Abstract
Recently, multiple spillover events between domesticated poultry and wild birds have been reported for several avian viruses. This phenomenon highlights the importance of the livestock-wildlife interface in the possible emergence of novel viruses. The aim of the current study was to investigate the potential spillover and epidemiological links of infectious bursal disease virus (IBDV) between wild birds and domestic poultry. To this end, twenty-eight cloacal swabs were collected from four species of free-living Egyptian wild birds (i.e. mallard duck, bean goose, white-fronted goose and black-billed magpie). Genetic and phylogenetic analysis of three positive isolates revealed that the IBDV/USC-1/2019 strain clustered with previously reported very virulent IBDV (vvIBDV) Egyptian isolates. Interestingly, two other wild bird-origin isolates (i.e. IBDV/USC-2/2019 and IBDV/USC-3/2019) grouped with a vaccine strain that is being used in commercial poultry. In conclusion, our results revealed the molecular detection of vaccine and vvIBDV-like strains in Egyptian wild birds and highlighted the potential role of wild birds in IBDV epidemiology in disease-endemic regions.
Keywords: Viruses, Spill over, Evolution, Poultry
Introduction
Infectious bursal disease (IBD) is an acute and highly contagious disease of chicks, and the clinical impact of IBD is mainly attributed to its severe immunosuppression especially in young chickens. The IBDV infection particularly targets and annihilates the precursors of antibody-producing B cells within the bursa of Fabricius (BF) [1]. Importantly, the damages to the BF are permanent, resulting in vaccination failure and expanded defencelessness to other diseases [2].
The IBDV is non-enveloped, icosahedral in shape, and carry double-stranded RNA genome within the genus Avibirnavirus of family Birnaviridae [3]. The IBDV is composed of segment A (~ 3.17 kb in length) and B (~ 2.8 kb in length) [3]. The segment A is comprised of two partially overlapping open reading frames (ORFs). The non-structural viral protein 5 (VP5) encoded by the first ORF, whereas the second ORF encodes a polyprotein. This polyprotein is eventually cleaved into two structural proteins known as VP2 and VP3, and a serine protease called VP4 [4–6]. The VP2 is the main structural protein and carries all the neutralizing epitopes, involved in virulence, cell tropism, and antigenic variation [7–9]. The RNA-dependent RNA polymerase (VP1) is encoded by Segment B [10], which plays critical functions in viral replication [11].
Out of two IBDV serotypes (i.e. I and II), only serotype I strains of IBDV are virulent in chickens. These strains are grouped into four characteristic pathotypes including classical, attenuated, antigenic variant, and very virulent strains [12–14]. Nearly 60–76% of IBDV isolates across four continents can be grouped as very virulent based on the global molecular epidemiological investigations [15]. Since the first report of the very virulent IBDV (vvIBDV) in the USA in 1957 [12, 16], the disease has been spreading worldwide [15] including Egypt [17–19] and has undergone a complex evolution. In Egypt, the vvIBDVs were first reported in 1989 [19]. To contain the infection, live-attenuated, intermediate plus, and classical strain-based vaccines are currently being used in the Egyptian poultry industry [20]. Despite mass vaccination regimes, Egypt is experiencing repeated IBDV outbreaks with high mortality rates since last two decades [17–19].
A relatively recent area of research at livestock-species interface is the spillover of viruses from the fared poultry into wild birds that can risk the wild birds' welfare. In commercial poultry farms, vaccination may have a significant effect on virus evolution [20] and possible spread to wild birds in vicinity [21, 22]. Several spillover events of vaccine viruses from domestic poultry to wild birds have been reported such as Newcastle disease virus and infectious bronchitis virus [21, 22]. Owing to high demands for free-range and backyard poultry production, the direct interaction between wild birds and farmed poultry is increasing [23]. Furthermore, massive size of the industrialized poultry production may risk the environment contamination with infectious materials through activities such as reuse of poultry litters [23].
This study was designed to investigate the potential spillover of infectious bursal disease virus (IBDV) between wild birds and domestic poultry. A total of 28 cloacal swabs were collected from Egyptian free-living wild birds during 2019, and genetics and transmission risks were assessed for the IBDV in Egypt.
