Abstract
Oxaliplatin resistance is a major challenge in the clinical treatment for advanced colorectal cancer (CRC). Long non-coding RNAs (lncRNAs) are involved in tumorigenesis and progression as critical regulators, while their potential roles in chemoresistance are poorly understood. In this study, we report that the LINC00460-miR-149-5p/miR-150-5p-mutant p53 feedback loop is responsible for oxaliplatin resistance in CRC. First, LINC00460 was found to exhibit higher expression in oxaliplatin-resistant CRC (CRC/OxR) cells compared with parental oxaliplatin-sensitive ones, and this expression pattern depends on mutant p53 (SW480/OxR), not wild-type p53 (HCT116/OxR). Oxaliplatin-induced LINC00460 in SW480/OxR cells was mainly located in the cytoplasm and was associated with AGO2 protein. LINC00460 functions as a competing endogenous RNA (ceRNA) to promote oxaliplatin resistance through sequestering miR-149-5p/miR-150-5p and upregulating the expression of the microRNA (miRNA) target p53. Knockdown of LINC00460 sensitized SW480/OxR cells to oxaliplatin by modulating p53 in vitro and in vivo. In turn, mutant p53 positively regulated the expression of LINC00460, thus forming a feedback loop. Clinical data showed that LINC00460 was upregulated in CRC tissues compared with paired normal tissues and was significantly correlated with clinical stage and node (N) status. Our findings uncover a mechanism for the LINC00460-miR-149-5p/miR-150-5p-mutant p53 feedback loop in oxaliplatin resistance of CRC, and they provide potential therapeutic targets for tumor chemoresistance.
Keywords: colorectal cancer, oxaliplatin resistance, LINC00460, miR-149-5p, miR-150-5p, ceRNA, mutant p53
Graphical Abstract
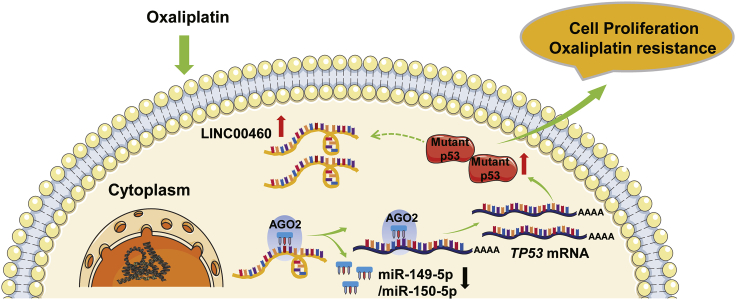
The functions of lncRNAs in oxaliplatin resistance of colorectal cancer (CRC), especially with TP53 mutations, are not clear. Wang and colleagues report that LINC00460 promotes oxaliplatin resistance through sequestering miR-149-5p/miR-150-5p and upregulating the expression of mutant p53, which provides a potential therapeutic target for tumor chemoresistance.
Introduction
Colorectal cancer (CRC) is the third most common type of cancer and the second leading cause of cancer death worldwide.1 Region, sex, and age differences have been documented in the incidence of CRC. Globally, CRC ranks the third in men and the second in women in incidence, and it is more likely to occur in people over the age of 50.1,2 Although the incidence of CRC is relatively high in developed countries, with the rapid development of economies and the improvement of living standards, it is also on the rise in developing countries due to changing diets. Early CRC without obvious symptoms is generally difficult to diagnose, and current treatment of CRC is mainly based on surgery and chemotherapy.
Oxaliplatin, sometimes combined with other drugs (e.g., 5-fluorouracil [5-FU], leucovorin), is one of the most effective first-line chemotherapeutic reagents for CRC.3 It is the first platinum drug proven to be active against CRC and is also used for treatment of gastric and pancreatic cancers.4 Unfortunately, intrinsic or acquired resistance to oxaliplatin alone or in combination will cause treatment failure, and almost all metastatic CRC eventually develops resistance.5,6 Currently, the molecular mechanism of oxaliplatin resistance has not been completely studied, and the cause is still elusive. Long non-coding RNAs (lncRNAs) are transcripts with a length longer than 200 nt and low protein-coding potential. Recently, lncRNAs have been found to be involved in a series of biological processes such as genomic imprinting, chromatin modification, cell differentiation, and tumorigenesis.7, 8, 9 Some studies have shown that lncRNAs are related to chemoresistance, involved in transcriptional and post-transcriptional regulation, and subsequently affect cell proliferation and apoptosis.10,11
LINC00460 is a lncRNA involved in tumor progression, including epithelial-mesenchymal transition (EMT), cell proliferation and apoptosis, and cell migration and invasion.12, 13, 14 However, the effect of LINC00460 on oxaliplatin resistance in CRC is unknown.
The tumor suppressor p53 plays a fundamental and extensive role in the development of cancer and cancer therapy; however, about 50%–60% of human cancers contain mutations in the TP53 gene.15 Mutant (MUT) p53 loses the tumor-suppressive activity and acquires new carcinogenic characteristics that promote tumor growth and progression.16 Approximately half of all CRCs show TP53 gene mutations, which are associated with poor clinical outcome.17 p53 functions primarily as a transcription factor to regulate the expression of various target genes that participate in cell cycle arrest, programmed cell death, DNA repair, proliferation, apoptosis, angiogenesis inhibition, and cellular stress response.18,19 TP53 copies in the SW480 cell line carry two point mutations (R273H and P309S) that lead to abnormalities in p53 protein.20 MUT p53 is an important element for 5-FU resistance in CRC.21,22 However, the relationship between p53 and oxaliplatin resistance in CRC is not clear.
In this study, we demonstrate that LINC00460 is significantly upregulated in oxaliplatin-resistant CRC cells that harbor MUT TP53, and we show that the LINC00460-miR-149-5p/miR-150-5p-MUT p53 feedback loop plays an important role in oxaliplatin resistance of CRC. These findings might provide therapeutic targets for CRC patients that suffer from chemoresistance.
