Highlight
-
•
In this study, we designed a novel detection system to detect DNA based on DNA G-quadruplex/hemin enzyme. It consisted of DNA probes with hemin that can form a special structure via self-assembly after paring with target DNA. The special structure containing hemin showed enzyme activity catalyzing substrates for coloration or luminescence.
-
•
Based on this chemiluminescence DNA biosensor system using DNA G-quadruplex/hemin enzyme, we designed a pair of DNA probes targeting β-actin DNA to detect circulating free DNA (cfDNA) in human serum from patients with gall bladder cancer (GBC), patients with cholecystitis, and healthy donors and calculated concentrations of cfDNA according to a standard curve.
-
•
In the serum of patients with GBC, the level of cfDNA was higher than those in the serum of patients with cholecystitis and healthy donors.
-
•
This is a convenient, economic, and promising approach to aid the diagnosis of patients with GBC via detecting serum cfDNA.
Keywords: Circulating free dna (CFDNA), Gall bladder carcinoma (GBC), Chemiluminescence dna biosensor, DNA G-quadruplex/hemin enzyme, Diagnosis
Abstract
Gall bladder cancer (GBC) is an insidious but rapidly progressed disease with a poor prognosis and high mortality rate. To explore a novel method for GBC diagnosis, we quantified circulating free DNA (cfDNA) in serum samples from 228 participants, including 83 patients with GBC, 75 patients with cholecystitis, and 70 healthy donors, using a chemiluminescence DNA biosensor system based on DNA G-quadruplex/hemin enzyme. We measured β-actin gene expression to evaluate serum cfDNA levels representing as chemiluminescence intensity with the addition of sufficient probes. We analyzed associations of cfDNA quantities in serum samples and corresponding pathological stages and found that the concentration of cfDNA was significantly higher in GBC group than in the healthy control and cholecystitis groups. The levels of cfDNA were significantly associated with TNM stage, lymph node involvement, metastasis, and jaundice. The ROC curves showed that the diagnostic value of chemiluminescence DNA biosensor system was nearly equivalent to that of qPCR. Our method can distinguish patients with GBC from healthy donors and patients with cholecystitis clearly; however, this method was not available to distinguish patients with cholecystitis from the healthy controls. In summary, cfDNA maybe serve as a new diagnostic and noninvasive marker for the diagnosis of GBC using chemiluminescence DNA biosensor system based on DNA G-quadruplex/hemin enzyme.
Introduction
The most common malignant neoplasm of the biliary tract, gallbladder cancer (GBC), accounts for about 90% of all biliary tract cancers [1], [2], [3]. GBC is the sixth most common gastrointestinal cancer, with an annual global incidence of 1.5–27/100,000 [4,5]. The clinical symptoms of GBC are nonspecific, and may include fever, weight loss, jaundice, and abdominal pain. It is very difficult to differentiate GBC from cholecystitis, other benign gallbladder conditions, and other abdominal malignancies [6,7]. Because of the nonspecific clinical presentation, the diagnosis of GBC is usually delayed; therefore, patients have poor 5-year survival rates, ranging from 4% to 60% [8], [9], [10], [11], [12]. Surgical resection is the only most effective curative treatment with a long-term survival in patients with GBC. Patients with early-stage tumors are often curable after surgical resection; unfortunately, the majority of patients present late-stage disease at the time of diagnosis when surgical resection is no longer effective [13]. Therefore, early diagnosis and accurate assessment is crucial to optimize treatment schemes and to improve long-term survival in patients with GBC.
Circulating cell-free DNA (cfDNA), a extracellular nucleic acid, is present in plasma, serum, and other body fluids like saliva and urine [14, 15]. In peripheral blood of healthy individuals, cfDNA mainly derives from the apoptotic lymphocytes and other nucleated cells, while in cancer patients, it comes from tumor necrosis and lysis, micro-metastases, or active release of circulating malignant cells [16]. In general, the cfDNA concentration is very low in healthy individuals or those with various non-malignant diseases, whereas it can be sharply increased in cancer patients. Thus, cfDNA levels are useful to distinguish cancer patients from healthy subjects or patients with benign diseases. In recent years, several studies have confirmed that cfDNA concentrations are higher in patients with cancers such as colon [17], ovarian [18], non-small cell lung [19], breast [20], prostate [21], bladder [22], hepatocellular [23], and gastric cancer [24], than in normal controls. These findings suggest the possibility that cfDNA may be used as a biomarker for early cancer diagnosis, prediction of the response to therapy, and prognosis assessment.
Kumari et al. first reported that quantitative analysis of cfDNA may be used to distinguish GBC from cholecystitis and healthy individuals [25]. These authors measured serum cfDNA levels using quantitative polymerase chain reaction (qPCR) through amplification of the β-globin gene. The qPCR technique significantly improves the sensitivity of cfDNA detection; however, it relies heavily on template replication [26,27]. This undoubtedly increases the risk of cross-contamination from amplicons, resulting in the occurrence of false-positive results [27]. In addition, the qPCR technique requires expensive experimental apparatus, skilled technicians, and high costs of reagents. Therefore, it is necessary to develop a novel method with high sensitivity, high specificity, simplicity, speed, low costs, and easy manipulation for cfDNA measurement.
DNAzyme (also called as catalytical DNA or deoxyribozyme), a single-stranded oligodeoxynucleotide, mimics peroxidase activity and catalyzes many chemical reactions [28,29]. Compared with traditional protein enzymes, the flexibility of altering the recognition region of DNAzyme sequences, the ease of synthesizing of nucleic acids, and the high thermal stability make DNAzymes ideal candidates for biosensing applications [29], [30], [31]. In particular, the G-quadruplex DNAzyme has been widely utilized in several optical biosensors in recent years [32]. G-quadruplex sequences form DNA G-quadruplex/hemin enzymes with the presence of hemin, which can efficiently catalyze luminol-H2O2 system to stimulate the generation of chemiluminescence.
