habitat in which they are found67.
The most prevalent taxa were a Fusarium sp. found in 63.3% of the plants collected, and Pleosporales sp. A, found in 56.7%, both were present in all B. oleracea accessions (Table 1). Thanks to the creation of the phylogenetic tree and the comparison of the representative sequence of the proposed taxon Pleosporales sp. A, we have determined how this sequence represents a species not yet described, between the genus Edenia and Setophoma (called from now Setophoma/Edenia). The distribution of the fungal taxa showed few commonalities among the kale accessions (Fig. 1). MBG-BRS0426 and MBG-BRS0468 have 5 fungal taxa in common, 2 of which were not shared with any other accession. Several taxa occurred in only one accession: 10 taxa in MBG-BRS0426, 5 taxa in MBG-BRS0106, 4 taxa in MBG-BRS0292, 4 taxa in MBG-BRS0446, and 3 taxa in MBG-BRS0446. The similarity analysis based on the Jaccard index (JI) (Table 2), shows that only the accessions MBG-BRS0292 and MBG-BRS0446 present a similarity between their fungal taxa greater than 0.3, while above 0.25 we find MBG-BRS0292 and MBG-BRS0468, MBG-BRS0426 and MBG-BRS0468, and MBG-BRS0446 and MBG-BRS0106.
Figure 1.
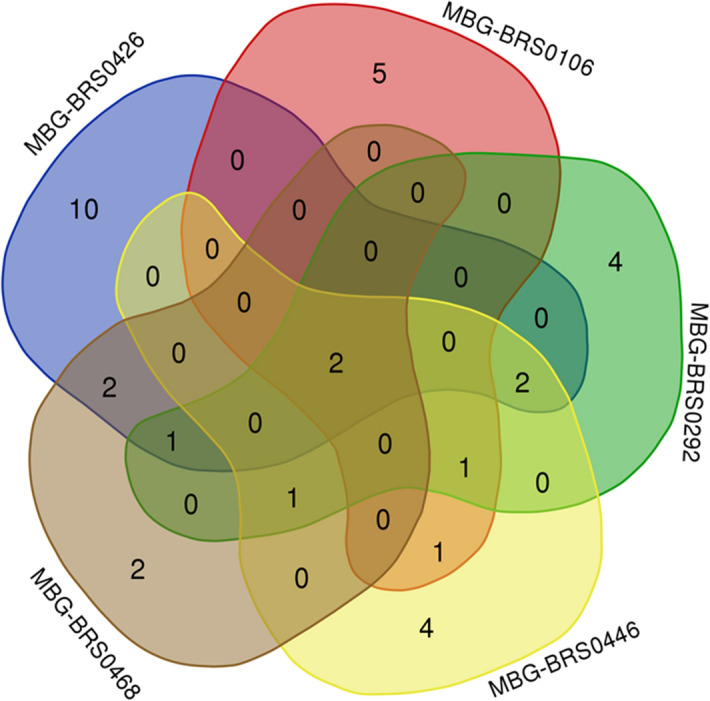
Venn diagram showing the distribution of fungal taxa isolated from the different accessions of B. oleracea (kale).
Table 2.
Jaccard index of similarity (bold) and total number of fungal taxons identified in roots of each pair of B. oleracea (kale) accessions.
| B. oleracea accession | MBG-BRS0426 | MBG-BRS0106 | MBG-BRS0292 | MBG-BRS0446 | MBG-BRS0468 |
|---|---|---|---|---|---|
| MBG-BRS0426 | 1.000 | 0.083 | 0.200 | 0.083 | 0.250 |
| MBG-BRS0106 | 24 | 1.000 | 0.176 | 0.250 | 0.133 |
| MBG-BRS0292 | 25 | 17 | 1.000 | 0.375 | 0.267 |
| MBG-BRS0446 | 24 | 16 | 16 | 1.000 | 0.188 |
| MBG-BRS0468 | 20 | 15 | 15 | 16 | 1.000 |
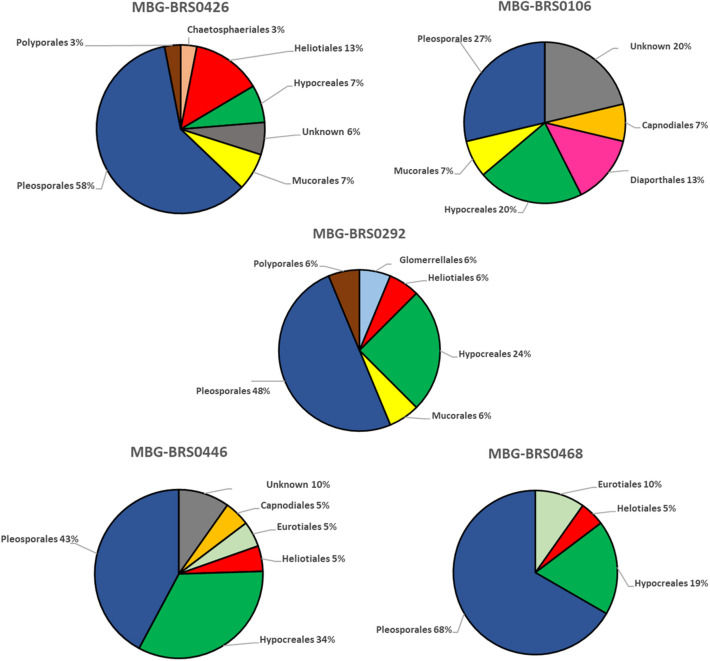
Pleosporales is the most representative order in terms of the number of taxa (32.3%) (Table 1). Most taxa are present only in one of the accessions (58.8%), with only 5.9% of the taxa present in all (Table 1). Taking into account the incidence of each order in each variety (Fig. 2), it can be seen that the most abundant order is Pleosporales (27–68%), followed by Hypocreales (7–34%), both present in all varieties. Helotiales is present in all accessions, except in MBG-BRS0106, where we find Diaporthales as an specific order. On the other hand, the distribution of taxa according to their incidence can be visualized in the rank-abundance curve shown in Fig. 3. Three taxa were present in more than 45% of the plants (Fusarium sp. [63.3%], Setophoma/Edenia [56.7%] and Acrocalymma sp.[46.7%]) while the remaining taxa were present in less than 15% of the plants (Table 1).
Figure 2.
Distribution of fungal taxa from roots of different accessions of B. oleracea (kale) according to orders.
Figure 3.
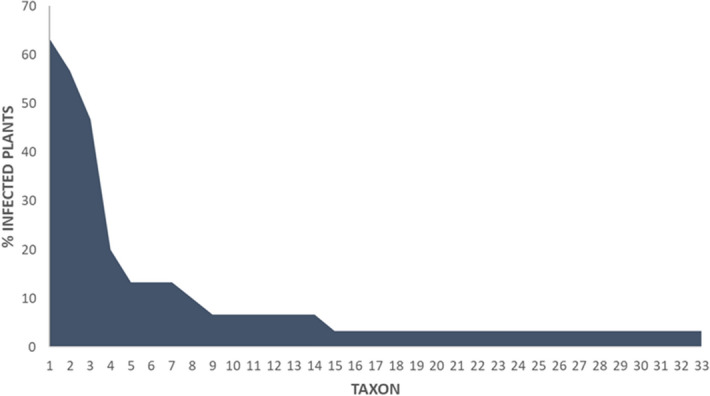
Rank-abundance plot showing the incidence in plants of each taxon identified in roots of B. oleracea (kale).
