ABSTRACT
Myeloid cells, including antigen-presenting cells (APCs) and myeloid-derived suppressor cells (MDSCs) play opposing roles to orchestrate innate and adaptive immune responses during physiological and pathological conditions. We investigated the role of DNA methylation in regulating the transcription of inhibitory/suppressive molecules in myeloid suppressive cells (identified as CD33+HLA-DR–) in comparison to APCs. We selected a number of immune checkpoints (ICs), IC ligands, and immunosuppressive molecules that have been implicated in MDSC function, including PD-L1, TIM-3, VISTA, galectin-9, TGF-β, ARG1 and MMP9. We examined their mRNA expression levels, and investigated whether DNA methylation regulates their transcription in sorted myeloid cell subpopulations. We found that mRNA levels of PD-L1, TIM-3, TGF-β, ARG1 and MMP9 in CD33+HLA-DR– cells were higher than APCs. However, VISTA and galectin-9 mRNA levels were relatively similar in both myeloid subpopulations. CpG islands in the promoter regions of TGF-β1, TIM-3 and ARG1 were highly unmethylated in CD33+HLA-DR–cells, compared with APCs, suggesting that DNA methylation is one of the key mechanisms, which regulate their expression. However, we did not find differences in the methylation status of PD-L1 and MMP9 between CD33+HLA-DR– and APCs, suggesting that their transcription could be regulated via other genetic and epigenetic mechanisms. The promoter methylation status of VISTA was relatively similar in both myeloid subpopulations. This study provides novel insights into the epigenetic mechanisms, which control the expression of inhibitory/suppressive molecules in circulating CD33+HLA-DR– cells in a steady-state condition, possibly to maintain immune tolerance and haemostasis.
KEYWORDS: MDSC, epigenetics, DNA methylation, immunosuppression
Introduction
Myeloid cells play important roles in regulating innate and adaptive immune responses during physiological and pathological conditions, including infections, inflammation, autoimmunity and cancer [1]. Under the influence of certain cytokines and growth factors, monocytes can differentiate into macrophages or monocyte-derived dendritic cells (Mo-DCs). These cells act as antigen-presenting cells (APCs) as they express the major histocompatibility complex class II (MHC II) molecule, HLA-DR, and secrete molecules to activate adaptive immunity [2]. In certain pathological conditions, these myeloid cells act as a double-edged sword and play deleterious or beneficial roles. They can exert suppressive/pro-tumorigenic or inflammatory/anti-tumour functions [3,4].
Securely regulated myelopoiesis is disrupted during a vast array of pathological conditions ranging from infectious diseases, trauma, transplantation, autoimmunity and cancer, leading to the expansion of a heterogeneous population of myeloid cells. This population comprises of myeloid cells halted at various stages of maturation/differentiation with an immunosuppressive function, referred to as myeloid-derived suppressor cells (MDSCs) [1,3]. MDSCs account for approximately 0.5% of peripheral blood mononuclear cells (PBMCs) in healthy individuals [5], suggesting their potential role in maintaining immune tolerance. However, studies have reported that the number of circulating MDSCs increases by 10-folds in pathological conditions such as cancer [5].
Although the role of MDSCs has been extensively studied in cancer, there is accumulating evidence suggesting the involvement of MDSCs in regulating the immune response following organ transplantation, and during various infectious, inflammatory and autoimmune diseases [1,6]. For instance, MDSCs maintain immune tolerance in organ transplantation [7], while in cancer, traumatic conditions and infectious diseases, their role is deleterious as they exert suppressive and anti-proliferative effects on the adaptive immune response [1,6,8]. On the other hand, MDSCs play a protective role in autoimmunity by suppressing autoreactive T cell responses, and enhancing T regulatory cell (Treg) survival and function to dampen inflammation [4].
MDSCs facilitate immunosuppression via the production of immunosuppressive mediators, such as transforming growth factor-β (TGF-β), secretion of proteases, for example matrix metalloproteinases (MMPs), and the expression of suppressive enzymes, such as arginase-1 (ARG1), indoleamine 2,3-dioxygenase (IDO) and inducible nitric oxide synthase (iNOS) [1,5]. This in turn causes APC dysfunction, reduces T effector cell (Teff) activation, and enhances Treg recruitment, survival and function [1,9–11]. Additionally, activated MDSCs induce T cell cycle arrest by expressing ARG1 that catabolizes L-arginine, an essential amino acid required for T cell proliferation, and they also release high levels of nitric oxide [6,12]. By a similar phenomenon, activated MDSCs express high levels of IDO, which catabolize tryptophan, another essential amino acid, and causes starvation leading to T cell cycle arrest and anergy [13].
MDSCs can inhibit Teff functions through the expression of inhibitory molecules, for example programmed death-ligand 1 (PD-L1), which interacts with its receptor programmed death-1 (PD-1) on Teffs, causing inhibitory signals that subsequently halt Teff activation and proliferation [12]. There is also a potential association between upregulated levels of PD-L1 on monocytes and the inhibition of T cell immune responses during viral infections [14]. Furthermore, the expression of immune checkpoints (ICs), such as T-cell immunoglobulin and mucin-domain containing-3 (TIM-3) [15,16] and V-domain Ig-containing Suppressor of T-cell Activation (VISTA) [17], have been detected in myeloid suppressive cell subsets. The expression of galectin-9, ligand of TIM-3, has also been detected in MDSCs, and the involvement of TIM-3/galectin-9 pathway in the suppression of anti-tumour immune response, cancer progression and resistance to immunotherapy has been reported [18,19]. Moreover, MDSC expansion and the suppression of Th1 immune responses in an autoimmune disease model, can be driven by TIM-3/galectin-9 pathway [20]. VISTA expression on APCs and/or MDSCs has been associated with the suppression of T cell responses in tumour models and resistance to immunotherapy [17,21–23]. Based on animal models of autoimmunity, MDSCs regulate T cell activation via cell-cell contact or by the expression of molecules such as ARG1, PD-L1 and TGF-β [24]. MDSCs are recruited to the inflamed sites, where they accumulate and become activated to exert suppressive functions and inhibit the activation of autoreactive T cells [25]. However, the suppressive function of MDSCs could be compromised due to the predominance of inflammatory cytokines in the inflamed sites, which subsequently alter the phenotype and function of MDSCs [4].
Epigenetics encompass gene regulation without altering the DNA sequence via DNA methylation, histone post-translational modifications and non-coding microRNAs (miRNAs) [26]. These mechanisms maintain transcriptional homoeostasis via favouring gene transcription or gene silencing under normal physiological conditions [27]. Epigenetic modifications have not only been implicated in immune tolerance, cancer pathogenesis/progression and autoimmunity, but also in MDSC function [28–30]. Studies have shown that epigenetic modification in MDSCs can alter their functional characteristics [28]. We have recently shown that epigenetic modifications play important roles in the transcriptional regulation of MDSCs in the tumour microenvironment of colorectal cancer patients [31]. Importantly, we found that blockade of histone deacetylase (HDAC) led to downregulation of genes related to immunosuppression in polymorphonuclear MDSCs [31].
