Abstract
Disruption of microbial communities within human hosts has been associated with infection, obesity, cognitive decline, cancer risk and frailty, suggesting that microbiome-targeted therapies may be an option for improving healthspan and lifespan. The objectives of this study were to determine the feasibility of delivering fecal microbiota transplants (FMTs) to marmosets via oral gavage and to evaluate if alteration of the gut microbiome post-FMT could be achieved. This was a prospective study of marmosets housed at the Barshop Institute for Longevity and Aging Studies in San Antonio, TX. Eligible animals included healthy young adult males (age 2-5 years) with no recent medication use. Stool from two donors was combined and administered in 0.5mL doses to five young recipients once weekly for 3 weeks. Safety outcomes and alterations in the gut microbiome composition via 16s rRNA sequencing were compared at baseline and monthly up to 6 months post-FMT. Overall, significant differences in the percent relative abundance was seen in FMT recipients at the phylum and family levels from baseline to 1 month and baseline to 6 months post-FMT. In PERMANOVA analyses, treatment status (donor versus recipient) (p=0.056) and time course (p=0.019) predicted beta diversity (p=0.056). The FMT recipients did not experience any negative health outcomes over the course of the treatment. FMT via oral gavage was safe to administer to young adult marmosets. The marmoset microbiome may be altered by FMT; however, progressive changes in the microbiome are strongly driven by the host and its baseline microbiome composition.
Keywords: fecal microbiota transplantation, aging, healthspan, gut microbiota
Background
In 2030, when the last baby boomer turns 65, one out of every five Americans (about 72 million people) will be 65 years or older (Centers for Disease & Prevention, 2003; Control & Prevention, 2015). This shift to an aged population is likely to increase overall economic burden, including costs of medical care and an increase in the percentage of the population not working (Dall et al., 2013; Weil, 1997). Due to the dynamic alteration of human population needs in the near future, aging researchers are no longer simply focused on extending human lifespan; they are interested in increasing quality of life, extending the length of time people are living independently, and overall healthspan for the aging population. Therefore, the focus of aging research has now shifted to treating aging as a systemic problem and determining whether an increase in lifespan alters the ability for that individual to remain disease free. The hope is that the treatment for multiple age-related diseases including diabetes, cardiovascular disease, Alzheimer’s disease, dementia, and sarcopenia could be discovered by understanding mechanisms that underlie the basic systemic aging process.
The microbiome is important in mediating human health and could impact biological aging; thus, microbiome-mediated therapies should be investigated as potential healthspan and lifespan interventions. Trillions of microorganisms live commensally within the gut and encode >150 times more genes than the human genome. These organisms serve several important physiological functions, including protection against pathogens, immune and inflammatory response regulation, nutrient and energy extraction, and hormone biosynthesis (Belkaid & Hand, 2014; Lynch & Pedersen, 2016). It is not surprising then that dysbiosis (i.e., disturbance or change in the microbiota composition or diversity) has been associated with increased inflammation and altered physiological homeostasis associated with more than two dozen comorbidities, including enteric infections, metabolic disorders (e.g., obesity, diabetes), neurological disorders (Alzheimer’s disease, Parkinson’s disease), cardiovascular disease, cancer, and frailty (Barlow, Yu, & Mathur, 2015; Cattaneo et al., 2017; Jackson et al., 2016; Komaroff, 2017). Notably, the majority of these conditions exhibit age-related increases in incidence, thus supporting the hypothesis that the gut microbiome may play a role in healthspan and physiological aging. Microbial communities have been found to differ between geriatric and young individuals in humans, mice, and other model species (Heintz & Mair, 2014).
Due to the connection between dysbiosis and disease, fecal microbiota transplantation (FMT) may be a possible therapeutic intervention for altering and sustaining microbiota homeostasis in aging individuals, but it has been scarcely investigated. FMT has been used successfully for decades to treat Clostridioides difficile infections (CDI) in geriatric people (Kassam, Lee, Yuan, & Hunt, 2013). Transfection from healthy donors via endoscopic or colonoscopic delivery results in reduction of the C. difficile load and recovery of the microbiome. Prior mouse models of FMT offer the best evidence of a causal relationship between microbial communities and disease. Evaluations of gnotobiotic mice recipients of human donor fecal material, as well as mouse-derived fecal material, result in weight gain and metabolic dysfunction if they received material from an obese donor (Boulangé, Neves, Chilloux, Nicholson, & Dumas, 2016; Ussar et al., 2015). Interestingly, transplanting fecal microbiota from lean individuals to obese individuals results in weight loss and rescued metabolic function (Di Luccia et al., 2015; Marotz & Zarrinpar, 2016). Furthermore, transplanting fecal microbiota from animals that were fed high fat diets resulted in increased anxiety, increased stereotypical behavior, and decreased cognition associated with increased neuro-inflammation in mice that did not yet show changes in obesity or metabolic function (Bruce-Keller et al., 2015). These findings in mice suggest that shifts in microbial communities, and the byproducts that they produce, are associated with inflammation and many disease states. They also suggest that FMT may be an interesting interventional treatment to stabilize microbial communities and restore homoeostasis (Jin Song et al., 2019; Liwinski & Elinav, 2019).