Results
LINC00460 Is Upregulated in SW480/OxR Cells and Is Responsible for Oxaliplatin Resistance in CRC with TP53 Mutations
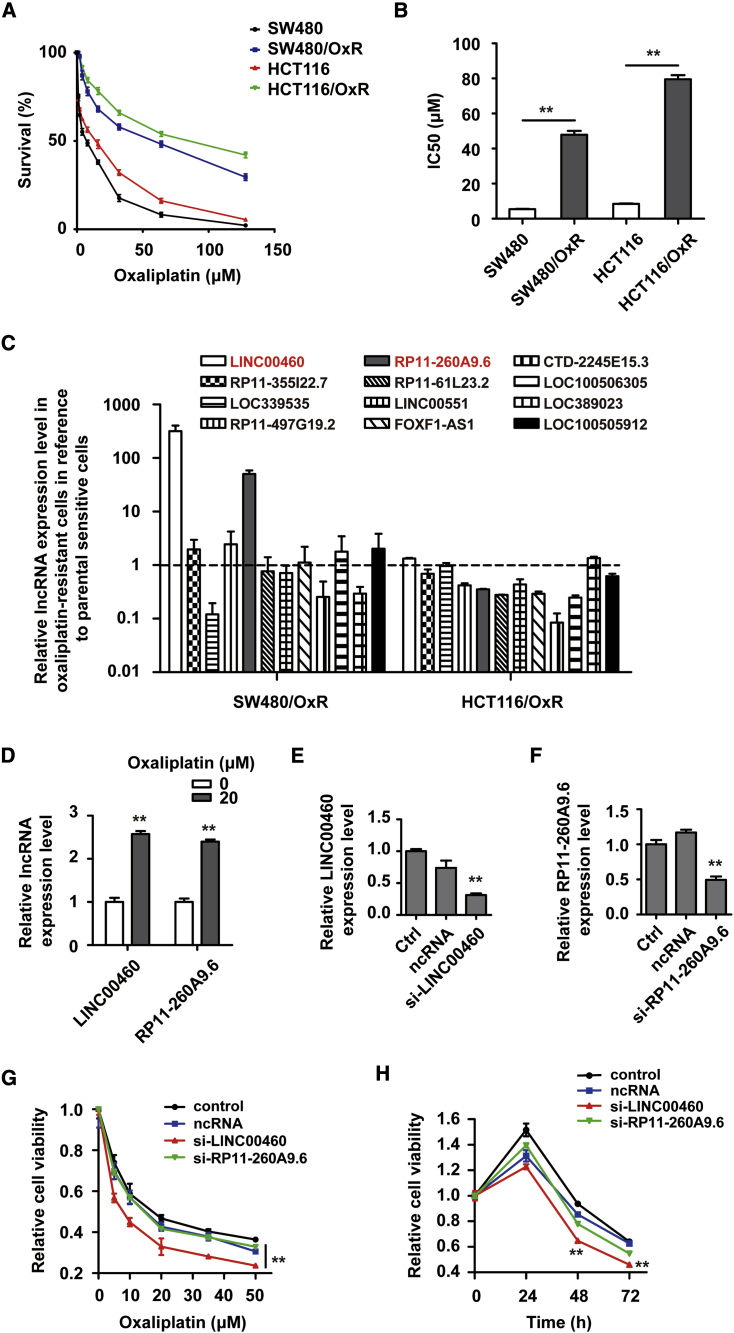
To investigate the role of lncRNAs in oxaliplatin resistance in CRC, we established oxaliplatin-resistant CRC (CRC/OxR) cell lines including SW480/OxR (MUT p53) and HCT116/OxR (wild-type [WT] p53) as described in the Materials and Methods. The resistance of SW480/OxR and HCT116/OxR cells to oxaliplatin was validated by measuring cell survival rate (Figure 1A) and half-maximal inhibitory concentration (IC50) (Figure 1B) in SW480/OxR, HCT116/OxR, and parental SW480 and HCT116 cell lines following oxaliplatin treatment. Differential expressions of lncRNAs, which may be associated with chemoresistance, were detected by quantitative real-time PCR in SW480/OxR and HCT116/OxR cells compared with parental sensitive ones. The data showed that LINC00460 and RP11-260A9.6 were significantly upregulated in SW480/OxR cells (>10-fold change), but not in HCT116/OxR cells (Figure 1C). Furthermore, we found that the expression levels of LINC00460 and RP11-260A9.6 in SW480/OxR cells upon oxaliplatin treatment were elevated (Figure 1D). To investigate the role of LINC00460 and RP11-260A9.6 in the chemoresistance of CRC, we knocked down the expressions of LINC00460 and RP11-260A9.6 in SW480/OxR cells using specific lncRNA smart silencers, validated by quantitative real-time PCR (Figures 1E and 1F). Cell Counting Kit-8 (CCK-8) assays showed that the knockdown of LINC00460 displayed a dose-dependent and time-dependent inhibition effect with significance on SW480/OxR cell survival following oxaliplatin treatment compared with controls, while this effect was not significant in RP11-260A9.6 knockdown cells (Figures 1G and 1H). Meanwhile, we silenced LINC00460 effectively in HCT116/OxR cells but found little effect on the cell viability following oxaliplatin treatment (Figure S1). These results indicate that LINC00460 is responsible for oxaliplatin resistance in CRC with TP53 mutations.
Figure 1.
LINC00460 Is Upregulated in SW480/OxR Cells and Is Responsible for Oxaliplatin Resistance in CRC
(A) Cell survival rates in SW480, SW480/OxR, HCT116, and HCT116/OxR cells treated with an increasing dose (0–128 μM) of oxaliplatin for 48 h. (B) The half-maximal inhibitory concentration (IC50) was calculated from (A) using GraphPad Prism v5.0 software. (C) Differential expressions of lncRNAs in SW480/OxR and HCT116/OxR cells relative to the parental-sensitive cells were determined by quantitative real-time PCR. (D) The expression levels of LINC00460 and RP11-260A9.6 in SW480/OxR cells after 20 μM oxaliplatin treatment for 48 h were compared with those from untreated cells. (E and F) Relative expression level of LINC00460 (E) or RP11-260A9.6 (F) in SW480/OxR cells transfected with si-LINC00460 or si-RP11-260A9.6. (G) CCK-8 assays of cell viability in LINC00460 or RP11-260A9.6 knockdown cells compared with the control cells following oxaliplatin treatment (5, 10, 20, 35, and 50 μM) for 48 h. (H) CCK-8 assays of cell viability in LINC00460 or RP11-260A9.6 knockdown cells following 20 μM oxaliplatin treatment for 24, 48, and 72 h. Data were shown as the mean ± SD; ∗∗p < 0.01.
LINC00460 Is Mainly Localized to the Cytoplasm and Is Associated with AGO2 in SW480/OxR Cells
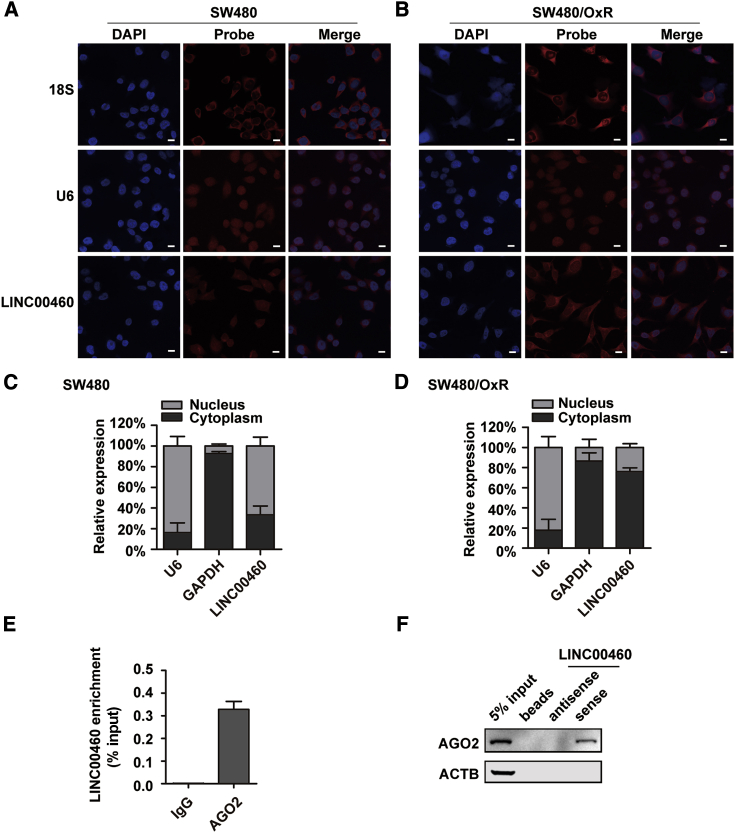
To explore the potential mechanism of LINC00460 for oxaliplatin resistance in CRC, we first performed fluorescence in situ hybridization (FISH) experiments in SW480 and SW480/OxR cells using Cy3-labeled probes that specifically recognized LINC00460. Interestingly, the results showed that LINC00460 was distributed throughout the SW480 cells, whereas it was predominantly localized to the cytoplasm in SW480/OxR cells (Figures 2A and 2B). These observations were confirmed by quantitative real-time PCR of LINC00460 following nucleus/cytoplasm separation (Figures 2C and 2D). The data suggest that induced expression of LINC00460 by oxaliplatin in SW480/OxR cells was mostly in the cytoplasm. Recently, lncRNAs have been reported to act as competing endogenous RNAs (ceRNAs), competitively bind miRNAs in vitro, and subsequently affect the expression of miRNA targets.23,24 The ceRNA network is dependent on AGO2, which physically interacts with miRNAs and ceRNAs.25 As oxaliplatin-induced LINC00460 was located in the cytoplasm, it might function as a ceRNA. To test this hypothesis, we conducted an anti-AGO2 RNA immunoprecipitation (RIP) experiment, which showed that LINC00460 was significantly enriched in the AGO2-immunoprecipitated complex of SW480/OxR cells (Figure 2E). RNA pull-down assays followed by western blotting also revealed the specific association of AGO2 with LINC00460 (Figure 2F). The protein band presented only in the LINC00460 sample but not in the antisense RNA or beads-only controls.
Figure 2.