In this study, we investigated the involvement of DNA methylation in regulating the expression of inhibitory/suppressive molecules in two myeloid cell subpopulations under normal physiological condition. We sorted CD33+HLA-DR– myeloid cell populations, which potentially comprise of MDSCs, and CD33+HLA-DR+ myeloid APCs, from peripheral blood mononuclear cells (PBMC) of 10 healthy individuals. We selected a number of molecules that play key roles in the function of myeloid suppressive cells, including PD-L1, TIM-3, galectin-9, VISTA, ARG1, MMP9 and TGF-β, to examine their mRNA expression levels in the sorted myeloid cell subsets, and to investigate whether DNA methylation regulates their transcription. We found increased mRNA expression of ten-eleven translocation (TET) enzymes and reduced levels of DNA methyltransferase (DNMT3a) in CD33+HLA-DR– cells, compared with CD33+HLA-DR+ myeloid APCs, suggesting a potential role for DNA unmethylation in regulating gene transcription. In addition, we investigated the promoter methylation status for PD-L1, TIM-3, ARG1, MMP9, TGF-β, VISTA and galectin-9 using CpG methylation analysis.
Results
Levels of circulating CD33+HLA-DR+ and CD33+HLA-DR – myeloid cells
It has been reported that MDSCs circulate at low levels in healthy individuals [5]. First, we investigated the levels of circulating CD33+ myeloid cells in 10 healthy donors (median age 33, 8 Male & 2 Female). We found that the level of circulating CD33+HLA-DR+ cells was significantly higher than CD33+HLA-DR – cells (92.4 ± 1.0 vs 6.1 ± 0.92, P = 0.001, Figure 1(a)). CD33+HLA-DR– myeloid cell population can represent heterogeneous populations of cells including immature myeloid cells (IMCs; identified as CD33+HLA-DR–CD15–CD14–), granulocytic myeloid cells (GMCs; identified as CD33+HLA-DR–CD15+CD14–) and monocytic myeloid cells (MMCs; identified as CD33+HLA-DR–CD15–CD14+) [32,33]. We investigated the percentage of each of these cell subsets in CD33+HLA-DR– cells. We found that the relative percentage of circulating MMCs was the highest (46.3 ± 7.9), followed by GMCs (29.8 ± 6.6) and finally IMCs (20.9 ± 2.3) (Figure 1(b)).
Figure 1.
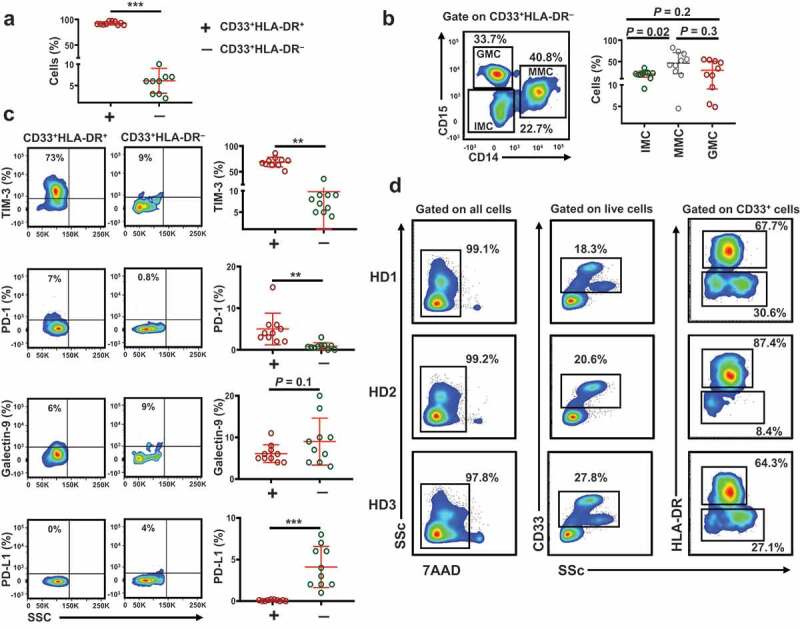
Levels of circulating CD33+HLA-DR+ and CD33+HLA-DR– myeloid cells and gating strategy for sorting
PBMC of 10 healthy donors were stained for CD33, HLA-DR, TIM-3, PD-1, galectin-9 and PD-L1. Scatter plot shows the levels of circulating CD33+HLA-DR+ and CD33+HLA-DR– cells (a). Representative flow cytometric and scatter plots show the levels of CD33+HLA-DR–CD15–CD14– immature myeloid cells (IMCs), CD33+HLA-DR–CD15–CD14+ monocytic myeloid cells (MMCs) and CD33+HLA-DR–CD15+CD14– granulocytic myeloid cells (GMCs) (b). Representative flow cytometric plots and scatter plots show the levels of circulating CD33+HLA-DR+ and CD33+HLA-DR– cells expressing TIM-3, PD-1, galectin-9 and PD-L1 (c). Representative flow cytometric plots from three donors show the gating strategy used to sort CD33+HLA-DR+ and CD33+HLA-DR– cells (D). Results obtained from 10 donors per myeloid cell subset, and expressed as mean ± SEM.
Next, we examined the expression levels of key ICs and IC ligands in the two myeloid subpopulations. We found that TIM-3 and PD-1 expression levels on CD33+HLA-DR+ cells were significantly higher than that of CD33+HLA-DR– cells (68.8 ± 2.9 vs 9.8 ± 2.9, P = 0.002, and 5.0 ± 1.2 vs 0.8 ± 0.2, P = 0.002, Figure 1(c)). In addition, there was a trend towards an increased level of galectin-9 expression on CD33+HLA-DR– cells, compared to CD33+HLA-DR+ cells (6.1 ± 2.1 vs 9.0 ± 1.7, P = 0.09, Figure 1(c)). The expression level of PD-L1 on CD33+HLA-DR– cells was significantly higher than that of CD33+HLA-DR+ cells (0.08 ± 0.02 vs 4.1 ± 0.78, P = 0.001, Figure 1(c)). Next, we sorted CD33+HLA-DR+ cells and CD33+HLA-DR– myeloid cells from the peripheral blood of 10 healthy donors to examine the mRNA expression of these ICs and IC ligands, in addition to other suppressive molecules, to investigate whether DNA methylation plays a role in their transcriptional regulation. The gating strategy employed for sorting is shown in Figure 1(d).
Genes encoding immune checkpoints, immune checkpoint ligands and suppressive molecules are upregulated in CD33+HLA-DR – myeloid cells
We examined the mRNA expression level of PD-L1, MMP9, galectin-9, TGF-β, TIM-3, ARG1 and VISTA mRNA in the two sorted myeloid cell subsets using RT-PCR. These molecules were selected due to their important roles in MDSC function. We found that PD-L1 (P = 0.007), MMP9 (P = 0.003), TGF-β (P = 0.003), TIM-3 (P = 0.04) and ARG1 (P = 0.009) mRNA expression levels were highly upregulated in CD33+HLA-DR– cells, compared with CD33+HLA-DR+ cells (Figure 2(a)). Galectin-9 and VISTA mRNA expression levels were comparable in both myeloid subpopulations (Figure 2(a)).
Figure 2.
