Abstract
In this paper, Doxil coupled with anti-CD133 monoclonal antibodies made by either routine or optimized post-insertion technique, were compared with respect to their size, drug leakage, release pattern and the number of antibodies conjugated per single liposome. The results demonstrated that the number of antibodies conjugated per liposome in the optimized post-insertion technique was almost two times more than those in the routine post-insertion method. However, the drug release and leakage pattern was almost similar between the two methods. Furthermore, anti-tumor activity and therapeutic efficacy of the preferred CD133–targeted Doxil with Doxil was compared in terms of their in vitro binding, uptake, internalization and cytotoxicity against HT-29 (CD133+) and CHO (CD133-) cells. Flow cytometry analyses and confocal laser scanning microscopy results exhibited a significantly higher cellular uptake, binding and internalization of CD133-targeted Doxil in CD+133 cells relative to Doxil. Cytotoxicity results revealed a lower in vitro inhibitory concentration for CD133–targeted Doxil compared to Doxil. However, CHO (CD133-) cells displayed a similar uptake and in vitro cytotoxicity for both CD133-Doxil and non-targeted Doxil. Therefore, the results of this study can exhibit that specific recognition and binding of antibodies with CD133 receptors on HT-29 cells can result in enhanced cellular uptake, internalization and cytotoxicity. The research suggests further investigation for in vivo studies and may offer proof-of-principle for an active targeting concept.
Keywords: Doxil, CD133–targeted Doxil, Routine post-insertion technique, Optimized post-insertion technique, Cellular uptake, Cellular binding, Cytotoxicity
1. Introduction
A recent advance in the nanocarriers has been proposed as a promising strategy to deliver anti-cancer drugs to cancer cells (Malam, Loizidou and Seifalian 2009). Leaky and tortuous tumor vessels allow nanoparticles to distribute through cancer cells more efficiently as compared to the healthy blood vessels in which their access has been limited (Xu, Paxton and Wu 2016). Liposomal formulations of anticancer drugs have been comprehensively considered as a tool to efficiently develop drug delivery systems (Malam et al. 2009) and the general therapeutic efficacy are applied to several types of cancer treatment (Malam et al., 2009, Corvo et al., 2016). However, in some cases, drug resistance in cancer chemotherapy has become a big issue being attributed to the existence of a small group of cancer cells called cancer stem cells (CSCs). They give rise to the tumor recurrence and ultimately metastasis as a result of their substantial chemo- and radio resistance (Mi, Huang and Deng 2018). Therefore, this scenario has requested further developments in the therapeutic index via ligand-targeted liposomes to selectively target cell surface receptors expressed on cancer cells. The choice of over- expressed cell surface receptors of cancer cells for active targeting is a key determinant for successful therapy. CD133 appears to be a potentially motivating ligand for targeted therapy that often over-expressed on CSC and was found in several kinds of cancer consisting of the breast; colon, prostate, liver, pancreatic, and lung cancer (Ferrandina et al. 2009). Previous results have shown that directly targeting CD133 with monoclonal antibody (Mi et al. 2018), aptamer (Gui et al. 2019) and antibody fragments (Olsen et al. 2019) in addition to biological drugs (Schmohl and Vallera 2016) might be an effective approach to eliminate cancer stem cells. CD133 plays an important role both in activating Wnt/β-catenin pathway (Evangelista, Tian and de Sauvage 2006) and cell growth and proliferation (Gong and Huang 2012), which may promote resistance to chemotherapy and radiotherapy procedures (Wu et al. 2015).
Results have been shown that ligand-targeted liposomal containing anticancer drugs could improve cellular binding and cytotoxicity relative to non-targeted liposomes. Therefore, a simple and flexible method is necessary to prepare ligand-targeted liposomes for clinical trials. Recently the post-insertion technique has been developed to prepare ligand-targeted liposomes (Iden and Allen, 2001). Principally, the technique involves linking ligands (antibodies, antibody fragments, peptides) to terminal groups of PEG, which can extend the circulation time, lessen undesirable side effects, and mainly target specific cells.
The purpose of this study was to optimize the conjugation of Doxil with anti CD133 antibody via post-insertion approach and then compare cellular uptake, binding, internalization, and cytotoxicity of the prefeered CD133–targeted Doxil with Doxil against HT-29 (CD133+) and CHO (CD133-) cell lines.
2. Materials and Methods:
2.1. Materials
Methoxy-polyethyleneglycol (MW 2000)–distearoylphosphatidyl- ethanoloamine (mPEG2000–DSPE) and 1,2-Distearoyl-sn-glycero-3-phospho-ethanolamine-N-[succinimidyl (polyethylene glycol)] (DSPE–PEG3400–NHS) was purchased from Nanocs Inc. (USA). Purified anti-human CD133 Antibody (clone 7) and PE anti-human CD133 Antibody and PE Mouse IgG1, κ Isotype Ctrl (FC) Antibody (clone MOPC-21) were obtained from Biolegend. HT-29 and CHO cell lines were prepared from the National Cell Bank of Iran (Pasteur Institute, Iran-Tehran). All the chemicals were used as usual without additional purification. Milli-Q water (18Mcm) was used for the rinsing and preparation of solutions.
2.2. Conjugation of Doxil with anti-CD133 monoclonal antibodies via post insertion method
2.2.1. Routine Post-Insertion method
At first, mPEG2000–DSPE, and DSPE–PEG3400–NHS as a coupling lipid was mixed at a 4:1 M ratio and dried under a stream of nitrogen gas until no liquid remained. The dried lipid films were hydrated in 1 ml of sterile water for injection at 47 °C, and then immediately was mixed with the aliquot of anti-CD133 antibody (0.5 mg/ml) at a molar ratio of 6:1 (micelle/antibody) in phosphate-buffered saline (0.1 M, pH 7.4). The solution was stirred for 4 h at room temperature. In order to quench the reaction, 5 mg glycine was added. In the end, the micellar dispersion was co-incubated with 0.4 ml Doxil at 60 °C for 2 h in a water bath with continuous stirring.
