ABSTRACT
Aim
In this study, we explored the ability of TAMs to affect the malignant phenotype of human hepatoma Huh-7 cells through the Gli2/IGF-II/ERK1/2 pathway.
Methods
The TAMs were characterized by flow cytometry and ELISA assays. Huh-7 cells were treated with conditioned medium of TAMs (TAMs-CM), and the proliferation, migration and invasion abilities were measured by CCK-8, Transwell and scratch assays. The levels of TGF-β1, Gli2, IGF-II and related proteins in the ERK1/2 pathway and the epithelial-mesenchymal transition (EMT) process were examined by RT-qPCR and western blot. Huh-7 cells were injected subcutaneously into nude mice with TAMs to explore the role of TAMs in tumor growth.
Results
The expression levels of TGF-β1, Gli2 and IGF-II and the cell proliferation, migration and invasion abilities were elevated in Huh-7 cells treated with TAMs-CM. TGF-β1 was upregulated in the conditioned medium and was found to be involved in the promotion of migration, invasion and the EMT of Huh-7 cells. The activation of TGF-β1 signaling increased the expression of Gli2. Knockdown of Gli2 decreased the expression of IGF-II and also reversed the promotional effect of the conditioned medium on migration, invasion and the EMT of Huh-7 cells. TGF-β1/Gli2/IGF-II signaling was shown to promote the malignant phenotype of Huh-7 cells by activating the ERK1/2 signaling pathway. Further, TGF-β1 knockdown attenuated the influence of TAMs on tumor growth in mouse model.
Conclusion
The TGF-β1 secreted by TAMs promotes the migration, invasion and EMT of human hepatoma Huh-7 cells through the Gli2/IGF-II/ERK1/2 pathway.
KEYWORDS: Hepatocellular carcinoma, tumor-associated macrophages, TGF-β1, Gli2, IGF-II, ERK1/2 signaling pathway, tumor growth
Introduction
Liver cancer is a common cause of cancer-related death and leads to 700,000 deaths globally every year. 1 . Hepatocellular carcinoma (HCC) is the predominant form of liver cancer, accounting for more than 90% of cases and is the leading cause of death in patients with cirrhosis.2 The underlying risk factors for HCC include chronic viral hepatitis, alcohol abuse and metabolic diseases.2,3 Although there are curative approaches for HCC treatment at the early stage, there are limited effective therapies for most patients diagnosed at the intermediate or advanced stages owing to the aggressive malignancy of HCC.4 Therefore, it is urgent to better understand the molecular mechanisms of HCC malignancy to identify more definitive therapeutic targets for HCC treatment.
The hedgehog (Hh)-glioma-associated oncogene (Gli) signaling pathway is implicated in embryo patterning, cell proliferation, survival and growth, and the dysregulation of this pathway has been associated with tumourigenesis and tumor development.5 Gli2 expression is activated by the Hh signaling pathway; however, it can also be stimulated by non-canonical pathways, including that of TGF-β.6 Gli2 acts as a transcription factor,7 and the expression of Gli2 was recently reported to be elevated in various cancers.6,8 Gli2 has also been shown to be upregulated in HCC cell lines and tissues, and its high expression is associated with poor prognosis in HCC patients.9 However, the mechanism of how Gli2 regulates HCC development has not been fully clarified.
Macrophages are a major immune cell component of the tumor microenvironment.10 Tumor-associated macrophages (TAMs) play a vital role in the balance between the immune response and tumor growth.11 Cancer stem cells (CSCs) are reportedly activated by TAMs in HCC and have been associated with the poor outcomes of HCC patients.11 TGF-β1, secreted by TAMs, has been found to mediate the promotion of CSC activation.12 TGF-β signaling has been reported to be a pivotal factor that stimulates various surrounding cells and participates in the invasion and migration of HCC cells.13 Gli2 is suggested to be a target gene of the TGF-β signaling pathway in human melanoma cells.14 However, the role of TGF-β1 secreted by TAMs in the tumor microenvironment in the expression of Gli2 and the role of the TGF-β1/Gli2 axis in modulating HCC progression remain uncertain.
Insulin-like growth factor II (IGF-II) regulates cell survival, growth and proliferation and is upregulated in various cancers.15,16 Upregulated IGF-II has been found in the tumors and blood of HCC patients and has been suggested as a diagnostic biomarker of HCC.17 IGF-II is regulated at the transcriptional level.15,18 The expression of IGF-II is reportedly induced by the Hh signaling pathway. However, whether Gli2 is involved in the regulation of IGF-II in HCC remains unclear.
Here, we hypothesized that Gli2 regulates IGF-II expression and that this process is modulated by TAMs through the secretion of TGF-β1, thus influencing the malignancy of human hepatoma Huh-7 cells. To test this hypothesis, human hepatoma cells (Huh-7) were cultured in conditioned medium (CM) from TAMs, and the expression levels of TGF-β1, Gli2 and IGF-II and the proliferation, migration, invasion and epithelial-mesenchymal transition (EMT) process of Huh-7 cells were found to be elevated. We also identified that the TGF-β1 presented in TAMs-CM positively regulated the expression of Gli2 and IGF-II and promoted the migration, invasion and EMT process of Huh-7 cells. In addition, extracellular-regulated protein kinases 1/2 (ERK1/2) was found to mediate the promotion of IGF-II in malignant phenotype of Huh-7 cells. TGF-β1 was necessary for the ability of TAMs to promote tumor growth in the mouse model. Our study is the first to identify that TGF-β1 secreted by TAMs can enhance the progression of HCC by stimulating the Gli2/IGF-II/ERK1/2 axis. These findings provide potential therapeutic targets for HCC treatment.
Materials and methods
Cell culture, differentiation and treatment
The human monocytic cell line THP-1 was purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in Roswell Park Memorial Institute medium (RPMI-1640, Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA), 100 U/mL penicillin and 100 μg/mL streptomycin (Beyotime, Shanghai, China). The human hepatocellular carcinoma cell line Huh-7 was purchased from the Chinese Academy of Sciences (Shanghai, China) and cultured in Dulbecco’s modified Eagle medium (DMEM, Gibco, Grand Island, NY, USA) containing 10% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin. The cells were cultured in a humidified incubator at 37°C with 5% CO2. Mycoplasma detection was tested using a Mycoplasma Stain Assay Kit (C0296, Beyotime, Shanghai, China). The cells were fixed with fixation buffer for 20 min at room temperature. The fixation buffer was then removed, and the cells were stained with Hoechst staining buffer for 30 min in the dark, followed by observation under a fluorescence microscope (Zeiss, Germany). In some cases, TGF-β1 inhibitor LY364947 (10, 50, 100 nM), recombinant human IGF-II (20 ng/mL), or ERK1/2 signaling pathway inhibitor PD98059 (50 µM, MedChem Express, Princeton, NJ, USA) were used to treat the Huh-7 cells.
