Abstract
Circulating tumor cells (CTCs) are rare cells that can be found in the peripheral blood of cancer patients. They have been demonstrated to be useful prognostic markers in many cancer types. Within the last decade various methods have been developed to detect rare cells within a liquid biopsy from a cancer patient. These methods have revealed the phenotypic diversity of CTCs and how they can represent the complement of cells that are found in a tumor. Single-cell proteogenomics has emerged as an all-encompassing next-generation technological approach for CTC research. This allows for the deconstruction of cellular heterogeneity, dynamics of metastatic initiation and progression, and response or resistance to therapeutics in the clinical settings. We take advantage of this opportunity to investigate CTC heterogeneity and understand their full potential in precision medicine.The high-definition single-cell analysis (HD-SCA) workflow combines detection of the entire population of CTCs and rare cancer related cells with single-cell genomic analysis and may therefore provide insight into their subpopulations based on molecular as well as morphological data. In this chapter we describe in detail the protocols from isolation of a candidate cell from a microscopy slide, through whole-genome amplification and library preparation, to CNV analysis of identified cells from the HD-SCA workflow. This process may also be applicable to any platform starting with a standard microscopy slide or isolated cell of interest.
Keywords: Circulating tumor cells, CTC, Copy number variation, CNV, Liquid biopsy, Precision medicine, Single-cell analysis
1. Introduction
Throughout the initiation and progression of carcinomas, cells are actively or passively released from a tumor into the blood stream. Often tumor associated cells detach during the process of vascularization, or tumor derived cells intravasate through destabilized cell junctions into a blood vessel [1–4]. These possible messengers of metastasis then circulate in the blood of patients and have been correlated with disease progression and patient outcome in numerous studies [5–7]. The variety of tumor and tumor associated cells and extracellular vesicles (EVs) that pass over into the blood stream include circulating tumor cells (CTCs), endothelial cells which line the tumor vasculature, stromal cells, platelets, exosomes, oncosomes (oncogene-containing EVs) and more [8–10].
The detection of CTCs can be achieved by various methods [11, 12] utilizing mostly specific biological or physical properties of known cell types, to enrich a subset of CTCs [13–15]. Such selection process can perform exceedingly well if the selection parameters are known with high accuracy and can be chosen with high precision. This process can be useful in a known diagnostic framework but can be challenging in a heterogeneous disease setting represented by carcinomas. A possible solution is the direct analysis of the entire population of peripheral blood mononuclear cells (PBMC) from a patient to allow the unbiased identification of all CTC types. Deeper molecular analysis of the identified candidate cells can follow without losing valuable information especially of rare subpopulations that might be responsible for relapse or disease progression.
One technical solution to the challenge of heterogeneity is the High-Definition Single-Cell Analysis (HD-SCA) workflow that has been developed to take the entire population of nucleated circulating cells in a peripheral blood sample into account [16, 17]. It offers an enrichment-free, high-throughput approach for rare cell detection and multimodal single cell analysis following the principle that “no cell is left behind” to be able to identify the one needle that matters in the haystack. For deeper insight about the impact of the detected rare cell, it is important to consider the spatiotemporal evolution of cancer; the migration of a CTC through the hematogenous and lymphatic system of the body and changes it might provoke within a new metastatic site [18]. Collecting various available spatial and temporal data provides the opportunity to detect patterns and may predict patient outcome.
Recently the HD-SCA workflow has been extended to a variety of tissue preparations such as “touch-preps” [19] where resected tumor tissue or biopsy specimens are lightly touched to the surface of a slide, leaving an imprint of the tissue comprising a layer of intact tumor and associated cells that maintains their positional information. Being able to not only investigate peripheral blood and bone marrow aspirate, but also solid tumor tissue cells from the primary tumor and metastases, allows the HD-SCA workflow to correlate high-resolution imaging and downstream molecular data of single cells of different spatiotemporal samples of the same patient to fully monitor and analyze disease progression.
This chapter aims to describe the methods for extracting individual cells of interest (COIs) from microscopy slides and to prepare them for genomic analysis, resulting in genome-wide copy number variation (CNV) profiles. This workflow is equally applicable not only to the PBMC fraction of a peripheral blood draw, but also to any slide-based cell preparation including “touch-preps.” The detailed methods for the handling of blood samples, preparation for immunofluorescent staining and characterization of COIs from blood samples have been previously published [16], but will be briefly described here for the better understanding of the analytical steps of the workflow.
2. Materials
All materials are listed in order of time of use for the corresponding part of the protocol listed below.
2.1. High-Definition Single-Cell Analysis (HD-SCA) Workflow
Blood collection tubes (BCTs) containing a cell fixation and DNA stabilization reagent (Streck Cell-Free DNA BCT®).
Customized adhesive glass slides (Marienfeld GmbH & Co. KG).
Fluorescently stained antibodies (Alexa Fluor©, ThermoFisher Scientific).
2.2. Single-Cell Isolation
2.2.1. Equipment
-
1
Inverted microscope, e.g., Olympus IX81, Nikon TE2000.
-
2
Micromanipulator, e.g., TransferMan® 4r (Eppendorf).
2.2.2. Reagents and Consumables
-
3
DNA AWAY® surface decontaminant (Molecular BioProducts).
-
4
Cell isolation coverslips: 25 × 75 mm (Electron Microscopy Sciences).
-
5
Glass capillaries (piezo drill tips): diameter of 15 μm, angle of 25°, length of 6000 μm, and a jagged front surface (Eppendorf).
-
6
Oil for hydraulic system: mineral oil.
-
7
Dry ice and wet ice to store cells and buffers (in thermal boxes).
-
8
Picking buffer PBS-T: 5 ml 1× PBS (pH = 7.4 ± 0.02) + 5 μl of Tween 20 (0.1% final concentration of Tween 20).