2.2.2. Optimization of Post-insertion method
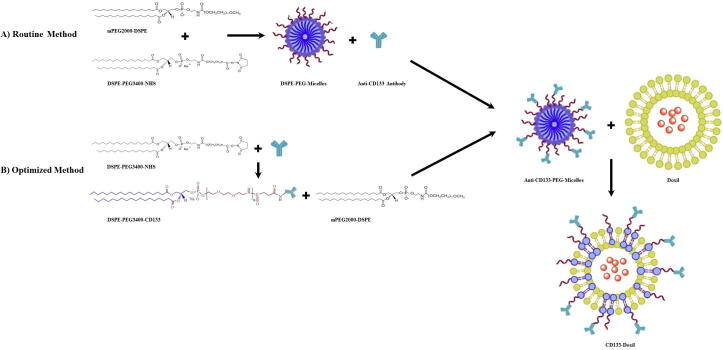
To this, DSPE–PEG3400–NHS as a coupling lipid was dissolved in 25 µl of chloroform at pH 7.4. After drying the chloroform, the antibody was immediately added (pH 7.4) to the dried film of DSPE-PEG (3400)-NHS at a molar ratio of 6:1 (micelle/antibody). After sonication for 5 min, the solution incubated for 4 h at room temperature. In order to quench the reaction, 5 mg glycine was added. Afterward, the lipid-conjugated antibody was then mixed with the mPEG2000–DSPE micelles for 2 h [mPEG2000–DSPE micelles were prepared by dissolving mPEG2000–DSPE lipid in 25 µl of chloroform. A dried lipid film was made by using a stream of nitrogen. Then 25 µl of PBS buffer was added to the dried lipid film and the hydrated lipid film was sonicated and stored in the fridge]. (the molar ratio of mPEG2000–DSPE to DSPE–PEG3400–NHS is 4:1). Finally, the micellar dispersion was co-incubated with Doxil being exactly the same as the routine post-insertion method. Fig. 1 shows the preparation of CD133-targeted Doxil using the post-insertion method.
Fig. 1.
Schematic of the route for the routine and optimized post-insertion method for preparation of CD133-Doxil.
2.3. Particle size and zeta potential
A dynamic light scattering (DLS, Malvern system ZS3600, Malvern, UK) was used to analyze the hydrodynamic size and zeta potential of the samples. Samples were freshly prepared by the addition of an aliquot of the buffer to 0.01 mg/ml lipid concentration. Measurements were obtained at 25 °C with a scattering angle of 173° and a wavelength of 633 nm.
2.4. Phospholipid concentration
Phospholipids concentration can be measured calorimetrically by Stewart assay. In this procedure, the red inorganic compound ammonium was produced by forming a complex between the phospholipid head groups and ammonium ferrothiocyanate which after extraction into chloroform could be quantified by spectrophotometry.
2.5. Doxorubicin concentration
Before and after the post-insertion method, Doxorubicin concentrations were calculated from the absorption at 480 nm in methanol by relationship to a standard curve. The percent of doxorubicin-loaded was determined using the following equation:
2.6. In vitro release of Doxorubicin from nanoparticles
Briefly, 250 µl of each sample was placed into dialysis membranes (membrane tubing; MWCO12-14 KDa, Sigma-Aldrich USA) and immersed in two different types of pH (5.5 and 7.4) at 25 ml phosphate-buffered saline. All samples were incubated at 37C° and rotated at 50 rpm in the dark to avoid Doxorubicin photodegradation. Then 1 ml of samples were collected at different time intervals (2, 4, 8, 12, 24, 48, and 72 h), and replaced with an equivalent volume of fresh buffer (37C°).
The absorbance of the withdrawn samples was measured at 480 nm using a UV–visible spectrophotometer and the drug concentration was calculated using the standard calibration curve. Drug release profiles from liposome were expressed according to the following equation:
Where [DXR]t is referred to the amount of Doxorubicin are released at time t, [DXR] total is the whole Doxorubicin existing in the Doxorubicin-loaded nanoparticles.
2.7. The amount of conjugated Anti-CD133 antibody
A 300-kDa molecular weight cut-off (MWCO) dialysis membrane (Spectrum) was used to separate the mAb conjugated micelles and free mAb. Intact antibodies were eliminated from liposomes via dialyzing four times against 10 wt% sucrose, 10 mM histidine buffer in pH 6.5 at 4 °C. BCA protein assay (Pierce) was used to quantify the amount of anti-CD133 antibody incorporation into the liposomes.
2.8. Cell culture
HT-29 and CHO cell lines were grown in DMEM and DMEM F12 medium respectively. All culture mediums supplemented with 10% (by volume) fetal bovine serum (FBS), penicillin 100 units/, 50 IU/ml streptomycin in an incubator with 5% CO2 (by volume).The cells were detached from the culture flasks by incubating in trypsin for 5 min.
2.8.1. Investigation of CD133 expression level
Aliquot containing approximately 1 × 106 (HT-29 and CHO) cells were suspended in 0.1 M PBS, including 2 mM Ethylene Diamine Tetra Acetic acid (EDTA) and 2% FBS. Then the cells were incubated with 2 μl of PE anti-human CD133 mAb and PE Mouse IgG2b, κ antibody as an isotype control at 4 °C for 30 min and washed three times with ice-cold PBS. The fluorescence intensity was analyzed using a flow Space Cytometer (Partec, Münster, Germany) supplied with a 488 laser in the FL2 channel. The analysis was performed using the cyflow software (Tree Star, San Carlos, CA).
2.8.2. Cellular binding using Flow Cytometry
Characteristic fluorescence spectra of doxorubicin (ex: 480 nm, em: 580 nm) was used to determine CD133-specific cellular binding. Approximately 1 × 106 (HT-29 and CHO cells) were suspended in 0.1 M PBS containing 2 mM EDTA and 2% FBS. The cells were then incubated with free doxorubicin, Doxil, and CD133-Doxil at 4 °C for 1 h. The fluorescence intensity was analyzed using the Cyflow Space Cytometer (Partec, Münster, Germany) equipped with a 488 laser in the FL2 channel.
2.8.3. Cellular uptake using Flow Cytometry
Approximately 2 × 105 HT-29 and CHO cells were added in 24-well plates in complete DMEM and DMEM F12 containing 10% FBS, respectively. At the indicated time (24 h), the cells were treated with free Doxorubicin, Doxil, and CD133-Doxil (10 μg Doxorubicin/ml,) for 3 h at 37 °C in a serum-free media. The cells then were washed twice with PBS and removed from the culture flasks by incubating in 0.25% trypsin-EDTA solution. After centrifugation of cells at 1500 rpm for 5 min, the cell pellets were washed three times with cold PBS followed by resuspending in PBS 0.1 M, containing 2 mM EDTA and 2% FBS. FL2 channel of FCM was used to analyses the Geometric mean fluorescence intensity (MFI) of doxorubicin in cells.