LINC00460 Is Mainly Localized to the Cytoplasm and Is Associated with AGO2 in SW480/OxR Cells
(A and B) FISH analysis of the distribution of LINC00460 in SW480 (A) and SW480/OxR (B) cells. LINC00460 was recognized by specific Cy3-labeled probes (red), followed by nucleic staining with DAPI (blue). 18S rRNA and U6 snRNA were used as cytoplasmic and nuclear markers, respectively. Scale bars, 10 μm. (C and D) Cell nucleus/cytoplasm fractionation followed by quantitative real-time PCR displayed the cellular distribution of LINC00460 in SW480 (C) and SW480/OxR (D) cells. (E) Anti-AGO2 RIP followed by quantitative real-time PCR to determine the association between LINC00460 and AGO2 in SW480/OxR cells. The RNA levels in the immunoprecipitation samples were normalized to the input samples. IgG was an isotype control. (F) Lysates from SW480/OxR cells were incubated with in vitro-synthesized, biotin-labeled sense or antisense LINC00460 for RNA pull-down followed by western blotting analysis of AGO2 protein. ACTB served as a control. Data were shown as the mean ± SD.
LINC00460 Functions as a ceRNA and Sponges miR-149-5p/miR-150-5p
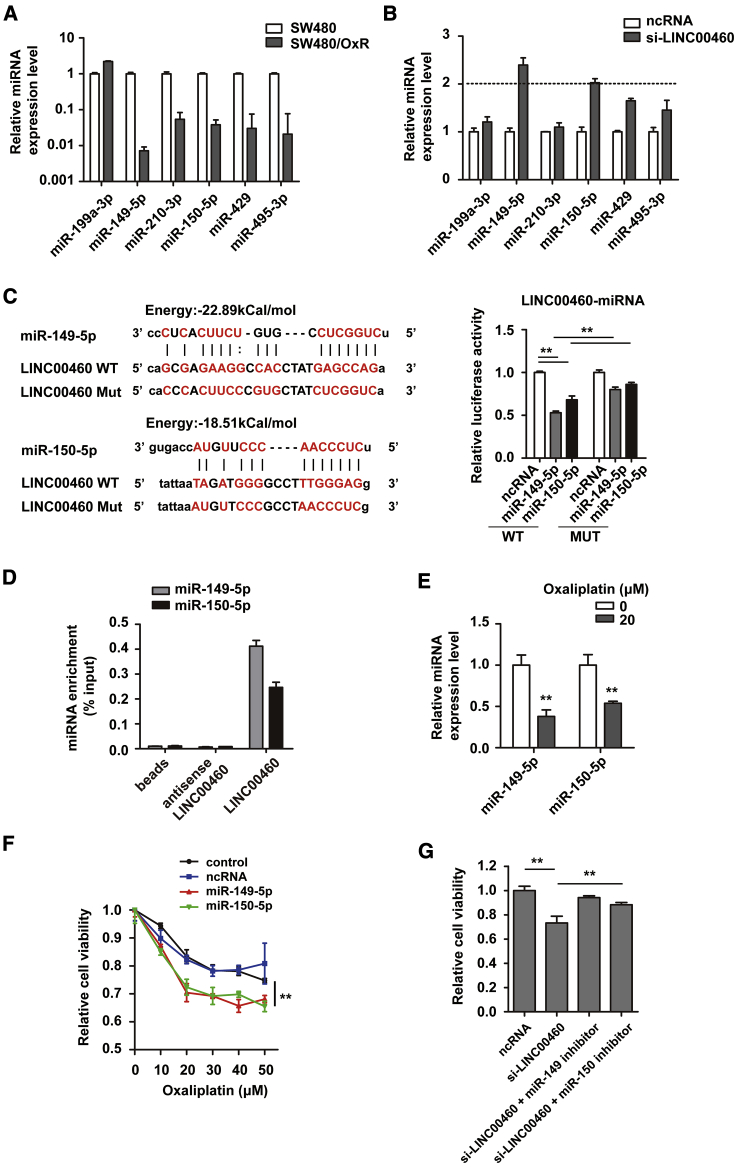
We predicted several miRNA target sites on LINC00460 by bioinformatics (lncRNASNP2 database, http://bioinfo.life.hust.edu.cn/lncRNASNP/#!/). Among these potential miRNA targets, miR-149-5p and miR-150-5p exhibited lower expression in SW480/OxR cells than in SW480 cells, and they were upregulated when LINC00460 was silenced in SW480/OxR cells (2-fold threshold) (Figures 3A and 3B). The direct interaction between LINC00460 and miR-149-5p/miR-150-5p was verified by luciferase reporter assays (Figure 3C) and RNA pull-down assays (Figure 3D). We further found that miR-149-5p/miR-150-5p were downregulated in SW480/OxR cells with the addition of oxaliplatin by quantitative real-time PCR (Figure 3E). CCK-8 assays showed that overexpression of miR-149-5p/miR-150-5p increased the sensitivity of SW480/OxR cells to oxaliplatin compared with controls (Figure 3F). The attenuated resistance by LINC00460 knockdown in SW480/OxR cells was restored with the supplement of miR-149-5p/miR-150-5p inhibitors (Figure 3G). These data demonstrate that LINC00460 may function as a ceRNA that physically interacts with miR-149-5p/miR-150-5p.
Figure 3.
LINC00460 Functions as a Competing Endogenous RNA and Sponges miR-149-5p/miR-150-5p
(A) Levels of predicted LINC00460-targeting miRNAs in SW480/OxR cells compared with SW480 cells. (B) Levels of predicted miRNAs in SW480/OxR cells when LINC00460 was knocked down. (C) Schematic diagram of putative base pairing between LINC00460 and miR-149-5p/miR-150-5p. Luciferase activity was measured in SW480/OxR cells co-transfected with pmirGLO-LINC00460-WT or pmirGLO-LINC00460-MUT and miR-149-5p/miR-150-5p mimics. (D) RNA pull-down assay followed by quantitative real-time PCR for detection of LINC00460-binding miRNAs. The miRNA levels in the pull-down samples were normalized to the input samples. (E) Expression levels of miR-149-5p and miR-150-5p in SW480/OxR cells after 20 μM oxaliplatin treatment compared with those of untreated cells. (F) SW480/OxR cells were transfected with miR-149-5p/miR-150-5p mimics, and cell viability following oxaliplatin treatment (10, 20, 30, 40, and 50 μM) compared with controls was assessed by CCK-8 assays. (G) The viability of SW480/OxR cells transfected with ncRNA, si-LINC00460, si-LINC00460 + miR-149-5p inhibitor, or miR-150-5p inhibitor and treated with 20 μM oxaliplatin was measured by CCK-8. Data were shown as the mean ± SD; ∗∗p < 0.01.
