Abstract
The emergence of Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has imposed a greater challenge for the world. Coronavirus has infected over 38.3 million people and caused millions of deaths worldwide. The COVID-19 outbreak has accentuated the need for additional efforts to develop broad-spectrum therapeutics to combat SARS-CoV-2 infection. In the current investigation, an attempt was made to design potential SARS-CoV PLpro inhibitors containing naphthalene and 3,4-dihydro-2H-pyran moieties connected via -NHCO- linker. The ligands obeyed Lipinski's rule and were found to have good drug-likeness and ADMET properties. Docking simulations confirmed strong binding affinity and inhibition potential of the designed ligands against the receptor SARS CoV-2 Papain-like protease (PLpro). LigandL10 incorporating the oxadiazole ring system displayed better binding affinity than the control 5-acetamido-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide. Further, the docked complex of LigandL10 was subjected to molecular dynamics (MD) simulation to examine the molecular mechanisms of protein-ligand interactions. The results of the present study are encouraging. Ligand L10 emerged as the most potent ligand in the series and could be considered for further research for the development of potential therapeutics for the treatment of COVID-19.
Keywords: COVID-19, SARS-CoV-2, Papain-like protease, Molecular docking, Molecular dynamics simulation, Computational chemistry, Organic chemistry, Theoretical chemistry, Pharmaceutical chemistry, Bioinformatics, Pharmaceutical science
COVID-19; SARS-CoV-2; papain-like protease; Molecular Docking; molecular dynamics simulation; Computational Chemistry; Organic chemistry; Theoretical chemistry; Pharmaceutical chemistry; Bioinformatics; Pharmaceutical Science
1. Introduction
The outbreak of coronavirus diseases (COVID-19) pandemic had influenced all the sections of the society and had put a great social economic and health care challenge. As of 14 September, there are 38,364,519 active cases and the number of deaths attributed to COVID-19 worldwide has already surpassed 1,090,811 [1]. The world is dealing with expeditious growth in the number of confirmed cases and deaths. At present no proven effective therapies available for COVID-19 treatment. SARS-CoV-2 belongs to the coronaviridiae family as a member of β-coronaviruses and has a positive-sense single-stranded RNA with the largest RNA genome possessing a helical nucleocapsid [2]. The understanding of virology and the mechanism underlying the SARS-CoV-2 replication and maturation could open up ways for the development of target-specific drug design and discovery. The scientists and pharmacists have a great interest in either repurposing the existing drugs or developing new drugs for the treatment of COVID-19. SARS-CoV-2 has zoonotic emergence and a high human-to-human transmission rate [3]. SARS coronavirus infections may develop the severe acute respiratory disease with multi-organ failure. Zou and co-workers in their study identified the organs such as lung, heart, esophagus, kidney, bladder, and ileum, and located specific cell types, at-risk and vulnerable to 2019-nCoV infection [4]. According to the pathogenesis of SARS-CoV-2, primary viral replication occurs in the mucosal epithelium of the upper respiratory tract and further multiplicated in the lower respiratory tract and gastrointestinal mucosa [5].
Two viral proteases, papain-like protease (PLpro) and 3C cleavage-like protease (3CLpro) (also known as Mpro) are correlated to viral transcription and replication in SARS-CoV-2 [6]. The processing of viral polyproteins is essential for the maturation and infectivity of the virus [7]. This protease cleave and transform, the polyproteins of coronaviridae genome pp1a and pp1ab in non-structural proteins (NSPs). The non-structural proteins play an important role in transcription/replication during the infection [8]. Therefore, antiviral drug candidates targeting these proteins may display anti-SARS-CoV-2 activity. The papain-like protease PLpro is an essential coronavirus enzyme that is required for processing viral polyproteins to generate a functional replicase complex and enable viral spread [9]. 3CLpro and PLpro mainly process the viral polyprotein, however, PLpro has the additional function of stripping ubiquitin and Interferon-stimulated gene 15 (ISG15) from host-cell proteins to aid coronaviruses in their evasion of the host innate immune responses. Inhibition of SAR-CoV2-PLpro with GRL-0617 impairs the virus-induced cytopathogenic effect, maintains the antiviral interferon pathway, and reduces viral replication in infected cells [10]. Targeting PLpro with antiviral drugs may have an advantage in not only inhibiting viral replication but also inhibiting the dysregulation of signaling cascades in infected cells [11]. Cho and coworkers [12] screened flavonoids that inhibited SARS-CoV PLpro with IC50 values between 5.0 and 14.4 μM, and it was shown that compounds containing the dihydro-2H-pyran moiety displayed better PLpro inhibition. Ratia and co-workers used a high-throughput screen (HTS) and reported two promising naphthalene inhibitors of SARS-CoV PLpro with IC50 values of 20.1 μM and 59 μM, respectively [13]. Naphthalene-based inhibitors are potent, competitive inhibitors, and bind within the active site of SARS-CoV PLpro [11]. In another study naphthalene-based PLpro inhibitors are shown to be effective at halting SARS-CoV-2 PLpro activity as well as SARS-CoV-2 replication [14]. PLpro is an indispensable enzyme in the process of coronavirus replication. It is very valuable for targeting PLpro to treat coronavirus infections, but no inhibitor has been approved by the FDA for marketing [15]. In light of the above findings, it was planned to design novel inhibitors targeting SARS-CoV-2 PLpro.
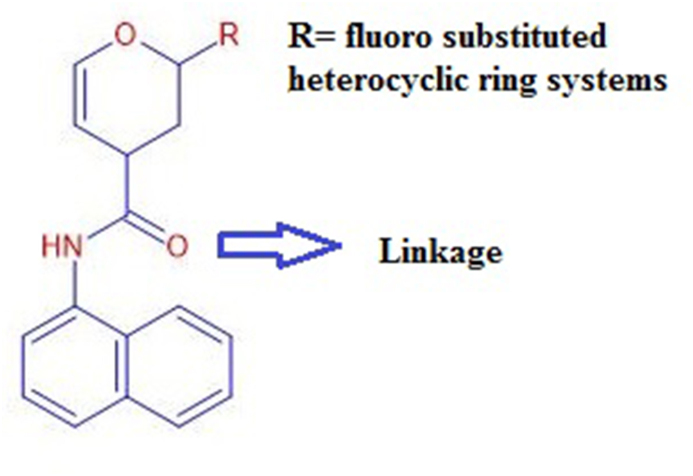
Structure-based drug designing and development of specific therapeutic drugs are critical for anti-SARS-CoV-2 drug discovery [16]. Computer-aided drug design expedites the drug designing process and has been a promising technique for designing novel and potent therapeutics [17, 18, 19]. In the present investigation, a series of naphthalene based SARS-CoV PLpro inhibitors were rationally designed by linking naphthalene scaffold to the 3,4-dihydro-2H-pyran moiety via -NHCO functional group. The pyran moiety tolerated different substituents, five and six-membered, and fused heterocyclic ring systems having oxygen and nitrogen hetero atoms, and fluoro substitution. Heterocyclic compounds possess a broad spectrum of antiviral activity [20, 21, 22, 23]. fluoro group results in a radical change in the biological activities of the molecules. Therefore the introduction of a fluoro substituent is an important strategy in the design and discovery of novel drug candidates [24]. The pharmacokinetic and pharmacodynamic properties of the designed inhibitors were examined through computational techniques. 5-acetamido-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide was used as a control that inhibits SARS-CoV-2 PLpro with an IC50 of 5.0 μM and an EC50 of 21.0 μM [13, 14]. The result obtained from the study will provide an insight into essential pharmacophoric features required to develop potential therapeutics for the treatment of COVID-19.