Evaluating the connections between the microbiome and aging has been difficult to interpret and evaluate from the human data due to broad variability between developmental environments, current environment, dietary choice, antibiotic exposure and a number of other factors. Furthermore, most microbiome aging studies have been cross-sectional in design, given the difficulty in following humans prospectively over years to decades of life. The use of an animal model to evaluate the interactions between the microbiome and phenotypic aging offers the advantage of being able to control current environment, diet, and exposure to antibiotics or probiotics. The common marmoset (Callithrix jacchus), a small Plattyrhine monkey from Brazil, has rapidly become an important nonhuman primate model in the study of age-related disease. The marmoset has a maximum lifespan of approximately 20 years, which is roughly half the maximum lifespan of the rhesus macaque (Ross, 2019; Ross & Salmon, 2019). Marmosets are described as displaying changes associated with geriatric decline beginning at around 10 years of age (Ross, 2019; Ross et al., 2019). Marmosets are easily group housed in closely controlled environmental settings that mirror their natural social arrangement (Ross et al., 2017), which is extremely difficult and expensive to accomplish with larger Cattarhine monkeys such as macaques. Marmosets have been found to display many aging phenotypes that mimic human aging, including increased risk of cardiovascular changes, inflammatory disease, metabolic impairment, suppressed immune function, and impaired cognition (Ross, 2019; Ross et al., 2019; Ross, Davis, Dobek, & Tardif, 2012). Moreover, marmosets have been found to exhibit significantly decreased microbial diversity with aging (Reveles, Patel, Forney, & Ross, 2019).
Given these findings, further testing of FMT as a potential bacteriotherapy option for healthy aging is needed. FMT may allow for the restoration of a stable, highly diverse gut community typically found in young adults consisting of Firmicutes and Bacteroidetes, but with high diversity in other phyla of bacteria (Wilson, Vatanen, Cutfield, & O'Sullivan, 2019). Given that FMT has become much more accessible to patients in recent years due to the availability of stool banks (e.g., OpenBiome) that provide rigorously screened donor stool, clinical application is likely. The objectives of this study were to 1) determine the feasibility and safety of delivering FMTs to marmosets via oral gavage and 2) to evaluate if alteration of the gut microbiome post-FMT could be achieved.
Methods
Marmoset selection and housing
This study used marmosets housed at the Barshop Institute for Longevity and Aging Studies colony in San Antonio, TX from January to July 2018. The marmosets are maintained on a daily standardized base diet consisting of a mix of commercial products (Harlan Teklad marmoset purified diet, Purina Mazuri). Marmosets in the colony receive limited dietary enrichment items consisting of irradiated primate enrichment mix (Harlan Teklad). The use of other enrichment including Cheerios, marshmallows, and peanuts is limited to behavioral training and assessment and was not provided to animals during the course of the study. Eligibility criteria for the study included animals not receiving interventions including probiotics, antibiotics, or rapamycin treatment, and had no recent medical concerns. The procedures for the study were reviewed and approved by the UT Health San Antonio Institutional Animal Care and Use Committee, and followed guidelines set forth by the American Society of Primatologists. The data that support the findings of this study are available on request from the corresponding author.
In a previously published study (Reveles et al., 2019), gut microbiome diversity was compared between 10 young adult (2-5 years old) and 10 older adult (8+ years old) marmosets. Animals from that study that were determined to have disparate microbiome composition were selected to assess the ability to administer FMT orally in the marmoset. A total of two young adult (2-5 years old) donors and five young adult (2-5 years old) recipients were selected from these preliminary studies. Selection was based on differences between donors and recipients using Bray-Curtis dissimilarity hierarchical clustering and the distinct high mean percent relative abundance of Bifidobacteriaceae in donors compared to recipients (33.5% versus 7.6%, respectively) (Reveles et al., 2019). Potential donors were screened for common pathogens, including Giardia, Cryptosporidium, and Clostridium perfringens prior to becoming donors. The two donor stool samples were pooled to ensure sufficient quantity for donation as previously described for rodents (Di Luccia et al., 2015). There is some evidence from human FMT treatment for C. difficile that success of transplant is affected not only by donor microbiome diversity but also composition leading to the hypothesis that super donors may exist (Wilson et al., 2019). However, there is currently no way to predict which donor may be a super donor and increase likelihood of success, so an accepted best strategy is to combine donor material into a single batch (Wilson et al., 2019). The combined donor sample was re-assessed for alpha and beta diversity metrics at baseline, prior to FMT. We chose only young adult male marmosets as donors and recipients (mean age = 3.18±0.6, mean weight = 436.6±45.2) to confirm the safety of the procedure prior to administering to older marmosets in future studies. Mean Shannon diversity did not significantly change in donors (3.30 vs. 3.27) or recipients (3.35 vs. 3.30) from the previously published data (Reveles et al., 2019) to the baseline sample collected as part of this study.
FMT procedures
Fecal material was collected from selected donors over several days and immediately frozen to obtain 50g of fecal material. Feces were then thawed and homogenized in sterile saline using a blender immediately prior to the procedure. The mixture was centrifuged at 800 rpm for 2 minutes and the supernatant removed. The supernatant was aliquoted into doses of 0.5 mL and frozen until treatment. Five unsedated recipient marmosets received an FMT via oral gavage of 0.5 mL once a week for three weeks following collection of a single stool sample at baseline. This dosing regimen was selected based on successful transplant in rat models of obesity (Di Luccia et al., 2015). Fecal samples were collected after the three weeks of treatment, and then again monthly for 6 months.