LINC00460 Promotes Oxaliplatin Resistance through Competitively Binding miR-149-5p/miR-150-5p and Upregulating the Expression of Corresponding miRNA target p53
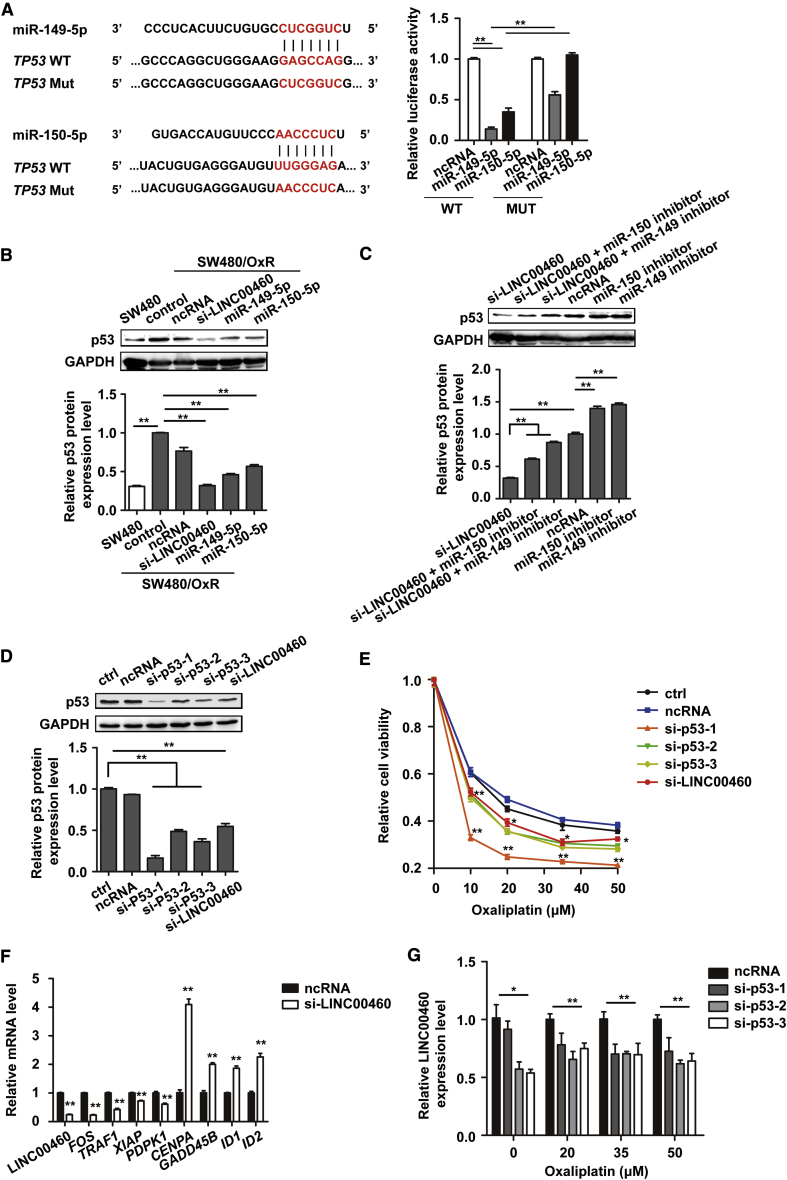
p53 was predicted to be a potential miR-149-5p/miR-150-5p target in SW480/OxR cells by TargetScan (http://www.targetscan.org/vert_72/), and luciferase reporter assays confirmed the direct binding of miR-149-5p/miR-150-5p with the 3′ untranslated region (UTR) of the TP53 mRNA (Figure 4A). Western blotting analysis further showed that the expression level of p53 protein was much higher in SW480/OxR cells than in parental SW480 cells; LINC00460 silencing, as well as miR-149-5p/miR-150-5p overexpression, reduced the expression of p53 in SW480/OxR cells to the level in SW480 cells (Figure 4B). This reduction in p53 protein level by silencing LINC00460 in SW480/OxR cells was restored by the action of miR-149-5p/miR-150-5p inhibitors (Figure 4C). These results demonstrate that LINC00460 promotes oxaliplatin resistance through competitively binding miR-149-5p/miR-150-5p and upregulating the expression of corresponding miRNA target p53 in vitro.
Figure 4.
LINC00460-miR-149-5p/miR-150-5p-Mutant p53 Feedback Loop Promoted Oxaliplatin Resistance in SW480/OxR Cells
(A) Schematic diagram of putative base pairing between miR-149-5p/miR-150-5p and the 3′ UTR of TP53 mRNA. Luciferase activity was measured in SW480/OxR cells co-transfected with pmirGLO-p53-3′ UTR-WT or pmirGLO-p53-3′ UTR-MUT and miR-149-5p/miR-150-5p mimics. (B) p53 protein levels in SW480 cells and in SW480/OxR cells transfected with ncRNA, si-LINC00460, miR-149-5p mimics, and miR-150-5p mimics were detected by a western blotting assay. GAPDH was used as a loading control. Normalized p53 protein level in these groups was compared with the SW480/OxR control group, as shown in the lower panel. (C) p53 protein levels in SW480/OxR cells transfected with ncRNA, si-LINC00460, si-LINC00460 + miR-150-5p inhibitor, miR-149-5p inhibitor, miR-150-5p inhibitor, or miR-149-5p inhibitor. Quantification of p53 was normalized by setting the ncRNA group as 1. (D) Effective knockdown of p53 by specific siRNAs (si-p53-1, -2, -3) was verified by western blotting. (E) CCK-8 assays of cell viability following oxaliplatin treatment in SW480/OxR cells with knockdown of p53 (si-p53-1, -2, -3) or with LINC00460. (F) mRNA expression level of genes, associated with mutant p53 and platinum resistance, was determined by quantitative real-time PCR in SW480/OxR cells after LINC00460 knockdown. (G) The expression levels of LINC00460 in SW480/OxR cells transfected with p53 siRNAs (si-p53-1, -2, -3) and treated with oxaliplatin (0, 20, 35, 50 μM) for 24 h were detected by quantitative real-time PCR. Data were shown as the mean ± SD; ∗p < 0.05, ∗∗p < 0.01.
LINC00460-MUT p53 Forms a Positive Feedback Loop in SW480/OxR Cells to Confer Oxaliplatin Resistance
Furthermore, we knocked down p53 in SW480/OxR cells by small interfering RNAs (siRNAs) against p53 (si-p53-1, -2, -3), which were verified by western blotting (Figure 4D). Then, the viability of SW480/OxR cells following oxaliplatin treatment in p53 or LINC00460 knockdown cells were examined by CCK-8 assays. The data showed that knockdown of p53 or LINC00460 enhanced the sensitivity to oxaliplatin in SW480/OxR cells (Figure 4E). The effective knockdown of p53 was also performed in HCT116/OxR cells. However, HCT116/OxR cells did not regain sensitivity to oxaliplatin after p53 knockdown; on the contrary, the resistance increased a little (Figure S2). We hypothesized that the effect of p53 knockdown on the sensitivity of HCT116/OxR cells and SW480/OxR cells to oxaliplatin varies with the status of p53. WT p53 in HCT116/OxR cells and MUT p53 in SW480/OxR cells (R273H/P309S mutations) were verified by sequencing the regions corresponding to TP53 mutations in SW480 (Figure S3). MUT p53 may function as a transcription factor with distinct targets. A list of gene promoters are shown to be potential targets of MUT p53 by chromatin immunoprecipitation (ChIP)-on-chip analysis, suggesting that MUT p53 could transcriptionally regulate these genes.26, 27, 28 As the MUT p53 protein level was positively correlated with LINC00460, we speculated that expression of MUT p53 target genes would be influenced upon LINC00460 knockdown. Therefore, we performed RNA sequencing to profile the gene expression of LINC00460-knockdown SW480/OxR cells compared with control cells. The differential expression of several MUT p53 target genes (FOS, TRAF1, XIAP, CENPA, GADD45B, ID1, ID2) was validated by quantitative real-time PCR (Figure 4F). We found that platinum resistance-related genes (XIAP, PDPK1) were downregulated while pro-apoptosis and aging-related genes (GADD45B, ID1, ID2) were upregulated in SW480/OxR cells after LINC00460 knockdown. Additionally, a p53 transcriptional factor binding site was predicted in the promoter region of LINC00460 by the PROMO database. Thus, we hypothesized that p53 might regulate the expression of LINC00460 as well. Quantitative real-time PCR results showed that the content of LINC00460 was significantly reduced by p53 knockdown in SW480/OxR cells following treatment with a gradient concentration of oxaliplatin for 24 h (Figure 4G). These data suggest that MUT p53 may function as a transcriptional activator to induce the expression of LINC00460, and that LINC00460 in turn promotes p53 expression through competitively binding miR-149-5p/miR-150-5p, thus forming a positive feedback loop that mediates oxaliplatin resistance in SW480/OxR cells.