In this study, we describe a simple, highly sensitive, sequence-specific DNA detection method, utilizing G-quadruplex/hemin complex as the critical detection component. We used the self-assembly system to quantify the amount of cfDNA in GBC patients as well as those with cholecystitis and healthy controls. The specificity and sensitivity of the method were also evaluated. To the best of our knowledge, this is the first time that the chemiluminescence DNA biosensor system based on DNA G-quadruplex/hemin enzyme was used to analyze levels of cfDNA in patients with GBC.
Materials and methods
Subjects and blood sample collection
This study included 228 participants from three groups as follows: Group I: 83 patients with histopathologic diagnosis of GBC; Group II: 75 patients with cholecystitis; and Group III: 70 gender- and age-matched healthy donors as the control group. All subjects were enrolled from Surgical Oncology, Gastrointestinal Surgery, or Physical Examination Center from The Second Hospital affiliated to Wannan Medical College in Wuhu, Anhui, R.P. China between February 2017 and February 2019. This study was approved by the ethics committee of The Second Hospital affiliated to Wannan Medical Collage before commencement. Written informed consent was obtained from all participants.
Peripheral blood (5 mL) was collected from patients and healthy donors before treatment or surgery in vacuum blood collection tubes without anticoagulants (BD Biosciences, Franklin Lakes, NJ, USA), and then serum was separated and collected within 1 h by centrifugation at 3500 × g for 10 min at 4 °C. The serum was transferred to clean tubes without nucleases (Eppendorf, Germany) and frozen at –80 °C until used.
DNA extraction
DNA was extracted from serum using the Charge SwitchⓇ gDNA 1 mL Serum Kit (Invitrogen, USA) according to the manufacturer's protocol. Briefly, the process consisted of several steps as follows: i) adding 560 μL of lysis buffer and 30 μL of Proteinase K to 1 mL of serum and incubating for 30 min at room temperature; ii) adding 25 μL of charge Switch magnetic beads and 200 μL of purification buffer to the above mixture and mixing by pipetting gently; iii) placing the tubes in the magna Rack™ for 3 min; iv) washing the tubes twice with 800 μL of wash buffer; v) adding 50 μL of Elution Buffer and leaving at room temperature for 2 min, followed by incubation in magna Rack™ for 1 min; and vi) eluting the purified cfDNA in a new tube and stored at –80 °C until further analysis.
CfDNA quantification using chemiluminescence DNA biosensor system
Human specific β-actin DNA sequences were selected as the target: 5′-ATGCCAACACAGTGCTGTCTGGTGGTACCACCATGTACCCTGGCATT-3′ (the italicized bases were the dividing point of two-part self-assembled luminous element; the underlined base was the assembly reservation site of self-assembly component). The primers for self-assembly chemiluminescence DNA biosensor system were designed according to the target indicator β-actin DNA sequences. Primer 1: 5′-TAGGGTTAGGGTTAGGGTCAGCACTGTGTTGGCAT-3′; Primer 2: 5′-AATGCCAGGGTACATGGTGGTACCACCAGTGGGAT-3′ (the underlined fragments were the complementary sequence of target DNA; the italicized fragments were two split G-rich segments).
The solution of detection system, containing various concentrations of DNA, 1 μM of primer 1, 1 μM of primer 2, and 2 μM hemin in 10 mM Tris-acetate buffer (pH 7.0) containing 5 mM KAc and 10 mM MgCl2, were mixed by pipetting gently and heating for 10 min at 95 °C, slowly cooling to room temperature and incubating for 30 min at 25 °C. Then, the solutions were sampled on PVDF membranes using a mechanized microarrayer and dried at room temperature. Finally, chemiluminescence measurements were performed by adding the mixture of luminol (0.1 mL) and H2O2 (0.1 mL) to the PVDF membranes and read using the SpectraMax M5e Multi-Mode Microplate Reader (Molecular Devices, USA). The standard curve of chemiluminescence intensities versus target β-actin DNA standards was plotted. Concentration of cfDNA in samples was calculated according to the chemiluminescence intensities, based on the standard curve.
CfDNA quantification using quantitative polymerase chain reaction (qPCR)
β-Actin primer sequences were as follows: forward primer, 5′-CCACACTGTGCCCATCTACG-3′; reverse primer, 5′-AGGATCTTCATGAGGTAGTCAGTCAG-3′. qPCR assay was performed on the ABI Step One Real-Time PCR System (Applied Bio systems; Thermo Fisher Scientific, Inc.) in a total volume of 20 μl, containing 2 μl of β-actin reference standard or cfDNA sample, 500 nM of each primer, 10 μl of 2 × SYBR-Green Supermix (Applied Biosystems, USA), and nuclease free water to a total volume of 20 μl. The amplification conditions were as follows: 95 °C for 5 min, followed by 30 cycles of 95 °C for 15 s, and extension at 58 °C for 30 s. Each assay was performed in duplicate. The standard curve was plotted by the machine, generating Ct values versus target β-actin DNA reference standards. Concentrations of cfDNA in samples were calculated according to the Ct values, based on the standard curve.
Statistical analysis
Data were expressed as mean ± standard deviation (X- ± SD) and were analyzed using SPSS software (version 20.0 SPSS Inc., Chicago, IL, USA). GraphPad Prism 6.0 software (GraphPad Software, Inc., La Jolla, CA, USA) was used to generate figures. One-way analysis of variance (ANOVA) and chi-square (χ2) tests were used to analyze differences regarding age and sex among the three groups, respectively. The serum cfDNA concentrations between groups were compared using non-parametric Mann–Whitney U tests and the Kruskal–Wallis H test. The diagnostic values of cfDNA to distinguish between the two groups were evaluated using the receiver operating characteristics (ROC) curves. p<0.05 was regarded as significantly different.
