Abstract
Background
Mycoplasma pneumoniae (MP) causes community-acquired pneumonia affecting mainly children, and tends to produce cyclic outbreaks. The widespread use of macrolides is increasing resistance rates to these antibiotics. Molecular tools can help in diagnosis, typing and resistance detection, leading to better patient management.
Objectives
To assess the MP genotypes and resistance pattern circulating in our area while comparing serological and molecular diagnosis of MP.
Methods
Molecular and serological diagnosis of MP was performed in 821 samples collected in Badalona (Barcelona, Spain) from 2013 to 2017. Multiple locus variable number tandem repeat analysis (MLVA) and macrolide resistance detection by pyrosequencing were performed in those cases positive by PCR. Presence of respiratory viruses and relevant clinical data were also recorded.
Results
MP was detected in 16.8% of cases by PCR, with an overall agreement with serology of 76%. Eleven different MLVA types were identified, with 4-5-7-2 (50.1%) and 3-5-6-2 (29.2%) being the most abundant, with the latter showing a seasonal increase during the study. A total of 8% of the strains harboured a point substitution associated with macrolide resistance, corresponding mainly to an A2063G 23S rRNA mutation and directly related to previous macrolide therapy. Analysis of respiratory viruses showed viral coinfections in most cases.
Conclusions
Serological and molecular tools combined could improve MP diagnosis and the analysis of its infection patterns. Macrolide resistance is associated with previous therapy. Given that MP pneumonia usually resolves spontaneously, it should be reconsidered whether antibiotic treatment is suitable for all cases.
Introduction
Mycoplasma pneumoniae (MP) is a prevalent respiratory pathogen that causes about 20%–40% of cases of community-acquired pneumonia (CAP) in children and young adults, with epidemic peaks at 3–7 year intervals worldwide.1 Serology is the gold standard, but molecular techniques offer a rapid and reliable alternative for MP diagnosis and epidemiological surveillance. Molecular typing by P1 restriction fragment length polymorphism analysis (RFLP)2 has been substituted by methods with higher discriminatory power, such as multiple locus variable number tandem repeat analysis (MLVA),3 allowing more accurate outbreak analysis and epidemic strain monitoring worldwide.
Macrolides (MLs) are the first-line treatment, due to their low side effects and convenient administration. MLs inhibit protein synthesis by binding to domain V of 23S rRNA.4 Mutations in domain II or V of 23S rRNA or in L4 and L22 ribosomal proteins have been described to be involved in ML resistance (MLr) in other species.5,6 In MP, the MLr in vivo is mainly driven by specific mutations in the V domain of 23S rRNA gene, leading either to a high level resistance profile (mutations at positions 2063 or 2064) or to a low level MLr (transition at position 2067 or a transversion at position 2617).4,7–11 However, although no mutations in domain II could be linked to MLr, mutations in ribosomal proteins L4 and L22 have been found both in vitro4 and in vivo,9 although their real impact in MLr is still unclear, so further research should be performed.
As MP culture is mainly performed in reference centres, MLr determination by broth dilution tests11 is not accessible to most clinical microbiology laboratories. Molecular techniques targeting the single-copy 23S rRNA gene (such as PCR followed by high resolution melting curve analysis,12–14 Sanger sequencing,15 RFLP8 or pyrosequencing analysis16–18) have been described. As a consequence of widespread ML use, resistance rates have risen from 40% in 2008 to 90%–100% in China recently.19,20 Concurrently, in Europe, rates vary from 1% in Slovenia to up to 26% in Italy.21–26 Due to the increase in the prevalence of MLr observed recently and the lack of data in Spain, it is imperative to improve technical tools for detecting resistant strains without delay and to adjust the patients’ therapy as necessary.
As there are no published data about the MP MLVA types circulating in our area, this study aimed to provide insights about the genotypes and yearly distribution found in Catalonia (Spain). Furthermore, the evaluation of the presence of single-base mutations in domain V of the 23S rRNA gene that provide MLr in our setting is described, in comparison with the results described in other countries. Additionally, the clinical performance of the molecular technique used for MP pneumonia diagnosis was compared with routine serological testing.
Materials and methods
Ethics, patients included, samples and collection
Written informed consent was obtained from patients’ guardians. This study was approved by the Clinical Research Ethics Committee at Germans Trias i Pujol University Hospital (‘Comité Ético de Investigación Clínica’, CEIC). Samples collected included nasopharyngeal aspirates and sera. A total of 821 nasopharyngeal samples from 774 individuals were obtained between April 2013 and January 2017 in the Pediatric Emergency room at the University Hospital Germans Trias i Pujol (Badalona, Spain). Samples were divided into different groups: children (<18 years old) clinically diagnosed with CAP, household or school contacts from a diagnosed pneumonia case and healthy volunteer subjects. Clinical data were registered by the Pediatric Department, and microbiological results were also recorded at the Microbiology Department.
M. pneumoniae serological and molecular diagnosis
Detection of IgG or IgM antibodies from sera was performed by a particle-agglutination assay (Serodia®-MYCO II, Fujirebio Europe) either singly or in two samples separated by at least 21 days in order to demonstrate a seroconversion. A positive result with clinical significance was defined as antibody levels ≥1/160 for single samples. For the molecular diagnosis, DNA was extracted from nasopharyngeal aspirates using the nucliSENS® easyMag® platform (bioMérieux, France). MP molecular diagnosis was performed using two different assays: during the first stage of the project, Real Time PCR Realcycler M. pneumoniae/Chlamydophila pneumoniae (Progenie molecular, Spain) was employed, whereas Anyplex® 2 RB5 Detection (Seegene Inc., Korea) was used later on. Those samples in which MP DNA was detected were further investigated for MLVA typing and MLr by pyrosequencing. Respiratory viruses were also tested for in all MP-positive samples, using three different methods: BinaxNOW® Influenza A&B and BinaxNOW® Respiratory Syncytial virus immunochromatographic assay (Abbott-Alere Healthcare, USA), direct immunofluorescence D3 Ultra8 DFA Respiratory virus Screening & Identification Kit (Quidel Corporation, USA), used until April 2014, and Anyplex II RV-16 Detection (Seegene Inc., Korea), used from May 2014, when Anyplex was integrated into our diagnosis routine for adults and also for children with negative immunochromatographic results.