Microbiome sample sequencing and analysis
Sequencing and analysis was performed by Second Genome, Inc. as described previously (Reveles et al., 2019). In brief, bacterial DNA was extracted using the DNeasy PowerSoil Pro kit (Qiagen, Hilden, Germany) per manufacturer’s instructions. The Illumina® HiSeq platform (San Diego, CA) was used for bacterial 250 base paired-end 16S V4 rRNA sequencing. Sequenced paired-end reads were compared to curated strains (StrainSelect) using USEARCH (Edgar, 2010). All sequences matching to a unique strain with an identity ≥99% were assigned a strain OTU. For each strain OTU, one of the matching reads was selected as representative and all sequences were mapped by USEARCH against the strain OTU representatives to calculate strain abundances. Resulting unique sequences were then clustered at 97% by UPARSE (de novo OTU clustering) and a representative consensus sequence per de novo OTU was determined (Edgar, 2013). All non-strain sequences that passed the quality filtering were mapped to the representative consensus sequences to generate an abundance table for de novo OTUs. Representative OTU sequences were assigned taxonomic classification via mothur’s bayesian classifier(Schloss et al., 2009), trained against the Greengenes reference database of 16S rRNA gene sequences clustered at 99% (McDonald et al., 2012).
Sample richness was estimated based on the number of OTUs present in a sample. Shannon diversity (Shannon, 1948) was also calculated, accounting for sample richness and relative abundance. Abundance-weighted sample pairwise differences (beta diversity) was calculated using the Bray-Curtis dissimilarity (Bray & Curtis, 1957). Univariate differential abundance of OTUs was tested using a negative binomial noise model for the overdispersion and Poisson process using the DESeq2 package (Love, Huber, & Anders, 2014), as described for microbiome applications (McMurdie & Holmes, 2014). The Wilcoxon signed rank test was used to compare diversity metrics between baseline and 1-month post-FMT and 6 months post-FMT. A PERMANOVA using distance matrices was performed for each variable of interest to determine if they significantly contributed to the beta diversity of the samples. False discovery rate-corrected p values (i.e., q-values) were calculated using the Benjamini-Hochberg procedure.
Safety endpoints
In order to evaluate the risk for potential side effects on marmoset health following transplant, several biological health parameters were assessed. Body mass was evaluated weekly by weighing the marmosets within the homecage. Body composition was assessed prior to dosing and at one month and 6 months post-dosing using quantitative magnetic resonance (Echo QMR), briefly the unsedated marmoset is placed in a cylinder which is inserted into the machine for a less than two minute scan (Tardif et al., 2009). Additionally, since the microbiome has been associated with insulin signaling and resistance, an oral glucose tolerance test was conducted prior to treatment and 6 months following treatment. Marmosets readily consume a dextrose solution and blood samples of < 50ul are taken at time points 0 (pre-dosing), and 15, 30, 60 and 120 minutes post-dosing via tail vein collection (Power, Ross, Schulkin, Ziegler, & Tardif, 2013). Blood was drawn from unsedated animals via the femoral vein for analysis of standard veterinary blood chemistry and complete blood count at baseline and at 6 months post FMT. All continuous variables were compared between baseline and post FMT using the paired t-test for matched comparisons. Given that this was a pilot efficacy study, we set an α value of 0.2 to determine statistically significant differences for all microbiome and safety comparisons, as advocated for clinical efficacy studies (Lee, Whitehead, Jacques, & Julious, 2014).
Results
Sample sequencing quality
A total of 355 OTUs were observed in 6,742,239 reads following independent filtering. Most sequences were classified at the phylum (99.68%), family (89.35%), and genus (74.36%) levels. Only 21.69% and 15.97% of sequences were classified at the species and strain level, respectively. Sequences per sample ranged from a minimum of 62,016 to a maximum of 243,118 filtered reads across samples. One sample from the recipient 2 months post-FMT group was removed from all month 2 analyses because it did not meet the required 50,000 total reads threshold for library size. Rarefaction curves approached saturation for all samples, indicating that samples were sequenced with sufficient depth to capture the whole microbiome composition.
Changes in alpha diversity and composition from baseline to 6 months post-FMT
Overall, there were no significant changes in alpha diversity estimates over time between donor and recipients. The mean OTU richness for the donor sample at baseline was 174 and recipient mean (SD) OTU richness ranged from 249 (15.6) at baseline to 239 (19.0) at 6 months follow-up (recipient baseline versus 6 months post-FMT p=0.42) (SI Fig 1). No significant changes were seen for Shannon diversity; donor Shannon diversity was 3.27 at baseline and mean (SD) Shannon diversity in recipients ranged from 3.50 (0.15) at baseline to 3.52 (0.08) at 6 months follow-up (recipient baseline versus 6 months post-FMT p=0.59) (Figure 1).
Figure 1.

Shannon diversity estimates for donor and recipient marmosets from baseline through the six-month follow-up post fecal microbiota transplant.
Figures 2a and 2b display the mean percent relative abundance of the most common bacterial taxa at the phylum and family levels. From baseline to 1 month, significant differences were noted at the phylum level for Firmicutes (19.9% vs. 25.6%, p=0.059), Bacteroidetes (39.2% vs. 36.3%, p=0.106), and Fusobacteria (7.9% vs. 6.0%, p=0.178). At the family level, significant differences were seen in Porphyromonadaceae (15.4% vs. 8.2%, p=0.059), Veillonellaceae (9.4% vs. 11.7%, p=0.059), and Fusobacteriaceae (7.9% vs. 6.0%, p=0.178). From baseline to 6 months, significant differences were noted in the phyla Actinobacteria (17.8% vs. 24.2%, p=0.059), Proteobacteria (13.9% vs. 9.3%, p=0.059), and Fusobacteria (7.9% vs. 10.9%, p=0.106). Interestingly, while Bifidobacteriaceae did not significantly increase from baseline to 1 month (9.2% vs. 9.5%, p=0.590), there was a significant increase from baseline to 6 months (9.2% vs. 11.8%, p=0.059). In general, there appeared to be a trend in microbial abundance between donors and recipients; if the donor abundance was higher than the recipients initially, the recipient abundances tended to increase post-FMT and vice versa. For example, the Firmicutes phyla abundance increased among recipients following FMT from the donor with a higher relative abundance of Firmicutes at baseline (40.5%). Alternatively, the Bacteroidetes phyla abundance decreased following FMT from the donor with lower Bacteroidetes at baseline (27.7%). Phylum and family differences for individual animals at baseline and each follow-up period can be found in SI Figs 2 and 3.