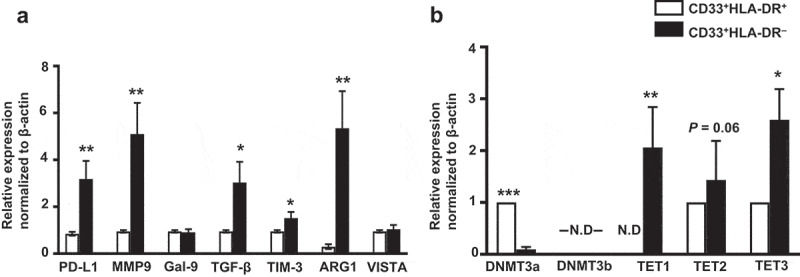
Relative gene expression of immune checkpoints, suppressive molecules, methyltransferases and demethylation enzymes in circulating CD33+HLA-DR– cells and antigen-presenting cells of healthy donors
The mRNA expression levels for PD-L1, MMP9, galectin-9, TGF-β1, TIM-3, ARG1 and VISTA in the sorted CD33+HLA-DR+ and CD33+HLA-DR– cells were determined by quantitative RT-PCR (a). The mRNA expression levels of DNMT3a, DNMT3b, TET1, TET2 and TET3 in CD33+HLA-DR+ and CD33+HLA-DR– cells were determined by quantitative RT-PCR (b). The relative gene expression was normalized to β-actin. Results obtained from 10 donors per myeloid cell subset, and expressed as mean ± SEM. N.D; not detected.
DNMT3a is downregulated and TET enzymes are upregulated in CD33+HLA-DR – myeloid cells
DNA methylation, mediated by DNA methyltransferases (DNMTs) such as DNMT3a and DNMT3b, induces transcriptional silencing and plays a major role in immune tolerance [30] and pathological conditions [34,35]. On the contrary, TET enzymes, including TET1, TET2, and TET3 [36], act as 5-methylcytosine oxidases to reverse DNA methylation and lead to transcriptional activation, for instance genes associated with cell growth and differentiation or immunosuppression [37–39]. It has been reported that promoter unmethylation/methylation could be maintained through the modulation of TETs and DNMTs [40]. This prompted us to investigate the gene expression of DNMT3a, DNMT3b and TET1, TET2 and TET3 enzymes in CD33+HLA-DR– and CD33+HLA-DR+ cells. Interestingly, we found that DNMT3a gene expression was significantly downregulated in CD33+HLA-DR– cells, compared to CD33+HLA-DR+ cells (P = 0.001, Figure 2(b)). However, DNMT3b gene expression was not detected in both myeloid subpopulations. TET1 gene was the most upregulated gene in CD33+HLA-DR– cells, followed by TET3 (P = 0.005 and P = 0.04, Figure 2(b)). Moreover, the mRNA level of TET2 was also high in CD33+HLA-DR– cells, compared to CD33+HLA-DR+ cells, but did not reach statistical significance (P = 0.06, Figure 2(b)). These data suggest that TET-mediated active demethylation could play a role in driving the upregulation of genes in CD33+HLA-DR– cells, compared to CD33+HLA-DR+ cells.
Galectin-9, TGF-β1, TIM-3 and ARG1 upregulation in CD33+HLA-DR – myeloid cells is associated with CpG promoter unmethylation
We then investigated the methylation status of PD-L1, MMP9, galectin-9, TGF-β1, TIM-3, ARG1 and VISTA promoters, in the two myeloid subpopulations. In order to investigate the promoter methylation profiles of ICs/ligands, we selected CpG islands in the promotor regions of PD-L1, TIM-3, and galectin-9 as we have previously described[39]. Additionally, we selected 5 CpG islands in the promoter regions of VISTA, 4 in ARG1, 12 in MMP9 and 16 in TGF-β1. We found that PD-L1 promoter was fully unmethylated and MMP9 promoter was moderately unmethylated in both myeloid cell subpopulations (Figure 3(a, b & h)). In contrast, galectin-9 promoter was highly unmethylated in CD33+HLA-DR– cells, compared to CD33+HLA-DR+ cells (P = 0.04, Figure 3(c) & (h)). TGF-β1 promoter showed a trend towards an increased percentage of unmethylation in CD33+HLA-DR – cells, but the data were not statistically significant (P = 0.08, Figure 3(d) & (h)). Moreover, TIM-3 and ARG1 promoters were significantly unmethylated in CD33+HLA-DR– cells (P = 0.006 and P = 0.04, Figure 3(e)–(f & h)). In contrast, VISTA promoter was equally unmethylated in both myeloid cell subpopulations (Figure 3(g) & (h)). These data suggest that DNA methylation is one of the key mechanisms regulating the transcription of TGF-β1, ARG1, TIM-3 and galectin-9 in CD33+HLA-DR– cells, while PD-L1 and MMP9 transcriptional regulation could be mediated by other epigenetic mechanisms. Interestingly, both myeloid subpopulations expressed VISTA at comparable levels and showed similar degrees of unmethylation within the promoter region.
Figure 3.
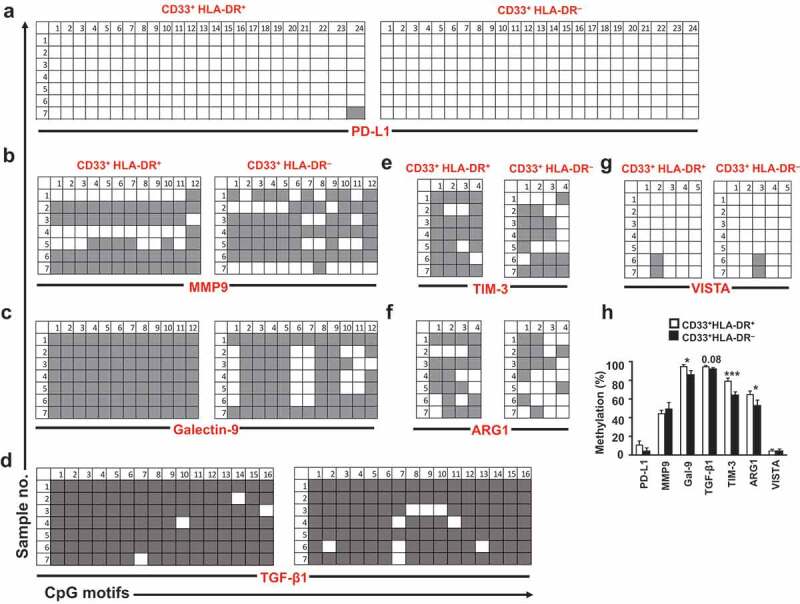
DNA methylation status of different genes in circulating CD33+HLA-DR– cells and antigen-presenting cells
Representative plots show the DNA methylation status in PD-L1 (a), MMP9 (b), galectin-9 (denoted as Gal-9) (c), TGF-β1 (d), TIM-3 (e), ARG1 (f) and VISTA (g) in the sorted CD33+HLA-DR+ and CD33+HLA-DR– cells as analysed by bisulphite sequencing of the gDNA. Methylation status of individual CpG motifs is shown in a colour-coded manner: white indicates unmethylation and grey indicates methylation. Bar plot shows the overall average methylation percentage of all genes investigated (h). Results obtained from 10 donors and expressed as mean ± SEM.
As shown in Figure 4(a) and (b), the unmethylation percentage of VISTA promoter was the highest in both myeloid subpopulations, followed by PD-L1, MMP9, ARG1, TIM-3, galectin-9 and TGF-β1. Additionally, we calculated the corrected unmethylation percentage by subtracting the unmethylation percentage of a particular promoter gene in CD33+HLA-DR+ cells from that of the corresponding CD33+HLA-DR– cells. TIM-3 showed the highest corrected percentage, followed by ARG1, galectin-9, PD-L1 and TGF-β1, while VISTA and MMP9 showed the least (Figure 4(c)).