2. Materials and methods
2.1. Generation of ligands
The ligands L1-L20 were built up and optimized using Advanced Chemistry Development (ACD) Labs ChemSketch 12.0 (http://www.acdlabs.com) software. Zinc compounds database (http://www.zinc.docking.org) and ChemSpider (http://www.chemspider.com) were employed to check the novelty of the designed ligands. Table1 summarizes the IUPAC (International Union of Pure and Applied Chemistry) names and molecular formula of newly designed SARS-CoV PLpro inhibitors. The general design of these inhibitors has been illustrated in Figure 1.
Table 1.
A summary of the designed PLpro inhibitors (L1-L20).
| Sl. No. | Compd No. | R | IUPAC name | Molecular Formula |
|---|---|---|---|---|
| 1 | L1 | 4-fluorophenyl | 2-(4-fluorophenyl)-N-(naphthalen-1-yl)-3,4-dihydro-2H-pyran-4-carboxamide | C22H18FNO2 |
| 2 | L2 | 4-fluoropyridin-2-yl | 2-(4-fluoropyridin-2-yl)-N-(naphthalen-1-yl)-3,4-dihydro-2H-pyran-4-carboxamide | C21H17FN2O2 |
| 3 | L3 | 2-fluoropyrimidin-4-yl | 2-(2-fluoropyrimidin-4-yl)-N-(naphthalen-1-yl)-3,4-dihydro-2H-pyran-4-carboxamide | C20H16FN3O2 |
| 4 | L4 | 4-fluoropiperidin-3-yl | 2-(4-fluoropiperidin-3-yl)-N-(naphthalen-1-yl)-3,4-dihydro-2H-pyran-4-carboxamide | C21H23FN2O2 |
| 5 | L5 | 2-fluoromorpholin-3-yl | 2-(2-fluoromorpholin-3-yl)-N-(naphthalen-1-yl)-3,4-dihydro-2H-pyran-4-carboxamide | C20H21FN2O3 |
| 6 | L6 | 4-fluoro-1H-pyrrol-3-yl | 2-(4-fluoro-1H-pyrrol-3-yl)-N-(naphthalen-1-yl)-3,4-dihydro-2H-pyran-4-carboxamide | C20H17FN2O2 |
| 7 | L7 | 2-fluoro-1H-imidazol-4-yl | 2-(2-fluoro-1H-imidazol-4-yl)-N-(naphthalen-1-yl)-3,4-dihydro-2H-pyran-4-carboxamide | C19H16FN3O2 |
| 8 | L8 | 4-fluorofuran-3-yl | 2-(4-fluorofuran-3-yl)-N-(naphthalen-1-yl)-3,4-dihydro-2H-pyran-4-carboxamide | C20H16FNO3 |
| 9 | L9 | 2-fluoro-1,3-oxazol-4-yl | 2-(2-fluoro-1,3-oxazol-4-yl)-N-(naphthalen-1-yl)-3,4-dihydro-2H-pyran-4-carboxamide | C19H15FN2O3 |
| 10 | L10 | 5-fluoro-1,3,4-oxadiazol-2-yl | 2-(5-fluoro-1,3,4-oxadiazol-2-yl)-N-(naphthalen-1-yl)-3,4-dihydro-2H-pyran-4-carboxamide | C18H14FN3O3 |
| 11 | L11 | 2-fluoro-2H-pyran | 2′-fluoro-N-(naphthalen-1-yl)-3,4-dihydro-2H,2′H-[2,4′-bipyran]-4-carboxamide | C21H18FNO3 |
| 12 | L12 | 3-fluoro-1,4-dioxan-2-yl | 2-(3-fluoro-1,4-dioxan-2-yl)-N-(naphthalen-1-yl)-3,4-dihydro-2H-pyran-4-carboxamide | C20H20FNO4 |
| 13 | L13 | 2-fluoro-2,3-dihydro-1H-indol-6-yl | 2-(2-fluoro-2,3-dihydro-1H-indol-6-yl)-N-(naphthalen-1-yl)-3,4-dihydro-2H-pyran-4-carboxamide | C24H21FN2O2 |
| 14 | L14 | 4-fluoroquinolin-7-yl | 2-(4-fluoroquinolin-7-yl)-N-(naphthalen-1-yl)-3,4-dihydro-2H-pyran-4-carboxamide | C25H19FN2O2 |
| 15 | L15 | 4-fluoro-2H-1-benzopyran-7-yl | 2-(4-fluoro-2H-1-benzopyran-7-yl)-N-(naphthalen-1-yl)-3,4-dihydro-2H-pyran-4-carboxamide | C25H20FNO3 |
| 16 | L16 | 6-fluoro-9H-purin-2-yl | 2-(6-fluoro-9H-purin-2-yl)-N-(naphthalen-1-yl)-3,4-dihydro-2H-pyran-4-carboxamide | C21H16FN5O2 |
| 17 | L17 | 2-fluoro-1-benzofuran-5-yl | 2-(2-fluoro-1-benzofuran-5-yl)-N-(naphthalen-1-yl)-3,4-dihydro-2H-pyran-4-carboxamide | C24H18FNO3 |
| 18 | L18 | 2-fluoro-1H-benzimidazol-5-yl | 2-(2-fluoro-1H-benzimidazol-5-yl)-N-(naphthalen-1-yl)-3,4-dihydro-2H-pyran-4-carboxamide | C23H18FN3O2 |
| 19 | L19 | 2-fluoro-1,3-benzoxazol-6-yl | 2-(2-fluoro-1,3-benzoxazol-6-yl)-N-(naphthalen-1-yl)-3,4-dihydro-2H-pyran-4-carboxamide | C23H17FN2O3 |
| 20 | L20 | 3-fluoro-2-oxo-2H-1-benzopyran-6-yl | 2-(3-fluoro-2-oxo-2H-1-benzopyran-6-yl)-N-(naphthalen-1-yl)-3,4-dihydro-2H-pyran-4-carboxamide | C25H18FNO4 |
Figure 1.