MLVA typing
DNA from PCR-positive samples was typed using the culture-independent MLVA approach described by Dumke and Jacobs,27 performing a nested PCR in order to increase the sensitivity. Products from the secondary PCR were analysed by high-resolution capillary electrophoresis (QIAxcel® Advanced System, Qiagen, Germany) and subsequently purified (QIAquick PCR Purification Kit, Qiagen). Purified PCR products were then Sanger sequenced and the number of repeats was found by uploading the FASTA file to the web resource Tandem Repeat Finder.28 Only four out of the five loci (Mp13, Mp14, Mp15 and Mp16) initially described by Dégrange et al.3 were analysed due to the high variability found in locus Mp1, as described elsewhere.29 The MLVA type is depicted as the number of repeats within the Mp13, Mp14, Mp15 and Mp16 loci, respectively, separated by a hyphen. Optimization of the protocol was performed using the DNA from M. pneumoniae M129, acquired from DSMZ (German Collection of Microorganisms and Cell Cultures, Germany).
Resistance detection by pyrosequencing
The pyrosequencing protocol was modified from Spuesens et al.16,17 Two different assays were performed in order to cover all known mutations (550 bp apart) conferring ML-resistant genotypes found in vivo: the first assay was able to detect single base mutations at positions 2063, 2064 and 2067, while the second detected the mutation at position 2617. The protocol described17 was modified by increasing the number of cycles to 45 for both primary and secondary PCR and by adding 2 μL of template for the secondary PCR. The presence of the targeted DNA fragment was detected by high-resolution capillary electrophoresis (QIAxcel® Advanced System, Qiagen), obtaining an approximately 165 bp fragment. Immobilization of the biotinylated product and the pyrosequencing reaction were also performed following the protocol of Spuesens et al.16 with the following modifications: a total of 4 μL of streptavidin beads and 26 μL of H2O per sample were used, and the plates were mixed for 10 min instead of 15 min. Pyrosequencing reactions and sequence analysis were also performed using the Pyromark™Q96MD sequencer (Biotage, Sweden) and associated software.
Statistical analysis
Qualitative and categorical data were summarized with number and percentage, and were compared using a χ2 test. Statistical analyses were performed with the statistical free software PSPP (http://www.gnu.org/software/pspp/, last accessed April 2020). Statistical significance was accepted at P < 0.05.
Results
Molecular detection of M. pneumoniae
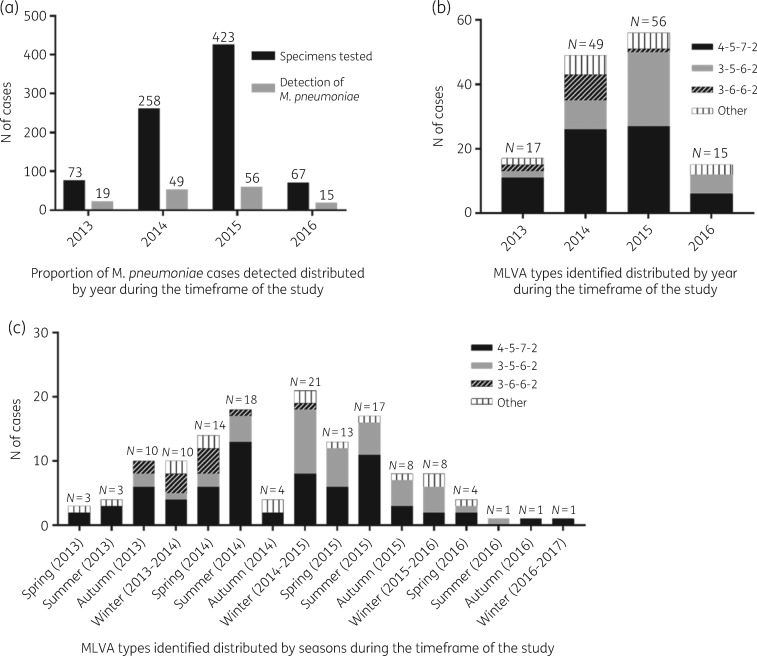
A total of 821 specimens (described in Table 1) were collected from 774 individuals, corresponding to children in 769 cases (766 patients with suspicion of pneumonia or relatives with pneumonia, and 3 healthy children) and adults in 52 cases (29 patients with a pneumonia diagnosis or their household contacts, and 23 volunteer health professionals). A positive detection of MP was obtained in 138 (16.8%) specimens, corresponding to 127 individuals. One positive specimen was excluded from the study due to insufficient DNA; thus, the final count of positive specimens was 137 (16.7%, 3 by Progenie, 134 by Anyplex). Positive detections were mostly from children (133, 97.1%), with only 4 (2.9%) from adults (close relatives). The median age of children was 6.6 years and the highest proportion of positive samples was detected in children aged >5 years (53.3%, 73/137), followed by children aged between 1 and 5 years (36.4%, 50/137) and ≤1 year old (7.3%, 10/137) (Table 2). The majority of specimens were collected during the middle years of the project (2014 and 2015), and the positivity rates per year were as follows: 26% in 2013 (19/73), 19% in 2014 (49/258), 13.2% in 2015 (56/423) and 22.4% in 2016 (15/67), as depicted in Figure 1(a).