Results
Clinical characteristics of participants in the study
Clinical characteristics of subjects in this study (83 GBC patients, 75 cholecystitis patients, and 70 healthy controls) are summarized in Table 1. The male-to-female (M/F) ratio was 42/41 in the GBC group, 42/33 in the cholecystitis group, and 35/35 in the healthy group. The age ranges of the GBC, cholecystitis and healthy control groups were 35–79 y, 32–81 y, and 29–75 y, respectively, with mean ± SD listed in Table 1. The gender and age distributions of study subjects in the three groups did not differ significantly (F = 1.37, p = 0.3624 and F = 0.6771, p = 0.5091, respectively). The majority of GBC patients were in stage IV (56.63%) with tumor stage of T3/T4 (83.14%). In GBC patients, regional lymph node involvement occurred in 80.72%, distant metastasis in 65.06%, and jaundice in 62.66%.
Table 1.
The clinical characteristics of participants in study.
| Clinical characteristics | Healthy control (n = 70)(%) | Cholecystitis (n = 75)(%) | Gall bladder cancer(n = 83)(%) | |
|---|---|---|---|---|
| Age (yrs) (Mean ± SD) | 63.61±15.69 | 62.59±15.34 | 65.53±17.25 | |
| Sex Male/ Female | 35(50)/35(50) | 42(56)/33(44) | 42(50.6)/41(49.4) | |
| Stage | Ⅱ | 15(18.07) | ||
| Ⅲ | 21(25.3) | |||
| Ⅳ | 47(56.63) | |||
| Primary tumor | T2 | 14(16.86) | ||
| T3 | 26(31.32) | |||
| T4 | 43(51.81) | |||
| Lymph node | N0 | 16(19.27) | ||
| N1 | 27(32.53) | |||
| N2 | 40(48.19) | |||
| Metastasis | M0 | 29(34.94) | ||
| M1 | 54(65.06) | |||
| Jaundice | No | 31(37.34) | ||
| Yes | 52(62.66) |
The principle of the autonomous assembly of chemiluminescence DNA biosensor system based on DNA G-quadruplex/hemin enzyme
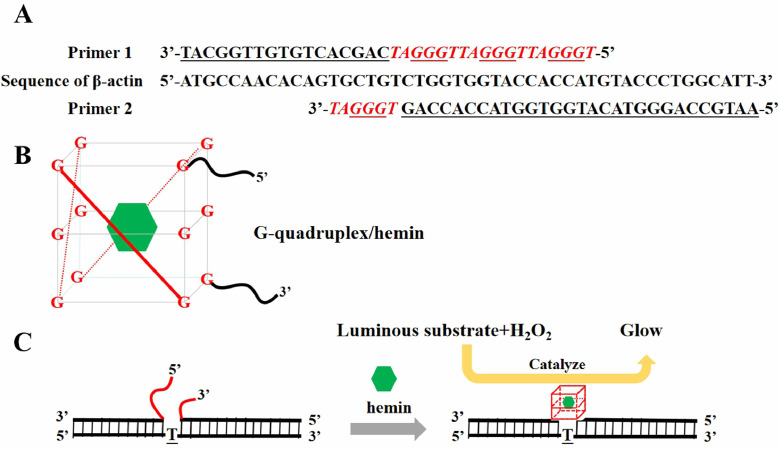
The basic operating principle of the chemiluminescence DNA sensing system, which utilizes G-quadruplex/hemin as the key catalytic unit, is depicted in Fig 1. A pair of primers with G-rich segments (P1 and P2) were designed to be complementary to the target β-actin DNA (Fig 1A). In the presence of target β-actin DNA, primers (P1 and P2) with G-rich segments were specifically bound to target DNA sequences by complementary base pairing. The G-rich segments at the 5′-end and 3′-end of primers form the active G-quadruplex structure, and then formed G-quadruplex/hemin complexes (Fig 1B) with the aid of hemin, which efficiently catalyzes the luminol-H2O2 system to stimulate the generation of chemiluminescence (Fig. 1C). The chemiluminescence intensity is related to the amount of G-quadruplex/hemin complex formed, and thus referred to the concentration of the target DNA.
Fig. 1.
(A) Primer 1 containing 3/4 fractional sequence of G-quadruplex and Primer 2 containing 1/4 fractional sequence of G-quadruplex were designed to pair the target sequence of β-actin DNA. The five bases, TAGGG, were to be a unit and such four units were constructed as a G-quadruplex with one “T” interlinked every units; four units and linker were in red italics, and the G framework was underlined; (B) The 3/4 fractional sequence of G-quadruplex at the 5′-end of primer 1 and 1/4 fractional sequence of the G-quadruplex 3′-end of primer 2 constructed the active G-quadruplex framework, and then formed G-quadruplex/hemin complex catalyzer; the green hexagon stands for hemin; (C) The process of enzymatic luminescence. Chemiluminescence intensity is related to the amount of G-quadruplex/hemin complex formed, and thus the concentration of the target DNA.
Detection sensitivity and specificity of the chemiluminescence DNA biosensor
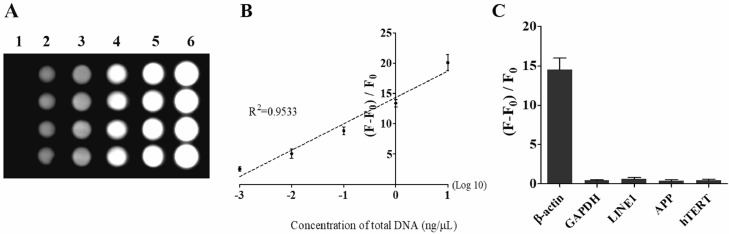
The sensitivity of the chemiluminescence DNA biosensor was evaluated by measuring the chemiluminescence intensities as a reflection of target DNA concentrations. The results displayed in Fig. 2A show that the chemiluminescence intensities increased with target β-actin DNA concentrations increasing from 0.001 ng/μL to 10 ng/μL when exposed continuously for 10 min. The relationship between the chemiluminescence intensities and the target β-actin DNA concentrations was investigated and shown in Fig. 2B. A linear relationship (R2=0.9533) was found existing between the chemiluminescence intensities and the target β-actin DNA concentrations. The detection limit of the chemiluminescence DNA biosensor was estimated and found under 0.001 ng/μL, which is satisfactory for cfDNA detection.