-
9
Cell deposition buffer (CDB): 10 mM Tris–HCl + EDTA pH 8.0 (TE): 10 mM Tris–HCl, 1 mM disodium EDTA, pH 8.0.
-
10
0.5 ml LoBind DNA tubes: sterile, with flat rim (Eppendorf).
-
11
70% ethanol.
-
12
Compressed air.
2.2.3. Software and Computing
-
13
Imaging Software: (ImagePro, MediaCybernetics Inc.).
-
14
Semiautomated custom ImagePro macros for relocation and imaging of cells.
-
15
Transformation matrix: Converts slide scanner coordinates for each cell of interest to reimaging/cell picking microscope coordinates.
2.3. Whole-Genome Amplification
2.3.1. Equipment
-
1
Thermal cycler (e.g., Mastercycler™ pro PCR System, Eppendorf).
-
2
PCR cooler (e.g., iceless cold storage system, Eppendorf®).
-
3
Equipment and chemicals for gel electrophoresis system: e.g., Quick-Load® 100 bp DNA Ladder (New England BioLabs), 10× SYBR® Safe DNA Gel Stain (Life Technologies), Gel Pilot Loading Dye 5× (Qiagen), GelPilot® Agarose (Qiagen).
-
4
DNA quantification instrument (e.g., Qubit, ThermoFisher Scientific).
2.3.2. Reagents and Consumables
-
5
DNA AWAY® surface decontaminant (Molecular BioProducts).
-
6
70% EtOH.
-
7
Lysis buffer: 1:1 dithiothreitol (DTT, 100 mM) + potassium hydroxide (KOH, 400 mM).
-
8
GenomePlex® Single Cell Whole-Genome Amplification Kit (WGA4, Sigma-Aldrich): 10× single cell lysis & fragmentation buffer, 1× single cell library preparation buffer, library stabilization solution, library preparation enzyme, 10× Amplification Master Mix, WGA DNA polymerase, Control gDNA (5 ng/ml), molecular grade water.
-
9
10 mM Tris–HCl + EDTA pH 8.0 (TE): 10 mM Tris–HCl, 1 mM disodium EDTA, pH 8.0.
-
10
QIAquick PCR Purification kit (Qiagen).
-
11
Qubit dsDNA HS Assay Kit (ThermoFisher Scientific).
-
12
Qubit assay tubes (ThermoFisher Scientific).
2.4. Library Construction
2.4.1. Equipment
-
1
Sonication device, e.g., Covaris S2 (COVARIS Inc.).
-
2
Thermal cycler (e.g., Mastercycler™ pro PCR System, Eppendorf).
-
3
DNA quantification instrument (e.g., Qubit, ThermoFisher Scientific).
-
4
Bioanalyzer; e.g., 2100 Bioanalyzer (Agilent Technologies).
-
5
Magnetic stands for PCR tubes/96-well plates (e.g., LifeTechnologies).
-
6
Magnetic stand for 1.5 ml tubes (e.g., DynaMag®, ThermoFisher Scientific).
2.4.2. Reagents and Consumables
-
7
Sonication tube for fragment size of 200–250 bp, e.g., Snap Cap microTUBE (COVARIS Inc.).
-
8
10 mM TE (pH = 7.5–8.0, RT): 10 mM Tris–HCl, 1 mM disodium EDTA.
-
9
Dnase-free 0.2 ml PCR 8-strips.
-
10
NEBNext® Ultra DNA Library Prep Kit for Illumina® (New England BioLabs Inc.).
-
11
NEBNext® Multiplex Oligos for Illumina®-Dual Index Primer Set 1 (New England BioLabs Inc.).
-
12
Agencourt® AMPure® XP Beads (Beckman Coulter).
-
13
100% EtOH.
-
14
DNA low-bind tubes (Eppendorf): PCR tubes, 1.5 ml tubes, 96-well plates.
-
15
Qubit dsDNA HS Assay Kit (ThermoFisher Scientific).
-
16
Qubit assay tubes (ThermoFisher Scientific).
-
17
High Sensitivity DNA Analysis Kit (Agilent Technologies).
2.5. Single-Cell CNV Profiling Analysis Software Tools
Bowtie (sequence analysis software; version 2.2.6 or later).
R: The R project for statistical computing (or equivalent).
University of California Santa Cruz (UCSC) Genome browser: https://genome.ucsc.edu/, hg19 reference genome.
3. Methods
The entire procedure from initial blood sample preparation to sequence-ready DNA library takes approximately 5 days of elapsed time, is visually summarized in Fig. 1 and consists of the following major steps (the steps described in detail in this chapter are marked in bold letters):
Fig. 1.
Overview of the complete HD-SCA workflow. The HD-SCA workflow from a blood draw to a single-cell CNV profile: Red blood cell (RBC) lysis is followed by plating of the entire PBMC fraction, fluorescent staining, full slide scanning and detection of COIs. The CTC detection is followed by single-cell extraction, next-generation sequencing and CNV profiling. In red: steps provided within this protocol: single-cell extraction through micromanipulation, WGA, library preparation (including sonication), and single-cell CNV profile analysis
Blood sample collection (time to sample process of up to 48 h to enable standard shipping conditions).
Blood sample processing (half day of elapsed time, 45 min of hands-on/sample).
Fluorescent staining (half day of elapsed time, 5 min of hands-on/slide).
Whole slide imaging (1.5 h of elapsed time/2 slides).
Technical analysis of COIs (5 min/slide).
Single-cell isolation (3 min hands-on/cell).
Whole-genome ampliftcation including cleanup (half day for 24 cells of elapsed time, 2 h of hands-on/24 cells).
