Abstract
Context.—
As circulating tumor cell (CTC) assays gain clinical relevance, it is essential to address preanalytic variability and to develop standard operating procedures for sample handling in order to successfully implement genomically informed, precision health care.
Objective.—
To evaluate the effects of blood collection tube (BCT) type and time-to-assay (TTA) on the enumeration and high-content characterization of CTCs by using the high-definition single-cell assay (HD-SCA).
Design.—
Blood samples of patients with early- and advanced-stage breast cancer were collected into cell-free DNA (CfDNA), EDTA, acid-citrate-dextrose solution, and heparin BCTs. Time-to-assay was evaluated at 24 and 72 hours, representing the fastest possible and more routine domestic shipping intervals, respectively.
Results.—
We detected the highest CTC levels and the lowest levels of negative events in CfDNA BCT at 24 hours. At 72 hours in this BCT, all CTC subpopulations were decreased with the larger effect observed in high-definition CTCs and cytokeratin-positive cells smaller than white blood cells. Overall cell retention was also optimal in CfDNA BCT at 24 hours. Whole-genome copy number variation profiles were generated from single cells isolated from all BCT types and TTAs. Cells from CfDNA BCT at 24-hour TTA exhibited the least noise.
Conclusions.—
Circulating tumor cells can be identified and characterized under a variety of collection, handling, and processing conditions, but the highest quality can be achieved with optimized conditions. We quantified performance differences of the HD-SCA for specific preanalytic variables that may be used as a guide to develop best practices for implementation into patient care and/or research biorepository processes.
Assessment of preanalytic variables is an important issue in diagnostic clinical laboratory medicine. While technical assay validation for interassay and intra-assay variability is routinely done, the conditions under which the sample is collected, the manner of transportation from the patient to the laboratory bench, and the preanalytic sample processing procedures are frequently insufficiently characterized and can lead to unstable clinical interpretation.1 Thus, assessing and controlling the preanalytic handling of biospecimens is essential for their optimal and routine use. Preanalytic variables can introduce in vitro modifications, either systematically or randomly, that adversely affect interpretation of results, thus putting the associated clinical decision-making at risk. When test results deviate from the expected, the analytic integrity is often questioned rather than the preanalytic integrity of the result. Yet, data show that most errors in a clinical chemistry laboratory are due to preanalytic factors.2,3
Liquid or fluid biopsies are minimally invasive blood tests that detect rare circulating tumor cells (CTCs) and circulating cell-free tumor nucleic acids. Thus, a blood sample is a biospecimen providing a source of rare cellular and acellular analytes that are relevant to various clinical contexts, and may be measured by using various platforms.
The utility of CTC and cellular or cell-free nucleic acid measurements rests on offering increased precision and timeliness of information. Precision medicine for oncology relies on the identification of the appropriate molecular tumor target, using validated assays. Currently, bulk tumor tissue derived from a biopsy, generally from the primary tumor, is used to determine molecular targets at a single time point, most often in treatment-naïve patients. Repeated biopsies cannot routinely be performed owing to the risks they pose for patients; they are often painful, costly, and take time to obtain. In addition, a bulk tissue sample may not accurately represent the complete, relevant molecular profile, owing to the complexities of tumor heterogeneity, both within a tumor and between a primary tumor and metastases.4,5 As a complementary assay, a single-cell–based liquid biopsy may better capture the heterogeneity of the disseminated disease, and most importantly, it can be repeated longitudinally to reflect the natural and treatment-induced evolution of the disease. Naturally, the ability of a liquid biopsy to represent the disseminated disease heterogeneity and its predictive value for specific therapeutic interventions has to be established for each particular clinical case. However, all cases are dependent on delivery of a high-quality sample to the analytic platform. Thus, characterization of the preanalytic variables of the liquid biopsy is an important parameter for human-scale spatiotemporal research and for companion diagnostic development. It is a prerequisite to allow for the clinical success of the implementation of the fluid biopsy approach in the clinical management of patients with cancer.6
The goal of this study was the evaluation of preanalytic variables on the identification of rare cells in blood for high-content analysis. The approach was to use a previously developed and technically validated high-definition single-cell analysis assay (HD-SCA)7–11 as a model platform to investigate the differences between 4 commonly used blood collection tubes (BCTs) and 2 time points from blood draw to assay. The effects of other preanalytic variables were not explored. We compared the performance of the assay in terms of cell enumeration and also evaluated the results of single-cell genomics, a downstream analysis intrinsic to the HD-SCA platform.
High-definition SCA is one of the second-generation platforms for CTC detection. It is a direct-analysis approach to identify rare cells and characterize them at the cellular, protein, and molecular level, developed at the Scripps Physics Oncology Center (San Diego, California) and licensed to Epic Sciences (San Diego, California) for commercial development.7–11 The HD-SCA directly converts a liquid sample to a cytology-type preparation of all nucleated cells on a glass slide. Each tube of blood can produce approximately 12 identical samples that are stored in a biorepository, hence allowing for repeated analyses. This direct-analysis assay has the advantage of enabling both enumeration and high-content characterization of specific subclasses of rare cells at the single-cell level, based on high-definition morphologic, genomic, and protein expression data for the characterization of the disease across a wide range of scales and applications.
MATERIALS AND METHODS
Study Design
Women with breast cancer participated in the ongoing prospective Physical Sciences in Oncology Study (PSOC-0068) entitled OPTICOLL (OPTImization of blood COLLection). A total of 100 patients were enrolled between April 2013 and June 2015 at 2 clinical sites in the United States, namely, Billings Clinic (Billings, Montana) and Duke University (Durham, North Carolina). Separate recruitment and study schedules for women with early-stage disease and women with metastatic disease were established. Patients with early-stage disease had 2 study-related blood draws: before any initial treatment and 3 to 8 weeks after starting primary therapy. Patients with metastatic disease were eligible for participation at the start of a new line of therapy, either as a first line of therapy or after experiencing progression while receiving therapy for treatment of metastatic disease. These patients had a potential for up to 12 study-related blood draws at intervals of 8 to 12 weeks, coordinated with routine clinic visits. This collection scheme was designed for screening and reproducible identification of circulating rare cells, and not intended for clinical outcome correlation. An institutional review board at each site approved all procedures, and all study participants provided written informed consent.
Study participants underwent phlebotomy of up to 40 mL of blood into a syringe, which was then redistributed into 8 BCTs. Four types of BCTs were used: cell-free DNA (CfDNA; Cat No. 218962, Streck Laboratories, Omaha, Nebraska), EDTA (Cat No. 367861, Becton, Dickinson and Company, Franklin Lakes, New Jersey), acid-citrate-dextrose (Cat No. VT4816, VWR, Radnor, Pennsylvania), and heparin (Cat No. 367884, Becton, Dickinson and Company). Blood was drawn into 2 duplicate sets of 4 BCTs; each set included all tube types. Tube brand and lot number were tracked throughout the study and between clinical sites. Expiration dates on tubes were checked, as the vacuum in evacuated systems decreases with tube age and can affect the blood draw and filling of the tube. Variables such as anatomic draw location, needle gauge, and tube fill number were tracked but not controlled for, with a standardized questionnaire.