2.8.4. Cellular uptake using confocal laser scanning microscopy (CLSM)
A Nikon confocal laser scanning microscopy was used to visualize the cellular internalization of formulations by HT-29 and CHO cells. The cells were placed in gelatin-coated cover-slips into 6-well plate overnight. The next day, approximately 2 × 105 cells were incubated with free Doxorubicin, Doxil, and CD133-Doxil (10 μg Doxorubicin/ml,) in a serum-free media for 3 h. To visualize the nucleus, the cells were washed three times with methanol before and after staining with DAPI-methanol (10 μg/ml) for 15 min. After fixing the cells with 75% ethanol at room temperature, the prepared slides were analyzed by CLSM.
2.8.5. Cytotoxicity study
Approximately 5 × 104 (HT-29 and CHO) cells per ml were added to each of the indicated samples in the 96-well plate. The cells were cultured overnight in DMEM and DMEM/F12 medium respectively, with growth medium, including 10% FBS at 37◦C in an incubator with 5% CO2. The formulations were serially diluted with serum-free media at eight doses in triplicate, following with incubating for 3 h at 37 °C. Complete culture media were used to wash the cells and remove the drug and then reincubated for an extra 48 h at 37 °C. Afterward, 10 µl of MTT (5 mg/mL) was added per well for 96 plates at 37 °C for 3 h. Then the media was replaced by 200 µl of DMSO. The absorbance was recorded at 570/630 nm.
Where blank contains no cells for measurement of the background, the absorbance of drug-treated cells and non-treated cells corresponded to sample and control respectively.
2.8.6. Competition assay
Competition assays were performed in cellular uptake, binding, and cytotoxicity to evaluate the specificity of CD133-targeted Doxil on HT-29 cells. To this, cells were pre-incubated with excess amounts of the free anti-CD133 antibody for 30 min before introducing CD133-Doxil to competitively inhibit the effect of CD133-Doxil.
2.9. Statistical analysis
Experiments were performed in triplicate. The results of each experimental group are stated as mean plus or minus standard deviation. Statistical analyses of the present study were completed using GraphPad Prism version 8.0.2.263 (GraphPad Software, San Diego, CA). Analysis comparisons were performed using a one-way analysis of variance (ANOVA). A p-value of less than < 0.05 is calculated to be statistically significant.
3. Results:
3.1. Size and charge of the nanoparticles
A dynamic light scattering was used to determine the mean liposome diameter and charge before and after insertion of an antibody to the formulation to ensure that CD133-Doxil had satisfactory parameters for improving cancer drug delivery based on EPR effects. Table 1 summarized size, zeta potential, and Polydispersity Indices (PDI) of Doxil before and after two post-insertion techniques. As can be seen in Table 1, after conjugation of Doxil to antibody, the size of CD133-Doxil in optimized and routine post-insertion techniques increased to 113 and 106 nm respectively. Polydispersity indices (PDI) could be an important parameter for assessing the quality of the nanoparticles in the systems. The PDI<0.10 could be a sign of the narrow particle size distribution (17). CD133-Doxil in both optimized and routine post-insertion methods showed a negative zeta potential of about −30 and −26 mv respectively. Zeta potentials with higher magnitude indicate improved stability, which is attributed to a greater electrostatic repulsion between nanoparticles.
Table 1.
Average size, Polydispersity Indices and zeta potential of CD133-Doxil of optimized and routine post-insertion method and non-targeted Doxil. Each value represents Mean ± standard deviation (n = 3). Particle sizes are presented as Z averages means ± standard deviation and Zeta potentials are reported as means ± zeta deviation. PDI values obtained from triplicate measurements for each formulation.
| Liposome | Z-Average (nm) | PDI | Zeta Potential (mv) |
|---|---|---|---|
| Doxil | 102.03 ± 0.680 | 0.036 ± 0.018 | −11.36 ± 0.23 |
| CD133-Doxil (Optimized method) | 113.86 ± 1.601 | 0.062 ± 0.025 | −30.7 ± 0.81 |
| CD133-Doxil (Routine method) | 106.01 ± 1.90 | 0.078 ± 0.057 | −26.9 ± 0.98 |
3.2. Doxorubicin Leakage:
The Doxorubicin concentration was calculated in the presence or absence of CD133-PEG3400-DSPE micelle to verify the change in the drug concentrations after the addition of antibody-micelles to the outer monolayer of liposomes. While in the absence of antibody-micelles, approximately 0.69% of Doxorubicin was released during 6 h incubation at 60 °C, this slightly increased to 7.69% and 8.98% in the presence of antibody-micelles in the optimized and routine post-insertion method respectively.
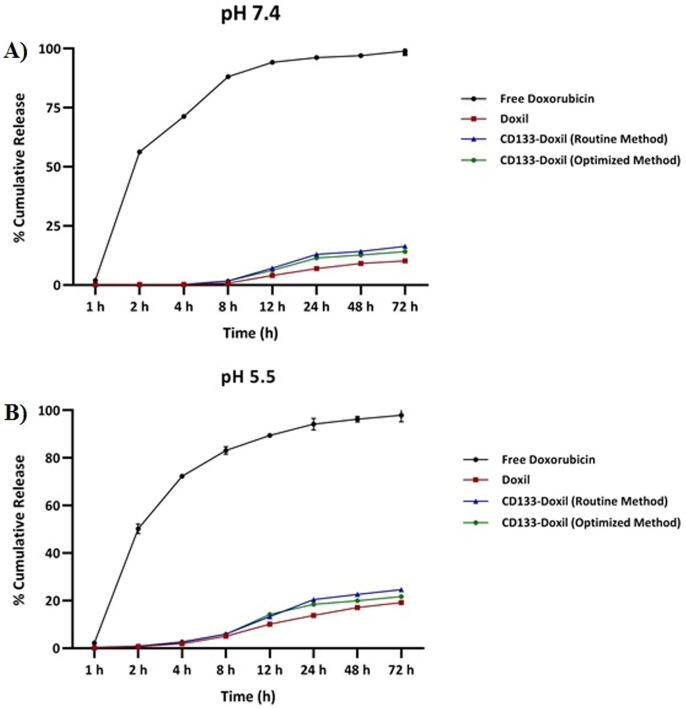
3.3. Doxorubicin release
The drug release profile of Doxorubicin from CD133-targeted Doxil was assessed to investigate whether the addition of anti CD133-PEG2000-DSPE micelles change the pattern release profile of CD133-targeted Doxil compared to that of Doxil. Fig. 2 shows the drug release profile of Doxorubicin from all formulations over a 72-hour period at pH 7.4 (corresponding to the pH of blood) and pH 5.5 (corresponding to the pH of the tumor extracellular). The Doxorubicin release for all formulations was linear and slow in physiological as well as the acidic medium. As can be seen in Fig. 2, doxorubicin was released at approximately the same rate for CD133-Doxil made by optimized and routine post-insertion method over a 72-hour period. However, the release rate of all formulations in acidic medium was faster and slightly higher than those in the physiologic one. The results also displayed that the doxorubicin release profile for all formulations slightly increased and then reached a plateau after 24 h and remained almost constant over 72 h. This is consistent with previous reports suggesting that doxorubicin release rate from liposomal doxorubicin in phosphate-buffered saline (PBS) at 37 is anticipated to be too low, because doxorubicin inside the liposome present as a precipitate which support the doxorubicin stability inside the liposome (Shibata et al. 2015). These results suggest that the change in the post-insertion method has almost no effect on the doxorubicin release pattern and slightly affect the release profile of Doxil.