Figure 4.
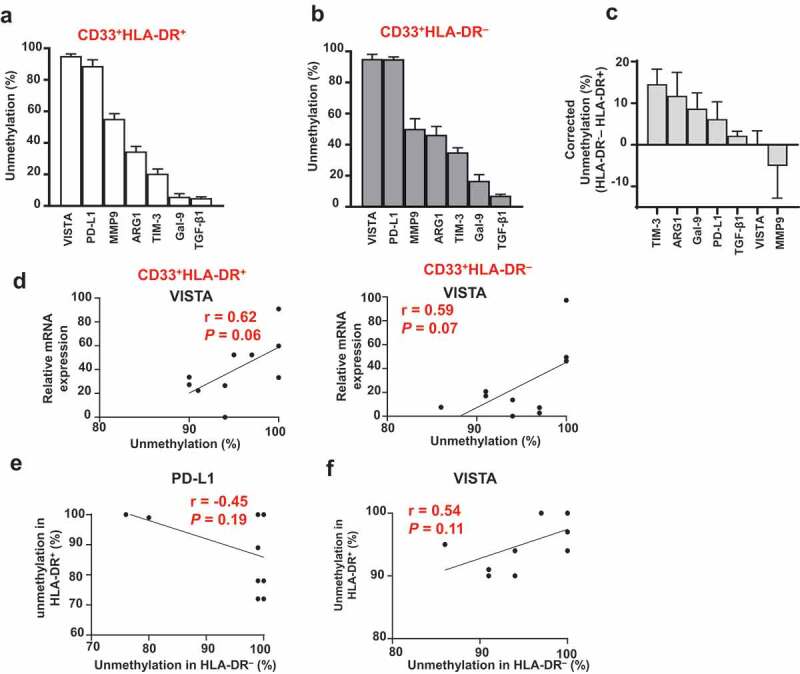
Hierarchical order of DNA methylation status and association with mRNA expression
Hierarchical order of DNA methylation status of different genes in circulating CD33+HLA-DR+ (a) and CD33+HLA-DR– cells (b). Corrected unmethylation percentage of different gene promoters showing the difference in unmethylation status between CD33+HLA-DR– cells and APCs (c). The association between DNA unmethylation status and relative mRNA expression of VISTA in CD33+HLA-DR+ and CD33+HLA-DR– cells (d). The association between PD-L1 promoter unmethylation status in CD33+HLA-DR+ and CD33+HLA-DR– cells (e). The association between VISTA promoter unmethylation status in CD33+HLA-DR+ and CD33+HLA-DR– cells (f). Results obtained from 10 donors and expressed as mean ± SEM.
Next, we investigated associations between mRNA expression and promoter DNA methylation status of a particular gene in the two myeloid subpopulations. We found a weak association between TIM-3, ARG1, galectin-9, PD-L1, MMP9 and TGF-β1 in both CD33+HLA-DR+ and CD33+HLA-DR– myeloid cells (data not shown). However, there was a moderate positive association between VISTA mRNA expression and promoter unmethylation in both myeloid subpopulations (P = 0.06 and P = 0.07, Figure 4(d)). In addition, we examined the association in gene promoter methylation status between CD33+HLA-DR+ and CD33+HLA-DR– myeloid cells. We found a weak association in the promoter unmethylation status of TIM-3, ARG1, galectin-9, MMP9 and TGF-β1 between CD33+HLA-DR+ and CD33+HLA-DR– myeloid cells (data not shown). Of note, PD-L1 promoter unmethylation in CD33+HLA-DR– cells was inversely correlated with that of CD33+HLA-DR+ cells (P = 0.19, Figure 4(e)), suggesting that transcriptional regulation of PD-L1 in both myeloid subpopulations is mediated via different mechanisms or that DNA methylation plays a distinct role in each subpopulation. On the other hand, VISTA promoter unmethylation in CD33+HLA-DR– cells was positively correlated with that of CD33+HLA-DR+ cells (P = 0.11, Figure 4(f)). This latter finding suggests that transcriptional regulation of VISTA in both myeloid subpopulations is mediated by DNA methylation in a similar manner.
Discussion
Cellular identity and functionality are determined by epigenetic modulations, including DNA methylation and post-translational histone modifications, which occur within the cellular environment. Aberrant modifications to the ‘normal’ epigenetic landscape of immune cells result in the disturbance of immune homoeostasis and lead to some significant clinical consequences [30]. MDSCs are one of the key immunoregulatory cells, which comprise of precursor and progenitor myeloid cells that ultimately differentiate into macrophages, granulocytes and dendritic cells upon their recruitment from the peripheral blood to organs/tissues. In physiological conditions, these immature precursor cells differentiate into mature dendritic cells, macrophages and granulocytes in peripheral lymphoid organs. However, a small proportion of these immature myeloid cells circulate at low levels in the peripheral blood of healthy individuals exhibiting a suppressive function. Consistent with this, we showed that the level of circulating CD33+HLA-DR– cells is significantly lower than CD33+HLA-DR+ cells in healthy donors. Moreover, it is noteworthy that due to the low levels of CD33+HLA-DR– cells in circulation, performing nucleic acid extractions from these cells was challenging and required additional amplification steps.
The suppressive capacity of CD33+HLA-DR– cells was indicated by the elevated level of ARG1 mRNA expression in these cells, compared to CD33+HLA-DR+ cells. Furthermore, we found that CD33+HLA-DR– cells express more PD-L1 and galectin-9 at the transcript (Figure 2(a)) and protein (Figure 1(b)) levels, suggesting their potentials to act as suppressive cells. The mRNA level of TIM-3 was higher in CD33+HLA-DR+ (Figure 2(b)), while the expression was lower in protein level (Figure 1(c)), compared with CD33+HLA-DR– cells. This discrepancy could be due to some other post-transcriptional modifications. It has been reported that many factors including RNA secondary structure, codon bias and amino acid usage, protein half-lives, ribosomal density and ribosome occupancy and regulatory RNAs could affect the translation of proteins [41]. Moreover, the correlation between mRNA and protein is reported to be ‘notoriously poor’ in biological samples [41].
We investigated the transcriptional regulation and DNA methylation of key genes involved in the suppressive function of myeloid cells including TGF-β1, ARG1 and MMP9, along with some ICs/ligands such as TIM-3, VISTA, galectin-9 and PD-L1. We found that mRNA expression levels of TGF-β1, TIM-3 and ARG1 were higher in CD33+HLA-DR– cells (potentially representing MDSCs) than APCs. CpG methylation analyses showed that the promoters of TGF-β1, TIM-3 and ARG1 genes were highly unmethylated in CD33+HLA-DR– cells, reflecting their increased expression at the transcript level. These data suggest that DNA methylation is one of the key epigenetic mechanisms, which controls the transcription of TGF-β1, TIM-3 and ARG1 genes in circulating CD33+HLA-DR– cells. On the other hand, PD-L1 and MMP9 showed a similar degree of unmethylation in both CD33+HLA-DR– and CD33+HLA-DR+ cells, which did not reflect RT-PCR data. This finding suggests that PD-L1 and MMP9 transcriptional regulation is mediated via different epigenetic mechanisms, perhaps histone post-translational modifications or microRNAs.