Identification and Characterization of Rare Cells
Blood samples were shipped at an average temperature of 22°C to the central laboratory via FedEx (Memphis, Tennessee) priority overnight. One set of 4 BCTs (1 CfDNA, 1 EDTA, 1 citrate, 1 heparin) was randomly selected and processed 24 hours after time of draw, that is, time-to-analysis (TTA) was 24 hours. The remaining set of 4 BCTs was stored at room temperature and processed 2 days later, producing a TTA of 72 hours.
All blood samples were processed as previously described.9 White blood cells (WBCs) were counted in whole blood by using the M series open vial sampling system analyzer (Cat No. 1400073, Clinical Diagnostics Solutions, Plantation, Florida). Each sample was treated independently regardless of tube type or TTA. The trained technologist processing the sample was randomly selected, not controlled but tracked. The WBC count was used to determine the volume of whole blood required to create slides, assuming a maximum deposition of 3 million WBCs per slide. The active area of a slide had a capacity of 750 μL. Red blood cells were lysed and nucleated cells were subsequently plated as a 750-μL monolayer on custom-made cell adhesion slides (Cat No. 0909100091, Marienfeld, Lauda, Germany). A minimum of 6 and as many as 12 replicate slides for rare cell identification and characterization were prepared and stored at −80°C until further analysis.9
For this study, we used a protocol based on the published high-definition circulating tumor cell (HD-CTC) assay that included a cocktail of pan-cytokeratin (CK) (Cat No. C2562, Sigma-Aldrich, St Louis, Missouri), CK19 (Cat No. M088801, Dako, Santa Clara, California), CD45 (Cat No. MCA87A-647, Bio-Rad, Hercules, California), and DAPI (4´,6-diamidino-2-phenylindole, dihydrochloride; Cat No. D1306, Invitrogen, Waltham, Massachusetts), plus antibody SP1 (Cat No. RM-9101-S, Thermo Scientific, Waltham, Massachusetts) for the evaluation of estrogen receptor (Janett Stoer, MS, unpublished data, 2013). Secondary antibodies were goat anti-mouse IgG1 Alexa Fluor 555 conjugate (Cat No. A-21127) and goat anti-rabbit IgG (H+L) Alexa Fluor 488 conjugate (Cat No. A-11034) from Life Technologies (Waltham, Massachusetts). The number of total retained cells was estimated by DAPI-stained nuclei counts. Cells that were CK+, CD45−, with intact DAPI staining, and generally larger and morphologically distinct from surrounding WBCs (annotated as “HD-CTCs”), as well as cells that only partially met these criteria (marginal CTC populations), were recorded.10 Marginal populations included the following: (1) “CTC-Small”: CK+, CD45−, intact DAPI-stained cells that were the same size or smaller than neighboring WBCs; (2) “CTC-LowCK”: CK+ cells with CK levels lower than those of HD-CTCs, CD45−, and intact DAPI staining; and (3) “CTC-Ap”: CK+ CD45− cells with a DAPI pattern of nuclear condensation and fragmentation and plasma membrane blebs that are common features of apoptotic cells.12,13
Positive events were defined as the total number of candidate cells (HD-CTC plus marginal populations) detected per total number of retained cells. Other components of tumor-produced material, or tumor-associated material, were not part of the scope of this work. Negative events were defined as the total number of events not associated with DAPI nuclear staining. Negative events are cellular and noncellular events that are identified by the system can be used to assess sample deterioration; they essentially represent debris of various types including cytoplasmic fragments, dust, and dye aggregates. These events were supplied to the analyst by the classification algorithm based on the intensity of the fluorescent signals. Once an event was classified as negative, it was removed from the cell classification algorithm as part of the dynamic analysis process. As both positive and negative events are expressed per total number of retained cells, we calculated the ratio of positive events to negative events per test for each BCT.
Slides generated from BCTs processed at 24 hours that had at least 1 HD-CTC in a test of 2 slides took part in analysis I (BCT comparison). Slides from the tube type with best HD-SCA performance at 24 hours and their matched slides from 72-hour TTA were then eligible for inclusion in analysis II (TTA comparison) with the goal of determining the best BCT and TTA for the HD-SCA platform. Intraday assay reproducibility at different time intervals or on multiple days was not addressed. Draws were excluded from analysis when fewer than 8 BCTs were collected, if scheduled TTAs were not met, or if samples were incorrectly processed.
All assay calculations were performed on a per test basis. One test corresponds to 2 slides per blood sample evaluated.10 The technologists, microscopes, and analysis systems were constant throughout this study.
Extraction of Single Cells and Whole-Genome Amplification
High-definition CTCs identified in the first draw of 1 patient with late-stage disease were selected for single-cell next-generation sequencing. Sixteen to 20 cells identified in slides generated from each BCT (CfDNA, EDTA, citrate, heparin) processed at 24-hour TTA and from CfDNA BCT processed at 72-hour TTA were relocated on the glass slides and reimaged at ×40 magnification for detailed cytomorphometric analysis.9 A total of 93 cells were captured by micromanipulation14 and subjected to whole-genome amplification (WGA) and sequencing library construction.15 Whole-genome amplification was carried out manually in a 96-well plate format with the WGA4 Genomeplex Single Cell Whole Genome Amplification Kit (Sigma-Aldrich), followed by purification using a QIAquick 96 PCR (polymerase chain reaction) Purification Kit (Qiagen, Hilden, Germany). Concentration of eluted DNA was measured by using a Nanodrop 8000 (Thermo Scientific).
For each well, amplification was considered successful if the resulting DNA concentration was 70 ng/μL or greater (elution volume of 50 μL), followed by further quality control to confirm the appropriate sample size distribution with the Agilent 2100 Bioanalyzer (High-Sensitivity DNA Assay and Kit, Agilent Technologies, Santa Clara, California). DNA samples were also loaded into 1% agarose gels, electrophoresed, and subsequently photographed under ultraviolet transillumination with a device camera. The GeneRuler DNA Ladder Mix (Cat No. SM0334, Thermo Scientific) was included in all gels and used to normalize band profiles between gels with the BioNumerics software (Applied Maths NV, Sint-Martens-Latem, Belgium). This software identified bands in the same position in different lanes of the gel and measured the intensity of the bands identified from digitized images.