For the differentiation of M2-polarized macrophages (also called TAMs), THP-1 cells were incubated with 300 nM phorbol 12-myristate 13-acetate (PMA, Sigma-Aldrich, Saint Louis, MO, USA) for 6 h, followed by incubation with PMA and IL-13 (20 ng/mL, STEMCELL Technologies Inc., Cambridge, MA, USA) and IL-4 (20 ng/mL, R&D Systems, Minneapolis, MN, USA) for an additional 18 h at 37°C. The conditioned medium (CM) from M2-polarized macrophages was co-cultured with the Huh-7 cells.
Flow cytometry analysis
After M2-polarized macrophage differentiation, the cells were washed twice and suspended in pre-chilled PBS containing 5% inactivated serum and 0.1% NaN3. After centrifuged at 200 g, 4°C for 5 minutes, the cells were incubated with FITC-conjugated anti-CD68 antibody (11–0689-42, eBioscience, San Diego, CA, USA) and PE-conjugated anti-CD206 antibody (12–2069-42, eBioscience, San Diego, CA, USA) diluted in PBS containing 5% inactivated serum and 0.1% NaN3. After washing three times with PBS containing 5% inactivated serum and 0.1% NaN3, the cells were fixed with 4% paraformaldehyde in cold PBS and analyzed by flow cytometry (BD Biosciences, Germany).
Enzyme-linked immunosorbent assay (ELISA)
After differentiation and polarization, the supernatant of the M2-polarized macrophages was collected and subjected to cytokine measurement using ELISA kits according to the manufacturer’s guidelines. TGF-β1 and IL-10 were detected by using ELISA kits from Abcam (Eugene, OR, USA).
Plasmid construction and cell transfection
The human Gli2 gene was cloned into the pcDNA3.1 vector (GenePharma, Shanghai, China) to achieve Gli2 overexpression (OE-Gli2). The pcDNA3.1 empty vector served as a negative control (NC). pGPH1 plasmid expressing short hairpin RNA (shRNA) targeting Gli2 (sh-Gli2) or IGF-II (sh-IGF-II) and scrambled control shRNA-expressing pGPH1 plasmid (sh-NC) were also ordered from GenePharma (Shanghai, China). Huh-7 cells were plated in 6-well plates and allowed to grow for 24 h. They were then transfected with OE-Gli2 vector, sh-Gli2 vector, or sh-IGF-II vector at 40–60% confluence using Lipofectamine 3000 reagent (Thermo Fisher Scientific, Waltham, MA, USA). The transfected cells were harvested 48 h after transfection.
Transwell invasion and migration assay
Transwell assays were used to measure the migration and invasion abilities of Huh-7 cells. The cells were first digested with trypsin, washed with PBS, suspended in serum-free medium, and then added into the upper chamber for migration measurement. Medium containing 20% serum was then added into the bottom chamber. Following an incubation of 24 h, the chamber was taken out and washed with PBS. The cells below the microporous membrane were fixed with 95% ethanol and stained with crystal violet solution. Finally, the cells that migrated through the membrane were observed under an optical microscope (Zeiss, Germany). The number of cells in 5 random fields was counted. For cell invasion measurement, the basement membrane of the upper chamber was pre-coated with Matrigel, and the cells were collected and treated similarly.
Scratch migration assay
Huh-7 cells were seeded into 6-well plates and cultured overnight. After the cells adhered, the cell monolayer was scratched gently and slowly with a sterile pipette tip across the center of the well in a straight line. Subsequently, to remove the detached cells, the well was gently washed twice with medium. The cells were cultured in fresh serum-free medium and incubated for 24 h, followed by observation under an optical microscope (Zeiss, Germany). Images were acquired and the migration distance of cells was measured by the following formula: (W0h–W24h)/W0h ×100%.
CCK-8 assay
Huh-7 cells were seeded into 96-well plates in 100 µL of culture medium and cultured for 12, 24 and 48 h. Cell Counting Kit-8 (C0038, Beyotime) was adopted to measure the proliferation of Huh-7 cells following the user’s guide. Briefly, 10 μL of the CCK-8 reagent was added into each well, and the cells were cultured for 1 h. The OD 450 value was detected, which indicated the cell proliferation rate.
RNA extraction, reverse transcription and quantitative polymerase chain reaction (qPCR)
Total RNA from cells or tissues was extracted with Trizol reagent (Sigma-Aldrich, St. Louis, MO, USA) by the regular method. The RNA was reversely transcribed into cDNA with a reverse transcription PrimeScript RT reagent Kit (Takara Biomedical Technology, Dalian, China). The obtained cDNA was used as template in the real-time qPCR using the SYBR Premix Ex TaqTM II (RNaseH Plus) Kit (Takara Biomedical Technology, Dalian, China). The qPCR reaction was performed in an ABI 7500HT real-time PCR system (Thermo Fisher Scientific). Primers for TGF-β1, Gli2, IGF-II, IL-10, Arg-1, CD206 and CD163 were purchased from Sangon Biotech (Shanghai, China). GAPDH was used as an internal control. The relative mRNA expression levels were calculated by the 2−ΔΔCt method.
Western blot analysis
Cells were washed with pre-chilled PBS and lysed in radio immunoprecipitation assay buffer (RIPA, Beyotime, Shanghai, China). Quantification of the protein concentration was performed using a BCA protein assay Kit (Beyotime). The protein samples were then separated by SDS-PAGE and then transferred onto a polyvinylidene fluoride (PVDF) membrane. After blocking with 5% skimmed milk at room temperature for 1 h, the membrane was incubated with primary antibodies diluted in Tris-buffered saline containing Tween-20 (TBST) at 4°C overnight. The following antibodies were used: anti-TGF-β1 (#3709), anti-Gli2 (#2585), anti-IGF-II (ab170304, Abcam), anti-E-cadherin (#3195), anti-vimentin (#5741), anti-MMP-9 (#13667), anti-p-ERK1/2 (#4370), anti-ERK1/2 (#4695) and anti-GAPDH (#5174). The membrane was then washed 3 times with TBST, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibody (#7074) at room temperature for 1 hour. After washing 3 times with TBST, the membrane was incubated with chemiluminescence reagent and scanned. The antibodies used for western blotting were obtained from Cell Signaling Technology (Danvers, MA, USA) unless indicated otherwise. The gray values of the bands were analyzed by ImageJ software.
Tumor xenograft in nude mice
40 BALB/c nude mice about 16–20 g were purchased from Shanghai SLAC Laboratory Animal Center (Shanghai, China) and housed in a pathogen-free environment. This animal experiment was approved by the Animal Care and Use Committee of the First Affiliated Hospital of Nanchang University (Nanchang, Jiangxi, China). To evaluate tumor growth, TAMs were stably transfected with lentiviral vectors to silence TGF-β1 or not, and 3 × 106 Huh-7 cells and 9 × 106 indicated TAMs were injected subcutaneously together into each mice in a total volume of 200 μL as previously reported.19 The tumor sizes were measured every 3 days. After 21 days, the mice were sacrificed and the tumor tissues were collected for size and weight measurement and gene expression analysis.
