ABSTRACT
Background
Colorectal cancer (CRC) is one of the most common digestive malignant tumors globally. Focally amplified lncRNA on chromosome 1 (FALEC) is a novel lncRNA that has been reported to be involved in many biological processes during carcinogenesis. However, its role in CRC remains poorly understood.
Methods
Gene expression at mRNA or protein level was measured by qRT-PCR or western blot, respectively. In vitro experiments including EdU, colony formation, flow cytometry, wound-healing and transwell assays, as well as in vivo xenograft experiment, were utilized to determine the functional role of FALEC in CRC. Relevant mechanical assays were performed to investigate the underlying molecular mechanism.
Results
FALEC was aberrantly up-regulated in CRC. FALEC knockdown could impair CRC cell proliferation, migration and invasion, whereas facilitate cell apoptosis. MiR-2116-3p was revealed to be sponged by FALEC. PIWIL1 was identified as the target of miR-2116-3p. Mechanically, FALEC restored the expression of PIWIL1 via absorbing miR-2116-3p. MiR-2116-3p inhibition and PIWIL1 enrichment could counteract the anti-tumor impact induced by silenced FALEC on the oncogenic behaviors of CRC cells.
Conclusion
Our study revealed that FALEC promoted CRC progression via restoring the expression of miR-2116-3p-targeted PIWIL1, suggesting the potential application of targeting FALEC in the treatment of CRC.
KEYWORDS: FALEC, miR-2116-3p, PIWIL1, colorectal cancer
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors in digestive system with a steadily rising incidence in United States.1 Notably, approximately 655,000 new cases are diagnosed each year. Although the five-year overall survival rate of patients with early CRC is nearly 90%, it decreases to 15% in those with metastases, especially metastasis in lymph nodes and major organs.2 So far, few research has revealed the molecular mechanism behind CRC carcinogenesis. Therefore, in-depth studies are required for exploring CRC pathogenesis, so as to provide theoretical supports for novel promising anti-tumor drugs targeting CRC.
Long non-coding RNAs (lncRNAs) are a class of non-coding transcripts longer than 200 nucleotides that participate in a wide array of physiological and pathological processes, such as cell proliferation, apoptosis, migration and invasion.3,4 LncRNAs have been identified as critical regulators in cancer biology through controlling gene expression via transcriptional or post-transcriptional manner.5 FALEC is a newly discovered intergenic non-coding RNA, which has been reported to exert oncogenic effects on the development and progression of prostate cancer (PCa).6 Yet its cellular role and potential mechanism in CRC remains largely obscure.
Notably, increasing lncRNAs have been revealed to act as competing endogenous RNAs (ceRNAs) to influence gene expression via sponging microRNAs (miRNAs) at post-transcriptional level, further affecting the initiation, development and progression of cancers.7,8 To date, a wide array of lncRNAs have been found to be dysregulated and exhibit oncogenic or anti-tumor properties in various cancers, including CRC. For instance, lncRNA NEAT1 has been found to play a carcinogenic role in CRC progression via regulating Akt signaling.9
MicroRNAs are a group of small non-coding RNAs (ncRNAs), and have been found to regulate disease development through mediating the silence of target genes at posttranscriptional level. The link between miRNAs and CRC has been uncovered. For instance, miRNA-708 has been found to act as a tumor suppressor in CRC by targeting ZEB1 through Akt/mTOR pathway.10 In this study, bioinformatics analysis predicted miR-2116-3p could interact with FALEC in CRC. Nevertheless, its function and potential downstream target are not clear in CRC.
Therefore, the aim of this study was to explore whether FALEC engaged in the carcinogenesis and progression of CRC via participating in the ceRNA network, which might help provide novel targets for anti-tumor drug research for CRC.
Materials and methods
Cell lines and cell culture
Human CRC cell lines (SW480, LoVo, HCT8 and DiFi) and normal colorectal epithelial cell line (NCM460) were propagated in the DMEM (Gibco, Grand Island, NY) with 10% FBS (Gibco) and 1% Pen/Strep solution under 5% CO2 and 37°C. All cell lines were procured from ATCC (Manassas, VA) and authenticated via STR profiling.
Cell transfection
Short hairpin RNAs (shRNAs) against FALEC and the negative control (NC) sh-NC were constructed by Genepharma (Shanghai, China), and then inserted into the Tet-pLKO-Puro plasmid (Addgene) to obtain the lentiviral particles as described11. Afterward, the lentiviral particles were transfected into LoVo and HCT8 cells via using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). The transfection duration was 2 days. At length, cells with stable transfections were collected after 48 h of selection via puromycin treatment. Meanwhile, miR-2116-3p mimics (5ʹ-CCUCCCAUGCCAAGAACUCCC-3ʹ), miR-2116-3p inhibitor (5ʹ-GGGAGUUCUUGGCAUGGGAGG-3ʹ), and corresponding negative controls NC mimics (5ʹ-CACACCCGCAUUACACCUCCG-3ʹ) and NC inhibitor (5ʹ-AGAUGUGGCGGGUCGGGAUUG-3ʹ) were also designed by Genepharma. To overexpress PIWIL1, the amplified cDNA sequence of PIWIL1 was cloned into pcDNA3.1 vector (Invitrogen) to obtain pcDNA3.1/PIWIL1, with the empty vector as the negative control. Then, the above plasmids were transfected into indicated CRC cells for 48 h with the aid of Lipofectamine 3000.