Fig. 2.
Release profile of Doxorubicin from CD133-targeted Doxil at 37 °C in (A) post-insertion method at pH 7.4 (B) post-insertion method at pH 5.5.The concentration of Doxil in the dispersion is 0.5 mg/mL. Each value and error bar represents the mean of triplicate samples and its standard deviation.
3.4. The number of Anti-CD133 antibodies conjugated per single liposome
BCA kit (Pierce) was used to determine the total antibody concentration per liposomes before and after purification using a 300-kDa molecular weight cut-off.
The average number of phospholipid molecules per mL of the liposome for the optimized post-insertion method was calculated by using the Avogadro’s number, 6.023 × 1023 molecules/mole. The total phospholipid concentration analyzed by Stewart assay was 15 mM which gives an average of 9.03 × 1022 lipid molecules/mL. Similarly, the average number of antibody molecules/mL of liposome after purification based on BCA kit assay was 2.7 × 10-5 mM. This offered an average of 1.63 × 1019 antibody molecules/mL of the liposome.
The total surface area of liposome includes external and internal surface area.
Equation1 is used to calculate the outer layer surface area of the liposome. Liposome average size is 100 nm in diameter.
| (1) |
The inner layer surface area is calculated from Equation (2).
| (2) |
Here (h) is the thickness of the bilayer and it is about 5 nm and (r) is the diameter of the liposome that here is = 50 nm. The head group area of phosphatidylcholine of the liposome was indicated as (a) with a value of 0.71 nm2 (Mittal et al. 2018). The surface areas of both monolayers are added together. Then the total lipid area is divided into the head group area of one lipid molecule.
| (3) |
The above equation for a unilamlellar liposome is simplified to Equation (4):
| (4) |
Therefore the number of lipids in a 100 nm size liposome is about 80047.
| (5) |
Number of average monoclonal antibody molecules per liposome was calculated from Equation (6).
| (6) |
Based on the calculations in Table 2, it is supposed that approximately 14–15 molecule of antibodies were conjugated on the surface of every single liposome in the optimized post-insertion method and this number reduced to almost 6 molecules for the routine post-insertion technique.
Table 2.
calculation of number of antibody molecules/liposome for optimized and post-insertion method.
| Factors | Optimized post-insertion method | Routine post-insertion method |
|---|---|---|
| Phospholipid concentration | 15 mM | 13 mM |
| Lipid molecules/mL | 9.03 × 1022 | 7.82 × 1022 |
| The average amount of antibody molecules/mL based on BCA kit assay* | 2.69 × 10-5 ± 0.135 × 10-5 mM | 0.973 × 10-5 ± 0.008 × 10-5 mM |
| The efficiency of CD133-Doxil | 79% | 32% |
| Antibody molecules/mL of liposome. | 1.63 × 1019 | 5.90 × 1018 |
| Number liposomes/mL | 1.12 × 1018 | 9.7 × 1018 |
| Number of antibody molecules/liposome | 14.55 | 6.08 |
The average amount of antibody molecules/mL based on BCA kit assay are the mean ± SD for triplicate samples.
3.5. Cellular Expression, binding and uptake with Flow Cytometry
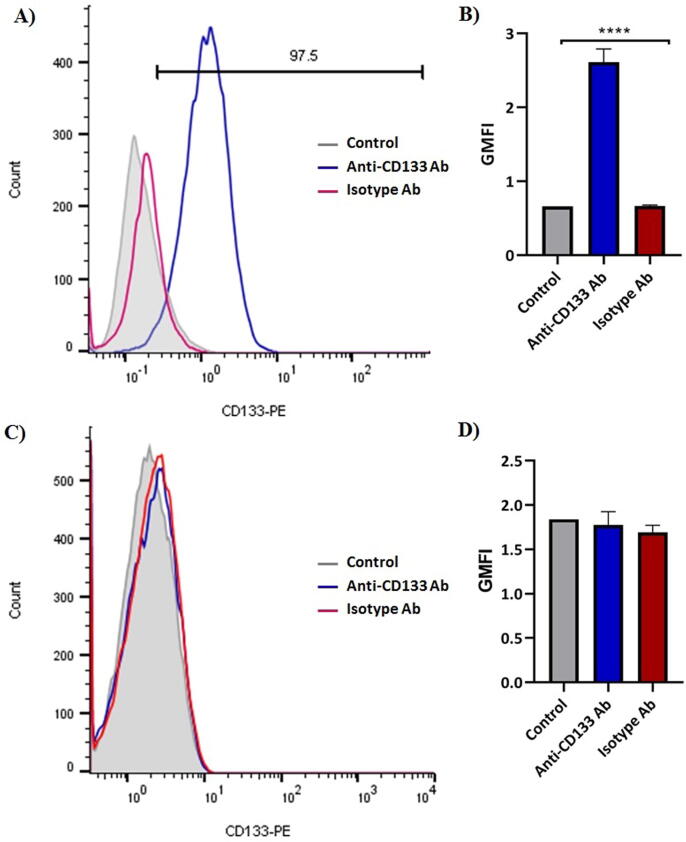