Designed SARS-CoV PLpro inhibitors.
2.2. Molecular descriptors, drug likeliness properties, and toxicity risk assessment
In the present study, Molinspiration online tool (http://www.molinspiration.com) was employed to predict molecular properties such as partition coefficient (Log P), Topological polar surface area (TPSA), number of atoms, hydrogen bond donors and acceptors, number of rotatable bonds, and molecular weight. Bioactivity scores for drug targets including G protein-coupled receptors (GPCR) ligands, ion channel modulators, kinase inhibitors, nuclear receptors ligands, protease inhibitors, and enzyme inhibitors were also computed. OSIRIS DataWarrior program [25] was employed to predict toxic properties such as mutagenicity, tumorogenicity, irritant, and reproductive effect. Synthetic accessibility (SA) of the ligands was computed using Ambit-SA (http://ambit.sourceforge.net/reactor.html) software tool. The model for SA uses four weighted molecular descriptors, which represent different structural and topological features, combined within an additive scheme [26]. The synthetic accessibility score of the ligands L1-L20 was found between the range 60.38–72.87, indicating that the ligands are easily synthesizable.
2.3. admetSAR predictions
The screening of the Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) profile of the designed inhibitors was done using the admetSAR tool [27]. This database is having 22 qualitative classification and 5 quantitative regression models with high predictive accuracy, used to estimate mammalian ADMET properties for novel chemicals. Various ADMET associated properties like Human Intestinal Absorption, blood-brain-barrier (BBB) penetration, Caco-2 permeability, CYP inhibitory promiscuity, AMES toxicity, carcinogenicity, and rate acute toxicity LD50 were determined for the designed ligands.
2.4. Docking and molecular dynamics simulations
The computation of molecular docking screening was performed using the Autodock 4.2 software [28]. The X-ray crystallographic structure of SARS CoV-2 papain-like protease (PDB deposited code: 6WUU), was retrieved from the protein data bank [29] and shown in Figure 2. Papain-Like cysteine protease (PLpro, NSP3) is essential for SARS-CoV-2 replication and represents a promising target for the development of antiviral drugs [30]. The drug-target interactions were analyzed by calculating binding energy (kcal/mol) and inhibition constant (nM).
Figure 2.
The three-dimensional structure of selected SARS CoV-2 papain-like protease (6WUU).
Preprocessing of the receptor was done using AutoDock Tools 4.2 program (ADT) (The Scripps research institute, La Jolla, California, USA). The receptor was processed by removing papain-like protease peptide inhibitors, metal ions, water molecules, and adding polar hydrogen atoms. Lamarckian genetic algorithm is an interesting application employed in the AutoDock program. In the present study for the Lamarckian genetic algorithm, the number of genetic algorithms (GA) runs, the maximum number of energy evaluations, the maximum number of generations, population size was set to 10, 2500000, 27000, and 150 respectively. To explore the most probable binding poses, the ligands were kept flexible while the receptor was set rigid. The grid box size was set to covers all the residues present inside the active site pocket with 126 × 126 × 126 points in x, y, and z directions, and 32.717, 77.276, 2.008 grid center was set for papain-like protease receptor. The box was centered based on the cognate ligand with a spacing of 0.375 Å. The binding energetics of the ligand-receptor complexes were determined by computing minimum binding energy, as well as inhibition constant values. “LigPlot+” is a graphical system that generates multiple two-dimensional (2D) diagrams of ligand-protein interactions from docked complexes. In the present study, the LigPlot+ program was employed to study the interacting residues, hydrogen bonds, and hydrophobic interactions of best docked pose for the selected ligand and papain-like protease receptor 6WUU [31]. Molecular dynamics simulations were performed using the program NAMD, NAnoscale Molecular Dynamics program [32], and all files were generated using visual molecular dynamics, VMD [33]. MD simulations were performed using the CHARMM36 force field. The protein-ligand complex was immersed in the center of a 50 Å box of water molecules, where all water molecule atoms (H-O-H) were closer than 1.5 Å. A 12 Å cutoff distance was used to calculate short-range nonbonded interactions. The solvated protein-ligand complex system was equilibrated with 1,000 minimization steps of the Powell algorithm at constant temperature (310 K), followed by a simulation of 5000000 runs for 10 ns.
3. Result and discussion
3.1. Molecular descriptors and drug likeliness properties
The molecular properties of designed SARS-CoV PLpro inhibitors were calculated by using Molinspiration cheminformatics software and are presented in Table2.
Table 2.
Molecular descriptors from molinspiration.