Increased plant growth, tolerance to cold and resistance to biotic stresses
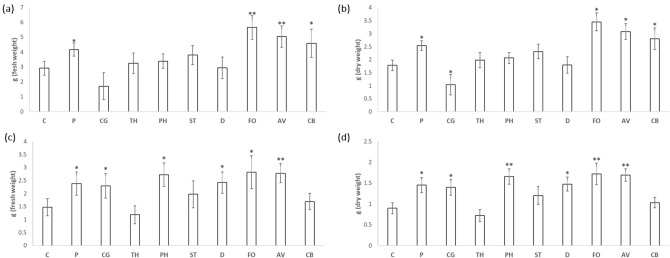
Nine fungal isolates were used to study beneficial effects on kale. Significant differences in the growth of the aerial part of kale were observed with the application of Fusarium sp. (5.66 g) and Acrocalymma sp. (4.83 g), being their weight almost double that observed for the control plants without inoculation (2.92 g). Growth of plants inoculated with Pleosporales sp. B next to Pyrenophora (called from now Pyrenophora) and Curvularia sp. was also significantly higher than that of the control plants. Likewise, although there was no significant difference respect to the control, we observed a reduction in the weight of the plants inoculated with Chaetomium sp. (1.71 g) (Fig. 4a); as well as dry weight (Fig. 4b).
Figure 4.
Mean of fresh (a,c) and dry weight (b,d) of kale plants grown in greenhouse (a,b) and cold (c,d) conditions. Plants without inoculation (C) and inoculated with unknown Pyrenophora (P), Chaetomium sp. (CG), Trichoderma sp. (TH), Phialocephala (PH), Setophoma/Edenia (ST), Diaporthe sp. (D), Fusarium sp. (FO), Acrocalymma sp. (AV) and Curvularia sp. (CB) were collected at 12-week-old and measured their fresh weight of the aerial part. Data are the mean of 25 plants for each inoculation with the corresponding standard deviation. Student’s t-test was performed. Asterisks denote significant differences at P ≤ 0.05 (*) and P ≤ 0.01 (**).
Under cold conditions (constant 12 °C) is observed how inoculation with Pyrenophora, Chaetomium sp., Helotiales sp.next to Phialocephala (called from now Phialocephala), Diaporthe sp., Fusarium sp. and Acrocalymma sp. caused a significant increase in growth, and therefore, of tolerance to abiotic stress. Inoculation with Phialocephala, Fusarium sp. and Acrocalymma sp. increased the plant weight almost twice (2.72 g, 2.82 g, and 2.78, respectively) respect to uninoculated control plants (1.47 g) (Fig. 4c); as in dry weight (Fig. 4d).
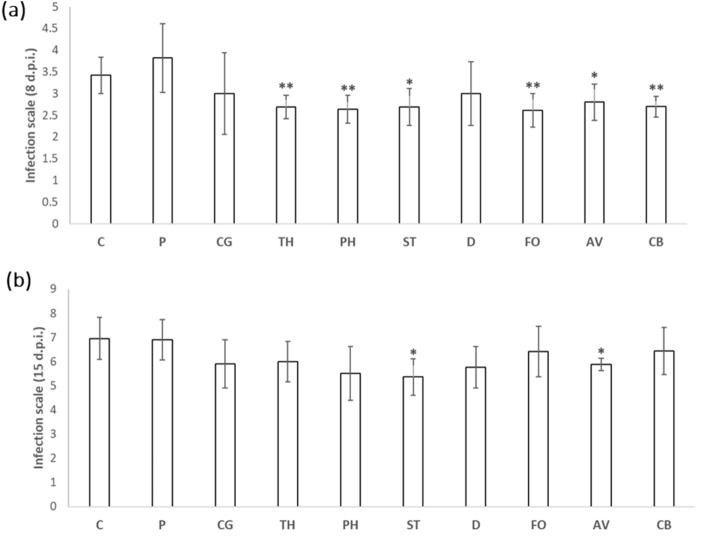
Regarding the activation of systemic resistance against biotic stressors, leaves of B. oleracea were inoculated with the pathogenic Xcc bacteria (Fig. 5). We observed a significant decrease in the damage caused by the bacteria at 8 d.p.i (Fig. 5a) in the plants inoculated with Trichoderma sp., Phialocephala, Fusarium sp., Curvularia sp., Setophoma/Edenia and Acrocalymma sp., being highly significant (P < 0.01) with the inoculation of the first four fungi indicated. On the other hand, at 15 d.p.i. (Fig. 5b) we only obtained a significant reduction in the incidence of the disease in the plants with the inoculation with the Setophoma/Edenia and Acrocalymma sp. strains.
Figure 5.
Effect of Xcc infection on kale plants inoculated with endophytic fungi. Plants without inoculation (C) and inoculated with unknown Pyrenophora (P), Chaetomium sp. (CG), Trichoderma sp. (TH), Phialocephala (PH), Setophoma/Edenia (ST), Diaporthe sp. (D), Fusarium sp. (FO), Acrocalymma sp. (AV) and Curvularia sp. (CB) were infected at 5–6 leaves stage and the leaves with lesions are classified according to an infection range from 1 to 5. Data are the mean 10 plants for each inoculation with the corresponding standard deviation. Student’s t-test was performed. Asterisks denote significant differences at P ≤ 0.05 (*) and P ≤ 0.01 (**).
Regarding the possible activation of systemic resistance in the inoculated plants against the insect pest Mb, the inoculation of kale with the fungi Setophoma/Edenia (DI 2.3) and Fusarium sp. (DI 2.2) strains supposed a significant decrease in the damage index, compared to the control plants without inoculation (DI 3.1). Furthermore, although no significant differences were observed with respect to the leaf area consumed by the larvae, both Setophoma/Edenia and Fusarium sp. had the lowest average leaf area consumed (Fig. 6).
Figure 6.