Materials and methods
Samples collection, virus isolation, and genetic characterization
Twenty-eight cloacal swabs were collected from three Egyptian provinces (Monofiya, Qaulubia and Sharkia) during 2019, which were considered wild birds-dense and IBDV-endemic areas in the Nile Delta region (Table 1). The Nile Delta of Northern Egypt is a crucial stopover for millions of birds migrating between the Palearctic and Afrotropical regions annually, and considered one of the most important migration routes for wild birds [24, 25].
Table 1.
Overview of wild bird samples involved in the study, and the prevalence of IBDVs in different species
| Order | Family | Genus | Species | Region/Governorate | Sampled (n) | Positive (n) |
|---|---|---|---|---|---|---|
| Anseriformes | Anatidae | Anas |
A. crecca (Green- winged teal) |
Monofiya, | 3 | 0 |
| Qaulubia | 2 | 0 | ||||
| Sharkia | 3 | 1 | ||||
|
A. platyrhynchos (Mallard) |
Monofiya, | 2 | 0 | |||
| Qaulubia | 3 | 0 | ||||
| Sharkia | 2 | 0 | ||||
| Pelecaniformes | Ardeidae | Bubulcus |
B. ibis (Cattle egret) |
Monofiya, | 3 | 1 |
| Qaulubia | 2 | 1 | ||||
| Sharkia | 1 | 0 | ||||
| Galliformes | Phasianidae | Coturnix |
C. coturnix (Common quail) |
Monofiya, | 2 | 0 |
| Qaulubia | 3 | 0 | ||||
| Sharkia | 2 | 0 |
n means: number
Capturing and sampling from live wild birds were carried out in accordance with all relevant guidelines, regulations and animal ethics permits issued by the Faculty of Veterinary Medicine, University of Sadat City, Egypt. The cloacal swabs were collected from each bird individually and placed in 1.5 ml of phosphate buffer saline (PBS) supplemented with 2000 unit/ml Penicillin G, 200 mg/ml Gentamicin, and 4 mg/ml Amphotericin B. The swab fluids were clarified by centrifugation at 1500 rpm for 10 min, and the supernatant was used for RNA extraction using TRIzol™ reagent as per manufacturer’s instructions. Using RT-PCR assays, the extracted RNA were screened for IBDV using a primer pair that amplifies a 743 bp region of VP2 gene, the forward primer was 5′-GCC CAG AGT CTA CAC CAT-3′ and the reverse primer was 5′-CCC GGA TTA TGT CTT TGA-3′ [26].
The RT-PCR-positive samples (n = 3) were inoculated on the chorioallantonic membrane (CAM) of specific pathogen free (SPF) embryonated chicken eggs following the standard procedures [27]. Five days post-inoculation, all embryos died. CAMs were harvested from dead embryos and screened by qRT-PCR for IBDV. The RNA was extracted from positive CAMs using TRIzol™ reagent as per manufacturer’s instructions (Invitrogen, USA). The extracted RNA treated with dimethylsulphoxide (DMSO) for 5 min at 98 °C and then snap chilled [27]. The synthesis of cDNA from the DMSO-treated RNA was performed using SuperScript™ IV Reverse Transcriptase (Thermo Scientific, USA) as per the manufacturer’s instruction. Polymerase chain reaction (PCR) was carried out using High Fidelity Q5 polymerase (NEB, UK), according to manufacturer’s instructions for the amplification of full length VP2 gene using the following primers; IBDVP2F-5′-ATG ACA AAC CTG CAA GAT CAA ACC CAA C-3′ and IBDVP2R-5′-TTA TGT CTT TGA AGC CAA ATG CTC CTG C-3′. These primers flank the conserved regions of VP2 ORF among IBDV serotype I strains. Briefly, a total of 50 μl reaction mixture contain 2 μl of cDNA, 10 μl of 5X Q5 Reaction Buffer, 10 ul of 5X Q5 High GC Enhancer 2.5 μl primer IBDVP2F, 2.5 μl primer IBDVP2R, 2 μl dNTPs mix, 0.5 μl of Q5 High-Fidelity DNA Polymerase and 20.5 μl nuclease free water. The PCR cycling protocol was as follows: 98 °C for 3 min followed by 40 three-step cycles of 98 °C for 30 s, 68 °C for 45 s and 72 °C for 2 min; then 72 °C for 10 min. The PCR products were analysed on electrophoreses using a 1% agarose gel containing ethidium bromide and were visualized under UV illumination. The QIAquick Gel Extraction Kit (Qiagen, Germany) was used to purify the PCR products. These products were sequenced bi-directionally with both sense (IBDVP2F) and antisense (IBDVP2R) primers that were used in the PCR amplification. The sequencing was performed utilizing BigDye terminator v3.1 cycle sequencing kit in an ABI 3100 genetic analyser (Applied Biosystems Foster City, California, USA).