| Sl.No. | Compd No. | LogP | TPSA | natom | MW | nON | nOHN | nVio | nRot | Volume |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | L1 | 4.68 | 38.33 | 26 | 347.39 | 3 | 1 | 0 | 3 | 311.78 |
| 2 | L2 | 3.85 | 51.22 | 26 | 348.38 | 4 | 1 | 0 | 3 | 307.62 |
| 3 | L3 | 3.32 | 64.12 | 26 | 349.37 | 5 | 1 | 0 | 3 | 303.46 |
| 4 | L4 | 3.15 | 50.36 | 26 | 354.43 | 4 | 2 | 0 | 3 | 325.99 |
| 5 | L5 | 2.82 | 59.59 | 26 | 356.40 | 5 | 2 | 0 | 3 | 318.17 |
| 6 | L6 | 3.46 | 54.12 | 25 | 336.37 | 4 | 2 | 0 | 3 | 293.76 |
| 7 | L7 | 3.23 | 67.02 | 25 | 337.35 | 5 | 2 | 0 | 3 | 292.61 |
| 8 | L8 | 3.56 | 51.47 | 25 | 337.35 | 4 | 1 | 0 | 3 | 293.35 |
| 9 | L9 | 3.34 | 64.36 | 25 | 338.34 | 5 | 1 | 0 | 3 | 289.19 |
| 10 | L10 | 2.31 | 77.26 | 25 | 339.33 | 6 | 1 | 0 | 3 | 285.03 |
| 11 | L11 | 3.68 | 47.57 | 26 | 351.38 | 4 | 1 | 0 | 3 | 310.17 |
| 12 | L12 | 2.83 | 56.80 | 26 | 357.38 | 5 | 1 | 0 | 3 | 314.76 |
| 13 | L13 | 4.62 | 50.36 | 29 | 388.44 | 4 | 2 | 0 | 3 | 346.97 |
| 14 | L14 | 4.90 | 51.22 | 30 | 398.44 | 4 | 1 | 0 | 3 | 351.61 |
| 15 | L15 | 4.88 | 47.57 | 30 | 401.44 | 4 | 1 | 0 | 3 | 354.14 |
| 16 | L16 | 2.80 | 92.80 | 29 | 389.39 | 7 | 2 | 0 | 3 | 328.29 |
| 17 | L17 | 5.11 | 51.47 | 29 | 387.40 | 4 | 1 | 1 | 3 | 337.34 |
| 18 | L18 | 4.47 | 67.02 | 29 | 387.41 | 5 | 2 | 0 | 3 | 336.60 |
| 19 | L19 | 4.58 | 64.36 | 29 | 388.40 | 5 | 1 | 0 | 3 | 333.18 |
| 20 | L20 | 3.69 | 68.54 | 31 | 415.42 | 5 | 1 | 0 | 3 | 356.32 |
| 21 | Control | 3.84 | 58.20 | 26 | 346.43 | 4 | 2 | 0 | 4 | 328.73 |
LogP, the logarithm of compound partition coefficient between n-octanol and water; TPSA, topological polar surface area; natom, number of atoms; MW, molecular weight; nON, number of hydrogen bond acceptors; nOHN, number of hydrogen bond donors; nVio, number of violations; Nrotb, number of rotatable bonds.
LogP, the logarithm of compound partition coefficient between n-octanol and water; TPSA, topological polar surface area; natom, number of atoms; MW, molecular weight; nON, number of hydrogen bond acceptors; nOHN, number of hydrogen bond donors; nVio, number of violations; Nrotb, number of rotatable bonds.
Lipinski's rule of five was used to analyze the drug-likeness of the designed ligands (L1-L20). The partition coefficient or Log P is a sum of fragment-based contributions and correlation factors and indicates molecular hydrophobicity or lipophilicity. Log P values of all the ligands were found below 5 (except ligand L17), suggesting good permeability across the cell membrane. In the series, the lowest value of log P was seen for ligand 10 (2.31). The topological polar surface area (TPSA) of the ligands was observed in the range of 38.33–92.80 Å and is well below the limit of 160 Å, suggesting good bioavailability of the ligands. The molecular weight of the derivatives was found to be less than 500. The molecular descriptors such as number of hydrogen bond acceptors (O and N atoms), number of hydrogen bond donors (NH and OH), and number of rotatable bonds (nRB) were in agreement with Lipinski's rule of five i.e. less than 10, 5, and 10 respectively. The designed PL pro inhibitors passed through the Lipinski filter and are anticipated to be orally active.
The drug likeliness properties of the designed ligands (L1-L20) concerning the prediction of bioactivity scores of the designed ligands (L1-L20) were analyzed by molinspiration and are reported in Table 3. The designed ligands demonstrated a bioactivity score of more than 0.00 for the three-drug targets namely GPCR ligand, protease inhibitor, and enzyme inhibitor. The predicted bioactivity score of the ligands is suggestive of considerable to moderate interaction with all drug targets. The ligands showed a better bioactivity score than the control for all drug targets.
Table 3.
Prediction of bioactivity score with molinspiration.
| Sl.No. | Compd No. | GPCR | ICM | KI | NRL | PI | EI |
|---|---|---|---|---|---|---|---|
| 1 | L1 | 0.27 | -0.08 | -0.04 | 0.05 | 0.16 | 0.10 |
| 2 | L2 | 0.23 | -0.04 | 0.11 | -0.11 | 0.17 | 0.11 |
| 3 | L3 | 0.36 | -0.02 | 0.05 | 0.20 | 0.12 | 0.21 |
| 4 | L4 | 0.38 | 0.07 | 0.15 | 0.00 | 0.25 | 0.22 |
| 5 | L5 | 0.20 | 0.17 | -0.02 | -0.08 | 0.29 | 0.06 |
| 6 | L6 | 0.38 | 0.07 | 0.15 | 0.00 | 0.25 | 0.22 |
| 7 | L7 | 0.35 | 0.02 | 0.06 | 0.04 | 0.20 | 0.26 |
| 8 | L8 | 0.28 | -0.06 | -0.04 | 0.02 | 0.15 | 0.12 |
| 9 | L9 | 0.18 | -0.07 | -0.09 | -0.09 | 0.06 | 0.04 |
| 10 | L10 | 0.14 | -0.13 | -0.17 | -0.16 | 0.12 | 0.02 |
| 11 | L11 | 0.14 | 0.17 | -0.07 | 0.13 | 0.16 | 0.18 |
| 12 | L12 | 0.20 | 0.17 | -0.01 | 0.18 | 0.28 | 0.13 |
| 13 | L13 | 0.21 | -0.11 | -0.08 | 0.04 | 0.13 | 0.04 |
| 14 | L14 | 0.35 | 0.01 | 0.22 | 0.08 | 0.21 | 0.21 |
| 15 | L15 | 0.24 | -0.13 | -0.09 | 0.11 | 0.09 | 0.18 |
| 16 | L16 | 0.37 | -0.03 | 0.19 | -0.41 | 0.16 | 0.34 |
| 17 | L17 | 0.22 | -0.12 | -0.09 | 0.06 | 0.15 | 0.10 |
| 18 | L18 | 0.36 | -0.01 | 0.08 | 0.16 | 0.15 | 0.22 |
| 19 | L19 | 0.20 | -0.09 | -0.08 | 0.03 | 0.11 | 0.09 |
| 20 | L20 | 0.08 | -0.23 | -0.20 | 0.03 | 0.07 | 0.12 |
| 21 | Control | -0.12 | -0.29 | -0.23 | -0.44 | -0.07 | -0.26 |
GPCR, GPCR ligand; ICM, Ion channel modulator, KI, Kinase inhibitor; NRL, Nuclear receptor ligand; PI, Protease inhibitor, EI, Enzyme inhibitor.
Solubility, Druglikeness, and toxicity of the designed ligands were assessed by Osiris property explorer and the results are presented in Table 4.
Table 4.