Library construction including sonication (2 days for 24 cells of elapsed time, 5 h hands-on/24 cells).
Single-Cell CNV proftling.
Details for required steps to detect COIs on glass slides (steps 1–5 above) are provided in Marrinucci et al. [16] and are discussed briefly in Subheading 3.1 below. The detailed protocol provided in Subheadings 3.2–3.5 below describes the necessary steps from isolation of a single candidate cell to genome-wide CNV analysis.
3.1. Summary of the HD-SCA Workflow
Blood is drawn in blood collection tubes (BCTs) containing a cell fixation and DNA stabilization reagent and then shipped in temperature controlled boxes to ensure preservation of intact cells and DNA. Samples are processed by plating of the PBMC fraction on customized adhesive glass slides after red blood cell (RBC) lysis.
Slides are then stained for immunofluorescent identification of rare cells of interest among the white blood cell (WBC) population. Briefly, cells are fixed, permeabilized, and afterward fluorescently stained with antibodies against a pan-cytokeratin panel (targeting an epithelial-specific intermediate filament), CD45 (a leukocyte specific marker) and DAPI (4′,6-diaminido-2-phenylindole, a nucleic acid stain). The markers for identification can be adapted depending on the COI, hence specialized assays, for example, for melanoma cells [20] and endothelial cells [21] are available. An additional disease-specific marker can be used as a forth channel marker to study the biology of the CTC. Fixation steps (chemically in the tube, physically on the slide and during staining) are not only necessary in order to identify potential CTCs, but also present a challenge, especially for the quality of downstream analysis [22]. The immunofluorescence staining is followed by a high-throughput digital imaging pipeline. All images are stored and analyzed by an R-based software routine.
The analysis algorithms take several features into consideration to identify cells distinct from a WBC including morphology data like nuclear size or shape, signal intensities, or absence of the epithelial and leukocyte marker. Finally the cells that have been calculated as rare events and probable high definition circulating tumor cells (HD-CTCs; CKpos/CD45neg with distinct nuclear shape) are presented to a specialist trained in hematopathology for final technical analysis and classification. An example for different categories of CTCs analyzed by the HD-SCA workflow are displayed in Fig. 2. The final result is an enumeration of all rare events within the entire PBMC cell population and a comprehensive morphometric data set for each cell.
Fig. 2.
Categories of detected potential CTCs. These CTCs are representing the pleomorphic character of CTCs detected in the blood of a metastatic prostate cancer patient. Cells have been stained for nuclei (DAPI; blue), CD45 (green) and CK (red). Displayed are the composite and the single channel images. HD-CTC: CKpos/CD45neg cells with a nuclear shape distinct from WBCs. CTC-Small: CKpos/CD45neg cells with a small nucleus (WBC-like). CTC-LowCK: CKneg/CD45neg cells with a nucleus at least double the size of a WBC. CTC-cfDNA producing: CTCs undergoing apoptosis (CKpos/CD45neg with irregular cytoplasmic or nuclear condensation) and about to release circulating tumor DNA. Scale bars equal 10 μm
For all cells on a slide, approximately 400 variables are detected and stored, including the position of each COI on the slide by their Cartesian coordinates. Two numerical coordinates define the exact position on the slide, using two perpendicular lines as reference axes. This coordinate system can be applied to any microscope without special instrumentation and allows the relocation of each cell further along the pipeline. In order to perform single-cell downstream analysis, COIs are relocated and imaged at high definition to enable subsequent correlation of genomic and morphological data. COIs are typically imaged at 400× magnification on a fluorescence microscope, but may also be analyzed through deconvolution or confocal microscopy. Each COI can then be isolated from the slide using the Cartesian coordinates and a micromanipulation station with a glass capillary and a hydraulic cylinder system.
3.2. Single-Cell Isolation
After a COI has been identified, cell isolation is started, but before isolation from the slide, reimaging of COIs at 400× magnification is recommended to achieve a high quality image. This enables future cytomorphologic downstream analysis of COIs.
Prepare all buffers and keep them on ice and check oil level in micromanipulation system (if you intend to proceed with cell isolation after relocation and reimaging of the cell).
Clean slide with 70% ethanol (do not disturb coverslip) and remove dust with compressed air spray.
COI is relocated using the imaging software ImagePro and a custom macro that uses a transformation matrix, which maps the coordinates between the scanning and reimaging microscope. 100× images from the scanning report are used to confirm the location. Detected offset can be applied to all other coordinates of COIs on this slide.
Each cell position is confirmed and images are taken in each fluorescent channel using a 400× magnification until all COIs of the slide are imaged.
Color composites and images of individual channels are stored in a database.
3.2.1. Pause Point: Slides Can Be Stored in a Dark and Dry Location for Future Downstream Analysis
To proceed with cell isolation, clean all areas around the microscope station with DNA AWAY® and 70% ethanol. Place a coverslip on the slide holder insert to provide PCR tube support. Prepare 0.5 ml PCR tubes using UV sterilization prior to use.
Peel off nail polish from slide carefully without moving the coverslip (a scalpel may be useful).
Place slide in 1× PBS in Coplin jar in a tilted position until coverslip comes off (~10 min).
Place slide (without coverslip) on microscope next to empty coverslip and add 1 ml of PBS-T to prevent cells from dehydration (refill if PBS-T starts to evaporate during longer cell extraction durations).
Insert glass capillary into the micromanipulation arm. Ensure straight orientation of capillary.
Use microscope software control box to turn on the live preview and choose brightfield with a 20× objective lens; use stored coordinates to navigate to cell.
Use control panels (one for oil pressure, one for capillary, one for microscopy stage) to navigate to the end of the capillary in field of vision, but keep the capillary above slide surface, but within PBS-T.