We then investigated the potential involvement of DNMTs and TETs in DNA methylation and regulating the transcription of MDSC-related genes and IC/ligands in CD33+HLA-DR– and CD33+HLA-DR+ cells. There are different isoforms of DNMTs, and amongst these are DNMT1, DNMT3a and DNMT3b [42,43]. DNMT1 is known to maintain DNA methylation pattern during replication, while DNMT3a/3b are responsible for the de novo methylation of DNA [42,43]. DNMT1 expression is essential for differentiated cells, and the expression pattern of DNMT isoforms in progenitor cells and differentiated cells is different [44]. Using RNA-Seq, we found that the expression of DNMT1 was higher in CD33+HLA-DR– cells, compared with CD33+HLA-DR+ cells (5.5 ± 0.3 vs. 4.8 ± 0.1, unpublished data). Moreover, we found the mRNA expression level of the DNMT3a was lower in CD33+HLA-DR–, compared with CD33+HLA-DR+ cells, while DNMT3b was not detected in both subsets (Figure 2). These results are consistent with other reports showing the opposite expression patterns of DNMT1 and DNMT3a/b [45–48]. Moreover, it has been shown that the silencing of both DNMT3a and DNMT3b upregulates DNMT1 as a compensatory mechanism [46]. In our data, we propose that the upregulation of DNMT1 can compensate the loss of DNMT3a, in addition to its role as a maintenance enzyme. In comparison to the low level of DNMT3a, we found that mRNA expression levels of TET1 and TET3 were significantly high in CD33+HLA-DR–. These data suggest that the upregulation of TGF-β1, TIM-3 and ARG1 in CD33+HLA-DR – could be due to the active demethylation of their promotors mediated by TET enzymes.
Although we showed that transcription of TGF-β1, ARG1 and TIM-3 in CD33+HLA-DR– cells is regulated by DNA methylation, this does not rule out the possible involvement of other epigenetic mechanisms. We found that galectin-9 mRNA level is relatively similar in CD33+HLA-DR– and CD33+HLA-DR+ cells. Conversely, this was not reflected in the CpG methylation analysis, which showed that galectin-9 promoter is highly unmethylated in CD33+HLA-DR– cells, compared to CD33+HLA-DR+ cells. This discrepancy may suggest that other mechanisms, such as histone modifications, are involved in the transcriptional regulation of galectin-9.
It is speculated that VISTA expression by APCs in a steady-state condition could act as a self-regulatory mechanism to inhibit T cell proliferation and activation, autoimmunity or an ongoing immune responses [23]. This might explain our results, which indicated that VISTA promoter is highly unmethylated in both CD33+HLA-DR+ and CD33+HLA-DR– cells, given that both subsets were isolated from the peripheral blood of healthy donors. This was also reflected in the RT-PCR data showing that VISTA mRNA expression level was comparable in both myeloid subpopulations. However, we hypothesize that the expression of VISTA in myeloid suppressive cells from cancer patients is higher than healthy donors based on pre-clinical and clinical data [21,22].
TGF-β released by myeloid suppressive cells can enhance the function and survival of Tregs, and suppress Teff proliferation [49]. Moreover, TGF-β also increases the suppressive function of MDSCs by upregulating the expression of immunosuppressive molecules such as iNOS, IL-10 and ARG1 [49]. In physiological conditions, mice lacking TGF-β1 develop inflammatory lesions in several organs as they age, suggesting the importance of TGF-β1 in regulating immune tolerance [50]. Mice lacking PD-L1 are susceptible to autoimmune diseases, suggesting the importance of PD-L1 in maintaining immune tolerance [51]. In vitro work showed that PD-L1 is expressed by APCs, in a steady-state condition, and serve as a self-regulatory mechanism to maintain haemostasis and suppress T cell activation [52].
The suppressive role of MDSCs and function of the molecules they express are well-studied in many pathological conditions (summarized in Figure 5(a) & (b)). ARG1+ MDSCs can downregulate CD3-ζ chain to alter T cell antigen recognition, and trigger T cell apoptosis via the production of NO and reactive oxygen species (ROS) [6,53,54]. TGF-β is an important mediator for MDSC generation [55,56], Treg differentiation and survival and epithelial-mesenchymal transition (EMT) [57,58]. TGF-β also influences protective immunity and inhibits the activation of Th1 response during bacterial infections [59] and chronic viral infections [59]. In autoimmunity and cancer settings, the activation of PD-1/PD-L1 and TIM-3/galectin-9 pathways results in the suppression of T cell immune responses associated with MDSC expansion [14,20,33,60]. In viral infection and cancer settings, VISTA+ MDSCs have been associated with T cell dysfunction and pro-tumorigenic effects [6,21,22]. Additionally, VISTA+ APCs were shown to be important for the regulation of inflammatory responses, including the expression of inflammatory cytokines [59]. MMP9 produced by MDSCs can also contribute to T cell dysfunction [61], tissue destruction and inflammation [62], as well as tumour invasion and metastasis [63]. Associations between MDSC expansion and the pathogenesis of various diseases are well-documented [4]. In cancer, several studies have shown correlations between MDSCs and poor disease prognosis. MDSCs exert their immunosuppressive effects through various molecules and pathways. The current FDA-approved or under development therapeutic agents target MDSCs through indirect approaches by targeting their suppressive mediators/molecules [33]. However, studies have shown that genes related to MDSC activity may be targeted by epigenetic modifiers, including histone deacetylases, to counter tumour growth and metastasis in various murine models [28]. In tumour models, 2′-deoxy-5-azacytidine (DNMT inhibitor) could downregulate percentage of MDSC and ARG1 expression within the TME, leading to favourable disease outcome [64]. We have recently reported that in CRC explant culture model, HDAC inhibitor treatment can downregulate ARG1 expression along with other myeloid chemotaxis markers including CCr2 and ITGAL [31]. In addition, pre-clinical and clinical studies have shown that combining epigenetic modifiers and immune checkpoint inhibitors is an effective anti-tumour therapy for various types of tumours [65].
Figure 5.