Sequencing and Bioinformatic Analysis
Low-depth sequencing was performed by multiplexing up to 50 amplified single copy DNAs per lane of an Illumina HiSeq instrument (San Diego, California) and data were analyzed as published.15,16 The copy number variation (CNV) profiles in this work are based on 20 000 variable length genome bins, averaging a length of ~150 kilobase pairs each and counting the total number of reads mapping in each bin. The bin count data were normalized for total read count and guanine and cytosine (GC) content of each bin and converted into copy number profiles by using the Circular Binary Segmentation algorithm. The data reported here had a median count of 1.78 million uniquely mapping reads, with a range from 244 190 (minimum cutoff 200 000) to 5.33 million. The variances associated with the copy number profiles were calculated as the ratio of the normalized bin counts in each bin divided by the mean ratio for the genome segment called by DNAcopy. To normalize this noise level by the number of reads, we multiply it by the square root of the number of reads obtained for each sample. For R = vector of normalized bin counts, S = vector of segment means computed by DNAcopy, TR = total reads, the normalized variance score is variance (R/S)*sqrt(TR). Cluster analysis and frequency analysis to define genomic alterations were performed as described.14
Statistical Analysis
Graphic representation and statistical analyses were performed with GraphPad Prism (GraphPad Software Inc, La Jolla, California). Friedman test was used to detect differences in BCTs across patient samples, followed by post hoc analysis using Dunn tests to decide which BCTs were significantly different from each other, based upon the mean rank differences of the groups. Wilcoxon matched-pairs signed rank test was used to compare TTA matched samples and to assess whether mean ranks were different.
DNA amplification success rates (dichotomous yes/no categorization) for single cells analyzed from matched samples at each of the 5 conditions under comparison were compared to detect any statistically significant differences by using χ2 test. Kruskal-Wallis test and Dunn multiple comparison tests were used to compare variance scores.
Multidimensional scaling of the WGA profiles was performed to visualize relationships in the data sets under comparison, based on the relative band intensity and positions, using the BioNumerics software.
RESULTS
Study Cohort
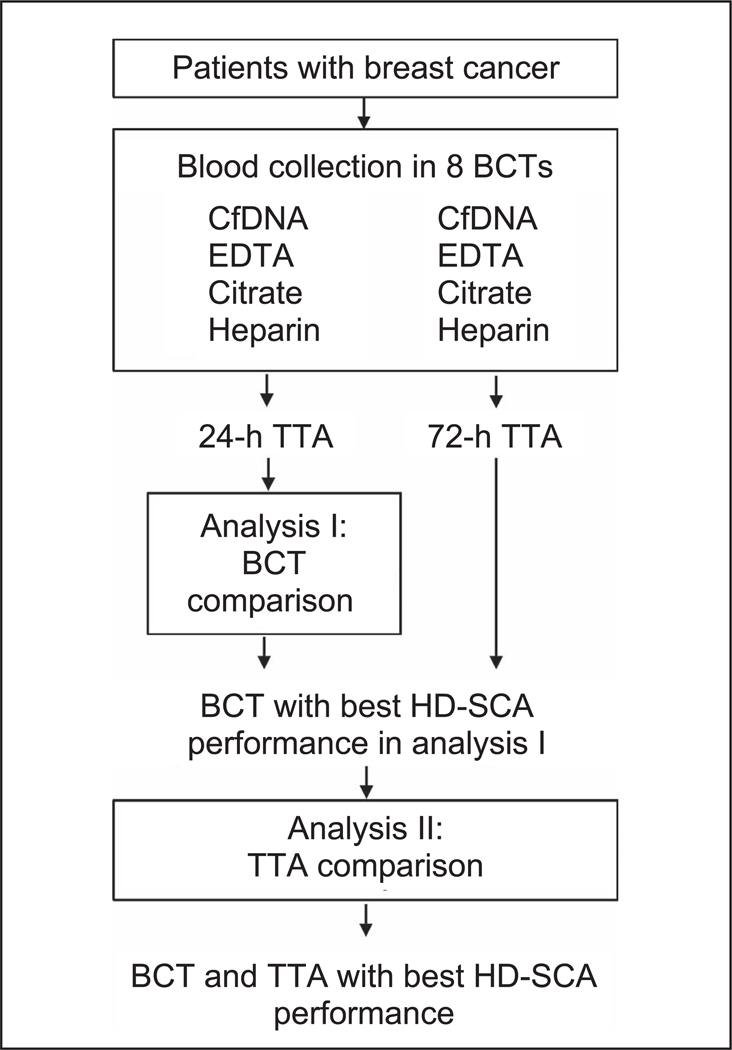
The experimental design of the study consisted of sample acquisition and 2 analyses (Figure 1). Analysis I consisted of various assay metrics for performance comparison among the 4 BCT types. Slides generated from BCTs processed at 24 hours that had 1 or more HD-CTCs in a test of 2 slides were included in analysis I (BCT comparison). A total of 50 CTC-positive draws were obtained from 119 complete sample sets of 4 BCTs. The positive draws represented 38 unique subjects: 27 early-stage and 11 late-stage cases. The best performing BCT was then the subject of analysis II comparing the 24-hour and 72-hour TTAs.
Figure 1.
Experimental design. Blood collected from patients with breast cancer was drawn into a syringe and distributed among 4 different blood collection tube (BCT) types. All BCTs were packaged and sent to University of Southern California (Los Angeles, California) on the same day of collection. The 24-hour time-to-analysis (TTA) tubes were processed and stored in the biorepository upon arrival. The 72-hour TTA tubes were stored at room temperature and processed 72 hours after time of draw. Slides generated from BCTs processed at 24 hours that had 1 or more high-definition circulating tumor cells (HD-CTCs) in a test of 2 slides were included in analysis I (BCT comparison). Slides from the tube type with best HD-CTC assay performance at 24 hours and their matched slides from 72-hour TTA were included in analysis II (TTA comparison) with the goal of determining the best BCT and TTA for the high-definition single-cell assay (HD-SCA) platform. Abbreviation: CfDNA, cell-free DNA.