Focus on the capillary tip and adjust oil so that air (visible as dark mass) in the capillary moves close to tip opening.
Focus on the cell and slowly navigate the capillary down and in front of the cell as described and illustrated in Fig. 3.
Dislodge the COI using the capillary tip or use the tip to push other cells out of the way first (see Note 1).
Position the capillary tip right in front of loose COI and use the oil pressure wheel to create suction and aspirate the COI into the capillary (make sure to ONLY aspirate the COI, not other cells!). Move the capillary up (out of the PBS-T); do NOT move the capillary in x and y direction!
Place a PCR tube with open lid (opaque label facing up) on the coverslip next to the slide and place a 1 μl drop of cell deposition buffer (CDB) between the first two lines behind the opening of the tube.
Now move the microscope stage to steer the tube to the capillary tip to position the CDB below the tip. Do not touch the capillary tip with the tube.
Focus on the buffer drop (the edge of the drop has to be a sharp, dark outline) and move the tip down until it is immersed in the drop.
Focus on the tip and use the oil wheel to slowly release the cell. Observe the tip opening, watch the cell moving through the capillary and make sure it is in the drop.
Move the tip up and the stage away from the tip. Take the tube, and tap it slightly on bench to let the drop with the cell move toward the bottom before closing the lid. Follow with a quick spin and store the tube directly on dry ice.
Mark the cell in your software or database as “isolated” and move to the next cell position; repeat steps 7–17.
Fig. 3.
The Single-Cell Isolation Process. (1) The capillary is positioned inside the buffer solution above the cell of interest and (1a) set into focus. (1b) The oil adjustment wheel is used to move the air close to the capillary opening and then stabilize it there. (2) The cell of interest (COI) is set into focus and the capillary tip moved down onto the slide (2a) to be in focus together with the COI. The capillary is used to loosen the COI and the oil wheel to create suction to (2b) aspirate the cell into the capillary. The capillary is moved up, out of the buffer. (3) A PCR tube is placed on a coverslip next to the sample slide. (3a) The edge of the 1 μl drop of TE buffer is set into focus and the capillary is lowered into the drop. (3b) The COI is released carefully by slowly turning the oil wheel to the “out” direction
3.3. Whole-Genome Amplification
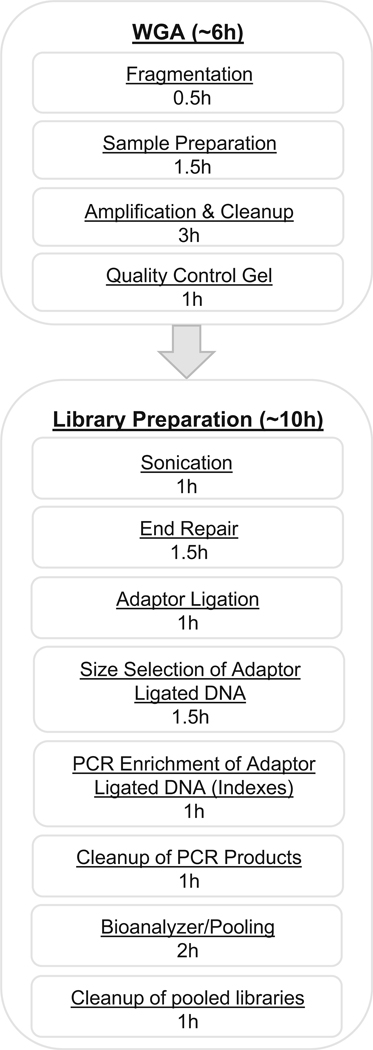
Following the extraction of single cells, whole-genome amplification (WGA) and fragmentation by sonication is used to reach a sufficient amount of DNA in the required fragment length for single-cell sequencing. Since a diploid human cell only contains about 7 pg of genomic DNA, amplification is necessary prior to genetic variation analyses [23]. An overview of all procedures involved in WGA and library preparation with time estimates are indicated in Fig. 4.
Fig. 4.
Whole-genome amplification (WGA) and library preparation workflow chart. Overview and approximate timing estimation of all steps included in the process from extracted single cell to ready-to-sequence library. Time estimates are based on processing of 24 samples
Clean all surfaces with DNA AWAY® and 70% EtOH.
Thaw PCR tubes containing the single cell in 1 μl of TE, always add a sample for negative control (TE only) and positive control (1.5 μl gDNA).
Add 1.5 μl of lysis buffer (1:1 DTT & KOH) and spin down (gently).
PCR program: 95 °C for 2 min.
-
Cool tubes on a PCR cooler and prepare master mix:
- Add 6.5 μl of 10 mM TE for each reaction.
- Add 1 μl 10× Single Cell Lysis & Fragmentation Buffer per reaction, mix thoroughly and spin down.
Add 7.5 μl of master mix to each reaction, including controls.
PCR program: 99 °C for 4 min (time sensitive: take samples out instantly and place on PCR cooler).
3.3.1. Pause Point: Store the Reactions at −20 °C or Keep on a PCR Cooler to Continue Immediately
-
Prepare master mix of:
- 2 μl of 1× Single Cell Library Preparation Buffer
- 1 μl of Library Stabilization Solution per reaction and mix thoroughly.
Add 3 μl of the mix to each reaction.
Mix thoroughly and place in thermal cycler at 95 °C for 2 min, cool samples on PCR cooler, spin down and store on PCR cooler.
Add 1 μl of Library Preparation Enzyme to each reaction, mix thoroughly and spin down.
Place samples in thermal cycler and incubate as follows: 16 °C for 20 min, 24 °C for 20 min, 37 °C for 20 min, 75 °C for 5 min, 4 °C hold.
Spin samples down.
