Abstract
Potassium (K+) acquisition, translocation and cellular homeostasis are mediated by various membrane transport systems in all organisms. We identified and described an ion channel in the ectomycorrhizal fungus Hebeloma cylindrosporum (HcSKC) that harbors features of animal voltage-dependent Shaker-like K+ channels, and investigated its role in both free-living hyphae and symbiotic conditions. RNAi lines affected in the expression of HcSKC were produced and used for in vitro mycorrhizal assays with the maritime pine as host plant, under standard or low K+ conditions. The adaptation of H. cylindrosporum to the downregulation of HcSKC was analyzed by qRT-PCR analyses for other K+-related transport proteins: the transporters HcTrk1, HcTrk2, and HcHAK, and the ion channels HcTOK1, HcTOK2.1, and HcTOK2.2. Downregulated HcSKC transformants displayed greater K+ contents at standard K+ only. In such conditions, plants inoculated with these transgenic lines were impaired in K+ nutrition. Taken together, these results support the hypothesis that the reduced expression of HcSKC modifies the pool of fungal K+ available for the plant and/or affects its symbiotic transfer to the roots. Our study reveals that the maintenance of K+ transport in H. cylindrosporum, through the regulation of HcSKC expression, is required for the K+ nutrition of the host plant.
Introduction
Potassium (K+) is an essential cation involved in many biological processes in plants, such as growth, photosynthesis and stress tolerance [1,2]. However, most soil K+ cations are complexed with minerals such as feldspars or micas, resulting in low mobility and availability. As a consequence, productivity of agricultural and agroforestry ecosystems is strongly dependent on large and regular addition of chemical fertilizers [3]. Plants developed various strategies to overcome this lack of K+ availability and to improve its uptake, such as expression of high-affinity transport systems, exudation of organic acids by the root system, and symbiotic associations with soil-living microbes (reviewed in [4–7]).
Ectomycorrhizal (ECM) symbiosis is an intimate relationship between the roots of woody plants and the mycelium of soil-borne fungi, and constitutes a major component of boreal and temperate forest ecosystems [8]. The main benefit of this association for the host plants is the improvement of macro- and micronutrient acquisition [9]. In ECM structures, nutrients are transferred from the soil-exploring extraradical mycelium to the hyphae developing in planta, through a fungal sheath surrounding the roots. The intra-radical colonization by the fungus forms a specific structure called the Hartig net. This net develops between the host root epidermis and cortex, where various transport proteins are expressed to drive nutrient exchanges between the two partners [9,10]. While most reports have focused on nitrogen and phosphorus nutrition in ECM plants, some studies have reported a significant transfer of fungal K+ towards the host, particularly in K+-limited and stressful conditions [11,12]. One hypothesis is that the ectomycorrhiza-mediated K+ nutrition could also be a strategy for the host plant to alleviate salt stress responses [13]. The transfer of K+ through the mycorrhizal pathway requires the expression and organization of fungal transport proteins specific to the soil-fungus or plant-fungus interfaces, allowing K+ movements from the soil into colonized roots. The fine tuning of this putative fungal hyphae polarization seems to be required for the distribution of K+ from the soil to plant cortical apoplasm [4].
Several proteins from ECM fungi involved in plant nutrient allocation have been identified and characterized [9,14–16], and some of these might be responsible for the maintenance of ECM symbiosis, in addition to exclusive involvement in trophic exchanges [17]. To our knowledge, only a handful of transport proteins involved in K+ transport has been reported in ECM fungi thus far. Among them, HcTrk1 from Hebeloma cylindrosporum [18] is specifically localized in extra-radical hyphae of H. cylindrosporum-Pinus pinaster ectomycorrhizas, suggesting a role in K+ uptake from the soil [12]. Genome sequencing of H. cylindrosporum [19] has allowed the identification of other putative transporters possibly involved in K+ acquisition from the soil, HcTrk2 and HcHAK [4], but their roles remain to be elucidated. Recently, we also described for the first time three fungus-specific tandem-pore outwardly rectifying K+ (TOK) channels from H. cylindrosporum [20,21]. Excitingly, we were able to use an overexpression approach to demonstrate that one of them, HcTOK2.2, participates in the release of K+ towards colonized P. pinaster roots.
In plants and animals, the efflux of K+ from cells is a crucial biological process involved in the electrical polarization and energization of the plasma membrane, as well as in adaptation to biotic and abiotic stresses. Potassium channels and non-selective cation channels are involved in such mechanisms [22]. The large family of tetrameric voltage-dependent K+ channels (KV), often called Shaker-like channels, constitutes a key component of the plasma membrane conductance. Many members are major contributors to the voltage-dependent influx or efflux of K+ in plants and animals [23–25]. To our knowledge, although some Shaker-like channels were identified in silico in ECM fungi, none has been studied so far [4]. Previous evidence in plants and animals establishes the relevance of this type of channels in the regulation of cellular K+ homeostasis, and suggests fungal Shaker-like channels are likely to have a similar function. Moreover, they are possible candidates for K+ allocation from ECM fungi towards the host plant at the symbiotic interface and/or K+ storage into the vacuole.
In this work, we provide the first description of the fungal Shaker-like KV channel family by in silico and phylogenetic analyses, and investigate the physiological role of one of them, HcSKC from H. cylindrosporum. To assess the role of this channel in the allocation of fungal K+ to the host plant P. pinaster, we generated transgenic fungal lines downregulated in HcSKC expression. We demonstrate that HcSKC silencing affects the fungal K+ homeostasis, alters the expression of other fungal K+ transport systems, and attenuates the K+ acquisition of ECM P. pinaster.
Materials and methods
Wild-type and transgenic fungal strains
The homokaryotic strain h7 of the ECM Basidiomycota H. cylindrosporum Romagnesi [26] was grown in the dark at 26°C in YMG medium (Yeast extract, Malt extract, Glucose) [27] either on agar-solidified Petri dishes or in liquid cultures.
The Agrobacterium tumefaciens-mediated genetic transformation process and production of the empty vector fungal strain (pPZP-133 control) have been detailed previously [12]. Using the same protocol and the bacterial strain LBA1126, carboxin-resistant RNAi-SKC fungal lines were produced.
Construction of RNAi-SKC plasmids
A 320 bp HcSKC-specific region was amplified on cDNA using RNAi-specific primers (S1 Table) for the construction of the silencing vector. The sense (S) and antisense (AS) fragments obtained were inserted into the two MCS of pSILBAγ vector used for gene silencing in Laccaria bicolor [28]. The silencing cassette was digested using XbaI enzyme and inserted in pPZP-133 plasmid in order to get the pPZP-RNAi-SKC vector.
Ectomycorrhiza production
Maritime pine seeds (P. pinaster Soland in Ait. from Medoc, Landes-Sore-VG source, France) were sterilized with 37% H2O2 [29] and sown on Petri dishes containing agarose (Eurobio Molecular Biology Grade) and 0.2% glucose. The co-culture method in glass tubes as well as the composition of standard K+ (SK, 1 mM K+) and low K+ (LK, 0.05 mM K+) liquid N1 media were described previously [12]. Culture conditions were 16 h-photoperiod (210 μmol.m-2.s-1), 20°C and 60% relative humidity. SK medium was used for in situ hybridization experiments, and both SK and LK media for assays analyzing the ECM phenotype of RNAi-SKC fungal lines. Six 2-month-old plants per condition were collected for each experiment.
In situ hybridization
Two successive PCR amplifications of HcSKC cDNA led to the synthesis of sense and antisense probes (300 bp). The first amplification was performed with ISHSKC-T7-S-F/ISHSKC-S-R or ISHSKC-AS-F/ISHSKC-T7-AS-R primers (S1 Table). A 1:100 dilution of the first PCR product was used for the second amplification with ISHT7-Prom and specific SKC-F or -R primers. Ectomycorrhizas were embedded in paraffin and 8 μm sections were obtained using a microtome. Sample preparation and hybridization were performed as previously described [20,30]. Slides were observed with a Leica DM6000 wide-field microscope (Montpelier RIO Imaging platform, www.mri.cnrs.fr) and pictures were analyzed by Volocity Acquisition 5.1.0 software (Perkin Elmer, www.perkinelmer.com).
Potassium shortage in Hebeloma cylindrosporum pure culture
Empty vector and RNAi-SKC fungal strains were cultivated in N6 liquid medium [31] containing 10 mM K+ (6 mM KNO3, 4 mM KCl). Fresh N6 was supplied on days 7 and 12. On day 14, thalli were washed five times with N6 medium without added K+ and cultivated in these media up to 48 h. In medium without K+, NO3 was added in the form of Ca(NO3)2 at the same concentration as in the standard K+ medium. Fungi were collected 0, 12, 24 and 48 h after K+ deprivation and washed twice in CaSO4 (0.2 mM)–glucose (5 gl-1) solution. Half of each sample was flash-frozen in liquid nitrogen for qRT-PCR analyses and the rest was dried for one week at 60°C, weighed and used for intracellular K+ content determination.
Additionally, four media with high or low concentrations of K+ (1 or 0.05 mM, respectively) and sodium (Na+; 1 or 0.2 mM, respectively) were used to assess fungal biomass of transgenic strains (S2 Table). These media were named: +K/-Na, +K/+Na, -K/-Na, and -K/+Na. Transgenic fungal strains were cultured on a solid version of these media for 1 week at 26°C, sub-cultured into 50 ml of their corresponding liquid media and placed at 26°C for 28 days. Thalli were collected, dried at 70°C, and dry weights were determined.