Fig. 2.
Performance of the chemiluminescence detection system for cf DNA.
(A) The microarray image of detection for 6 concentration gradients of target cfDNA by chemiluminescence detection system. Vertical lines 1 to 6 represent 0 ng/μL, 0.001 ng/μL, 0.01 ng/μL, 0.1 ng/μL, 1 ng/μL and 10 ng/μL of target cfDNA. Every concentration was measured four times; (B) The fitted curve of detection was constructed according to 6 concentration gradients using the chemiluminescence detection system (R2=0.9533). The abscissa took the logarithm 10 of concentration values. The chemiluminescence intensity of blank control (0 ng/μL) was recorded as F0, other groups were recorded as F. The ordinate displayed fold-magnitudes according to a formula: (F-F0)/F0; (C) The detection specificity of chemiluminescence detection system. This detection system distinguished target DNA from other DNA sequences (GAPDH, LINE1, APP and hTERT) prominently.
The selectivity of the chemiluminescence DNA biosensor detection system reflects the ability to distinguish target DNA from other non-target DNAs. Therefore, to determine the detection specificity of the DNA biosensor, we measured and compared the dose effect of non-target DNAs (glyceraldehyde-3-phosphate dehydrogenase (GAPDH), human telomerase reverse transcriptase (hTERT), long interspersed nuclear element-1 (LINE1) and amyloid precursor protein (APP)), artificially synthesized oligonucleotides, to induce chemiluminescence with that of target β-actin DNA. As shown in Fig. 2C, significant chemiluminescence intensity increases occurred in the presence of increased target β-actin DNA, whereas other non-target DNAs showed negligible responses. These results suggest that the constructed chemiluminescence DNA biosensor is highly sensitive and selective for the target DNA.
Serum cfDNA level
It should be noted that cfDNAs in human serum have attracted many attentions as novel non-invasive biomarkers for diagnosis of serious diseases. Therefore, cfDNA levels in our samples were measured using the chemiluminescence DNA biosensor system. As shown in Fig. 2B, the standard curve of chemiluminescence intensities versus target β-actin DNA standards had a high R2 value of 0.9533. Concentrations of cfDNA in healthy controls, cholecystitis patients, and cancer patients were calculated according to the standard curve of chemiluminescence intensities versus target β-actin DNA standards. As shown in Fig. 3, the cfDNA concentration of GBC patients was dramatically highest among three groups, GBC patients (1013.95±681.96 ng/mL) than those in cholecystitis patients (198.66±103.64 ng/ mL) and healthy controls (82.94±47.51 ng/mL).
Fig. 3.
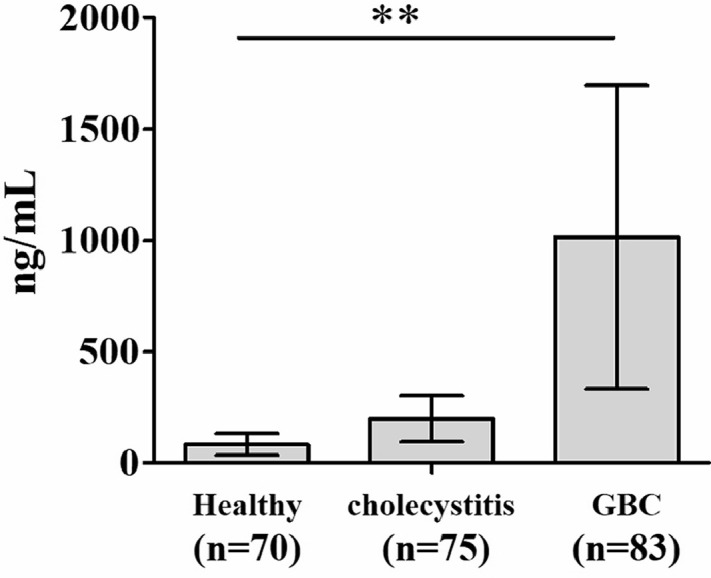
Comparison of the levels of cfDNA among the three groups using chemiluminescence DNA biosensor.
**p< 0.01 as compared to healthy group and cholecystitis group.
Serum cfDNA levels in normal controls, cholecystitis patients, and cancer patients were also measured using classical quantitative polymerase chain reaction (qPCR) through amplification of the β-actin gene. The concentrations of cfDNA in three groups are listed in Table 2. The cfDNA levels measured by these two methods in each of three groups did not show significant differences (p>0.05).
Table 2.
Cf DNA levels (ng/mL) measured by two methods in samples.
| Groups (n) | chemiluminescence DNA biosensor |
qPCR |
t | p | ||
|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | |||
| Healthy (70) | 82.94 | 47.51 | 71 | 31 | 1.761 | 0.0805 |
| Cholecystitis (75) | 198.66 | 113.64 | 168 | 96 | 1.785 | 0.0763 |
| GBC (83) | 1013.95 | 681.96 | 895 | 507 | 1.275 | 0.2040 |
Associations of serum cfDNA concentration with various clinical characteristics in GBC patients
The associations of serum cfDNA concentration with various clinical characteristics of GBC patients are listed in Table 3. Analysis of cfDNA levels in different subgroups of GBC patients showed that serum cfDNA level was independent of age and sex: there were no significant differences (p>0.05) in serum cfDNA concentration between GBC patients of different sexes (42 males and 41 females) or between GBC patients aged no less than 60 years and more than 60 years. The cfDNA concentrations were considerably higher in GBC patients with advanced stage disease (stage IV) than in those with early stage disease (stage II and III) (p<0.001). GBC patients with jaundice also had dramatically higher cfDNA concentrations than patients without jaundice (p<0.001). The cfDNA concentrations were higher in GBC patients with primary tumors of T4 than in those with primary tumors of T3 & T2, higher in patients with N1 and N2 lymph node status than in those with N0, and higher in patients with metastasis than in those without metastasis, the differences were statistically significant (p<0.01).