Table 1.
Classification of the patients and specimens included in this study
| Samples tested for MP | Age group | Group of individuals | Number of samples tested | Positive MP detection (number) | Complementary data |
|---|---|---|---|---|---|
| 821 specimens (774 individuals) | Children (<18 years old) | Children with suspicion of pneumonia or relatives with pneumonia | 766a | 17.5% (134)a |
|
| Healthy children | 3 | 0.0% (0) | |||
| Adults | Adults with diagnosis of pneumonia or close relatives with pneumonia | 29 | 13.8% (4) |
|
|
| Healthy adults (volunteer healthcare professionals) | 23 | 0.0% (0) |
For one specimen where MP was detected by PCR, it was not possible to perform the MLVA typing due to insufficient DNA. Thus, the total number of specimens from children typed was 133.
Table 2.
Characteristics and clinical symptoms distributed by MLVA type identified
| MLVA type |
% (n) from total of positive specimens (N = 137) | P value | ||||
|---|---|---|---|---|---|---|
| Characteristic or clinical symptoms | 4-7-5-2 (N = 70) | 3-5-6-2 (N = 40) | 3-6-6-2 (N = 11) | Other (N = 16) | ||
| Sex | ||||||
| male (n) | 58.6% (41) | 52.5% (21) | 45.5% (5) | 56.3% (9) | 55.5% (76) | 0.834 |
| female (n) | 41.4% (29) | 47.5% (19) | 54.5% (6) | 43.7% (7) | 44.53% (61) | |
| Age group | ||||||
| ≤1 year (n) | 5.7% (4) | 5.0% (2) | 0.0% (0) | 25.0% (4) | 7.3% (10) | 0.252 |
| 1–5 years (n) | 38.6% (27) | 40.0% (16) | 36.4% (4) | 18.6% (3) | 36.4% (50) | |
| >5 years (n) | 51.4% (36) | 52.5% (21) | 63.6% (7) | 56.2% (9) | 53.3% (73) | |
| adult (n) | 2.2% (3) | 0.73% (1) | 0.0% (0) | 0.0% (0) | 2.9% (4) | |
| Body temperature | ||||||
| afebrile (n) | 54.3% (38) | 57.5% (23) | 81.8% (9) | 50.0% (8) | 57.0% (78) | 0.348 |
| fever/slight fever (n) | 45.7% (32) | 42.5% (17) | 18.2% (2) | 50.0% (8) | 43.0% (59) | |
| Respiratory rate | ||||||
| eupnoea (n) | 70% (49) | 72.5% (29) | 81.82% (9) | 68.75% (11) | 71.5% (98) | 0.857 |
| tachypnoea (n) | 30% (21) | 27.5% (11) | 18.18% (2) | 31.25% (5) | 28.5% (39) | |
| Lung auscultation | ||||||
| normal (n) | 45.7% (32) | 47.5% (19) | 72.73% (8) | 37.5% (6) | 47.5% (65) | 0.316 |
| altered (n) | 54.29% (38) | 52.5% (21) | 27.27% (3) | 62.5% (10) | 52.5% (72) | |
| Chest X-ray | ||||||
| normal (n) | 2.9% (2) | 10.0% (4) | 0.0% (0) | 6.2% (1) | 5.1% (7) | 0.655 |
| pathological (n) | 80.0% (56) | 72.5% (29) | 90.91% (10) | 81.3% (13) | 78.8% (108) | |
| not performed (n) | 17.1% (12) | 17.5% (7) | 9.1% (1) | 12.5% (2) | 16.1% (22) | |
| Adenopathies | ||||||
| absent (n) | 97.1% (68) | 97.5% (39) | 100.0% (11) | 100.0% (16) | 97.8% (134) | 0.857 |
| present (n) | 2.9% (2) | 2.5% (1) | 0.0% (0) | 0.0% (0) | 2.2% (3) | |
| Arthralgia | ||||||
| absent (n) | 97.1% (68) | 97.5% (39) | 100.0% (11) | 100.0% (16) | 97.8% (134) | 0.857 |
| present (n) | 2.9% (2) | 2.5% (1) | 0.0% (0) | 0.0% (0) | 2.2% (3) | |
| Stevens–Johnson syndrome | ||||||
| absent (n) | 98.57% (69) | 97.5% (39) | 100.0% (11) | 100.0% (16) | 98.5% (135) | 0.873 |
| present (n) | 1.43% (1) | 2.5% (1) | 0.0% (0) | 0.0% (0) | 1.5% (2) | |
| Viral coinfection | ||||||
| absent (n) | 58.6% (41) | 42.5% (17) | 72.7% (8) | 56.25% (9) | 54.7% (75) | 0.232 |
| present (n) | 41.4% (29) | 57.5% (23) | 27.3% (3) | 43.75% (7) | 45.3% (62) | |
Figure 1.
Molecular detection and typing of M. pneumoniae. (a) Detected cases of M. pneumoniae distributed by year. (b) MLVA types identified distributed by year. (c) MLVA types identified distributed by season during the time frame of the study.