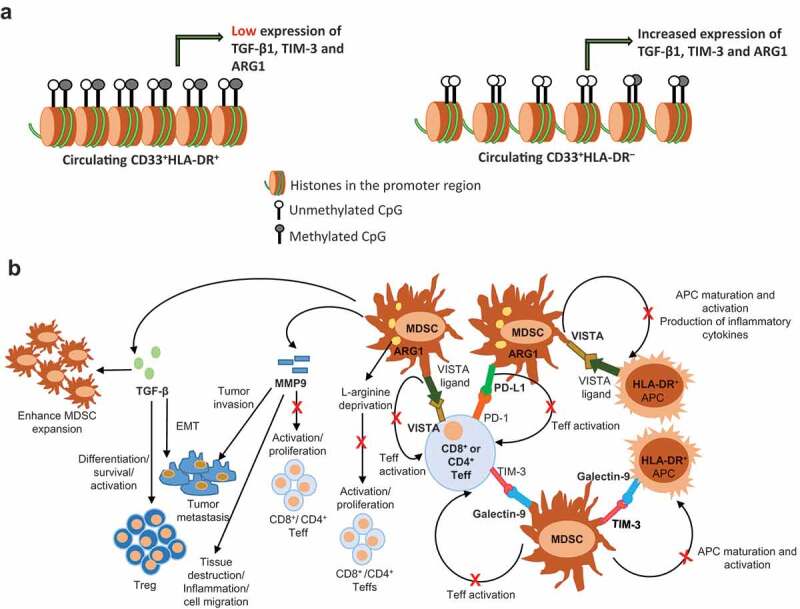
Epigenetic and suppressive mechanisms of MDSCs
DNA unmethylation alters chromatin compaction and increases the transcription of TGF-β1, TIM-3 and ARG1 in circulating CD33+HLA-DR– cells of healthy donors, compared to CD33+HLA-DR+ cells (a). Schematic summary for the suppressive mechanisms that could be mediated by CD33+HLA-DR– cells in pathological conditions (b). The interaction between PD-L1 on MDSCs and PD-1 on CD8+ or CD4+ Teffs restrains T cell activation. Similarly, the interaction between galectin-9 on MDSCs and TIM-3 on Teffs inhibits T cell activation, and may cause T cell apoptosis. Alternatively, the interaction between TIM-3 on MDSCs and galectin-9 on APCs may result in the inhibition of APC maturation and activation, and ultimately prevents antigen presentation and T cell activation. The binding of VISTA on MDSCs or Teffs to its ligand may also result in the inhibition of T cell activation, APC activation and production of inflammatory cytokines. Via other means, MDSCs express elevated levels of ARG1, which catabolizes L-arginine, and inhibits Teff proliferation. MDSCs release high levels of MMP9, which could suppress anti-tumour T cell responses, and promote tumour invasion and metastasis. In autoimmunity, MMP9 promotes inflammation by the generation of signals to trigger tissue destruction and recruit inflammatory cells into inflamed lesions. MDSCs also release high levels of TGF-β, which in turn enhance Treg differentiation, survival and function, and induce EMT to promote tumour metastasis. TGF-β also acts as an important mediator for MDSC expansion.
Our findings suggest that the transcription of TGF-β1, TIM-3, galectin-9 and ARG1 genes in circulating CD33+HLA-DR– cells can be regulated by DNA methylation, while PD-L1 and MMP9 gene transcription could be mediated via different epigenetic mechanisms. Our goal was to report the epigenetic control of these genes under steady state conditions in order to identify their regulatory mechanisms, which could be targeted in other physiological conditions in which these genes are elevated and contribute to disease progression. Our data provide novel insights into the epigenetic mechanisms, which control the expression of suppressive molecules in circulating CD33+HLA-DR– cells in a steady-state condition and provide potential rationale for using DNA hypermethylating agents (e.g. TET inhibitors) for counteracting the upregulation of these suppressive molecules in various disease conditions for therapeutic benefits. However, we did not perform intervention studies using epigenetic modifiers to investigate the effects on gene/protein expression. Further studies could be performed to examine whether DNA methylation regulates the transcription of the same investigated genes in circulating myeloid cells of patients with cancer, infectious or autoimmune diseases.
Materials and methods
PBMC isolation
PBMCs were isolated from peripheral blood samples from 10 healthy individuals by density gradient centrifugation using Histopaque-1077 (Sigma-Aldrich, Missouri, USA). PBMCs were frozen in cryovials at a density of 10 × 106 cells in 1 ml of freezing medium [(10% dimethylsulphoxide (DMSO; Sigma-Aldrich), 50% foetal calf serum (FCS; Hyclone, GE Healthcare Life Sciences, Utah, USA) and 40% RPMI-1640 medium (Life Technologies, New York, USA)], and stored in liquid nitrogen to be used in batches in subsequent experiments. This study was executed under ethical approval from Qatar Biomedical Research Institute, Doha, Qatar (protocol no. 2017-006).
Multi-parametric flow cytometry and cell sorting
PBMCs were washed with PBS and re-suspended in 100 µl flow cytometry staining buffer (PBS with 1% FCS and 0.1% sodium azide). Fc receptors (FcR) were first blocked using FcR Blocking Reagent (Miltenyi Biotech, Bergisch Gladbach, Germany). 7-AAD viability dye or Fixable Viability Dye eFluor™ 780 (eBioscience, San Diego, USA) was used to gate live cells. Cells were then stained with cell surface antibodies against CD33-FITC (clone HIM3-4; BD Biosciences, Oxford, UK), HLA-DR-PE (clone G46-6; BD Biosciences), TIM-3-BV711 (clone 7D3; BD Horizon), PD-1-PE-CF594 (clone EH12.1; BD Pharmingen), galectin-9-PerCP (clone 9M1-3; Biolegend) and PD-L1-APC (clone MIH1; BD Pharmingen), and incubated at 4°C for 30 min. Cells were then washed twice with flow cytometry staining buffer and data were acquired using BD LSRFortessa X-20 SORP flow cytometer (BD Biosciences).
For sorting, cells stained with 7-AAD, CD33 and HLA-DR were re-suspended in Pre-Sort buffer (BD Biosciences). Cell sorting was performed on BD FACSAria III SORP cell sorter with BD FACSDiva software (BD Biosciences). Applicable measures were taken to ensure minimal sorter-induced cell stress (SICS). Data analyses were performed on FlowJo V10 software (FlowJo, Ashland, USA).
DNA and RNA extraction
Total DNA and RNA were extracted from sorted, pure CD33+HLA-DR– and CD33+HLA-DR+ myeloid cell populations, isolated from ten healthy donors, using Norgen RNA/DNA/protein purification Plus Micro Kit (Norgen Bioteck Corporation, Ontario, Canada) according to the manufacture’s protocol. RNA was then amplified using 5X MessageAmp™ II aRNA Amplification Kit (Invitrogen). RNA concentrations were determined by Qubit RNA HS and Broad Range Assay Kits, respectively (Invitrogen).
Quantitative real-time PCR
Extracted RNA was reverse transcribed into cDNA using QuantiTect Reverse Transcription Kit (Qiagen). Real-time (RT)-PCR was performed using QuantStudio 6/7 Flex Real-time PCR system (Applied Biosystems, California, USA) for PD-L1, TIM-3, galectin-9, VISTA, TGF-β, MMP9, ARG1 and β-actin genes using PowerUp SYBR Green Master Mix (Applied Biosystems). All samples were assayed in duplicate. Quantification of relative gene expression was determined, using 2−ΔΔCT and normalized to β-actin. Sequences for the primers are shown in Supplementary Table 1a. The RT primers were all obtained from PrimerBank database (https://pga.mgh.harvard.edu/primerbank/).
CpG methylation analysis by bisulphite sequencing
Extracted DNA from sorted myeloid cells was subjected to bisulphite treatment via EZ DNA Methylation Gold Kit (Zymo Research, California, USA), followed by PCR amplification using TaKaRa Taq polymerase (TaKaRa Bio Inc., Shiga Prefecture, Japan) for PD-L1, TIM-3, galectin-9, VISTA, ARG1, MMP9 and TGF-β1. The PCR products were cloned into pGEM-T-vector (Promega Corporation, Wisconsin, USA) using DNA Ligation Mighty Mix (TaKaRa Bio). Methylation primers were designed by MethPrimer database (https://www.urogene.org/methprimer/index1.html). Sequences are listed in Supplementary Table 1b. Seven colonies were picked from each sample and plasmid extraction was performed using GENEJET Plasmid miniprep Kit (Thermo Fisher Scientific). M13 reverse primer was used for Sanger sequencing (Supplementary Table 1c).
Sanger sequencing
Purified plasmid DNA samples were subjected to sequencing using 3130X Genetic Analyser (Applied Biosystems) as described previously [39]. Sequencing data were analysed using a software for Bisulphite Sequencing DNA Methylation Analysis (BISMA, Jacobs University, Vegesack, Germany) and Quantification tool for Methylation Analysis (QUMA, RIKEN, Wako, Japan).