Knockdown of LINC00460 Sensitizes SW480/OxR Cells to Oxaliplatin by Modulating p53 In Vivo
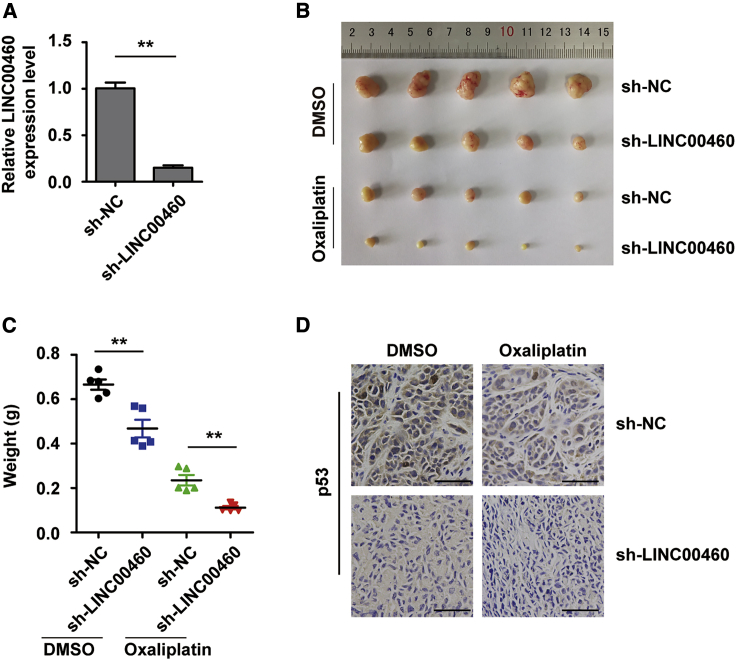
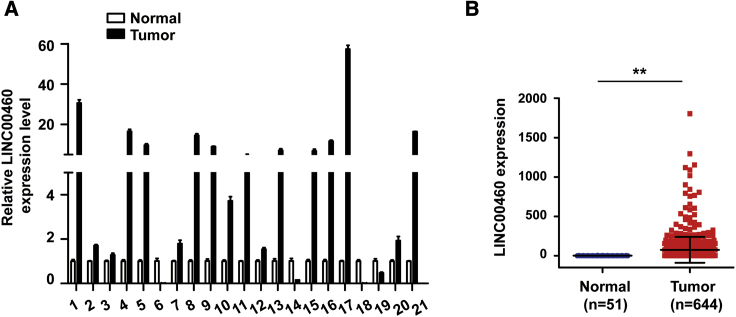
To explore whether LINC00460 mediates oxaliplatin resistance by modulating the expression of p53 in vivo, we used a xenograft mouse model. Briefly, LINC00460-knockdown SW480/OxR cells were established using short hairpin (sh-)LINC00460 lentiviruses and validated by quantitative real-time PCR compared with sh-negative control (NC) cells (Figure 5A). The cells were then subcutaneously injected into the right flanks of nude mice. On day 28 after inoculation, the tumor volume and weight were measured to be smaller and lighter in the sh-LINC00460 group than in the sh-NC group, especially when oxaliplatin was administered (Figures 5B and 5C). Immunohistochemistry (IHC) analysis further displayed that the expression level of p53 was much lower in the sh-LINC00460 group than in the sh-NC group (Figure 5D). Additionally, we determined the expression of LINC00460 in 21 pairs of CRC tissues and paired normal tissues and found that LINC00460 was upregulated in CRC tissues (Figure 6A). The clinical characteristics of the patients are listed in Table 1. LINC00460 expression was significantly associated with clinical stage (p = 0.008) and node (N) status (p = 0.033). LINC00460 expression data downloaded from The Cancer Genome Atlas (TCGA) database also showed that LINC00460 was upregulated in human CRC tissues compared with normal tissues (Figure 6B). Based on all of the findings above, we proposed a schematic model of LINC00460 regulating oxaliplatin resistance in CRC (Figure 7).
Figure 5.
Knockdown of LINC00460 Sensitizes SW480/OxR Cells to Oxaliplatin by Modulating Mutant p53 In Vivo
(A) Quantitative real-time PCR validation of LINC00460 knockdown in SW480/OxR cells infected with sh-LINC00460 lentiviruses. (B and C) Tumors generated by xenograft sh-NC or sh-LINC00460 cells treated with or without oxaliplatin on day 28 were dissected, photographed (B), and weighed (C). (D) IHC analysis of tumor p53 expression in the LINC00460 knockdown group compared with the control group. Scale bars, 50 μm. Data were shown as the mean ± SD; ∗∗p < 0.01.
Figure 6.
LINC00460 Expression Is Higher in CRC Tissues
(A) Quantitative real-time PCR of LINC00460 expression normalized by GAPDH mRNA in 21 cases of CRC tissues (tumor) compared with paired normal tissues (normal). (B) Data of LINC00460 expression level in CRC tissues and normal tissues were downloaded from TCGA database. Data were shown as the mean ± SD; ∗∗p < 0.01.
Table 1.
Association between Clinical Features and LINC00460 Expression
| Variable | n = 21 | LINC00460 Expression |
p Valueb | |
|---|---|---|---|---|
| High (n = 11)a | Low (n = 10) | |||
| Age | ||||
| <60 | 13 | 6 | 7 | 0.659 |
| ≥60 | 8 | 5 | 3 | |
| Sex | ||||
| Male | 12 | 7 | 5 | 0.670 |
| Female | 9 | 4 | 5 | |
| Cancer Subtype | ||||
| Colon | 16 | 8 | 8 | 1.000 |
| Rectal | 5 | 3 | 2 | |
| Clinical Stage | ||||
| II | 12 | 3 | 9 | 0.008∗∗ |
| III | 9 | 8 | 1 | |
| T Status | ||||
| T1+T2 | 3 | 2 | 1 | 1.000 |
| T3+T4 | 18 | 9 | 9 | |
| N Status | ||||
| N0 | 13 | 4 | 9 | 0.033∗ |
| N1 | 4 | 3 | 1 | |
| N2 | 4 | 4 | 0 | |
The median expression level was used as the cutoff. Low expression of LINC00460 in 10 patients was classified as values below the 50th percentile. High LINC00460 expression in 11 patients was classified as values at or above the 50th percentile.
For analysis of association between LINC00460 levels and clinical features, Fisher’s exact tests were used. ∗p < 0.05, ∗∗p < 0.01, for association among the variables.
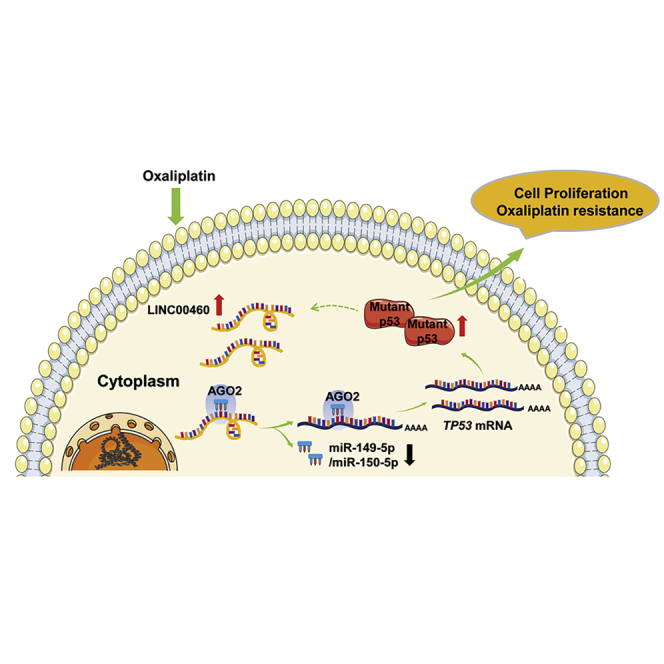
Figure 7.
A Proposed Schematic Model of LINC00460 Regulating Oxaliplatin Resistance in CRC
Discussion
Currently, lncRNAs have been reported as key regulators in tumorigenesis and progression, including cell proliferation and invasion, DNA damage response, and apoptosis.29, 30, 31 As an oncogenic lncRNA, LINC00460 is found to be upregulated in non-small-cell lung cancer (NSCLC) and esophageal squamous cell carcinoma (ESCC).32,33 In this study, we report that LINC00460 is significantly upregulated in oxaliplatin-resistant SW480 cells and promotes oxaliplatin resistance in vitro and in vivo. We further found that LINC00460 mediates oxaliplatin resistance by competitively binding miR-149-5p/miR-150-5p and thus upregulates the expression of MUT p53 in SW480/OxR cells. Additionally, LINC00460 expression was downregulated after p53 knockdown in SW480/OxR cells, suggesting that LINC00460-miR-149-5p/miR-150-5p-MUT p53 form a positive feedback loop that is responsible for oxaliplatin resistance in CRC.
Recent studies have reported the ceRNA-crosstalk network where lncRNAs function via competitively binding miRNAs and further regulate the expression of downstream proteins.34,35 Herein, we found that oxaliplatin-induced LINC00460 in SW480/OxR cells was mainly localized to the cytoplasm and was associated with AGO2 protein, suggesting that LINC00460 may function as a miRNA sponge. By bioinformatics analysis, luciferase reporter assays, and quantitative real-time PCR, we verified that miR-149-5p/miR-150-5p were physically associated with LINC00460, as well as TP53 mRNA. Furthermore, we observed that the reduced resistance by LINC00460 knockdown in SW480/OxR cells was restored by miR-149-5p/miR-150-5p inhibitors, indicating that LINC00460 functions as a ceRNA to mediate oxaliplatin resistance through sequestering miR-149-5p/miR-150-5p.