Quantificative real-time PCR (qRT-PCR) analysis
Total RNAs from LoVo and HCT8 cells (underwent 24 h of incubation) were obtained by TRIzol reagent (Invitrogen), and then converted into cDNA for quantitative PCR experiment performed using SYBR Green PCR Master Mix (Takara, Kyoto, Japan). Relative gene expression was standardized to GAPDH or U6, and fold-change was calculated by 2−ΔΔCT method. The primer sequences are shown in Supplementary Table 1.
Colony formation assay
Clonogenic LoVo and HCT8 cells in the 6-well plates were seeded at the density of 500 cells per well for 14 days. Colonies were then fixed in 4% paraformaldehyde and dyed with 0.5% crystal violet. Finally, colonies containing more than 50 cells were counted manually.
EdU staining assay
Cell proliferation was assessed by use of EdU kit (RiboBio, Guangzhou, China). After 48 h of transfection, LoVo and HCT8 cells were further cultured for 24 h and then put into the 24-well plates, followed by processing with 100 μL of EdU reagent (50 μM) for 2 hours. The nuclei were double-dyed with DAPI (Beyotime, Shanghai, China) after fixing cells by 4% paraformaldehyde. After that, EdU-positive cells were captured and analyzed under Laser confocal microscopy (Olympus, Tokyo, Japan).
Flow cytometry analysis of cell apoptosis
Following 24 h of incubation, LoVo and HCT8 cells (2 × 105) after transfection were collected and rinsed in phosphate-buffered saline (PBS) for re-suspending in the 500 μL of binding buffer. 10 μL of Annexin V-FITC and 10 μL of PI were added to the solution for 15 min of incubation, followed by analysis of the apoptotic cells via flow cytometry (BD Biosciences, San Jose, CA).
Wound-healing assay
After further incubation for 24 h after transfection, cells were collected and re-suspended, and then cell resuspension was added into 24-well plates (with 1 × 105 cells per well). After cells reaching 90% confluence, wounds were produced by 200 μl tip on the cell monolayer. Then, the wounds at 0 h and 24 h after produced were imaged using inverted light microscopy.
Cell invasion assay
After further incubation for 24 h after transfection, cells were collected and re-suspended, and then cell resuspension was plated into the upper Matrigel coated chamber (BD Biosciences), while the lower chamber was filled with 500 μl of culture medium containing 10% FBS. Following 24 h of incubation, cells on the lower filter were dyed with 0.5% crystal violet solution after fixation, and then observed with inverted microscope.
Tube formation assay
The transfected LoVo and HCT8 cells were cultured for 48 h, and then the culture medium was collected as conditional medium (CM) for incubating human umbilical vein endothelial cells (HUVECs, Corning Incorporated, N.Y., USA) which was seeded into 48-well plates in 5% CO2 at 37°C. The 48-well plates were solidified with 150 μL of Matrigel for 30 min. After incubation with indicated CM for 8 h, tubes formed by HUVECs were observed and analyzed under microscope.
Subcellular fractionation
Subcellular fractionation was performed in CRC cell lines to separate cell nucleus and cell cytoplasm in line with the protocol of PARIS™ Kit (Invitrogen). Expression levels of FALEC, U6 (nuclear control) and GAPDH (cytoplasmic control) were measured by qRT-PCR.
Fluorescence in situ hybridization (FISH) assay
The RNA FISH probe synthesized for recognizing FALEC was produced by RiboBio (Guangzhou, China) and used following the guidebook. After 48 h of incubation, cells were subjected to incubation with FALEC probes, counterstaining with DAPI and then visualization via Laser confocal microscopy.
Dual luciferase reporter assay
LoVo and HCT8 cells were prepared in the 48-well plates (5 × 103 cells per well) and co-transfected with the 100 ng of pmirGLO-PIWIL1 WT/MUT or pmirGLO-PIWIL1 WT/MUT luciferase reporter vectors, and miR-2116-3p mimics or NC mimics. 48 h later, luciferase activities were tested by Dual-Luciferase Reporter Assay System (Promega Corporation, Fitchburg, WI, USA). The relative activities were standardized to Renilla luciferase activity.
RNA pull down assay
The wild type or mutant FALEC sequence covering the putative miR-2116-3p binding sites were biotin-labeled into Biotin FALEC WT/MUT, and then incubated with cell lysates (lysed after 48 h of culturing) for 1 h. After adding beads with streptavidin, the pull-downs were acquired for qRT-PCR analysis.
RNA Immunoprecipitation (RIP)
RIP assay was carried out as per the protocol of EZ-Magna RIP RNA Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA). In brief, cells were lysed via RIP lysis buffer after 48 h of incubation. Then, cell lysates were treated with magnetic beads coupled with human Ago2 or control IgG antibody (Millipore). Finally, RNAs in immunoprecipitates were analyzed by qRT-PCR.
Western blot
Following 24 h of incubation after transfection, CRC cells were lysed via RIPA lysis buffer (Upstate Biotechnology, New York, USA) to collect total protein. After that, proteins were subjected to electrophoresis by 12% SDS-PAGE, and then shifted to PVDF membranes. Following sealing with 5% skimmed milk for 2 h, the samples were incubated with the primary antibodies against GAPDH (the loading control; ab181602, 1:10000 dilution) and PIWIL1 (ab12337, 1:500 dilution) all night, followed by processing with the secondary antibodies tagged with HRP (ab205718, 1:10000 dilution) for 1 h. All antibodies were procured from Abcam (Cambridge, MA).