3.3.2. Pause Point: Store at −20 °C for Up to 3 Days or Amplify Immediately
-
Prepare master mix of:
- 7.5 μl of 10× Amplification master mix.
- 48.5 μl of molecular grade water (provided in the kit).
- 5 μl of WGA DNA Polymerase.
Mix thoroughly. Add 61 μl of the master mix to each reaction.
Mix thoroughly, spin down and place in thermal cycler: 95 °C for 3 min, 24 cycles of: 94 °C for 30 s, 65 °C for 5 min; then hold at 4 °C.
3.3.3. Pause Point: Store the Reactions at −20 °C (Pause Point) or Keep at 4 °C to Continue Immediately
Prepare a 1.5% agarose gel including gel stain. Mix 2 μl 5 × loading dye with 8 μl DNA sample and load samples onto agarose gel. Add Quick-Load® 100 bp DNA Ladder in one well and run for 35 min at 90 V (for 100 ml gel; adjust accordingly for other gel volumes).
Take image of agarose gel and evaluate samples: Successful WGA produces a continuous smear mainly between 150 and 1000 bp. If no smear is visible, WGA was not successful and sample cannot be used for library preparation (see Note 2 and Fig. 7).
Use QIAquick PCR Purification kit according to manufacturer instructions.
Keep eluted DNA on PCR cooler and quantify all samples using for example Qubit quantification system according to manufacturer instructions; note DNA concentrations in an excel sheet or similar form.
Fig. 7.
Whole-genome amplification (WGA) control gel. On the left is the DNA ladder, followed by 17 samples, the gDNA control (C+) and the negative control (C−). The samples labeled in red (2, 13, 15) have no or only a very dim smear and therefore no sufficient amplification. In cells that show no smear, like in this example cells 2, 13, 15, cell transfer during isolation might have been incomplete or DNA quality of those cells insufficient
3.4. Library Construction
Before starting library construction all samples have to be adjusted to same concentrations to obtain equal number of reads per sample during sequencing.
For each sample use 185 ng DNA input in 55.5 μl total volume; calculate your sample volume and add 10 mM TE buffer accordingly.
Transfer all samples to a sonication tube suitable for your equipment, volume, and fragment size (here: a COVARIS Snap Cap microTUBE).
Follow the manufacturers protocol to reach a fragment size of approximately 200–250 bp and continue library preparation with fragmented DNA.
Transfer the fragmented DNA samples to strips of 0.2 ml PCR tubes.
-
Prepare a master mix using the NEBNext End Prep Kit:
- 3 μl End Prep Enzyme Mix.
- 6.5 μl 10× End Repair Reaction Buffer.
Add 9.5 μl of master mix to each 55.5 μl fragmented DNA, mix by pipetting and spin down.
-
Place in a thermal cycler and incubate:
20 °C for 30 min, 65 °C for 30 min, 4 °C hold
-
Prepare master mix:
- 15 μl Blunt/TA Ligase Master Mix.
- 2.5 μl NEBNext Adaptor for Illumina.
- 1 μl Ligation enhancer.
Add the 18.5 μl master mix immediately to the 65 μl of cooled down reaction mix. Total volume is 83.5 μl.
Mix by pipetting and spin down.
Place in a thermal cycler and incubate at 20 °C for 15 min, then place on PCR cooler.
Add 3 μl of USER enzyme to each sample reaction mix.
Spin down and incubate at 37 °C for 15 min.
3.4.1. Pause Point: Store the Reactions at −20 °C or Keep on PCR Cooler to Continue
Thaw reagents for the next step (AMPure XP bead size selection) for 30 min, keep DNA on PCR coolers until use.
-
For AMPure XP bead size selection of adaptor ligated DNA, follow the manufacturer instructions to select fragments between 200 and 300 bp. Let beads and TE come to RT, vortex beads, prepare fresh 80% ethanol.
At the end of the AMPure XP bead size selection protocol, elute in 17 μl TE and finally transfer 15 μl to a new PCR tube for amplification.
- For PCR Enrichment of Adaptor Ligated DNA mix the following components in fresh sterile nuclease-free tubes (no master mix here!). Make sure to record index numbers for each sample and to assign to each reaction a unique combination of indexes within one set of libraries that is about to get pooled:
- 15 μl Adaptor Ligated DNA Fragments.
- 25 μl NEBNext Q5 Hot Start HiFi PCR Master Mix.
- 5 μl Index Primer/i7 Primer.
- 5 μl Universal PCR Primer/i5 Primer.
- Total reaction volume: 50 μl.
-
Place in thermal cycler and run the following program:
98 °C for 30 s; 7 cycles of: 98 °C for 10 s, 65 °C for 75 s; then final extension: 65 °C for 5 min and hold at 4 °C.
Use AMPure XP beads for PCR product cleanup: let beads and TE come to RT, vortex beads, prepare fresh 80% ethanol.
Transfer library into low-binding 1.5 ml tube, add 0.8 volume of beads (for 50 μl sample: 40 μl beads) to sample, mix thoroughly, but gently by pipetting up and down and incubate 5 min at RT.
Spin down, place in magnet stand with open lid, incubate 5 min at RT, discard supernatant.
Wash twice with 200 μl of 80% EtOH, incubate 30 s, discard supernatant.
Air-dry beads for exactly 2 min with lid open on magnet.
Remove tube from magnet and elute DNA in 35 μl TE, mix well, incubate for 3 min at RT, spin down, place in magnet, and let sit for 5 min.
Collect 32 μl supernatant that contains the DNA, and discard beads.
Measure DNA quantity (e.g., Qubit); following the manufacturer’s protocol.