Quantification of potassium contents
Plant tissue and mycelia were collected and weighed to determine the dry biomass (DW). Acid extraction of tissue ion contents and assays of K+ content by flame atomic absorption spectrophotometry were performed as previously described [12].
qRT-PCR analyses
H. cylindrosporum RNA extraction, cDNA synthesis and qRT-PCR protocol were described [30]. Expression levels of the transport systems HcSKC (protein ID 79961; https://mycocosm.jgi.doe.gov/Hebcy2/Hebcy2.home.html), HcTOK1 (31571), HcTOK2.1 (129509), HcTOK2.2 (127201), HcTrk1 (445173), HcTrk2 (176376), and HcHAK (435192) were determined relatively to the internal control α-tubulin (24108) on mycelium samples (for primers see S1 Table).
In silico analysis of HcSKC
Hydrophobicity profiles of HcSKC (H. cylindrosporum), LbSKC (L. bicolor), and XlKV2.1 (Xenopus laevis) subunits were obtained using Kyte-Doolittle algorithm with a window size of 11 amino acids [32] (http://gcat.davidson.edu/DGPB/kd/kyte-doolittle.htm).
Amino acid alignment of HcSKC, LbSKC, and XlKV2.1 subunits was performed using Clustal Omega program (http://www.ebi.ac.uk/Tools/msa/) and formatted with BoxShade program (http://www.ch.embnet.org/software/BOX_form.html).
Homology structure of the HcSKC subunit was modeled by Swiss-Model server using the template structure of Rattus norvegicus RnKV2.1 subunit [33] (http://swissmodel.expasy.org/). Amino acid alignment between these two subunits was obtained using the same program.
Phylogenetic tree construction
The protein sequences of SKC subunits of 185 fungi, 4 animals and 2 plants were collected from JGI [34], NCBI (NCBI Resource Coordinator 2015), UniProt (The UniProt Consortium 2015) or TAIR (www.arabidopsis.org) databases by BLASTP analysis based on HcSKC of H. cylindrosporum (S2 Table). All fungal databases used here are publicly available. All sequences without a predicted G[YF]G[DE] pore motif were removed manually. Protein sequences were aligned with MUSCLE algorithm v3.8.31 [35] and the alignment was cured by Gblocks program [36]. Phylogenetic analysis was performed by PhyML [37] using the maximum likelihood method (1000 bootstraps) and the tree was visualized by Dendroscope software v3.2.10 [38].
Construction of the HcSKC-EGFP fusion and expression in Saccharomyces cerevisiae
The HcSKC-EGFP C-terminal cassette was created with a PCR fusion approach, amplifying separately HcSKC from H. cylindrosporum cDNA and EGFP from a preexisting construct, with primers overlapping the fusion region containing both HcSKC and EGFP sequences (S1 Table). A third PCR with the flanking primers was required to amplify the whole construct, which was later digested with restriction enzymes and inserted in the corresponding vectors. Subcellular localization of the HcSKC-EGFP construct in the yeast strain ply232 was attempted with two approaches, first a galactose-induced expression in the vector pYES2 (ThermoFisher Scientific) and later under the constitutive promoter PGK in the pFL61 vector [39].
Statistical analyses
Data normality was checked using the Wilk-Shapiro test. Differences between averages were analyzed by one-way ANOVA followed by Tukey HSD post-hoc tests, or by Student’s test, depending on the experiment. Mean comparisons were carried out using Dunnett’s test. Dunnett’s test was also used to search for significant differences in pairwise comparisons. All statistical analyses were performed with R software at the 5% level of statistical significance.
Results
Identification of a Shaker-like KV channel in Hebeloma cylindrosporum
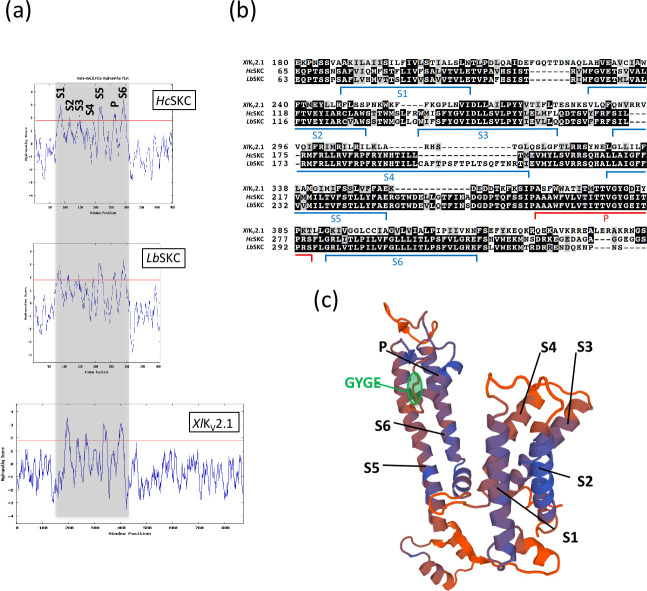
We previously identified a putative K+ channel (STC no.: 009A07R1.0A1.1) in EST resources obtained from H. cylindrosporum pure cultures [40]. BLASTP analysis of 009A07R1.0A1.1 protein sequence against the L. bicolor S238N genomic database allowed the identification of a unique orthologue (protein ID 297800; https://mycocosm.jgi.doe.gov/Lacbi2/Lacbi2.home.html). The hydrophobicity profile of these two proteins generated by the Kyte-Doolittle algorithm revealed the presence of 7 hydrophobic domains, similar to the XlKV2.1 Shaker channel from X. laevis (Fig 1A). The amino acid alignment of these fungal Shaker-like proteins with XlKV2.1 confirmed the presence of 6 conserved transmembrane domains and one pore domain characterized by the GYGE motif ensuring K+ selectivity (Fig 1B). Consequently, we named these proteins SKC (Shaker-like K+-Channel). The S4 domain (constituting the voltage sensor in Shaker channels) of HcSKC and LbSKC harbored several arginine residues (R). Such positively charged residues have been shown in other KV channels to contribute to the channel voltage sensitivity and gating charge [41]. When compared with XlKV2.1, HcSKC and LbSKC have a shorter C-terminal region, suggesting reduced "built-in" regulation mechanisms (Fig 1A). Based on these in silico analyses, we used the R. norvegicus RnKV2.1 channel (NP_037318.1; S3 Table) as a template to propose a structural model for the fungal Shaker-like proteins (Fig 1C, S1 Fig).
Fig 1. In silico prediction of a Shaker-like channel in the fungus Hebeloma cylindrosporum.
The hydrophobicity profiles of the Shaker subunits from H. cylindrosporum (HcSKC), L. bicolor (LbSKC), and X. laevis (XlKV2.1) (a) and their partial amino acid sequence alignment (b) predicted six transmembrane domains (S1 to S6). One pore domain in HcSKC and LbSKC subunits containing the conserved G[YF]G[DE] motif of the Shaker channel selectivity filter was also detected. The S4 transmembrane domain corresponds to the predicted voltage-sensor domain. (c) The plausible 3-D model of the fungal HcSKC subunit showing the transmembrane (S1 to S6) and pore (P) domains was generated by homology with the 3-D structure of RnKV2.1 K+ channel from R. norvegicus (NP_037318.1; S2 Table) as a template using Swiss-Model server [33] (http://swissmodel.expasy.org/). The GYGE signature motif of the pore region was highlighted in green. The sequence alignment of HcSKC and RnKV2.1 polypeptides is displayed in S1 Fig.
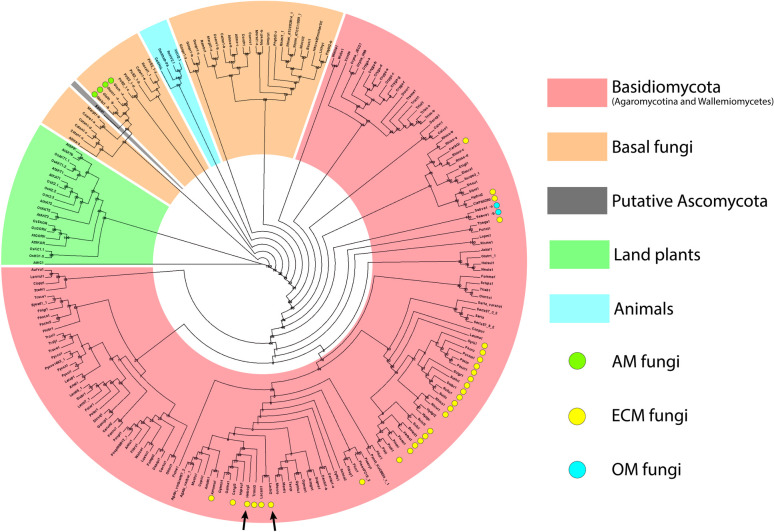
Distribution of Shaker-like channels in the fungal kingdom
Using HcSKC as a reference, we performed BLASTP analysis on publicly available sequenced fungi found on NCBI and JGI fungal portals (http://genome.jgi-psf.org/programs/fungi/index.jsf). In addition, a cure based on the presence of a putative G[YF]G[DE] pore motif was achieved. In the Basidiomycota phylum, the presence of putative SKC proteins was detected only in the Agaricomycotina subphylum, as is defined in the JGI phylogenetic tree (Fig 2). In addition, putative SKC proteins were found in Wallemia melicola, which belongs to Wallemiomycetes, a sister group to Agaricomycotina [42]. Two putative Shaker-like channels were also detected in the arbuscular mycorrhizal (AM) fungus Rhizophagus irregularis (Glomeromycotina subphylum) and other basal fungi belonging to Cryptomycota, Chytridiomycota, Blastocladiomycota, Zoopagomycota and Mucoromycota phyla (Fig 2). Surprisingly, no SKC was found in the Pucciniomycotina and Ustilaginomycotina subphyla of Basidiomycota, and in the Ascomycota phylum. Moreover, proteins similar to KV channels, but without the G[YF]G[DE] motif, were not found in these fungi, indicating the complete loss of Shaker-like genes in Pucciniomycotina, Ustilaginomycotina and Ascomycota species, with the exception in the Ascomycota phylum of Saitoella complicata, which presents one putative SKC channel (Fig 2).