Figure 2.
Mean percent relative abundance changes from baseline to follow-up for common microbial taxa a) Phylum-level differences b) Family-level differences.
In PERMANOVA analyses, treatment status (donor versus recipient) was significantly associated with beta diversity (p=0.056). Time course also significantly predicted beta diversity (p=0.019). In paired analyses, there was a significant difference between recipient baseline beta diversity compared to 1-month follow-up (p=0.111) (Fig 3) and compared to 6-month follow-up (p=0.164) (Fig 4).
Figure 3.
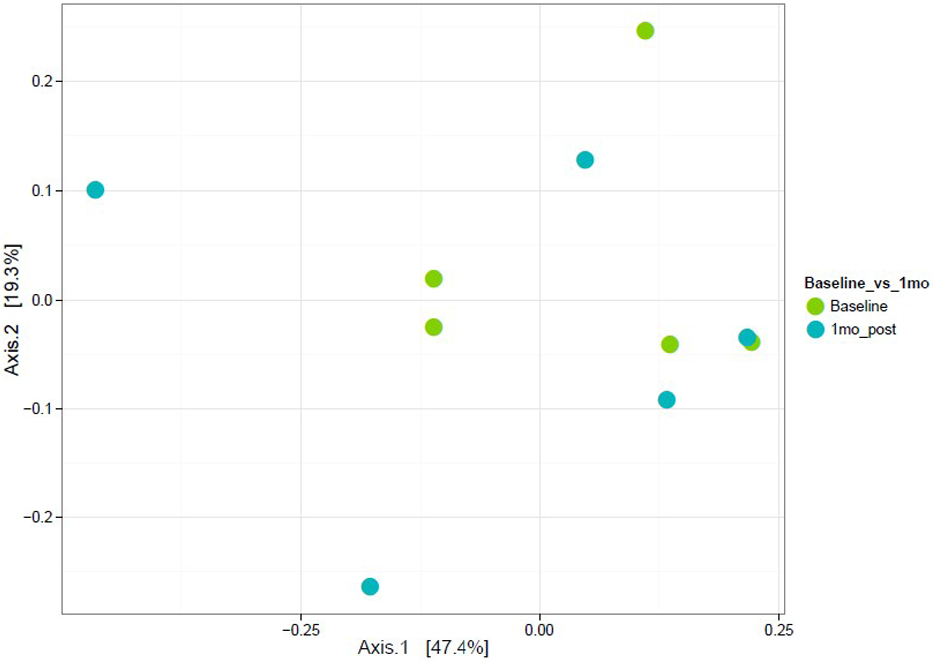
Weighted ordination using abundance in recipients from baseline to 1 month post FMT
Figure 4.

Weighted ordination using abundance in recipients from baseline to 6 months post FMT
Safety endpoints
The FMT recipients did not experience any negative health outcomes over the course of the treatment. Table 1 displays safety endpoints among recipients from baseline to follow-up. There were no significant differences from baseline to follow-up in complete blood count endpoints, oral glucose tolerance test area under the curve, or body composition. There were statistically significant decreases in fasting serum glucose and anion gap, and significant increases in total carbon dioxide and cholesterol in subjects from baseline to 6-months. These laboratory values still fell within normal clinical limits.
Table 1.