In vivo experiment
Male nude mice, aged 6 weeks, were acquired commercially from the National Laboratory Animal Center (Beijing, China). All mice were housed at room temperature in SPF-grade lab with enough feed and water under 12 h of lighting every day. Three mice were placed in a cage and the cages were changed every 2 days. The animal experiment was conducted with the approval from the Animal Research Ethics Committee of Shanxi Cancer Hospital. 5 × 106 LoVo cells, underwent 24 h of incubation following transfection with sh-NC, sh-FALEC#1, or sh-FALEC#1+ pcDNA3.1/PIWIL1, were inoculated subcutaneously to mice (three mice per group). Tumor volume was examined every fourth day. Tumors were dissected from mice that were sacrificed after 28 days, and then the tumors were weighed.
Statistical analyses
Three biological repeats were included in all assays, and results were exhibited as the mean ± SD. GraphPad Prism 7.0 was used to process data analyses through t-test or One-way ANOVA, with p value less than 0.05 as the threshold of significance.
Results
Silencing FALEC could inhibit the malignant behaviors of CRC cells
performed qRT-PCR analysis to detect the expression pattern of FALEC in CRC cell lines (SW480, LoVo, HCT8, DiFi) and normal colorectal epithelial cell line (NCM460). Results showed an aberrant up-regulation of FALEC in CRC cell lines in contrast to NCM460 cells (Figure 1A). To determine the role of FALEC, we performed loss-of-function assays by knocking down FALEC expression in LoVo and HCT8 cells via using sh-FALEC#1/2. The outcomes of qRT-PCR demonstrated that the level of FALEC was nearly unaffected by transfection with sh-NC compared to the untreated mock groups, while FALEC expression was notably reduced in response to the transfection with sh-FALEC#1/2 relative to the sh-NC groups (Figure 1B). Colony formation assay showed that the number of colonies was significantly reduced after knocking down FALEC, whereas that in sh-NC transfected groups was similar to that in mock groups (Figure 1C). EdU assay outcomes further confirmed that silencing FALEC dampened the proliferation of LoVo and HCT8 cells, while transfecting with sh-NC had no apparent influence on cell proliferation (Figure 1D). Flow cytometry assay indicated that FALEC knockdown could significantly strengthen cell apoptosis ability, but the transfection with sh-NC could not (Figure 1E). Results of wound-healing and transwell assays showed that FALEC knockdown could inhibit the migration and invasion of LoVo and HCT8 cells, but transfecting with sh-NC had no evident impact on such processes (figure 1F-G). Meanwhile, we also wondered the impact of FALEC on angiogenesis, an important process for tumor growth and migration. Hence, tube formation assays were performed in human umbilical vein endothelial cells (HUVECs) incubated with the conditional medium (CM) of LoVo and HCT8 cells. The results demonstrated that the number of tube branches formed by HUVECs was obviously declined by transfecting sh-FALEC#1/2 but not sh-NC (Figure 1H). Importantly, based on the above data, we observed that compared with mock groups, the transfection of sh-NC had no effect on FALEC expression or CRC cell function, so we only applied groups with sh-NC as the negative controls subsequently.
Figure 1.
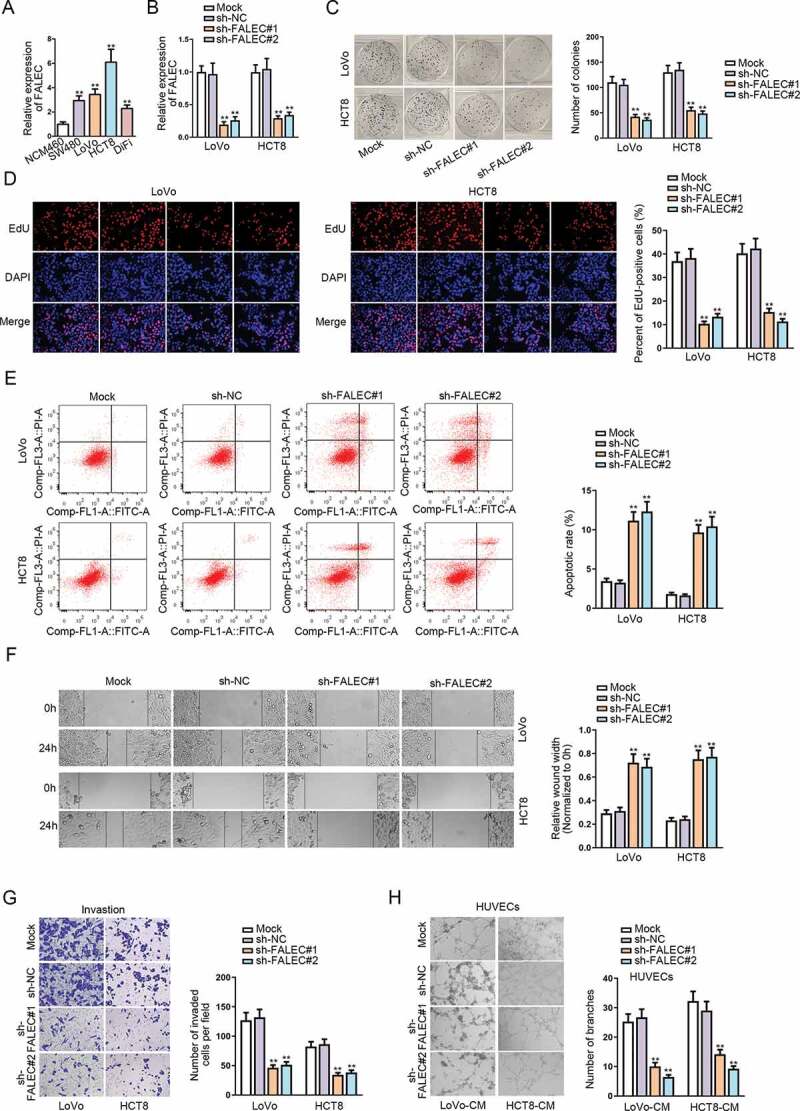
Silencing FALEC could inhibit cell proliferation, motility and angiogenesis in CRC
A. The expression of FALEC in CRC cell lines and normal NCM460 cells was evaluated by qRT-PCR analysis. B. The inhibitory efficiency of sh-FALEC#1/2 was guaranteed by qRT-PCR assay. C–D. Colony formation and EdU assays were implemented to investigate the proliferation ability of LoVo and HCT8 cells. E. Flow cytometry analysis was conducted to assess cell apoptosis rate. F–G. Wound-healing and transwell assays were conducted to determine cell migration and invasion abilities after FALEC knockdown. H. Tube formation assays were performed to determine the impact of FALEC knockdown on angiogenesis. **P < .01.