In the present study, p53 was identified as a target of miR-149-5p/miR-150-5p, competing with LINC00460 for miR-149-5p/miR-150-5p binding sites. The tumor suppressor gene TP53 plays a critical role in multiple biological processes, including proliferation, cell cycle arrest, DNA replication and repair, apoptosis, cellular stress response, as well as in tumor suppression.36,37 However, mutations in the TP53 gene are the most frequent genetic alterations found in more than 50% of human cancers.38 Most TP53 mutations in cancers are missense mutations, leading to the expression of full-length MUT p53 protein.39 Many tumor-associated MUT p53 proteins not only lose their original tumor suppressive functions, but they also acquire new functions (“gain of function”) that promote tumorigenesis and progression independent of WT p53.40,41 In our study, compared with parental oxaliplatin-sensitive ones, the upregulated expression of LINC00460 in the CRC cell line SW480/OxR, but not HCT116/OxR, depends on MUT p53. In addition, knockdown of LINC00460 or p53 increased the sensitivity of SW480/OxR cells to oxaliplatin, while similar effects were not observed in HCT116/OxR cells (WT p53).
p53 works primarily as a transcriptional activator, inducing the expression of its downstream target genes.36,37 MUT p53 can regulate the expression of downstream targets that differ from WT p53. In this study, we examined the expression of genes related to MUT p53 and platinum drug resistance in LINC00460-knockdown SW480/OxR cells compared with control cells. The differential expression of several MUT p53 target genes was validated by quantitative real-time PCR (Figure 4F). For example, the expression of growth arrest and DNA damage-inducible beta (GADD45B), which participates in the DNA damage response of the p53 signaling pathway and promotes apoptosis, was upregulated in SW480/OxR cells after LINC00460 knockdown. Aging-related gene ID2, the promoter of which responds to MUT but not WT p53,42 was also upregulated. Meanwhile, platinum-resistant genes (XIAP, PDPK1) were downregulated. Additionally, by bioinformatics analysis, we hypothesized that LINC00460 might be a target of p53. When p53 was knocked down in SW480/OxR cells, the expression of LINC00460 was reduced as expected (Figure 4G). These data indicate that LINC00460-MUT p53 forms a positive feedback in SW480/OxR cells to confer oxaliplatin resistance.
Collectively, our results show that LINC00460 is upregulated in SW480/OxR cells and CRC tissues, and that it is responsible for oxaliplatin resistance of CRC with TP53 mutations. We propose the functional mechanism for LINC00460 (Figure 7). Oxaliplatin-induced LINC00460 in the cytoplasm promotes MUT p53 expression through competitively binding miR-149-5p/miR-150-5p, and MUT p53 in turn induces the expression of LINC00460, thus forming a positive feedback loop that drives oxaliplatin resistance in SW480/OxR cells. Our findings report the LINC00460-miR-149-5p/miR-150-5p-MUT p53 feedback loop in CRC and provide potential therapeutic targets for tumor chemoresistance.
Materials and Methods
Clinical Tissue Samples
Twenty-one samples of CRC tissues and paired normal tissues were obtained from patients undergoing surgery at the Drum Tower Hospital Affiliated to the Medical School of Nanjing University (Nanjing, China). Written consent was obtained from all patients, and all experiments in this study were conducted in accordance with the Chinese Ethical Regulations and Guidelines approved by the Ethics Committee of Nanjing University. Clinical stage and the tumor-node-metastasis (TNM) classification in all 21 cases were determined by diagnosis and histological analysis. During surgery, the tissue samples were immediately frozen in liquid nitrogen and stored at −80°C. The clinical characteristics of the patients are listed in Table 1.
Cell Culture and Establishment of CRC/OxR Cell Lines
SW480 and HCT116 cell lines were purchased from the Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Science, Chinese Academy of Sciences (Shanghai, China). All cells were cultured in RPMI 1640 medium (Gibco, CA, USA) supplemented with 10% fetal bovine serum (FBS, Gibco) and 100 U/mL of penicillin and streptomycin (Gibco) in a cell incubator at 37°C with 5% CO2.
CRC/OxR cell lines including SW480/OxR and HCT116/OxR were established by intermittently and gradually increasing the concentration of oxaliplatin in the culture medium of parental oxaliplatin-sensitivity CRC cell lines (SW480 and HCT116) for a 10-month period as described previously.43 To maintain the oxaliplatin resistance of the CRC/OxR cell lines, oxaliplatin at a final concentration of 2 μM was added to the culture medium until 1 month before the study began. CRC/OxR cells were used at no more than 10 passages following their establishment.
Mimics or Inhibitors of miRNA and siRNA Oligonucleotides
Synthetic miR-149-5p, miR-150-5p mimics or inhibitors, siRNAs against p53 (si-p53-1, -2, -3), and the scrambled NC RNAs (ncRNAs) were purchased from RiboBio (Guangzhou, China). lncRNA Smart Silencer for LINC00460 (si-LINC00460), RP11-260A9.6 (si-RP11-260A9.6), and the corresponding ncRNAs were synthesized by RiboBio. Cell transfection was performed using the Lipofectamine 2000 kit (Invitrogen, CA, USA) as instructed by the supplier.
CCK-8 Assays
Cell viability was determined using CCK-8 assays (CK04-500, Dojindo, Japan) following the manufacturer’s instructions. To detect the cell viability in the presence of oxaliplatin at different concentrations, cells were seeded into 96-well plates at a density of 1 × 104 cells/well, followed by exposure to a gradient concentration of oxaliplatin for 48 h. To detect the cell viability at different time points, cells were seeded into 96-well plates at a density of 5 × 103 cells/well followed by exposure to oxaliplatin at 20 μM, and they were cultivated for 24, 48, and 72 h. Then, 100 μL/well RPMI 1640 medium containing 10% CCK-8 was added to the above 96-well plates for a 1-h incubation. The optical density (OD) was then measured at 450 nm.
RNA Extraction and Quantitative Real-Time PCR
Total RNA was isolated from cultured cells and tissue samples by TRIzol reagent (Invitrogen, CA, USA) according to the manufacturer’s instructions. To quantify mature miRNAs, quantitative real-time PCR was performed using TaqMan miRNA assay probes (Applied Biosystems, Foster City, CA, USA), a TaqMan PCR kit, and an Applied Biosystems 7500 sequence detection system. To quantify lncRNAs and mRNAs, total RNA was converted to cDNA using oligo(dT) and random primers (Takara, Japan), followed by PCR using SYBR Green (Takara) and gene-specific primers. All of the reactions were run in triplicate. The expression levels of miRNAs were normalized to U6 small nuclear RNA (snRNA), while those of lncRNAs and mRNAs were normalized to GAPDH mRNA using the 2−ΔΔCt method. The primers used for quantitative real-time PCR were provided in Table S1.
Nuclear and Cytoplasmic Separation
The nuclear and cytoplasmic fractions from SW480 and SW480/OxR cells were isolated by a Paris kit (Invitrogen). The total, nuclear, and cytoplasmic RNAs were extracted using TRIzol to detect the content of U6, GAPDH, and LINC00460 by quantitative real-time PCR.
FISH Assay
FISH experiments were performed using Cy3 fluorescence-labeled probes specific for LINC00460, U6 snRNA (nuclear marker), and 18S rRNA (cytoplasmic marker), which were designed and synthesized by RiboBio, and a Ribo FISH kit (RiboBio). Briefly, cells were fixed with 4% paraformaldehyde for 15 min, penetrated with 0.25% Triton X-100 for 5 min, and after pre-hybridization at 37°C for 30 min, cells were then in situ hybridized at 37°C overnight. After six washes with 1× PBS for 5 min, cell nuclei were stained with DAPI and images were taken with a confocal microscope.
RIP Assay
For anti-AGO2 RIP, SW480/OxR cell lysates were incubated with human anti-AGO2 antibody (ab57113, Abcam, Cambridge, UK) or control immunoglobulin G (IgG) (catalog no. 1265, BioVision, Milpitas, CA, USA) on a rotary shaker at 4°C overnight, followed by protein A/G-agarose bead (sc-2003, Santa Cruz, USA) incubation for 4 h. The immunoprecipitated RNA was examined by quantitative real-time PCR.