Analysis I: Effect of Blood Collection Tube Type in Rare Cell Enumeration
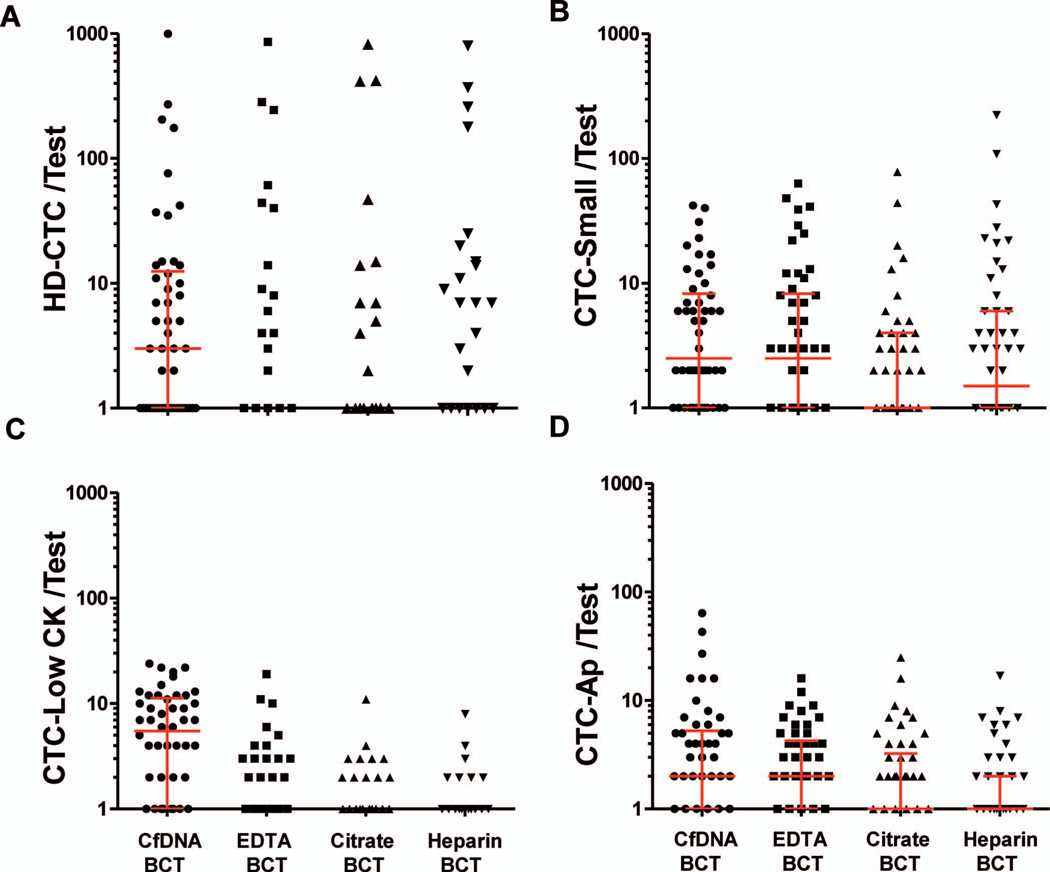
Fifty sets of 4 BCTs were subjected to quantitative comparisons (Figure 2, A through D). The highest numbers of CTC candidate cells (HD-CTCs and marginal populations) were detected in the CfDNA BCT. No differences were found between EDTA, citrate, and heparin BCTs. Citrate and heparin BCTs had comparable levels of total negative events detected per test, whereas the CfDNA BCT had the lowest level. As a result, CfDNA BCT exhibited the highest ratio of positive to negative events detected. Cell retention (million cells per test) was not different between CfDNA (median, 4.33; interquartile range [IQR], 0.58), EDTA (median, 4.42; IQR, 0.77), and citrate (median, 4.00; IQR, 0.85) BCTs, whereas heparin BCT (median, 3.40; IQR, 1.10) resulted in 21.4% fewer cells than CfDNA BCT.
Figure 2.
Analysis I: blood collection tube (BCT) comparison. A, Circulating tumor cell (CTC) candidate cells enumerated per test included high-definition CTC and marginal CTC populations as defined in Materials and Methods: CTC-Small, CTC-LowCK, and CTC-Ap. P < .001: Cell-free DNA (CfDNA) BCT versus EDTA, citrate, or heparin BCTs. B, Total negative events detected per test: total number of negative events per total number of retained cells in 1 test of 2 slides per blood sample. P<.001: CfDNA BCT versus EDTA, citrate, or heparin BCTs; EDTA BCT versus citrate BCT; EDTA BCT versus heparin BCT. C, Number of total retained cells as estimated by the DAPI nuclear stain. P < .001: heparin BCT versus CfDNA, EDTA, or citrate BCTs. D, Ratio of positive to negative events per test. Positive events were defined as the total number of candidate cells detected per total number of retained cells. P < .001: CfDNA BCT versus EDTA, citrate, or heparin BCTs. Friedman test was used to detect differences in BCTs across patient samples, followed by post hoc analysis using Dunn tests. Median and interquartile range are indicated. Abbreviation: DAPI, 4´,6-diamidino-2-phenylindole, dihydrochloride.
Figure 3, A through D, shows the distribution across different subtypes of candidate cell populations detected in the 4 BCT types. Higher numbers of HD-CTCs and CTC-LowCK were detected in CfDNA BCT than in the other 3 BCTs. No differences between EDTA, citrate, and heparin BCTs were observed. The CTC-LowCK subpopulation was mostly detected in CfDNA BCT; its level was significantly reduced in the other 3 BCTs. CTC-Small and CTC-Ap subpopulations exhibited less variation between BCTs. Citrate BCT had the lowest levels of CTC-Small, whereas CfDNA, EDTA, and heparin BCTs had comparable levels. Heparin BCT had the lowest levels of detected CTC-Ap, whereas CfDNA, EDTA, and citrate BCTs had comparable levels.
Figure 3.
Cell enumeration in analysis I. Circulating tumor cell (CTC) candidate cells identified included high-definition circulating tumor cell (HD-CTC) (A) and marginal CTC populations as defined in Materials and Methods: CTC-Small (B), CTC-LowCK (C), and CTC-Ap (D). One test consisted of 2 slides per blood sample. Friedman test was used to detect differences in blood collection tubes (BCTs) across patient samples, followed by post hoc analysis using Dunn tests. P values: <.001 (A), .007 (B), <.001 (C), .001 (D). Median and interquartile range are indicated. Abbreviation: CfDNA, cell-free DNA.
The results of analysis I indicated that the CfDNA BCT showed the best performance at 24 hours. Therefore, we thawed and evaluated slides generated from 50 drawmatched CfDNA BCTs that were processed at 72-hour TTA and included them in analysis II.