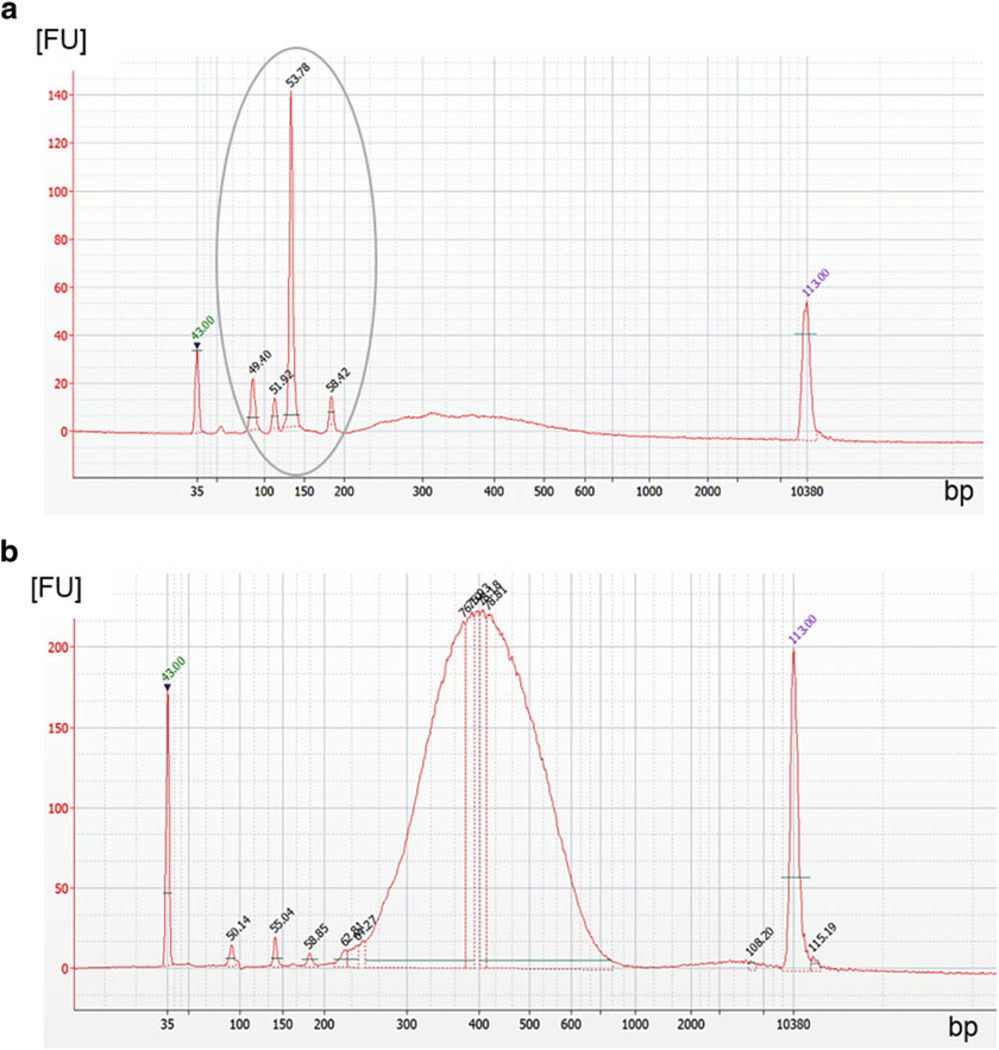
Use a Bioanalyzer for analysis of size distribution by following the manufacturer’s instructions.
-
Quality control of Bioanalyzer size distribution check: If the library preparation has been successful, the lower and upper markers frame a normal curve of distribution with a peak around 300–400 bps (see Note 3). A low concentration of primer dimers and adaptor dimers (<130 bps) might be observed right after the lower marker, but these low concentrations will be eliminated during the final cleanup.
If single peaks are observed, especially in the area of short fragments (primer dimer: ~80 bps or adaptor dimer: ~130 bps) and no fragment distribution with a peak around 380 bps, library preparation has not been successful. Examples for library control results are displayed in Fig. 5.
-
Calculate the molarity of each library:
Pool all libraries at a final concentration of 10 mM using a volume of 5 μl/sample (dilute with TE buffer).
Perform one last cleanup using the AMPure XP beads (repeat steps 5–8) with the total volume of all pooled libraries: Total Vol. = number of libraries × 5 μl.
Air-dry beads for 2 min with lid open, elute DNA in the same volume of TE as the volume you started the cleanup with (Total Vol.). Mix well, incubate for 3 min at RT, spin down, place in magnet, and let sit for 5 min.
Collect as much supernatant (that contains the DNA) as possible without touching the beads, and discard beads.
Pooled library is ready for sequencing. Protocols for next generation-sequencing of libraries for CNV analysis are in detail described in Baslan et al. (here, DNA libraries were sequenced on the HiSeq2500 platform (Illumina) using single end read 50 base pair protocol (SR50)) [24].
Fig. 5.
Quality control of size distribution after library preparation using the Bioanalyzer. (a) An example of a poor quality library with primer dimers (~80 bp) and a high number of adaptor dimers (~130 bp), both within grey encircled area. The concentration of the library is very low (represented by a fluorescent signal <10 FU), but the size (~350 bp) is correct. Even after an additional cleanup this library would not be sufficient for sequencing. (b) Adaptor dimers and primer dimers are of very low concentration. Library shows a perfect size distribution with a peak around 380 bp and a high concentration (>200 FU). After the final cleanup, this library will be sufficient for next-generation sequencing
3.5. Single-Cell CNV Profiling
Single-cell CNV profiling has become a valuable tool in CTC research to monitor the evolution of disease during the course of therapy [25–27]. It enables detection of new lineages or subclones appearing in the circulation that reflect changes in the tumor load that would not be easily detected by bulk based applications [27]. As described by Dago et al. [25] such rare subclones may play an important role in chemotherapy resistance and disease progression and their early detection could thus greatly improve therapeutic outcomes.