Solubility, Drug-likeness and toxicity prediction through OSIRIS Property Explorer.
| Sl.No. | Compd No. | ClogS | Drug-likeness | Mutagenic | Tumorigenic | Reproductive | Irritant |
|---|---|---|---|---|---|---|---|
| 1 | L1 | -5.633 | 1.0646 | High | High | None | None |
| 2 | L2 | -4.862 | 1.0646 | High | High | None | None |
| 3 | L3 | -4.924 | 1.0646 | High | High | None | None |
| 4 | L4 | -5.087 | 2.2466 | High | High | None | None |
| 5 | L5 | -4.629 | 2.1525 | High | High | None | None |
| 6 | L6 | -4.657 | 1.4464 | High | High | None | None |
| 7 | L7 | -4.743 | 1.5828 | High | High | None | None |
| 8 | L8 | -5.291 | 1.4464 | High | High | None | None |
| 9 | L9 | -5.377 | 1.4937 | High | High | None | None |
| 10 | L10 | -4.772 | 2.0104 | High | High | None | None |
| 11 | L11 | -5.396 | 1.6189 | High | High | None | None |
| 12 | L12 | -4.765 | -5.6984 | High | High | None | Low |
| 13 | L13 | -6.124 | 1.2642 | High | High | None | None |
| 14 | L14 | -6.339 | 1.0646 | High | High | None | None |
| 15 | L15 | -6.206 | 1.2261 | High | High | Low | None |
| 16 | L16 | -5.494 | 1.5509 | High | High | None | None |
| 17 | L17 | -7.317 | 0.9672 | High | High | None | None |
| 18 | L18 | -6.115 | 1.2445 | High | High | None | None |
| 19 | L19 | -6.749 | 1.1528 | High | High | None | None |
| 20 | L20 | -6.586 | -7.0596 | High | High | None | None |
| 21 | Control | -5.307 | 2.5325 | Low | High | None | High |
The ClogS value indicated that the designed ligands possess good solubility. A positive drug-likeness value (0.1–10) suggests that a molecule contains fragments that are commonly present in commercial drugs. The drug-likeness score of the ligands was found to be positive and significant in comparison to the control except for ligand L12 and L20. The toxicity calculations revealed that the ligands possess high mutagenic and tumorigenic toxicity, but are safe regarding the reproductive and irritant effect.
3.2. admetSAR predictions
The ADMET profile of the designed PLpro inhibitors was studied using the admetSAR tool. The pharmacokinetic properties such as Absorption, Distribution, Metabolism, Excretion, Toxicity were predicted and results are presented in Table 5. Blood-Brain Barrier (BBB) penetration, HIA (Human Intestinal Absorption), Caco-2 cell permeability, CYP inhibitory promiscuity, AMES toxicity, and rat acute toxicity were calculated. The results revealed that ligands possess a blood-brain barrier (BBB) and human intestinal absorption (HIA). The ligands showed permeability through human colon epithelial cancer cells (Caco-2) except ligands L4, L6, L7, L10, L16, and L18. The AMES test indicated the compounds are non-toxic except ligands L1 and L15. The rat acute toxicity LD50 of the tested ligands was found between 2.3373-2.6917 mol/Kg. Furthermore, the designed inhibitors possess substantial CYP450 inhibitory promiscuity and are non-carcinogenic.
Table 5.
ADMET predictions using admetSAR.
| Sl.No. | Compd No. | Blood-Brain Barrier (BBB) | Human Intestinal Absorption (HIA) | Caco-2 Permeability | CYP Inhibitory Promiscuity | AMES toxicity | Carcinogenicity | Rat Acute ToxicityLD50 mol/Kg |
|---|---|---|---|---|---|---|---|---|
| 1 | L1 | BBB+ | HIA+ | Caco2+ | High | Toxic | Non- Carcinogenic | 2.5083 |
| 2 | L2 | BBB+ | HIA+ | Caco2+ | High | Non-Toxic | Non- Carcinogenic | 2.3373 |
| 3 | L3 | BBB+ | HIA+ | Caco2+ | High | Non- Toxic | Non- Carcinogenic | 2.4076 |
| 4 | L4 | BBB+ | HIA+ | Caco2- | Low | Non-Toxic | Non- Carcinogenic | 2.6657 |
| 5 | L5 | BBB+ | HIA+ | Caco2+ | High | Non- Toxic | Non- Carcinogenic | 2.5854 |
| 6 | L6 | BBB+ | HIA+ | Caco2- | High | Non- Toxic | Non- Carcinogenic | 2.4940 |
| 7 | L7 | BBB+ | HIA+ | Caco2- | High | Non- Toxic | Non- Carcinogenic | 2.5120 |
| 8 | L8 | BBB+ | HIA+ | Caco2+ | High | Non- Toxic | Non- Carcinogenic | 2.6917 |
| 9 | L9 | BBB+ | HIA+ | Caco2+ | High | Non- Toxic | Non- Carcinogenic | 2.4729 |
| 10 | L10 | BBB+ | HIA+ | Caco2- | High | Non- Toxic | Non- Carcinogenic | 2.4344 |
| 11 | L11 | BBB+ | HIA+ | Caco2+ | High | Non- Toxic | Non- Carcinogenic | 2.5645 |
| 12 | L12 | BBB+ | HIA+ | Caco2+ | High | Non- Toxic | Non- Carcinogenic | 2.4801 |
| 13 | L13 | BBB+ | HIA+ | Caco2+ | High | Non- Toxic | Non- Carcinogenic | 2.5201 |
| 14 | L14 | BBB+ | HIA+ | Caco2+ | High | Non- Toxic | Non- Carcinogenic | 2.4166 |
| 15 | L15 | BBB+ | HIA+ | Caco2+ | High | Toxic | Non- Carcinogenic | 2.6190 |
| 16 | L16 | BBB+ | HIA+ | Caco2- | High | Non- Toxic | Non- Carcinogenic | 2.5580 |
| 17 | L17 | BBB+ | HIA+ | Caco2+ | High | Non- Toxic | Non- Carcinogenic | 2.5982 |
| 18 | L18 | BBB+ | HIA+ | Caco2- | High | Non- Toxic | Non- Carcinogenic | 2.6036 |
| 19 | L19 | BBB+ | HIA+ | Caco2+ | High | Non- Toxic | Non- Carcinogenic | 2.4261 |
| 20 | L20 | BBB+ | HIA+ | Caco2+ | Low | Non- Toxic | Non- Carcinogenic | 2.4973 |
| 21 | Control | BBB+ | HIA+ | Caco2+ | High | Toxic | Non- Carcinogenic | 1.6699 |
3.3. Molecular docking studies
The designed ligands were docked into the active site of SARS CoV-2 Papain-like protease. The results for the binding free energies of the designed ligands and inhibition constants are reported in Table 6.