Analysis II: Effect of Time-To-Analysis in CTC Enumeration
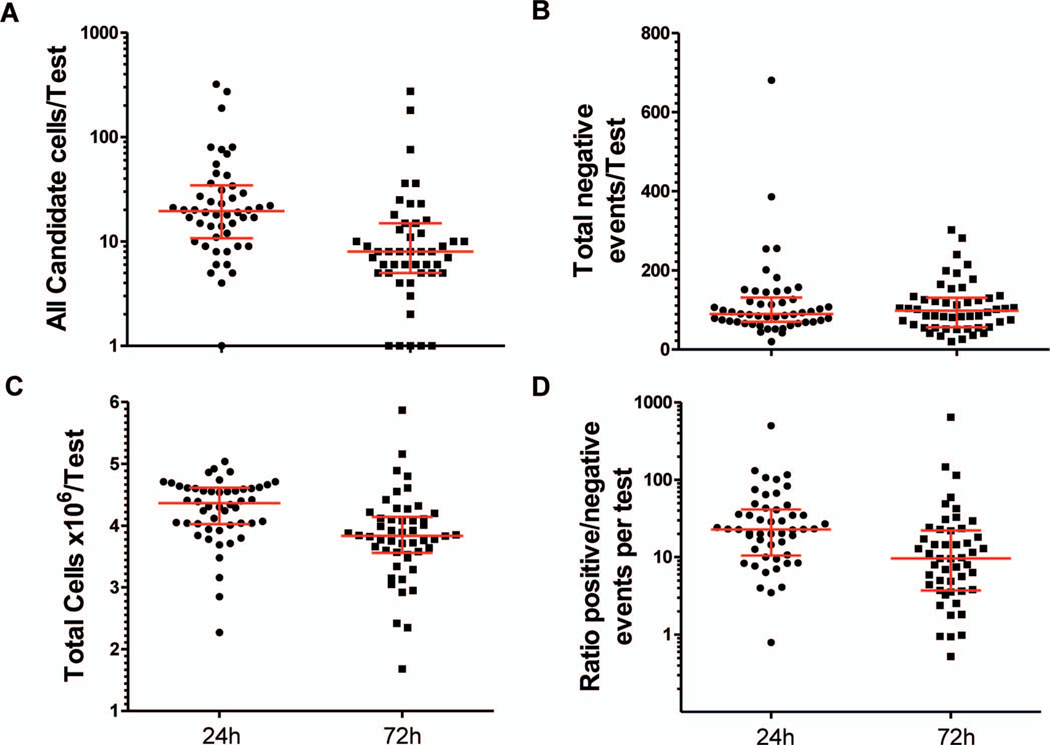
Blood samples collected in CfDNA BCT exhibited differences in various assay metrics at the 2 TTAs evaluated (Figure 4, A through D). The total number of candidate cells detected was reduced at 72 hours. No differences between TTAs were found in the total number of negative events detected but cell retention was decreased at 72 hours. The 24-hour TTA exhibited a higher ratio of positive to negative events.
Figure 4.
Analysis II: time-to-assay comparison. A, Circulating tumor cell (CTC) candidate cells enumerated per test included high-definition CTC and marginal CTC populations as defined in Materials and Methods: CTC-Small, CTC-LowCK, and CTC-Ap. B, Total negative events detected per test: total number of negative events per total number of retained cells in 1 test of 2 slides per blood sample. C, Number of total retained cells as estimated by the DAPI nuclear stain. D, Ratio of positive to negative events per test. Positive events were defined as the total number of candidate cells detected per total number of retained cells. Wilcoxon matched-pairs signed rank test was used to compare matched samples and to assess whether mean ranks were different.P values: <.001 (A), .61 (B), <.001 (C), .001 (D). Median and interquartile range are indicated. Abbreviation: DAPI, 4´,6-diamidino-2-phenylindole, dihydrochloride.
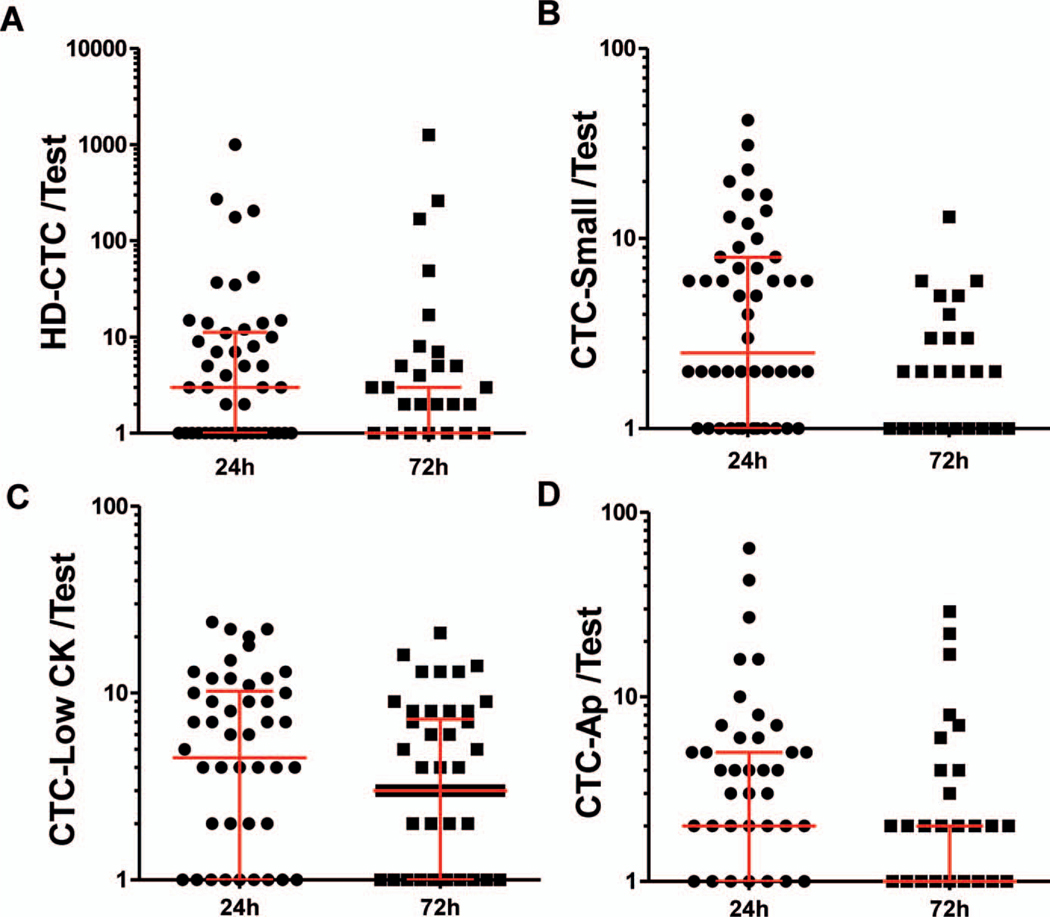
Figure 5, A through D, indicates the distribution across different subtypes of candidate cells that were detected at 24-hour and 72-hour TTAs in matched CfDNA BCTs. At 72 hours, reduced numbers of all CTC subpopulations were detected with the larger effect observed in HD-CTC and CTC-Small subpopulations.
Figure 5.
Cell enumeration in analysis II. Circulating tumor cell (CTC) candidate cells identified included high-definition circulating tumor cell (HD-CTC) (A) and marginal CTC populations as defined in Materials and Methods: CTC-Small (B), CTC-LowCK (C), and CTC-Ap (D). One test consisted of 2 slides per blood sample. Wilcoxon matched-pairs signed rank test was used to compare matched samples and to assess whether mean ranks were different. P values: <.001 (A), <.001 (B), .04 (C), .009 (D). Median and interquartile range are indicated.