Fig 2. Phylogenetic tree of KV channels in fungi, and representative animal and plant species.
The Shaker-like K+ channel tree of publicly available sequenced fungi from the Basidiomycota phylum and from basal fungi (Cryptomycota, Chytridiomycota, Blastocladiomycota, Zoopagomycota and Mucoromycota) was calculated by the maximum likelihood method (1000 bootstraps). Shaker-like proteins were found only in one putative Ascomycota species (Saitoella complicata) and not in the Pucciniomycotina and Ustilaginomycotina subphyla of Basidiomycota. Ectomycorrhizal (ECM), arbuscular mycorrhizal (AM), and orchid mycorrhizal (OM) fungi are indicated by yellow, green, and blue dots, respectively. KV channels from representative land plant and animal species were included in the tree. All species displayed in this tree are listed in the S2 Table. Shaker-like proteins from H. cylindrosporum (HcSKC) and L. bicolor (LbSKC) used in this study are indicated by black arrows.
The functional properties of HcSKC could not be revealed in various heterologous systems
Functional characterization is an important step for the analysis of K+ channel activity to predict transport features, such as selectivity and direction of rectification. Many strategies were applied to characterize HcSKC, using multiple heterologous systems (X. laevis oocytes and yeast) and co-injection of putative regulatory proteins (protein kinase K and putative channel β-subunits from H. cylindrosporum: HcKCNAB1 and HcKCNAB2) (S4 Table). No significant result was obtained from any of these experiments. Moreover, the expression of HcSKC triggers endogenous currents in X. laevis oocytes, making the observation of specific exogenous currents tricky (S4 Table). In addition, the previously identified HcSKC channel in the H. cylindrosporum h1 strain [40] has been extensively tested by voltage-clamp experiments in X. laevis oocytes, without any conclusive result (S4 Table). As an alternative, a functional characterization of the homologous channel in L. bicolor LbSKC was also attempted in X. laevis oocytes, but unsuccessfully (S4 Table). Finally, attempts to perform subcellular localizations in yeast using HcSKC-EGFP fused constructs driven by a constitutive or a galactose-inducible promoter failed too (S2 Fig). None of the experiments yielded a clear localization pattern of HcSKC, probably due to misprocessing of the protein product, reinforcing the issues in expressing HcSKC in the tested heterologous systems.
HcSKC transcripts are localized in all types of ectomycorrhizal hyphae
To elucidate the role of HcSKC channels in the symbiotic association with P. pinaster, the localization of corresponding transcripts was analyzed in 2-month-old ectomycorrhizas through in situ hybridization experiments. Cross-section and probe hybridization processes resulted in the detection of HcSKC transcripts in extra-radical hyphae, the hyphal mantle, and the Hartig net, indicating no specific expression of this channel in ectomycorrhizas (Fig 3). Only the antisense probe provided signals indicating the presence of HcSKC transcripts (Fig 3B). Since the sense probe sequence is not complementary to the HcSKC mRNA sequence, it cannot hybridize, and, therefore, serves as a negative control. Consequently, it did not give rise to any signal (Fig 3A). A construct harboring the EGFP gene marker fused to the HcSKC promoter was cloned to show that HcSKC was well expressed in free-living conditions (S3 Fig).
Fig 3. HcSKC transcript localization in Hebeloma cylindrosporum—Pinus pinaster ectomycorrhizas.
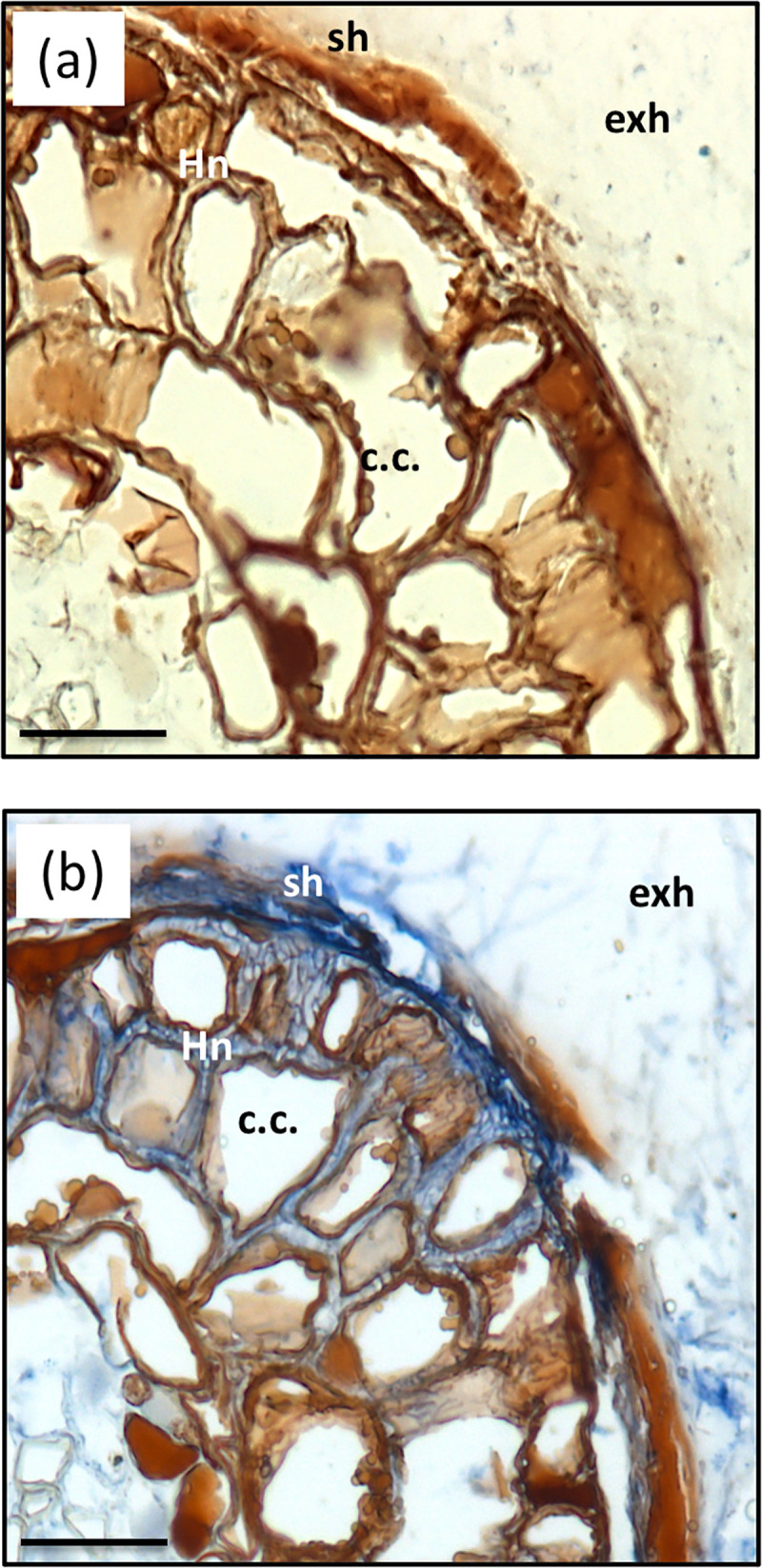
In situ hybridization in ectomycorrhizas from 2-month-old co-cultures of P pinaster–H. cylindrosporum with HcSKC-specific probes. (a) Control with a sense probe did not show any signal. (b) Using the antisense probe, HcSKC transcripts were detected in extra-radical hyphae, the fungal sheath and the Hartig net of ectomycorrhizas. Bars, 50 μm. c.c., cortical cell; exh, extraradical hyphae; Hn, Hartig net; sh, fungal sheath.
Silencing of HcSKC affects Hebeloma cylindrosporum potassium homeostasis
Decoding the role of HcSKC in axenic conditions was attempted by RNAi downregulation of the corresponding gene. HcSKC-silencing lines of H. cylindrosporum were produced by Agrobacterium tumefaciens–mediated transformation. The expression level of HcSKC was determined by qRT-PCR in fungal mycelia growing on selection medium. Two isolates displaying a five-fold decrease in HcSKC expression, named RNAi-SKC-5 and RNAi-SKC-7, were selected for ECM assays (S4 Fig). We then investigated the K+ accumulation and expression of K+ transport-related genes in RNAi-SKC-5 and RNAi-SKC-7 lines. In fungi grown in standard medium (i.e., in presence of a relatively high external K+, 1 mM), internal K+ content was significantly higher in RNAi-SKC transformants than in the control line, indicating an increase in K+ acquisition and/or a decrease in K+ efflux (Fig 4). Once these fungal isolates were transferred into the K+-free medium, no difference in K+ content between RNAi-SKC and control lines could be detected at 12, 24 and 48 h after the transfer (Fig 4). In addition, we assessed the effect of high and low K+ (1 and 0.05 mM, respectively) and Na+ (1 or 0.2 mM, respectively) availability on fungal growth in all transgenic strains (S5 Fig). All media were prepared on the basis of the media used for the subsequent ECM assay (S4 Table). After 28 days of culture in axenic condition, dry weights were recorded. Although the biomass of both RNAi-SKC transgenic lines differed in all conditions, they did not significantly differ from the control line, except for RNAi-SKC-5 that showed a slightly reduced biomass in +K/-Na condition compared to the control line (S5 Fig).