Statistical analysis
All experiments were performed at least three times, and the data were analyzed using GraphPad prism 6.0 software. The results are presented as the mean ± standard deviation (SD). Data between two groups were compared using unpaired Student’s t-test, and data among multiple groups were compared using one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test. A P value less than 0.05 was considered to indicate statistical significance.
Results
Conditioned medium from TAMs upregulates the expression of Gli2 and IGF-II and promotes migration and invasion of Huh-7 cells
TAMs are characterized as an M2-polarized subtype.20,21 To explore the role of TAMs in regulating hepatocellular carcinoma cells, THP-1 cells were differentiated to the M2 phenotype, which was indicated by an increased percentage of CD68+CD206+ macrophages detected by flow cytometry (Figure 1A-B). ELISA assays were performed to assess the level of anti-inflammatory cytokine IL-10 (Figure 1C). The results suggested that THP-1 cells were polarized into M2 macrophages, as previously reported.22 Surprisingly, the level of TGF-β1 was also markedly increased (Figure 1C). Human hepatoma Huh-7 cells were then treated with CM from M2 macrophages. Compared with control group, the mRNA levels of TGF-β1, Gli2 and IGF-II were increased in Huh-7 cells treated with CM from M2 macrophages (Figure 1D). Furthermore, western blotting showed that the protein expressions of TGF-β1, Gli2 and IGF-II were also elevated (Figure 1E). Next, we detected whether the malignant phenotype of Huh-7 cells could be altered during this process. We adopted Transwell and scratch assays to examine the migration and invasion abilities of Huh-7 cells. As shown in figure 1F, the migration and invasion abilities were significantly enhanced when treated with CM from M2 macrophages. The enhanced migration ability of the Huh-7 cells was also confirmed by scratch assay (Figure 1G). Further, we performed CCK-8 assays to examine the effect of the CM from M2 macrophages on Huh-7 cell proliferation. As shown in Figure 1H, the proliferation of Huh-7 cells was increased upon treatment with CM from M2 macrophages compared with the control group. Altogether, these data suggest that conditioned medium from TAMs (TAMs-CM) upregulates the expression levels of Gli2 and IGF-II in Huh-7 cells and promotes the malignant phenotypes.
Figure 1.
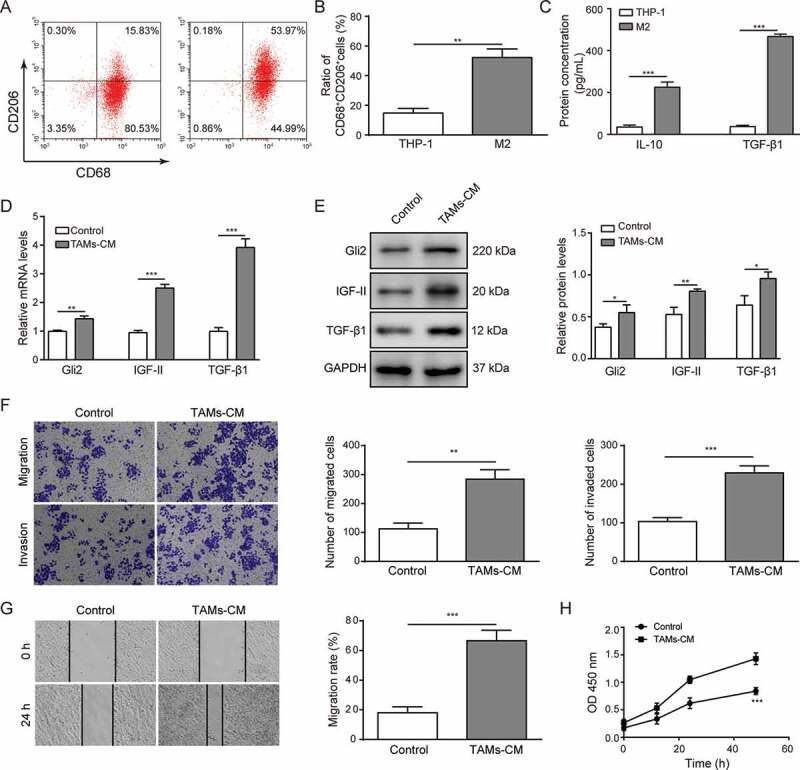
Conditioned medium from TAMs upregulates the expression of Gli2 and IGF-II and promotes migration and invasion of Huh-7 cells
(A-B) THP-1 cells were differentiated and polarized into M2 macrophages, and the expression levels of CD68 and CD206 were detected by flow cytometry (A) and quantified (B). (C) After differentiation and polarization, the medium of THP-1 cells was collected, and the concentrations of IL-10 and TGF-β1 were measured by ELISA. (D-E) After treatment with conditioned medium of TAMs (TAMs-CM), the Huh-7 cells were collected, and the levels of TGF-β1, Gli2 and IGF-II were examined by qPCR (D) and western blotting (E). (F) The migration and invasion abilities of Huh-7 cells treated with TAMs-CM were detected by Transwell assay. (G) The migration of Huh-7 cells treated with TAMs-CM was detected by scratch assay. (H) The proliferation of Huh-7 cells treated with TAMs-CM was detected by CCK-8 assay. *** p < .001, ** p < .01, * p < .05.
TGF-β1 signaling is responsible for the effects of TAMs-CM on Gli2 expression and Huh-7 cell migration and invasion
Gli2 is reportedly ubiquitously regulated by the TGF-β pathway. We next tested whether TGF-β1 was involved in the process whereby TAMs-CM modulated Gli2 expression in Huh-7 cells. The TGF-β1 inhibitor LY364947 showed a dosage-dependent suppressing effect on the Gli2 expression upon TAMs-CM treatment at both the mRNA and protein levels in Huh-7 cells (Figure 2A-B). The inhibited TGF-β1 expression was also confirmed by western blotting (Figure 2B). Accordingly, we observed that the migration and invasion abilities of Huh-7 cells treated with TAMs-CM were suppressed by LY364947 treatment in a dosage-dependent manner (Figure 2C-D). Further, a scratch assay was performed and confirmed the reduced migration ability of the Huh-7 cells upon LY364947 treatment (Figure 2E-F). Taken together, our data suggest that TGF-β1 participates in the regulation of Gli2 expression and in cell migration and invasion in Huh-7 cells.
Figure 2.