Performance of molecular detection in comparison with serological diagnosis
In parallel to PCR, serological diagnosis was also performed in 376 cases, including 105 patients with a positive MP detection by PCR and 271 with a negative MP result. Among the 105 MP PCR-positive subjects, serology results in the acute phase (first serum sample) were also positive (antibody levels ≥1/160) in 75 (71.4%) cases, whereas in the remaining 30 cases with negative serology (antibody levels <1/160), seroconversion was observed in 13 (43.3%) patients in convalescence phase (second serum sample), leading to 88 out of 105 patients (83.8%) with both positive serology and PCR, and an overall agreement between techniques of 76%. Among the 271 MP PCR-negative subjects, serology was also negative in 199 cases (73.4%). The remaining 72 cases showed a positive serology result in a single sample. Thus, sensitivity and specificity of the PCR compared with serology was 55% and 92%, respectively (Table 3).
Table 3.
Performance of molecular diagnosis compared with serology for the detection of Mycoplasma pneumoniae
| Serology |
||||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| PCR | Positive | 75/88a | 30/17a | 105 |
| Negative | 72 | 199 | 271 | |
|
| ||||
| Total | 147/160a | 229/216a | 376 | |
|
| ||||
| Sensitivity | 51%/55%a | |||
| Specificity | 87%/92%a | |||
| Overall agreement | 73%/76%a | |||
Patients that showed seroconversion (second serology with a positive result). PCR performed from nasopharyngeal aspirates.
M. pneumoniae MLVA typing
Eleven different MLVA types were identified; the most abundant was 4-5-7-2 (70/137, 50.1%), followed by 3-5-6-2 (40/137, 29.2%), 3-6-6-2 (11/137, 8%), 4-5-6-2 (4/137, 2.9%), 3-5-7–2 (3/137, 2.2%), 4-5-5-2 (2/137, 1.5%), 4-5-7-3 (2/137, 1.5%), 4-6-7-2 (2/137, 1.5%), 3-3-6-2 (1/137, 0.7%), 3-4-6-2 (1/137, 0.7%) and 3-4-7-2 (1/137, 0.7%). The proportions of the types varied widely between the years of the study (Figure 1b). Comparing the 2 years with higher numbers of positive cases (2014 and 2015, Figure 1a), the proportion of type 4-5-7-2 during those years was similar, while an increase of type 3-5-6-2 and a decrease of type 3-6-6-2 was observed from 2014 to 2015. Interestingly, the distribution of the types also varied depending on the season (Figure 1c), with genotype 4-5-7-2 mostly detected during summer (both 2014 and 2015) in contrast to type 3-5-6-2, which was more frequent from winter 2014 to spring 2015. Genotype 3-6-6-2 also showed a peak during spring 2014, whereas the other minority genotypes were detected in similar numbers among all seasons from spring 2013 to spring 2016.
The four positive specimens detected from adults had the same MLVA profile as the children, suggesting transmission of MP within the same household.
Twelve out of the 137 cases were reinfections after 1 or 2 months from the primary specimen. In 9 specimens, the MLVA type detected was the same as the initial sample; however, for 2 patients, the MLVA changed. In one case, the initial MLVA was 3-6-6-2 (March 2015) but the MLVA from the specimens obtained in April and May 2015 was 3-5-6-2. For the other case, the initial type was 3-5-6-2 (March 2016) whereas the specimen obtained on April 2016 was 3-5-7-2. Given that both isolates had only a single repeat change from the initial profile, the second detected specimen could be a variant of the first isolate.
Clinical findings
Several parameters such as chest radiography, axillary temperature, respiratory rate, presence of adenopathy, arthralgia and pleural effusion were recorded, and a statistical analysis was performed to determine if there was a relationship to the MLVA type. Results are shown in Table 2; however, none of the clinical symptoms evaluated was related to a specific MLVA type.
Viral infections were also evaluated in MP-positive patients. Respiratory viruses were found in 45.3% of patients (N = 62), with two or more viruses detected in 12.4% of the cases. A total of 79 virus-positive samples were detected, listed in Table 4. The most frequently detected was Rhinovirus (37.9%, 30) followed by Parainfluenza virus (15.2%, 12) and Respiratory Syncytial Virus (13.9%, 11). The genotypes 4-5-7-2 and 3-5-6-2 were the most frequently co-infected with virus (P = 0.033).
Table 4.
Virus detecteda distributed by MLVA type identified
| MLVA type |
|||||
|---|---|---|---|---|---|
| Virus | 4-7-5-2 (N = 70) | 3-5-6-2 (N = 40) | 3-6-6-2 (N = 11) | other (N = 16) | Total |
| Influenza virus | 28.6% (2) | 28.6% (2) | 0.0% (0) | 42.8% (3) | 8.9% (7) |
| Metapneumovirus | 0.0% (0) | 0.0% (0) | 0.0% (0) | 100% (1) | 1.3% (1) |
| Parainfluenza virus | 66.7% (8) | 25.0% (3) | 0.0% (0) | 8.3% (1) | 15.2% (12) |
| Respiratory Syncytial Virus | 27.3% (3) | 36.3% (4) | 18.2% (2) | 18.2% (2) | 13.9% (11) |
| Rhinovirus | 53.3% (16) | 36.7% (11) | 3.3% (1) | 6.7% (2) | 37.9% (30) |
| Adenovirus | 50.0% (5) | 50.0% (5) | 0.0% (0) | 0.0% (0) | 12.7% (10) |
| Bocavirus | 0.0% (0) | 50.0% (1) | 50.0% (1) | 0.0% (0) | 2.5% (2) |
| Coronavirus | 0.0% (0) | 100% (2) | 0.0% (0) | 0.0% (0) | 2.5% (2) |
| Enterovirus | 50.0% (2) | 0.0% (0) | 25.0% (1) | 25.0% (1) | 5.1% (4) |
|
| |||||
| Total | 45.6% (36) | 35.4% (28) | 6.3% (5) | 12.7% (10) | 100% (79) |
Results are shown as the percentage with virus detected (n).