Fig 4. Potassium content in Hebeloma cylindrosporum HcSKC downregulated lines.
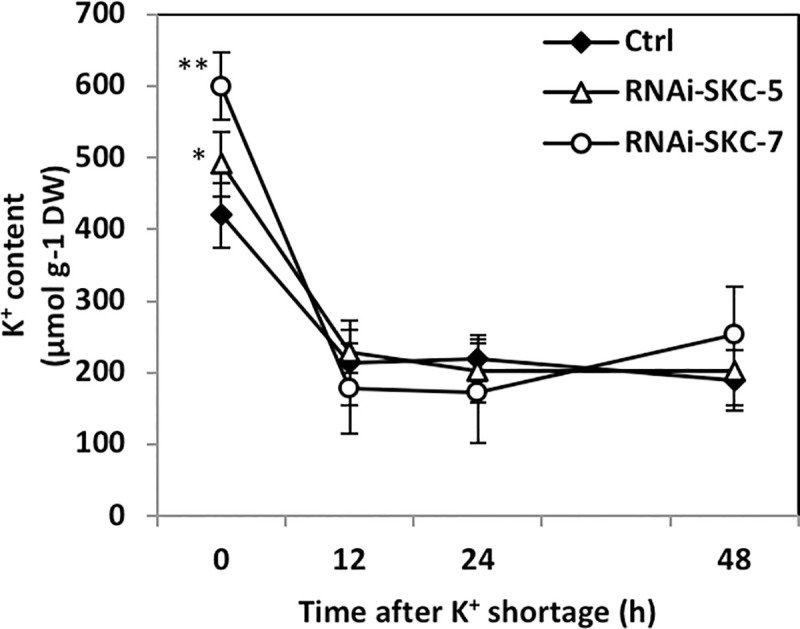
K+ content was measured in fungal lines transformed with the empty (Ctrl) or HcSKC-silencing vectors (RNAi-SKC-5 and RNAi-SKC-7) using an atomic absorption flame spectrophotometer. Statistical differences were evaluated using the Student’s test with respect to the Ctrl strain (*, P < 0.05; **, P < 0.01). n = 6.
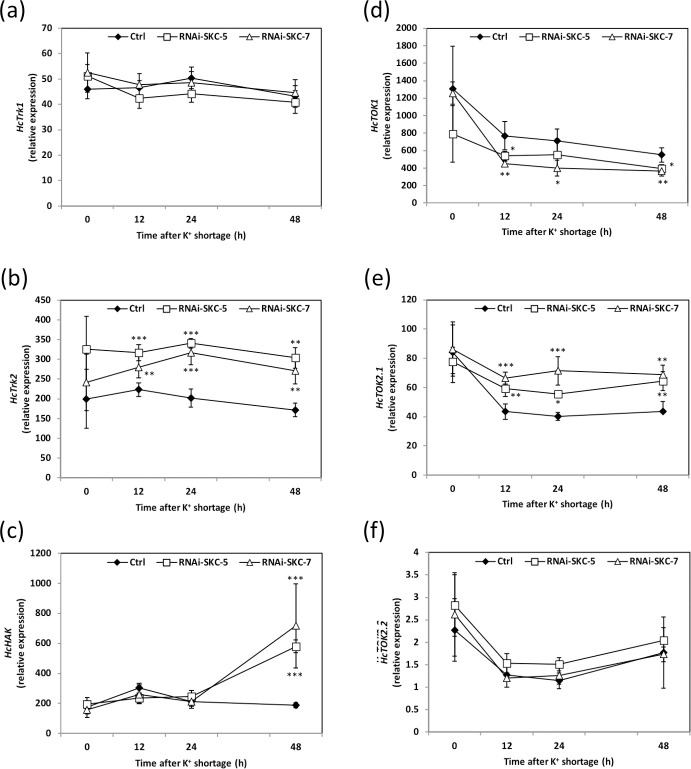
Besides HcSKC, six other putative K+ transport-related genes can be identified in the H. cylindrosporum genomic database [4,19]. To figure out whether their expression levels were affected by the downregulation of HcSKC, the same fungal samples as those used for K+ content measurements were analyzed by qRT-PCR. When grown in K+ standard pure culture conditions (SK medium), none of the six targeted K+ transport system genes was significantly differentially expressed in the RNAi-SKC lines compared to the control (Fig 5). In contrast, 12, 24 and 48 h after the transfer into the K+-free medium, HcTrk2 and HcTOK2.1 were upregulated and HcTOK1 was downregulated in the two RNAi-SKC lines (Fig 5B, 5D and 5E). Interestingly, HcHAK was upregulated in both RNAi lines only 48 h after the transfer into the K+-free medium (Fig 5C). The expression in the transgenic lines of the other two transport systems, HcTrk1 and HcTOK2.2, was not significantly affected by the transfer into the K+-free medium (Fig 5A–5F). Taken together, these data indicate that the downregulation of HcSKC is sufficient to modify the K+ homeostasis in H. cylindrosporum at standard K+. However, in conditions of K+ shortage, the modification of the expression of three K+ transport-related genes compensated for the effect of HcSKC downregulation and restored a wild-type K+ content in the fungus.
Fig 5. Relative expression of putative K+ channels and transporters in Hebeloma cylindrosporum control and HcSKC-silencing lines.
Relative expressions of the K+ transporters HcTrk1 (a), HcTrk2 (b), HcHAK (c) and the K+ channels HcTOK1 (d), HcTOK2.1 (e) and HcTOK2.2 (f) were analyzed in dependence on K+ shortage in control and RNAi fungal lines. The relative expressions of the K+ transporter HcTrk2 (b) and the K+ channels HcTOK1 (d) and HcTOK2.1 (e) were altered in RNAi-SKC-5 and RNAi-SKC-7 lines in comparison to the transgenic control line (Ctrl) under K+ limiting conditions. The relative expressions of HcTrk1 (a) and HcHAK (c) K+ transporters, and of the K+ channel HcTOK2.2 (f) were not affected by K+ shortage in RNAi-SKC-5 and RNAi-SKC-7 lines in comparison to the transgenic control line (Ctrl). Mean values are provided with the standard error (n = 6). Statistical differences were determined using a one-way ANOVA followed by Dunnett’s test relative to Ctrl strain for each data point P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***).
Downregulation of HcSKC affects K+ transfer to the host plant at standard K+ conditions
To investigate the role of HcSKC in symbiotic conditions, P. pinaster seedlings were inoculated with control, RNAi-SKC-5, or RNAi-SKC-7 lines, or kept non-inoculated, and grown in SK and LK conditions (Fig 6). Mycorrhization with the control fungal strain (transformed with the empty plasmid) resulted in statistically significant higher K+ contents in the plant roots and shoots when grown in LK, but not in SK, conditions. In comparison with these control plants, mycorrhizal association with the SKC-silencing fungal lines resulted in a significant decrease of K+ contents in shoots and roots in SK conditions, indicating a less cooperative behavior of H. cylindrosporum that affected K+ transfer to P. pinaster (Fig 6A). Surprisingly, this negative effect of HcSKC silencing on K+ contents in the host plant was not observed in LK conditions (Fig 6B). Hence, in low K+ conditions, the mycorrhizal pathway for plant K+ nutrition was not affected by the reduced expression of HcSKC. This result suggests that, in this LK condition, other underlying mechanisms are at work to achieve an efficient mycorrhizal K+ transport towards the host plant. In other words, HcSKC seems to play a significant role in K+ delivery toward the plant in SK conditions (1 mM), and not in conditions of K+ shortage (0.05 mM).
Fig 6. Potassium content in 2-month-old Pinus pinaster plants growing alone or in co-culture with Hebeloma cylindrosporum under K+-sufficient or -deficient conditions.