Table 6.
Docking results of the designed ligands (L1-L20) and control.
| Sl.No. | Compd No. | Binding Energy (kcal/mol) | Inhibition Constant (nM) |
|---|---|---|---|
| 1 | L1 | -7.98 | 1420 |
| 2 | L2 | -8.42 | 667.48 |
| 3 | L3 | -8.49 | 598.76 |
| 4 | L4 | -8.62 | 483.78 |
| 5 | L5 | -8.25 | 900.61 |
| 6 | L6 | -8.17 | 1020 |
| 7 | L7 | -8.76 | 378.34 |
| 8 | L8 | -8.52 | 570.19 |
| 9 | L9 | -8.50 | 589.73 |
| 10 | L10 | -8.81 | 349.00 |
| 11 | L11 | -8.77 | 375.00 |
| 12 | L12 | -8.30 | 820.54 |
| 13 | L13 | -8.33 | 780.60 |
| 14 | L14 | -8.2 | 976.69 |
| 15 | L15 | -8.27 | 868.09 |
| 16 | L16 | -8.15 | 1060 |
| 17 | L17 | -8.62 | 478.72 |
| 18 | L18 | -8.04 | 1290 |
| 19 | L19 | -8.35 | 757.89 |
| 20 | L20 | -8.41 | 681.61 |
| 21 | Control | -8.77 | 372.57 |
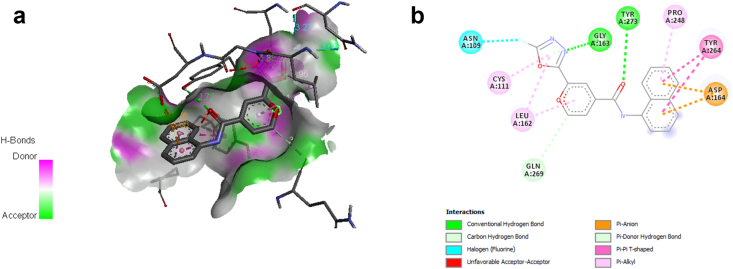
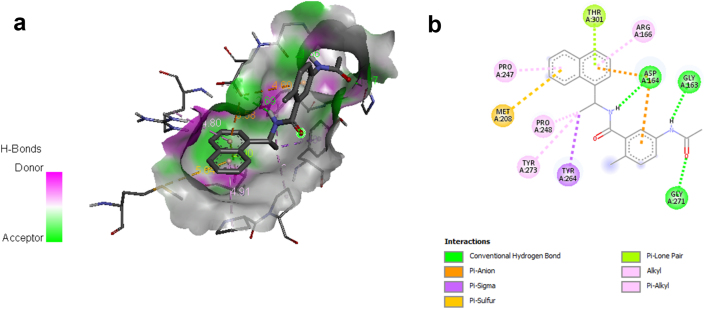
Binding free energies are comprised in the range -7.8 to -8.81 kcal/mol and are significant in comparison to the control. Ligand L10 is the most potent docked ligand in the series with the highest binding energy of -8.81 kcal/mol followed by ligand L11, which displayed a similar value of binding energy as the control i.e. -8.77 kcal/mol. The 2D and 3D interaction of ligand L10 and control 5-acetamido-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide with SARS CoV-2 papain-like protease 6WUU visualized using Discovery Studio Visualizer and are depicted in Figure 3(a)-(b), and Figure 4(a)-(b) respectively. The inhibition constant of the designed ligands was found between 349.00 nM to 1420 nM. The results indicated that the designed ligands can firmly bind to the active site of SARS CoV-2 Papain-like protease. Ligand L10 incorporating a 1,3,4-oxadiazole ring system displayed better affinity than the control 5-acetamido-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide. 1,3,4- oxadiazole exhibits broad and potent biological activities [34, 35, 36, 37, 38].
Figure 3.
(a) and (b) show the 3D and 2D interactions between LigandL10 and SARS CoV-2 papain-like protease 6WUU.
Figure 4.
(a) and (b) show the 3D and 2D interactions between control and SARS CoV-2 papain-like protease 6WUU.
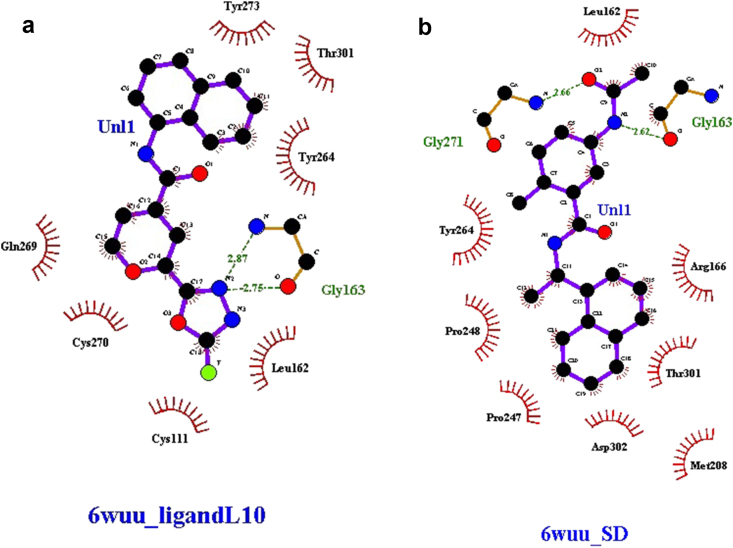
The interactions of ligand L10 and control with the functional residues of 6WUU were studied using the Ligplot+ program and are depicted in Figure 5 (a) and (b) respectively. A Docking study of SARS CoV-2 papain-like protease and ligandL10, revealed the participation of hydrogen bonding between ligandL10 and amino acid residues of the binding site of the receptor. LigandL10 stabilized through a pair of hydrogen bonds and a series of hydrophobic interactions stemming from residues lining the binding pocket of SARS CoV-2 Papain-like protease. The oxadiazole ring of ligandL10 formed two hydrogen bonds with active site amino acid residue Gly163 at a distance of 2.75 and 2.87 respectively. On the other hand, the hydrophobic interactions were observed with Tyr273, Thr301, Tyr264, Leu162, Cys112, Cys270, and Gln269 residues. However, the control 5-acetamido-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide was found to show two hydrogen bond interactions with amino acid residue Gly163 and Gly271 at a distance of 2.62 and 2.66 respectively, and hydrophobic interactions with Leu162, Tyr264, Pro248, Pro247, Asp302, Met208, Thr301, and Arg166 residues. The results suggested that the Gly, Thr, Tyr, and Leu are the most common amino acid residues in the docking site and have a significant role in the formation of the hydrogen bond. These interactions contributed to the ligand-receptor complex stability and maintaining a stable conformation in the active site of 6WUU.