Sequence analysis, phylogeny, and selective pressure analysis
Nucleotide sequences were aligned with ClustalW [28] and analysed using the BioEdit 5.0 package [29]. The obtained nucleotide sequences were submitted to GenBank and are available under the accession numbers; MT304668-MT304670. Sequence Demarcation Tool (SDT) was used to display the nucleotide pairwise identity scores through a color-coded matrix [30]. Phylogenetic analyses were carried out using general time-reversible (GTR) model [31], which was selected using jModelTest [32], and maximum-likelihood trees were constructed using RaxML version 8.2.11 [33] with 1000 bootstrap replicates.
The VP2 gene-specific estimates of dN/dS were predicted using the Synonymous-Non-Synonymous Analysis Program (SNAP) [34]. The number of potential synonymous and non-synonymous changes were counted as well as the number of actual synonymous and non-synonymous changes in codon between each pair. The dN/dS ratio was calculated by comparing the proportion of observed non-synonymous substitutions over the proportion of observed synonymous substitutions. These were then adjusted for multiple hits using the Jukes–Cantor correction [34].
Results and discussion
Understanding the epidemiology of vvIBDV is important to underpin the viral evolution, virus spread and up-to-date field status for effective control strategies. Previous studies have reported a widespread usage of live vaccines help in the spread of IBDVs with emergence of vaccine escape mutant strains [35–38]. Based on serological evidences of IBDV serotype I in wild birds, it has been suggested that wild birds may be critical player in the epidemiology of IBDV and may act as reservoir for the IBDV [39–43].
Usage of live vaccines is blamed to be responsible for spillover of viral vaccines from poultry into wild birds [22, 23] The safety of attenuated IBDV vaccines that are commonly used in the Egyptian poultry sectors might be examined systematically within the commercial avian species but not in wild birds that might be susceptible to infection [24]. In spite of restricted epidemiological studies for viruses in wild birds, spilling over of poultry vaccines has been documented in wild birds [24]. Despite the direct impacts of the attenuated viral vaccines on wild birds, the potential for these vaccines to develop significant levels of pathogenicity in wild birds is a major challenge [44]. These findings highlight the potential roles of wild birds in the spread of IBDV. In the current study, twenty-eight samples were collected from randomly selected wild birds from three Egyptian Governorates These samples were individually screened for IBDV by the RT-PCR targeting the VP2 gene. Three samples (3 out of 28) were identified positive among the tested cloacal samples (Table 1). The sampled wild birds were classified into four different families; Anatidae (A. crecca species, n = 8 and A. platyrhynchos species, n = 7), Ardeidae (B. ibis species, n = 6) and Phasianidae (C. coturnix species, n = 7) based on their taxonomy (Table 1). The vvIBDV isolate Egypt-USC-IBD-1-2019 was collected from B. ibis species of Qaulubia Governorate while the IBDV vaccine-like strains Egypt-USC-IBD-2-2019 and IBDV isolate Egypt-USC-IBD-3-2019 were isolated from A. crecca species, Sharkia Governorate and B. ibis species, Monofiya Governorate, respectively. Identification of these IBDV in birds from the Nile Delta of Northern Egypt is of particular concern. The Nile Delta is historically a crucial stopover for millions of birds. These birds migrate between the Palearctic and Afrotropical regions every year. Therefore, the Nile Delta is considered one of the most important migration routes for wild birds [24, 25]. Circulation of IBDV in these wild birds could pose a risk of infection to other birds migrating through multiple routes.