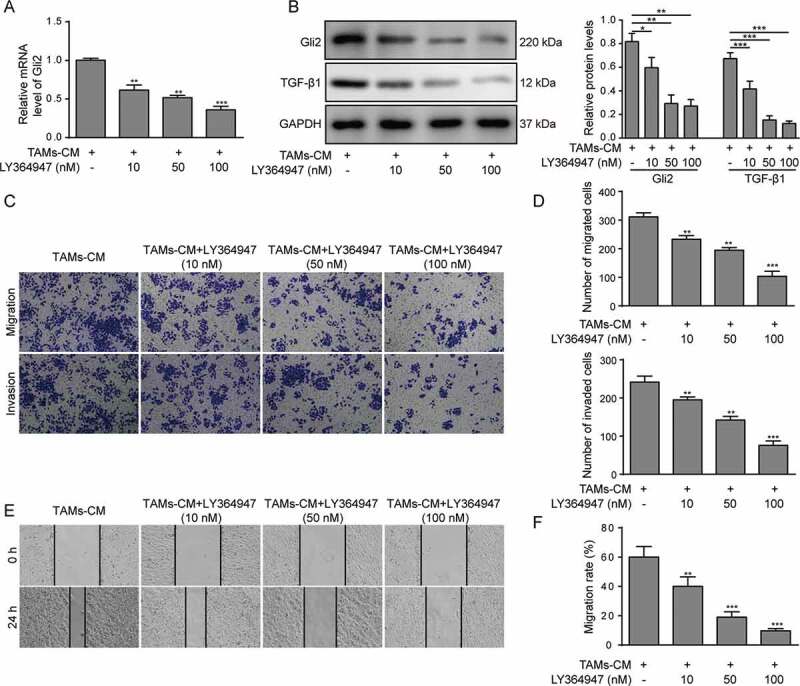
TGF-β1 signaling mediates the effect of TAMs-CM on Gli2 expression and Huh-7 cell migration and invasion
Huh-7 cells were treated with the conditioned medium of TAMs and with different concentrations of the TGF-β1 inhibitor LY364947. (A) The level of Gli2 mRNA was measured by qPCR. (B) The levels of Gli2 and TGF-β1 protein were measured by western blotting. (C-D) The invasion and migration abilities were measured by Transwell assays; the quantification is shown in (D). (E-F) The migration of Huh-7 cells was measured by scratch assay; the quantification is shown in (F). *** p < .001, ** p < .01, * p < .05.
Gli2 induces IGF-II expression to promote Huh-7 cell migration and invasion
As Gli2 is a known transcription factor, we next measured whether the expression of IGF-II was regulated by Gli2. Gli2 shRNA (sh-Gli2) was introduced into Huh-7 cells, and the mRNA and protein expression of Gli2 was markedly reduced (Figure 3A and C). Meanwhile, the mRNA and protein expression of IGF-II was observed to be decreased compared with untreated control and sh-NC transfection control (Figure 3B-C). These data suggest that the expression of IGF-II can be regulated by Gli2.
Figure 3.
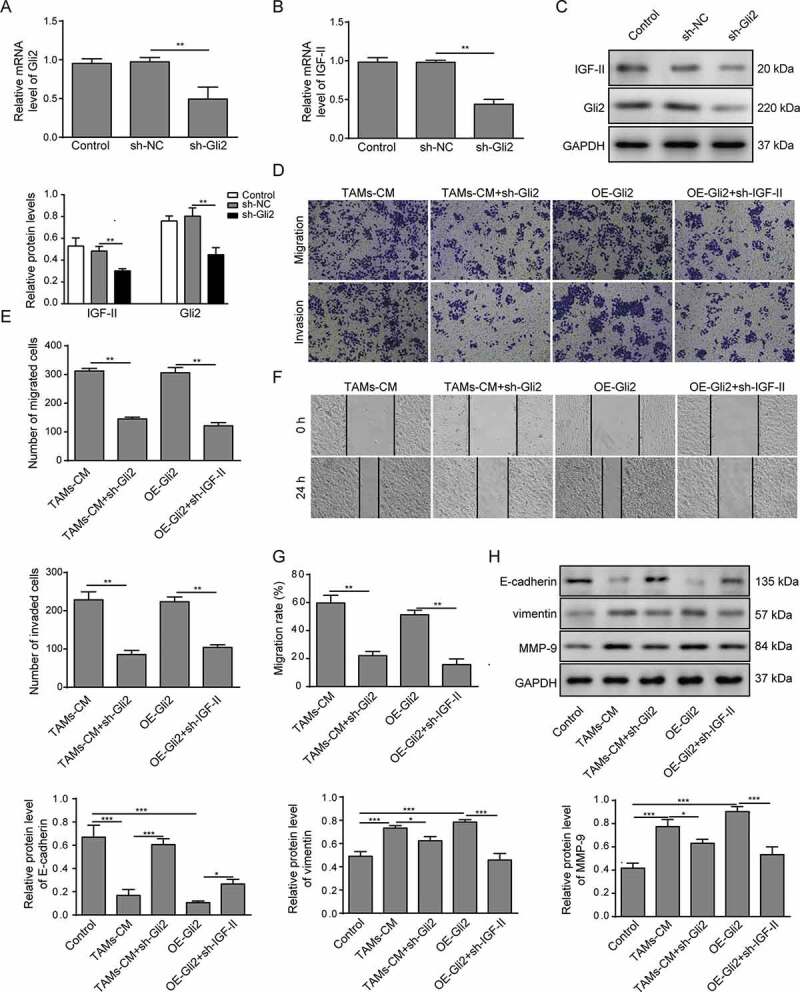
Gli2 induces IGF-II expression to promote Huh-7 cell migration and invasion
(A-C) Huh-7 cells were transfected with sh-Gli2, and the levels of Gli2 and IGF-II mRNA were measured by qPCR (A-B); the levels of Gli2 and IGF-II protein were measured by western blotting (C). (D-E) The migration and invasion abilities of Huh-7 cells were measured by Transwell assay. (F-G) The migration of Huh-7 cells was confirmed by scratch assay. (H) The protein levels of E-cadherin, vimentin and MMP-9 were detected by western blotting. *** p < .001, ** p < .01, * p < .05.
We next examined whether Gli2 and the subsequent regulated IGF-II expression have any influence on the migration and invasion of Huh-7 cells induced by TAMs-CM. The expression of Gli2 in Huh-7 cells upon overexpression (Supplemental Figure 1A-B) and the expression of IGF-II (Supplemental Figure 1C-D) upon knockdown were confirmed by qPCR and western blotting. As shown in Figure 3D-E, Gli2 knockdown in Huh-7 cells (TAMs-CM+sh-Gli2) attenuated the migration and invasion abilities induced by TAMs-CM. In addition, Huh-7 cells transfected with Gli2 (OE-Gli2) showed increased migration and invasion abilities that were comparable with those treated with TAMs-CM (Figure 3D-E). Moreover, the effects of Gli2 overexpression could be considerably suppressed by treatment with IGF-II shRNA (OE-Gli2+ sh-IGF-II, Figure 3D-E). Likewise, the scratch assay also showed that the migration of Huh-7 cells induced by TAMs-CM treatment was restrained by sh-Gli2 treatment, while downregulation of IGF-II reduced the migration of Huh-7 cells that was induced by Gli2 overexpression (figure 3F-G). These data suggest that the IGF-II expression regulated by Gli2 may be involved in the migration and invasion promoted by TAMs-CM in Huh-7 cells.