MiR-2116-3p could directly bind to FALEC
LncRNAs could play different roles depending on their sub-cellular distribution. We found that FALEC predominantly located in the cytoplasm of LoVo and HCT8 cells according to the results of subcellular fractionation assay (Figure 2A). FISH assay further confirmed the cytoplastic location of FALEC in these two CRC cells (Figure 2B). The findings highlighted the miRNA sponging role of FALEC in CRC. Therefore, we utilized DIANA database (http://carolina.imis.athena-innovation.gr/diana_tools/) to screen potential miRNAs that could bind to FALEC. As a result, eight potential miRNA candidates were screened out based on the condition: prediction score >0.85. Subsequent qRT-PCR analysis revealed that the expression of miR-2116-3p was enhanced most markedly in LoVo cells with FALEC inhibition (Figure 2C). Next, we examined an abnormal down-regulation of miR-2116-3p in CRC cells relative to NCM460 cells (Figure 2D), which was contrary to the expression status of FALEC in CRC. Then, the putative miR-2116-3p binding site in the sequence of FALEC was predicted by DIANA, and we mutated the putative sequence accordingly, as shown in Figure 2E. The outcomes of luciferase reporter assays manifested that miR-2116-3p mimics could significantly decrease the luciferase activity of pmirGLO vector containing FALEC-WT, while no effects on the luciferase activity of FALEC-MUT (Figure 2F). RNA pull down data demonstrated that miR-2116-3p was pulled down only by Bio-FALEC-WT (Figure 2G), which verified the physical binding between FALEC and miR-2116-3p in CRC. Together, these data indicated that FALEC directly bind to miR-2116-3p in CRC.
Figure 2.
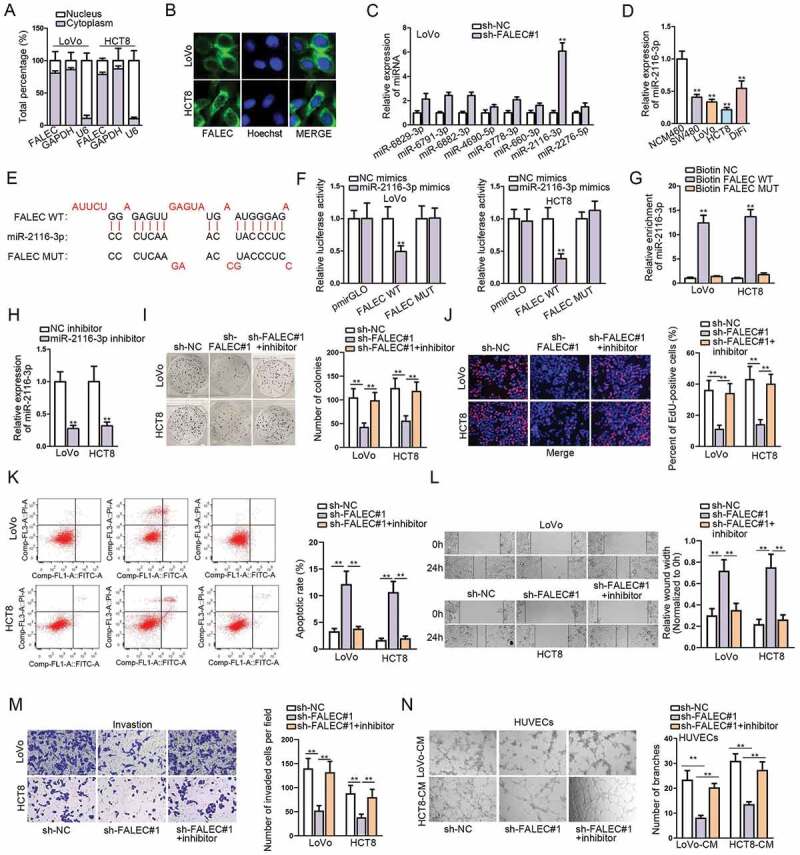
MiR-2116-3p could directly bind to FALEC
A–B. The subcellular localization of FALEC in LoVo and HCT8 cells was detected by subcellular fractionation and FISH assays. C. qRT-PCR analysis was performed to evaluate the expression of candidate miRNAs in LoVo cells after knockdown of FALEC. D. The expression of miR-2116-3p in CRC cell lines and normal NCM460 cells was detected by qRT-PCR. E. Putative binding sequences between miR-2116-3p and FALEC predicted by DIANA, as well as the sequence of FALEC-MUT with such binding sites mutated. F. Dual luciferase reporter assays demonstrated that miR-2116-3p overexpression successfully decreased the luciferase activity of FALEC-WT. G. RNA pull down assay showed that miR-2116-3p was preferentially enriched by biotin-labeled FALEC-WT. H. The expression of miR-2116-3p was evaluated via qRT-PCR in CRC cells with the transfection of miR-2116-3p inhibitor. I–N. Rescue experiments were performed in LoVo and HCT8 cells to explore the effects of miR-2116-3p inhibitor on sh-FALEC#1-transfected cell proliferation, apoptosis, migration, invasion and angiogenesis. **P < .01.