MLr detection
The MP PCR-positive samples were further screened for point mutations known to confer MLr, and they were detected in 11 samples (8%, 10 patients), showing four different types of mutation, three of them in region 1 and one in region 2: A2063G (N = 7), A2063T (N = 1), A2064G (N = 2) (region 1) and C2617A (N = 1) (region 2).
Among the resistant strains, the most frequent MLVA type was 4-5-7-2 (N = 8), followed by 3-5-6-2 (N = 3), and all of them were detected between 2014 and 2016. All the resistant strains detected were linked to a previous treatment with MLs. In six cases, patients developed the resistance during the treatment of the studied episode, with a specimen before treatment being susceptible to MLs and having the same MLVA profile as the after-treatment and resistant specimen. In four cases, resistant strains were collected after ML treatment during the ongoing or a previous CAP episode. The last case was a patient whose brother previously had CAP caused by MP and was subsequently treated with MLs.
Discussion
M. pneumoniae is known to cause between 20% and 40%1 of CAP in children and young adults, with increasing resistance to MLs,13–20 which are the first-line treatment. Prevalence of MP CAP in our region has not been widely studied yet, and, to the best of our knowledge, this is the first study to document the MLVA types circulating in our area as well as the MLr prevalence. During the study period, a total of 137 (16.7%) specimens were positive and could be further analysed for MLVA typing and MLr detection. The proportion of positive samples detected is similar to that in other European countries.21,30
During our study, a higher number of positive cases was detected during 2014 and 2015 (Figure 1a), probably related to a higher number of specimens processed. In accordance with similar studies found in the literature,21 the highest proportion of positive cases was in children older than 5 years (N = 73).
Culturing MP is usually performed in reference centres. Given the prompt presence of IgM antibodies, especially in children, serological tests are the most often applied methods for MP diagnosis. Different techniques have been used, such as complement fixation tests, enzyme immunoassays or passive agglutination with latex or gelatin particles, which have been extensively reviewed.1 Usually, two samples (from initial and convalescent phases) are required to demonstrate the seroconversion and to increase the diagnostic sensitivity, although some authors have found that using only an acute phase sample could lead to a diagnosis in up to 50% of cases.31,32 In our comparison of serology and molecular methods, we found an overall agreement of 73% between both techniques during the acute phase (first serum sample positive, antibody titre ≥1/160). After the seroconversion of 13 patients, the overall agreement increased to 76%. Discrepancies between these methods can be explained due the nature of the techniques.33–35 Immunosuppressed patients or collecting the serology specimen too early during the acute phase could lead to false-negative results.36 On the other hand, in the event of a previous MP infection, IgM can be detected for months or even years.34,35 Kim et al.36 suggested to set the positivity cut-off to a titre ≥1/640 for the diagnosis of acute-phase CAP. In our study, in 61 out of the 72 serology-positive–PCR-negative cases, their antibody titres were really close to the cut-off value (22 with a titre of 1/160 with no increase during the convalescent phase and 46 cases with a titre of 1/320), which could indicate previous contact with MP and thus be false positives for the current acute infection. On the other hand, 17 patients had negative serology but positive molecular detection of MP. It has been reported that MP can colonize the nasopharynx for a long time without any clinical manifestation. Meyer Sauteur et al.37 detected MP DNA in 48% of healthy children tested as controls during their study, and Nilsson et al.33 were able to detect MP DNA for 7 weeks after the clinical symptoms, even though the patients were correctly treated with MLs. Further research would be necessary to clarify the impact of carriers in the community on infection transmission. Molecular techniques can be valuable tools for MP diagnosis, and further allow the study of epidemic clusters by performing molecular typing or the detection of resistance patterns. Thus, combining serological methods with molecular tools should improve MP diagnosis and avoid misinterpretation.
MLVA is a powerful genotyping tool that can be used to better distinguish among strains.38 The genotypes 4-5-7-2 and 3-5-6-2 were the most abundant, as has been described in other geographic areas such as Europe,39 the USA40 and China.10 Furthermore, those genotypes were seasonal, with 4-5-7-2 mainly detected during summer whereas 3-5-6-2 was mainly circulating during winter and spring. Minority genotypes were evenly distributed within seasons and the years of the study.
Clinical aspects in MP pneumonia can be subtle and shared with other viral or bacterial entities. However, some clinical findings or an altered X-ray test, showing only bilateral pneumonia (20% of cases), could help in its diagnosis.1,41 Thus, the most reliable diagnostic is direct or indirect pathogen detection. Respiratory viruses were also tested for in patients with a confirmed MP pneumonia. In most cases we detected multiple viral coinfections, which complicated distinguishing whether the observed clinical symptoms were due to the virus or MP. This has been observed before, as some studies have shown that different viruses, such as Rhinovirus, could be found in children without any symptoms;42 thus their detection should be taken into consideration with some caution. In our study, this virus was the most frequently detected. However, it must be kept in mind that infections with other viruses such as Parainfluenza or Respiratory Syncytial Virus could be related to the clinical symptoms shared with MP.
In our study, no correlation was found between the MLVA type and the different symptomatology, in contrast to what was described by Yan et al.,43 where the MLVA was linked to the clinical outcome, although this group also found that some MLVA types, irrespective of their susceptibility profile, showed a higher risk of progression to severe MP pneumonia.