Figure 5.
(a) and (b) The schematic diagrams of protein-ligand interactions of ligandL10 and control respectively with SARS CoV-2 papain-like protease 6WUU.
3.4. Molecular dynamic simulation of the docked complex
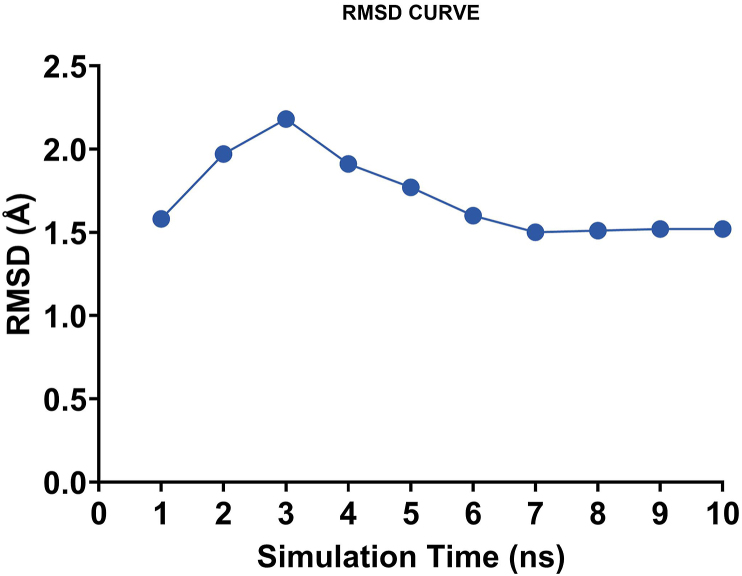
The docked receptor-ligand complex of ligandL10 was subjected to molecular dynamic (MD) simulations using NAMD software to validate the intrinsic atomic interaction and binding conformation [39]. Visualizations and data analysis were performed with VMD software. The psf protein structure file was created by an automatic psf generation plugin within the VMD program. The integrator parameter included 2fs/step for all rigid bonds. The simulation was performed for 10 ns to study the conformational stability of the complex. Figure 6 shows a water box simulations using molecular dynamics simulation of the papain-like protease of SARS-CoV-2 and ligandL10. The root mean square deviation (RMSD) of the protein backbone and ligandL10 was calculated during a 10 ns MD trajectory, as shown in Figure 7. The results revealed that the RMSD values increased in the beginning from 1-3 ns, then started converging, the system equilibrated after 6ns. The RMSD values showed that the docked receptor-ligandL10 complex was stable, the RMSD value was found to be 1.52 Å at 10 ns.
Figure 6.
The molecular dynamics simulation box of the SARS-CoV PLpro and ligandL10 complex.
Figure 7.
RMSD studies the SARS-CoV PLpro and ligandL10 complex.
4. Conclusion
In the present work, several naphthalenes-based SARS-CoV PLpro inhibitors incorporating substituted 3,4-dihydro-2H-pyran moiety were rationally designed and evaluated for their pharmacokinetic and pharmacodynamic properties employing computational tools. The ligands confirmed Lipinski's rule of five and had good bioavailability and drug-likeness. The ADMET profile of the ligands was very promising. The results of the docking study confirmed that the ligands possess a strong affinity towards SARS CoV-2 papain-like protease. Ligand10 i.e. 2-(5-fluoro-1,3,4-oxadiazol-2-yl)-N-(naphthalen-1-yl)-3,4-dihydro-2H-pyran-4-carboxamide displayed better binding efficiency than that of control 5-acetamido-2-methyl-N-[(1R)-1-naphthalen-1-yl ethyl]benzamide. The docked complex of ligandL10 was submitted to 10 ns molecular dynamics (MD) simulations to examine the stability of ligand binding modes. The molecular dynamics simulation study suggested that the receptor-ligand complex might be stable over time of the simulation. Ligand L10 emerged as a lead compound in the series. However, further in-vitro and in-vivo validation is needed to examine their potential of developing into the therapeutic agents for the treatment of COVID-19.
Declarations
Author contribution statement
S. Bhati: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The author would like to thank the Management, The Oxford Group of institutions for their support and providing the necessary facilities to conduct this study.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.World Health Organization-Coronavirus disease 2019 (COVID-19): situation report. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports [PubMed]
- 2.Orhan I.E., Senol Deniz F.S. Natural products as potential leads against coronaviruses: could they Be encouraging structural models against SARS-CoV-2. Nat. Prod. Bioprospect. 2020;10:171–186. doi: 10.1007/s13659-020-00250-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Metcalf C.J.E., Lessler J. Opportunities and challenges in modeling emerging infectious diseases. Science. 2017;357:149–152. doi: 10.1126/science.aam8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14(2):85–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao F., Tang M., Zheng X., Li C., He J., Hong Z. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses, drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92(4):418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weißbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 9.Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 2004;78:13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shin D., Mukherjee R., Grewe D., Bojkova D., Baek K., Bhattacharya A. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature. 2020 doi: 10.1038/s41586-020-2601-5. Advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Báez-Santos Y.M., St John S.E., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho J.K., Curtis-Long M.J., Lee K.H., Kim D.W., Ryu H.W., Yuk H.J., Park K.H. Geranylated flavonoids displaying SARS-CoV papain-like protease inhibition from the fruits of Paulownia tomentosa. Bioorg. Med. Chem. 2013;21(11):3051–3057. doi: 10.1016/j.bmc.2013.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ratia K., Pegan S., Takayama J., Sleeman K., Coughlin M., Baliki S., Chaudhuri R., Fu W., Prabhakar B.S., Johnson M.E. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. Unit. States Am. 2008;105(42):16119–16124. doi: 10.1073/pnas.0805240105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freitas B.T., Durie I.A., Murray J., Longo J.E., Miller H.C., Crich D., Hogan R.J., Tripp R.A., Pegan S.D. Characterization and noncovalent inhibition of the deubiquitinase and deISGylase activity of SARS-CoV-2 papain-like protease. ACS Infect. Dis. 2020;6(8):2099–2109. doi: 10.1021/acsinfecdis.0c00168. [DOI] [PubMed] [Google Scholar]