To further prove that FALEC contributed to CRC progression via sponging miR-2116-3p, we performed rescue experiments in subsequence. Firstly, we detected that the expression of miR-2116-3p was notably decreased in both LoVo and HCT8 cells after transfecting with miR-2116-3p inhibitor (Figure 2H). As anticipated, we observed that miR-2116-3p inhibitor could significantly reverse the repressive effect exerted by silencing FALEC on CRC cell proliferation through colony formation and EdU assays (Figure 2I, 2J). The promoting effect of FALEC knockdown on CRC cell apoptosis was abolished by inhibiting miR-2116-3p, as illustrated in Figure 2K. Additionally, miR-2116-3p inhibition markedly offset the restraining impacts of FALEC depletion on CRC cell migration (Figure 2L), invasion (Figure 2M) and tube formation (Figure 2N).
PIWIL1 is the direct target of miR-2116-3p
MiRNAs have been extensively reported to regulate disease development via targeting specific downstream genes. We utilized DIANA to find out downstream targets of miR-2116-3p, and found only two mRNA candidates (INPP5A and PIWIL1) with prediction score >0.6. Subsequently, we observed that PIWIL1 expression was significantly abrogated in miR-2116-3p-elevated LoVo and HTC8 cells, while INPP5A level was not evidently affected in such cells (Figure 3A). Furthermore, we unveiled that PIWIL1 expression was remarkably higher in CRC cell lines than that in normal NCM460 cells (Figure 3B). Thus, we recommended PIWIL1 as the downstream of miR-2116-3p in CRC. The putative miR-2116-3p binding site in the 3ʹUTR sequence of PIWIL1 was found by browsing TargetScan (http://www.targetscan.org/vert_72/), and the binding site was also mutated, as shown in Figure 3C. Dual luciferase reporter assays demonstrated that the luciferase activity of PIWIL1-WT, instead of PIWIL1-MUT, was obviously declined in response to co-transfection with miR-2116-3p mimics (Figure 3D). The results of RIP assays manifested that miR-2116-3p, PIWIL1 and FALEC were enriched only in Ago2 group but not IgG group in both LoVo and HCT8 cells (Figure 3E). Importantly, we proved that the expression of PIWIL1 was depleted by silencing FALEC, yet recovered by co-transfection of miR-2116-3p inhibitor in LoVo and HCT8 cells (Figure 3F). These observations added additional evidence for the ceRNA role of FALEC in CRC by targeting miR-2116-3p/PIWIL1 axis.
Figure 3.
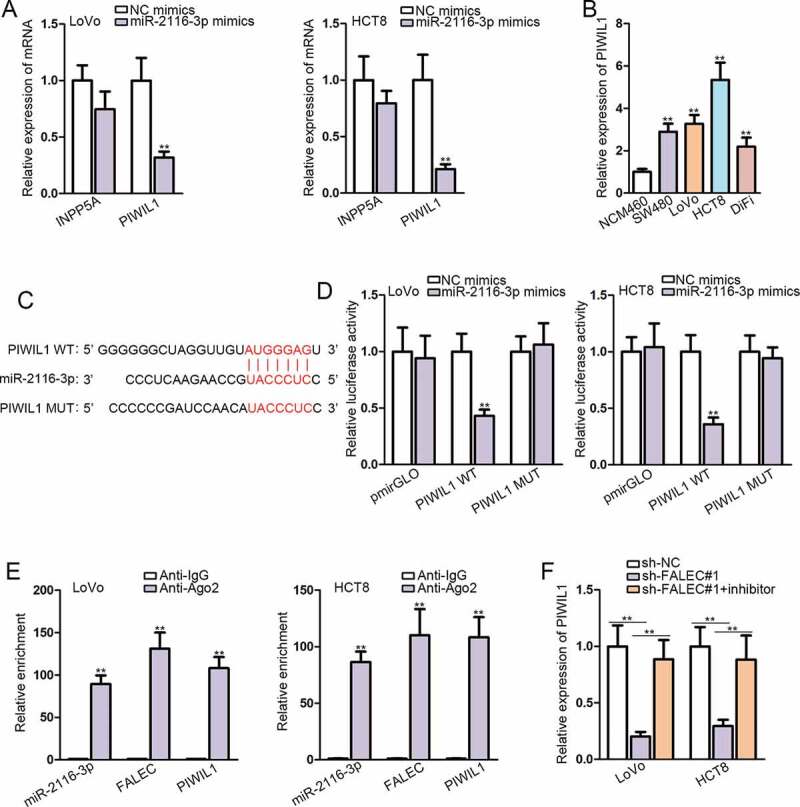
PIWIL1 is the target of miR-2116-3p
A. The expression of potential target genes was assessed by qRT-PCR in LoVo and HCT8 cells after overexpressing miR-2116-3p. B. The expression level of PIWIL1 in CRC cell lines and normal NCM460 controls was determined by qRT-PCR. C. Putative binding sites between miR-2116-3p and PIWIL1 predicted by TargetScan. D. Dual luciferase reporter assays were performed in LoVo and HCT8 cells to determine the physical interaction between miR-2116-3p and PIWIL1. E. RIP assays revealed the significant enrichment of FALEC, miR-2116-3p and PIWIL1 by anti-Ago2. F. qRT-PCR was conducted to study the expression of PIWIL1 in CRC cells with FALEC silence or together with miR-2116-3p inhibition. **P < .01.