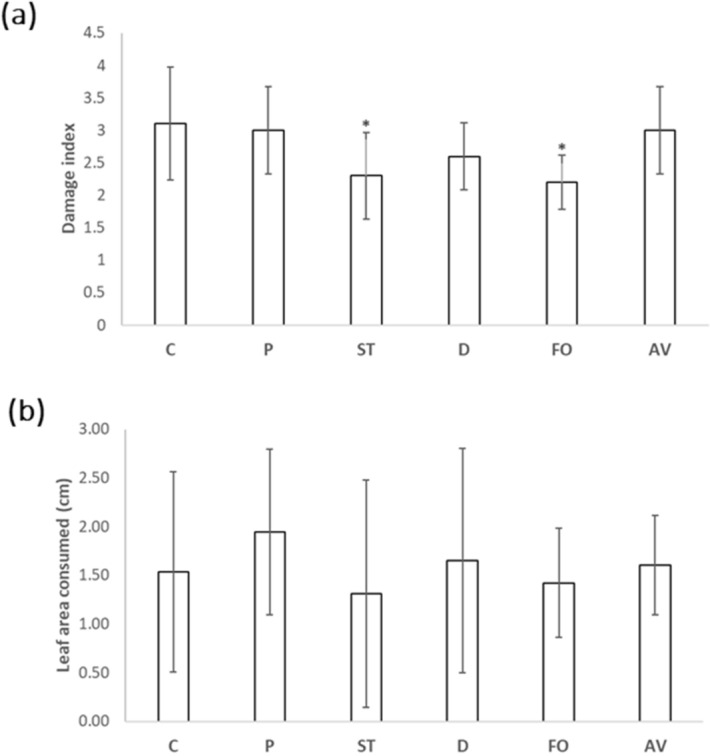
Effect of infestation with M. brassicae on kale plants inoculated with endophytic fungi. Plants without inoculation (C) and inoculated with Pyrenophora (P), Setophoma/Edenia (ST), Diaporthe sp. (D), Fusarium sp. (FO) and Acrocalymma sp. (AV) were attacked at 12-week-old, obtaining 5 d.p.a. the damage index in a range from 1 to 5 and the leaf area consumed by the larvae. Data are the mean of 10 plants for each inoculation with the corresponding standard deviation, and for each biological replicate and condition, three leaves/plants were used. Student’s t-test was performed. Asterisks denote significant differences at P ≤ 0.05 (*).
Discussion
Few studies have investigated fungal diversity in varieties of the same plant species within a study site. In other words, whether genotypic variation affects the structure of the root microbiome. Regarding the B. oleracea accessions of our study, we have reported a possible specificity of accession. Most of the determined fungal taxa are only present in one accession and the similarity indices give great differences despite the fact that all the plants are in the same geographical plot. These differences could be due to differences in the root exudates produced by each kale accession, which could affect the ability of endophytic fungi to colonize the roots, just as it has been observed in different tomato genotypes and their rhizospheric microbiota19. Despite this, it is very difficult to consider the existence of such specificity in a study like this, since the possibility of detecting those fungi with low prevalence is more difficult than those that are present to a greater extent, also the great differences existing at the level of alpha (Table 1, Fig. 2) and beta (Table 2) microbial diversity. On the other hand, the presence in all accessions of Fusarium sp. and Setophoma/Edenia, suggests that those two taxa could be components of a core microbiome of kale. This could be confirmed by surveying kale root endophytes at different locations.
The diversity of endophytic fungi present in the roots of plants of the Brassica genus has been scarcely studied so far, as has the diversity of fungi between different varieties of the same species. In our study, most isolated fungi belonged to the Pleosporales order. To our knowledge, only one species of the Pleosporales order, Leptosphaeria biglobosa, has been found in B. napus13. Besides, order Hypocreales was found in all accessions. This order is more common in Brassicaceae, and taxa such as Fusarium sp.20, C. rosea13 and M. anisopliae15 have been reported in B. napus. Mucorales order was found in 3 of the 5 accessions, having previously being reported Mucor sp. in B. campestris21 and B. chinensis22. Sordariales order was only found in MBG-BRS0292 and MBG-BRS0468 accessions, having previously being described the fungus Chaetomium globosum in B. napus roots13. And Eurotiales order was only found in the MBG-BRS0446 accession, having previously reported the fungus Penicillium sp.20, A. flavipes13 and A. capensis14 in B. napus roots. Therefore, our study reports the first description of the Helotiales, Capnodiales, Diaporthales, Chaetosphaeriales and Minutisphaerales orders in roots of a plant of the Brassica genus; Heliotales order was widely present in 4 of the 5 accessions.
In our study carried out with different accessions of B. oleracea (kale), we have been able to determine the presence of two dominant taxa in all the accessions examined (Fusarium sp. and Setophoma/Edenia). Species of the genus Fusarium, such as F. oxysporum can be neutral, beneficial, or detrimental for host plants. Fusarium wilt is one of the most devastating diseases in agriculture, since some strains can also cause vascular wilts resulting in serious yield losses in affected crops. Despite this, numerous strains of F. oxysporum behave as endophytes capable of activating plant systemic resistance against the pathogenic strains of the fungus23, or produce protective secondary metabolites24, like other species such as F. tricinctum25. Nevertheless, all the existing studies with F. oxysporum f. sp. conglutinans describe it as a pathogen of the Brassica genus, for example, studying the transcriptome profiles in different lines of B. rapa after its inoculation26, or transcriptomic and proteomic studies in B. oleracea27,28. So, this is the first study that describes Fusarium as an endophyte of brassica roots, although Fusarium oxysporum and other species of the genus have been described as dominant components of the microbiome of several plant species29.
As regards the taxa Setophoma/Edenia, there is information about pathogenic and endophytic species in both genus. Setophoma includes disease-causing pathogenic fungal species such as leaf spot in Camellia sinensis (S. antiqua, S. longinqua, S. yingyisheniae and S. yunnanensis)30, but mainly to root level. S. terrestris (formerly called Phoma terrestris) causes pink root rot on members of the Allium genus and other crops, such as tomato, eggplant, pepper, maize and carrot. In 2017, the first case of pink root rot in the Brassica genus was described in B. napus31, being the only report of S. terrestris-Brassica interaction described so far. On the other hand, S. terrestris has been described as an endophyte of other plant species such as Gloriosa superba32 and horseradish (Armoracia rusticana), where is able to decompose various glucosinolates33. In turn, Edenia is a well-known genus of endophytic fungi that produce compounds of high medical interest such as anti-inflammatories34 or antileshmanials35. Furthermore, the E. gomezpompae species is capable of producing highly toxic compounds for plants, which are used as natural herbicides36. Both within the Setophoma genus and the Edenia genus, our isolate represents the first endophyte identified in roots of Brassica plants.
An Acrocalymma sp. was present in 3 of the 5 accessions studied. A. medicaginis has been described as a causal agent of crown and root rot in Medicago plants37. However, A. vagum is a dark septate endophyte able to decrease heavy metal content in tobacco38 and to promote plant growth in Glycyrrhiza uralensis39, but never before reported in Brassica plants.
Therefore, in the present study we have been able to identify several taxa of endophytic fungi in kale roots, which had not been described so far as endophytic fungi (although in some cases as pathogens) within the Brassica genus.
Once the different endophytic fungi in the kale roots were identified, we made a selection of them to determine their possible biological role in the plant. To do this, we inoculated kale plants and tried to determine if the endophytes were capable of promoting plant growth, increasing cold tolerance, or inducing systemic plant resistance against pathogens and/or pests.