- 15.Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panda P.K., Arul M.N., Patel P., Verma S.K., Luo W., Rubahn H.G., Mishra Y.K., Suar M., Ahuja R. Structure-based drug designing and immunoinformatics approach for SARS-CoV-2. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abb8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah R., Panda P.K., Patel P., Panchal H.K. Pharmacophore based virtual screening and molecular docking studies of inherited compounds against Ebola virus receptor proteins. World J. Pharm. Sci. 2015;4:1268–1282. [Google Scholar]
- 18.Bhati S., Kaushik V., Singh J. In Silico identification of piperazine linked thiohydantoin derivatives as novel androgen antagonist in prostate cancer treatment. Int. J. Pept. Res. Ther. 2018;25:845–860. [Google Scholar]
- 19.Ray N.M., Singh R., Singh J., Bhati S., Kaushik V. Computational screening of Thiohydantoin Derivatives for antitumor activity. Res. J. Pharm. Technol. 2020;13:795–800. [Google Scholar]
- 20.Zhang D.J., Sun W.F., Zhong Z.J., Gao R.M., Yi H., Li Y.H., Peng Z.G., Li Z.R. Synthesis and broad-spectrum antiviral activity of some novel benzo-heterocyclic amine compounds. Molecules. 2014;19(1):925–939. doi: 10.3390/molecules19010925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saudi M., Zmurko J., Kaptein S., Rozenski J., Gadakh B., Chaltin P., Marchand A., Neyts J., Van Aerschot A. Synthetic strategy and antiviral evaluation of diamide containing heterocycles targeting dengue and yellow fever virus. Eur. J. Med. Chem. 2016;121:158–168. doi: 10.1016/j.ejmech.2016.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhi S., Li Y., Qiang J., Hu J., Song W., Zhao J. Synthesis and anticancer evaluation of benzo-N-heterocycles transition metal complexes against esophageal cancer cell lines. J. Inorg. Biochem. 2019;201:110816. doi: 10.1016/j.jinorgbio.2019.110816. [DOI] [PubMed] [Google Scholar]
- 23.Rashamuse T.J., Njengele Z., Coyanis E.M., Sayed Y., Mosebi S., Bode M.L. Design, synthesis and biological evaluation of novel 2-(5-aryl-1H-imidazol-1-yl) derivatives as potential inhibitors of the HIV-1 Vpu and host BST-2 protein interaction. Eur. J. Med. Chem. 2020;190:112111. doi: 10.1016/j.ejmech.2020.112111. [DOI] [PubMed] [Google Scholar]
- 24.Liu P., Sharon A., Chu C.K. Fluorinated nucleosides: synthesis and biological implication. J. Fluor. Chem. 2008;129(9):743–766. doi: 10.1016/j.jfluchem.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sander T., Freyss J., Von Korff M., Rufener C. DataWarrior: an open-source program for chemistry aware data visualization and analysis. J. Chem. Inf. Model. 2015;55(2):460–473. doi: 10.1021/ci500588j. [DOI] [PubMed] [Google Scholar]
- 26.Kochev N., Avramova S., Jeliazkova N. Ambit-SMIRKS: a software module for reaction representation, reaction search and structure transformation. J. Cheminf. 2018;10:42. doi: 10.1186/s13321-018-0295-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng F., Yu Y., Zhou Y., Shen Z., Xiao W., Liu G., Li W., Lee P.W., Tang Y. Insights into molecular basis of cytochrome p450 inhibitory promiscuity of compounds. J. Chem. Inf. Model. 2011;51(10):2482–2495. doi: 10.1021/ci200317s. [DOI] [PubMed] [Google Scholar]
- 28.Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. Automated docking using a lamarckian genetic algorithm and empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 29.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rut W., Lv Z., Zmudzinski M., Patchett S., Nayak D., Snipas S.J., El Oualid F., Huang T.T., Bekes M., Drag M., Olsen S.K. Activity profiling and structures of inhibitor-bound SARS-CoV-2-PLpro protease provides a framework for anti-COVID-19 drug design. bioRxiv. 2020 doi: 10.1126/sciadv.abd4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laskowski R.A., Swindells M.B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 32.Nelson M.T., Humphrey W., Gursoy A., Dalke A., Kalé L.V., Skeel R.D., Schulten K. NAMD: a parallel, object-oriented molecular dynamics program. Int. J. Supercomput. Appl. High Perform. Comput. 1996;10(4):251–268. [Google Scholar]
- 33.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14(1):28–33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 34.Li Z., Zhan P., Liu X. 1,3,4-oxadiazole: a privileged structure in antiviral agents. Mini Rev. Med. Chem. 2011;11(13):1130–1142. doi: 10.2174/138955711797655407. [DOI] [PubMed] [Google Scholar]
- 35.Bhati S. Microwave assisted synthesis, antimicrobial activity and in silico pharmacokinetic study of some novel 1', 3', 4'-oxadiazole derivatives. Rasayan J. Chem. 2018;11(3):1366–1375. [Google Scholar]
- 36.Caneschi W., Enes K.B., Carvalho de Mendonça C., de Souza Fernandes F., Miguel F.B., da Silva Martins J., Le Hyaric M., Pinho R.R., Duarte L.M., Leal de Oliveira M.A., Dos Santos H.F., Paz Lopes M.T., Dittz D., Silva H., Costa Couri M.R. Synthesis and anticancer evaluation of new lipophilic 1,2,4 and 1,3,4-oxadiazoles. Eur. J. Med. Chem. 2019;165:18–30. doi: 10.1016/j.ejmech.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 37.Bhati S., Kumar V., Singh S., Singh J. Synthesis, biological activities and docking studies of piperazine incorporated 1, 3, 4-oxadiazole derivatives. J. Mol. Struct. 2019;1191:197–205. [Google Scholar]
- 38.S. Bhati, V. Kumar, S. Singh, J. Singh, Synthesis, characterization, antimicrobial, anti-tubercular, antioxidant activities and docking simulations of derivatives of 2-(pyridin-3-yl)-1H benzo [ d] imidazole and 1,3,4-oxadiazole analogy, Lett. Drug Des. Discov., 17, 1047-1059.
- 39.Laxmikant K., Robert S., Milind B., Robert B., Attila G., Neal K., James P., Aritomo S., Krishnan V., Klaus S. NAMD2: greater scalability for parallel molecular dynamics. J. Comp. Phys. 1999;151:283–312. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.