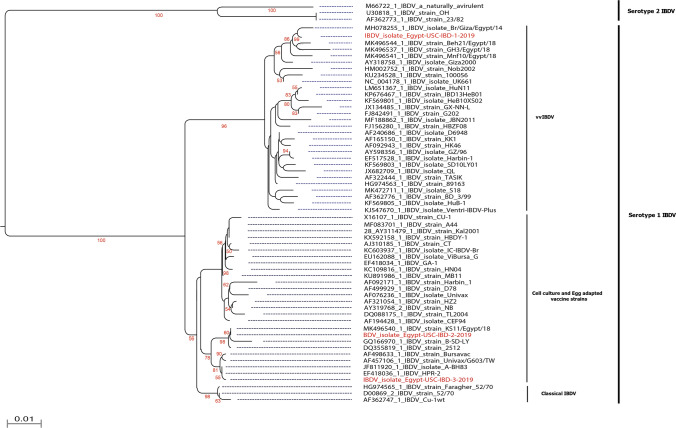
The phylogenetic analysis based on VP2 sequences revealed that IBDV isolate Egypt-USC-IBD-1-2019 clustered with vvIBDV (Fig. 1) whereas two other isolates (IBDV isolate Egypt-USC-IBD-2-2019 and IBDV isolate Egypt-USC-IBD-3-2019) clustered with cell-culture adapted IBDV vaccine strains (Fig. 1). The highly variable domain of VP2 protein carries the antigenic region which is accountable for neutralizing antibody as well as virulence [9]. Genetic analysis of the highly variable domain of VP2 may help to identify the genetic relationship among IBDV strains [9]. Previous studies have demonstrated that there are two major and three minor hydrophilic regions within the VP2 [45]. The major hydrophilic regions are represented by peak A (aa 212–224) and B (aa 314–324) while the three minor hydrophilic regions ranged from aa 248–252, 279–290 and 299–305 [45]. Likewise, there is a serine-rich heptapeptide 326SWSASGS332 sequence close to the second major hydrophilic region, was found in virulent strains and it might be the virulence marker for IBDV [45] which is detected in the isolated vvIBDV characterized in this study. Previous reports demonstrated the structural conformation of the major hydrophilic peaks A and B as critical in determining the IBDV antigenicity. Overall, finding revealed high selection pressures in peak A and B, and highlight key amino acids that can play critical roles in preserving the structural confirmation of the VP2 protein and decide the magnitude of virulence, pathogenicity and characterization of IBV.
Fig. 1.
Phylogenetic analysis of studied isolates and their clustering patterns with representative IBDVs. Full length VP2 gene based phylogenetic analysis of three wild-bird origin IBDV isolates with representative strains of currently circulating IBDVs in Egypt. One of the reported isolates clustered within vvIBDVs with close relationship with the previously characterized strains from commercial poultry while the other one clustered vaccine strains. The reported isolated marked with red colour
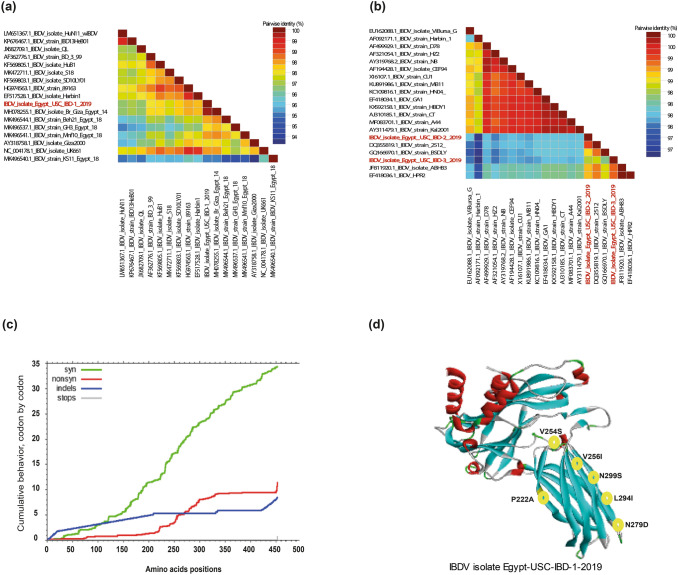
Our analysis of VP2 gene sequences indicated that wild bird-origin IBDV isolates carried high similarity with vvIBDV (Fig. 2a) and vaccine strains (Fig. 2b) previously reported from domestic chickens in Egypt [17]. Moreover, the presence of IBDV in the cloacal swabs of the wild birds suggested that these birds can shed the virus without developing disease, which may have implications in the IBDV epidemiology. These data suggest an epidemiological link between domestic chickens and wild birds in the epidemiology of IBDVs. Previous studies have demonstrated the serological presence of IBDV in multiple wild bird species [39–43]. Since serotype I of IBDV is known to be a pathogenic in avian species other than chicken, it become clear that IBDV didn't assume a significant role in the bird’s deaths [46]. Our results revealed that such isolates are most likely spilt over from previous outbreaks in vaccinated poultry and are carried by free-living wild birds, which may be playing a role in their dissemination. It has been well recognized the spillover of wild birds’ viruses to domesticated poultry causing disease and also in the other direction (from poultry to wild birds) [21, 22]. Previous studies have reported that passaging of NDV vaccine strains in wild bird species may provide selective pressures that could lead to antigenic variabilities or an increase in virulence [47–49]. The VP2 gene-specific estimates of dN/dS were predicted using SNAP and the number of potential synonymous and non-synonymous changes were counted. The sites under positive or negative selection were mapped and outlined in Fig. 2c.