The ability of different species of endophytes to promote plant growth as well as tolerance to abiotic stresses such as cold, or resistance to pathogens has been described40. In our study, we have observed that the Pyrenophora strain was capable of promoting the growth of the plant and its cold tolerance. This would be the first time that a benefit of the Pyrenophora-plant interaction has been described, this genus is best known for pathogens, such as Pyrenophora teres, causal agent of net blotch of barley41. The beneficial role observed in B. oleracea by Pyrenophora could be due to the defensive capacity of cruciferous plants through secondary metabolites such as glucosinolates, not present in other plant groups, which is why a possible pathogen in a species may be a harmless symbiont in Brassica. Moreover, inoculation with Fusarium sp. in B. oleracea also increased its growth and cold tolerance, more significantly than with Pyrenophora. Although there are numerous studies that demonstrate the ability of non-pathogenic strains of Fusarium to increase the resistance of plants against various pathogens, such as F. oxysporum23, their role as a possible promoter of plant growth or cold tolerance has not been studied.
The interaction B. oleracea-Acrocalymma sp. resulted in a significant increase in plant growth, something previously observed in other plants such as Glycyrrhiza uralensis39, Medicago sativa and Ammopiptanthus mongolicus42 in interaction with A. vagum. Similar results have been reported in B. oleracea-Curvularia sp. interaction, fungal genus that, although it includes important plant pathogens43, also has species classified as endophytes that promote plant growth, such as C. geniculata in Parthenium hysterophorus through phosphate solubilization and phytohormone production44.
Regarding the increase in cold tolerance, we have observed that the B. oleracea-Chaetomium sp. interaction results in a significant benefit for the plant. In this sense, several authors have described different mechanisms such as heat shock proteins or antioxidant compounds that, for example, the endophyte C. globosum has to tolerate cold, some of which may also be beneficial for the plant thanks to the interaction45–47. The interaction of B. oleracea with Phialocephala also produced an increase in plant tolerance to cold, as has been verified in different subarctic herbaceous plants by Phialocephala fortinii48. Also, we have observed the capacity of Diaporthe sp. and Acrocalymma sp. to increase the tolerance of B. oleracea to cold, being the first time, even within their fungal genera, that these beneficial capacities for plants have been described.
After infection with Xcc, we quantified a decrease in the harmful effect of the bacteria on B. oleracea plants preinoculated with Trichoderma sp. The ability of different Trichoderma species to activate plant systemic resistance against different pathogens has been widely proven in various plant species49, including Brassica plants as B. napus with T. harzianum50,51 or T. viride52, B. rapa with T. pseudokoningii53, and even B. oleracea var. capitate with T. harzianum against Rhizoctonia solani54.
As far as the Fusarium sp. strain is concerned, the ability of F. oxysporum endophytic strains to induce systemic plant resistance has been described in zucchini plants (Cucurbita pepo) against the insect Aphis gossypii55, in banana plants (Musa spp.) against the nematode Radopholus similis56, in Asparagus officinalis against the pathogen F. oxysporum f. sp. asparagi57, and in tomato plants against F. oxysporum f. sp. lycopersici58. While in the case of inoculation with Phialocephala, Curvularia sp., Setophoma/Edenia and Acrocalymma sp., the data obtained represents the first report of activation of plant systemic resistance by these fungi as root endophytes.
As conclusions, the diversity of endophytic fungi found in the kale roots of various accessions is lesser compared to non-brassicaceae species, where up to 49 fungal taxa can be found per plant59. In addition, several of the isolated fungal taxa have been widely described as pathogens of different crops, but never as endophytes, as species of the genus Pyrenophora, an aspect that occurs in kale probably due to their powerful defensive capacity through the use of glucosinolates against fungi60. On the other hand, we find taxa present in several of the accessions and even in all of them, as is the case of Fusarium sp. and Setophoma/Edenia, representing a possible core microbiome for kale. It is precisely these taxa that have shown in our preliminary studies a greater capacity to promote the productivity of kale, by promoting its growth, increasing its tolerance to cold and increasing its defensive capacity against pathogens and pests.
Materials and methods
Plant material, crop and sampling
Five kale accessions were used in this study. These are local populations from Galicia (Northwestern Spain), kept in the Brassica Germplasm Collection of the Misión Biológica de Galicia (MBG-CSIC). The codes for these accessions are MBG-BRS0106, MBG-BRS0292, MBG-BRS0426, MBG-BRS0446 and MBG-BRS0468.
The study was conducted in 2016 at MBG-CSIC (Galicia, NW Spain). Two hundred seeds of each accession were sown in trays in May 2016, and kept in a greenhouse under environmental conditions. After 50 days, when most seedlings were at the 4–5 leaf stage, one hundred and fifty plants of each accession were transplanted into an experimental plot randomly distributed. The distance was 0.8 m between rows and 0.6 m between plants. The experimental plot was established in a slightly acidic soil (pH close to 5.5), organic matter content of 6.3%, 100 ppm available P, 335 ppm assimilable K, 133 ppm changeable Mg, and 7.83 cmol/kg Ca2+. As management practices, no fungicide was applied, or any other pesticide, or any form of fertilization. To prevent the development of weeds, the soil was covered with a mesh that prevents their presence in the plot.
Sampling was carried out in 30-week-old plants, randomly choosing 6 plants per accession. After digging out the entire root system, samples of the roots were collected, and stored in cold for a few hours until processing in the laboratory.
Isolation of fungi
A subsample of 30 root fragments of 4–5 cm per plant was collected. Disinfection of roots and fungal isolation was done following the procedure described by Ref.29. Each root sample was washed with tap water and then surface-disinfected with a solution of 20% commercial bleach (1% active chlorine) containing 0.02% Tween 80 (v:v) for 6 min, followed by treatment with an aqueous solution of 70% ethanol for 30 s. Finally, the roots were rinsed with sterile water and cut into pieces about 5 cm long. Thirty root pieces of each sample were plated in three Petri plates (10 pieces/plate) with potato dextrose agar (PDA) (Sigma-Aldrich, St. Louis, USA) containing 200 mg/L of chloramphenicol (the antibiotic was used to avoid the isolation of endophytic bacteria), and kept in the dark at room temperature. As mycelium emerged from a root fragment into the agar, a small piece of the mycelium from the leading edge of the colony was transferred to a new PDA plate and maintained at room temperature. The root fragment and remaining mycelium were taken out of the original plate to avoid overgrowth. The plates with root samples were checked daily for the presence of fungi for about 4 weeks.
Identification of fungi and diversity analysis
The fungal isolates obtained from roots were first grouped into different morphotypes according to morphological characteristics such as colony color, exudate production, mycelium appearance, and growth rate29. One or a few isolates of each morphotype were used for further classification and identification based on rDNA nucleotide sequences. Fungal DNA was extracted from a small amount of mycelium scraped from a PDA culture using the Phire Plant Direct PCR Kit (Thermo Fisher Scientific). A ribosomal DNA region including the internal transcribed spacer 1 (ITS1), 5.8S rDNA, and ITS2 was amplified by PCR using primers ITS1 and ITS461. Amplification conditions were: 98 °C for 5 min, followed by 35 cycles of 98 °C for 5 s, 54 °C for 5 s, and 72 °C for 20 s; after that the reaction was kept at 72 °C for 1 min. PCR amplicons were cleaned (MSB Spin PCRapace, Stratec biomedical, Germany) and sequenced at the Genomics Service in the CACTI, University of Vigo, Spain (https://cactiweb.webs.uvigo.es/).