Table 3.
Association between clinical characteristics of 83 GBC patients and cfDNA levels.
| Characteristic | n | cf DNA (ng/mL) | t / F value | P value |
|---|---|---|---|---|
| Age (years) : | ||||
| ≤60 | 36 | 1258.71±754.65 | 1.877 | 0.0656 |
| >60 | 47 | 986.56±494.07 | ||
| Sex: | ||||
| Male | 42 | 1276.38±865.99 | 1.791 | 0.0777 |
| Female | 41 | 996.36±519.27 | ||
| Stage: | ||||
| Ⅱ+Ⅲ | 36 | 798.45±168.32 | 4.996 | 0.000* |
| Ⅳ | 47 | 1381.48±776.54 | ||
| Primary tumor: | ||||
| T2 | 14 | 811.48±226.35 | 5.327 | 0.0067* |
| T3 | 26 | 985.26±209.35 | ||
| T4 | 43 | 1351.47±826.33 | ||
| Lymph node: | ||||
| N0 | 16 | 664.72±135.67 | 5.828 | 0.0043* |
| N1 | 27 | 1047.64±678.59 | ||
| N2 | 43 | 1411.25±944.35 | ||
| Metastasis: | ||||
| M0 | 29 | 887.31±361.92 | 3.62 | 0.0005* |
| M1 | 54 | 1375.76±859.65 | ||
| Jaundice: | ||||
| No | 31 | 786.33±269.26 | 4.356 | 0.000* |
| Yes | 52 | 1398.94±952.36 |
Diagnostic utility of serum cfDNA detection in GBC patients
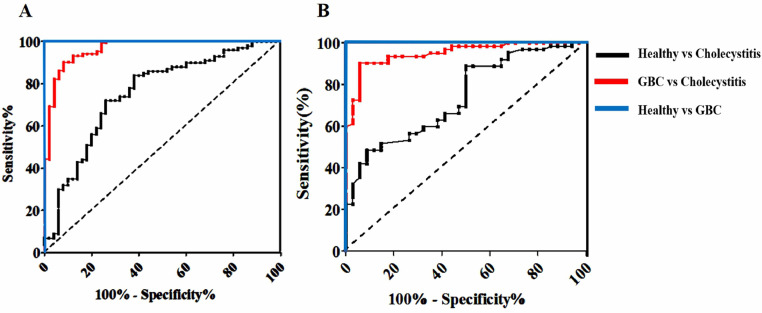
ROC curves were performed to evaluate the diagnostic accuracy of serum cfDNA detection to distinguish cancer patients, i.e., GBC patients in this case, from healthy controls and from cholecystitis patients and to distinguish cholecystitis patients from healthy controls. As shown in Fig. 4 and Table 4, the cutoff value of cfDNA at 113.82 ng/mL dramatically distinguished GBC patients from healthy donors controls with 100% sensitivity (95% CI = 88.78%–100%) and 100% specificity (95% CI = 87.66%–100%). The cfDNA value at 403.65 ng/mL dramatically distinguished GBC patients from cholecystitis patients with 93.52% sensitivity (95% CI = 86.54%–97.83%) and 96.81% specificity (95% CI =85.92%–98.31%) (Table 5). The cfDNA value at 89.56 ng/mL dramatically distinguished cholecystitis patients from normal controls with 75.32% sensitivity (95% CI = 49.41–88.32) and 69.14% specificity (95% CI = 42.37%–84.99%) (Table 6). qPCR was the classical method for cfDNA detection with high sensitivity and specificity in routine laboratory practice. We compared the diagnostic accuracy of the chemiluminescence DNA biosensor system for GBC with that of qPCR for GBC. As shown in Table 4, for distinguishing GBC patients from healthy donors, the AUCs of the chemiluminescence DNA biosensor and of the qPCR were 1 (95% CI: 0.864–1) and 1 (95% CI: 0.829–1), respectively. For distinguishing GBC patients from cholecystitis patients, the AUCs of these two methods were 0.972 (95% CI: 0.771–0.995) and 0.965 (95% CI: 0.753–0.989), respectively (Table 5). For distinguishing cholecystitis patients form normal controls, the AUCs of these two methods were 0.643 (95% CI: 0.394–0.805) and 0.611 (95% CI: 0.368–0.785), respectively (Table 6). No obvious differences were found between AUC values of these two methods for cfDNA detection in GBC diagnosis. These results suggest that the novel method could be effectively used for the detection of cfDNA for GBC diagnosis.
Fig. 4.
ROC curves of detection cfDNA among three groups using two methods.
(A) ROC curves of detection cfDNA among three groups using chemiluminescence DNA biosensor; (B) ROC curves of detection cfDNA among three groups using qRT-PCR.
Table 4.
Diagnostic accuracy of cfDNA to discriminate GBC patients from healthy donors using ROC curve analysis.
| Methods | AUC (95% CI) | Sensitivity (95% CI) | Specifcity (95% CI) | Cutoff value (ng/ml) | Positive predictive value (%) | Negative predictive value (%) |
|---|---|---|---|---|---|---|
| chemilumines-cence DNA biosensor | 1 (0.864–1.000) |
100 (88.78–100) |
100 (87.66–100) |
113.82 | 100% | 100% |
| qPCR | 1 (0.829–1.000) |
100 (85.66–100) |
100 (89.72–100) |
96 | 100% | 100% |
Table 5.
Diagnostic accuracy of cfDNA to discriminate GBC patients from cholecystitis patients using ROC curve analysis.
| Methods | AUC (95% CI) | Sensitivity (95% CI) | Specifcity (95% CI) | Cutoff value (ng/ml) | Positive predictive value (%) | Negative predictive value (%) |
|---|---|---|---|---|---|---|
| Chemiluminesc-ence DNA biosensor | 0.972 (0.771–0.995) |
93.52 (86.54–97.83) |
96.81 (85.92–98.31) |
403.65 | 94.31 | 93.33 |
| qPCR | 0.965 (0.753–0.989) |
92.36 (82.36–95.37) |
95.24 (87.51–98.29) |
364 | 92.13 | 90.66 |
Table 6.