Safety endpoints changes in FMT recipients from baseline to 6 months follow-up
| Safety endpoint | Baseline (n=5) |
6 Month Follow-Up (n=5) |
P-value |
|---|---|---|---|
| Blood chemistry, mean ± SD | |||
| Blood urea nitrogen (mg/dL) | 25.6 ± 4.5 | 26.4 ± 5.0 | 0.7142 |
| Serum creatinine (mg/dL) | 0.32 ± 0.04 | 0.30 ± 0.00 | 0.3739 |
| Glucose (mg/dL) | 147.6 ± 46.2 | 99.4 ± 23.6 | 0.0996 |
| Alkaline phosphatase (U/L) | 68.4 ± 17.6 | 73.4 ± 16.9 | 0.4795 |
| AST (U/L) | 131.0 ± 40.3 | 125.4 ± 29.9 | 0.7661 |
| ALT (U/L) | 7.6 ± 2.3 | 6.8 ± 3.0 | 0.6842 |
| Total protein (mg/dL) | 6.7 ± 0.8 | 7.0 ± 1.0 | 0.4075 |
| Total CO2 (mEq/L) | 16.6 ± 6.0 | 22.6 ± 2.9 | 0.0905 |
| Sodium (mEq/L) | 149.6 ± 4.7 | 151.6 ± 3.0 | 0.3262 |
| Potassium (mEq/L) | 4.0 ± 1.0 | 3.4 ± 0.2 | 0.2259 |
| Chloride (mEq/L) | 104.0 ± 4.5 | 106.0 ± 1.9 | 0.4085 |
| Anion gap | 33.0 ± 6.4 | 26.4 ± 4.8 | 0.0339 |
| C-reactive protein (ng/mL) | 1.4 ± 0.5 | 1.5 ± 0.4 | 0.6010 |
| Cholesterol (mmol/L) | 140.4 ± 13.6 | 167.4 ± 15.4 | 0.0229 |
| Albumin (g/dL) | 4.1 ± 0.6 | 4.0 ± 0.6 | 0.9255 |
| Globulin (g/dL) | 2.6 ± 0.3 | 3.0 ± 0.5 | 0.2386 |
| Albumin/globulin ratio | 1.6 ± 0.2 | 1.4 ± 0.2 | 0.2488 |
| Complete blood count, mean ± SD | |||
| White blood cells (x 103 cells/μL) | 6.4 ± 3.9 | 6.3 ± 2.1 | 0.8931 |
| Red blood cells (x 103 cells/μL) | 7.2 ± 0.3 | 7.3 ± 0.5 | 0.6350 |
| Hemoglobin (g/dL) | 16.6 ± 0.9 | 17.1 ± 0.9 | 0.5811 |
| Hematocrit (%) | 51.7 ± 3.1 | 52.2 ± 2.3 | 0.8357 |
| Mean cell volume (μm3) | 72.0 ± 2.8 | 71.3 ± 1.8 | 0.2943 |
| Mean cell hemoglobin (pg) | 23.2 ± 0.5 | 23.4 ± 0.5 | 0.3792 |
| MCHC (g/dL) | 32.2 ± 0.8 | 32.8 ± 0.3 | 0.2682 |
| Red blood cell distribution width (%) | 15.5 ± 0.9 | 15.9 ± 1.8 | 0.4575 |
| Platelets (x 103 cells/μL) | 544.8 ± 78.3 | 608.0 ± 247.6 | 0.6333 |
| Measure platelet volume (fL) | 8.9 ± 0.7 | 9.7 ± 0.9 | 0.2157 |
| Oral glucose tolerance test, mean ± SD | |||
| Glucose (mg/dL), 0 minutes | 98.0 ± 17.3 | 109.6 ± 22.8 | 0.3970 |
| Glucose (mg/dL), 15 minutes | 110.6 ± 45.6 | 147.0 ± 51.9 | 0.3248 |
| Glucose (mg/dL), 30 minutes | 135.2 ± 66.7 | 127.6 ± 35.0 | 0.8411 |
| Glucose (mg/dL), 60 minutes | 163.0 ± 40.9 | 130.0 ± 30.7 | 0.2371 |
| Glucose (mg/dL), 120 minutes | 91.0 ± 24.0 | 127.8 ± 48.8 | 0.0379 |
| Area under the curve | 15501 ± 4500 | 15582 ± 2296 | 0.9677 |
| Body composition, mean ± SD | |||
| Weight, grams | 435.8 ± 46.28 | 433.1 ± 46.2 | 0.8212 |
| Fat weight, grams | 49.7 ± 12.2 | 35.1 ± 20.7 | 0.2056 |
| Lean weight, grams | 374.0 ± 27.5 | 367.2 ± 28.8 | 0.6895 |
| Water weight, grams | 2.9 ± 0.4 | 2.9 ± 0.4 | 0.7324 |
ALT: alanine aminotransferase; AST: aspartate aminotransferase; MCHC: mean corpuscular hemoglobin concentration
Note: Bolding indicates statistical significance at p<0.2
Discussion
Studies in other model animal species suggest that FMT may be an effective treatment to alter disease states. Prior mouse models of FMT offer the best evidence of a causal relationship between microbial communities and disease to date, and have mostly focused upon characteristics and transmission of obesity (Boulangé et al., 2016; Ussar et al., 2015). The studies of FMT in mice suggest that shifts in microbial communities are associated with inflammation and downstream inflammatory cascades, making them of particular interest for studies regarding stabilization of microbial communities to restore homoeostasis, potentially decreasing inflammatory states. If FMT can be used to stabilize microbial diversity and decrease inflammatory states then they are a potential intervention to prevent disease states.
To our knowledge, this is the first study to attempt FMT in marmosets and aimed to determine the safety and feasibility of FMT via oral gavage in marmosets. While the overall alpha diversity was not significantly different in recipients six months following the transfer, there were initial shifts in microbial taxa that reflected the donor diversity. These results suggest that FMT is a feasible procedure to be used to alter gut microbiota in the marmosets. Importantly, there were no clinical changes in the recipients that resulted in poor health outcomes. While, due to small sample size in this initial efficacy study, the analyses were not powered to detect significant differences in microbiome composition post-FMT at an alpha <0.05, we were able to detect shifts in the microbiome that reflected the donor diversity. We started with a relatively homogenous sample of young adult male marmosets to assess safety of the procedure prior to attempting FMT in older marmosets. Because of this, there was less variation in microbiome composition at baseline; thus, limiting the potential effect size of FMT in altering the gut microbiome composition. We only collected a single baseline sample from donors and recipients, thus longitudinal variability in microbiome structure prior to FMT was not captured. Despite this, prior studies have found gut microbiome structure to be relatively consistent over time (Caporaso et al., 2011; Faith et al., 2013; Mehta et al., 2018) and any changes would not have impacted the safety and microbiome effects of the FMT. Furthermore, we found that mean Shannon diversity at baseline for this study did not significantly change in donors from the previously published data (Reveles et al., 2019).