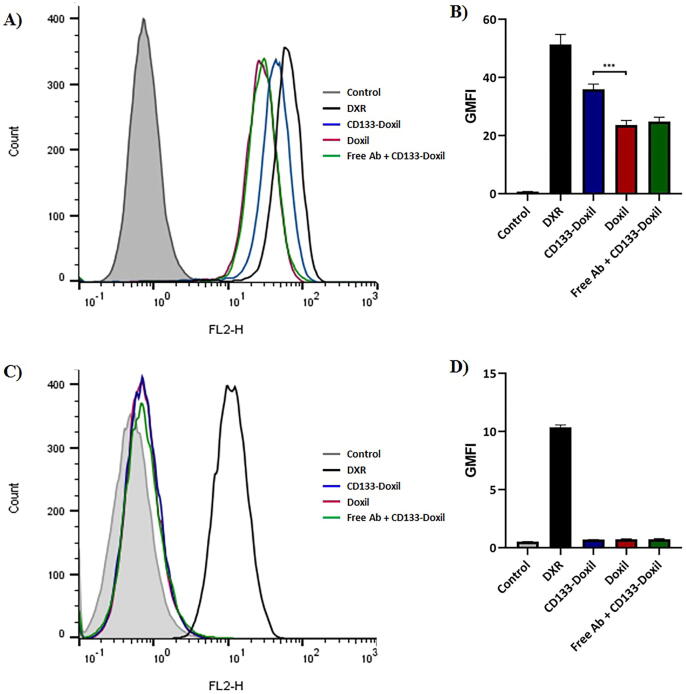
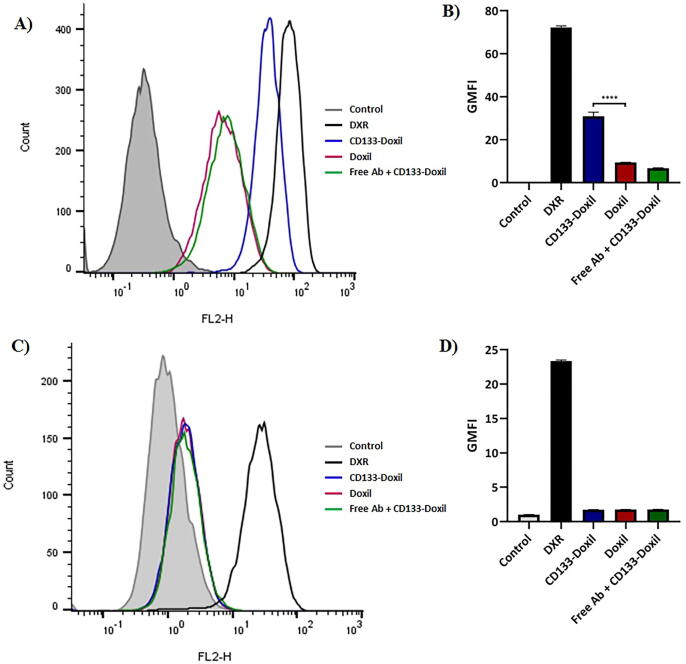
The relative percentages of cells expressing a single surface marker were determined by Flow Cytometric analysis. HT-29 and CHO cells were directly stained with PE anti-human CD133 mAb and PE Mouse IgG2b, κ antibody as an isotype control to determine the expression of CD133. While HT-29 cells showed a CD133 + population of greater than 97%, CD133+ cells were not detectable in CHO cells (Fig. 3). The cellular binding and uptake of doxorubicin in different liposome formulations by HT-29 (CD133+) and CHO (CD133-) cells were quantified using Flow Cytometry. According to the MFI of the cells (Fig. 4), the in vitro results indicated that CD133-Doxil bound to HT-29 cells were significantly (p < 0.001) higher than those for Doxil. Also, the specific uptake of monoclonal antibody conjugated Doxil by HT-29 cells was approximately 3-fold higher than those for the Doxil solution (Fig. 5). In contrast, no obvious difference was observed in cellular binding and uptake between Doxil and CD133-Doxil in CHO cells. Results from the competition study revealed that the HT-29 cells pretreated with the free anti-CD133 antibody were not different from those of cells treated with Doxil. This indicates that the excess amount of free anti-CD133 can block the interaction of HT-29 cell receptors with CD133-Doxil.
Fig. 3.
Expression level of CD133 receptor in HT-29 cell line (A&B) and CHO cell line (C&D) by Flow Cytometry Analysis. Cells were stained with PE-labeled anti-CD133 antibodies. A nonspecific binding of PE-labeled isotype antibody to the cells is pointed to the red peak and a specific binding of PE-labeled anti-CD133 antibody to the cells are shown by the blue peak. The data, expressed as Geometric mean fluorescence intensity (GMFI), represented mean ± S.E.M. (n = 3).
Fig. 4.
Cellular binding of CD133-Doxil, Doxil and free Doxorubicin to CD133-positive HT-29 cells (A&B) and CD133-negative CHO cells (C&D) at 4 °C in vitro. Doxil was used as control. Results are representative of three independent experiments. *** (p = 0.0002) represents significant differences of CD133-Doxil compared to Doxil in cellular binding.
Fig. 5.
Cellular uptake of CD133-Doxil, Doxil and free doxorubicin to CD133-positive HT-29 cells (A&B) and CD133-negative CHO cells (C&D) at 37 °C in vitro. Doxil was used as control. Results are representative of three independent experiments. **** (p < 0.0001) represents significant differences of CD133-Doxil compared to Doxil in cellular uptake.
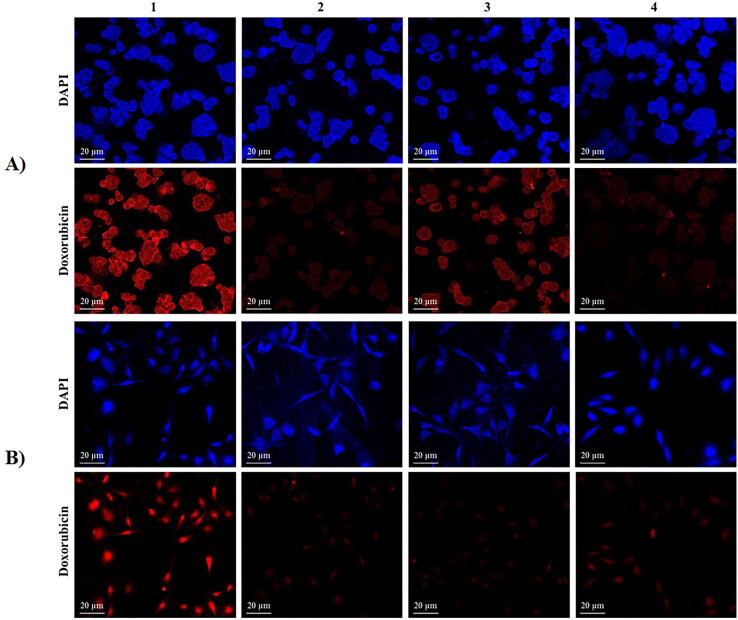
3.6. Cellular uptake of Doxorubicin using CLSM
CLSM was used to assess the relative internalization of immonoliposome formulations after incubation with HT-29 and CHO cells in a serum-free media. The CLSM images displayed a significantly higher accumulation of doxorubicin in HT-29 cells incubated with CD133-Doxil relative to those treated with Doxil (Fig. 6). A result of competition studies was not different from HT-29 cells treated with Doxil, suggesting the role of CD133 antibody in facilitating the cellular uptake of the liposome.