To further confirm the malignant phenotype of Huh-7 cells in different conditions, we measured the expression of tumor malignancy-related proteins. As shown in Figure 3H, E-cadherin was downregulated in TAMs-CM-treated Huh-7 cells, while vimentin, a marker of epithelial-to-mesenchymal transition (EMT),12 and MMP-9, a metalloproteinase that facilitates tumor progression,23 were upregulated. These results indicate that TAMs-CM regulates the expression of EMT-related proteins to promote the migration and invasion of Huh-7 cells. In accordance with the above findings, Gli2 knockdown was able to partially rescue the alterations induced by TAMs-CM, further supporting the ability of Gli2 to mediate the influence of TAMs-CM on Huh-7 cells. In addition, the reduced expression of E-cadherin and the elevated expression of vimentin and MMP-9 were observed in Huh-7 cells overexpressing Gli2 (Figure 3H). However, knockdown of IGF-II rescued the altered expression levels of malignancy-related proteins that were induced by Gli2 overexpression, further conforming that IGF-II is involved in the regulation of Gli2 in the migration and invasion of Huh-7 cells.
IGF-II promotes migration and invasion of Huh-7 cells via ERK1/2 signaling
The ERK signaling pathway has been reported as being strongly related to tumor progression.24 We observed that the phosphorylation of ERK1/2 was enhanced in Huh-7 cells treated with IGF-II compared with untreated or vehicle-treated cells (Figure 4A), suggesting that the ERK1/2 signaling pathway was activated by IGF-II in Huh-7 cells. Next, Huh-7 cells were treated with the ERK1/2 inhibitor PD98059 in the presence of IGF-II. As shown in Figure 4F, PD98059 treatment attenuated the phosphorylation of ERK1/2. Accordingly, Huh-7 cells treated with IGF-II and PD98059 showed decreased migration and invasion abilities compared to the Huh-7 cells treated with IGF-II alone, which was detected by both Transwell assay (Figure 4B-C) and scratch assay (Figure 4D-E). In addition, PD98059 treatment also promoted the expression of E-cadherin and reduced the expression of vimentin and MMP-9 (figure 4F), further indicating that the activation of the ERK1/2 signaling pathway may have a role in facilitating the IGF-II-regulated migration and invasion of Huh-7 cells.
Figure 4.
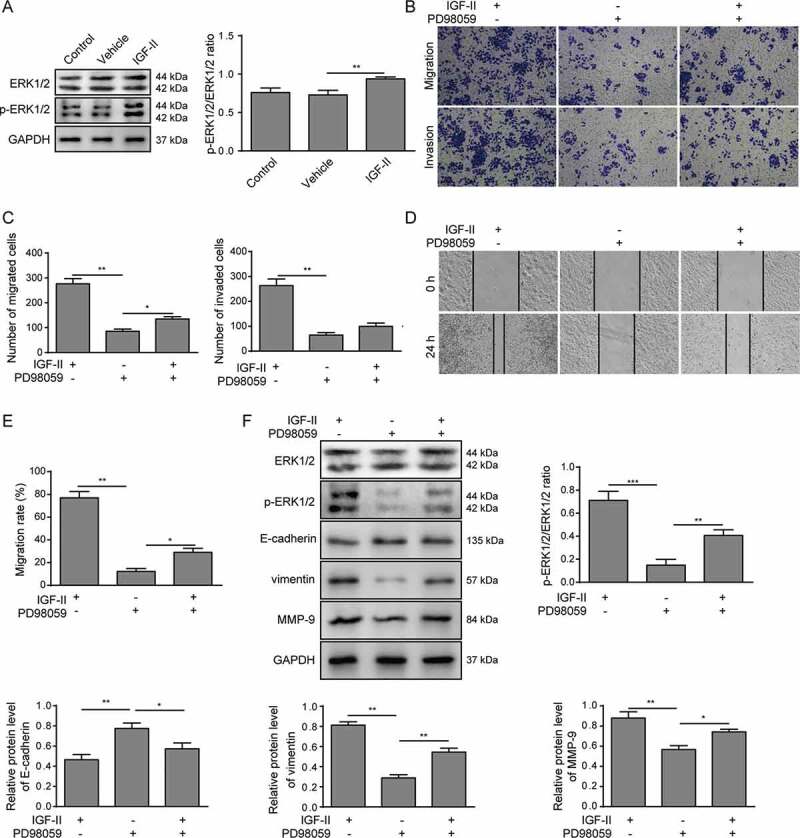
IGF-II promotes migration and invasion of Huh-7 cells via the ERK1/2 signaling pathway
(A) Huh-7 cells were treated with IGF-II or its vehicle, and the expression and phosphorylation levels of ERK1/2 were measured by western blotting. (B-E) Huh-7 cells were treated with IGF-II or the ERK1/2 inhibitor PD98059 or with both IGF-II and PD98059. The invasion and migration of Huh-7 cells were measured by Transwell assay (B, C), and the migration was measured by scratch assay (D, E). (F) The activation of ERK1/2 and the expression of E-cadherin, vimentin and MMP-9 were analyzed by western blotting. *** p < .001, ** p < .01, * p < .05.
TAMs-CM promotes the migration and invasion of Huh-7 cells through Gli2/IGF-II/ERK1/2 axis
We further investigated whether the ERK1/2 signaling pathway was involved in the ability of TAMs-CM and Gli2 to promote Huh-7 cell migration and invasion. Huh-7 cells grown in TAMs-CM were treated with PD98059, and dramatic decrease of migration and invasion were observed (TAMs-CM vs TAMs-CM+PD98059, Figure 5A-D). Furthermore, the migration and invasion abilities of Huh-7 cells overexpressing Gli2 were also markedly suppressed by PD98059 treatment (OE-Gli2 vs OE-Gli2+ PD98059, Figure 5A-D). In accordance with the suppressed migration and invasion abilities upon PD98059 treatment, the expression of E-cadherin was increased, while the phosphorylation of ERK1/2 and the expression of vimentin and MMP-9 were decreased in PD98059-treated Huh-7 cells compared to those treated with TAMs-CM or Gli2 alone (Figure 5E). These findings suggest that the ability of TAMs-CM and Gli2 to promote Huh-7 cell migration and invasion is dependent on ERK1/2 phosphorylation.
Figure 5.