Fig. 2.
Pairwise identity, localization of specific mutations in the VP2 protein of the newly identified vvIBDV strain and IBDVs selective pressure. The pairwise identities plot of VP2 gene for a Egypt-USC-IBD-1-2019 compared to vvIBDVs and b Egypt-USC-IBD-2-2019 and Egypt-USC-IBD-3-2019 compared to IBDV vaccine-like strains aligned by ClustalW and displayed by Sequence Demarcation Tool (SDT) software. c Cumulative behaviour of the average synonymous and non-synonymous substitutions moving codon by codon across VP2 gene. d 3D structure template for IBDV isolate IBDV/USC-3/2019 showed the localization of specific mutations in the VP2 protein for IBDV isolate IBDV/USC-1/2019. The 3D was visualized by PyMOL software
Interestingly, the VP2 gene of Egypt-USC-IBD-1-2019 vvIBDV isolated from wild bird gained specific amino acid mutations (P222A, V256I, N279D, L294I, and N299S) (Fig. 2d), which are conserved among all Egyptian vvIBDV strains. However, a unique amino acid mutation (G254S) was observed in the studied isolates (Fig. 2d). These results suggested an existing close link between the IBDV epidemiology in both domesticated chickens and wild birds. The IBDV strains characterized from wild birds may be infectious and virulent in chickens and warrant future investigations. Although the number of samples analysed in this study were limited, it is plausible that the circulation of IBDVs among wild birds is much higher than previously thought. Continuous disease monitoring, surveillance, and subsequent complete viral genome characterization is advisable in case of spillover from wild birds to commercial poultry and/or reverse spillover from commercial poultry to wild birds.
Future investigations are warranted to underpin the proposed virulence markers as guidelines for the cataloguing of IBDV strains into diverse pathotypes. Additional animal trials of the currently used commercial inactivated IBDV vaccines are needed to confirm their effectiveness against field IBDV strains without the use of live IBDV vaccines. To further understand the transmissibility of the wild bird-origin IBDV strains, additional experiments such as assessment of the minimum infectious and lethal doses need to be performed. Thus, further research is needed to investigate the pathobiology of wild bird-origin IBDVs that might help to explore the pathobiology and immunosuppressive impacts of IBDV isolates and tracking their evolutionary changes to better assess the nature of recently circulating strains of IBDV.
Acknowledgements
This work was financed by International Foundation for Science (IFS), project No. I-3-B-6270-1), The Organisation of Islamic Cooperation’s Standing Committee on Scientific and Technological Cooperation (COMSTECH). Additionally, this study was supported by the British Council under Grants Numbers: 172710323 and 332228521. The funding sources had no role in the study design, collection, or analysis of the data, writing of the manuscript, or in the decision to submit the manuscript for publication.
Author contributions
RFE and MM: designed the study; RFE and MAR: performed experiments; RFE, MAR and MM: wrote the manuscript; and RFE and MAR: provided the virus samples.
Data availability
All sequence data are available in GenBank, and their accession numbers are MT304668-MT304670.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
International, national, and/or institutional guidelines for collection of cloacal swabs from wild birds were followed. Samples collection and experiments were conducted with the approval of the Local Ethics Committee on Animal Experimentation at Faculty of Veterinary Medicine, University of Sadat City, Egypt.
Footnotes
The original online version of this article was revised: Fig. 1 and Fig. 2 has interchanged their position in the article and it has been corrected.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rania F. El Naggar and Mohammed A. Rohaim have contributed equally.
Change history
1/30/2021
The original version of this article unfortunately contained an error in figure.