Diagnostic accuracy of cfDNA to discriminate cholecystitis patients from healthy donors using ROC curve analysis.
| Methods | AUC (95% CI) | Sensitivity (95% CI) | Specifcity (95% CI) | Cutoff value (ng/mL) | Positive predictive value (%) | Negative predictive value (%) |
|---|---|---|---|---|---|---|
| Chemilumin-escence DNA biosensor | 0.643 (0.394–0.805) |
75.32 (49.41–88.32) |
69.14 (42.37–84.99) |
89.56 | 78.37 | 76.05 |
| qPCR | 0.611 (0.368–0.785) |
73.69 (45.27–83.65) |
62.92 (42.33–81.52) |
81 | 74.32 | 74.02 |
Discussion
G-quadruplex is a G-rich oligonucleotide that can fold into unique four-stranded structure and the structure can associate with the co-factor hemin to form DNA G-quadruplex/hemin enzymes [33, 34]. Because of its outstanding characteristics, including low cost, greater resistance to hydrolysis and heat, low nonspecific adsorption, relative ease of labeling, and typical catalytic ability, DNA G-quadruplex/hemin enzymes have been widely utilized in biosensing [35], [36], [37], [38]. In the present study, we developed a simple yet ultrasensitive chemiluminescence DNA biosensor based on the G-quadruplex/hemin complex. Taking human β-actin DNA sequences as the target, two primers with G-rich segments were designed to pair target DNA. The rationally designed primers formed G-quadruplexes in the presence of target DNA, and further formed G-quadruplex/hemin complexes with the aid of hemin, which can efficiently catalyze the luminol-H2O2 system to stimulate the generation of chemiluminescence. Chemiluminescence intensity is related to the amount of G-quadruplex/hemin complex, and therefore, to the concentration of target DNA. The detection limit of the chemiluminescence DNA biosensor is estimated to be 3.8 fM. For specificity, an obvious chemiluminescence intensity increase occurred in the presence of increased target β-actin DNA but did not in the presence of increased other non-target DNAs. Our results suggest that the chemiluminescence DNA biosensor system is highly sensitive and selective for the target DNA, which is satisfactory for cfDNA detection.
Gallbladder cancer (GBC) is the most common malignant neoplasm of the biliary tract and is usually diagnosed at advanced stages due to the nonspecific clinical presentation [25]. At present, in clinical practice, the diagnosis of GBC is based on ultrasound, computed tomography, magnetic resonance imaging, endoscopy, and biopsy. Of these, tissue biopsy is the gold standard for the diagnosis of GBC, which is invasive and expensive and increases risk and pain for patients. These limitations restrict its application in clinical practice. With the development of medical science and liquid biopsy, cfDNA has rapidly emerged as a novel biomarker with promising clinical applications because of features such as non-invasive nature, low cost, and lack of risk [39]. In recent years, many studies have demonstrate the diagnostic value of cfDNA detection in breast cancer [20], prostate cancer [21], and gastric cancer [24]. In the present study, we used a chemiluminescence DNA biosensor system to quantify the amount of cfDNA in GBC patients as compared to cholecystitis patients and healthy controls. We found that levels of cfDNA were higher in GBC patients than in cholecystitis patients and healthy donors. This is consistent with the results of qPCR analysis. We also found that cfDNA levels in GBC patients did not correlate with age and sex; however patients with more advanced stage disease (stage IV) and jaundice had considerably higher cfDNA levels, which was consistent with findings of previous reports [25]. ROC curve analysis showed that cfDNA levels measured by our chemiluminescence DNA biosensor obtained both sensitivity and specificity of 100% to discriminate GBC patients in this study, from normal controls and with the sensitivity and specificity of 93.52% and 96.81%, respectively to differentiate GBC patients from cholecystitis patients. For qPCR assay, ROC analysis for GBC group versus healthy group and versus cholecystitis groups had sensitivities of 100% and 92.36%, respectively, and specificities of 100% and 95.24%, respectively. The diagnostic accuracy of quantification cfDNA for GBC via chemiluminescence DNA biosensor system based on DNA G-quadruplex/hemin enzyme was slightly higher than that of qPCR; however, there was no significant difference (p>0.05).
There are also some limitations for the present study. For example, whether this chemiluminescence DNA biosensor system also works well for other kinds of cancers was not analyzed and we did not study whether it could distinguish patients with early stage GBC from patients with cholecystitis and from healthy controls. Moreover, only one cohort was used and a validation cohort is needed to confirm the diagnostic value of this chemiluminescence DNA biosensor system. That is our future research work and this study is only to proof the partial detection performance of method.
In conclusion, a simple yet ultrasensitive chemiluminescence DNA biosensor system based on DNA G-quadruplex/hemin enzyme was designed to quantify the concentration of cfDNA in serum samples. The levels of cfDNA were dramatically greater in the serum of GBC patients than in those of cholecystitis patients and healthy controls, consistent with the results of qPCR analysis. These cfDNA levels might be a valuable biomarker for diagnosing GBC. Although the diagnostic accuracy of the chemiluminescence DNA biosensor system is almost equal with that of qPCR, it is important to note that the new approach is operationally simple, highly sensitive, and does not require expensive analytical instruments.
Declaration of Competing Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 81601806).
Compliance with ethical standards
This study was complied with the World Medical Association Declaration of Helsinki and approved by the Ethical committees of the Second affiliated hospital of Wannan Medical College (Wuhu, China).
Author Contributions Section
We confirm that all methods were performed in accordance with the relevant guidelines and regulations in this work.All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission. Ying Hua and Feng Hu completed this experiment and wrote this manuscript. Yan-hong Wu and Feng-ying Sun prepared the figures and helped to do experiment, Xian-ru Xia and Xiao-lei Tang provided idea and expenditure for this experiment.