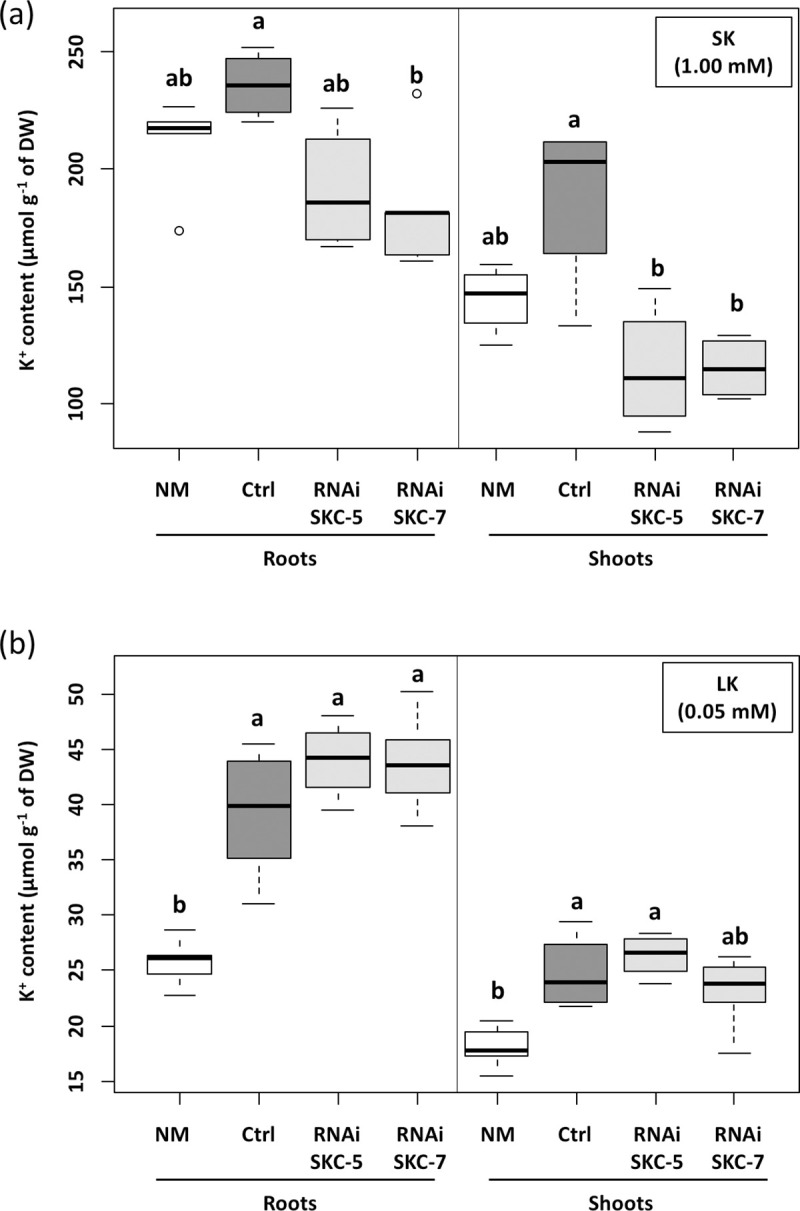
Root and shoot contents were measured in non-mycorrhizal plants (NM) and plants inoculated with fungi transformed with the empty (Ctrl) or HcSKC-silencing vectors (RNAi-SKC-5 and RNAi-SKC-7), and growing at standard K+ (SK medium, 1 mM K+, a) or low K+ (LK medium, 0.05 mM K+, b). Mean values are provided with the standard error (n = 5–6). Different letters indicate significant differences between treatments according to one-way ANOVA followed by Tukey HSD post-hoc tests (P < 0.05).
Discussion
As has recently been revealed, K+ acquisition in higher fungi seems to be mediated by membrane transport proteins belonging to the Trk/Ktr/HKT and KT/KUP/HAK families [4,43]. In addition, ACU ATPases have been reported as being involved in K+ and Na+ uptake in Ustilago maydis, and more generally envisioned as key players of K+ transport in fungi [7,44]. PAT ATPases could emerge as mechanisms for absorption of these cations as well, but they have not been studied thoroughly [45,46]. Regarding outward K+ transport systems, yeast and filamentous fungi developed TOK and Shaker-like K+ channels, from which only some members have been investigated thus far [20,21]. To our knowledge, this work reports the first description of a Shaker-like KV channel in higher fungi and highlights its role in both K+ homeostasis and mycorrhizal symbiosis.
In silico analysis of HcSKC revealed features from animal Shaker K+ channels
The initial identification of the HcSKC Shaker-like channel was performed from an EST library of H. cylindrosporum produced in free-living conditions [40]. In this study, one putative Shaker-like K+ channel was found to be highly expressed (STC no.: 009A07R1.0A1.1), suggesting an important role of this transport system in the fungus. Thanks to the release of H. cylindrosporum h7 genomic data [19,47,48], in silico analyses confirmed the homology of this fungal candidate to animal and plant KV channels (Fig 1). Moreover, phylogenetic analyses provided additional evidence on the proximity of fungal Shaker channels to animal ones (Fig 2). The major difference between HcSKC/LbSKC and animal channels is the length of their C-terminal regions. Several studies have evidenced a crucial role of this C-terminal region in the regulation of plant and animal Shaker channel activity [49,50]. In plants, it was reported that the C-terminal regions are involved in intracellular K+ sensing, heteromerization or voltage-dependent gating mechanisms [51–53]. In animal Shaker-type channels, the C-terminus can play multiple roles e.g. in regulation by β-subunits [54], by voltage [55], or in localization [56]. The fact that the C-terminal domains of H. cylindrosporum and L. bicolor are significantly shorter than those of the animal Shaker channels (Fig 1) suggests differences in channel activity regulation. Moreover, LbSKC seems to diverge by an additional sequence in the region of the S4 domain (Fig 1B) with unknown function.
Shaker-like channels were lost in Pucciniomycotina, Ustilaginomycotina and Ascomycota phyla
Although well-described in plants and animals, the presence of KV channels in the fungal kingdom has been hypothetical until now. Indeed, no Shaker channel gene could be detected in the Saccharomyces cerevisiae genome sequence. Our phylogenetic analysis based on many fungal genomes belonging to Basidiomycota and Ascomycota phyla, as well as basal fungi (Cryptomycota, Chytridiomycota, Blastocladiomycota, Zoopagomycota and Mucoromycota phyla) revealed the absence of putative SKC proteins in Pucciniomycotina and Ustilaginomycotina, which are sub-phyla of Basidiomycota, and the entire Ascomycota phylum. Interestingly, one SKC protein was found in S. complicata, which is related to the Ascomycota phylum. It is, however, worth noting that the affiliation of S. complicata to Ascomycota remains unclear [57,58], and its SKC protein sequence clusters with those of basal fungi, in agreement with the idea that this species might belong to a more basal phylum than previously thought. Given that Shaker-like channels were found in basal fungi and in the most recent subphylum of Badisiomycota (Agaricomycotina), their absence in Pucciniomycotina, Ustilaginomycotina, and Ascomycota strongly suggests that they have been lost in the ancestors of these phyla. This indicates that other transport systems can fill the roles of Shaker channels and play a central role in the maintenance of K+ homeostasis in these organisms. The survey and analysis of other K+ channels found in fungi, such as the TOK channels [4], have been integral in describing other key regulators of K+ transport and homeostasis in fungi (e.g. [20,21]).
HcSKC cannot be functionally characterized in heterologous systems
Although several attempts using different setups, strategies and heterologous systems were performed, the functional characterization of H. cylindrosporum and L. bicolor Shaker-like channels is still missing (S4 Table). Similarly, no conclusive data were obtained after expressing a HcSKC-EGFP construct driven by a constitutive or a galactose-inducible promoter in S. cerevisiae (S2 Fig). This might result from the fact that conditions or proteins required for the expression, function, localization, and regulation of fungal SKC channels were missing in the tested heterologous systems. Recently, the similar failed expression of a zinc transporter from the ECM fungus Suillus luteus was reported, showing the risky nature of these experiments [59]. It is well-known that animal and plant Shaker channels can physically interact with modulatory β-subunits [60]. For example, rat KV1 Shaker channels are able to form stable complexes with cytosolic β-subunits (KVβ), which modify the channel activity [61,62]. In the model plant Arabidopsis thaliana, KAB1 is a protein related to animal β-subunits that interacts with the K+ channel KAT1 [63]. We identified ten putative β-subunits in the genomic database of H. cylindrosporum and two of them, HcKCNAB1 and HcKCNAB2, were co-expressed with HcSKC in X. laevis oocytes, resulting in no exogenous current detection. At this stage we cannot conclude whether fungal Shaker channels require β-subunits or other types of peptides for their activation. It is known that some compounds can alter the activity of voltage-dependent K+ channels, such as human hormones that inhibit KV1.3 and KV1.5 [64]. Thus, the presence in X. laevis oocytes of undefined compounds that would inhibit fungal SKC currents might be possible. Another possibility would also be a regulation by other membrane proteins. For instance, in the nematode Caenorhabditis elegans, the two-pore K+ channel SUP-9 forms a complex with two proteins, UNC-93 (Major Facilitator Superfamily) and SUP-10 (The Potassium Channel Regulatory Protein Sup-10 Family), which coordinate muscle contraction [65]. In A. thaliana, the homolog of UNC-93 regulates K+ translocation from the root to the aerial parts of the plant. Disruption of this gene leads to a phenotype similar to that of knockout mutants of the AtSKOR Shaker-like channel, indicating that they may function together, although no physical interactions have yet been observed [66,67]. There is one UNC-93 gene in the genome of H. cylindrosporum which is induced in ectomycorrhizas, compared to free-living mycelium, and could be a promising candidate to test as a regulator of the activity of HcSKC.
Calcium signaling pathways are well-known regulators of K+ transport systems in plants and animals. In plants, calcium mediates signals during biotic interactions and environmental stress, and regulates a series of proteins that can regulate the activity of K+ channels through phosphorylation, dephosphorylation, structural modification, and physical interaction [68]. In animals these pathways are different, but there is also evidence of the modulation of K+ channel transport by calcium-dependent phosphorylation and dephosphorylation [69]. In filamentous fungi, there are few links between the regulation of K+ transport and calcium signaling, probably because most of the studies have been carried on the model Ascomycota yeast S. cerevisiae [70]. However, the first description of Shaker-like K+ channels in fungi may lead to the discovery of new pathways. Supporting this hypothesis, a rapid survey of the genome of H. cylindrosporum yields many putative calcium-regulated genes: three calmodulins (Protein ID 385514, 443591 and 443596), one calcineurin B (444455) and a wide range of Ca2+/calmodulin-dependent and serine/threonine protein kinases. It is also tempting to imagine that the most promising heterologous system for the characterization of fungal transport systems would be S. cerevisiae, which is phylogenetically closer to higher fungi than animal cells. In general, the use of yeast seems more adapted for the characterization of inward transport systems than of outward systems, but so far it has not provided encouraging results in our uptake experiments (S4 Table). Direct transport measurements by the electrophysiological patch-clamp method might provide missing insights in fungal SKC functional properties.