References
- 1.Sharma JM, Kim IJ, Rautenschlein S, Yeh HY. Infectious bursal disease virus of chickens: pathogenesis and immunosuppression. Dev Comp Immunol. 2000;24:223–235. doi: 10.1016/s0145-305x(99)00074-9. [DOI] [PubMed] [Google Scholar]
- 2.Withers DR, Young JR, Davison TF. Infectious bursal disease virus-induced immunosuppression in the chick is associated with the presence of undifferentiated follicles in the recovering bursa. Viral Immunol. 2005;18(1):127–137. doi: 10.1089/vim.2005.18.127. [DOI] [PubMed] [Google Scholar]
- 3.Müller H, Scholtissek C, Becht H, Muller H. The genome of infectious bursal disease virus consists of two segments of double-stranded RNA. J Virol. 1979;31:584–589. doi: 10.1128/jvi.31.3.584-589.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manning DS, Leong JC. Expression in Escherichia coli of the large genomic segment of infectious pancreatic necrosis virus. Virology. 1990;179:16–25. doi: 10.1016/0042-6822(90)90268-v. [DOI] [PubMed] [Google Scholar]
- 5.Manning DS, Mason CL, Leong JC. Cell-free translational analysis of the processing of infectious pancreatic necrosis virus polyprotein. Virology. 1990;179:9–15. doi: 10.1016/0042-6822(90)90267-u. [DOI] [PubMed] [Google Scholar]
- 6.Lejal N, Da Costa B, Huet JC, Delmas B. Role of Ser-652 and Lys-692 in the protease activity of infectious bursal disease virus VP4 and identification of its substrate cleavage sites. J Gen Virol. 2000;81:983–992. doi: 10.1099/0022-1317-81-4-983. [DOI] [PubMed] [Google Scholar]
- 7.Brandt M, Yao K, Liu M, Heckert RA, Vakharia VN. Molecular determinants of virulence, cell tropism, and pathogenic phenotype of infectious bursal disease virus. J Virol. 2001;75:11974–11982. doi: 10.1128/JVI.75.24.11974-11982.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coulibaly F, Chevalier C, Gutsche I, Pous J, Navaza J, Bressanelli S, Delmas B, Rey FA. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell. 2005;120:761–772. doi: 10.1016/j.cell.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 9.Letzel T, Coulibaly F, Rey FA, Delmas B, Jagt E, van Loon AAMW, Mundt E. Molecular and structural bases for the antigenicity of VP2 of infectious bursal disease virus. J Virol. 2007;81:12827–12835. doi: 10.1128/JVI.01501-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Einem UI, Gorbalenya AE, Schirrmeier H, Behrens SE, Letzel T, Mundt E. Vp1 of infectious bursal disease virus is an RNA-dependent RNA polymerase. J Gen Virol. 2004;85:2221–2229. doi: 10.1099/vir.0.19772-0. [DOI] [PubMed] [Google Scholar]
- 11.Yu F, Ren X, Wang Y, Qi X, Song J, Gao Y, Qin L, Gao H, Wang X. A single amino acid V4I substitution in VP1 attenuates virulence of very virulent infectious bursal disease virus (vvIBDV) in SPF chickens and increases replication in CEF cells. Virology. 2013;440:204–209. doi: 10.1016/j.virol.2013.02.026. [DOI] [PubMed] [Google Scholar]
- 12.Cosgrove AS. An apparently new disease of chickens-avian nephrosis. Avian Dis. 1962;6:385–389. [Google Scholar]
- 13.Jackwood DH, Saif YM. Antigenic diversity of infectious bursal disease viruses. Avian Dis. 1987;31:766–770. [PubMed] [Google Scholar]
- 14.Chettle N, Stuart JC, Wyeth PJ. Outbreak of virulent infectious bursal disease in East Anglia. Vet Rec. 1989;125:271–272. doi: 10.1136/vr.125.10.271. [DOI] [PubMed] [Google Scholar]
- 15.Alkie TN, Rautenschlein S. Infectious bursal disease virus in poultry: current status and future prospects. Vet Med (Auckl) 2016;7:9–18. doi: 10.2147/VMRR.S68905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eterradossi N, Saif YM. Infectious bursal disease. In: Swayne DE, Glisson JR, Mcdougald LR, Nolan LK, Suarez DL, Nair V, editors. Diseases of poultry. 13. Hoboken: Wiley-Blackwell Publishing; 2013. pp. 219–246. [Google Scholar]
- 17.Samy A, Courtillon C, Briand F, Khalifa M, Selim A, Arafa A, Hegazy A, Eterradossi N, Soubies SM. Continuous circulation of an antigenically modified very virulent infectious bursal disease virus for fifteen years in Egypt. Infect Genet Evol. 2020;78:104099. doi: 10.1016/j.meegid.2019.104099. [DOI] [PubMed] [Google Scholar]
- 18.Shehata AA, Sultan H, Halami MY, Talaat S, Vahlenkamp TW. Molecular characterization of very virulent infectious bursal disease virus strains circulating in Egypt from 2003 to 2014. Arch Virol. 2017;162(12):3803–3815. doi: 10.1007/s00705-017-3554-3. [DOI] [PubMed] [Google Scholar]
- 19.El-Batrawy A (1990) Studies on severe outbreaks of infectious bursal disease. Proceedings of the 2nd Scientific Conference of the Egyptian Veterinary Poultry Association (pp. 239–252). Cairo, Egypt.