While FMT has been conducted in humans via either lower (e.g., colonoscopy) or upper (e.g., capsule, suspension) gastrointestinal delivery, we chose the oral route of administration for this study for several reasons. First, from a safety and ethics perspective, oral administration does not require surgical intervention, thus limiting the risk for adverse events in the animals. Second, rodent FMT has been traditionally conducted using oral gavage and was the basis of our design in this marmoset study. Mouse studies have indicated successful microbiome engraftment post-FMT. For example, studies by (Staley et al., 2017; Wrzosek et al., 2018) evaluated mouse microbiome engraftment following human microbiota transfer and improved engraftment after sequential antibiotic treatment. In each of these studies, microbiome engraftment was successful and stable over the study period. Finally, we aimed to produce study findings that will be most translatable to humans in the future. Oral FMT is likely more translatable to human use, as capsule formulations are already readily available from stool banks (e.g., OpenBiome) and does not require surgical intervention. While there is some concern that the microbiome might not fully engraft following passage through the acidic stomach, recent studies have indicated this might not be of significant clinical concern. In a phase 3 clinical trial, a new oral FMT suspension for human use in CDI resulted in successful microbiome engraftment and continued stability over the study period (60 days). Participant microbiomes progressively became more similar to the intervention post-transplant (Blount, Shannon, Deych, & Jones, 2019). Similarly, recent meta-analyses indicate no significant clinical differences between upper and lower FMT delivery in the treatment of CDI (Ramai, Zakhia, Ofosu, Ofori, & Reddy, 2019). A recent study comparing oral FMT capsules to colonoscopy delivery found that FMT by oral capsule was non-inferior to delivery by colonoscopy in preventing future CDI recurrences (Kao et al., 2017). This study also found that Shannon diversity in recipients significantly increased following FMT and that the increased diversity was maintained up to 12 weeks post-FMT, suggesting that oral administration of FMT does have the ability to alter gut microbiome composition. Some prior studies in humans and rodents have administered antibiotics prior to FMT to deplete the host’s commensal microbiota and promote uptake of the donor microorganisms. There is limited evidence to support this practice, and further studies are needed to evaluate the impact of antibiotic depletion of the marmoset microbiome prior to FMT on FMT engraftment and long-term stability and health outcomes. In addition, the use of antibiotics for FMT delivery may ultimately limit translation to clinical use in humans.
Donor selection for FMT and processing of the samples are of critical importance for positively altering the gut microbiome. There is some evidence from human FMT treatment of CDI that success of transplant is affected not only by donor microbiome diversity, but also composition, leading to the hypothesis that super donors may exist (Wilson et al., 2019). Since it is not currently possible to predict which donor may be a super donor, or which donor material may increase likelihood of success, most FMT studies currently use an accepted best strategy combining donor material into a single batch (Wilson et al., 2019). This is the technique that we used in this efficacy study, however, this method could introduce niche competition between microbiota resulting in altered implantation (Zlitni et al., 2020). Future studies of FMT should include a comparison of benefits gained through combination of donor materials versus the risk of potential niche competition. Another potential limitation of our procedures was that recent studies have suggested that preparation of donor material in an anaerobic environment may better preserve anaerobic bacteria and thus increase the likelihood of survival and engraftment of these species (Chu, Smith, Perrotta, Kassam, & Alm, 2017). However, to date this method has not been assessed in animal models of FMT and is not included in the recent guidelines for human FMT procedures (Sokol et al., 2016), and therefore was not included in our procedural methods. Furthermore, the use of two freeze-thaw cycles (original stool collection and supernatant post-processing) could have resulted in some microbial degradation, though we expect this effect to be minimal (Shkoporov et al., 2018). Future studies should continue to characterize conditions of donor material and microbiome composition most predictive of positive health outcomes and the ability of the donor microbiome to engraft in a recipient. These studies should also incorporate functional analyses to determine if structural microbiome changes contribute to byproduct production and functional changes as well. This initial efficacy study in marmosets is the first step in evaluating the use of microbiome-targeted therapies, such as FMT, in a nonhuman primate. Future studies may evaluate the use of these therapies in treatments of diseases such as obesity, metabolic syndrome, aging, frailty or marmoset wasting syndrome.
Conclusions
In this study, FMT via oral gavage was safe to administer to young adult marmosets. Overall, the data suggest that the marmoset microbiome may be altered by FMT; however, the progressive changes in the microbiome are strongly driven by the host and its baseline microbiome composition. Further studies are needed to evaluate the safety and effectiveness of FMT as a potential intervention to improve healthspan and lifespan, particularly focusing on optimizing dose, route of administration, and donor selection.
Supplementary Material
Research Highlights.
Fecal microbiota transplantation (FMT) via oral gavage had no negative impact on clinical measures in young adult marmosets after 6 months.
The data suggest that the marmoset microbiome may be altered by oral delivery of FMT.
Acknowledgements
The investigators would like to thank Dr. Larry Forney at the University of Idaho for microbiome expertise during study design. We would also like to thank Aubrey Sills and Joselyn Artavia for their dedicated care of the marmosets, the staff at the Barshop Colony for Longevity and Aging Studies, UT Health San Antonio and at Second Genome, Inc. for their assistance with study procedures.
Funding
This work was supported by the National Institutes of Health/National Institute on Aging San Antonio Claude D. Pepper Older Americans Independence Center (1P30AG044271-01A1).
Footnotes
Conflicts of Interest
The authors report no competing interests related to the content of this manuscript.