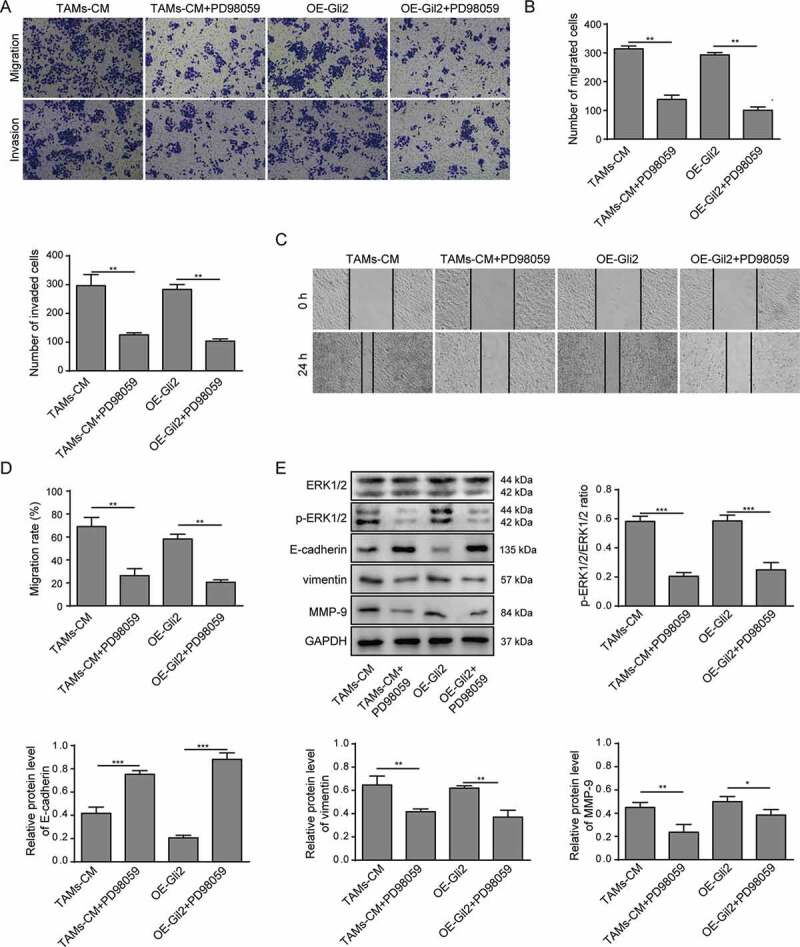
TAMs-CM promotes the migration and invasion of Huh-7 cells through the Gli2/IGF-II/ERK1/2 axis
(A-B) Huh-7 cells were treated with TAMs-CM, TAMs-CM and PD98059, Gli2 overexpression, or Gli2 overexpression and PD98059. Cell invasion and migration were measured by Transwell assay. (C-D) Cell migration was confirmed by scratch assay. (E) The activation of ERK1/2 and the expression of E-cadherin, vimentin and MMP-9 were analyzed by western blotting. *** p < .001, ** p < .01, * p < .05.
TAMs promote Huh-7 tumor growth in vivo mediated by TGF-β1
We also evaluated whether TGF-β1 secreted by TAMs was involved in tumor growth in vivo. Lentivirus was employed to knockdown the expression of TGF-β1 in TAMs. As shown in Figure 6A-B, compared with control TAMs or TAMs infected with lentivirus harboring scrambled control shRNA (sh-NC), the TAMs infected with lentivirus harboring shRNA targeting TGF-β1 (sh-TGF-β1) showed robustly decreased TGF-β1 expression, as demonstrated by qPCR (Figure 6A) and western blotting (Figure 6B). Huh-7 cells were then injected subcutaneously with different TAMs into nude mice. The tumor sizes were measured, intriguingly, the TAMs with TGF-β1 knockdown showed a reduced capability to promote tumor growth compared with control TAMs (sh-NC TAMs, Figure 6C-D). Likewise, the weights of tumors induced by TAMs were decreased upon TGF-β1 knockdown in TAMs (Figure 6D).
Figure 6.
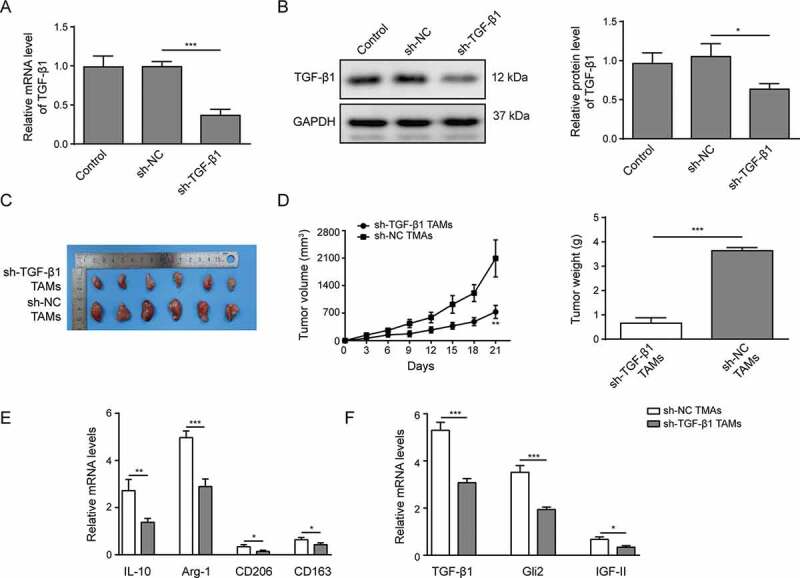
TAMs promote Huh-7 tumor growth in vivo mediated by TGF-β1
(A-B) TAMs were infected with lentivirus harboring scrambled control shRNA (sh-NC) or shRNA targeting TGF-β1 (sh-TGF-β1). The expression of TGF-β1 was examined by qPCR (A) and western blotting (B). (C) An image of the tumors collected after the mice were sacrificed. (D) The growth rate and weight of tumors in each group. (E) The expression levels of M2 macrophage markers IL-10, Arg-1, CD206 and CD163 in isolated tumors, as detected by qPCR. (F) The expression levels of TGF-β1, Gli2 and IGF-II in tumors detected by qPCR. *** p < .001, ** p < .01, * p < .05.
Furthermore, we isolated the tumors and probed their characteristics. As shown in Figure 6E, the expression levels of M2 macrophage markers IL-10, Arg-1, CD206 and CD163 were reduced upon TGF-β1 knockdown, indicating that the activation of TAMs was suppressed. In addition, we observed that TGF-β1, Gli2 and IGF-II were downregulated in the tumor tissues from mice that were co-injected with sh-TGF-β1 TAMs (figure 6F). Altogether, our data demonstrate that TAMs promote the tumor growth in vivo mediated by TGF-β1.
Discussion
Increasing evidence suggests that TAMs participate in tumor invasion and metastasis.25,26 Here, we showed that TAMs promoted the proliferation, migration and invasion of human hepatoma Huh-7 cells in vitro, as well as tumor growth in vivo. This promotional ability was mediated by the secretion of TGF-β1, which induced the expression of Gli2 and IGF-II, and then activated ERK1/2 signaling pathway in tumor cells. Our study elucidated the molecular mechanism by which TAMs promote the malignant phenotype of Huh-7 cells. These findings identify potential therapeutic targets for HCC treatment.