Statistical analyses
Statistical analyses were performed using GraphPad Prism 8 software (GraphPad Software, California, USA). Paired t-tests were performed on samples that passed the Shapiro-Wilk normality test, and Wilcoxon matched-pairs signed rank tests were performed for samples that did not show normal distribution. A P value of > 0.05 was considered statistically non-significant. The P values are represented as follows; ***P < 0.001, **P < 0.01, *P < 0.05. Data are presented as mean ± standard error of the mean (SEM).
Supplementary Material
Acknowledgments
We would like to thank the staff of the Genomics Core at Qatar Biomedical Research Institute for performing Sanger sequencing. The publication of this article was funded by the Qatar National Library.
Funding Statement
This work was supported by a start-up grant [VR04] for Professor Eyad Elkord from Qatar Biomedical Research Institute, Qatar Foundation.
List of abbreviations
| APC | antigen-presenting cells |
| ARG1 | arginase-1 |
| DNMT | DNA methyltransferase |
| EMT | epithelial-mesenchymal transition |
| GMC | granulocytic myeloid cells |
| IMC | immature myeloid cells |
| MDSC | myeloid-derived suppressor cells |
| MMC | monocytic myeloid cells |
| MMP9 | Matrix metalloproteinase-9 |
| PBMC | peripheral blood mononuclear cells |
| PD-L1 | programmed cell death-ligand1 |
| ROS | reactive oxygen species |
| Teff | T effector cell |
| TET | ten-eleven translocation |
| TGF-β | transforming growth factor-β |
| TIM-3 | T-cell immunoglobulin and mucin-domain containing-3 |
| TME | tumor microenvironment |
| Treg | T regulatory cell |
| VISTA | V-domain Ig-containing Suppressor of T-cell Activation |
Disclosure statement
The authors declare no conflicts of interest.
Complying with Ethics of experimentation
This study was executed under ethical approval from Qatar Biomedical Research Institute, Doha, Qatar (protocol no. 2017-006).
Supplementary material
Supplemental data for this article can be accessedhere.
References
- [1].Gabrilovich DI, Nagaraj S.. Myeloid-derived suppressor cells as regulators of the immune system. Nat Rev Immunol. 2009;9(3):162–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ennaciri J, Girard D.. Immune system: maturation of myeloid cells. Methods Mol Biol. 2009;550:195–203. [DOI] [PubMed] [Google Scholar]
- [3].Awad RM, De Vlaeminck Y, Maebe J, et al. Turn back the time: targeting tumor infiltrating myeloid cells to revert cancer progression. Front Immunol. 2018;9:1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cripps JG, Gorham JD. MDSC in autoimmunity. Int Immunopharmacol. 2011;11(7):789–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Khaled YS, Ammori BJ, Elkord E. Myeloid-derived suppressor cells in cancer: recent progress and prospects. Immunol Cell Biol. 2013;91(8):493–502. [DOI] [PubMed] [Google Scholar]
- [6].O’Connor MA, Rastad JL, Green WR. The role of myeloid-derived suppressor cells in viral infection. Viral Immunol. 2017;30(2):82–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yang T, Li J, Li R, et al. Correlation between MDSC and immune tolerance in transplantation: cytokines, pathways and cell-cell interaction. Curr Gene Ther. 2019;19(2):81–92. [DOI] [PubMed] [Google Scholar]
- [8].Makarenkova VP, Bansal V, Matta BM, et al. CD11b + /Gr-1 + myeloid suppressor cells cause T cell dysfunction after traumatic stress. J Immunol. 2006;176(4):2085–2094. [DOI] [PubMed] [Google Scholar]
- [9].Lindau D, Gielen P, Kroesen M, et al. The immunosuppressive tumour network: myeloid-derived suppressor cells, regulatory T cells and natural killer T cells. Immunology. 2013;138(2):105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Schlecker E, Stojanovic A, Eisen C, et al. Tumor-infiltrating monocytic myeloid-derived suppressor cells mediate CCR5-dependent recruitment of regulatory T cells favoring tumor growth. J Immunol. 2012;189(12):5602–5611. [DOI] [PubMed] [Google Scholar]
- [11].Bernal-Estevez DA, Garcia O, Sanchez R, et al. Monitoring the responsiveness of T and antigen presenting cell compartments in breast cancer patients is useful to predict clinical tumor response to neoadjuvant chemotherapy. BMC Cancer. 2018;18(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Weber R, Fleming V, Hu X, et al. Myeloid-derived suppressor cells hinder the anti-cancer activity of immune checkpoint inhibitors. Front Immunol. 2018;9:1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Platten M, Wick W, Van den Eynde BJ. Tryptophan catabolism in cancer: beyond IDO and tryptophan depletion. Cancer Res. 2012;72(21):5435–5440. [DOI] [PubMed] [Google Scholar]
- [14].Meier A, Bagchi A, Sidhu HK, et al. Upregulation of PD-L1 on monocytes and dendritic cells by HIV-1 derived TLR ligands. AIDS. 2008;22(5):655–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Anderson AC, Anderson DE, Bregoli L, et al. Promotion of tissue inflammation by the immune receptor Tim-3 expressed on innate immune cells. Science. 2007;318(5853):1141–1143. [DOI] [PubMed] [Google Scholar]
- [16].Das M, Zhu C, Kuchroo VK. Tim-3 and its role in regulating anti-tumor immunity. Immunol Rev. 2017;276(1):97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gao J, Ward JF, Pettaway CA, et al. VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer. Nat Med. 2017;23(5):551–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Limagne E, Richard C, Thibaudin M, et al. Tim-3/galectin-9 pathway and mMDSC control primary and secondary resistances to PD-1 blockade in lung cancer patients. Oncoimmunology. 2019;8(4):e1564505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Goncalves Silva I, Yasinska IM, Sakhnevych SS, et al. The Tim-3-galectin-9 secretory pathway is involved in the immune escape of human acute myeloid leukemia cells. EBioMedicine. 2017;22:44–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dardalhon V, Anderson AC, Karman J, et al. Tim-3/galectin-9 pathway: regulation of Th1 immunity through promotion of CD11b+Ly-6G+ myeloid cells. J Immunol. 2010;185(3):1383–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Deng J, Li J, Sarde A, et al. Hypoxia-induced VISTA promotes the suppressive function of myeloid-derived suppressor cells in the tumor microenvironment. Cancer Immunol Res. 2019;7(7):1079–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang L, Jia B, Claxton DF, et al. VISTA is highly expressed on MDSCs and mediates an inhibition of T cell response in patients with AML. Oncoimmunology. 2018;7(9):e1469594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang L, Rubinstein R, Lines JL, et al. VISTA, a novel mouse Ig superfamily ligand that negatively regulates T cell responses. J Exp Med. 2011;208(3):577–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Crook KR. Role of myeloid-derived suppressor cells in autoimmune disease. World J Immunol. 2014;4(1):26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Li M, Zhu D, Wang T, et al. Roles of myeloid-derived suppressor cell subpopulations in autoimmune arthritis. Front Immunol. 2018;9:2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Putiri EL, Robertson KD. Epigenetic mechanisms and genome stability. Clin Epigenetics. 2011;2(2):299–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mazzio EA, Soliman KF. Basic concepts of epigenetics: impact of environmental signals on gene expression. Epigenetics. 2012;7(2):119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zhang C, Wang S, Liu Y, et al. Epigenetics in myeloid derived suppressor cells: a sheathed sword towards cancer. Oncotarget. 2016;7(35):57452–57463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lu Q. The critical importance of epigenetics in autoimmunity. J Autoimmun. 2013;41:1–5. [DOI] [PubMed] [Google Scholar]