FALEC promotes CRC progression via targeting miR-2116-3p/PIWIL1 axis
Thereafter, we intended to further elucidate that FALEC regulate CRC progression via regulating miR-2116-3p/PIWIL1 axis. Firstly, we observed that the transfection of pcDNA3.1/PIWIL1 led to PIWIL1 overexpression in two CRC cell lines (Figure 4A). Further, it manifested that both the mRNA and protein levels of PIWIL1 in LoVo and HTC8 cells were depleted after knockdown of FALEC, while restored by co-transfection with pcDNA3.1/PIWIL1 (Figure 4B, 4C). Colony formation and EdU assays showed that PIWIL1 up-regulation promoted cell proliferation that was inhibited by silencing FALEC (Figure 4D, 4E). Flow cytometry analysis indicated that PIWIL1 overexpression suppressed CRC cell apoptosis that was strengthened by FALEC depletion (figure 4F). Besides, PIWIL1 overexpression counteracted the blocking impact of FALEC inhibition on CRC cell migration and invasion (Figure 4G, 4H). Moreover, the angiogenesis restrained by FALEC silence was restored after overexpressing PIWIL1 (Figure 4I). To further testify the role of FALEC in CRC, we also performed rescue assays in vivo. Consistently, silencing FALEC could inhibit tumor growth in vivo, but this inhibitory effect was counteracted by PIWIL1 overexpression (Figure 4J, 4K). Moreover, the tumor volume and weight were lessened after silencing FALEC, but increased again under PIWIL1 overexpression (Figure 4L, 4M). Taken together, these results further confirmed the oncogenic function of FALEC in CRC via regulating miR-2116-3p/PIWIL1 axis.
Figure 4.
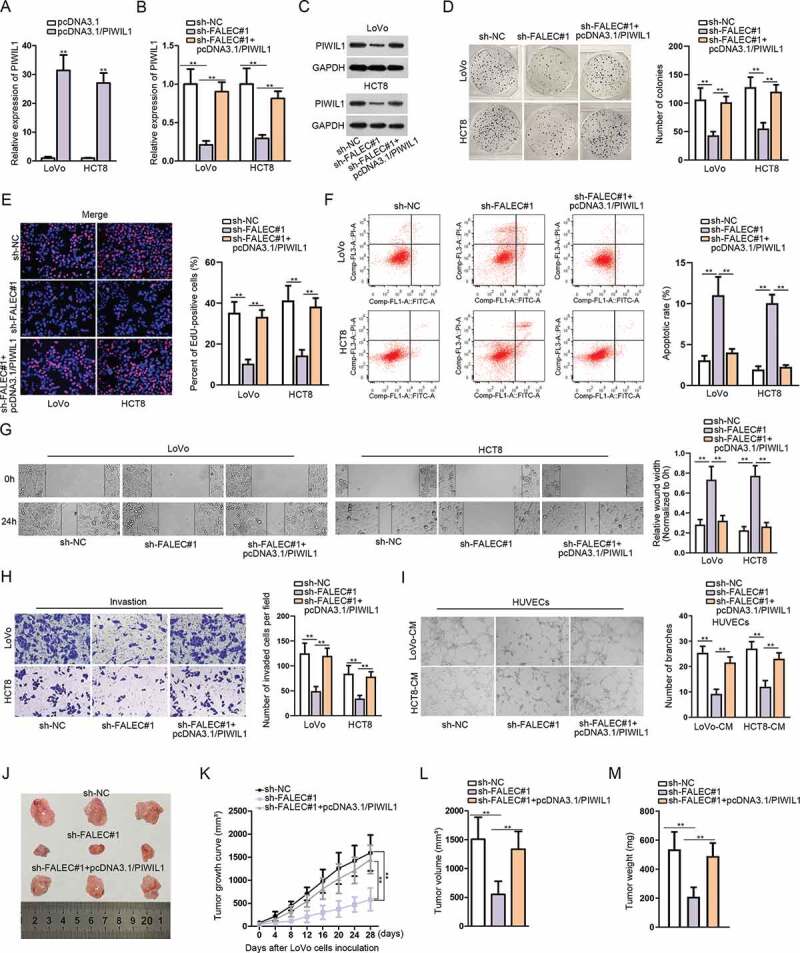
FALEC promotes CRC progression via targeting miR-2116-3p/PIWIL1 axis
A. The expression of PIWIL1 after transfecting with pcDNA3.1/PIWIL1 was detected by qRT-PCR analysis. B–C. The mRNA and protein levels of PIWIL1 after silencing FALEC were detected by qRT-PCR and western blot, respectively. D–E. Colony formation and EdU assays were performed to study the impact of overexpressing PIWIL1 on the proliferation of FALEC-silenced LoVo and HCT8 cells. F. Flow cytometry analysis was performed to assess the percent of apoptotic CRC cells under different conditions. G–I. Wound-healing, transwell and tube formation assays were performed to examine cell migration, invasion and angiogenesis under diverse contexts. J. Representative images of tumors excised from mice in sh-NC, sh-FALEC#1 or sh-FALEC#1+ pcDNA3.1/PIWIL1 group. K. The growth curve of above tumors recorded every 4 days during 4 weeks. L–M. Tumor volume and weight were measured and compared among sh-NC, sh-FALEC#1 and sh-FALEC1+ pcDNA3.1/PIWIL1 groups. **P < .01.