- 20.Read AF, Baigent SJ, Powers C, Kgosana LB, Blackwell L, Smith LP, Kennedy DA, Walkden-Brown SW, Nair VK. Imperfect vaccination can enhance the transmission of highly virulent pathogens. PLoS Biol. 2015;13(7):e1002198. doi: 10.1371/journal.pbio.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rohaim MA, El Naggar RF, Helal AM, Hussein HA, Munir M. Reverse spillover of avian viral vaccine strains from domesticated poultry to wild birds. Vaccine. 2017;35(28):3523–3527. doi: 10.1016/j.vaccine.2017.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collet SR. Principles of disease prevention, diagnosis and control introduction. In: Swayne DE, Glisson JR, McDougald LR, Nolan LK, Suarez DL, Nair VL, editors. Diseases of poultry. 13. Hoboken: Wiley-Blackwell; 2013. pp. 4–40. [Google Scholar]
- 23.Kelleher BP, Leahy JJ, Henihan AM, O'Dwyer TF, Sutton D, Leahy MJ. Advances in poultry litter disposal technology—a review. Bioresour Technol. 2002;83:27–36. doi: 10.1016/s0960-8524(01)00133-x. [DOI] [PubMed] [Google Scholar]
- 24.Denny P. Africa. In: Finlayson M, Moser M, editors. Wetlands. London: International Waterfowl and Wetlands Research Bureau; 1991. pp. 115–148. [Google Scholar]
- 25.Gerloff NA, Jones J, Simpson N, Balish A, Elbadry MA, Baghat V, Rusev I, de Mattos CC, de Mattos CA, Zonkle LE, Kis Z, Davis CT, Yingst S, Cornelius C, Soliman A, Mohareb E, Klimov A, Donis RO. A high diversity of Eurasian lineage low pathogenicity avian influenza A viruses circulate among wild birds sampled in Egypt. PLoS ONE. 2013;8(7):e68522. doi: 10.1371/journal.pone.0068522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackwood DJ, Sommer-Wagner S. Genetic characteristics of infectious bursal disease viruses from four continents. Virology. 2007;365:369375. doi: 10.1016/j.virol.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 27.OIE (2012) Infectious bursal disease. In OIE Biological Standards Commission (Ed). Manual of diagnostic tests and vaccines for terrestrial animals (Mammals, Birds and Bees) 7th edn (pp. 549–565). Paris: Office international des epizooties
- 28.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 30.Muhire BM, Varsani A, Martin DP. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE. 2014;9:e108277. doi: 10.1371/journal.pone.0108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tavaré S (1986) Some probabilistic and statistical problems in the analysis of DNA sequences, In: Miura RM (eds) Some mathematical questions in biology—DNA sequence analysis, (Providence, RI: Amer Math Soc), pp. 57–86.