MLs are the first-line treatment for children with MP pneumonia in our country.44 Fluoroquinolones and tetracyclines are generally avoided in this population due to their side effects,45 although countries with high rates of ML-resistant strains (such as Japan) have approved tosufloxacin or minocycline as second-line drugs for children with MP pneumonia who respond poorly to MLs.46 Several studies reporting MLr have been published. The first resistant strains were detected in Asia and were linked to broad ML use,47 with resistance rates that vary between 13% and 100% in China19,20,48–50 or Japan.8,51–53 Similarly, antibiotic pressure seems to be the cause of the high resistance rate detected in Italy (25.6%).26 Lower resistance rates have been described in other European countries, such as 9.8% in France,13 9.3% in England,23 1.2% in Germany24 and 1% in Slovenia.21 Interestingly, no resistant strains were detected in northern countries, such as Sweden.25
Eight percent of the strains tested had a point substitution in domain V of the 23S rRNA gene that conferred resistance to MLs. Interestingly, in 6 out of 11 patients, a shift to a resistant pattern was detected from the initial to the second sample collected for the clinical follow-up after ML treatment. It is important to highlight that one of the resistant cases was resistant only after two doses of MLs, as the treatment had to be interrupted due to an allergic event. MLVA typing confirmed that it was probably the same strain and not a reinfection with an already resistant strain. Given that the MLr found in vivo is mainly linked to point mutations in domain V of the 23S rRNA,45 only that region was analysed. However, the real impact on the MLr phenotype of mutation on L4 and L22 ribosomal proteins remains unclear,4,9 and it should be addressed with further studies.
The rising incidence of ML-resistant strains favoured by a previous ML treatment is a significant finding. Interestingly, symptom clearance was observed in all ML-resistant CAP cases even after maintaining ML as antibiotic therapy.
This study had several limitations. First, sera and nasopharyngeal aspirate were simultaneously collected for MP serological and molecular diagnosis, respectively. Thus, when the CAP was suspected to be caused by a virus, there were no sera available to perform a serological diagnosis of MP, which would have given a more accurate comparison between the two techniques. Secondly, even though the number of specimens investigated was high (N = 821), the final number of specimens positive for MP was relatively small for statistical purposes (N = 137). Thirdly, this study only covers a small area of Spain. It would be interesting to perform a multicentre study to include several regions of our country in order to corroborate our findings, preferably during an extended period to determine the occurrence of infection peaks in our area and to confirm the distribution of the observed MLVA types.
In conclusion, we have described the first evaluation of M. pneumoniae MLVA types circulating in our area. The most abundant types found were 4-5-7-2 and 3–5-6-2, with an MLr rate of 8%. Given that the clinical course of CAP usually resolves spontaneously, and MLs are extensively employed in children, it should be reconsidered whether the treatment is suitable in all cases, as it may unnecessarily increase MLr.
Acknowledgements
We thank Dr Elisa Martró for helping with the research project design and serology, and molecular biology technicians for their collaboration. Subsets of this work have been presented at the European Congresses of Clinical Microbiology and Infectious Diseases (ECCMID) in the years 2015 (P1150 and EV0883), 2016 (P0779) and 2017 (P15549).
Funding
This work was supported by a Health Research Funding—Carlos III Health Institute (FIS-ISCIII) grant (PI12/02298). The funding body had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Transparency declarations
None to declare.
References
- 1. Atkinson TP, Balish MF, Waites KB. Epidemiology, clinical manifestations, pathogenesis and laboratory detection of Mycoplasma pneumoniae infections. FEMS Microbiol Rev 2008; 32: 956–73. [DOI] [PubMed] [Google Scholar]
- 2. Cousin-Allery A, Charron A, de Barbeyrac B et al. Molecular typing of Mycoplasma pneumoniae strains by PCR-based methods and pulsed-field gel electrophoresis. Application to French and Danish isolates. Epidemiol Infect 2000; 124: 103–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dégrange S, Cazanave C, Charron A et al. Development of multiple-locus variable-number tandem-repeat analysis for molecular typing of Mycoplasma pneumoniae. J Clin Microbiol 2009; 47: 914–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pereyre S, Guyot C, Renaudin H et al. In vitro selection and characterization of resistance to macrolides and related antibiotics in Mycoplasma pneumoniae. Antimicrob Agents Chemother 2004; 48: 460–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Davies TA, Dewasse BE, Jacobs MR et al. In vitro development of resistance to telithromycin (HMR 3647), four macrolides, clindamycin, and pristinamycin in Streptococcus pneumoniae. Antimicrob Agents Chemother 2000; 44: 414–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Prunier AL, Malbruny B, Tande D et al. Clinical isolates of Staphylococcus aureus with ribosomal mutations conferring resistance to macrolides. Antimicrob Agents Chemother 2002; 46: 3054–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pereyre S, Gonzalez P, De Barbeyrac B et al. Mutations in 23S rRNA account for intrinsic resistance to macrolides in Mycoplasma hominis and Mycoplasma fermentans and