Potassium homeostasis mediated by HcSKC is a prerequisite for K+ allocation to pine
In order to investigate in vivo the role of the KV channel SKC in both free-living and ECM conditions, we decided to generate RNAi fungal lines affected in HcSKC expression. Analysis of the phenotype of these transgenic fungal lines revealed a close relationship between HcSKC expression, K+ accumulation in the fungus, and improvement of K+ nutrition of the colonized plants. Indeed, HcSKC silencing resulted in contrasting phenotypes of both the fungus and the mycorrhizal plant, depending on external K+ availability. At 1 mM K+, the downregulation of HcSKC expression altered K+ accumulation in the free-living mycelia and the ECM plants. It should be noted that, in our previous report [12], H. cylindrosporum had a significant effect on K+ allocation to P. pinaster only in K+ limiting conditions and not in sufficient conditions. Such a phenotype is further confirmed in the present report by the comparison between plants inoculated by the control transformed fungus harboring the empty vector and the control non-mycorrhizal plants (Fig 4). However, this does not mean that K+ transfer from the fungus to the plant did not occur in standard (SK) conditions. Indeed, the direct uptake of K+ from the medium by the root system may be reduced upon mycorrhizal association, and this reduction could be compensated by fungal K+ allocation to the plants. Parts of the ECM root system are somehow isolated from the external medium due to a loss of root hairs and to the presence of the fungal mantle surrounding the roots, and thus are dependent on the ECM symbiont to acquire nutrients. Given that HcSKC RNAi transformants accumulated more K+ than the control strain at standard external K+, it may be assumed that, when such transformants were grown in association with P. pinaster, their reduced expression of HcSKC either modified the pool of fungal K+ available for the plant and/or affected its symbiotic transfer to host roots. This would result in impaired K+ nutrition of the host. It is worth noting that varying external concentrations of K+ and Na+ did not drastically impact the growth of the RNAi-SKC transgenic fungal lines (S5 Fig). This indicates that the alteration of K+ nutrition observed in plants colonized by the RNAi-SKC lines at standard K+ cannot be explained by a modification of fungal growth and sensitivity to external conditions, but by the actual downregulation of HcSKC. Interestingly, the over-accumulation of K+ in the RNAi-SKC transformants was not found to involve the upregulation of putative inward K+ transporters, and thus, might result from a decrease in K+ efflux from the fungus. In contrast, in low K+ conditions, three other putative K+ transport systems were up- or downregulated in the RNAi lines. We postulate that the altered expression of HcTrk2, HcHAK, HcTOK1 and HcTOK2.1 restored the overall fungal K+ transport activities and homeostasis, as suggested by the recovery of the fungal K+ content observed in free-living conditions (Fig 4). This could explain the restoration of the K+ contents displayed by the mycorrhizal plants, suggesting that efficient ectomycorrhiza-mediated K+ nutrition occurred again. Another hypothesis, that HcSKC is involved in K+ movements from/to the vacuole, cannot be completely excluded, even though only plasma membrane localization of this type of ion channel has been described in any organism so far. Subcellular localization at either the plasma membrane or at the tonoplast should allow us to fully appreciate the role of HcSKC, regarding its manifest importance in H. cylindrosporum biology under axenic and symbiotic conditions.
To summarize, homology of HcSKC with animal outward KV channels and the observed K+ nutrition-related mycorrhizal and fungal phenotypes lead to the simplest hypothesis: that the fungal Shaker-like channel HcSKC may be involved in K+ efflux from fungal cells, allowing K+ translocation towards the host plant. Thus, the role played by HcSKC in K+ homeostasis in H. cylindrosporum seems crucial for fungal K+ availability and/or release to the host plant.
Supporting information
Structure-based sequence alignment and sequence conservation between HcSKC and RnKV2.1 K+ channel from Rattus norvegicus. The GYGE motif of the pore domain was highlighted in green.
(TIF)
The HcSKC-EGFP construct was inserted into the pYES2 vector, with galactose-induced expression (a,b), or into the pFL61 vector, under the PGK constitutive promoter (c,d). None of these attempts yielded a clear localization pattern of HcSKC, probably due to misprocessing of the protein product.
(TIF)
EGFP expression was observed in fungal lines harboring the EGFP cassette under control of the promoters of HcSKC (pPZP-PSKC1-E). Fungal isolates were grown on YMG medium for 2–3 weeks before analysis. (a) bright field image (b) EGFP image (GFP filter 505–530 nm). Scale bar: 100 μm.
(TIF)
The relative expression of HcSKC was determined in empty vector (Ctrl), RNAi-SKC-5 and RNAi-SKC-7 fungal lines by qRT-PCR. Mean values are provided with the standard deviation (n = 6). Statistical differences were evaluated using the Student’s test with respect to E.V. strain (**, P < 0.01).
(TIF)
Dry weights of empty vector (Ctrl), RNAi-SKC-5 and RNAi-SKC-7 fungal lines were determined after 28 days of culture in media containing high (+K and +Na) or low concentration (-K and -Na) of potassium (K+) and sodium (Na+), respectively. +K corresponds to 1 mM of K+, -K to 0.05 mM of K+, +Na to 1 mM of Na+, and -Na to 0.2 mM of Na+. Full recipes are provided in S1 Table. Mean values are provided with the standard deviation (n = 4–6). Different letters indicate significant differences between treatments according to one-way ANOVA followed by Tukey HSD post-hoc tests (P < 0.05).
(TIF)
Restriction enzyme sites indicated in primer name, were underlined in the corresponding 5’-3’ sequence.
(TIF)
Final concentrations of components constituting the +K/-Na, +K/+Na, -K/-Na, and -K/+Na media used in S5 Fig were presented in this table. pH was adjusted at 5.5 with Ca(OH)2 before autoclave.
(TIF)
(XLS)
Whole-cell current recordings in Xenopus oocytes expressing HcSKC alone or with two β-subunits or a kinase as well as expression of LbSKC did not give evidence for K+-dependent currents. Growth of a triple yeast mutant PLY246 (Δtrk1Δtrk2Δtok1; Bertl et al., 2003) was not complemented by expression of HcSKC.
(TIF)
Acknowledgments
We would like to thank Dr. A.G. Pardo and Dr. M.J. Kemppainen (Laboratorio de Micología Molecular; Universidad Nacional de Quilmes, Argentina) for kindly providing the vector pSILBAγ. We also thank Dr. A. Kafle for assistance with some statistical analyses.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
RL and CGG were financially supported by grants from the French Minister of Research and Technology, AD by funding of the ANR project "TRANSMUT" 2010 BLAN 1604 03. KG acknowledges support of the North Carolina Agriculture Research Service (NCARS) and the North Carolina Soybean Producers Association (2019-1656). SDZ is supported by the French ANR project “MYCOTRANS”. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zimmermann SD, Chérel I. Potassium. In: Broadley MR, White PJ, editors. Plant Nutritional Genomics. Blackwell; Oxford, UK; 2005. pp. 26–65. [Google Scholar]
- 2.Anschütz U, Becker D, Shabala S. Going beyond nutrition: Regulation of potassium homoeostasis as a common denominator of plant adaptive responses to environment. J Plant Physiol. 2014;171(9): 670–687. 10.1016/j.jplph.2014.01.009 [DOI] [PubMed] [Google Scholar]
- 3.Zörb C, Senbayram M, Peiter E. Potassium in agriculture—Status and perspectives. J Plant Physiol. 2014;171(9): 656–669. 10.1016/j.jplph.2013.08.008 [DOI] [PubMed] [Google Scholar]
- 4.Garcia K, Zimmermann SD. The role of mycorrhizal associations in plant potassium nutrition. Front Plant Sci. 2014;5(337): 1–9. 10.3389/fpls.2014.00337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meena VS, Maurya BR, Verma JP. Does a rhizospheric microorganism enhance K+ availability in agricultural soils? Microbiological Research. 2014;169: 337–347. 10.1016/j.micres.2013.09.003 [DOI] [PubMed] [Google Scholar]
- 6.Shin R. Strategies for improving potassium use efficiency in plants. Mol Cells. 2014;37(8): 575–584. 10.14348/molcells.2014.0141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haro R, Benito B. The role of soil fungi in K+ plant nutrition. Int J Mol Sci. 2019;20(13): 3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith SE, Read D. Mycorrhizal Symbiosis. 3rd ed. Smith SE, Read D, editors. London: Academic Press; 2008. [Google Scholar]