REFERENCES CITED
- Barlow GM, Yu A, & Mathur R (2015). Role of the Gut Microbiome in Obesity and Diabetes Mellitus. Nutr Clin Pract, 30(6), 787–797. doi: 10.1177/0884533615609896 [DOI] [PubMed] [Google Scholar]
- Belkaid Y, & Hand TW (2014). Role of the microbiota in immunity and inflammation. Cell, 157(1), 121–141. doi: 10.1016/j.cell.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount KF, Shannon WD, Deych E, & Jones C (2019). Restoration of bacterial microbiome composition and diversity among treatment responders in a phase 2 trial of RBX2660: an investigational microbiome restoration therapeutic. Paper presented at the Open forum infectious diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulangé CL, Neves AL, Chilloux J, Nicholson JK, & Dumas M-E (2016). Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome medicine, 8(1), 42. doi: 10.1186/s13073-016-0303-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray JR, & Curtis JT (1957). An ordination of the upland forest communities of southern Wisconsin. Ecological monographs, 27(4), 325–349. doi: 10.2307/1942268 [DOI] [Google Scholar]
- Bruce-Keller AJ, Salbaum JM, Luo M, Blanchard E IV, Taylor CM, Welsh DA, & Berthoud H-R (2015). Obese-type gut microbiota induce neurobehavioral changes in the absence of obesity. Biological psychiatry, 77(7), 607–615. doi: 10.1016/j.biopsych.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, … Fierer N (2011). Moving pictures of the human microbiome. Genome biology, 12(5), 1–8. doi: 10.1186/gb-2011-12-5-r50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo A, Cattane N, Galluzzi S, Provasi S, Lopizzo N, Festari C, … Muscio C (2017). Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiology of aging, 49, 60–68. doi: 10.1016/j.neurobiolaging.2016.08.019 [DOI] [PubMed] [Google Scholar]
- Centers for Disease, C., & Prevention. (2003). Trends in aging--United States and worldwide. MMWR Morb Mortal Wkly Rep, 52(6), 101–104, 106. [PubMed] [Google Scholar]
- Chu ND, Smith MB, Perrotta AR, Kassam Z, & Alm EJ (2017). Profiling living bacteria informs preparation of fecal microbiota transplantations. PLoS One, 12(1), e0170922. doi: 10.1371/journal.pone.0170922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Control, C. f. D., & Prevention. (2015). The State of Aging and Health in America 2013. Atlanta, GA: Centers for Disease Control and Prevention, US Department of Health and Human Services; 2013. In. [Google Scholar]
- Dall TM, Gallo PD, Chakrabarti R, West T, Semilla AP, & Storm MV (2013). An aging population and growing disease burden will require alarge and specialized health care workforce by 2025. Health affairs, 32(11), 2013–2020. doi: 10.1377/hlthaff.2013.0714 [DOI] [PubMed] [Google Scholar]
- Di Luccia B, Crescenzo R, Mazzoli A, Cigliano L, Venditti P, Walser J-C, … Iossa S (2015). Rescue of fructose-induced metabolic syndrome by antibiotics or faecal transplantation in a rat model of obesity. PLoS One, 10(8), e0134893. doi: 10.1371/journal.pone.0134893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26(19), 2460–2461. doi: 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Edgar RC (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods, 10(10), 996–998. doi: 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, … Leibel RL (2013). The long-term stability of the human gut microbiota. Science, 341(6141). doi: 10.1126/science.1237439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz C, & Mair W (2014). You are what you host: microbiome modulation of the aging process. Cell, 156(3), 408–411. doi: 10.1016/j.cell.2014.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MA, Jeffery IB, Beaumont M, Bell JT, Clark AG, Ley RE, … Steves CJ (2016). Signatures of early frailty in the gut microbiota. Genome Med, 8(1), 8. doi: 10.1186/s13073-016-0262-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Song S, Woodhams DC, Martino C, Allaband C, Mu A, Javorschi-Miller-Montgomery S, … Knight R (2019). Engineering the microbiome for animal health and conservation. Experimental Biology and Medicine, 244(6), 494–504. doi: 10.1177/1535370219830075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao D, Roach B, Silva M, Beck P, Rioux K, Kaplan GG, … Xu H (2017). Effect of oral capsule–vs colonoscopy-delivered fecal microbiota transplantation on recurrent Clostridium difficile infection: a randomized clinical trial. JAMA, 318(20), 1985–1993. doi: 10.1001/jama.2017.17077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassam Z, Lee CH, Yuan Y, & Hunt RH (2013). Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. The American journal of gastroenterology, 108(4), 500. doi: 10.1038/ajg.2013.59 [DOI] [PubMed] [Google Scholar]
- Komaroff AL (2017). The microbiome and risk for obesity and diabetes. JAMA, 317(4), 355–356. doi: 10.1001/jama.2016.20099 [DOI] [PubMed] [Google Scholar]
- Lee EC, Whitehead AL, Jacques RM, & Julious SA (2014). The statistical interpretation of pilot trials: should significance thresholds be reconsidered? BMC medical research methodology, 14, 41–41. doi: 10.1186/1471-2288-14-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liwinski T, & Elinav E (2019). Harnessing the microbiota for therapeutic purposes. American Journal of Transplantation. doi: 10.1111/ajt.15753 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, & Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology, 15(12), 550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch SV, & Pedersen O (2016). The human intestinal microbiome in health and disease. New England Journal of Medicine, 375(24), 2369–2379. doi: 10.1056/NEJMra1600266 [DOI] [PubMed] [Google Scholar]