Discussion
CRC is one of the most prevalent malignant tumors with increasing morbidity and mortality worldwide. Numerous studies have revealed the important regulatory role of lncRNAs in the tumorigenesis of multiple human cancers. For example, LINC00645 was found to be significantly up-regulated in glioblastoma tissues and cell lines, and its upregulation was associated with poor overall survival for glioblastoma patients.12 Similarly, FALEC has been reported to exhibit the oncogenic property in cervical cancer.13 Here, we detected that FALEC was aberrantly highly expressed in CRC cell lines, which was similar to its expression pattern in endometrial cancer,14 but contrary to the findings in tongue squamous cell carcinoma.15 The different phenomena might be attributed to tissue specificity. Furthermore, knockdown of FALEC could inhibit CRC cell proliferation, migration and invasion while facilitating apoptosis, consistent with the effects of FALEC silence on gastric cancer cells.16
Previous reports have revealed that lncRNAs can serve as key regulators in cancer mainly through transcriptional or post-transcriptional control of genes.17,18 In recent years, lncRNAs have been found to be involved in ceRNA network to function in various cancers.19–21 Present study evidenced the cytoplastic localization of FALEC in CRC cells, and revealed miR-2116-3p as the factor downstream of FALEC in CRC. Currently, few research pay attention to miR-2116-3p, and our work was the first to uncover the downregulation of miR-2116-3p in CRC. Moreover, we identified intimate interaction between FALEC and miR-2116-3p via molecular mechanical experiments. In addition, miR-2116-3p inhibition could rescue the oncogenic effects of FALEC in CRC progression. These findings suggested that FALEC could function in CRC by sponging miR-2116-3p, while the sponging of miR-2116-3p by lncRNA LINC01433 was previously reported by Wu, M., et al.22
MiRNAs could suppress the expression of target genes and alter the production of corresponding proteins. We found that PIWIL1 was the potential downstream target of miR-2116-3p by utilizing TargetScan database. PIWIL1 has been reported to be an oncogene that is positively correlated with factors related to molecular subtype in pancreatic cancer.23 Herein, we verified the physical interaction between miR-2116-3p and PIWIL1, and that miR-2116-3p could negatively regulate PIWIL1 in CRC. Intriguingly, protein encoded by PIWIL1 belongs to PIWI protein family, which could interact with PIWI-interacting RNAs (piRNAs) and finally lead to gene silencing.24,25 This provided a potential that PIWIL1 might regulate miR-2116-3p expression in turn via a piRNAs-mediated manner. Besides, we performed rescue functional experiments and found that PIWIL1 overexpression reversed the biological effects of FALEC knockdown both in vitro and in vivo, indicating that FALEC facilitated CRC progression via regulating miR-2116-3p/PIWIL1 axis.
In a nutshell, our findings suggested that FALEC contributed to CRC progression via serving as a ceRNA of PIWIL1 by sequestering miR-2116-3p, which might provide some evidence for targeting FALEC in future research on CRC treatment.
Supplementary Material
Acknowledgments
This study was supported by research members.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A.. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–193. [DOI] [PubMed] [Google Scholar]
- 2.Jones P, Cade JE, Evans CEL, Hancock N, Greenwood DC.. Does adherence to the World Cancer Research fund/American Institute Of Cancer Research cancer prevention guidelines reduce risk of colorectal cancer in the UK women’s cohort study? Br J Nutr. 2018;119:340–348. doi: 10.1017/S0007114517003622. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Y, Chen Y, Ding W, Hua Z, Wang L, Zhu Y, Qian H, Dai T.. LncRNA UCA1 impacts cell proliferation, invasion, and migration of pancreatic cancer through regulating miR-96/FOXO3. IUBMB Life. 2018;70:276–290. doi: 10.1002/iub.1699. [DOI] [PubMed] [Google Scholar]
- 4.Chen Z, Hu X, Wu Y, Cong L, He X, Lu J, Feng J, Liu D. Long non-coding RNA XIST promotes the development of esophageal cancer by sponging miR-494 to regulate CDK6 expression. Biomed Pharmacother. 2019;109:2228–2236. doi: 10.1016/j.biopha.2018.11.049. [DOI] [PubMed] [Google Scholar]
- 5.Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9:703–719. doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao R, Sun F, Bei X, Wang X, Zhu Y, Jiang C, Zhao F, Han B, Xia S. Upregulation of the long non-coding RNA FALEC promotes proliferation and migration of prostate cancer cell lines and predicts prognosis of PCa patients. Prostate. 2017;77:1107–1117. doi: 10.1002/pros.23367. [DOI] [PubMed] [Google Scholar]