- 9.Becquer A, Guerrero-Galán C, Eibensteiner JL, Houdinet G, Bücking H, Zimmermann SD, et al. The ectomycorrhizal contribution to tree nutrition. In: Advances in Botanical Research 89. Academic Press Inc.; 2019. pp. 77–126. [Google Scholar]
- 10.Guerrero-Galán C, Houdinet G, Calvo-Polanco M, Bonaldi KE, Garcia K, Zimmermann SD. The role of plant transporters in mycorrhizal symbioses. In: Advances in Botanical Research 87. Academic Press Inc.; 2018. pp. 303–342. [Google Scholar]
- 11.Jentschke G, Brandes B, Kuhn AJ, Schröder WH, Godbold DL. Interdependence of phosphorus, nitrogen, potassium and magnesium translocation by the ectomycorrhizal fungus Paxillus involutus. New Phytol. 2001;149(2): 327–337. [DOI] [PubMed] [Google Scholar]
- 12.Garcia K, Delteil A, Conéjéro G, Becquer A, Plassard C, Sentenac H, et al. Potassium nutrition of ectomycorrhizal Pinus pinaster: Overexpression of the Hebeloma cylindrosporum HcTrk1 transporter affects the translocation of both K+ and phosphorus in the host plant. New Phytol. 2014;201: 951–960. 10.1111/nph.12603 [DOI] [PubMed] [Google Scholar]
- 13.Guerrero-Galán C, Calvo-Polanco M, Zimmermann SD. Ectomycorrhizal symbiosis helps plants to challenge salt stress conditions. Mycorrhiza. 2019;29: 291–301. 10.1007/s00572-019-00894-2 [DOI] [PubMed] [Google Scholar]
- 14.Casieri L, Ait Lahmidi N, Doidy J, Veneault-Fourrey C, Migeon A, Bonneau L, et al. Biotrophic transportome in mutualistic plant-fungal interactions. Mycorrhiza. 2013;23: 597–625. 10.1007/s00572-013-0496-9 [DOI] [PubMed] [Google Scholar]
- 15.Courty PE, Doidy J, Garcia K, Wipf D, Zimmermann SD. The transportome of mycorrhizal systems. In: Molecular Mycorrhizal Symbiosis. Wiley Blackwell; 2016. pp. 239–256. [Google Scholar]
- 16.Garcia K, Doidy J, Zimmermann SD, Wipf D, Courty PE. Take a trip through the plant and fungal transportome of mycorrhiza. Trends Plant Sci. 2016;21(11): 937–950. 10.1016/j.tplants.2016.07.010 [DOI] [PubMed] [Google Scholar]
- 17.Garcia K, Delaux PM, Cope KR, Ané JM. Molecular signals required for the establishment and maintenance of ectomycorrhizal symbioses. New Phytol. 2015;208(1): 79–87. 10.1111/nph.13423 [DOI] [PubMed] [Google Scholar]
- 18.Corratgé C, Zimmermann S, Lambilliotte R, Plassard C, Marmeisse R, Thibaud JB, et al. Molecular and functional characterization of a Na+-K+ transporter from the Trk family in the ectomycorrhizal fungus Hebeloma cylindrosporum. J Biol Chem. 2007;282: 26057–26066. 10.1074/jbc.M611613200 [DOI] [PubMed] [Google Scholar]
- 19.Kohler A, Kuo A, Nagy LG, Morin E, Barry KW, Buscot F, et al. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat Genet. 2015;47(4): 410–415. 10.1038/ng.3223 [DOI] [PubMed] [Google Scholar]
- 20.Guerrero-Galán C, Delteil A, Garcia K, Houdinet G, Conéjéro G, Gaillard I, et al. Plant potassium nutrition in ectomycorrhizal symbiosis: properties and roles of the three fungal TOK potassium channels in Hebeloma cylindrosporum. Environ Microbiol. 2018;20(5): 1873–1887. 10.1111/1462-2920.14122 [DOI] [PubMed] [Google Scholar]
- 21.Guerrero-Galán C, Garcia K, Houdinet G, Zimmermann SD. HcTOK1 participates in the maintenance of K+ homeostasis in the ectomycorrhizal fungus Hebeloma cylindrosporum, which is essential for the symbiotic K+ nutrition of Pinus pinaster. Plant Signal Behav. 2018; 13(6): e1480845 10.1080/15592324.2018.1480845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gambale F, Uozumi N. Properties of shaker-type potassium channels in higher plants. J Membr Biol. 2006;210(1): 1–19. 10.1007/s00232-006-0856-x [DOI] [PubMed] [Google Scholar]
- 23.Zimmermann S, Sentenac H. Plant ion channels: from molecular structures to physiological functions. Curr Opin Plant Biol. 1999;2(6): 477–482. 10.1016/s1369-5266(99)00020-5 [DOI] [PubMed] [Google Scholar]
- 24.Hedrich R. Ion channels in plants. Physiol Rev. 2012;92(4): 1777–1811. 10.1152/physrev.00038.2011 [DOI] [PubMed] [Google Scholar]
- 25.Jegla T, Busey G, Assmann SM. Evolution and structural characteristics of plant voltage-gated K+ channels. Plant Cell. 2018;30(12): 2898–2909. 10.1105/tpc.18.00523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Debaud JC, Gay G. In vitro fruiting under controlled conditions of the ectomycorrhizal fungus Hebeloma cylindrosporum associated with Pinus pinaster. New Phytol. 1987;105: 429–435. [DOI] [PubMed] [Google Scholar]
- 27.Rao PS, Niederpruem DJ. Carbohydrate metabolism during morphogenesis of Coprinus lagopus (sensu Buller). J Bacteriol. 1969;100(3): 1222–1228. 10.1128/JB.100.3.1222-1228.1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kemppainen MJ, Pardo AG. pHg/pSILBAγ vector system for efficient gene silencing in homobasidiomycetes: optimization of ihpRNA–triggering in the mycorrhizal fungus Laccaria bicolor. Microb Biotechnol. 2010;3(2): 178–200. 10.1111/j.1751-7915.2009.00122.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ali MA, Louche J, Legname E, Duchemin M, Plassard C. Pinus pinaster seedlings and their fungal symbionts show high plasticity in phosphorus acquisition in acidic soils. Tree Physiol. 2009;29(12): 1587–1597. 10.1093/treephys/tpp088 [DOI] [PubMed] [Google Scholar]
- 30.Garcia K, Haider MZ, Delteil A, Corratgé-Faillie C, Conéjero G, Tatry M-V, et al. Promoter-dependent expression of the fungal transporter HcPT1.1 under Pi shortage and its spatial localization in ectomycorrhiza. Fungal Genet Biol. 2013;58–59: 53–61. 10.1016/j.fgb.2013.06.007 [DOI] [PubMed] [Google Scholar]
- 31.Louche J, Ali MA, Cloutier-Hurteau B, Sauvage FX, Quiquampoix H, Plassard C. Efficiency of acid phosphatases secreted from the ectomycorrhizal fungus Hebeloma cylindrosporum to hydrolyse organic phosphorus in podzols. FEMS Microbiol Ecol. 2010;73(2): 323–335. 10.1111/j.1574-6941.2010.00899.x [DOI] [PubMed] [Google Scholar]
- 32.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157(1): 105–132. 10.1016/0022-2836(82)90515-0 [DOI] [PubMed] [Google Scholar]
- 33.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42(1): 252–258. 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grigoriev IV, Cullen D, Goodwin SB, Hibbett D, Jeffries TW, Kubicek CP, et al. Fueling the future with fungal genomics. Mycology. 2011;2(3): 192–209. [Google Scholar]
- 35.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5): 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17(4): 540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- 37.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59: 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 38.Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8(1): 460 10.1186/1471-2105-8-460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minet M, Dufour M-E, Lacroute F. Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant Journal. 1992;2: 417–422. 10.1111/j.1365-313x.1992.00417.x [DOI] [PubMed] [Google Scholar]
- 40.Lambilliotte R, Cooke R, Samson D, Fizames C, Gaymard F, Plassard C, et al. Large-scale identification of genes in the fungus Hebeloma cylindrosporum paves the way to molecular analyses of ectomycorrhizal symbiosis. New Phytol. 2004;164: 505–513. [Google Scholar]
- 41.Seoh SA, Sigg D, Papazian DM, Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. 1996;16(6): 1159–1167. 10.1016/s0896-6273(00)80142-7 [DOI] [PubMed] [Google Scholar]
- 42.Matheny PB, Gossmann JA, Zalar P, Kumar TKA, Hibbett DS. Resolving the phylogenetic position of the Wallemiomycetes: An enigmatic major lineage of Basidiomycota. Can J Bot. 2006;84(12): 1794–1805. [Google Scholar]
- 43.Corratgé-Faillie C, Jabnoune M, Zimmermann S, Véry AA, Fizames C, Sentenac H. Potassium and sodium transport in non-animal cells: The Trk/Ktr/HKT transporter family. Cell Mol Life Sci. 2010;67(15): 2511–2532. 10.1007/s00018-010-0317-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benito B, Garciadeblás B, Schreier P, Rodríguez-Navarro A. Novel P-type ATPases mediate high-affinity potassium or sodium uptake in fungi. Eukaryot Cell. 2004;3(2): 359–368. 10.1128/ec.3.2.359-368.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fietto LG, Pugliese L, Gomes SL. Characterization and expression of two genes encoding isoforms of a putative Na, K-ATPase in the Chytridiomycete Blastocladiella emersonii. Biochim Biophys Acta. 2002;1576(1–2): 59–69. 10.1016/s0167-4781(02)00297-x [DOI] [PubMed] [Google Scholar]