for acquired resistance to macrolides in M. hominis. Antimicrob Agents Chemother 2002; 46: 3142–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Matsuoka M, Narita M, Okazaki N et al. Characterization and molecular analysis of macrolide-resistant Mycoplasma pneumoniae clinical isolates obtained in Japan. Antimicrob Agents Chemother 2004; 48: 4624–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu X, Jiang Y, Chen X et al. Drug resistance mechanisms of Mycoplasma pneumoniae to macrolide antibiotics. Biomed Res Int 2014; 2014: 320801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao F, Li J, Liu J et al. Antimicrobial susceptibility and molecular characteristics of Mycoplasma pneumoniae isolates across different regions of China. Antimicrob Resist Infect Control 2019; 8: 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lucier TS, Heitzman K, Liu SK et al. Transition mutations in the 23S rRNA of erythromycin-resistant isolates of Mycoplasma pneumoniae. Antimicrob Agents Chemother 1995; 39: 2770–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wolff BJ, Thacker WL, Schwartz SB et al. Detection of macrolide resistance in Mycoplasma pneumoniae by real-time PCR and high-resolution melt analysis. Antimicrob Agents Chemother 2008; 52: 3542–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peuchant O, Menard A, Renaudin H et al. Increased macrolide resistance of Mycoplasma pneumoniae in France directly detected in clinical specimens by real-time PCR and melting curve analysis. J Antimicrob Chemother 2009; 64: 52–8. [DOI] [PubMed] [Google Scholar]
- 14. Li X, Atkinson TP, Hagood J et al. Emerging macrolide resistance in Mycoplasma pneumoniae in children: detection and characterization of resistant isolates. Pediatr Infect Dis J 2009; 28: 693–6. [DOI] [PubMed] [Google Scholar]
- 15. Yoo SJ, Kim HB, Choi SH et al. Differences in the frequency of 23S rRNA gene mutations in Mycoplasma pneumoniae between children and adults with community-acquired pneumonia: clinical impact of mutations conferring macrolide resistance. Antimicrob Agents Chemother 2012; 56: 6393–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Spuesens EB, Hoogenboezem T, Sluijter M et al. Macrolide resistance determination and molecular typing of Mycoplasma pneumoniae by pyrosequencing. J Microbiol Methods 2010; 82: 214–22. [DOI] [PubMed] [Google Scholar]
- 17. Spuesens EB, Meijer A, Bierschenk D et al. Macrolide resistance determination and molecular typing of Mycoplasma pneumoniae in respiratory specimens collected between 1997 and 2008 in The Netherlands. J Clin Microbiol 2012; 50: 1999–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chan KH, To KK, Chan BW et al. Comparison of pyrosequencing, Sanger sequencing, and melting curve analysis for detection of low-frequency macrolide-resistant Mycoplasma pneumoniae quasispecies in respiratory specimens. J Clin Microbiol 2013; 51: 2592–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou Z, Li X, Chen X et al. Macrolide-resistant Mycoplasma pneumoniae in adults in Zhejiang, China. Antimicrob Agents Chemother 2015; 59: 1048–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Y, Ye X, Zhang H et al. Characterization of macrolide resistance in Mycoplasma pneumoniae isolated from children in Shanghai, China. Diagn Microbiol Infect Dis 2010; 67: 355–8. [DOI] [PubMed] [Google Scholar]
- 21. Kogoj R, Mrvic T, Praprotnik M et al. Prevalence, genotyping and macrolide resistance of Mycoplasma pneumoniae among isolates of patients with respiratory tract infections, Central Slovenia, 2006 to 2014. Euro Surveill 2015; 20: pii=30018. [DOI] [PubMed] [Google Scholar]
- 22. Pereyre S, Touati A, Petitjean-Lecherbonnier J et al. The increased incidence of Mycoplasma pneumoniae in France in 2011 was polyclonal, mainly involving M. pneumoniae type 1 strains. Clin Microbiol Infect 2013; 19: E212–7. [DOI] [PubMed] [Google Scholar]
- 23. Brown RJ, Macfarlane-Smith L, Phillips S et al. Detection of macrolide resistant Mycoplasma pneumoniae in England, September 2014 to September 2015. Euro Surveill 2015; 20: 30078. [DOI] [PubMed] [Google Scholar]
- 24. Dumke R, von Baum H, Luck PC et al. Occurrence of macrolide-resistant Mycoplasma pneumoniae strains in Germany. Clin Microbiol Infect 2010; 16: 613–6. [DOI] [PubMed] [Google Scholar]
- 25. Gullsby K, Bondeson K. No detection of macrolide-resistant Mycoplasma pneumoniae from Swedish patients, 1996–2013. Infect Ecol Epidemiol 2016; 6: 31374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chironna M, Sallustio A, Esposito S et al. Emergence of macrolide-resistant strains during an outbreak of Mycoplasma pneumoniae infections in children. J Antimicrob Chemother 2011; 66: 734–7. [DOI] [PubMed] [Google Scholar]
- 27. Dumke R, Jacobs E. Culture-independent multi-locus variable-number tandem-repeat analysis (MLVA) of Mycoplasma pneumoniae. J Microbiol Methods 2011; 86: 393–6. [DOI] [PubMed] [Google Scholar]
- 28. Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 1999; 27: 573–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Diaz MH, Winchell JM. The evolution of advanced molecular diagnostics for the detection and characterization of Mycoplasma pneumoniae. Front Microbiol 2016; 7: 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meyer Sauteur PM, Bleisch B, Voit A et al. Survey of macrolide-resistant Mycoplasma pneumoniae in children with community-acquired pneumonia in Switzerland. Swiss Med Wkly 2014; 144: w14041. [DOI] [PubMed] [Google Scholar]