All the sequences obtained were grouped into operational taxonomic units (OTU), considering that groups of sequences with a similarity greater than 97% belonged to the same OTU. This clustering operation was done using CD-Hit-Est software62. For taxonomic identification, a representative sequence of each OTU was used to search the GenBank nucleotide database using BLAST. This search was limited to sequences belonging to the ITS region from fungi type and reference material. Taxonomic identification was limited to genus rank because often occurs that ITS sequences are not accurate for species rank identification (i.e. Ref.30). When the identity between our sequences and that of a type strain was lower than 95%, the genus of our sequence was considered to be unknown. Further taxonomic information was obtained by means of a phylogenetic tree made with representative sequences from each taxon. This tree was made with MEGA6 software using the maximum likelihood method with distances calculated according to the Tamura 3 parameter model63. Tree branch confidence values were estimated by bootstrapping with 1000 replications.
For each kale accession, the incidence of each fungal taxon in plants was calculated, relating the number of plants in which it was found against the total of plants sampled (30 root fragments per plant, 6 plants per accession, 30 plants in total), as alpha diversity analysis. Additionally, the distribution of the relative abundance of each taxon was observed with a rank-abundance curve. Furthermore, beta diversity was analyzed by comparing between accessions, using the Jaccard index.
In planta assays
Plant–fungus interaction
One of the local varieties previously used in the diversity study (MBG-BRS0106, from here on referred to as kale) was used for the reinoculation tests and the study of the biological activity of the selected fungi.
Different fungal isolates (Table 3) were chosen based on their presence in all accessions, or their proven biological activity in other plant species (bibliographic search). To inoculate plants, one part of beet pulp inoculum was mixed with seven parts (w/w) of a substrate consisting of peat moss (Profi-Substract, Gramoflor, Valencia, Spain) previously treated at 80 °C for 24 h. The fungal inoculum was a 4-week-old culture of each fungus grown in sugar beet pulp medium64. Uninoculated controls were transplanted to soil mixed with uninoculated beet pulp medium. By qPCR, the ability of fungi to effectively colonize kale roots could be verified (data not shown).
Table 3.
Fungal isolates used in kale inoculation.
| Proposed taxon | Name in the text | Abbreviation | Presence in kale accessions |
|---|---|---|---|
| Pleosporales sp. B (next to Pyrenophora) | Pyrenophora | P |
MBG-BRS0292 MBG-BRS0446 MBG-BRS0468 |
| Chaetomium sp. | Chaetomium sp. | CG | MBG-BRS0292 |
| Trichoderma sp. | Trichoderma sp. | TH | MBG-BRS0446 |
| Helotiales sp. (next to Phialocephala) | Phialocephala | PH |
MBG-BRS0426 MBG-BRS0468 |
| Pleosporales sp. A (next to Setophoma and Edenia) | Setophoma/Edenia | ST | All |
| Diaporthe sp. | Diaporthe sp. | D | MBG-BRS0106 |
| Fusarium sp. | Fusarium sp. | FO | All |
| Acrocalymma sp. | Acrocalymma sp. | AV |
MBG-BRS0426 MBG-BRS0292 MBG-BRS0468 |
| Curvularia sp. | Curvularia sp. | CB | MBG-BRS0426 |
Growth promotion
For the analysis of plant growth promotion kale plants were inoculated in 20 L pots. The plants were kept under controlled greenhouse conditions until their aerial part was harvested in 8-week-old-plants to record fresh weight. The plants were watered 2–3 times a week, according to the observed needs, always with the same amount of water in all the plants. Exogenous fertilization was not used. Greenhouse conditions were 14 h photoperiod, environmental temperature (12–30 °C) and relative humidity above 80%. A total of 25 plants were used for each fungal isolate inoculated.
Cold tolerance
For the cold tolerance assay, we stablished the same conditions reported in Ref.65. Seeds of kale were planted in 1 L pots and grown under fluorescent light (228 μmol m−2 s−1) in a 14 h light/10 h dark photoperiod regime and watered as needed. The temperature in the cold-exposure treatment was set at 12 ± 1 °C, since lower temperatures reduced seed germination and seedling survival dramatically. A total of 25 plants were used for each inoculation.
Biotic resistance
Inoculation of kale leaves with the bacterial plant pathogen Xanthomonas campestris pv. campestris (Xcc) was made according to Ref.66. Bacterial cultures were grown in PDA at 30 °C for 48 h. A bacterial suspension was diluted with sterile water to reach a turbidity value of 0.5, which corresponds to a concentration of 5 × 108 cfu/mL. Turbidity of the suspension was measured with a microplate spectrophotometer (Spectra MR; DynexTechnologies, Chantilly, VA) at a wavelength of 600 nm. The second leaf counting from the apex was inoculated when plants were at a 5–6 leaf stage. Inoculation was made by biting on three veins located on the edge of the leaf with a mouse-tooth forceps wrapped in cotton submerged in the inoculum. Xcc race 1 type strain HRI3811, provided by Joana Vicente (University of Warwick, UK), was used to inoculate the plants. After the inoculation, plants were kept under controlled greenhouse conditions (20 °C at night and 28 °C during the day, and relative humidity greater than 80%). Damage was measured using a visual damage index (DI) scale ranging from 1 to 9, where 1 = no visible symptoms and 9 = severely diseased with typical V-shaped chlorotic leaf edge lesions presenting blackened veins areas. Data were taken 8 and 11 days post infection (d.p.i.). A total of 10 plants were used for each fungal strain.
Infestation with the insect pest Mamestra brassicae (Mb) was carried out under the controlled greenhouse conditions above described using 12-week-old-plants. Eggs were provided by the Laboratory of Entomology, Wageningen University, The Netherlands. Egg hatching larvae were fed with fresh leaves of B. oleracea (MBG-BRS0106) ad libitum. Mb inoculation was carried out depositing 5 7–10 days old larvae on the 5–6th true leaf of the plant. After 5 days, data on leaf area consumed per plant and the damage index were recorded: 1 = no damage, 2 = 1–10% leaf consumed, 3 = 11–20% leaf consumed, 4 = 21–30% leaf consumed, 5 = more than 30% of the leaf consumed. A total of 10 plants were used for each fungal strain.
Statistical analysis
For each trait (i.e. growth promotion, tolerance to cold, inoculation with Xcc and infestation with Mb) a Student’s t-test was used to compare the means of each fungal inoculation treatment with the control at P < 0.05 and P < 0.01; significant differences are denoted using asterisks.
Supplementary information
Acknowledgements
This research was financially supported by project RTI2018-096591-B-I00 34 (MCIU/AEI/FEDER, UE). JP has a contract from the Xunta de Galicia IN607A2016/13. Authors want to thank Dr. Rieta Gols (Laboratory of Entomology, Wageningen University, Wageningen, The Netherlands) for providing us with Mamestra eggs and for the unvaluable inputs in this manuscript.