Procedures based on the protocols of Baslan et al. [24] are then performed to allow CNV profiling of single CTCs, and this reference provides method details for single-cell CNV profiling including all processing steps, software details, and scripts. An overview of the single-cell CNV profiling process is provided here. After sequencing, results are transferred as a fastq-file, the open-source software “Bowtie” can be used to align the sequences to the reference genome “hg19,” which can be acquired from the UCSC Genome Browser. Before the sequences can be mapped, a set with an arbitrary number of “bins” is created across the genome with each bin containing the same number of mappable positions (intervals of sequence reads). A high number of bins will allow for higher resolution but is also more sensitive for noise, which will lower confidence in observed events. A good balance between resolution and noise has to be determined and will differ on a cell to cell basis due to differences in whole-genome amplification and sequencing efficiency. This concept of optimized sparse sequencing of single cells for copy number analysis has been developed by Navin et al. [28]. In 2012 Baslan et al. [24] improved the method for high-throughput yet low-cost single-cell sequencing. The optimized method of sparse sequencing allows for accurate, but cost-efficient acquisition of CNV profiles even from single-cells. It requires 20 times fewer reads compared to Navin et al. [28] and only about 75 million bases instead of over 100 billion as is standard for whole-genome sequencing [29].
Once the binned data has been acquired, it can be simplified using mathematical segmentation [30]. Segmentation quantifies the amplitude and location of copy number gains and losses across the genome, creating a genome-wide, numerical “CNV profile” of each cell. CNV profiles, in form of numerical vectors, can be clustered to identify the lineage relationships among cells in a blood draw or to compare and contrast cells from different time points or different patients. The degree of similarity among groups of individual CNV profiles can reveal clonal and subclonal structures in the population that are typical of cancer. Among CTCs, the identification of complex clonal CNV profiles distinguishes the cancer cells from white blood cells or other cells in circulation that exhibit “flat” profiles typical of normal diploid genomes with no significant copy number alterations (see Fig. 6).
Fig. 6.
Copy Number Variation (CNV) profile examples. (a) An example of a complex genome with multiple copy number alterations consistent with a malignant CNV profile of a COI. (b) The profile of a diploid genome derived from a white blood cell (WBC) with no large copy number variation. (c) Various graphic programs can be used to visually display CNV profiles. Here the software CIRCOS [31] has been used to illustrate CNVs in each chromosome of the cell displayed in the center, which is the same cell-profile as shown in (a)
Most importantly, the tracking of single-cell CNV profiles across multiple time points during therapy provides an important “window” into the genetic response of the cancer to treatment. Dago et al. [25] demonstrated that a cancer that was genetically stable for months can rapidly change its genomic makeup in response to treatment pressure. Although more work is clearly needed to understand how to translate these genomic changes into improved outcomes, we believe that a combined approach of proteomorphometric and genomic analysis on the rare cells identified in the HD-SCA workflow will be a key component of real-time cancer diagnosis and targeted therapies.
Acknowledgment
Disclosure
James Hicks: Epic Sciences, Inc.—Clinical Advisory Board
Peter Kuhn: Epic Sciences, Inc.—Stock and Ownership Interests, Advisory Role, Patents, and Royalties
4. Notes
- While using the capillary to push other cells out of the way or scraping the COI off, ensure to not have a cell stick to the capillary or aspirate loosened cells together with the COI.
- When aspirating the COI into the capillary, ensure to turn the oil wheel until the cell is all the way out of the field of vision (about two full turns), then one quarter-turn backward to stop the oil flow.
- Not moving the capillary in x or y direction helps keeping the capillary in focus, enabling a faster process for depositing the COI in the CDB drop.
- If the cell “disappeared” in the capillary during the attempt to place it in the drop, it can be caused by one of three issues:
-
-Low oil level in the system.
-
-Air bubbles in the hose of the oil system.
-
-Oil and air bubbles in the capillary.
-
-
Check oil level before isolating cells. Remove air bubbles from oil supply hose and make sure to have no air trapped in the hose while inserting the capillary. Change capillary after a maximum of about ten cells to prevent oil moving into the capillary (danger of contaminating the sample).
- If most samples show a smear, but a few do not (see Fig. 7), it might be due to a loss of the cell during cell isolation. Make sure to observe the deposition of the cell into the buffer.
- If no smear is visible, but primer dimers are: PCR has worked, but cells have not been lysed properly. Ensure that 10× Single Cell Lysis & Fragmentation Buffer is thawed and mixed well before use until no precipitation is visible.
-
If neither smear nor primer dimers appear, the enzyme might not have worked sufficiently. Repeat four cycles of amplification PCR:95 °C for 3 min, four cycles of: 94 °C for 30 s, 65 °C for 5 min; hold at 4 °C.If still neither smear nor primer dimers are visible, PCR has not worked and new reagents are required.
- If the negative control shows a smear, water and TE have to be discarded immediately. Other components of the kit should be tested (buffers and enzymes) again with a new negative and positive control to make sure the contamination is eliminated before moving forward with valuable cells. If contamination continues, all used components have to be discarded.
- If the positive control shows no smear, test the gDNA concentration using the Qubit. Concentration should be 5 pg/μl. Mix gDNA well before use.
- If amount of library is insufficient it could be caused by two things: Low quality or concentration of starting material. DNA input can be increased up to the maximum input in 55 μl sample volume (without additional water). Due to some volume loss during sonication, 60 μl can be used for sonication if the sonication protocol and tube size allows it. The quality of the starting DNA (WGA products) can be determined via Bioanalyzer. In addition, make sure calculations were correct, and that magnetic beads and TE were brought to RT.