RNA Pull-Down Assay
RNA pull-down assays were carried out as described previously.44 Biotinylated LINC00460 or antisense RNA was incubated with SW480/OxR whole-cell lysates, which were prepared by briefly sonicating 10 million cells in 1 mL IP buffer (20 mM Tris [pH 7.4], 150 mM NaCl, 1% Nonidet P-40 [NP-40], 1 mM EDTA, 0.5 mM DTT, 1 mM NaF, protease inhibitors, and RNase inhibitor). After incubation, RNA-protein complexes were retrieved by streptavidin beads, washed five times in IP buffer, and eluted in SDS loading buffer for western blotting or TRIzol reagent for quantitative real-time PCR.
Plasmid Construction and Luciferase Reporter Assay
A fragment of LINC00460 or the 3′ UTR of TP53 mRNA containing predicted binding sites for miR-149-5p and miR-150-5p was inserted into pMIR-REPORT luciferase to construct WT luciferase reporter plasmids (pmirGLO-LINC00460-WT and pmirGLO-p53-3′ UTR-WT). Additionally, potential binding sites were mutated into complementary sequences to construct MUT luciferase reporter plasmids (pmirGLO-LINC00460-MUT and pmirGLO-p53-3′UTR-MUT). All of the pMIR-REPORT luciferase plasmids and β-galactosidase (β-gal) control plasmid were provided by GenScript (Nanjing, China).
For luciferase reporter assays, 1.0 μg of firefly luciferase reporter plasmid, 1.0 μg of β-gal expression plasmid, and 100 pmol of miR-149-5p, miR-150-5p mimics, or ncRNAs were co-transfected into SW480/OxR cells seeded in 24-well plates. 24 h later, cells were lysed and the firefly luciferase activities were detected using a luciferase reporter assay kit (Promega, Madison, WI, USA). Relative luciferase activity was normalized to β-gal.
Western Blotting
Cells were lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, Shanghai, China) on ice for 30 min. After centrifugation at 16,000 × g for 10 min, the supernatant was quantified for protein concentration using a bicinchoninic acid (BCA) kit (Pierce, CA, USA). Total cell lysates supplemented with 5× loading buffer were boiled at 99°C for 5 min. Equal amounts of protein were separated by a SDS-PAGE gel and blotted onto a polyvinylidene fluoride (PVDF) membrane. The membrane was then incubated with specific primary antibodies for p53 (1:1,000, 21891-1-AP, Proteintech, USA) and GAPDH (1:2,000, MA5-15738, Invitrogen, USA), followed by secondary horseradish peroxidase (HRP)-conjugated antibodies. For detection of LINC00460-binding proteins in RNA pull-down assays, antibodies for AGO2 (1:1,000, #2897, Cell Signaling Technology, USA) and ACTB (1:2,000, MA5-15739, Invitrogen, USA) were used. After washes with 1× Tris-buffered saline with Tween 20 (TBST), blots were reacted with enhanced chemiluminescence (ECL) reagent (Pierce) and protein bands were detected by a chemiluminescence system (Tanon-5200, Shanghai, China).
Lentivirus Infection and Stable Cell Line Establishment
sh-LINC00460 lentiviruses were generated by GenePharma (Shanghai, China). According to the manufacturer’s instructions, SW480/OxR cells were seeded at 1 × 105 per well on six-well plates, and 100 μL of sh-LINC00460 lentiviruses or sh-NC lentiviruses was added to 900 μL of RPMI 1640 medium supplemented with 2% inactivated FBS and 5 μg/mL Polybrene. 24 h later, 1 mL of RPMI 1640 medium containing 2% inactivated FBS was supplemented to six-well plates, and 48 h post-infection, transduced cells were selected by 1 μg/mL puromycin for 48 h. Stable cell lines were established by adding 1 μg/mL puromycin to the culture medium for three generations. The efficient knockdown of LINC00460 by sh-LINC00460 lentiviruses was verified by quantitative real-time PCR.
Nude Mouse Xenograft Model and IHC Analysis
5-week-old BALB/c nude mice were purchased from GemPharmatech (Nanjing, China) and housed under specific pathogen-free (SPF) conditions. All animal experimental procedures were approved by the Animal Care and Use Committee of Nanjing University. Mice were randomly divided into four groups (n = 5 per group) and injected subcutaneously with 5 × 106 SW480/OxR cells stably expressing sh-LINC00460 or sh-NC, followed by intraperitoneal injection of oxaliplatin at 5 mg/kg/day or DMSO twice a week when xenograft tumor volume reached 200 mm3. On day 28 post-inoculation, tumors were dissected, weighed, and subjected to IHC analysis of p53 protein. Briefly, the formalin-fixed, paraffin-embedded samples were sliced, dewaxed, and rehydrated, followed by incubation with anti-p53 (1:200, 21891-1-AP, Proteintech, USA) antibody and secondary antibody. After that, samples were incubated with diaminobenzidine (DAB) (Dako, USA) and restained with hematoxylin (Sigma, USA).
Statistical Analysis
Data from three independent experiments are shown as the mean ± the standard deviation (SD). GraphPad Prism v5.0 software and SPSS 25.0 statistical software were used for statistical analysis. The associations between LINC00460 expression and clinical features of CRC patients were determined using the Fisher’s exact test. Statistical differences between groups were analyzed using a Student’s t test or ANOVA. A p value <0.05 was considered statistically significant (∗p < 0.05, ∗∗p < 0.01).
Author Contributions
C.W., X.M., and J.Y. conceived and designed the experiments; X.M., W.S., J.Y., Y.Z., Y.G., J.H., and L.Z. performed the experiments; C.W., X.J., X.M., and J.Y. analyzed the data; W.S. provided the clinical samples; C.W. and X.M. wrote the paper. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31601057) and the Fundamental Research Funds for the Central Universities (14380138).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.10.018.
Supplemental Information
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Rejhová A., Opattová A., Čumová A., Slíva D., Vodička P. Natural compounds and combination therapy in colorectal cancer treatment. Eur. J. Med. Chem. 2018;144:582–594. doi: 10.1016/j.ejmech.2017.12.039. [DOI] [PubMed] [Google Scholar]
- 3.Kalofonos H.P., Aravantinos G., Kosmidis P., Papakostas P., Economopoulos T., Dimopoulos M., Skarlos D., Bamias A., Pectasides D., Chalkidou S. Irinotecan or oxaliplatin combined with leucovorin and 5-fluorouracil as first-line treatment in advanced colorectal cancer: a multicenter, randomized, phase II study. Ann. Oncol. 2005;16:869–877. doi: 10.1093/annonc/mdi193. [DOI] [PubMed] [Google Scholar]
- 4.Meyerhardt J.A., Mayer R.J. Systemic therapy for colorectal cancer. N. Engl. J. Med. 2005;352:476–487. doi: 10.1056/NEJMra040958. [DOI] [PubMed] [Google Scholar]
- 5.Martinez-Balibrea E., Martínez-Cardús A., Ginés A., Ruiz de Porras V., Moutinho C., Layos L., Manzano J.L., Bugés C., Bystrup S., Esteller M., Abad A. Tumor-related molecular mechanisms of oxaliplatin resistance. Mol. Cancer Ther. 2015;14:1767–1776. doi: 10.1158/1535-7163.MCT-14-0636. [DOI] [PubMed] [Google Scholar]
- 6.Goldberg R.M., Sargent D.J., Morton R.F., Fuchs C.S., Ramanathan R.K., Williamson S.K., Findlay B.P., Pitot H.C., Alberts S.R. A randomized controlled trial of fluorouracil plus leucovorin, irinotecan, and oxaliplatin combinations in patients with previously untreated metastatic colorectal cancer. J. Clin. Oncol. 2004;22:23–30. doi: 10.1200/JCO.2004.09.046. [DOI] [PubMed] [Google Scholar]
- 7.Fatica A., Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 8.Geisler S., Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou G., Chen X. Emerging role of extracellular microRNAs and lncRNAs. ExRNA. 2019;1:10. [Google Scholar]