Author contributions
P.V. conceived the study. P.V., J.P. and I.Z. discussed the results. P.S. carried out the Xcc experiment. M.E.C. carried out the Mb experiment. V.M.R. carried out the cold experiment and the molecular analyses. R.A. maintained the fungal collection. J.P. and P.V. performed statistical analysis of the data. I.Z. identified the fungal sequences. J.P. drafted the initial manuscript. All authors read, edited and approved the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-77215-7.
References
- 1.Francisco M, Tortosa M, Martínez-Ballesta MDC, Velasco P, García-Viguera C, Moreno DA. Nutritional and phytochemical value of Brassica crops from the agri-food perspective. Ann. Appl. Biol. 2017;170:273–285. doi: 10.1111/aab.12318. [DOI] [Google Scholar]
- 2.Šamec D, Urlić B, Salopek-Sondi B. Kale (Brassica oleracea var. acephala) as a superfood: Review of the scientific evidence behind the statement. Crit. Rev. Food Sci. Nutr. 2019;59:2411–2422. doi: 10.1080/10408398.2018.1454400. [DOI] [PubMed] [Google Scholar]
- 3.Velasco P, Cartea ME, González C, Vilar M, Ordás A. Factors affecting the glucosinolate content of kale (Brassica oleracea acephala group) J. Agric. Food Chem. 2017;55:955–962. doi: 10.1021/jf0624897. [DOI] [PubMed] [Google Scholar]
- 4.Schouten, A. Endophytic fungi: Definitions, diversity, distribution and their significance in plant life. In Endophyte Biotechnology: Potential for Agriculture and Pharmacology. 6–31 (CABI, 2019).
- 5.Compant S, Saikkonen K, Mitter B, Campisano A, Mercado-Blanco J. Editorial special issue: Soil, plants and endophytes. Plant Soil. 2016;405:1–11. doi: 10.1007/s11104-016-2927-9. [DOI] [Google Scholar]
- 6.Kumar V, Soni R, Jain L, Dash B, Goel R. Endophytic Fungi: Recent Advances in Identification and Explorations. In Advances in Endophytic Fungal Research. Cham: . Springer; 2019. pp. 267–281. [Google Scholar]
- 7.Busby PE, Ridout M, Newcombe G. Fungal endophytes: Modifiers of plant disease. Plant Mol. Biol. 2016;90:645–655. doi: 10.1007/s11103-015-0412-0. [DOI] [PubMed] [Google Scholar]
- 8.Poveda J, Abril-Urias P, Escobar C. Biological control of plant-parasitic nematodes by filamentous fungi inducers of resistance: Trichoderma, mycorrhizal and endophytic fungi. Front. Microbiol. 2020;11:992. doi: 10.3389/fmicb.2020.00992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quesada-Moraga, E., Herrero, N., Zabalgogeazcoa, Í. Entomopathogenic and nematophagous fungal endophytes. In Advances in Endophytic Research. 85–99 (Springer, New Delhi, 2014).
- 10.Lata R, Chowdhury S, Gond SK, White JF., Jr Induction of abiotic stress tolerance in plants by endophytic microbes. Lett. Appl. Microbiol. 2018;66:268–276. doi: 10.1111/lam.12855. [DOI] [PubMed] [Google Scholar]
- 11.Card SD, Hume DE, Roodi D, McGill CR, Millner JP, Johnson RD. Beneficial endophytic microorganisms of Brassica—A review. Biol. Control. 2015;90:102–112. doi: 10.1016/j.biocontrol.2015.06.001. [DOI] [Google Scholar]
- 12.Su ZZ, Wang T, Shrivastava N, Chen YY, Liu X, Sun C, Yin Y, Gao QK, Lou BG. Piriformospora indica promotes growth, seed yield and quality of Brassica napus L. Microbiol. Res. 2017;199:29–39. doi: 10.1016/j.micres.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Q, Zhang J, Yang L, Zhang L, Jiang D, Chen W, Li G. Diversity and biocontrol potential of endophytic fungi in Brassica napus. Biol. Control. 2014;72:98–108. doi: 10.1016/j.biocontrol.2014.02.018. [DOI] [Google Scholar]
- 14.Qin J, Lyu A, Zhang QH, Yang L, Zhang J, Li GQ. Strain identification and metabolites isolation of Aspergillus capensis CanS-34A from Brassica napus. Mol. Biol. Rep. 2019;46:3451–3460. doi: 10.1007/s11033-019-04808-5. [DOI] [PubMed] [Google Scholar]
- 15.Batta YA. Efficacy of endophytic and applied Metarhizium anisopliae (Metch.) Sorokin (Ascomycota: Hypocreales) against larvae of Plutella xylostella L. (Yponomeutidae: Lepidoptera) infesting Brassica napus plants. Crop Prot. 2013;44:128–134. doi: 10.1016/j.cropro.2012.11.001. [DOI] [Google Scholar]
- 16.Hiruma K, Kobae Y, Toju H. Beneficial associations between Brassicaceae plants and fungal endophytes under nutrient-limiting conditions: Evolutionary origins and host–symbiont molecular mechanisms. Curr. Opin. Plant Biol. 2018;44:145–154. doi: 10.1016/j.pbi.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Ilyas M, Kanti A, Jamal Y, Hertina H, Agusta A. Biodiversity of endophytic fungi associated with Uncaria gambier Roxb. (Rubiaceae) from west Sumatra. Biodiversitas. 2009;10:2009. doi: 10.13057/biodiv/d100105. [DOI] [Google Scholar]
- 18.Zhao Y, Xiong Z, Wu G, Bai W, Zhu Z, Gao Y, Parmar S, Sharma VK, Li H. Fungal endophytic communities of two wild Rosa varieties with different powdery mildew susceptibilities. Front. Microbiol. 2018;9:2462. doi: 10.3389/fmicb.2018.02462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovacs ED, Rusu T, Lech WS, Kovacs MH, Di T, Roman C. Rhizosphere microbiota profile changes with different genetic types of tomato species. Agricultura. 2019;109:140–150. [Google Scholar]
- 20.Shi Y, Xie H, Cao L, Zhang R, Xu Z, Wang Z, Deng Z. Effects of Cd-and Pb-resistant endophytic fungi on growth and phytoextraction of Brassica napus in metal-contaminated soils. Environ. Sci. Pollut. Res. 2017;24:417–426. doi: 10.1007/s11356-016-7693-y. [DOI] [PubMed] [Google Scholar]
- 21.Zahoor M, Irshad M, Rahman H, Qasim M, Afridi SG, Qadir M, Hussain A. Alleviation of heavy metal toxicity and phytostimulation of Brassica campestris L. by endophytic Mucor sp. MHR-7. Ecotox. Environ. Saf. 2017;142:139–149. doi: 10.1016/j.ecoenv.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Deng Z, Cao L, Huang H, Jiang X, Wang W, Shi Y, Zhang R. Characterization of Cd-and Pb-resistant fungal endophyte Mucor sp. CBRF59 isolated from rapes (Brassica chinensis) in a metal-contaminated soil. J. Hazard. Mater. 2011;185:717–724. doi: 10.1016/j.jhazmat.2010.09.078. [DOI] [PubMed] [Google Scholar]
- 23.de Lamo FJ, Takken FL. Biocontrol by Fusarium oxysporum using endophyte-mediated resistance. Front. Plant Sci. 2020;11:37. doi: 10.3389/fpls.2020.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caicedo NH, Davalos AF, Puente PA, Rodríguez AY, Caicedo PA. Antioxidant activity of exo-metabolites produced by Fusarium oxysporum: An endophytic fungus isolated from leaves of Otoba gracilipes. Microbiol. Open. 2019;8:e903. doi: 10.1002/mbo3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ola AR, Thomy D, Lai D, Brötz-Oesterhelt H, Proksch P. Inducing secondary metabolite production by the endophytic fungus Fusarium tricinctum through coculture with Bacillus subtilis. J. Nat. Prod. 2013;76:2094–2099. doi: 10.1021/np400589h. [DOI] [PubMed] [Google Scholar]