High expression levels of Gli2 have been shown to be associated with metastasis in many tumor types.5,27 We found that TAMs promoted the proliferation, migration and invasion of Huh-7 cells by elevating the expression of Gli2, this promotion was suppressed when Gli2 was downregulated. Furthermore, the expression of Gli2 was reduced by TGF-β1 inhibitor, which was consistent with the findings of previous reports showing that Gli2 is a direct target of TGF-β in various cancers.14,28–30 We also found that the migration and invasion abilities of Huh-7 cells promoted by the CM of TAMs could be reduced by the TGF-β1 inhibitor. Accordingly, we found that the capability of TAMs to induce tumor growth was diminished by TGF-β1 suppression in the mouse model. The expression of E-cadherin in HCC cells was decreased upon TAMs-CM treatment or Gli2 overexpression. These results are in accordance with previous reports showing that the expression of E-cadherin is repressed by Gli2 in human melanoma cells.14 In addition, vimentin and MMP-9 in HCC cells were upregulated by TAMs-CM treatment.31,32 Although the regulation of Gli2 by TGF-β1 has been reported in many kinds of cancers, we explain the mechanism by which TAMs enhance the migration and invasion of Huh-7 cells and provide the first evidence that TGF-β1 regulates the migration and invasion of human hepatoma cells through regulating Gli2 expression.
The mitogenic peptide IGF-II is well known to exert critical roles in oncogenesis.33,34 In this study, we found that the expression of IGF-II was regulated by Gli2 and could promote the migration and invasion of Huh-7 cells. The downregulation of IGF-II enhanced the survival of HCC mice.16 Curcumin was reported to inhibit IGF-II expression and suppress tumor development in a rat model of bladder cancer.15 Together, these findings suggest that IGF-II could be a potential therapeutic target for cancer treatment, and the current study is the first to report that Gli2 might promote HCC progression by upregulating IGF-II.
Many signaling pathways have been found to be dysregulated in cancer. In particular, the ERK pathway is reportedly involved in various cancers.35 Our data showed ERK1/2 activation upon the upregulation of IGF-II, and the inhibition of ERK1/2 phosphorylation reduced the migration and invasion of Huh-7 cells that was induced by Gli2 or IGF-II overexpression, further confirming the central role of ERK1/2 in cancer progression.36 In addition, the expression of E-cadherin was increased and the expression of vimentin and MMP-9 were decreased by ERK1/2 inhibitor, suggesting that ERK1/2 signaling might regulate the expression of cancer-related genes.
In conclusion, the present study explored the molecular mechanism by which TAMs influence the migration and invasion abilities of human hepatoma Huh-7 cells. TGF-β1 secreted by TAMs was found to promote HCC cell migration and invasion by elevating the expression of Gli2 and IGF-II, which then induced ERK1/2 activation. Although we elucidated the mechanism with in vitro and in vivo experiments, it would be more solid if we use multiple hepatoma cell lines. Nevertheless, our study provides novel insights into the molecular mechanisms of HCC malignancy and offers potential therapeutic targets for HCC treatment.
Supplementary Material
Acknowledgments
This work is supported by National Natural Science Foundation of China (No. 81660467); Youth Science Foundation of Jiangxi Province (20171BAB215081)
Funding Statement
This work was supported by the National Natural Science Foundation of China [81660467]; Natural Science Fund project of Jiangxi province [20171BAB215081].
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Disclosure statement
The authors declare no conflict of interests.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Cancer Genome Atlas Research Network . Electronic address wbe, cancer genome atlas research N. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017;169(7):1327–1341e1323. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forner A, Llovet JM, Bruix J.. 2012. Hepatocellular carcinoma. Lancet. 379(9822):1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 3.Kulik L, El-Serag HB.. 2019. Epidemiology and management of hepatocellular carcinoma. Gastroenterology. 156(2):477–491 e471. doi: 10.1053/j.gastro.2018.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruix J, Gores GJ, Mazzaferro V. 2014. Hepatocellular carcinoma: clinical frontiers and perspectives. Gut. 63(5):844–855. doi: 10.1136/gutjnl-2013-306627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruiz I Altaba A, Sanchez P, Dahmane N. 2002. Gli and hedgehog in cancer: tumours, embryos and stem cells. Nat Rev Cancer. 2(5):361–372. doi: 10.1038/nrc796. [DOI] [PubMed] [Google Scholar]
- 6.Javelaud D, Alexaki VI, Dennler S, Mohammad KS, Guise TA, Mauviel A. 2011. TGF-beta/SMAD/GLI2 signaling axis in cancer progression and metastasis. Cancer Res. 71(17):5606–5610. doi: 10.1158/0008-5472.CAN-11-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thiyagarajan S, Bhatia N, Reagan-Shaw S, Cozma D, Thomas-Tikhonenko A, Ahmad N, Spiegelman VS. 2007. Role of GLI2 transcription factor in growth and tumorigenicity of prostate cells. Cancer Res. 67(22):10642–10646. doi: 10.1158/0008-5472.CAN-07-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narita S, So A, Ettinger S, Hayashi N, Muramaki M, Fazli L, Kim Y, Gleave ME. 2008. GLI2 knockdown using an antisense oligonucleotide induces apoptosis and chemosensitizes cells to paclitaxel in androgen-independent prostate cancer. Clin Cancer Res. 14(18):5769–5777. doi: 10.1158/1078-0432.CCR-07-4282. [DOI] [PubMed] [Google Scholar]
- 9.Zhang D, Cao L, Li Y, Lu H, Yang X, Xue P. 2013. Expression of glioma-associated oncogene 2 (Gli 2) is correlated with poor prognosis in patients with hepatocellular carcinoma undergoing hepatectomy. World J Surg Oncol. 11:25. doi: 10.1186/1477-7819-11-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. 2002. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 23(11):549–555. doi: 10.1016/S1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 11.Wan S, Zhao E, Kryczek I, Vatan L, Sadovskaya A, Ludema G, Simeone DM, Zou W, Welling TH. 2014. Tumor-associated macrophages produce interleukin 6 and signal via STAT3 to promote expansion of human hepatocellular carcinoma stem cells. Gastroenterology. 147(6):1393–1404. doi: 10.1053/j.gastro.2014.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan QM, Jing YY, Yu GF, Kou XR, Ye F, Gao L, Li R, Zhao QD, Yang Y, Lu ZH, et al. 2014. Tumor-associated macrophages promote cancer stem cell-like properties via transforming growth factor-beta1-induced epithelial-mesenchymal transition in hepatocellular carcinoma. Cancer Lett. 352(2):160–168. DOI: 10.1016/j.canlet.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Gupta DK, Singh N, Sahu DK. 2014. TGF-beta mediated crosstalk between malignant hepatocyte and tumor microenvironment in hepatocellular carcinoma. Cancer Growth Metast. 7:1–8. doi: 10.4137/CGM.S14205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perrot CY, Gilbert C, Marsaud V, Postigo A, Javelaud D, Mauviel A. 2013. GLI2 cooperates with ZEB1 for transcriptional repression of CDH1 expression in human melanoma cells. Pigment Cell Melanoma Res. 26(6):861–873. doi: 10.1111/pcmr.12149. [DOI] [PubMed] [Google Scholar]