Single-Cell Next-Generation Sequencing and Copy Number Profiling
The HD-SCA platform has the ability to isolate single and grouped cells for further analysis including single-cell genomics. Thus, we can then integrate phenotypic and genotypic data to understand the alterations that occur in the tumor tissue as it evolves in the spatiotemporal dimensions.14 We isolated HD-CTCs from 1 draw of 1 patient with late-stage disease. A total of 93 HD-CTCs were selected from 5 evaluated slides, 1 slide from each of 5 conditions under comparison: 19 cells from CfDNA BCT, 24-hour TTA; 19 cells from EDTA BCT, 24-hour TTA; 17 cells from citrate BCT, 24-hour TTA; 20 cells from heparin BCT, 24-hour TTA; and 18 cells from CfDNA BCT, 72-hour TTA. All cells were subjected to WGA. The fraction of cells that was successfully amplified ranged from 89% to 100% and did not differ across the 5 conditions under comparison (P = .68, x2 test). Eighty-seven single cells successfully underwent amplification: 17 cells from CfDNA BCT, 24-hour TTA; 17 cells from EDTA BCT, 24-hour TTA; 17 cells from citrate BCT, 24-hour TTA; 19 cells from heparin BCT, 24-hour TTA; and 17 cells from CfDNA BCT, 72-hour TTA. From each successful amplification, a CNV profile was generated—with the exception of 1 HD-CTC from citrate BCT, 24-hour TTA—resulting in 86 CNV profiles available.
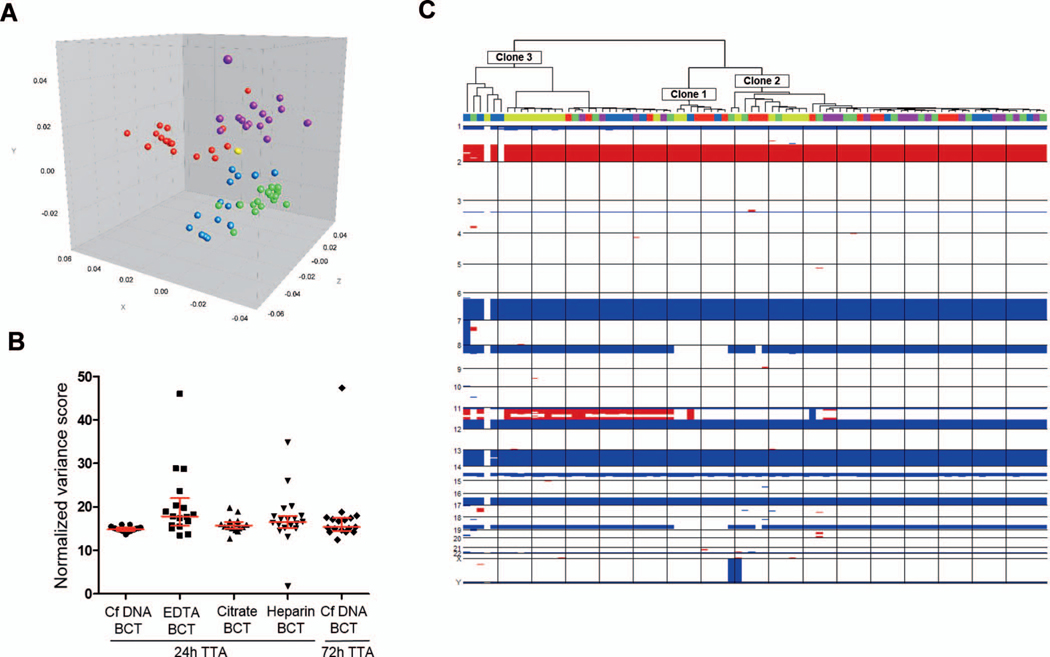
Processing of digitized images generated profiles for all lanes of the 5 different WGA gels. Multidimensional scaling analysis was performed on the composite data set derived from the combination of gel strips from successful WGA. This analysis indicated a specific clustering pattern, depending on the BCT type and TTA. As the distance between 2 data points in the 3-dimensional multidimensional scaling plot is a visual representation of the difference in the WGA composition of the different isolated cells, the multidimensional scaling analysis indicated that WGA profiles from the cells isolated from the CfDNA BCT processed at 24-hour TTA revealed the highest level of similarity, indicated by less distance, when compared to the profiles from the other 4 conditions under comparison (Figure 6, A).
Figure 6.
Single-cell genomics. A, Three-dimensional multidimensional scaling analysis conducted on the whole-genome amplification (WGA) profiles composite data set: yellow= cell-free DNA (CfDNA) blood collection tube (BCT), 24 hours (17); blue= EDTA BCT, 24 hours (17); purple=citrate BCT, 24 hours (16); green=heparin BCT, 24 hours (19); red= CfDNA BCT, 72 hours (17). Dimensions x, y, and z are arbitrary scales that represent the first, second, and third components defined by the method to compare distances between entries of the similarity matrix. Less distance indicates higher level of similarity. B, Variance of copy number variation (CNV) profiles (n= 86). The noise associated with the copy number profiles was calculated for each cell picked and amplified from each of the 5 conditions under comparison as the variance of the ratio of the normalized bin counts in each bin divided by the mean ratio for the genome segment called by DNAcopy as defined in Materials and Methods. Kruskal-Wallis test and Dunn multiple comparison tests were used to compare variance scores (P =.001). Median and interquartile range are indicated. C, Three different clonal lineages, represented as clones 1, 2, and 3, were identified by comparison of single-cell CNV profiles in an unsupervised hierarchical clustering (n= 86). The BCT and time-to-analysis (TTA) from which each cell was isolated is indicated with colors as in (A).
The noise associated with the copy number profiles was normalized to account for differences in read depth, hence allowing sample-to-sample comparison. Samples with a high noise would indicate a nonuniform distribution of reads stemming from uneven amplification during WGA. As this field is currently in development there is no historical gold standard data for a calculation of this type. It is therefore impossible to predict a priori the exact threshold for what would constitute a significant difference in these parameters. A larger number of cells would probably be required for this analysis. However, we here show that the CNV profiles from the cells isolated from the CfDNA BCT processed at 24-hour TTA exhibited less noise with the lowest dispersion (interquartile) range when compared with the other 4 sets of samples (Figure 6, B).
Complex patterns of genomic rearrangements were identified from 86 CNV profiles from all 5 conditions that were characterized by the presence of 3 different clonal lineages: clones 1, 2, and 3 (Figure 6, C). Clones 2 and 3 exhibited deletions on chromosomes 8 and 18, which were absent in clone 1. Clone 1 exhibited an amplification on chromosome 11, which was absent in clones 1 and 2.
Overall, these results suggest that these 4 BCT types when processed at 24 hours, and CfDNA BCT when processed at 72 hours, enable single cell genomics. However, CfDNA BCT at 24-hour TTA allows for the most robust and reproducible single-cell genomic results.
DISCUSSION
The goal of the study was the evaluation of 2 preanalytic variables on the enumeration and downstream characterization of CTCs as an essential step toward clinical implementation of fluid or liquid biopsies that seek to recover and characterize whole cells. The approach was to use the previously developed and validated HD-SCA as a model platform to investigate the differences between 4 commonly used BCTs and 2 time points from blood draw to assay.