References
- 1.Dutta U. Gallbladder cancer: can newer insights improve the outcome. J. Gastroenterol. Hepatol. 2012;27:642–653. doi: 10.1111/j.1440-1746.2011.07048.x. [DOI] [PubMed] [Google Scholar]
- 2.Montalvo-Jave E.E., Rahnemai-Azar A.A., Papaconstantinou D., Deloiza M.E., Tsilimigras D.I., Moris D. Molecular pathways and potential biomarkers in gallbladder cancer: a comprehensive review. Surg. Oncol. 2019;31:83–89. doi: 10.1016/j.suronc.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Hundal R., Shaffer E.A. Gallbladder cancer: epidemiology and outcome. Clin. Epidemiol. 2014;6:99–109. doi: 10.2147/CLEP.S37357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanthan R., Senger J.L., Ahmed S., Kanthan S.C. Gallbladder cancer in the 21st century. J. Oncol. 2015;2015 doi: 10.1155/2015/967472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stinton L.M., Shaffer E.A. Epidemiology of gallbladder disease: cholelithiasis and cancer. Gut Liver. 2012;6:172–187. doi: 10.5009/gnl.2012.6.2.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furlan A., Ferris J.V., Hosseinzadeh K., Borhani A.A. Gallbladder carcinoma update: multimodality imaging evaluation, staging, and treatment options. AJR Am. J. Roentgenol. 2008;191:1440–1447. doi: 10.2214/AJR.07.3599. [DOI] [PubMed] [Google Scholar]
- 7.van der Horst M.P., Hendriks E.R., Blok P., Brouwers M.A., Steup W.H. [Diversity of complaints in manifesting carcinoma of the gallbladder] Ned. Tijdschr. Geneeskd. 2007;151:1083–1086. PMID: 17552418. [PubMed] [Google Scholar]
- 8.Singh S.K., Talwar R., Kannan N., Tyagi A.K., Jaiswal P., Kumar A. Patterns of presentation, treatment, and survival rates of gallbladder cancer: a prospective study at a tertiary care centre. J. Gastrointest. Cancer. 2018;49:268–274. doi: 10.1007/s12029-017-9940-y. [DOI] [PubMed] [Google Scholar]
- 9.Moris D., Tsilimigras D.I., Lim J., Camastra D., Nanavati A., Knechtle S.J. Neuroendocrine carcinomas of the gallbladder: lessons learnt from cases at opposite ends of the spectrum. J. BUON. 2018;23:1922–1926. PMID: 30610822. [PubMed] [Google Scholar]
- 10.Dixon E., Vollmer C.M., Jr, Sahajpal A., Cattral M., Grant D., Doig C. An aggressive surgical approach leads to improved survival in patients with gallbladder cancer: a 12-year study at a North American Center. Ann. Surg. 2005;241:385–394. doi: 10.1097/01.sla.0000154118.07704.ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fong Y., Jarnagin W., Blumgart L.H. Gallbladder cancer: comparison of patients presenting initially for definitive operation with those presenting after prior noncurative intervention. Ann. Surg. 2000;232:557–569. doi: 10.1097/00000658-200010000-00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsilimigras D.I., Hyer J.M., Paredes A.Z., Moris D., Beal E.W., Merath K. The optimal number of lymph nodes to evaluate among patients undergoing surgery for gallbladder cancer: correlating the number of nodes removed with survival in 6531 patients. J. Surg. Oncol. 2019;119:1099–1107. doi: 10.1002/jso.25450. [DOI] [PubMed] [Google Scholar]
- 13.Miller G., Jarnagin W.R. Gallbladder carcinoma. Eur. J. Surg. Oncol. 2008;34:306–312. doi: 10.1016/j.ejso.2007.07.206. [DOI] [PubMed] [Google Scholar]
- 14.Yao W., Mei C., Nan X., Hui L. Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: a qualitative study. GeneGene. 2016;590:142–148. doi: 10.1016/j.gene.2016.06.033. [DOI] [PubMed] [Google Scholar]
- 15.Su Y.H., Wang M., Brenner D.E., Norton P.A., Block T.M. Detection of mutated K-ras DNA in urine, plasma, and serum of patients with colorectal carcinoma or adenomatous polyps. Ann. N. Y. Acad. Sci. 2008;1137:197–206. doi: 10.1196/annals.1448.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stroun M., Maurice P., Vasioukhin V., Lyautey J., Lederrey C., Lefort F. The origin and mechanism of circulating DNA. Ann. N. Y. Acad. Sci. 2000;906:161–168. doi: 10.1111/j.1749-6632.2000.tb06608.x. [DOI] [PubMed] [Google Scholar]
- 17.Vietsch E.E., Graham G.T., McCutcheon J.N., Javaid A., Giaccone G., Marshall J.L. Circulating cell-free DNA mutation patterns in early and late stage colon and pancreatic cancer. Cancer Genet. 2017;218-219:39–50. doi: 10.1016/j.cancergen.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng X., Zhang L., Chen Y., Qing C. Circulating cell-free DNA and circulating tumor cells, the "liquid biopsies" in ovarian cancer. J. Ovarian Res. 2017;10:75. doi: 10.1186/s13048-017-0369-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ulivi P., Silvestrini R. Role of quantitative and qualitative characteristics of free circulating DNA in the management of patients with non-small cell lung cancer. Cell. Oncol. (Dordr) 2013;36:439–448. doi: 10.1007/s13402-013-0155-3. [DOI] [PubMed] [Google Scholar]
- 20.Miao Y., Fan Y., Zhang L., Ma T., Li R. Clinical value of plasma cfDNA concentration and integrity in breast cancer patients. Cell. Mol. Biol. (Noisy-le-grand) 2019;65:64–72. [PubMed] [Google Scholar]