Fig. 6.
Fluorescent Confocal Microscopy images of HT-29 (A) and CHO (B) cells treated with free Doxorubicin (1), Doxil (2), CD133-Doxil (3) and free anti-CD133 antibody + CD133-Doxil (4) for 3 h at 37 °C. Cells were stained with DAPI to determine nuclei. Scale bar is 20 μm.
3.7. Cytotoxicity of CD133-targeted Doxil against HT-29 cells in vitro
MTT assay was measured to further explore the in vitro anti-tumor activity and IC50 of free Doxorubicin, CD133-Doxil, Doxil, and free anti-CD133 antibody against HT-29 and CHO cells. Among all the formulations, except Doxorubicin, CD133-Doxil displayed considerably higher cytotoxicity and a significantly lower IC50 against HT-29 cells because of the greater uptake of CD133-Doxil by the target cells (Table 3). In order to clarify whether Ab-liposomes were precisely bound to HT29 cells, the liposome formulations were incubated with the CHO cell line, which lacks antigens recognized by anti CD133 antibodies. It can be seen from Table 3 that there was almost no observable difference in cytotoxicity evaluation results between the nanoparticle formulations against CHO cells. This can be attributed to the fact that CHO cells do not express CD133 surface markers. Indeed, results of the competitive assay showed that incubation of HT-29 cells with Ab-liposomes in the presence of excess free Ab caused a reduction in the cytotoxicity of CD133-Doxil to an extent that was comparable to that of Doxil. This suggests that the higher cytotoxicity of CD133-Doxil was attributed to the specific ligand-receptor interaction. These results are consistent with what has been found in a previous study, suggesting the significant uptake of targeted-liposomes via receptor-mediated endocytosis is responsible for the cytotoxicity of encapsulated drugs (Sawant and Torchilin 2012).
Table 3.
Cytotoxicity effect (IC50) of different Doxorubicin formulations against HT-29 and CHO cancer cells in vitro. Data represented as μg/ml ± standard deviation (n = 3).
| IC50 |
||
|---|---|---|
| Cell Line | HT-29 | CHO |
| DXR | 0.293 ± 0.029 | 0.484 ± 0.041 |
| CD133-Doxil | 0.663 ± 0.092 | 4.087 ± 0.518 |
| Doxil | 2.031 ± 0.311 | 4.263 ± 0.53 |
| FreeAb + CD133-Doxil | 1.953 ± 0.213 | 3.996 ± 0.446 |
4. Discussion
The research was motivated to optimize post-insertion method to modify the average number of conjugated antibody molecules per single liposome. Then, we investigated the effect of preferred targeted Doxil on cellular uptake, binding, internalization and cytotoxicity against two different cell types. Our data showed that CD133-targeted Doxil could efficiently bind to HT-29 cells and result in considerably higher anti-cancer effects, relative to unmodified Doxil. These results are in agreement with previous studies which reported that specific interaction of antibody/antigen is responsible for higher therapeutic efficacy of targeted Doxil (Qhattal and Liu 2011).
Initially, the routine post-insertion method was used for antibody-conjugated Doxil; however, the number of conjugated antibodies per single liposome was not enough (<10). Then we decided to optimize post-insertion techniques as the same molar ratio as in the routine post-insertion method with the intention of increasing the number of antibodies conjugated per every single liposome. Our results exhibited that the drug release and leakage profiles of the two methods were almost similar. However, the surface size of the nanoliposomes in optimized post-insertion method (113 nm) was slightly bigger than those for the routine post-insertion method (106 nm). Of course, it has been reported that nanoparticles with diameters around 150 nm could facilitate receptor mediated endocytosis and cause the high accumulation in the tumor site (He et al. 2010).
Besides, surprisingly the number of conjugated antibodies per single liposome in the optimized post-insertion method was almost two times more than those in the routine post-insertion method. A possible explanation for less conjugated antibodies in the routine post-insertion method compared to an optimized post-insertion method could be the insufficient transfer of the antibodies from micells into liposomes. This could be attributed to either hydrolysis of the NHS ester or sensitivity of the reaction to the temperature variation (Psarra et al. 2017). The primary amine group of the antibodies reacts with the N-Hydroxysuccinimide (NHS) esters of DSPE-PEG-NHS liposomes resulting in the formation of an amide linkage. In the routine post-insertion method, direct exposure of DSPE-PEG-NHS lipid to either the sterile water or an elevated temperature of 47 °C may initiate the hydrolysis of the NHS group. This can subsequently diminish the coupling yield before the reaction with the antibody. However, in the optimized post-insertion method, the antibody was immediately added to the DSPE–PEG3400–NHS lipid after drying the film. As a result, in order to reduce efficiency losses, DSPE-PEG-NHS lipid needs to be used directly for conjugation to antibodies to prevent the hydrolysis of NHS ester. Of course, in most reactions, some NHS ester hydrolyzes which relies on protein structure, solubility, and reagent, etc. Therefore, comparing the results of the two post-insertion methods, it could be seen that almost 15 and 6 molecule antibodies have been coupled to the surface of each single liposome in the optimized and the routine post-insertion method respectively. It has been reported that relatively (10–20) ligand molecules per liposome are required to selectively increase the number of drugs accumulated within target cells through the mechanism of receptor mediated internalization (Sapra and Allen, 2003, Sapra and Allen, 2002). In fact, there is a positive correlation between the number of conjugated antibodies on the liposome surface and cellular uptake, binding and cytotoxicity. Previous research demonstrated the importance of the ligand density in cellular responses (Shamsi, 2016, Shamsi, 2017, Shamsi et al., 2011). Furthermore, other studies reported that a variation in the amount of antibodies at the liposome surfaces affects the pharmacokinetics and accelerate autoantibody clearance [12]. Of course, there are optimal values for the surface ligand density and beyond that could lead to a decrease in the overall cellular uptake and binding (Elias et al. 2013). Therefore, based on the above explanations, CD133-Doxil made by the optimized post-insertion method was relatively suitable for further experiments.