- 46.Benito B, Garciadeblás B, Fraile-Escanciano A, Rodríguez-Navarro A. Potassium and sodium uptake systems in fungi. The transporter diversity of Magnaporthe oryzae. Fungal Genet Biol. 2011;48(8): 812–822. 10.1016/j.fgb.2011.03.002 [DOI] [PubMed] [Google Scholar]
- 47.Doré J, Perraud M, Dieryckx C, Kohler A, Morin E, Henrissat B, et al. Comparative genomics, proteomics and transcriptomics give new insight into the exoproteome of the basidiomycete Hebeloma cylindrosporum and its involvement in ectomycorrhizal symbiosis. New Phytol. 2015;208: 1169–1187. 10.1111/nph.13546 [DOI] [PubMed] [Google Scholar]
- 48.Doré J, Kohler A, Dubost A, Hundley H, Singan V, Peng Y, et al. The ectomycorrhizal basidiomycete Hebeloma cylindrosporum undergoes early waves of transcriptional reprogramming prior to symbiotic structures differentiation. Environ Microbiol. 2017;19: 1338–1354. 10.1111/1462-2920.13670 [DOI] [PubMed] [Google Scholar]
- 49.Chérel I. Regulation of K+ channel activities in plants: from physiological to molecular aspects. J Exp Bot. 2004;55(396): 337–351. 10.1093/jxb/erh028 [DOI] [PubMed] [Google Scholar]
- 50.Li Y, Um SY, Mcdonald TV. Voltage-gated potassium channels: regulation by accessory subunits. Neurosci. 2006;12(3): 199–210. [DOI] [PubMed] [Google Scholar]
- 51.Dreyer I, Porée F, Schneider A, Mittelstädt J, Bertl A, Sentenac H, et al. Assembly of plant Shaker-like Kout channels requires two distinct sites of the channel α-subunit. Biophys J. 2004;87(2): 858–872. 10.1529/biophysj.103.037671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu K, Li L, Luan S. Intracellular K+ sensing of SKOR, a Shaker-type K+ channel from Arabidopsis. Plant J. 2006;46(2): 260–268. 10.1111/j.1365-313X.2006.02689.x [DOI] [PubMed] [Google Scholar]
- 53.Naso A, Dreyer I, Pedemonte L, Testa I, Gomez-Porras JL, Usai C, et al. The role of the C-terminus for functional heteromerization of the plant channel KDC1. Biophys J. 2009;96(10): 4063–4074. 10.1016/j.bpj.2009.02.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sokolova O, Accardi A, Gutierrez D, Lau A, Rigney M, Grigorieff N. Conformational changes in the C terminus of Shaker K+ channel bound to the rat Kvβ2-subunit. Proc Natl Acad Sci. 2003;100(22): 12607–12612. 10.1073/pnas.2235650100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zandany N, Marciano S, Magidovich E, Frimerman T, Yehezkel R, Shem-Ad T, et al. Alternative splicing modulates Kv channel clustering through a molecular ball and chain mechanism. Nat Commun. 2015;6(1): 6488 10.1038/ncomms7488 [DOI] [PubMed] [Google Scholar]
- 56.Ruiz-Cañada C, Koh YH, Budnik V, Tejedor FJ. DLG differentially localizes Shaker K+-channels in the central nervous system and retina of Drosophila. J Neurochem. 2002;82(6): 1490–1501. 10.1046/j.1471-4159.2002.01092.x [DOI] [PubMed] [Google Scholar]
- 57.Goto S, Sugiyama J, Hamamoto M, Komagata K. Saitoella, a new anamorph genus in the Cryptococcaceae to accommodate two himalayan yeast isolates formerly identified as Rhodotorula glutinis. J Gen Appl Microbiol. 1987;33(1): 75–85. [Google Scholar]
- 58.Nishida H, Hamamoto M, Sugiyama J. Draft genome sequencing of the enigmatic yeast Saitoella complicata. J Gen Appl Microbiol. 2011;57(4): 243–246. 10.2323/jgam.57.243 [DOI] [PubMed] [Google Scholar]
- 59.Ruytinx J, Coninx L, Nguyen H, Smisdom N, Morin E, Kohler A, et al. Identification, evolution and functional characterization of two Zn CDF-family transporters of the ectomycorrhizal fungus Suillus luteus. Environ Microbiol Rep. 2017;9(4): 419–427. 10.1111/1758-2229.12551 [DOI] [PubMed] [Google Scholar]
- 60.Torres YP, Morera FJ, Carvacho I, Latorre R. A marriage of convenience: β-subunits and voltage-dependent K+ channels. J Biol Chem. 2007;282(34): 24485–24489. 10.1074/jbc.R700022200 [DOI] [PubMed] [Google Scholar]
- 61.Rettig J, Heinemann SH, Wunder F, Lorra C, Parcej DN, Oliver Dolly J, et al. Inactivation properties of voltage-gated K+ channels altered by presence of β-subunit. Nature. 1994;369(6478): 289–294. 10.1038/369289a0 [DOI] [PubMed] [Google Scholar]
- 62.Pan Y, Weng J, Cao Y, Bhosle RC, Zhou M. Functional coupling between the Kv1.1 channel and aldoketoreductase Kvbeta1. J Biol Chem. 2008;283(13): 8634–8642. 10.1074/jbc.M709304200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang H, Vasconcelos AC, Berkowitz GA. Physical association of KAB1 with plant K+ channel alpha subunits. Plant Cell. 1996;8(9): 1545–1553. 10.1105/tpc.8.9.1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu J, Park M-H, Jo S-H. Inhibitory effects of cortisone and hydrocortisone on human Kv1.5 channel currents. Eur J Pharmacol. 2015;746: 158–166. 10.1016/j.ejphar.2014.11.007 [DOI] [PubMed] [Google Scholar]
- 65.de la Cruz IP, Levin JZ, Cummins C, Anderson P, Horvitz HR. sup-9, sup-10, and unc-93 may encode components of a two-pore K+ channel that coordinates muscle contraction in Caenorhabditis elegans. J Neurosci. 2003;23(27): 9133–9145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xiang J, Zhou X, Zhang X, Liu A, Xiang Y, Yan M, et al. The Arabidopsis AtUNC-93 acts as a positive regulator of abiotic stress tolerance and plant growth via modulation of ABA signaling and K+ homeostasis. Front Plant Sci. 2018;9: 718 10.3389/fpls.2018.00718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gaymard F, Pilot G, Lacombe B, Bouchez D, Bruneau D, Boucherez J, et al. Identification and disruption of a plant Shaker-like outward channel involved in K+ release into the xylem sap. Cell. 1998;94(5): 647–655. 10.1016/s0092-8674(00)81606-2 [DOI] [PubMed] [Google Scholar]
- 68.Chérel I, Gaillard I. The complex fine-tuning of K+ fluxes in plants in relation to osmotic and ionic abiotic stresses. Int J Mol Sci. 2019;20(3): 715 10.3390/ijms20030715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roeper J, Lorra C, Pongs O. Frequency-dependent inactivation of mammalian A-type K+ channel KV1.4 regulated by Ca2+/Calmodulin-dependent protein kinase. J Neurosci. 1997;17(10): 3379–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Casado C, Yenush L, Melero C, del Carmen Ruiz M, Serrano R, Pérez-Valle J, et al. Regulation of Trk-dependent potassium transport by the calcineurin pathway involves the Hal5 kinase. FEBS Lett. 2010;584(11): 2415–2420. 10.1016/j.febslet.2010.04.042 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structure-based sequence alignment and sequence conservation between HcSKC and RnKV2.1 K+ channel from Rattus norvegicus. The GYGE motif of the pore domain was highlighted in green.
(TIF)
The HcSKC-EGFP construct was inserted into the pYES2 vector, with galactose-induced expression (a,b), or into the pFL61 vector, under the PGK constitutive promoter (c,d). None of these attempts yielded a clear localization pattern of HcSKC, probably due to misprocessing of the protein product.
(TIF)
EGFP expression was observed in fungal lines harboring the EGFP cassette under control of the promoters of HcSKC (pPZP-PSKC1-E). Fungal isolates were grown on YMG medium for 2–3 weeks before analysis. (a) bright field image (b) EGFP image (GFP filter 505–530 nm). Scale bar: 100 μm.
(TIF)
The relative expression of HcSKC was determined in empty vector (Ctrl), RNAi-SKC-5 and RNAi-SKC-7 fungal lines by qRT-PCR. Mean values are provided with the standard deviation (n = 6). Statistical differences were evaluated using the Student’s test with respect to E.V. strain (**, P < 0.01).
(TIF)
Dry weights of empty vector (Ctrl), RNAi-SKC-5 and RNAi-SKC-7 fungal lines were determined after 28 days of culture in media containing high (+K and +Na) or low concentration (-K and -Na) of potassium (K+) and sodium (Na+), respectively. +K corresponds to 1 mM of K+, -K to 0.05 mM of K+, +Na to 1 mM of Na+, and -Na to 0.2 mM of Na+. Full recipes are provided in S1 Table. Mean values are provided with the standard deviation (n = 4–6). Different letters indicate significant differences between treatments according to one-way ANOVA followed by Tukey HSD post-hoc tests (P < 0.05).
(TIF)
Restriction enzyme sites indicated in primer name, were underlined in the corresponding 5’-3’ sequence.
(TIF)
Final concentrations of components constituting the +K/-Na, +K/+Na, -K/-Na, and -K/+Na media used in S5 Fig were presented in this table. pH was adjusted at 5.5 with Ca(OH)2 before autoclave.
(TIF)
(XLS)
Whole-cell current recordings in Xenopus oocytes expressing HcSKC alone or with two β-subunits or a kinase as well as expression of LbSKC did not give evidence for K+-dependent currents. Growth of a triple yeast mutant PLY246 (Δtrk1Δtrk2Δtok1; Bertl et al., 2003) was not complemented by expression of HcSKC.
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.