- 21.Choudhury A.D., Werner L., Francini E., Wei X.X., Ha G., Freeman S.S. Tumor fraction in cell-free DNA as a biomarker in prostate cancer. JCI Insight. 2018;3 doi: 10.1172/jci.insight.122109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thoma C. Bladder cancer: the promise of liquid biopsy ctDNA analysis. Nat. Rev. Urol. 2017;14:580–581. doi: 10.1038/nrurol.2017.138. [DOI] [PubMed] [Google Scholar]
- 23.Li Z., Xiao D., Li X., Zhan P., Wang J., Zhang H. Early recurrence detected in hepatocellular carcinoma patients after transcatheter arterial chemoembolization treatment with plasma cell-free DNA. Eur. J. Gastroenterol. Hepatol. 2019;31:885–892. doi: 10.1097/MEG.0000000000001373. [DOI] [PubMed] [Google Scholar]
- 24.Sai S., Ichikawa D., Tomita H., Ikoma D., Tani N., Ikoma H. Quantification of plasma cell-free DNA in patients with gastric cancer. Anticancer Res. 2007;27:2747–2751. PMID: 17695442. [PubMed] [Google Scholar]
- 25.Kumari S., Tewari S., Husain N., Agarwal A., Pandey A., Singhal A. Quantification of circulating free DNA as a diagnostic marker in gall bladder cancer. Pathol. Oncol. Res. 2017;23:91–97. doi: 10.1007/s12253-016-0087-0. [DOI] [PubMed] [Google Scholar]
- 26.Tan Y., Guo Q., Zhao X., Yang X., Wang K., Huang J. Proximity-dependent protein detection based on enzyme-assisted fluorescence signal amplification. Biosens. Bioelectron. 2014;51:255–260. doi: 10.1016/j.bios.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 27.Dong J., Cui X., Deng Y., Tang Z. Amplified detection of nucleic acid by G-quadruplex based hybridization chain reaction. Biosens. Bioelectron. 2012;38:258–263. doi: 10.1016/j.bios.2012.05.042. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L., Zhu J., Li T., Wang E. Bifunctional colorimetric oligonucleotide probe based on a G-quadruplex DNAzyme molecular beacon. Anal. Chem. 2011;83:8871–8876. doi: 10.1021/ac2006763. [DOI] [PubMed] [Google Scholar]
- 29.Gao Y., Li B. G-quadruplex DNAzyme-based chemiluminescence biosensing strategy for ultrasensitive DNA detection: combination of exonuclease III-assisted signal amplification and carbon nanotubes-assisted background reducing. Anal. Chem. 2013;85:11494–11500. doi: 10.1021/ac402728d. [DOI] [PubMed] [Google Scholar]
- 30.Zhao X.H., Gong L., Zhang X.B., Yang B., Fu T., Hu R. Versatile DNAzyme-based amplified biosensing platforms for nucleic acid, protein, and enzyme activity detection. Anal. Chem. 2013;85:3614–3620. doi: 10.1021/ac303457u. [DOI] [PubMed] [Google Scholar]
- 31.Zhang P., Wu X., Yuan R., Chai Y. An "off-on" electrochemiluminescent biosensor based on DNAzyme-assisted target recycling and rolling circle amplifications for ultrasensitive detection of microRNA. Anal. Chem. 2015;87:3202–3207. doi: 10.1021/ac504455z. [DOI] [PubMed] [Google Scholar]
- 32.Huang P.J., Lin J., Cao J., Vazin M., Liu J. Ultrasensitive DNAzyme beacon for lanthanides and metal speciation. Anal. Chem. 2014;86:1816–1821. doi: 10.1021/ac403762s. [DOI] [PubMed] [Google Scholar]
- 33.Zou P., Liu Y., Wang H., Wu J., Zhu F., Wu H. G-quadruplex DNAzyme-based chemiluminescence biosensing platform based on dual signal amplification for label-free and sensitive detection of protein. Biosens. Bioelectron. 2016;79:29–33. doi: 10.1016/j.bios.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 34.Sun X., Chen H., Wang S., Zhang Y., Tian Y., Zhou N. Electrochemical detection of sequence-specific DNA based on formation of G-quadruplex-hemin through continuous hybridization chain reaction. Anal. Chim. Acta. 2018;1021:121–128. doi: 10.1016/j.aca.2018.02.076. [DOI] [PubMed] [Google Scholar]
- 35.Sharon E., Freeman R., Willner I. CdSe/ZnS quantum dots-G-quadruplex/hemin hybrids as optical DNA sensors and aptasensors. Anal. Chem. 2010;82:7073–7077. doi: 10.1021/ac101456x. [DOI] [PubMed] [Google Scholar]
- 36.Sun A., Qi Q., Wang X., Bie P. Porous platinum nanotubes labeled with hemin/G-quadruplex based electrochemical aptasensor for sensitive thrombin analysis via the cascade signal amplification. Biosens. Bioelectron. 2014;57:16–21. doi: 10.1016/j.bios.2014.01.040. [DOI] [PubMed] [Google Scholar]
- 37.Yuan Y., Chai Y., Yuan R., Zhuo Y., Gan X. An ultrasensitive electrochemical aptasensor with autonomous assembly of hemin-G-quadruplex DNAzyme nanowires for pseudo triple-enzyme cascade electrocatalytic amplification. Chem. Commun. (Camb.) 2013;49:7328–7330. doi: 10.1039/c3cc42874e. [DOI] [PubMed] [Google Scholar]
- 38.Nakatsuka K., Shigeto H., Kuroda A., Funabashi H. A split G-quadruplex-based DNA nano-tweezers structure as a signal-transducing molecule for the homogeneous detection of specific nucleic acids. Biosens. Bioelectron. 2015;74:222–226. doi: 10.1016/j.bios.2015.06.055. [DOI] [PubMed] [Google Scholar]
- 39.Cervena K., Vodicka P., Vymetalkova V. Diagnostic and prognostic impact of cell-free DNA in human cancers: systematic review. Mutat. Res. 2019;781:100–129. doi: 10.1016/j.mrrev.2019.05.002. [DOI] [PubMed] [Google Scholar]