The issue that needs to be taken into consideration is that conjugation of the full-size monoclonal antibodies to the liposome surface could increase the liposome uptake because of the recognition of Fc portion of antibodies during circulation (Joshi, Butchar and Tridandapani 2006). However, it could also trigger antibody-dependent cell-mediated cytotoxicity and might result in synergistic anti-tumor effects in combination with the liposomal anticancer drugs (Baselga et al. 1998). Prior studies have noted the importance of the coupling method, the length and amount of PEG chain in the clearance rate of liposome when whole antibodies (Ab) are used as targeting ligands (Sapra and Allen 2003). As mentioned in the previous research, long-chain PEGs such as DSPE-PEG liposomes with higher molecular weight PEG (i.e., PEG 2000 and PEG 5000) produced higher prolonged circulation time compared with liposomes containing PEG-lipid with a lower molecular weight PEG (Gabizon 2001). Furthermore, while the ligand is attached at the distal terminal of the PEG chain either reduce the steric hindrances for target binding or prolong the circulation time (Klibanov et al. 1991). Moreover, the addition of mPEG-DSPE with long circulation time has proved to increase targeted delivery (Maruyama 2002).
Another factor which can affect cellular uptake, binding and cytotoxicity is the receptor density on the cancer cells (Sapra and Allen 2003). Population of CD133+ of HT-29 cells was more than 97%, which is a good indication of significant expression of CD133 receptors on the cancer cells. The results of competition assays could prove that superior cellular uptake, binding, and cytotoxicity was through specific binding of antibodies with CD133 receptors on HT-29 cells. Therefore, upon binding of a ligand to a specific receptor on the cancer cells, the receptor-mediated endocytosis pathway could efficiently internalize the liposome-drug package into cancer cells which could result in either higher drug delivery into HT-29 cells or greater overall efficacy due to the decrease in the drug diffusion back from the tumor. Following internalization of the liposomal drug and degradation of the drug-liposome package, the rate of drug release from the liposomes, determines the cytotoxicity of targeted liposomes (Sapra and Allen, 2004, Allen et al., 2005). Furthermore, in regard to antibody targeted liposomes, the cytotoxicity is proportionally associated with the antigen site density (Hosokawa et al. 2003). A reason for this could be that CHO cells with no or relatively low CD133 receptors were negligibly affected by CD133-doxil.
Receptor-mediated internalization remain an alternative route to the plasma membrane and could harness the activity of multidrug-resistant (MDR) transporters like P glycoprotein that carry out a significant role in pumping drugs like Doxorubicin out of cells (Sapra and Allen 2002). It is therefore likely that targeted Daxil with anti CD133 antibody could result in binding and subsequently increasing drugs into tumor cells via receptor-mediated endocytosis. This might lead to either significantly increasing anti-tumor efficacy or partially overcoming drug resistance.
5. Conclusion
A modified post-insertion technique was used to conjugate Doxil with a monoclonal antibody (mAb) against CD133, which is one of the recognized surface markers linked to cancer stem cells. Then the therapeutic efficacy of the CD133-targeted Doxil against the colorectal cancer cells, which highly express CD133 receptor markers was examined. Our findings confirmed higher cellular uptake and binding of CD133-Doxil relative to Doxil. This could suggest that CD133 targeting ligand is responsible for high drug concentrations inside the CD133+ tumor cells, leading to significantly improved either cytotoxicity or therapeutic efficacy of CD133-Doxil against HT-29 cell. Although in vitro studies prove high therapeutic efficacy for targeting CD133+ cells, in vivo examinations are necessary to ensure that they are suitable as CSC-targeting therapies that express high level of CD133 surface markers.
Acknowledgement
The award of a grant to authors from SUMS is gratefully acknowledged.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Fahimeh Shamsi, Email: fahimeh.shamsi@ymail.com.
Ali Khaleghian, Email: Biotechsemnan@gmail.com.
References
- Allen T.M., Mumbengegwi D.R., Charrois G.J. Anti-CD19-targeted liposomal doxorubicin improves the therapeutic efficacy in murine B-cell lymphoma and ameliorates the toxicity of liposomes with varying drug release rates. Clin Cancer Res. 2005;11:3567–3573. doi: 10.1158/1078-0432.CCR-04-2517. [DOI] [PubMed] [Google Scholar]
- Baselga J., Norton L., Albanell J., Kim Y.M., Mendelsohn J. Recombinant humanized anti-HER2 antibody (Herceptin) enhances the antitumor activity of paclitaxel and doxorubicin against HER2/neu overexpressing human breast cancer xenografts. Cancer Res. 1998;58:2825–2831. [PubMed] [Google Scholar]
- Corvo M.L., Mendo A.S., Figueiredo S., Gaspar R., Larguinho M., Guedes da Silva M.F.C., Baptista P.V., Fernandes A.R. Liposomes as Delivery System of a Sn(IV) Complex for Cancer Therapy. Pharm. Res. 2016;33:1351–1358. doi: 10.1007/s11095-016-1876-6. [DOI] [PubMed] [Google Scholar]
- Elias D.R., Poloukhtine A., Popik V., Tsourkas A. Effect of ligand density, receptor density, and nanoparticle size on cell targeting. Nanomedicine. 2013;9:194–201. doi: 10.1016/j.nano.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M., Tian H., de Sauvage F.J. The hedgehog signaling pathway in cancer. Clin Cancer Res. 2006;12:5924–5928. doi: 10.1158/1078-0432.CCR-06-1736. [DOI] [PubMed] [Google Scholar]
- Ferrandina G., Petrillo M., Bonanno G., Scambia G. Targeting CD133 antigen in cancer. Expert Opin Ther Targets. 2009;13:823–837. doi: 10.1517/14728220903005616. [DOI] [PubMed] [Google Scholar]
- Gabizon A.A. Pegylated Liposomal Doxorubicin: Metamorphosis of an Old Drug into a New Form of Chemotherapy. Cancer Invest. 2001;19:424–436. doi: 10.1081/cnv-100103136. [DOI] [PubMed] [Google Scholar]