- 26.Miyaji N, Shimizu M, Miyazaki J, Osabe K, Sato M, Ebe Y, et al. Comparison of transcriptome profiles by Fusarium oxysporum inoculation between Fusarium yellows resistant and susceptible lines in Brassica rapa L. Plant Cell. Rep. 2017;36:1841–1854. doi: 10.1007/s00299-017-2198-9. [DOI] [PubMed] [Google Scholar]
- 27.Pu Z, Ino Y, Kimura Y, Tago A, Shimizu M, Natsume S, et al. Changes in the proteome of xylem sap in Brassica oleracea in response to Fusarium oxysporum stress. Front. Plant Sci. 2016;7:31. doi: 10.3389/fpls.2016.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xing M, Lv H, Ma J, Xu D, Li H, Yang L, et al. Transcriptome profiling of resistance to Fusarium oxysporum f. sp. conglutinans in cabbage (Brassica oleracea) roots. PLoS ONE. 2016;11:e0148048. doi: 10.1371/journal.pone.0148048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pereira E, Vázquez de Aldana BR, San Emeterio L, Zabalgogeazcoa I. A survey of culturable fungal endophytes from Festuca rubra subsp. pruinosa, a grass from marine cliffs, reveals a core microbiome. Front. Microbiol. 2019;9:3321. doi: 10.3389/fmicb.2018.03321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu F, Wang J, Li H, Wang W, Cai L. Setophoma spp. on Camellia sinensis. Fungal Syst. Evol. 2019;4:43–57. doi: 10.3114/fuse.2019.04.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y, Zuzak K, Harding M, Neilson E, Feindel D, Feng J. First report of pink root rot caused by Setophoma (Pyrenochaeta) terrestris on canola. Can. J. Plant. Pathol. 2017;39:354–360. doi: 10.1080/07060661.2017.1355849. [DOI] [Google Scholar]
- 32.Shobha M, Kumara KS, Prakash HS. Fungal endophytes associated with Gloriosa superba (L.) Proc. Natl Acad. Sci. India Sect. B Biol. Sci. 2019;89:1335–1342. doi: 10.1007/s40011-018-1053-2. [DOI] [Google Scholar]
- 33.Szűcs Z, Plaszkó T, Cziáky Z, Kiss-Szikszai A, Emri T, Bertóti R, et al. Endophytic fungi from the roots of horseradish (Armoracia rusticana) and their interactions with the defensive metabolites of the glucosinolate-myrosinase-isothiocyanate system. BMC Plant Biol. 2018;18:85. doi: 10.1186/s12870-018-1295-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan Y, Guo Z, Zhu M, Shi J, Li W, Jiao R, et al. Anti-inflammatory spirobisnaphthalene natural products from a plant-derived endophytic fungus Edenia gomezpompae. Chin. Chem. Lett. 2020;31:1406–1409. doi: 10.1016/j.cclet.2020.03.059. [DOI] [Google Scholar]
- 35.Martínez-Luis S, Della-Togna G, Coley PD, Kursar TA, Gerwick WH, Cubilla-Rios L. Antileishmanial constituents of the Panamanian endophytic fungus Edenia sp. J. Nat. Prod. 2008;71:2011–2014. doi: 10.1021/np800472q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Macias-Rubalcava ML, Ruiz-Velasco Sobrino ME, Melendez-Gonzalez C, Hernandez-Ortega S. Naphthoquinone spiroketals and organic extracts from the endophytic fungus Edenia gomezpompae as potential herbicides. J. Agric. Food Chem. 2014;62:3553–3562. doi: 10.1021/jf500965k. [DOI] [PubMed] [Google Scholar]
- 37.Irwin JAG, Mackie JM, Marney TS, Musial JM, Roberts S. Incidence of Stagonaspora meliloti and Acracalymma medicaginis in lucerne crowns and roots in eastern Australia, their comparative aggressiveness to lucerne and inheritance of reaction to S. meliloti in lucerne. Aust. Plant Pathol. 2004;33:61–67. doi: 10.1071/AP03083. [DOI] [Google Scholar]
- 38.Jin HQ, Liu HB, Xie YY, Zhang YG, Xu QQ, Mao LJ, et al. Effect of the dark septate endophytic fungus Acrocalymma vagum on heavy metal content in tobacco leaves. Symbiosis. 2018;74:89–95. doi: 10.1007/s13199-017-0485-4. [DOI] [Google Scholar]
- 39.He C, Wang W, Hou J. Characterization of dark septate endophytic fungi and improve the performance of liquorice under organic residue treatment. Front. Microbiol. 2019;10:1364. doi: 10.3389/fmicb.2019.01364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh BP. Advances in Endophytic Fungal Research. New York: Springer; 2019. [Google Scholar]
- 41.Clare SJ, Wyatt NA, Brueggeman RS, Friesen TL. Research advances in the Pyrenophora teres-barley interaction. Mol. Plant. Pathol. 2020;21:272–288. doi: 10.1111/mpp.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou L, Yu J, Zhao L, He X. Dark septate endophytes improve the growth and tolerance of Medicago sativa and Ammopiptanthus mongolicus under cadmium stress. Front. Microbiol. 2019;10:3061. doi: 10.3389/fmicb.2019.03061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bengyella L, Iftikhar S, Nawaz K, Fonmboh DJ, Yekwa EL, Jones RC, et al. Biotechnological application of endophytic filamentous bipolaris and curvularia: A review on bioeconomy impact. World J. Microbiol. Biotechnol. 2019;35:69. doi: 10.1007/s11274-019-2644-7. [DOI] [PubMed] [Google Scholar]
- 44.Priyadharsini P, Muthukumar T. The root endophytic fungus Curvularia geniculata from Parthenium hysterophorus roots improves plant growth through phosphate solubilization and phytohormone production. Fungal Ecol. 2017;27:69–77. doi: 10.1016/j.funeco.2017.02.007. [DOI] [Google Scholar]
- 45.Liu ZH, Yang Q, Nie YH. Cloning and expression of Hsp22.4 gene from Chaetomium globosum. J. For. Res. 2006;17:259–262. doi: 10.1007/s11676-006-0059-4. [DOI] [Google Scholar]
- 46.Liu ZH, Yang Q, Ma J. A heat shock protein gene (hsp22.4) from Chaetomium globosum confers heat and Na2 CO3 tolerance to yeast. Appl. Microbiol. Biotechnol. 2007;77:901–908. doi: 10.1007/s00253-007-1226-z. [DOI] [PubMed] [Google Scholar]
- 47.Wang Z, Jia S, Cui J, Qu J, Yue Y, Sun Q, Zhang H. Antioxidant activity of a polysaccharide produced by Chaetomium globosum CGMCC 6882. Int. J. Biol. Macromol. 2019;141:955–960. doi: 10.1016/j.ijbiomac.2019.09.069. [DOI] [PubMed] [Google Scholar]
- 48.Acuña-Rodríguez IS, Newsham KK, Gundel PE, Torres-Díaz C, Molina-Montenegro MA. Functional roles of microbial symbionts in plant cold tolerance. Ecol. Lett. 2020;23:1034–1048. doi: 10.1111/ele.13502. [DOI] [PubMed] [Google Scholar]
- 49.Poveda, J., Eugui, D. & Abril-Urías, P. Could be Trichoderma a plant pathogen? Succesful of colonization. In: Trichoderma: Host Pathogen Interactions and Applications. 35–39 (Springer, New York, 2020).