- Marotz CA, & Zarrinpar A (2016). Focus: Microbiome: Treating Obesity and Metabolic Syndrome with Fecal Microbiota Transplantation. The Yale journal of biology and medicine, 89(3), 383. [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, … Hugenholtz P (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J, 6(3), 610–618. doi: 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, & Holmes S (2014). Waste not, want not: why rarefying microbiome data is inadmissible. PLoS computational biology, 10(4), e1003531. doi: 10.1371/journal.pcbi.1003531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta RS, Abu-Ali GS, Drew DA, Lloyd-Price J, Subramanian A, Lochhead P, … Brown GT (2018). Stability of the human faecal microbiome in a cohort of adult men. Nature microbiology, 3(3), 347–355. doi: 10.1038/s41564-017-0096-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power ML, Ross CN, Schulkin J, Ziegler TE, & Tardif SD (2013). Metabolic consequences of the early onset of obesity in common marmoset monkeys. Obesity (Silver Spring), 21(12), E592–598. doi: 10.1002/oby.20462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramai D, Zakhia K, Ofosu A, Ofori E, & Reddy M (2019). Fecal microbiota transplantation: donor relation, fresh or frozen, delivery methods, cost-effectiveness. Annals of gastroenterology, 32(1), 30. doi: 10.20524/aog.2018.0328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reveles KR, Patel S, Forney LJ, & Ross CN (2019). Age-related changes in the marmoset gut microbiome. American Journal of Primatology, 81(2), e22960. doi: 10.1002/ajp.22960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CN (2019). Marmosets in Aging Research In The Common Marmoset in Captivity and Biomedical Research (pp. 355–376): Elsevier. [Google Scholar]
- Ross CN, Adams J, Gonzalez O, Dick E, Giavedoni L, Hodara VL, … Tardif SD (2019). Cross-sectional comparison of health-span phenotypes in young versus geriatric marmosets. American Journal of Primatology, 0(0), e22952. doi:doi: 10.1002/ajp.22952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CN, Austad S, Brasky K, Brown CJ, Forney LJ, Gelfond JA, … Tardif SD (2017). The development of a specific pathogen free (SPF) barrier colony of marmosets (Callithrix jacchus) for aging research. Aging (Albany NY), 9(12), 2544–2558. doi: 10.18632/aging.101340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CN, Davis K, Dobek G, & Tardif SD (2012). Aging phenotypes of common marmosets (Callithrix jacchus). Journal of Aging Research, 2012, 567143. doi: 10.1155/2012/567143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CN, & Salmon AB (2019). Aging research using the common marmoset: Focus on aging interventions. Nutrition and Healthy Aging, 5(2), 97–109. doi: 10.3233/NHA-180046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, … Weber CF (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol, 75(23), 7537–7541. doi: 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon CE (1948). A mathematical theory of communication. Bell system technical journal, 27(3), 379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x [DOI] [Google Scholar]
- Shkoporov AN, Ryan FJ, Draper LA, Forde A, Stockdale SR, Daly KM, … Dalmasso M (2018). Reproducible protocols for metagenomic analysis of human faecal phageomes. Microbiome, 6(1), 68. doi: 10.1186/s40168-018-0446-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Galperine T, Kapel N, Bourlioux P, Seksik P, Barbut F, … Joly F (2016). Faecal microbiota transplantation in recurrent Clostridium difficile infection: Recommendations from the French Group of Faecal microbiota Transplantation. Digestive and Liver Disease, 48(3), 242–247. doi: 10.1016/j.dld.2015.08.017 [DOI] [PubMed] [Google Scholar]
- Staley C, Hamilton MJ, Vaughn BP, Graiziger CT, Newman KM, Kabage AJ, … Khoruts A (2017). Successful resolution of recurrent Clostridium difficile infection using freeze-dried, encapsulated fecal microbiota; pragmatic cohort study. The American journal of gastroenterology, 112(6), 940. doi: 10.1038/ajg.2017.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif SD, Power ML, Ross CN, Rutherford JN, Layne-Colon DG, & Paulik MA (2009). Characterization of obese phenotypes in a small nonhuman primate, the common marmoset (Callithrix jacchus). Obesity (Silver Spring), 17(8), 1499–1505. doi: 10.1038/oby.2009.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ussar S, Griffin NW, Bezy O, Fujisaka S, Vienberg S, Softic S, … Kahn CR (2015). Interactions between gut microbiota, host genetics and diet modulate the predisposition to obesity and metabolic syndrome. Cell Metabolism, 22(3), 516–530. doi: 10.1016/j.cmet.2015.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil DN (1997). The economics of population aging. Handbook of population and family economics, 1(2), 967–1014. [Google Scholar]
- Wilson BC, Vatanen T, Cutfield WS, & O'Sullivan JM (2019). The Super-Donor Phenomenon in Fecal Microbiota Transplantation. Frontiers in Cellular and Infection Microbiology, 9, 2. doi: 10.3389/fcimb.2019.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrzosek L, Ciocan D, Borentain P, Spatz M, Puchois V, Hugot C, … Cassard A-M (2018). Transplantation of human microbiota into conventional mice durably reshapes the gut microbiota. Scientific reports, 8(1), 1–9. doi: 10.1038/s41598-018-25300-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlitni S, Bishara A, Moss EL, Tkachenko E, Kang JB, Culver RN, … Handy C (2020). Strain-resolved microbiome sequencing reveals mobile elements that drive bacterial competition on a clinical timescale. Genome medicine, 12, 1–17. doi: 10.1186/s13073-020-00747-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