- [30].Suarez-Alvarez B, Baragano Raneros A, Ortega F, et al. Epigenetic modulation of the immune function: a potential target for tolerance. Epigenetics. 2013;8(7):694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sasidharan Nair V, Saleh R, Toor SM, et al. Transcriptomic profiling disclosed the role of DNA methylation and histone modifications in tumor-infiltrating myeloid-derived suppressor cell subsets in colorectal cancer. Clin Epigenetics. 2020;12(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Toor SM, Syed Khaja AS, El Salhat H, et al. Myeloid cells in circulation and tumor microenvironment of breast cancer patients. Cancer Immunol Immunother. 2017;66(6):753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Toor SM, Elkord E. Therapeutic prospects of targeting myeloid-derived suppressor cells and immune checkpoints in cancer. Immunol Cell Biol. 2018;96(9):888–897. [DOI] [PubMed] [Google Scholar]
- [34].Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21(35):5427–5440. [DOI] [PubMed] [Google Scholar]
- [35].Sun B, Hu L, Luo Z-Y, et al. DNA methylation perspectives in the pathogenesis of autoimmune diseases. Clin Immunol. 2016;164:21–27. [DOI] [PubMed] [Google Scholar]
- [36].Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14(6):341–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Huang Y, Rao A. Connections between TET proteins and aberrant DNA modification in cancer. Trends Genet. 2014;30(10):464–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Elashi AA, Sasidharan Nair V, Taha RZ, et al. DNA methylation of immune checkpoints in the peripheral blood of breast and colorectal cancer patients. Oncoimmunology. 2019;8(2):e1542918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sasidharan Nair V, El Salhat H, Taha RZ, et al. DNA methylation and repressive H3K9 and H3K27 trimethylation in the promoter regions of PD-1, CTLA-4, TIM-3, LAG-3, TIGIT, and PD-L1 genes in human primary breast cancer. Clin Epigenetics. 2018;10(1):78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nair VS, Song MH, Ko M, et al. DNA demethylation of the Foxp3 enhancer is maintained through modulation of ten-eleven-translocation and DNA methyltransferases. Mol Cells. 2016;39(12):888–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583(24):3966–3973. [DOI] [PubMed] [Google Scholar]
- [42].Turek-Plewa J, Jagodzinski PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett. 2005;10(4):631–647. [PubMed] [Google Scholar]
- [43].Bestor TH. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. Embo J. 1992;11(7):2611–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Hopfer OJ, Komor M, Koehler IS, et al. Expression of DNMT isoforms is differentially associated with aberrant promotor methylation in MDS hematopoietic progenitor cells during lineage specific differentiation. Blood. 2006;108(11):2628. [Google Scholar]
- [45].Li S, Chiang T-C, Richard-Davis G, et al. DNA hypomethylation and imbalanced expression of DNA methyltransferases (DNMT1, 3A, and 3B) in human uterine leiomyoma. Gynecol Oncol. 2003;90(1):123–130. [DOI] [PubMed] [Google Scholar]
- [46].Gustafsson JR, Katsioudi G, Degn M, et al. DNMT1 regulates expression of MHC class I in post-mitotic neurons. Mol Brain. 2018;11(1):36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zhong W, Li B, Xu Y, et al. Hypermethylation of the micro-RNA 145 promoter is the key regulator for NLRP3 inflammasome-induced activation and plaque formation. JACC Basic Transl Sci. 2018;3(5):604–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mishra M, Kowluru RA. Epigenetic modification of mitochondrial DNA in the development of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2015;56(9):5133–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lee C-R, Kwak Y, Yang T, et al. Myeloid-derived suppressor cells are controlled by regulatory T cells via TGF-β during murine colitis. Cell Rep. 2016;17(12):3219–3232. [DOI] [PubMed] [Google Scholar]
- [50].Aoki CA, Borchers AT, Li M, et al. Transforming growth factor beta (TGF-beta) and autoimmunity. Autoimmun Rev. 2005;4(7):450–459. [DOI] [PubMed] [Google Scholar]
- [51].Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity. 2018;48(3):434–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Sato R, Imamura K, Sakata S, et al. Disorder of coagulation-fibrinolysis system: an emerging toxicity of anti-PD-1/PD-L1 monoclonal antibodies. J Clin Med. 2019;8(6):6. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Bogdan C. Nitric oxide and the immune response. Nat Immunol. 2001;2(10):907–916. [DOI] [PubMed] [Google Scholar]
- [54].Nagaraj S, Gupta K, Pisarev V, et al. Altered recognition of antigen is a mechanism of CD8+ T cell tolerance in cancer. Nat Med. 2007;13(7):828–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Budhwar S, Verma P, Verma R, et al. The Yin and Yang of myeloid derived suppressor cells. Front Immunol. 2018;9:2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Gonzalez-Junca A, Driscoll KE, Pellicciotta I, et al. Autocrine TGFβ is a survival factor for monocytes and drives immunosuppressive lineage commitment. Cancer Immunol Res. 2019;7(2):306–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Shvedova AA, Kisin ER, Yanamala N, et al. MDSC and TGFβ are required for facilitation of tumor growth in the lungs of mice exposed to carbon nanotubes. Cancer Res. 2015;75(8):1615–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Saleh R, Elkord E. Acquired resistance to cancer immunotherapy: role of tumor-mediated immunosuppression. Semin Cancer Biol. 2019. DOI: 10.1016/j.semcancer.2019.07.017 [DOI] [PubMed] [Google Scholar]
- [59].Sanjabi S, Oh SA, Li MO. Regulation of the immune response by TGF-β: from conception to autoimmunity and infection. Cold Spring Harb Perspect Biol. 2017;9(6):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Wegner A, Verhagen J, Wraith DC. Myeloid-derived suppressor cells mediate tolerance induction in autoimmune disease. Immunology. 2017;151(1):26–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Lee S-E, Lim J-Y, Kim TW, et al. Matrix metalloproteinase-9 in monocytic myeloid-derived suppressor cells correlate with early infections and clinical outcomes in allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2018;24(1):32–42. [DOI] [PubMed] [Google Scholar]
- [62].Ram M, Sherer Y, Shoenfeld Y. Matrix metalloproteinase-9 and autoimmune diseases. J Clin Immunol. 2006;26(4):299–307. [DOI] [PubMed] [Google Scholar]
- [63].Fleming V, Hu X, Weber R, et al. Targeting myeloid-derived suppressor cells to bypass tumor-induced immunosuppression. Front Immunol. 2018;9:398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Mikyskova R, Indrova M, Vlkova V, et al. DNA demethylating agent 5-azacytidine inhibits myeloid-derived suppressor cells induced by tumor growth and cyclophosphamide treatment. J Leukoc Biol. 2014;95(5):743–753. [DOI] [PubMed] [Google Scholar]
- [65].Terranova-Barberio M, Thomas S, Munster PN. Epigenetic modifiers in immunotherapy: a focus on checkpoint inhibitors. Immunotherapy. 2016;8(6):705–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