- 50.Alkooranee JT, Yin Y, Aledan TR, Jiang Y, Lu G, Wu J, Li M. Systemic resistance to powdery mildew in Brassica napus (AACC) and Raphanus alboglabra (RRCC) by Trichoderma harzianum TH12. PLoS ONE. 2015;10:e0142177. doi: 10.1371/journal.pone.0142177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poveda J, Hermosa R, Monte E, Nicolás C. The Trichoderma harzianum Kelch protein ThKEL1 plays a key role in root colonization and the induction of systemic defense in Brassicaceae plants. Front. Plant Sci. 2019;10:1478. doi: 10.3389/fpls.2019.01478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alkooranee JT, Aledan TR, Xiang J, Lu G, Li M. Induced systemic resistance in two genotypes of Brassica napus (AACC) and Raphanus oleracea (RRCC) by Trichoderma isolates against Sclerotinia sclerotiorum. Am. J. Plant Sci. 2015;6:1662. doi: 10.4236/ajps.2015.610166. [DOI] [Google Scholar]
- 53.Li HY, Luo Y, Zhang XS, Shi WL, Gong ZT, Shi M, et al. Trichokonins from Trichoderma pseudokoningii SMF2 induce resistance against Gram-negative Pectobacterium carotovorum subsp. carotovorum in Chinese cabbage. FEMS Microbiol. Lett. 2014;354:75–82. doi: 10.1111/1574-6968.12427. [DOI] [PubMed] [Google Scholar]
- 54.Shibu MA, Lin HS, Yang HH, Peng KC. Trichoderma harzianum ETS 323-mediated resistance in Brassica oleracea var capitata to Rhizoctonia solani involves the novel expression of a glutathione S-transferase and a deoxycytidine deaminase. J. Agric. Food Chem. 2012;60:10723–10732. doi: 10.1021/jf3025634. [DOI] [PubMed] [Google Scholar]
- 55.Martinuz A, Schouten A, Menjivar RD, Sikora RA. Effectiveness of systemic resistance toward Aphis gossypii (Hom., Aphididae) as induced by combined applications of the endophytes Fusarium oxysporum Fo162 and Rhizobium etli G12. Biol. Control. 2012;62:206–212. doi: 10.1016/j.biocontrol.2012.05.006. [DOI] [Google Scholar]
- 56.Paparu P, Dubois T, Coyne D, Viljoen A. Defense-related gene expression in susceptible and tolerant bananas (Musa spp.) following inoculation with non-pathogenic Fusarium oxysporum endophytes and challenge with Radopholus similis. Physiol. Mol. Plant Pathol. 2007;71:149–157. doi: 10.1016/j.pmpp.2007.12.001. [DOI] [Google Scholar]
- 57.He CY, Hsiang T, Wolyn DJ. Induction of systemic disease resistance and pathogen defence responses in Asparagus officinalis inoculated with nonpathogenic strains of Fusarium oxysporum. Plant Pathol. 2002;51:225–230. doi: 10.1046/j.1365-3059.2002.00682.x. [DOI] [Google Scholar]
- 58.Constantin ME, de Lamo FJ, Vlieger BV, Rep M, Takken FL. Endophyte-mediated resistance in tomato to Fusarium oxysporum is independent of ET, JA, and SA. Front. Plant Sci. 2019;10:79. doi: 10.3389/fpls.2019.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vandenkoornhuyse P, Baldauf SL, Leyval C, Straczek J, Young JPW. Extensive fungal diversity in plant roots. Science. 2002;295:2051–2051. doi: 10.1126/science.295.5562.2051. [DOI] [PubMed] [Google Scholar]
- 60.Poveda J, Eugui D, Velasco P. Natural control of plant pathogens through glucosinolates: An effective strategy against fungi and oomycetes. Phytochem. Rev. 2020;161:1–15. [Google Scholar]
- 61.White, T. J., Bruns, T., Lee, S. J. W. T., Taylor, J. Amplification and direct sequencing of fungal ribosomal rna genes for phylogenetics. In PCR Protocols: a Guide to Methods and Applications. 315–322 (Academic Press, Cambridge, 1990).
- 62.Li, W. & Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006) https://weizhongli-lab.org/cd-hit/. [DOI] [PubMed]
- 63.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vázquez-de-Aldana BR, Cuesta MJ, Zabalgogeazcoa I. Cultivation and growth dynamics of endophytic fungi in a solid culture medium based on sugar beet pulp. J. Sci. Food Agric. 2020;100:441–446. doi: 10.1002/jsfa.10030. [DOI] [PubMed] [Google Scholar]
- 65.Rodríguez VM, Soengas P, Alonso-Villaverde V, Sotelo T, Cartea ME, Velasco P. Effect of temperature stress on the early vegetative development of Brassica oleracea L. BMC Plant. Biol. 2015;15:145. doi: 10.1186/s12870-015-0535-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iglesias-Bernabé L, Madloo P, Rodríguez VM, Francisco M, Soengas P. Dissecting quantitative resistance to Xanthomonas campestris pv. campestris in leaves of Brassica oleracea by QTL analysis. Sci. Rep. 2019;9:1–11. doi: 10.1038/s41598-019-38527-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hernandez-Agreda A, Gates RD, Ainsworth TD. Defining the core microbiome in corals’ microbial soup. Trends Microbiol. 2017;25:125–140. doi: 10.1016/j.tim.2016.11.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.