- 31. Talkington DF, Shott S, Fallon MT et al. Analysis of eight commercial enzyme immunoassay tests for detection of antibodies to Mycoplasma pneumoniae in human serum. Clin Diagn Lab Immunol 2004; 11: 862–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Matas L, Dominguez J, De Ory F et al. Evaluation of Meridian ImmunoCard Mycoplasma test for the detection of Mycoplasma pneumoniae-specific IgM in paediatric patients. Scand J Infect Dis 1998; 30: 289–93. [DOI] [PubMed] [Google Scholar]
- 33. Nilsson AC, Bjorkman P, Persson K. Polymerase chain reaction is superior to serology for the diagnosis of acute Mycoplasma pneumoniae infection and reveals a high rate of persistent infection. BMC Microbiol 2008; 8: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Copete AR, Vera C, Herrera M et al. Mycoplasma pneumoniae in children with and without community-acquired pneumonia. What do PCR and serology say? Pediatr Infect Dis J 2020; doi:10.1097/INF.0000000000002636. [DOI] [PubMed] [Google Scholar]
- 35. Wang L, Feng Z, Zhao M et al. A comparison study between GeXP-based multiplex-PCR and serology assay for Mycoplasma pneumoniae detection in children with community acquired pneumonia. BMC Infect Dis 2017; 17: 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim NH, Lee JA, Eun BW et al. Comparison of polymerase chain reaction and the indirect particle agglutination antibody test for the diagnosis of Mycoplasma pneumoniae pneumonia in children during two outbreaks. Pediatr Infect Dis J 2007; 26: 897–903. [DOI] [PubMed] [Google Scholar]
- 37. Meyer Sauteur PM, Krautter S, Ambroggio L et al. Improved diagnostics help to identify clinical features and biomarkers that predict Mycoplasma pneumoniae community-acquired pneumonia in children. Clin Infect Dis 2019; doi:10.1093/cid/ciz1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Benitez AJ, Diaz MH, Wolff BJ et al. Multilocus variable-number tandem-repeat analysis of Mycoplasma pneumoniae clinical isolates from 1962 to the present: a retrospective study. J Clin Microbiol 2012; 50: 3620–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun H, Xue G, Yan C et al. Multiple-locus variable-number tandem-repeat analysis of Mycoplasma pneumoniae clinical specimens and proposal for amendment of MLVA nomenclature. PLoS One 2013; 8: e64607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Diaz MH, Benitez AJ, Winchell JM. Investigations of Mycoplasma pneumoniae infections in the United States: trends in molecular typing and macrolide resistance from 2006 to 2013. J Clin Microbiol 2015; 53: 124–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Waites KB, Talkington DF. Mycoplasma pneumoniae and its role as a human pathogen. Clin Microbiol Rev 2004; 17: 697–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. El Kholy AA, Mostafa NA, Ali AA et al. The use of multiplex PCR for the diagnosis of viral severe acute respiratory infection in children: a high rate of co-detection during the winter season. Eur J Clin Microbiol Infect Dis 2016; 35: 1607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yan C, Xue G, Zhao H et al. Molecular and clinical characteristics of severe Mycoplasma pneumoniae pneumonia in children. Pediatr Pulmonol 2019; 54: 1012–21. [DOI] [PubMed] [Google Scholar]
- 44. Moreno-Perez D, Andres Martin A, Tagarro Garcia A et al. Community acquired pneumonia in children: treatment of complicated cases and risk patients. Consensus statement by the Spanish Society of Paediatric Infectious Diseases (SEIP) and the Spanish Society of Paediatric Chest Diseases (SENP). An Pediatr (Barc) 2015; 83: 217 e1–11. [DOI] [PubMed] [Google Scholar]
- 45. Lee H, Yun KW, Lee HJ, Choi EH. Antimicrobial therapy of macrolide-resistant Mycoplasma pneumoniae pneumonia in children. Expert Rev Anti Infect Ther 2018; 16: 23–34. [DOI] [PubMed] [Google Scholar]
- 46. Mikasa K, Aoki N, Aoki Y et al. JAID/JSC Guidelines for the Treatment of Respiratory Infectious Diseases: the Japanese Association for Infectious Diseases/Japanese Society of Chemotherapy—The JAID/JSC Guide to Clinical Management of Infectious Disease/Guideline-preparing Committee Respiratory Infectious Disease WG. J Infect Chemother 2016; 22: S1–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bébéar C, Pereyre S, Peuchant O. Mycoplasma pneumoniae: susceptibility and resistance to antibiotics. Future Microbiol 2011; 6: 423–31. [DOI] [PubMed] [Google Scholar]
- 48. Zhao F, Liu G, Wu J et al. Surveillance of macrolide-resistant Mycoplasma pneumoniae in Beijing, China, from 2008 to 2012. Antimicrob Agents Chemother 2013; 57: 1521–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ho PL, Law PY, Chan BW et al. Emergence of macrolide-resistant Mycoplasma pneumoniae in Hong Kong is linked to increasing macrolide resistance in multilocus variable-number tandem-repeat analysis type 4-5-7-2. J Clin Microbiol 2015; 53: 3560–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhao F, Liu J, Shi W et al. Antimicrobial susceptibility and genotyping of Mycoplasma pneumoniae isolates in Beijing, China, from 2014 to 2016. Antimicrob Resist Infect Control 2019; 8: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Matsuda K, Narita M, Sera N et al. Gene and cytokine profile analysis of macrolide-resistant Mycoplasma pneumoniae infection in Fukuoka. BMC Infect Dis 2013; 13: 591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Okada T, Morozumi M, Tajima T et al. Rapid effectiveness of minocycline or doxycycline against macrolide-resistant Mycoplasma pneumoniae infection in a 2011 outbreak among Japanese children. Clin Infect Dis 2012; 55: 1642–9. [DOI] [PubMed] [Google Scholar]
- 53. Tanaka T, Oishi T, Miyata I et al. Macrolide-resistant Mycoplasma pneumoniae infection, Japan, 2008–2015. Emerg Infect Dis 2017; 23: 1703–6. [DOI] [PMC free article] [PubMed] [Google Scholar]