- Gong A., Huang S. FoxM1 and Wnt/beta-catenin signaling in glioma stem cells. Cancer Res. 2012;72:5658–5662. doi: 10.1158/0008-5472.CAN-12-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui K., Zhang X., Chen F., Ge Z., Zhang S., Qi X., Sun J., Yu Z. Lipid-polymer nanoparticles with CD133 aptamers for targeted delivery of all-trans retinoic acid to osteosarcoma initiating cells. Biomed Pharmacother. 2019:751–764. doi: 10.1016/j.biopha.2018.11.118. [DOI] [PubMed] [Google Scholar]
- He C., Hu Y., Yin L., Tang C., Yin C. Effects of particle size and surface charge on cellular uptake and biodistribution of polymeric nanoparticles. Biomaterials. 2010;31:3657–3666. doi: 10.1016/j.biomaterials.2010.01.065. [DOI] [PubMed] [Google Scholar]
- Hosokawa S., Tagawa T., Niki H., Hirakawa Y., Nohga K., Nagaike K. Efficacy of immunoliposomes on cancer models in a cell-surface-antigen-density-dependent manner. Br. J. Cancer. 2003;89:1545–1551. doi: 10.1038/sj.bjc.6601341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iden D.L., Allen T.M. In vitro and in vivo comparison of immunoliposomes made by conventional coupling techniques with those made by a new post-insertion approach. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2001;1513:207–216. doi: 10.1016/s0005-2736(01)00357-1. [DOI] [PubMed] [Google Scholar]
- Joshi T., Butchar J.P., Tridandapani S. Fcγ receptor signaling in phagocytes. Int. J. Hematol. 2006;84:210–216. doi: 10.1532/IJH97.06140. [DOI] [PubMed] [Google Scholar]
- Klibanov, A. L., K. Maruyama, A. M. Beckerleg, V. P. Torchilin & L. Huang (1991) Activity of amphipathic poly(ethylene glycol) 5000 to prolong the circulation time of liposomes depends on the liposome size and is unfavorable for immunoliposome binding to target. Biochimica et Biophysica Acta (BBA) - Biomembranes, 1062, 142-148. [DOI] [PubMed]
- Malam Y., Loizidou M., Seifalian A.M. Liposomes and nanoparticles: nanosized vehicles for drug delivery in cancer. Trends Pharmacol Sci. 2009;30:592–599. doi: 10.1016/j.tips.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Maruyama K. PEG-immunoliposome. Biosci. Rep. 2002;22:251–266. doi: 10.1023/a:1020138622686. [DOI] [PubMed] [Google Scholar]
- Mi Y., Huang Y., Deng J. The enhanced delivery of salinomycin to CD133(+) ovarian cancer stem cells through CD133 antibody conjugation with poly(lactic-co-glycolic acid)-poly(ethylene glycol) nanoparticles. Oncol Lett. 2018;15:6611–6621. doi: 10.3892/ol.2018.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal N.K., Mandal B., Balabathula P., Setua S., Janagam D.R., Lothstein L., Thoma L.A., Wood G.C. Formulation, Development, and In Vitro Evaluation of a CD22 Targeted Liposomal System Containing a Non-Cardiotoxic Anthracycline for B Cell Malignancies. Pharmaceutics. 2018;10 doi: 10.3390/pharmaceutics10020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen C.E., Cheung L.H., Weyergang A., Berg K., Vallera D.A., Rosenblum M.G., Selbo P.K. Design, Characterization, and Evaluation of scFvCD133/rGelonin: A CD133-Targeting Recombinant Immunotoxin for Use in Combination with Photochemical Internalization. J Clin Med. 2019;9 doi: 10.3390/jcm9010068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psarra E., König U., Müller M., Bittrich E., Eichhorn K.-J., Welzel P.B., Stamm M., Uhlmann P. In Situ Monitoring of Linear RGD-Peptide Bioconjugation with Nanoscale Polymer Brushes. ACS Omega. 2017;2:946–958. doi: 10.1021/acsomega.6b00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qhattal H.S., Liu X. Characterization of CD44-mediated cancer cell uptake and intracellular distribution of hyaluronan-grafted liposomes. Mol Pharm. 2011;8:1233–1246. doi: 10.1021/mp2000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapra P., Allen T.M. Internalizing antibodies are necessary for improved therapeutic efficacy of antibody-targeted liposomal drugs. Cancer Res. 2002;62:7190–7194. [PubMed] [Google Scholar]
- Sapra P., Allen T.M. Ligand-targeted liposomal anticancer drugs. Prog. Lipid Res. 2003;42:439–462. doi: 10.1016/s0163-7827(03)00032-8. [DOI] [PubMed] [Google Scholar]
- Sapra P., Allen T.M. Improved outcome when B-cell lymphoma is treated with combinations of immunoliposomal anticancer drugs targeted to both the CD19 and CD20 epitopes. Clin Cancer Res. 2004;10:2530–2537. doi: 10.1158/1078-0432.ccr-03-0376. [DOI] [PubMed] [Google Scholar]
- Sawant R.R., Torchilin V.P. Challenges in development of targeted liposomal therapeutics. Aaps j. 2012;14:303–315. doi: 10.1208/s12248-012-9330-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmohl, J. U. & D. A. Vallera (2016) CD133, Selectively Targeting the Root of Cancer. Toxins (Basel), 8. [DOI] [PMC free article] [PubMed]
- Shamsi F. Investigation of human cell response to covalently attached RADA16-I peptide on silicon surfaces. Colloids Surf B Biointerfaces. 2016;145:470–478. doi: 10.1016/j.colsurfb.2016.05.030. [DOI] [PubMed] [Google Scholar]
- Shamsi F. Investigation of cellular response to covalent immobilization of peptide and hydrophobic attachment of peptide amphiphiles on substrates. Biochem. Eng. J. 2017;117:82–88. [Google Scholar]
- Shamsi F., Coster H., Jolliffe K.A. Characterization of peptide immobilization on an acetylene terminated surface via click chemistry. Surf. Sci. 2011;605:1763–1770. [Google Scholar]
- Shibata H., Izutsu K., Yomota C., Okuda H., Goda Y. Investigation of factors affecting in vitro doxorubicin release from PEGylated liposomal doxorubicin for the development of in vitro release testing conditions. Drug Dev Ind Pharm. 2015;41:1376–1386. doi: 10.3109/03639045.2014.954582. [DOI] [PubMed] [Google Scholar]
- Wu B., Sun C., Feng F., Ge M., Xia L. Do relevant markers of cancer stem cells CD133 and Nestin indicate a poor prognosis in glioma patients? A systematic review and meta-analysis. J Exp Clin Cancer Res. 2015;34:44. doi: 10.1186/s13046-015-0163-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Paxton J.W., Wu Z. Development of Long-Circulating pH-Sensitive Liposomes to Circumvent Gemcitabine Resistance in Pancreatic Cancer Cells. Pharm. Res. 2016;33:1628–1637. doi: 10.1007/s11095-016-1902-8. [DOI] [PubMed] [Google Scholar]