- 15.Tian B, Zhao Y, Liang T, Ye X, Li Z, Yan D, Fu Q, Li Y. 2017. Curcumin inhibits urothelial tumor development by suppressing IGF2 and IGF2-mediated PI3K/AKT/mTOR signaling pathway. J Drug Target. 25(7):626–636. doi: 10.1080/1061186X.2017.1306535. [DOI] [PubMed] [Google Scholar]
- 16.Yao X, Hu JF, Daniels M, Shiran H, Zhou X, Yan H, Lu H, Zeng Z, Wang Q, Li T, et al. 2003. A methylated oligonucleotide inhibits IGF2 expression and enhances survival in a model of hepatocellular carcinoma. J Clin Invest. 111(2):265–273. DOI: 10.1172/JCI15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tai BJ, Yao M, Zheng WJ, Shen YC, Wang L, Sun JY, Wu MN, Dong ZZ, Yao DF. 2019. Alteration of oncogenic IGF-II gene methylation status associates with hepatocyte malignant transformation. Hbpd Int. 18(2):158–163. doi: 10.1016/j.hbpd.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 18.Brouwer-Visser J, Huang GS. 2015. IGF2 signaling and regulation in cancer. Cytokine Growth Factor Rev. 26(3):371–377. doi: 10.1016/j.cytogfr.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Huang R, Wang S, Wang N, Zheng Y, Zhou J, Yang B, Wang X, Zhang J, Guo L, Wang S, et al. 2020. CCL5 derived from tumor-associated macrophages promotes prostate cancer stem cells and metastasis via activating β-catenin/STAT3 signaling. Cell Death Dis. 11(4):234. DOI: 10.1038/s41419-020-2435-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang L, Zhang Y. 2017. Tumor-associated macrophages: from basic research to clinical application. J Hematol Oncol. 10:58. doi: 10.1186/s13045-017-0430-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tao YSG, Zhu Q, Dai D, Li N, Wang H, Chen X, Hou D, Wang Y, Pan Q, Xu J, et al. 2019. Modulation of M2 macrophage polarization by the crosstalk between Stat6 and Trim24. Nat Commun. 10:4353. doi: 10.1038/s41467-019-12384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genin M, Clement F, Fattaccioli A, Raes M, Michiels C. 2015. M1 and M2 macrophages derived from THP-1 cells differentially modulate the response of cancer cells to etoposide. BMC Cancer. 15:577. doi: 10.1186/s12885-015-1546-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malik R, Lelkes PI, Cukierman E. 2015. Biomechanical and biochemical remodeling of stromal extracellular matrix in cancer. Trends Biotechnol. 33(4):230–236. doi: 10.1016/j.tibtech.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji S, Qin Y, Shi S, Liu X, Hu H, Zhou H, Gao J, Zhang B, Xu W, Liu J, et al. 2015. ERK kinase phosphorylates and destabilizes the tumor suppressor FBW7 in pancreatic cancer. Cell Res. 25(5):561–573. DOI: 10.1038/cr.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Yao W, Yuan Y, Chen P, Li B, Li J, Chu R, Song H, Xie D, Jiang X, et al. 2017. Targeting of tumour-infiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut. 66(1):157–167. DOI: 10.1136/gutjnl-2015-310514. [DOI] [PubMed] [Google Scholar]
- 26.Komohara Y, Jinushi M, Takeya M. 2014. Clinical significance of macrophage heterogeneity in human malignant tumors. Cancer Sci. 105(1):1–8. doi: 10.1111/cas.12314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen F, Mo J, Zhang L. 2016. Long noncoding RNA BCAR4 promotes osteosarcoma progression through activating GLI2-dependent gene transcription. Tumour Biol. 37(10):13403–13412. doi: 10.1007/s13277-016-5256-y. [DOI] [PubMed] [Google Scholar]
- 28.Islam SS, Mokhtari RB, Noman AS, Uddin M, Rahman MZ, Azadi MA, Zlotta A, van der Kwast T, Yeger H, Farhat WA. 2016. Sonic hedgehog (Shh) signaling promotes tumorigenicity and stemness via activation of epithelial-to-mesenchymal transition (EMT) in bladder cancer. Mol Carcinog. 55(5):537–551. doi: 10.1002/mc.22300. [DOI] [PubMed] [Google Scholar]
- 29.Dennler S, André J, Alexaki I, Li A, Magnaldo T, Ten Dijke P, Wang XJ, Verrecchia F, Mauviel A. 2007. Induction of sonic hedgehog mediators by transforming growth factor-beta: smad3-dependent activation of Gli2 and Gli1 expression in vitro and in vivo. Cancer Res. 67(14):6981–6986. doi: 10.1158/0008-5472.CAN-07-0491. [DOI] [PubMed] [Google Scholar]
- 30.Shao JB, Gao ZM, Huang WY, Lu ZB. 2017. The mechanism of epithelial-mesenchymal transition induced by TGF-β1 in neuroblastoma cells. Int J Oncol. 50(5):1623–1633. doi: 10.3892/ijo.2017.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gan L, Qiu Z, Huang J, Li Y, Huang H, Xiang T, Wan J, Hui T, Lin Y, Li H, et al. 2016. Cyclooxygenase-2 in tumor-associated macrophages promotes metastatic potential of breast cancer cells through Akt pathway. Int J Biol Sci. 12(12):1533–1543. DOI: 10.7150/ijbs.15943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu CY, Xu JY, Shi XY, Huang W, Ruan TY, Xie P, Ding JL. 2013. M2-polarized tumor-associated macrophages promoted epithelial-mesenchymal transition in pancreatic cancer cells, partially through TLR4/IL-10 signaling pathway. Lab Invest. 93(7):844–854. doi: 10.1038/labinvest.2013.69. [DOI] [PubMed] [Google Scholar]
- 33.Diaz LE, Chuan YC, Lewitt M, Fernandez-Perez L, Carrasco-Rodriguez S, Sanchez-Gomez M, Flores-Morales A. 2007. IGF-II regulates metastatic properties of choriocarcinoma cells through the activation of the insulin receptor. Mol Hum Reprod. 13(8):567–576. doi: 10.1093/molehr/gam039. [DOI] [PubMed] [Google Scholar]
- 34.Moorehead RA, Sanchez OH, Baldwin RM, Khokha R. 2003. Transgenic overexpression of IGF-II induces spontaneous lung tumors: a model for human lung adenocarcinoma. Oncogene. 22(6):853–857. doi: 10.1038/sj.onc.1206188. [DOI] [PubMed] [Google Scholar]
- 35.Montagut C, Settleman J. 2009. Targeting the RAF-MEK-ERK pathway in cancer therapy. Cancer Lett. 283(2):125–134. doi: 10.1016/j.canlet.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 36.Bao L, Yan Y, Xu C, Ji W, Shen S, Xu G, Zeng Y, Sun B, Qian H, Chen L, et al. 2013. MicroRNA-21 suppresses PTEN and hSulf-1 expression and promotes hepatocellular carcinoma progression through AKT/ERK pathways. Cancer Lett. 337(2):226–236. DOI: 10.1016/j.canlet.2013.05.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