- To ensure that the final cleanup has successfully removed most primer and adaptor dimers, retest all final libraries on a Bioanalyzer.
- If the Bionalayzer does not show a peak size of 300–400 bp (with a total library fragment range between 100 and 600 bp) the magnetic bead cleanup was not successful. During the cleanup ensure that all components are brought to RT and that beads are vortexed well right before use.
References
- 1.Ashworth TR (1869) A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Med J Aust 14:146–147 [Google Scholar]
- 2.Salsbury AJ (1975) The significance of the circulating cancer cell. Cancer Treat Rev 2:55–72 [DOI] [PubMed] [Google Scholar]
- 3.Chang YS, Tomaso E d, McDonald DM et al. (2000) Mosaic blood vessels in tumors: frequency of cancer cells in contact with flowing blood. Proc Natl Acad Sci U S A 97:14608–14613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Massagué J, Obenauf AC (2016) Metastatic colonization by circulating tumour cells. Nature 529:298–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cristofanilli M, Budd GT, Ellis MJ et al. (2004) Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 351:781–791 [DOI] [PubMed] [Google Scholar]
- 6.Miller MC, Doyle GV, Terstappen LWMM (2010) Significance of circulating tumor cells detected by the cellsearch system in patients with metastatic breast colorectal and prostate cancer. J Oncol 2010:617421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aktas B, Kasimir-Bauer S, Heubner M et al. (2011) Molecular profiling and prognostic relevance of circulating tumor cells in the blood of ovarian cancer patients at primary diagnosis and after platinum-based chemotherapy. Int J Gynecol Cancer 21:822–830 [DOI] [PubMed] [Google Scholar]
- 8.Minciacchi VR, Zijlstra A, Rubin MA et al. (2017) Extracellular vesicles for liquid biopsy in prostate cancer: where are we and where are we headed? Prostate Cancer Prostatic Dis 20:251–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thiele J-A, Bethel K, Králĺčková M et al. (2017) Circulating tumor cells: fluid surrogates of solid tumors. Annu Rev Pathol 12:419–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hesari A, Moghadam SAG, Siasi A et al. (2017) Tumor-derived exosomes: potential biomarker or therapeutic target in breast cancer?: exosomes in breast cancer. J Cell Biochem 119 (6):4236–4240 [DOI] [PubMed] [Google Scholar]
- 11.Alix-Panabieres C, Pantel K (2016) Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov 6(5):479–491 [DOI] [PubMed] [Google Scholar]
- 12.Cho WCS (2014) Emerging techniques in molecular detection of circulating tumor cells. Expert Rev Mol Diagn 14:131–134 [DOI] [PubMed] [Google Scholar]
- 13.Alix-Panabières C (2012) EPISPOT assay: detection of viable DTCs/CTCs in solid tumor patients. Recent Results Cancer Res 195:69–76 [DOI] [PubMed] [Google Scholar]
- 14.Tewes M, Kasimir-Bauer S, Welt A et al. (2015) Detection of disseminated tumor cells in bone marrow and circulating tumor cells in blood of patients with early-stage male breast cancer. J Cancer Res Clin Oncol 141:87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagrath S, Sequist LV, Maheswaran S et al. (2007) Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 450:1235–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marrinucci D, Bethel K, Kolatkar A et al. (2012) Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Phys Biol 9:016003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlsson A, Nair VS, Luttgen MS et al. (2014) Circulating tumor microemboli diagnostics for patients with non–small-cell lung cancer. J Thorac Oncol 9:1111–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newton PK, Mason J, Bethel K et al. (2013) Spreaders and sponges define metastasis in lung cancer: a Markov chain Monte Carlo mathematical model. Cancer Res 73:2760–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malihi PD, Morikado M, Welter L et al. (2018) Clonal diversity revealed by morphoproteomic and copy number profiles of single prostate cancer cells at diagnosis. Converg Sci Phys Oncol 4:015003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruiz C, Li J, Luttgen MS et al. (2015) Limited genomic heterogeneity of circulating melanoma cells in advanced stage patients. Phys Biol 12:016008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bethel K, Luttgen MS, Damani S et al. (2014) Fluid phase biopsy for detection and characterization of circulating endothelial cells in myocardial infarction. Phys Biol 11:016002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xuan J, Yu Y, Qing T et al. (2013) Next-generation sequencing in the clinic: Promises and challenges. Cancer Lett 340:284–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ranek L (1976) Cytophotometric studies of the DNA, nucleic acid and protein content of human liver cell nuclei. Acta Cytol 20:151–157 [PubMed] [Google Scholar]
- 24.Baslan T, Kendall J, Rodgers L et al. (2012) Genome-wide copy number analysis of single cells. Nat Protoc 7:1024–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dago AE, Stepansky A, Carlsson A et al. (2014) Rapid phenotypic and genomic change in response to therapeutic pressure in prostate cancer inferred by high content analysis of single circulating tumor cells. PLoS One 9: e101777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ni X, Zhuo M, Su Z et al. (2013) Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc Natl Acad Sci U S A 110:21083–21088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navin N, Hicks J (2011) Future medical applications of single-cell sequencing in cancer. Genome Med 3:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Navin N, Kendall J, Troge J et al. (2011) Tumour evolution inferred by single-cell sequencing. Nature 472:90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baslan T, Kendall J, Ward B et al. (2015) Optimizing sparse sequencing of single cells for highly multiplex copy number profiling. Genome Res 25:714–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venkatraman ES, Olshen AB (2007) A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics 23:657–663 [DOI] [PubMed] [Google Scholar]
- 31.Krzywinski M, Schein J, Birol I et al. (2009) Circos: An information aesthetic for comparative genomics. Genome Res 19:1639–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]