Anticoagulants are commonly added to collection tubes either to maintain blood in the fluid state for hematologic testing or to obtain suitable plasma for coagulation and clinical chemistry analyses. Depending on the expected downstream analyses, multiple collection tube types involving different additives may be needed according to the anticipated immediate and future analyses.17 The general recommendation is to follow a special order of draw dependent on the additive in the tube to avoid cross-contamination among the tubes.18 Historically, EDTA has been recommended as the anticoagulant of choice for hematologic testing because it allows the best preservation of cellular components and morphology of blood cells.19 Citrate has been the standard anticoagulant used by hematologists and blood transfusion services for stored blood products and also as an extracorporeal anticoagulant for centrifugal platelet and leucopheresis techniques and plasma exchange.20 Plasma isolated from heparin BCTs is suitable for a wide range of determinations in chemistry. CfDNA BCT is preferred for stabilization of cell-free plasma DNA. It contains EDTA as anticoagulant, a formaldehyde-free preservative that may be selected from the group consisting of diazolidinyl urea and imidazolidinyl urea, and a quenching agent for substantially abating any free aldehyde from reacting with DNA within a sample. This cell-stabilizing reagent mix has an advantage over aldehyde-based stabilizing agents because it prevents the release of genomic DNA, thus allowing isolation of high-quality cell-free DNA and it has no negative effect on DNA amplification by polymerase chain reaction.21,22 CfDNA BCT is also known to stabilize cell-free RNA in blood during storage and transport at room temperature for 3 days.23–25 Previous studies also found that CfDNA BCT preserves and stabilizes spiked cancer cell lines in donor blood samples for at least 4 days at room temperature.26
Results from analysis I indicated that the CfDNA BCT performs significantly better than the other 3 (EDTA, citrate, heparin) with the highest detection levels of rare cells and the lowest of total negative events. As the slides used by the HD-SCA are customized for retention of intact cells, we hypothesize that cellular negative events resulting from the destabilization of the cells within the tube over time may be washed away before plating, and are thus not present on the slide. However, the excessive cellular negative events present in EDTA, citrate, and heparin BCTs that lack the protective agent mix in CfDNA BCT may not have been completely washed away. Noncellular negative events will be detected regardless of BCT or TTA, as they include dust particles or dye aggregates.
Blood samples collected in CfDNA BCT exhibited differences in various assay metrics at the 2 TTAs evaluated (analysis II). Storage (sample age-related) artifacts can result in many false changes in hematology results. When blood was collected in the same BCT, namely, CfDNA BCT, but processed at 72 hours instead of at 24 hours, we detected fewer candidate cells, with the larger effect observed in HD-CTC and CTC-Small, and lower total number of cells retained on the slides. We hypothesize this is the result of the destabilization of the cells within the tube over time, possibly due to a reduced effect of the protective agent mix. No differences between TTAs were found in the total number of negative events detected because the cellular negative events are equally washed away before plating at both TTAs.
It is known that aldehydes induce oxidative stress and apoptosis. The protective agent mix in the CfDNA BCT claims to protect the CTCs by inhibiting all cellular metabolic activity in CTCs in blood. As a result of metabolic inhibition, all apoptotic and necrotic pathways are inhibited and CTCs are protected from cell degradation. Thus, we did not anticipate increased levels of CTC-Ap at 72 hours. However, it should be noted that CTC-Ap, as cataloged by the HD-SCA, comprises CK+ CD45− cells with cytoplasmic blebbing and/or nuclear fragmentation. This categorization is based on coarse morphologic features rather than a biological characterization. Further investigation is needed to evaluate the expression of specific apoptosis markers in this CTC-Ap population.
Low CTC incidence may limit the clinical utility of CTC assays. Therefore, increasing assay sensitivity without increasing false-positive results in healthy controls is important. Here, we have demonstrated that HD-SCA performance is optimized for high detection levels of rare tumor cells when blood is collected into CfDNA BCT and processed within 24 hours of collection. This BCT type and time from blood collection to assay also enabled robust single-cell genomic downstream applications. Although other CTC platforms may have specific features that require different collection methodologies, we argue that the basic stability of the cells in the blood sample is a universal need, and that absent other specific requirements, this work, using the HD-SCA platform as a model and surrogate for other general CTC platforms, suggests an optimal blood collection tube, namely, CfDNA, and an optimal time to assay, namely, 24 hours. Although we have demonstrated that both BCT type and TTA significantly impact CTC detection, we cannot comment on whether and how these variables may impact clinical utility. Ongoing studies aim to define the best analytic approach to maximize clinical correlation. However, the results presented here provide an important framework for understanding how preanalytic variables can impact the fluid biopsy test results. The HD-SCA performance differences we have quantified may be used as a best practices guide for site-specific implementation into patient care and/or research biorepository processes.
Acknowledgments
We thank our patients who consented to this study; we thank you for advancing our understanding of the preanalytic variables of the fluid biopsy. We also thank the clinical research staff who contributed to the study: Tauna Jeffery, RN, and Mary Branger at Billings Clinic; Kristen Brannock, BA, MPH, Wendy Lorizio, MD, MPH, Michelle Parks, BS, BSChE, Kenya Stringfellow, AAS, and the Breast Clinical Trials team at Duke University Medical Center. We are grateful to past and current technical staff at the Kuhn Laboratory for processing of the blood samples: Julia Li, MS, Natalie Felch, BS, Luisa Fernandez Altuna, PhD, Nadia Ebrahim, BS, Janett Stoehr, MS, Newsha Sahaf, BS, Samantha Reiss, MS, Lisa Welter, MS, and Xiomara Villasenor, BS. In addition, we thank Janett Stoer, MS, for the development of the estrogen receptor staining protocol; Lisa Welter, MS, for imaging supporting work; and the staff at the Cold Spring Harbor Laboratory (CSHL) Cancer Center Genomics Core Facility for the sequencing work performed. The authors would like to acknowledge the support of the National Cancer Institute (NCI) and Leidos Biomedical Research, Inc (contract No. HHSN261200800001E, contract agreements 12XS527 and 15X003) and of the NCI’s USC Norris Comprehensive Cancer Center (CORE) Support (grant No. 5P30CA014089-40).
Footnotes
The high-definition single-cell assay (HD-SCA) technology described here is licensed to Epic Sciences, San Diego, California. Dr Kolatkar, Ms McCormick (Luttgen), Dr Bethel, Dr Hicks, and Dr Kuhn have stock ownership in Epic Sciences. Dr Bethel is also a consultant and laboratory director for Epic Sciences, Inc. Dr Waitman is a speaker for Novartis, East Hanover, New Jersey, and Genentech, South San Francisco, California. The other authors have no relevant financial interest in the products or companies described in this article.