- 10.Xue X., Yang Y.A., Zhang A., Fong K.W., Kim J., Song B., Li S., Zhao J.C., Yu J. lncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016;35:2746–2755. doi: 10.1038/onc.2015.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yue B., Cai D., Liu C., Fang C., Yan D. linc00152 functions as a competing endogenous RNA to confer oxaliplatin resistance and holds prognostic values in colon cancer. Mol. Ther. 2016;24:2064–2077. doi: 10.1038/mt.2016.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang Y., Cao W., Wu K., Qin X., Wang X., Li Y., Yu B., Zhang Z., Wang X., Yan M. lncRNA LINC00460 promotes EMT in head and neck squamous cell carcinoma by facilitating peroxiredoxin-1 into the nucleus. J. Exp. Clin. Cancer Res. 2019;38:365. doi: 10.1186/s13046-019-1364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lian Y., Yan C., Xu H., Yang J., Yu Y., Zhou J., Shi Y., Ren J., Ji G., Wang K. A novel lncRNA, LINC00460, affects cell proliferation and apoptosis by regulating KLF2 and CUL4A expression in colorectal cancer. Mol. Ther. Nucleic Acids. 2018;12:684–697. doi: 10.1016/j.omtn.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li K., Sun D., Gou Q., Ke X., Gong Y., Zuo Y., Zhou J.K., Guo C., Xia Z., Liu L. Long non-coding RNA linc00460 promotes epithelial-mesenchymal transition and cell migration in lung cancer cells. Cancer Lett. 2018;420:80–90. doi: 10.1016/j.canlet.2018.01.060. [DOI] [PubMed] [Google Scholar]
- 15.Baugh E.H., Ke H., Levine A.J., Bonneau R.A., Chan C.S. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ. 2018;25:154–160. doi: 10.1038/cdd.2017.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yue X., Zhang C., Zhao Y., Liu J., Lin A.W., Tan V.M., Drake J.M., Liu L., Boateng M.N., Li J. Gain-of-function mutant p53 activates small GTPase Rac1 through SUMOylation to promote tumor progression. Genes Dev. 2017;31:1641–1654. doi: 10.1101/gad.301564.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petitjean A., Mathe E., Kato S., Ishioka C., Tavtigian S.V., Hainaut P., Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 18.Aubrey B.J., Strasser A., Kelly G.L. Tumor-suppressor functions of the TP53 pathway. Cold Spring Harb. Perspect. Med. 2016;6:a026062. doi: 10.1101/cshperspect.a026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J. The cell-cycle arrest and apoptotic functions of p53 in tumor initiation and progression. Cold Spring Harb. Perspect. Med. 2016;6:a026104. doi: 10.1101/cshperspect.a026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rochette P.J., Bastien N., Lavoie J., Guérin S.L., Drouin R. SW480, a p53 double-mutant cell line retains proficiency for some p53 functions. J. Mol. Biol. 2005;352:44–57. doi: 10.1016/j.jmb.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 21.Dominijanni A., Gmeiner W.H. Improved potency of F10 relative to 5-fluorouracil in colorectal cancer cells with p53 mutations. Cancer Drug Resist. 2018;1:48–58. doi: 10.20517/cdr.2018.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han J., Li J., Tang K., Zhang H., Guo B., Hou N., Huang C. miR-338-3p confers 5-fluorouracil resistance in p53 mutant colon cancer cells by targeting the mammalian target of rapamycin. Exp. Cell Res. 2017;360:328–336. doi: 10.1016/j.yexcr.2017.09.023. [DOI] [PubMed] [Google Scholar]
- 23.Qu L., Ding J., Chen C., Wu Z.J., Liu B., Gao Y., Chen W., Liu F., Sun W., Li X.F. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29:653–668. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 24.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bosson A.D., Zamudio J.R., Sharp P.A. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol. Cell. 2014;56:347–359. doi: 10.1016/j.molcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freed-Pastor W.A., Prives C. Mutant p53: one name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dell’Orso S., Fontemaggi G., Stambolsky P., Goeman F., Voellenkle C., Levrero M., Strano S., Rotter V., Oren M., Blandino G. ChIP-on-chip analysis of in vivo mutant p53 binding to selected gene promoters. OMICS. 2011;15:305–312. doi: 10.1089/omi.2010.0084. [DOI] [PubMed] [Google Scholar]
- 28.Brosh R., Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat. Rev. Cancer. 2009;9:701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 29.Hu Y., Lin J., Fang H., Fang J., Li C., Chen W., Liu S., Ondrejka S., Gong Z., Reu F. Targeting the MALAT1/PARP1/LIG3 complex induces DNA damage and apoptosis in multiple myeloma. Leukemia. 2018;32:2250–2262. doi: 10.1038/s41375-018-0104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu P., Wu J., Wang Y., Zhu X., Lu T., Liu B., He L., Ye B., Wang S., Meng S. lncGata6 maintains stemness of intestinal stem cells and promotes intestinal tumorigenesis. Nat. Cell Biol. 2018;20:1134–1144. doi: 10.1038/s41556-018-0194-0. [DOI] [PubMed] [Google Scholar]
- 31.Zou Y., Xu S., Xiao Y., Qiu Q., Shi M., Wang J., Liang L., Zhan Z., Yang X., Olsen N. Long noncoding RNA LERFS negatively regulates rheumatoid synovial aggression and proliferation. J. Clin. Invest. 2018;128:4510–4524. doi: 10.1172/JCI97965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang Y., Wu Y., Chen X., Zhang S., Wang K., Guan X., Yang K., Li J., Bai Y. A novel long noncoding RNA linc00460 up-regulated by CBP/P300 promotes carcinogenesis in esophageal squamous cell carcinoma. Biosci. Rep. 2017;37 doi: 10.1042/BSR20171019. BSR20171019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yue Q.Y., Zhang Y. Effects of linc00460 on cell migration and invasion through regulating epithelial-mesenchymal transition (EMT) in non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2018;22:1003–1010. doi: 10.26355/eurrev_201802_14382. [DOI] [PubMed] [Google Scholar]
- 34.Bak R.O., Mikkelsen J.G. miRNA sponges: soaking up miRNAs for regulation of gene expression. Wiley Interdiscip. Rev. RNA. 2014;5:317–333. doi: 10.1002/wrna.1213. [DOI] [PubMed] [Google Scholar]
- 35.Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tokino T., Nakamura Y. The role of p53-target genes in human cancer. Crit. Rev. Oncol. Hematol. 2000;33:1–6. doi: 10.1016/s1040-8428(99)00051-7. [DOI] [PubMed] [Google Scholar]
- 37.Hafner A., Bulyk M.L., Jambhekar A., Lahav G. The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 2019;20:199–210. doi: 10.1038/s41580-019-0110-x. [DOI] [PubMed] [Google Scholar]
- 38.Hainaut P., Pfeifer G.P. Somatic TP53 mutations in the era of genome sequencing. Cold Spring Harb. Perspect. Med. 2016;6:a026179. doi: 10.1101/cshperspect.a026179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bouaoun L., Sonkin D., Ardin M., Hollstein M., Byrnes G., Zavadil J., Olivier M. TP53 variations in human cancers: new lessons from the IARC TP53 database and genomics data. Hum. Mutat. 2016;37:865–876. doi: 10.1002/humu.23035. [DOI] [PubMed] [Google Scholar]
- 40.Alexandrova E.M., Mirza S.A., Xu S., Schulz-Heddergott R., Marchenko N.D., Moll U.M. p53 loss-of-heterozygosity is a necessary prerequisite for mutant p53 stabilization and gain-of-function in vivo. Cell Death Dis. 2017;8:e2661. doi: 10.1038/cddis.2017.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alexandrova E.M., Yallowitz A.R., Li D., Xu S., Schulz R., Proia D.A., Lozano G., Dobbelstein M., Moll U.M. Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature. 2015;523:352–356. doi: 10.1038/nature14430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan W., Liu G., Scoumanne A., Chen X. Suppression of inhibitor of differentiation 2, a target of mutant p53, is required for gain-of-function mutations. Cancer Res. 2008;68:6789–6796. doi: 10.1158/0008-5472.CAN-08-0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howells L.M., Sale S., Sriramareddy S.N., Irving G.R., Jones D.J., Ottley C.J., Pearson D.G., Mann C.D., Manson M.M., Berry D.P. Curcumin ameliorates oxaliplatin-induced chemoresistance in HCT116 colorectal cancer cells in vitro and in vivo. Int. J. Cancer. 2011;129:476–486. doi: 10.1002/ijc.25670. [DOI] [PubMed] [Google Scholar]
- 44.Lan Y., Xiao X., He Z., Luo Y., Wu C., Li L., Song X. Long noncoding RNA OCC-1 suppresses cell growth through destabilizing HuR protein in colorectal cancer. Nucleic Acids Res. 2018;46:5809–5821. doi: 10.1093/nar/gky214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.