- 7.Zeng Y, Gao T, Huang W, Yang Y, Qiu R, Hou Y, Yu W, Leng S, Feng D, Liu W, et al. The microRNA-455-3p mediates GATA3 tumor suppression in mammary epithelial cells by inhibiting TGF-beta signaling. J Biol Chem. 2019;294(43):15808–15825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou G, Xu W, Jin Y, Wu J, Pan Y, Zhou F. MiRNA-217 accelerates the proliferation and migration of bladder cancer via inhibiting KMT2D. Biochem Biophys Res Commun. 2019;519:747–753. doi: 10.1016/j.bbrc.2019.09.029. [DOI] [PubMed] [Google Scholar]
- 9.Peng W, Wang Z, Fan H. LncRNA NEAT1 impacts cell proliferation and apoptosis of colorectal cancer via regulation of akt signaling. POR. 2017;23:651–656. doi: 10.1007/s12253-016-0172-4. [DOI] [PubMed] [Google Scholar]
- 10.Sun S, Hang T, Zhang B, Zhu L, Wu Y, Lv X, Huang Q, Yao H. miRNA-708 functions as a tumor suppressor in colorectal cancer by targeting ZEB1 through Akt/mTOR signaling pathway. Am J Transl Res. 2019;11:5338–5356. [PMC free article] [PubMed] [Google Scholar]
- 11.Lin C, Garruss AS, Luo Z, Guo F, Shilatifard A. The RNA Pol II elongation factor Ell3 marks enhancers in ES cells and primes future gene activation. Cell. 2013;152:144–156. doi: 10.1016/j.cell.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C, Zheng H, Hou W, Bao H, Xiong J, Che W, Gu Y, Sun H, Liang P. Long non-coding RNA linc00645 promotes TGF-beta-induced epithelial-mesenchymal transition by regulating miR-205-3p-ZEB1 axis in glioma. Cell Death Dis. 2019;10:717. doi: 10.1038/s41419-019-1948-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Naizhaer G, Kuerban A, Meilipa KR, Zhou P. Up-regulation of lncRNA FALEC indicates prognosis and diagnosis values in cervical cancer. Pathol Res Pract. 2019;215:152495. doi: 10.1016/j.prp.2019.152495. [DOI] [PubMed] [Google Scholar]
- 14.Zheng QH, Shi L, Li HL. FALEC exerts oncogenic properties to regulate cell proliferation and cell-cycle in endometrial cancer. Biomed Pharmacother. 2019;118:109212. doi: 10.1016/j.biopha.2019.109212. [DOI] [PubMed] [Google Scholar]
- 15.Jia B, Xie T, Qiu X, Sun X, Chen J, Huang Z, Zheng X, Wang Z, Zhao J. Long noncoding RNA FALEC inhibits proliferation and metastasis of tongue squamous cell carcinoma by epigenetically silencing ECM1 through EZH2. Aging. 2019;11:4990–5007. doi: 10.18632/aging.102094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H, Qiao F, Zhao Y, Wu S, Hu M, Wu T, Huang F, Chen W, Sun D, Liu M, et al. Downregulation of long non-coding RNA FALEC inhibits gastric cancer cell migration and invasion through impairing ECM1 expression by exerting its enhancer-like function. Front Genet. 2019;10:255. doi: 10.3389/fgene.2019.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genom Proteom Bioinform. 2017;15:177–186. doi: 10.1016/j.gpb.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong Y, Lu Z, Liu P, Liu Y, Wang F, Liang EY, Hou FF, Liang M. Long noncoding RNA: genomics and relevance to physiology. Compr Physiol. 2019;9:933–946. [DOI] [PubMed] [Google Scholar]
- 19.Hu J, Ni G, Mao L, Xue X, Zhang J, Wu W, Zhang S, Zhao H, Ding L, Wang L. LINC00565 promotes proliferation and inhibits apoptosis of gastric cancer by targeting miR-665/AKT3 axis. Onco Targets Ther. 2019;12:7865–7875. doi: 10.2147/OTT.S189471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Z, Mi M, Li X, Zheng X, Wu G, Zhang L. lncRNA OSTN-AS1 may represent a novel immune-related prognostic marker for triple-negative breast cancer based on integrated analysis of a ceRNA network. Front Genet. 2019;10:850. doi: 10.3389/fgene.2019.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu X, Li J, Ren Y, Zuo Z, Ni S, Cai J. MEG3 can affect the proliferation and migration of colorectal cancer cells through regulating miR-376/PRKD1 axis. Am J Transl Res. 2019;11:5740–5751. [PMC free article] [PubMed] [Google Scholar]
- 22.Wu M, Wu W, Ding J, Yang J. LINC01433/miR-2116-3p/MYC feedback loop promotes cell proliferation, migration, and the epithelial-mesenchymal transition in breast cancer. Cancer Biother Radiopharm. 2019;34:388–397. doi: 10.1089/cbr.2019.2772. [DOI] [PubMed] [Google Scholar]
- 23.Li W, Martinez-Useros J, Garcia-Carbonero N, Fernandez-Acenero MJ, Ortega-Medina L, Garcia-Botella S, Perez-Aguirre E, Diez-Valladares L, Garcia-Foncillas J. The prognosis value of PIWIL1 and PIWIL2 expression in pancreatic cancer. J Clin Med. 2019:8: 1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rojas-Ríos P, Simonelig M. piRNAs and PIWI proteins: regulators of gene expression in development and stem cells. Development (Cambridge, England). 2018;145(17):dev161786. doi: 10.1242/dev.161786. [DOI] [PubMed] [Google Scholar]
- 25.Ozata DM, Gainetdinov I, Zoch A, O’Carroll D, Zamore PD. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet. 2019;20:89–108. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