- 32.Guindon S, Dufayard J, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 33.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kryazhimskiy S, Plotkin JB. The population genetics of dN/dS. PLoS Genet. 2008;4(12):e1000304. doi: 10.1371/journal.pgen.1000304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adamu J, Owoade AA, Abdu PA, Kazeem HM, Fatihu MY. Characterization of field and vaccine infectious bursal disease virus from Nigeria revealing possible virulence and regional markers in the VP2 minor hydrophilic peaks. Avian Pathol. 2013;42(5):420–433. doi: 10.1080/03079457.2013.822055. [DOI] [PubMed] [Google Scholar]
- 36.Habiba U, Maqbool A, Safdar M, Zia N, Mehmood A, Usman M, Sharif M, Khan A, Umar S. Detection and phylogeny of infectious bursal disease virus (IBDV) during field outbreaks in broilers. Pak J Zool. 2020;52(2):659–667. [Google Scholar]
- 37.Imran MS, Aslam A, Yasin M, Yaqoob T, Zahid B. A Comparative assessment of efficacy of currently applied vaccines in broiler chicken against individual and co-infection with field prevailing newcastle disease and infectious bronchitis viruses. Pak J Zool. 2020;52(5):1895–1901. [Google Scholar]
- 38.Arshad RW, Aslam A, Imran MS, Ashraf K, Akhtar R. Comparative evaluation of live vector, immune complex and intermediate plus vaccines of infectious bursal disease in broiler chicken. Pak J Zool. 2019;51(5):1837–1841. [Google Scholar]
- 39.Nawathe DR, Onunkwo O, Smith IM. Serological evidence of infection with the virus of infectious bursal disease in wild and domestic birds in Nigeria. Vet Rec. 1978;102:444. doi: 10.1136/vr.102.20.444. [DOI] [PubMed] [Google Scholar]
- 40.Wilcox G, Flower R, Baxendale W, Mackenzie J. Serological survey of wild birds in Australia for the prevalence of antibodies to egg drop syndrome 1976 (EDS-76) and infectious bursal disease viruses. Avian Pathol. 1983;12:135–139. doi: 10.1080/03079458308436155. [DOI] [PubMed] [Google Scholar]
- 41.Jeon WJ, Lee EK, Joh SJ, Kwon Jh, Yang CB, Yoon YS, Choi KS. Very virulent infectious bursal disease virus isolated from wild birds in Korea: epidemiological implications. Virus Res. 2008;137:153–156. doi: 10.1016/j.virusres.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 42.Kasanga CJ, Yamaguchi T, Wambura PN, Munang’andu HM, Ohya K, Fukushi H. Detection of infectious bursal disease virus (IBDV) genome in free-living pigeon and guinea fowl in Africa suggests involvement of wild birds in the epidemiology of IBDV. Virus Genes. 2008;36:521–529. doi: 10.1007/s11262-008-0219-z. [DOI] [PubMed] [Google Scholar]
- 43.Ogawa M, Wakuda T, Yamaguchi T, Murata K, Setiyono A, Fukushi H, Hirai K. Seroprevalence of infectious bursal disease virus in free-living wild birds in Japan. J Vet Med Sci. 1998;60:1277–1279. doi: 10.1292/jvms.60.1277. [DOI] [PubMed] [Google Scholar]
- 44.Chong YL, Padhi A, Hudson PJ, Poss M. The effect of vaccination on the evolution and population dynamics of avian paramyxovirus-1. PLoS Pathog. 2010;6(4):e1000872. doi: 10.1371/journal.ppat.1000872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van den Berg TP, Gonze M, Morales D, Meulemans G. Acute infectious bursal disease in poultry: immunological and molecular basis of antigenicity of a highly virulent strain. Avian Pathol. 1996;25:751–768. doi: 10.1080/03079459608419179. [DOI] [PubMed] [Google Scholar]
- 46.van den Berg TP, Morales D, Enterradossi N, Rivallan G, Toquin D, Raue R, Zierenberg K, Zhang MF, Zhu YP, Wang CQ, Zheng HJ, Wang X, Chen GC, Lim BL, Muller H. Assessment of genetic, antigenic and pathotypic criteria for the characterization of IBDV strains. Avian Pathol. 2004;33(5):470–476. doi: 10.1080/03079450400003650. [DOI] [PubMed] [Google Scholar]
- 47.Gould AR, Kattenbelt JA, Selleck P, Hansson E, Della-Porta A, Westbury HA. Virulent Newcastle disease in Australia: molecular epidemiological analysis of viruses isolated prior to and during the outbreaks of 1998–2000. Virus Res. 2001;77:51–60. doi: 10.1016/s0168-1702(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 48.Zahid B, Qazi IJ, Zohaib A, Aslam A, Akhter R, Sadia H, Ulain Q, Sultana R, Irshad I, Alyas S. Detection and molecular characterization of virulent newcastle disease virus in ducks (Anas platyrhynchos domesticus) Pak J Zool. 2019;52(1):369–372. [Google Scholar]
- 49.Brown VR, Bevins SN (2017) A review of virulent newcastle disease viruses in the United States and the role of wild birds in viral persistence and spread [published correction appears in Vet Res 48(1):77 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequence data are available in GenBank, and their accession numbers are MT304668-MT304670.