The content of this article is solely the responsibility of the authors and does not necessarily reflect the views or polices of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Contributor Information
Mariam Rodríguez-Lee, Department of Biological Sciences, Bridge Institute, University of Southern California, Los Angeles.
Anand Kolatkar, Department of Biological Sciences, Bridge Institute, University of Southern California, Los Angeles.
Madelyn McCormick (Luttgen), Department of Cell and Molecular Biology, The Scripps Research Institute, La Jolla, California.
Angel D. Dago, Department of Cell and Molecular Biology, The Scripps Research Institute, La Jolla, California.
Jude Kendall, Division of Cancer Genomics, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York.
Nils Anders Carlsson, Department of Biological Sciences, Bridge Institute, University of Southern California, Los Angeles.
Kelly Bethel, Department of Pathology, Scripps Clinic, La Jolla, California.
Emily J. Greenspan, Center for Strategic Scientific Initiatives (CSSI), Office of the Director, National Cancer Institute, National Institutes of Health, Bethesda, Maryland.
Shelley E. Hwang, Department of Surgery, Duke University Medical Center, Durham, North Carolina.
Kathryn R. Waitman, Medical Oncology, Billings Clinic, Billings, Montana.
Jorge J. Nieva, Department of Medicine, Keck School of Medicine, University of Southern California, Los Angeles.
James Hicks, Department of Biological Sciences, Bridge Institute, University of Southern California, Los Angeles.
Peter Kuhn, Department of Biological Sciences, Bridge Institute, University of Southern California, Los Angeles.
References
- 1.Ellervik C, Vaught J. Preanalytical variables affecting the integrity of human biospecimens in biobanking. Clin Chem. 2015;61(7):914–934. [DOI] [PubMed] [Google Scholar]
- 2.Carraro P, Zago T, Plebani M. Exploring the initial steps of the testing process: frequency and nature of pre-preanalytic errors. Clin Chem. 2012;58(3): 638–642. [DOI] [PubMed] [Google Scholar]
- 3.Lippi G, Guidi GC, Mattiuzzi C, Plebani M. Preanalytical variability: the dark side of the moon in laboratory testing. Clin Chem Lab Med. 2006;44(4):358–365. [DOI] [PubMed] [Google Scholar]
- 4.Allison KH, Sledge GW. Heterogeneity and cancer. Oncology. 2014;28(9): 772–778. [PubMed] [Google Scholar]
- 5.Kleppe M, Levine RL. Tumor heterogeneity confounds and illuminates: assessing the implications. Nat Med. 2014;20(4):342–344. [DOI] [PubMed] [Google Scholar]
- 6.Parkinson DR, Dracopoli N, Petty BG, et al. Considerations in the development of circulating tumor cell technology for clinical use. J Transl Med. 2012;10:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho EH, Wendel M, Luttgen M, et al. Characterization of circulating tumor cell aggregates identified in patients with epithelial tumors. Phys Biol. 2012;9(1): 016001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lazar DC, Cho EH, Luttgen MS, et al. Cytometric comparisons between circulating tumor cells from prostate cancer patients and the prostate-tumor-derived LNCaP cell line. Phys Biol. 2012;9(1):016002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marrinucci D, Bethel K, Kolatkar A, et al. Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Phys Biol. 2012;9(1):016003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nieva J, Wendel M, Luttgen MS, et al. High-definition imaging of circulating tumor cells and associated cellular events in non-small cell lung cancer patients: a longitudinal analysis. Phys Biol. 2012;9(1):016004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wendel M, Bazhenova L, Boshuizen R, et al. Fluid biopsy for circulating tumor cell identification in patients with early-and late-stage non-small cell lung cancer: a glimpse into lung cancer biology. Phys Biol. 2012;9(1):016005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tone S, Sugimoto K, Tanda K, et al. Three distinct stages of apoptotic nuclear condensation revealed by time-lapse imaging, biochemical and electron microscopy analysis of cell-free apoptosis. Exp Cell Res. 2007;313(16):3635–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wickman G, Julian L, Olson MF. How apoptotic cells aid in the removal of their own cold dead bodies. Cell Death Differ. 2012;19(5):735–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dago AE, Stepansky A, Carlsson A, et al. Rapid phenotypic and genomic change in response to therapeutic pressure in prostate cancer inferred by high content analysis of single circulating tumor cells. PLoS One. 2014;9(8):e101777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baslan T, Kendall J, Rodgers L, et al. Genome-wide copy number analysis of single cells. Nat Protoc. 2012;7(6):1024–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer LR, Zweig AS, Hinrichs AS, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41(Database issue):D64–D69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shabihkhani M, Lucey GM, Wei B, et al. The procurement, storage, and quality assurance of frozen blood and tissue biospecimens in pathology, biorepository, and biobank settings. Clin Biochem. 2014;47(4–5):258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowen RA, Remaley AT. Interferences from blood collection tube components on clinical chemistry assays. Biochem Med. 2014;24(1):31–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banfi G, Salvagno GL, Lippi G. The role of ethylenediamine tetraacetic acid (EDTA) as in vitro anticoagulant for diagnostic purposes. Clin Chem Lab Med. 2007;45(5):565–576. [DOI] [PubMed] [Google Scholar]
- 20.Davenport A, Tolwani A. Citrate anticoagulation for continuous renal replacement therapy (CRRT) in patients with acute kidney injury admitted to the intensive care unit. NDT Plus. 2009;2(6):439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das K, Dumais J, Basiaga S, Krzyzanowski GD. Carbon-13 nuclear magnetic resonance analysis of formaldehyde free preservatives. Acta Histochem. 2013;115(5):481–486. [DOI] [PubMed] [Google Scholar]
- 22.Das K, Fernando MR, Basiaga S, Wigginton SM, Williams T. Effects of a novel cell stabilizing reagent on DNA amplification by PCR as compared to traditional stabilizing reagents. Acta Histochem. 2014;116(1):55–60. [DOI] [PubMed] [Google Scholar]
- 23.Das K, Norton SE, Alt JR, Krzyzanowski GD, Williams TL, Fernando MR. Stabilization of cellular RNA in blood during storage at room temperature: a comparison of cell-free RNA BCT((R)) with K3EDTA tubes. Mol Diagn Ther. 2014; 18(6):647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernando MR, Norton SE, Luna KK, Lechner JM, Qin J. Stabilization of cell-free RNA in blood samples using a new collection device. Clin Biochem. 2012; 45(16–17):1497–1502. [DOI] [PubMed] [Google Scholar]
- 25.Qin J, Williams TL, Fernando MR. A novel blood collection device stabilizes cell-free RNA in blood during sample shipping and storage. BMC Res Notes. 2013;6:380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin J, Alt JR, Hunsley BA, Williams TL, Fernando MR. Stabilization of circulating tumor cells in blood using a collection device with a preservative reagent. Cancer Cell Int. 2014;14(1):23. [DOI] [PMC free article] [PubMed] [Google Scholar]